ABSTRACT
Neobacillus cucumis T4S4 was isolated from montane rain forest soil in Thailand. This strain possesses the ability to hydrolyze stevioside and rebaudioside A, the two major steviol glycosides found in the stevia plant. The draft genome sequence of T4S4 yielded a circular chromosome of 5,978,437 bases with 38.9% GC content.
ANNOUNCEMENT
A facultative anaerobic, Gram-positive, rod-shaped soil bacterium, designated strain T4S4, was isolated in Nan Province, Thailand (19°8′14″N, 100°57′25″E). T4S4 exhibits unique hydrolytic activities against stevioside and rebaudioside A (1). 16S rRNA sequence analysis of the PCR amplicon (1,416 bp) from T4S4 genomic DNA (gDNA) suggested that T4S4 belongs to the species Neobacillus cucumis, marking it as the first of the genus Neobacillus to harbor such activities. Here, we report the draft genome sequence of N. cucumis T4S4.
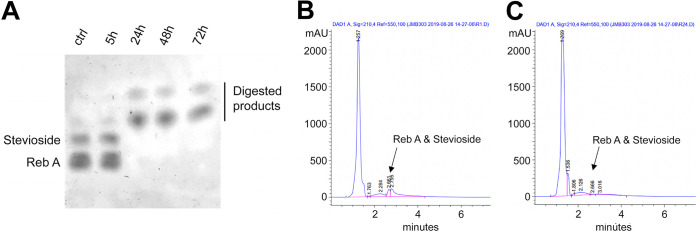
A soil specimen was sampled from at least 10 cm underneath the topsoil layer. The soil was mixed with minimal salt medium supplemented with 0.1% stevia extract (MSS) (per liter, 1 g stevia extract [Sugavia Co., Ltd., Thailand], 0.2 g yeast extract, 2 g NaNO3, 0.5 g MgSO4·7H2O, 0.05 g K2HPO4, 0.01 g FeSO4·7H2O, 0.02 g CaCl2, and 0.02 g MnSO4) + 25 μg/ml of amphotericin-B at a soil/medium ratio of 1:9. The mixture was microaerobically incubated at 40°C for 7 days. The culture was serially diluted and spread onto MSS plates. Hydrolytic activities against stevioside and rebaudioside A were detected using thin-layer liquid chromatography (TLC) (Fig. 1A) (2, 3). High-performance liquid chromatography (HPLC) was utilized to confirm substrate degradation (Fig. 1B and C) (2, 3).
FIG 1.
Hydrolytic activities of T4S4 against stevioside and rebaudioside A (Reb A). (A) Shifts in the stevioside and Reb A bands were observed after a 24 h cultivation. ctrl (control) = MSS. Chromatograms of stevioside and Reb A revealed hydrolytic activities by T4S4 1 h postincubation (B) and 24 h postincubation. (C) Reductions in stevioside and Reb A were observed at 24 h.
A single colony of T4S4 was cultivated in LB broth at 37°C, with agitation at 200 rpm. Genomic DNA was extracted using the E.Z.N.A. bacterial DNA kit (Omega Bio-tek, GA) according to the manufacturer’s recommendations. The quantity and quality of the extracted DNA were evaluated using NanoDrop. Analysis of the 16S rRNA sequence (using the primers 27F and 1492R) (4) and a BLASTn search (5) identified T4S4 as N. cucumis with 98.66% identity. The genomic DNA was submitted for whole-genome sequencing at the Omics Center of Chulalongkorn University.
Sequencing libraries were prepared according to the Qiagen FX protocol (USA). The libraries were analyzed using the Agilent 2100 Bioanalyzer and DeNovix fluorometer before pooling at equimolar quantities. The Illumina MiSeq sequencer was used in 2 × 250-bp paired-end format, resulting in 8,645,960 raw reads (genome coverage, 250×). FastQC was used to check the raw read quality (6). Adaptors and poor-quality reads were trimmed using Trim Galore v. 0.4.4 (7). Genome assembly was conducted using SPAdes v. 3.11.1 (8). The assembled genome was annotated using Prokka v. 1.13 (9). Default parameters were used for all software.
The draft genome sequence of N. cucumis T4S4 consists of 52 contigs and has a total length of 5,978,437 bp, a GC content of 38.9%, and an N50 value of 358,581 bp. Genome annotation revealed a total of 5,908 genes, with 5,781 protein-coding sequences and 123 tRNAs.
Data availability.
This whole-genome sequencing (WGS) project has been deposited at GenBank under accession number JAEHGC000000000. The version described in this paper is version JAEHGC010000000. The raw Illumina data have been submitted under SRA accession number SRR12699712 under the GenBank BioProject number PRJNA663754.
ACKNOWLEDGMENT
This project was funded by the Center of Excellence on Biodiversity (BDC), Office of the Higher Education Commission, Thailand (BDC-PG1-160004).
Contributor Information
S. Chankhamhaengdecha, Email: surang.cha@mahidol.ac.th.
P. Ounjai, Email: puey.oun@mahidol.edu.
Kenneth M. Stedman, Portland State University
REFERENCES
- 1.Ceunen S, Geuns JMC. 2013. Steviol glycosides: chemical diversity, metabolism, and function. J Nat Prod 76:1201–1228. doi: 10.1021/np400203b. [DOI] [PubMed] [Google Scholar]
- 2.Udompaisarn S, Arthan D, Somana J. 2017. Development and validation of an enzymatic method to determine stevioside content from Stevia rebaudiana. J Agric Food Chem 65:3223–3229. doi: 10.1021/acs.jafc.6b05793. [DOI] [PubMed] [Google Scholar]
- 3.Boonkaew B, Udompaisarn S, Arthan D, Somana J. 2019. Expression and characterization of a recombinant stevioside hydrolyzing β-glycosidase from Enterococcus casseliflavus. Protein Expr Purif 163:105449. doi: 10.1016/j.pep.2019.105449. [DOI] [PubMed] [Google Scholar]
- 4.Miller CS, Handley KM, Wrighton KC, Frischkorn KR, Thomas BC, Banfield JF. 2013. Short-read assembly of full-length 16S amplicons reveals bacterial diversity in subsurface sediments. PLoS One 8:e56018. doi: 10.1371/journal.pone.0056018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGinnis S, Madden TL. 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 7.Krueger F. 2015. Trim Galore: a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore.
- 8.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This whole-genome sequencing (WGS) project has been deposited at GenBank under accession number JAEHGC000000000. The version described in this paper is version JAEHGC010000000. The raw Illumina data have been submitted under SRA accession number SRR12699712 under the GenBank BioProject number PRJNA663754.