Abstract
Genomic and transcriptomic analyses have well established that the major fraction of the mammalian genome is transcribed into different classes of RNAs ranging in size from a few nucleotides to hundreds of thousands of nucleotides, which do not encode any protein. Some of these noncoding RNAs (ncRNAs) are directly or indirectly linked to the regulation of expression or functions of ∼25,000 proteins coded by <2% of the human genome. Among these regulatory RNAs, microRNAs are small (21–25 nucleotides) RNAs that are processed from precursor RNAs that have stem–loop structure, whereas noncoding RNAs >200 nucleotides are termed long noncoding RNAs (lncRNAs). Circular RNAs (circRNAs) are newly identified lncRNA members that are generated by back-splicing of primary transcripts. The functions of ncRNAs in modulating liver toxicity of xenobiotics are emerging only recently. Acetaminophen (N-acetyl-para-aminophenol, paracetamol or APAP) is a safe analgesic and antipyretic drug at the therapeutic dose. However, it can cause severe liver toxicity that may lead to liver failure if overdosed or combined with alcohol, herbs, or other xenobiotics. This review discusses the role of ncRNAs in acetaminophen metabolism, toxicity, and liver regeneration after APAP-induced liver injury (AILI).
Key words: Noncoding RNAs, Acetaminophen metabolism, Acetaminophen-induced liver injury (AILI), Toxicity, Liver regeneration
ACETAMINOPHEN-INDUCED LIVER INJURY (AILI)
Acetaminophen (APAP), first used in 1893 by von Mering, is the most commonly used over-the-counter antipyretic and analgesic drug. It has a large therapeutic window, and only at a high dose or in combination with alcohol or other xenobiotics, it causes centrilobular hepatic necrosis, resulting in acute liver failure (ALF)1,2. APAP accounts for ∼46% of all ALF cases annually and is the leading suicide drug in the US and UK.
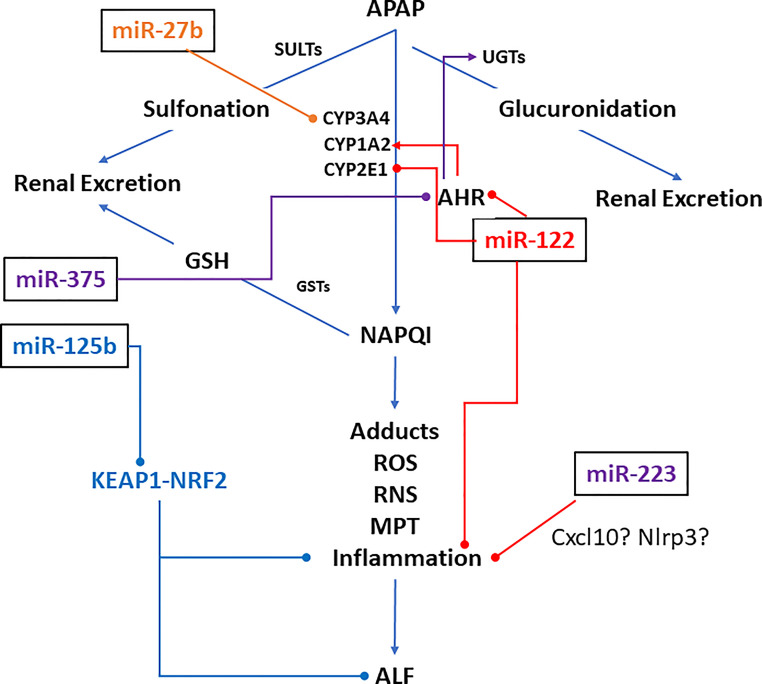
At regular doses, APAP is mostly converted to APAP-sulfate by sulfotransferases and APAP-glucuronide by glucuronidases and excreted in the urine (Fig. 1). Only ∼5% of APAP is oxidized by CYPP450s (CYP1A2, CYP2E1, and CYP3A4 in humans, and CYP2E1 and CYP1A2 in mice) into the highly reactive metabolite, N-acetyl-para-benzoquinone imine (NAPQI or NAPBQI)3, which is detoxified to APAP-GSH upon conjugation with glutathione (GSH). However, at a toxic dose, when the GSH pool is exhausted, NAPQI covalently binds to cellular proteins forming adducts4,5, the amount of which is directly proportional to hepatotoxicity6. Administration of N-acetyl-cysteine (NAC) that restores cellular GSH level was developed as an antidote against APAP-induced hepatotoxicity7,8. Although it is still being used, it is not very effective in enhancing survival by alleviating toxicity, especially after 24 h of overdose1,2.
Figure 1.
MicroRNAs that are known to modulate acetaminophen metabolism in animal models (miR-122, miR-125b, and miR-22) and in cell (hepatocytes and HepaRG) cultures (miR-27b and miR-375). Arrows and pins denote activation and repression, respectively.
The mechanism of hepatocellular death after the formation of NAPQI-protein adducts is not well understood. Initially, it was thought that the inactivation of essential cellular proteins due to NAPQI-adduct formation causes hepatic death. However, studies from different labs have shown that additional mechanisms such as mitochondrial and nuclear ion imbalance following APAP overdose are also involved in hepatocyte necrosis. This ion imbalance elevates cytoplasmic Ca2+ ion levels activating various proteases and endonucleases to cause DNA strand break and apoptosis9.
The immune system involving a complex interplay of immune cell subsets plays a critical role in ALF pathogenesis10,11. APAP overdose-induced hepatocyte death releases danger-associated molecular patterns (DAMPs) such as HMGB1 and nuclear and mitochondrial DNA fragments that bind to pattern recognition receptors on the macrophage. This interaction induces cytokines and chemokines from the macrophage, causing an influx of neutrophils and monocytes into the liver and sterile inflammation. Whether inflammation aggravates the liver injury or helps in the recovery and regeneration of hepatocytes is not fully understood. Additional studies are required to fully elucidate the role of various immune components in AILI1,11.
NONCODING RNAs
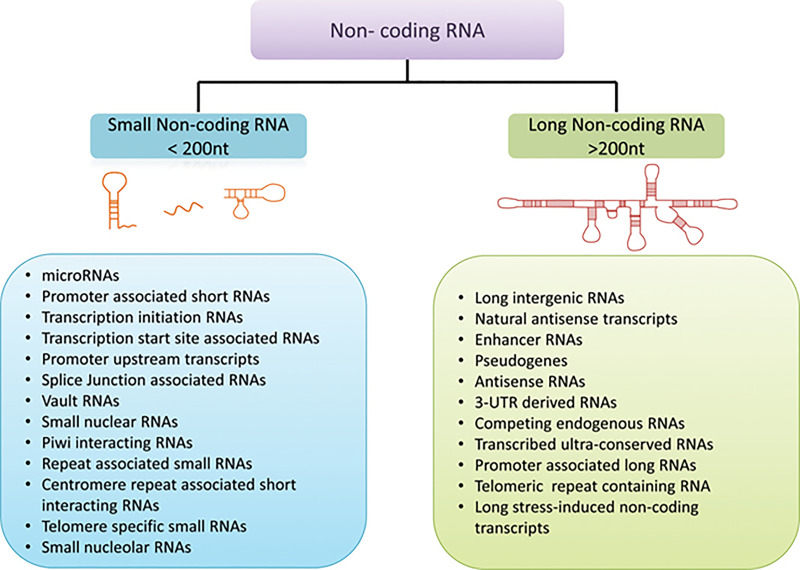
One surprising revelation of high-throughput genomic and transcriptomic sequencing studies is that the major fraction of the mammalian genome, once thought of as “junk DNA,” is transcribed into RNAs that do not code for any protein12,13. Compared to lower eukaryotes, noncoding regions of the genome have expanded enormously in mammals, which is thought to contribute to the complexity of higher organisms14. Among diverse types of non-protein-coding transcripts, those <200 nucleotides are classified as small noncoding RNAs (ncRNAs), whereas those >200 nucleotides are classified as long ncRNAs (lncRNAs) (Fig. 2).
Figure 2.
Classification of noncoding RNAs. From Takahashi et al.14 [Hepatology, 60(2), 744-753, 2014 (10.1002/hep.27043)], reproduced with permission from the publisher.
MicroRNAs
MicroRNAs (miRNAs) are small RNAs (21–25 nucleotides) that are generated by multistep processing of imperfectly base-paired precursor stem–loop region (pre-miRNA) of primary transcripts (pri-miRNA) predominantly coded by RNA polymerase II15. Each pri-miRNA is processed in the nuclei by the microprocessor complex, generating a ∼60-nucleotide pre-miRNA that is then transported to the cytoplasm and further processed, generating the guide RNA–passenger RNA duplex. The guide strand (mature miRNA) is then incorporated into the miRNA-induced silencing complex (mi-RISC) to interact with target RNAs. In general, miRNAs regulate gene expression by complementary base pairing with the target mRNAs through the seed sequence located at the 3′-end of miRNAs, causing mRNA decay and blocking their translation. However, emerging studies have shown that miRNAs can also regulate gene expression by interacting with the seed match located in the exons and 5′-UTRs of protein-coding transcripts16. On the other hand, specific target mRNAs can also induce degradation of certain targeting miRNAs with extensive complementarity, a phenomenon called target-directed miRNA decay (TDMD)17.
Hundreds of distinct miRNAs have been identified in different organisms, including humans. Many of these miRNAs are conserved among animals, whereas others are conserved among vertebrates or mammals, and others are species specific15. Expressions of >60% of protein-coding genes in mammals are repressed by miRNAs.
lncRNAs
lncRNAs are a heterogeneous class of RNAs consisting of intergenic transcripts, chromatin-associated RNAs, promoter-associated RNA, enhancer RNAs (eRNAs), sense or antisense transcripts overlapping with certain protein-coding genes, and many more (Fig. 2). The functions of most of these RNAs are mostly unknown. Emerging studies have shown that specific lncRNAs exhibit multifarious functions, including transcriptional regulation in the cis or trans, organization of nuclear structures, and regulation of expressions and/or functions of RNAs and proteins. Some transcripts annotated as lncRNAs actually encode for small proteins18. It is approximated that the numbers of lncRNA genes in mammals range from <20,000 to >100,000, many of which are not conserved12. Elucidating the biological functions of these RNAs, and their potential role in disease pathogenesis, although a monumental challenge in the field, is an exciting research area to pursue.
CIRCULAR RNAs (circRNAs)
circRNAs lacking 5′-Cap and 3′ poly-A tail are newly discovered lncRNAs19. These covalently closed RNAs are formed by back-splicing of downstream 5′ to upstream 3′ splice sites, a noncanonical splicing mechanism. They are evolutionarily conserved and play a pivotal role in regulating vital biological functions by interacting with target RNAs or proteins. circRNAs are widely expressed in animals, and some are expressed tissue specifically independent of respective linear mRNAs generated by conventional forward splicing20. Because of their covalently closed structures, circRNAs are incredibly stable in body fluids. Many circRNAs have already been identified in human peripheral whole blood, and levels of many circRNAs are much higher than corresponding linear mRNAs20. These can be used as noninvasive biomarkers for different diseases because they are highly resistant to nucleases. CircRNAs can modulate gene expression by acting as competing endogenous RNA (ceRNA) for miRNAs, and some are translated into polypeptides.
In the following sections, we have comprehensively summarized the literature on noncoding RNAs in APAP metabolism and AILI.
miRNA-MEDIATED REGULATION OF PHASE I AND PHASE II ENZYMES INVOLVED IN APAP METABOLISM
miRNAs that are reported to target different factors involved in APAP metabolism is shown in Figure 1. Several in vitro studies have demonstrated the regulation of CYP450s by various miRNAs. miR-27b-mediated regulation of CYP3A4 by binding to its 3′-untranslated region (3′-UTR) was shown in HEK293 cells21. Gill et al. found significant elevation of serum miR-122-5p, miR-378a-5p, miR-125b-5p, and miR-27b-3p levels in children with APAP overdose compared to healthy controls21. To understand the significance of this observation, they treated HepaRG cells with APAP (20 mM) and found that expressions of CYP1A2, CYP3A4, and CYP2E1 were decreased, whereas miR-122-5p, miR-378a-5p, and miR-27b-3p were increased at 6 h, miR-125b-5p at 12 h, and miR-27b-3p at 24 h. Moreover, overexpression of miR-122-5p and miR-378a-5p suppressed CYP1A2, CYP3A4, and CYP2E1 protein levels. Among these, miR-122 was found to target CYP1A2 and CYP3A4 3′-UTRs. Using bioinformatic analysis and luciferase reporter assay, Papageorgiou et al. demonstrated that miR-375 regulates expression of UGT1A1, involved in APAP glucuronidation, by targeting AHR, its transcriptional activator22. These results indicate the miRNA-mediated regulation of APAP metabolizing enzymes.
ROLE OF miR-122 IN AILI
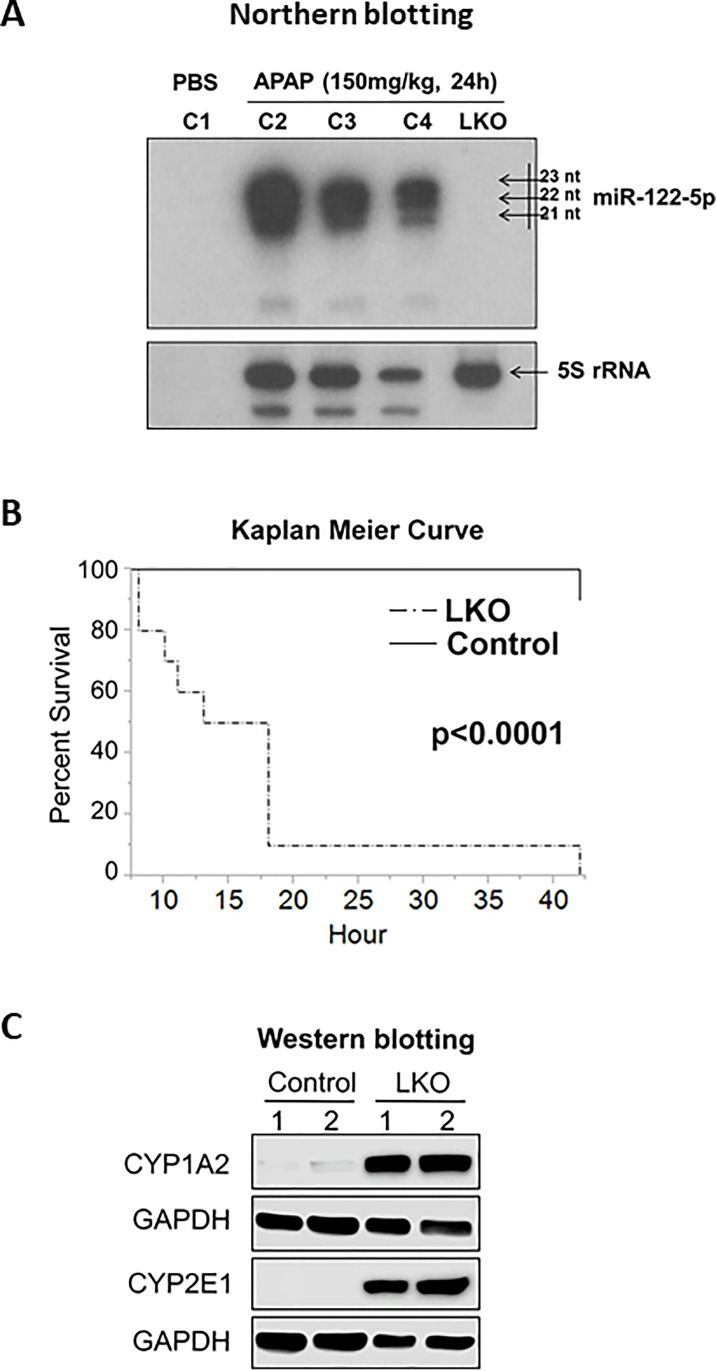
miR-122 is a liver-specific miRNA that constitutes ∼52% and ∼70% of miRNAs in human23 and mouse livers24,25, respectively. Notably, mature miR-122 is conserved from zebrafish to humans, suggesting this miRNA is critical for vertebrates. miR-122, although not essential for liver development, is required for maintenance of hepatic differentiation state, liver metabolic homeostasis, and tumor suppression26,27. In 2017, we reported that mice subjected to APAP overdose (300 mg/kg) repressed production of pri-miR-122 that correlated with reduced expression of HNF6 and HNF4α, two key trans-activators of Mir-122 gene28. By Northern blotting analysis, we also demonstrated that all three forms (21–23 nucleotides) of miR-122 were released in the serum of the wild-type (WT) mice after APAP overdose, which is absent in miR-122 knockouts (Fig. 3A). More importantly, we found that the liver-specific miR-122 knockout (LKO) mice were significantly more sensitive to APAP (500 mg/kg) and died within 24 h (Fig. 3B), which correlated with increased formation of the toxic metabolite, NAPQI28. Higher basal hepatic CYP2E1 and CYP1A2 enzymes contributed to the higher NAPQI formation in the mutant mice (Fig. 3C). We also showed that miR-122 indirectly suppressed Cyp1a2 gene expression in mouse liver by targeting Med1 and Ctcf 28. We found MED1 activates Cyp1a2 expression directly, whereas CTCF regulates the expression of Ahr, which encodes for AHR, a known Cyp1a2 trans-activator. In contrast, loss of miR-122 increased basal hepatic CYP2E1 protein level (Fig. 3C) without altering its gene expression28. Thus, loss of miR-122 facilitates NAPQI formation by upregulating the Cyp1a2 gene and CYP2E1 protein. We also showed that adeno-associated virus-mediated miR-122 delivery in miR-122 LKO mice before APAP treatment partially ameliorated AILI28.
Figure 3.
(A) Northern blot analysis of serum miR-122 and 5S rRNA isolated from the control (miR-122fl/fl and miR-122 LKO mice injected with APAP or PBS). (B) Survival of control and miR-122 LKO mice injected with APAP (500 mg/kg). (C) Western blot analysis of CYP1A2 and CYP2E1 in the hepatocytes isolated from the control and miR-122 LKO mice. From Chowdhary et al.28 [Am J Pathol. 187(12), 2758–2774, 2017 (doi: 10.1016/j.ajpath.2017.08.026)], with permission from the publisher.
We earlier reported that activation of the CCL2–CCR2 axis is causally linked to massive liver inflammation in miR-122-depleted mice due to infiltration of monocytes and neutrophils that produce proinflammatory cytokines [e.g., interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α)]27. Accumulation of neutrophils has been reported to promote AILI29, and pharmacological inhibition of CCL2 or CCR2 could attenuate APAP toxicity by impeding infiltration of macrophages in the liver30. Thus, the accelerated formation of NAPQI and a strong sterile inflammatory milieu in the liver of miR-122 LKO mice aggravate APAP toxicity in these mice. It is expected that blocking of the CCL2–CCR2 axis along with inhibition of these CYP450s will act in concert to alleviate APAP toxicity in miR-122 LKO mice.
ROLE OF miR-125b IN AILI
By transfecting 302 conserved miRNA mimic library in mouse and human hepatocytes followed by APAP treatment, Yang et al. identified seven miRNAs (miR-194-5p, miR-125b-5p, miR-21-5p, let-7a-5p, miR-122-5p, miR-30c-5p, and miR-193a-3p) attenuated APAP-induced cell death in these cells31. Among these, blocking functions of let-7a-5p, miR-125b-5p, and miR-122-5p, by transfecting anti-miRs, reduced hepatocyte viability compared with the scramble miRNA control, suggesting a protective role of these miRNAs against APAP toxicity. Furthermore, delivery of miR-125b mimic or AAV8-miR-125b virus impeded AILI and improved survival in mice31. Mechanistically, miR-125b-5p inhibited ALF by directly targeting KEAP1, thereby stabilizing its partner NRF2 and its downstream targets that protect livers from ALF. NRF2 has also been shown to inhibit Fas-induced ALF in mice32. Thus, miR-125b-5p delivery could attenuate ALF induced by multiple insults.
ROLE OF miR-223 IN AILI
He et al. investigated the role of miR-223, abundantly expressed in neutrophils in AILI by using genetically mutant mice (GEMMs) and cultured neutrophils33. They found that neutrophil-specific miR-223 knockout (KO) mice are more susceptible to AILI due to enhanced neutrophil infiltration, oxidative stress, hepatocyte injury, and induction proinflammatory mediators such as ICAM1 in neutrophils. Notably, Icam-1 gene disruption reversed APAP-induced neutrophil infiltration, and AILI in miR-223 depleted mice33.
miRNAs AS BIOMARKERS FOR AILI
A seminal study by Wang et al. demonstrating elevation of liver-specific miR-122-5p (miR-122) even before the increase in the ALT levels in the sera of APAP overdosed mice34 has generated impetus in its potential use as a diagnostic marker for AILI. However, subsequent studies from different labs have shown that the circulating miR-122 is elevated in almost all types of liver injury and disease, including nonalcoholic steatohepatitis (NASH) and hepatocellular carcinoma (HCC)35. According to a recent report, glutamate dehydrogenase (GLDH), high-mobility group box 1 (HMGB-1), Keratin-18 (K18), miR-122, and ornithine carbamoyltransferase (OCT) are more sensitive markers of hepatotoxicity than serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), and total bilirubin36.
Efficacy of the combination of specific miRNAs as biomarkers for AILI has been tested in various laboratories. Vliegenthart et al. reported that miR-122-5p, miR-885-5p, and miR-151-3p demonstrated the most significant fold change between patients who developed organ toxicity and those who did not upon APAP overdose37. The specificity of miRNAs is organ specific but not etiology specific. For example, in acute liver injury, miR-122-5p, miR-885-5p, miR-151-3p, and miR-382-5p showed a significant change in serum but no detectable difference in kidney injury. However, the levels of these miRNAs could not differentiate among liver injuries due to APAP or some other causes37. Since the circulating miR-122 level is elevated in any condition associated with hepatocyte injury and liver disease (e.g., HBV and HCV infection, nonalcoholic or alcoholic liver disease, and HCC)38, miR-122 alone is no longer considered as a specific biomarker for AILI.
Yang et al. analyzed circulating miRNAs in the serum and urine in children in three groups: 1) healthy children, 2) hospitalized children receiving therapeutic doses of APAP, and 3) children hospitalized for APAP overdose39. Among 147 miRNAs detected in the APAP overdose group, median levels of miR-122, miR-375, miR-423-5p, miR-30d-5p, miR-125b-5p, miR-4732-5p, miR-204-5p in serum and miR-574-3p and of miR-375, miR-940, miR-9-3p, and miR-302a in urine were increased compared to the other two groups. Serum miR-122 and miR-375 and urinary miR-375 and miR-940 levels were significantly increased in children with APAP overdose. There was a significant correlation between peak serum miR-122 and APAP-protein adducts39. In another study, 47 miRNAs were identified by miRNA-seq in HepaRG cells treated with APAP, among which four miRNAs (miR-224-5p, miR-320a, miR-449a, and miR-877-5p) were found to suppress drug-metabolizing enzymes involved in AILI by downregulating liver-enrich transcription factors (e.g., HNF1A, HNF4A, and NR1I2)40. Ectopic expression of these miRNAs protected HepaRG cells from APAP toxicity, confirming their protective role against AILI. More importantly, miR-320a and miR-877-5p levels were upregulated in sera of children with APAP overdose relative to healthy controls. Interestingly, these two studies identified entirely different sets of miRNAs in the sera of APAP-overdosed children. The reason for this incongruity is likely due to the use of different model systems to identify APAP-modulated miRNAs, serum for the first one and HepaRG cells for the other.
Recently, Carreiro et al. profiled 327 miRNAs by real-time reverse transcription-polymerase chain reaction (RT-PCR) in the sera of 27 DILI patients, among which 21 were diagnosed with AILI41. They identified five clusters, two of which showed clear clinical patterns, and included specific elevated miRNAs, as previously reported42. Serum miRNA signatures associated with clusters 1 and 5 confirmed APAP toxicity, high ALT, and late appearance, whereas those of clusters 2–4 exhibited a lower increase in miRNA and ALT levels and heterogeneous clinical manifestations. A set of seven miRNAs (miR-194-5b, miR-125b-5b, miR-193a-5b, miR-122-5p, miR-21-5p, miR-27b-3p, and miR-1290) were able to differentiate AILI from other forms of liver injury (ischemic hepatitis). In contrast, those with less pronounced AILI were difficult to distinguish from non-APAP-associated liver injury. Six among this group had the highest levels of the seven APAP-associated miRNAs with high ALT. The second set clustered 12 miRNAs (miR-3646-3p, miR-412, miR-2467-3p, miR-1207-5p, miR-138-1-3p, miR-605, miR-4258, miR-372, miR-4524a-3p, miR-19b-1-5p, miR-122-5p, and miR-483-5p) that are associated with the subjects exhibiting highest AILI. Elevation of circulating miR-122 was maximal in cluster 5, followed by cluster 1 without significant changes in clusters 2–4. miR-194-5p and miR-483-5 associated with AILI were most significantly elevated only in cluster 5. It has been reported that the expression profile of a set of 11 circulating miRNAs during NAC treatment can discriminate AILI from ischemic hepatitis42. These findings suggest that profiling of these miRNA clusters instead of miR-122, which is released into circulation in response to almost all types of hepatic injury, would be better diagnostic and prognostic biomarkers for AILI.
Recently, serum miRNA profiles were identified in one patient with AILI using next-generation sequencing (NGS)43. The patient showed elevated levels of AST, ALT, and GLDH. NAC treatment was initiated within 24 h after the APAP overdose. Circulating miRNA sequences from 24 consecutive serum specimens collected at different periods from the time of admission to death were compared with those from participants with nonlethal APAP overdose and healthy volunteers. Thirty-six APAP-inducible miRNAs, including several isomiRs (with the same seed sequence) and some novel miRNAs, were identified in the patients who succumbed to AILI. These results demonstrated that the miRNA profile could better distinguish the lethal outcome than the conventional serum biochemical markers. However, additional studies are needed to draw any conclusion.
Surprisingly, the elevation of circulating miR-122 has also been reported in different types of heart disease44. Since miR-122 is almost exclusively expressed in the liver, cardiac failure likely causes liver ischemia, resulting in the release of miR-122 from damaged hepatocytes. Rodenburg et al. analyzed serum miRNA levels in a large number of critically ill patients and found that serum miR-122 levels were associated only with hepatic injury45.
ROLE OF lncRNAs IN AILI
Dysregulation of lncRNAs in AILI and its consequence are emerging only recently. By gene expression profiling, Zheng et al. identified the expression of four lncRNAs (NONMMUT023651.2, NONMMUT029382.2, NONMMUT029383.2, and NONMMUT102053.1) is altered in the livers of mice treated with APAP (300 mg/kg)46. However, whether there are human orthologs of these lncRNAs and how their expression was affected in human AILI patients remain unknown. Another study identified two lncRNAs, HNF1α antisense RNA 1 and HNF 4α antisense RNA 1, which affected APAP-induced cell death in HepaRG cells probably by regulating CYP450s46,47.
Khanal et al. found that NR2E3 KO mice are highly susceptible to AILI that correlated with the suppression of DINO (Damage-induced lncRNA) and p53 activation48. They found NR2E3 transcriptionally activated DINO to directly interact with p53, enhancing p53 stability and activity to cope with various stresses, including AILI.
Recently, Pei et al. examined the role of KCNQ1OT1 in APAP toxicity in mice and HepaRG cells by knockdown and overexpression studies49. They showed that KCNQ1OT1 exhibited a protective role in mice by acting as an miR-122 sponge, thereby upregulating its target CES2 that detoxifies APAP. It is hard to comprehend that CES2 upregulation is mediated through sponging miR-122 since the loss of miR-122 enhances the susceptibility of human hepatocytes and mice APAP toxicity28,31.
ROLE OF circRNAs IN AILI
Recently, Wang et al. reported that circCBFB, generated from the CBFB primary transcript, was abundantly expressed in mouse livers upon APAP overdose50. They also found miR-185-5p targets P66Shc, an adapter protein that acts as a regulator of mitochondrial oxidative stress. Moreover, knockdown of circ-CBFB blocked APAP-induced mitochondrial damage and hepatocyte injury by acting as an miR-185-5p sponge, resulting in depression of P66Shc50.
miRNAs IN LIVER REGENERATION AFTER AILI
The liver is the only organ that can regenerate to carry on its function even after excessive loss of the liver mass after surgery or due to toxic insults51. Recovery depends on how fast the liver can generate after ALF. A liver transplant is the only option for survival if its regenerative capacity is compromised. However, due to the scarcity of transplantable livers, there is an impetus to develop novel regenerative therapy.
John et al. identified circulating miRNAs associated with liver regeneration in 63 ALF patients52. They found significantly higher miR-21 and miR-221 levels in the sera but reduced levels in liver biopsies in patients who recovered spontaneously from ALF compared to those who failed to recover. In contrast, miR-122 was upregulated both in the sera and livers of spontaneous survivors. As expected, expression of genes that promote hepatocyte proliferation was upregulated, whereas those that impede liver regeneration, such as HO-1, PDCD4, and the cyclin-dependent kinase inhibitors p21, p27, and p57, were downregulated in spontaneously recovered livers52.
Recently, Salehi et al. analyzed 18 patients admitted with APAP-induced ALF to the liver intensive therapy unit at King’s College Hospital in the UK53. Among these, five received liver transplants, seven recovered spontaneously, and six died soon after hospitalization. The median time to hospitalization after APAP overdose was 2 days. They found serum miR-30a and miR-26a levels were lower, whereas the miR-29b level was higher in the patients who recovered spontaneously than the deceased and transplanted cohort53. Identifying entirely different serum miRNAs by different groups is primarily due to a small number of patients analyzed, although different serum collection methods, miRNA isolation, and analysis could also be contributing factors. Studies in primary human hepatocytes and animal models might unravel the significance of these findings and the therapeutic potential of some of these miRNAs.
FUTURE PERSPECTIVE
Because of the higher stability of miRNAs and circRNAs in body fluids, these RNAs hold promise as noninvasive biomarkers for early detection of AILI and monitoring patients’ prognosis. Although these ncRNAs could be specific and more sensitive biomarkers than serum ALT, their application in the clinic has not yet been possible due to time-consuming detection methods. In this era of novel technology development for faster detection of circulating nucleic acids, it will be possible to image and quantify these RNAs directly in body fluid and biopsied tissues in the near future.
Emerging studies have shown that ncRNAs, especially those conserved, play an essential role in liver biology, and their dysregulation is associated with different disease processes. Except for a few, the role of ncRNAs in drug metabolism and drug toxicity is mostly unknown. Generation of GEMMs or cell lines deleted of specific ncRNA will be pivotal to address their functions. Availability of CRISPR/Cas9 technology will accelerate this process, especially in knocking out isomiR genes with identical/overlapping functions, and some are located in close proximity in the mammalian genome.
APAP-induced liver toxicity is widely used as a model to study the molecular mechanism for drug-induced liver injury because its metabolism is well defined. Since many drugs share common phase I and phase II metabolic enzymes, ncRNAs modulating the expression or function of these enzymes are likely to alter the metabolism and toxicity of many drugs.
Alternative approaches for AILI treatment are urgently needed as NAC is effective only at very early stages. Because most AILI patients are hospitalized long after APAP overdose, there is a significant impetus in identifying novel therapy for liver regeneration. It has been reported that CD1 mice pretreated with replication-defective human type 5 adenovirus vector containing a rodent MIP-2 cDNA insert (AdMIP-2) conferred protection against AILI54. Similarly, AAV-Nrf2 pretreatment protected Nrf2 −/− and WT mice against APAP-induced hepatotoxicity55. We have also shown AAV-DJ-mediated overexpression of miR-122 in Mir-122 −/− mice protected against APAP-induced necrosis of hepatocytes28. Though gene therapies hold promise for treating AILI, we are far from implementing them in the clinic. Alternatively, the drug-induced hepatotoxicity model is effectively used in selecting genetically targeted/modified hepatocytes in therapy applications. The therapeutic transgene is expressed with a short hairpin RNA (shRNA) that imparts modified hepatocytes with resistance to drug-induced toxicity56. After drug challenge, modified hepatocytes are likely to survive and regenerate, replacing unmodified hepatocytes. A better option was demonstrated by Li et al.: CRISPR/Cas9-mediated correction in newborn rabbits with hereditary tyrosinemia type I57. A limitation of AAV-mediated protection against AILI can be an immune response to capsid or transgene. Severe toxicity associated with high-doses of AAV in nonhuman primates and piglets may prove to be counterintuitive to using this method for treating AILI58.
RNA-based therapeutics (e.g., miRNA mimic, anti-miRs) are also being considered, but their effectiveness so far has only been shown as a preventive measure in animal models. It has been reported that a phase 1 clinical trial of miR-34a mimic was abandoned in HCC patients due to severe adverse immune response that resulted in patient deaths59. Oligonucleotides, especially double-stranded miRNA mimics, might cause interferon response in the host, aggravating AILI in patients. Nevertheless, a fundamental understanding of the regulation and mechanism of action of ncRNAs in APAP metabolism and toxicity will be pivotal, as some novel regulators of these pathways could be exploited for the future development of therapy.
ACKNOWLEDGMENTS
This study was supported in part by National Institutes of Health grants DK088076 (K.G.) and R01A193244 (K.G.). Vivek Chowdhary wrote the first draft of the review, and Pipasha Biswas and Kalpana Ghoshal critically read and edited it.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Ramachandran A, Jaeschke H. Acetaminophen toxicity: Novel insights into mechanisms and future perspectives. Gene Expr. 2018;18(1):19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stravitz RT, Lee WM. Acute liver failure. Lancet 2019;394(10201):869–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dahlin DC, Miwa GT, Lu AY, Nelson SD. N-acetyl-p-benzoquinone imine: A cytochrome P-450-mediated oxidation product of acetaminophen. Proc Natl Acad Sci USA 1984;81(5):1327–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mitchell JR, Jollow DJ, Potter WZ, Gillette JR, Brodie BB. Acetaminophen-induced hepatic necrosis. IV. Protective role of glutathione. J Pharmacol Exp Ther 1973;187(1):211–7. [PubMed] [Google Scholar]
- 5. Jollow DJ, Thorgeirsson SS, Potter WZ, Hashimoto M, Mitchell JR. Acetaminophen-induced hepatic necrosis. VI. Metabolic disposition of toxic and nontoxic doses of acetaminophen. Pharmacology 1974;12(4–5):251–71. [DOI] [PubMed] [Google Scholar]
- 6. Jollow DJ, Mitchell JR, Potter WZ, Davis DC, Gillette JR, Brodie BB. Acetaminophen-induced hepatic necrosis. II. Role of covalent binding in vivo. J Pharmacol Exp Ther 1973;187(1):195–202. [PubMed] [Google Scholar]
- 7. Prescott LF, Park J, Ballantyne A, Adriaenssens P, Proudfoot AT. Treatment of paracetamol (acetaminophen) poisoning with N-acetylcysteine. Lancet 1977;2(8035):432–4. [DOI] [PubMed] [Google Scholar]
- 8. Peterson RG, Rumack BH. Treating acute acetaminophen poisoning with acetylcysteine. JAMA 1977;237(22):2406–7. [PubMed] [Google Scholar]
- 9. James LP, McCullough SS, Knight TR, Jaeschke H, Hinson JA. Acetaminophen toxicity in mice lacking NADPH oxidase activity: Role of peroxynitrite formation and mitochondrial oxidant stress. Free Radic Res. 2003;37(12):1289–97. [DOI] [PubMed] [Google Scholar]
- 10. Jaeschke H, Ramachandran A. Pleiotropic roles of platelets and neutrophils in cell death and recovery during acetaminophen hepatotoxicity. Hepatology 2020;72(5):1873–6. [DOI] [PubMed] [Google Scholar]
- 11. Krenkel O, Mossanen JC, Tacke F. Immune mechanisms in acetaminophen-induced acute liver failure. Hepatobiliary Surg Nutr. 2014;3(6):331–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018;172(3):393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ulitsky I, Bartel DP. lincRNAs: Genomics, evolution, and mechanisms. Cell 2013;154(1):26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takahashi K, Yan I, Haga H, Patel T. Long noncoding RNA in liver diseases. Hepatology 2014;60(2):744–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bartel DP. Metazoan microRNAs. Cell 2018;173(1):20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luna JM, Barajas JM, Teng KY, Sun HL, Moore MJ, Rice CM, Darnell RB, Ghoshal K. Argonaute CLIP defines a deregulated miR-122-bound transcriptome that correlates with patient survival in human liver cancer. Mol Cell 2017;67(3):400–10 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fuchs Wightman F, Giono LE, Fededa JP, de la Mata M. Target RNAs strike back on microRNAs. Front Genet. 2018;9:435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tajbakhsh S. lncRNA-encoded polypeptide SPAR(s) with mTORC1 to regulate skeletal muscle regeneration. Cell Stem Cell 2017;20(4):428–30. [DOI] [PubMed] [Google Scholar]
- 19. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. [DOI] [PubMed] [Google Scholar]
- 20. Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One 2015;10(10):e0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gill P, Bhattacharyya S, McCullough S, Letzig L, Mishra PJ, Luo C, Dweep H, James L. MicroRNA regulation of CYP 1A2, CYP3A4 and CYP2E1 expression in acetaminophen toxicity. Sci Rep. 2017;7(1):12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Papageorgiou I, Freytsis M, Court MH. Transcriptome association analysis identifies miR-375 as a major determinant of variable acetaminophen glucuronidation by human liver. Biochem Pharmacol. 2016;117:78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122—A key factor and therapeutic target in liver disease. J Hepatol. 2015;62(2):448–57. [DOI] [PubMed] [Google Scholar]
- 24. Girard M, Jacquemin E, Munnich A, Lyonnet S, Henrion-Caude A. miR-122, a paradigm for the role of microRNAs in the liver. J Hepatol. 2008;48(4):648–56. [DOI] [PubMed] [Google Scholar]
- 25. Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–9. [DOI] [PubMed] [Google Scholar]
- 26. Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science 2005;309(5732):310–1. [DOI] [PubMed] [Google Scholar]
- 27. Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122(8):2871–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chowdhary V, Teng KY, Thakral S, Zhang B, Lin CH, Wani N, Bruschweiler-Li L, Zhang X, James L, Yang D, et al. miRNA-122 protects mice and human hepatocytes from acetaminophen toxicity by regulating cytochrome P450 family 1 subfamily A member 2 and family 2 subfamily E member 1 expression. Am J Pathol, 2017;187(12):2758–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Woolbright BL, Jaeschke H. Mechanisms of inflammatory liver injury and drug-induced hepatotoxicity. Curr Pharmacol Rep. 2018;4(5):346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mossanen JC, Krenkel O, Ergen C, Govaere O, Liepelt A, Puengel T, Heymann F, Kalthoff S, Lefebvre E, Eulberg D, et al. Chemokine (C-C motif) receptor 2-positive monocytes aggravate the early phase of acetaminophen-induced acute liver injury. Hepatology 2016;64(5):1667–82. [DOI] [PubMed] [Google Scholar]
- 31. Yang D, Yuan Q, Balakrishnan A, Bantel H, Klusmann JH, Manns MP, Ott M, Cantz T, Sharma AD. MicroRNA-125b-5p mimic inhibits acute liver failure. Nat Commun. 2016;7:11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morito N, Yoh K, Itoh K, Hirayama A, Koyama A, Yamamoto M, Takahashi S. Nrf2 regulates the sensitivity of death receptor signals by affecting intracellular glutathione levels. Oncogene 2003;22(58):9275–81. [DOI] [PubMed] [Google Scholar]
- 33. He Y, Feng D, Li M, Gao Y, Ramirez T, Cao H, Kim SJ, Yang Y, Cai Y, Ju C and others. Hepatic mitochondrial DNA/Toll-like receptor 9/microRNA-223 forms a negative feedback loop to limit neutrophil overactivation and acetaminophen hepatotoxicity in mice. Hepatology 2017;66(1):220–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA 2009;106(11):4402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thakral S, Ghoshal K. miR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Curr Gene Ther. 2015;15(2):142–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tajima S, Yamamoto N, Masuda S. Clinical prospects of biomarkers for the early detection and/or prediction of organ injury associated with pharmacotherapy. Biochem Pharmacol. 2019;170:113664. [DOI] [PubMed] [Google Scholar]
- 37. Vliegenthart AD, Shaffer JM, Clarke JI, Peeters LE, Caporali A, Bateman DN, Wood DM, Dargan PI, Craig DG, Moore JK, et al. Comprehensive microRNA profiling in acetaminophen toxicity identifies novel circulating biomarkers for human liver and kidney injury. Sci Rep. 2015;5:15501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gloor Y, Schvartz D, C FS. Old problem, new solutions: Biomarker discovery for acetaminophen liver toxicity. Expert Opin Drug Metab Toxicol. 2019;15(8):659–69. [DOI] [PubMed] [Google Scholar]
- 39. Yang X, Salminen WF, Shi Q, Greenhaw J, Gill PS, Bhattacharyya S, Beger RD, Mendrick DL, Mattes WB, James LP. Potential of extracellular microRNAs as biomarkers of acetaminophen toxicity in children. Toxicol Appl Pharmacol. 2015;284(2):180–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yu D, Wu L, Gill P, Tolleson WH, Chen S, Sun J, Knox B, Jin Y, Xiao W, Hong H and others. Multiple microRNAs function as self-protective modules in acetaminophen-induced hepatotoxicity in humans. Arch Toxicol. 2018;92(2):845–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Carreiro S, Marvel-Coen J, Lee R, Chapman B, Ambros V. Circulating microRNA profiles in acetaminophen toxicity. J Med Toxicol. 2020;16(2):177–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ward J, Kanchagar C, Veksler-Lublinsky I, Lee RC, McGill MR, Jaeschke H, Curry SC, Ambros VR. Circulating microRNA profiles in human patients with acetaminophen hepatotoxicity or ischemic hepatitis. Proc Natl Acad Sci USA 2014;111(33):12169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Krauskopf J, Gosink MM, Schomaker S, Caiment F, Warner R, Johnson K, Kleinjans J, Aubrecht J. The microRNA-based liquid biopsy improves early assessment of lethal acetaminophen poisoning: A case report. Am J Case Rep. 2020;21:e919289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peterlin A, Pocivavsek K, Petrovic D, Peterlin B. The role of microRNAs in heart failure: A systematic review. Front Cardiovasc Med. 2020;7:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roderburg C, Benz F, Vargas Cardenas D, Koch A, Janssen J, Vucur M, Gautheron J, Schneider AT, Koppe C, Kreggenwinkel K, et al. Elevated miR-122 serum levels are an independent marker of liver injury in inflammatory diseases. Liver Int. 2015;35(4):1172–84. [DOI] [PubMed] [Google Scholar]
- 46. Zheng JX, Tang YJ, Yang TH, Qin T, Liu JC, Gu XQ, Xue F, Xia Q. Long non-coding RNAs play regulatory roles in acetaminophen-induced liver injury. J Dig Dis. 2019;20(6):308–317. [DOI] [PubMed] [Google Scholar]
- 47. Chen L, Wang P, Manautou JE, Zhong XB. Knockdown of long noncoding RNAs hepatocyte nuclear factor 1alpha antisense RNA 1 and hepatocyte nuclear factor 4alpha antisense RNA 1 alters susceptibility of acetaminophen-induced cytotoxicity in HepaRG cells. Mol Pharmacol. 2020;97(4):278–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Khanal T, Leung YK, Jiang W, Timchenko N, Ho SM, Kim K. NR2E3 is a key component in p53 activation by regulating a long noncoding RNA DINO in acute liver injuries. FASEB J. 2019;33(7):8335–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pei J, Sun X, Yang G, Zhang S. LncRNA KCNQ1OT1 ameliorates the liver injury induced by acetaminophen through the regulation of miR-122-5p/CES2 axis. Mol Cell Biochem. 2020;475(1–2):107–18. [DOI] [PubMed] [Google Scholar]
- 50. Wang Z, Zhao Y, Sun R, Sun Y, Liu D, Lin M, Chen Z, Zhou J, Lv L, Tian X, et al. circ-CBFB upregulates p66Shc to perturb mitochondrial dynamics in APAP-induced liver injury. Cell Death Dis. 2020;11(11):953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bhushan B, Gunewardena S, Edwards G, Apte U. Comparison of liver regeneration after partial hepatectomy and acetaminophen-induced acute liver failure: A global picture based on transcriptome analysis. Food Chem Toxicol. 2020;139:111186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. John K, Hadem J, Krech T, Wahl K, Manns MP, Dooley S, Batkai S, Thum T, Schulze-Osthoff K, Bantel H. MicroRNAs play a role in spontaneous recovery from acute liver failure. Hepatology 2014;60(4):1346–55. [DOI] [PubMed] [Google Scholar]
- 53. Salehi S, Tavabie OD, Verma S, McPhail MJW, Farzaneh F, Bernal W, Menon K, Agarwal K, Aluvihare VR. Serum microRNA signatures in recovery from acute and chronic liver injury and selection for liver transplantation. Liver Transpl. 2020;26(6):811–22. [DOI] [PubMed] [Google Scholar]
- 54. Hogaboam CM, Bone-Larson CL, Steinhauser ML, Lukacs NW, Colletti LM, Simpson KJ, Strieter RM, Kunkel SL. Novel CXCR2-dependent liver regenerative qualities of ELR-containing CXC chemokines. FASEB J. 1999;13(12):1565–74. [DOI] [PubMed] [Google Scholar]
- 55. Liang KJ, Woodard KT, Weaver MA, Gaylor JP, Weiss ER, Samulski RJ. AAV-Nrf2 promotes protection and recovery in animal models of oxidative stress. Mol Ther. 2017;25(3):765–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nygaard S, Barzel A, Haft A, Major A, Finegold M, Kay MA, Grompe M. A universal system to select gene-modified hepatocytes in vivo. Sci Transl Med. 2016;8(342):342ra79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li N, Gou S, Wang J, Zhang Q, Huang X, Xie J, Li L, Jin Q, Ouyang Z, Chen F, et al. CRISPR/Cas9-mediated gene correction in newborn rabbits with hereditary tyrosinemia type I. Mol Ther. 2021;29(3):1001–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hinderer C, Katz N, Buza EL, Dyer C, Goode T, Bell P, Richman LK, Wilson JM. Severe toxicity in nonhuman primates and piglets following high-dose intravenous administration of an adeno-associated virus vector expressing human SMN. Hum Gene Ther. 2018;29(3):285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hong DS, Kang YK, Borad M, Sachdev J, Ejadi S, Lim HY, Brenner AJ, Park K, Lee JL, Kim TY, et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br J Cancer 2020;122(11):1630–7. [DOI] [PMC free article] [PubMed] [Google Scholar]