Significance
Long interspersed element class 1 (LINE1) elements are transposable elements that comprise ∼20% of mammalian genomes. Their activity must be tightly controlled or genome integrity will be compromised, leading to DNA damage and cell death. Here, we report that several members of a rapidly evolving X-linked homeobox gene cluster suppress LINE1 transposition. One family member of this Rhox gene cluster—Rhox10—silences LINE1 expression and transposition in the male germline when it is hypomethylated and thus highly susceptible to LINE1 activation. Rhox10 acts by driving the expression of Piwil2, which encodes a key component in the Piwi-interacting RNA pathway. The ability of Rhox genes to suppress LINE1 elements is evolutionarily conserved and perturbed by mutation in infertility patients.
Keywords: LINE1, piRNA, transposon, RHOX10, Piwil2
Abstract
Transposable elements (TEs) are mobile sequences that engender widespread mutations and thus are a major hazard that must be silenced. The most abundant active class of TEs in mammalian genomes is long interspersed element class 1 (LINE1). Here, we report that LINE1 transposition is suppressed in the male germline by transcription factors encoded by a rapidly evolving X-linked homeobox gene cluster. LINE1 transposition is repressed by many members of this RHOX transcription factor family, including those with different patterns of expression during spermatogenesis. One family member—RHOX10—suppresses LINE1 transposition during fetal development in vivo when the germline would otherwise be susceptible to LINE1 activation because of epigenetic reprogramming. We provide evidence that RHOX10 suppresses LINE transposition by inducing Piwil2, which encodes a key component in the Piwi-interacting RNA pathway that protects against TEs. The ability of RHOX transcription factors to suppress LINE1 is conserved in humans but is lost in RHOXF2 mutants from several infertile human patients, raising the possibility that loss of RHOXF2 causes human infertility by allowing uncontrolled LINE1 expression in the germline. Together, our results support a model in which the Rhox gene cluster is in an evolutionary arms race with TEs, resulting in expansion of the Rhox gene cluster to suppress TEs in different biological contexts.
Mammalian genomes are dominated by transposable elements (TEs)—parasitic genetic units that can reach copy numbers in the hundreds of thousands (1). The most abundant active class of TEs in mammals is long interspersed element class 1 (LINE1), which comprises ∼20% of mammalian genomes (2). LINE1 elements are autonomous TEs that propagate in the genome by a copy-and-paste mechanism through retrotransposition. LINE1 propagation depends on the LINE1-encoded proteins, ORF1p and ORF2p, which can also mobilize nonautonomous retrotransposons, other noncoding RNAs, and messenger RNAs, leading to the generation of processed pseudogenes (3, 4). All told, LINE1-mediated transposition has been estimated to have generated at least a third of the human genome (5).
LINE1 elements are detrimental to cells. One obvious negative consequence of LINE1 elements is they engender mutations when they undergo transposition to a new genomic site. Indeed, insertional mutagenesis elicited by LINE1 and other TEs has been shown to cause more than 65 human genetic diseases (6). In addition, the LINE1 open reading frames (ORFs) are themselves known to encode deleterious endonuclease and reverse transcriptase activities (7–9). The ultimate effect of overly active LINE1s is accumulation of DNA damage, checkpoint activation, and cell death (8, 9). Thus, limiting LINE1 expression and translocation is fundamental to genome integrity and health.
To protect against these negative effects, multiple epigenetic and RNA-mediated mechanisms have been postulated to have evolved (10). Such silencing mechanisms are particularly critical for the germline, as they reduce the transmission of TE-induced mutations to subsequent generations. A major opportunity for TEs to undergo transposition is when the male germline undergoes genome-wide demethylation (between embryonic day [E]7.5 and E12.5) in primordial germ cells (PGCs) (11). This hypomethylated state is maintained when PGCs become nonproliferative cells called prospermatogonia (ProSG, also known as gonocytes) at ∼E13.5 (12). While genome-wide DNA demethylation provides the benefit of reprogramming the genome for the next generation, it opens the door for a major hazard, as it allows an opportunity for TE activation and transposition. Thus, mechanisms have evolved to defend against TEs becoming active when PGCs and ProSG are in this hypomethylated state. For example, evidence suggests that PGCs and ProSG make use of the histone modification, H2A/H4R3me2, to suppress TE expression and protect genomic integrity (13). Another major mechanism that protects against TEs in PGCs and ProSG is the Piwi-interacting (pi) RNA pathway. This pathway is mediated by short (24 to 31 nt) noncoding RNAs called “piRNAs” that associate with members of the PIWI RNA-binding Argonaute protein family to suppress transposons and maintain germline genome integrity (14). In mice, there are three PIWI family members—PIWIL1, PIWIL2, and PIWIL4 (also known as MIWI, MILI, and MIWI2, respectively)—the latter two of which are expressed in PGCs and/or ProSG. Piwil2 is first detected in gonadal tissue at E12.5, around the PGC-to-ProSG transition (15), whereas Piwil4 is not detectable until E15.5 (16). Both PIWIL2 and PIWIL4 are critical for suppression of LINE1 elements and other TEs in ProSG, based on analysis of Piwil2- and Piwl4-knockout (KO) mice (16, 17). Both KO strains suffer from male sterility (16, 17).
In this communication, we report evidence for a new line of defense against LINE1 elements: the RHOX homeobox transcription factor family. Our laboratory identified a member of this family—Pem (Rhox5)—as an oncofetal gene expressed during normal embryogenesis and in diverse tumors (18). Later, we and others discovered that Rhox5 is part of a very large (33-member) homeobox gene cluster on the X chromosome in mice (19–23). All known members of this Rhox gene cluster are primarily expressed in the placenta and in the male and female reproductive tracts (24). By definition, all homeobox genes encode a DNA-binding homeodomain, suggesting that the RHOX proteins are transcription factors devoted to reproductive functions. Indeed, this possibility has been supported by several studies, including those showing that loss of Rhox5, Rhox8, Rhox10, or Rhox13 in mice causes spermatogenic defects (25–28). Relevant to this communication, we found that knock out of the entire Rhox cluster (either globally or specifically in germ cells) causes progressive spermatogenic decline, consistent with a defect in spermatogonial stem cells (SSCs) (26). We then demonstrated that one particular Rhox gene—Rhox10—is critical for this defect, as we found that Rhox10-null mice have the same progressive spermatogenic decline as Rhox cluster KO mice (26). Through a battery of assays, including germ cell transplantation and single-cell RNA sequencing (RNA-seq) analyses, the defect in Rhox10-null mice was pinpointed to be an inability of ProSG to efficiently undergo differentiation and thereby generate SSCs. Thus, Rhox10 functions in the “gateway cells” that drive the initiation of spermatogenesis: ProSG.
In the present study, we demonstrate that RHOX transcription factors have an unexpected role in TE defense in the germline. We initially postulated this possibility for several reasons: First, all Rhox genes are expressed in the testis and ovary (19, 24). In humans, the RHOX genes are expressed exclusively in germ cells in the testes and ovary, suggesting they have specific roles in the germline (29). Second, the Rhox genes encode DNA-binding homeobox proteins (19, 24), suggesting obvious mechanisms by which they could act on TEs. Third, Rhox genes are dramatically transcriptionally derepressed in response to DNA hypomethylation, based on many lines of evidence both in vitro and in vivo (30–35). We reasoned that this regulatory trigger would generate RHOX transcription factors precisely when needed for TE defense—during genome-wide demethylation of the germline. Consistent with this reasoning, most Rhox genes are induced in PGCs precisely when they undergo genome-wide hypomethylation (36, 37).
In this communication, we report our experiments to directly test whether RHOX transcription factors have a role in TE defense. Our experiments revealed that, indeed, several members of the mouse RHOX homeobox family are capable of suppressing LINE1 transposition. We demonstrate that one RHOX family member—RHOX10—strongly suppresses LINE1 transposition during the critical period when the germline is largely hypomethylated and thus highly susceptible to LINE1 aggression. We determined the mechanism by which this suppression occurs and then asked whether LINE1 defense extends to the human RHOX cluster. Together, our results suggest that the Rhox gene cluster is engaging in an evolutionary arms race with LINE1 elements to suppress the expression and expansion of these parasitic TEs.
Results
Rhox10 Represses LINE1 Transposition in the Male Germline.
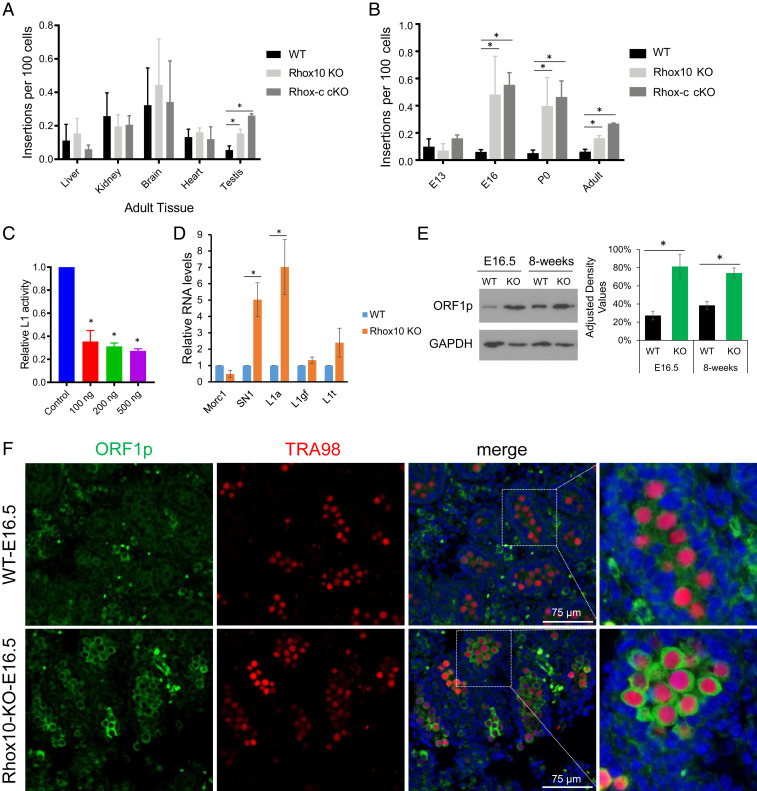
To determine whether the Rhox cluster represses LINE1 transposition, we crossed SN1 LINE1 reporter mice (38) with Rhox cluster (Rhox-c)fl/fl;Vasa-Cre mice, which delete the entire Rhox cluster in germ cells (26). We chose to use the SN1 single-copy LINE1 transgene reporter mouse line for several reasons, including that it contains codon-optimized mouse ORF1 and ORF2 for improved translation and an intron-containing split EGFP for assaying retrotransposition (38). Furthermore, unlike past LINE1 reporters (39–45), SN1 is under the control of an endogenous mouse LINE1 promoter, allowing for more physiological evaluation of LINE1 regulation (38). We used the digital droplet (dd) PCR assay to assay reporter genomic copy number. This assay showed that SN1 LINE1 reporter copy number was high in the brains of wild-type (WT) mice (Fig. 1A), consistent with previous reports that LINE1 elements undergo high rates of transposition in the brain and neural progenitor cells (42, 46, 47). ddPCR analysis of Rhox-c conditional KO (cKO) mice revealed SN1 LINE1 copy number was significantly higher in the testes from these cKO mice than control (Vasa-Cre;SN1+/−) mice, indicative of greatly increased LINE1 transposition as a result of Rhox cluster loss (Fig. 1A). In contrast, SN1 copy number was not significantly increased in other adult tissues we tested, including brain (Fig. 1A). Time-course analysis showed that these Rhox-c cKO mice exhibited a dramatic increase in SN1 copy number in testes between E13.5 and E16.5, which was sustained at postnatal (P) day 0 and adult stages (Fig. 1B). This indicated that the Rhox cluster suppresses LINE1 transposition in ProSG, as this is the only germ cell type present from ∼E13.5 to birth (12).
Fig. 1.
Rhox10 represses LINE1 transposition in the male germline. (A) ddPCR analysis of SN1 LINE1 copy number in adult tissues from SN1 reporter mice mated with Rhox cluster (c) cKO (Rhox-cfl/Y;Vasa-Cre;SN1+/−), Rhox10-null (Rhox10-/Y;SN1+/−) (KO), or WT (Vasa-Cre;SN1+/−) mice (n = 6). *P < 0.05. (B) SN1 LINE1 copy number in testes from mice of the ages indicated (adult testis is the same as in A). The genotypes and analysis are described in A (n = 3). *P < 0.05. (C) Luciferase analysis of LINE1 transposition activity in the GC1 spermatogonia cell line cotransfected with the mouse ORFeus LINE1 reporter and a Rhox10-expression vector or an empty expression vector (n = 3). *P < 0.05. (D) qPCR analysis of testes from E16.5 Rhox10-null (KO) and littermate WT mice. The primers used are specific for the indicated LINE1 subfamilies (n = 3). *P < 0.05. (E) Western blot analysis of LINE1 ORF1p protein expression in testes lysates from Rhox10-null (KO) and littermate WT mice of the indicated ages. (Right) ORF1p quantification when normalized to GAPDH (n = 3). *P < 0.05. (F) Immunofluorescence analysis of ORF1p (green) in Rhox10-null (KO) and littermate WT testes from E16.5 mice. The sections were costained with an antibody against TRA98 to detect all germ cells (red) and DAPI to detect all cells (blue). n = 3.
The Rhox cluster contains 33 homeobox genes (19, 24), any of whose loss could potentially contribute to LINE1 suppression. Since Rhox10 is expressed in the germ cell stage in which the Rhox cluster acts to suppress LINE1 transposition—ProSG (26)—we examined the role of Rhox10. To this end, we generated Rhox10-null;SN1 transgenic mice. Like Rhox-c cKO mice, these Rhox10-null mice had greatly elevated SN1 transgene copy number compared to control (SN1+/−) mice. Furthermore, both Rhox10-null and Rhox-c cKO mice had elevated SN1 transgene copy number in precisely the same contexts: E16.5, P0, and adult testes but not in E13.5 testes or nontesticular adult tissues (Fig. 1 A and B). This finding suggested that Rhox10 is largely responsible for the ability of the Rhox cluster to suppress LINE1 transposition during fetal germ cell development in vivo.
The notion that Rhox10 suppresses LINE1 transposition was validated by several lines of evidence. First, using the ORFeus dual luciferase reporter (48), we confirmed the suppressive effect of Rhox10 on LINE1 transposition in the GC1 spermatogonial cell line (Fig. 1C). Second, qPCR analysis showed that SN1 LINE1 transgene expression was elevated in Rhox10-null mice testes (Fig. 1D). Third, to examine whether regulation extends to endogenous LINE1 elements, we used primer pairs specific for several different LINE1 subfamilies. This analysis revealed that the L1a LINE1 subfamily exhibited strongly elevated expression in Rhox10-null mouse testes (Fig. 1D). Specificity was demonstrated by the finding that L1gf and L1t subfamilies did not exhibit statistically significant changes in expression (Fig. 1D). Fourth, to assess whether Rhox10 might suppress LINE1 transposition by repressing LINE1 ORF1p protein, we examined ORF1p expression and found it was significantly up-regulated in Rhox10-null mouse testes, as detected by Western blot analysis of both fetal and adult testes (Fig. 1E). Fifth, we confirmed ORF1 up-regulation using immunofluorescence analysis: at E16.5, most germ cells (TRA98+ cells) in WT testes had a very dim anti-ORF1p signal, whereas most germ cells in Rhox10-null mice testes had a strong signal (Fig. 1F). In adult WT mice, all germ cells in all seminiferous tubules examined had undetectable or weak expression of ORF1p, while in adult Rhox10-null mice, 18% of tubules (11 of 61) had germs cells with a strong ORF1p signal (SI Appendix, Fig. S1).
Evidence for a Rhox10–Piwil2–LINE1 Circuit.
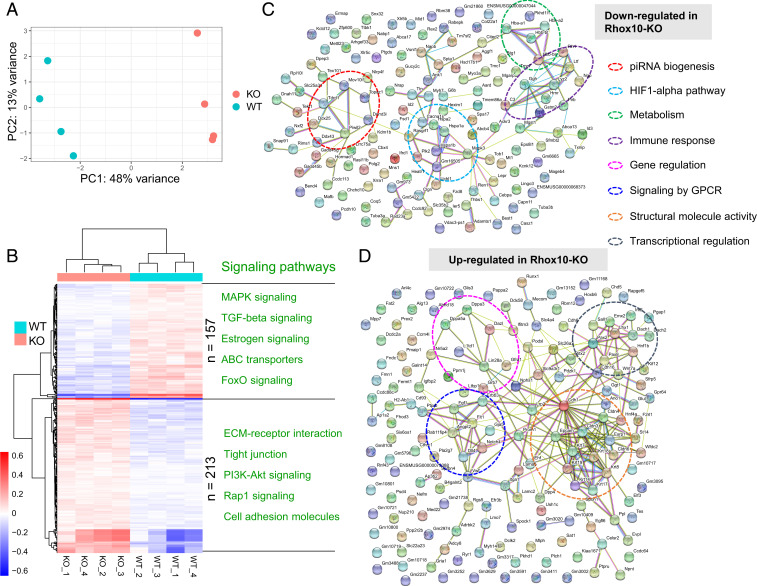
To identify candidate genes that act downstream of the RHOX10 transcription factor to suppress LINE1 elements in the male germline, we performed RNA-seq analysis on fluorescence-activated cell sorting (FACS)-purified germ cells from fetal Rhox10-null;Oct4-eGFP+/+ (KO) and littermate Oct4-eGFP+/+ (control) mice. Principal component analysis of quadruplicate replicates of Rhox10-/Y;Oct4-eGFP and Rhox10+/Y;Oct4-eGFP germ cells showed that their transcriptomes segregated separately, indicative of a significant change in gene expression elicited by loss of Rhox10 (Fig. 2A). In total, 370 genes exhibited significantly altered expression in Rhox10-null germ cells compared to control germ cells (q < 0.05, Basemean > 10, |Log2FC| > 0.25). Of these 370 differentially expressed genes (DEGs), 213 were up-regulated and 157 were down-regulated (Dataset S1). Fig. 2B shows signaling pathways associated with these DEGs.
Fig. 2.
RHOX10-regulated genes in fetal germ cells. (A) Principal component analysis plot of the RNA-seq datasets from quadruplicate testicular germ cell samples obtained from E16.5 Rhox10-null; Oct4-eGFP+/+ (KO) and Oct4-eGFP+/+ (WT) mice. (B) Heatmap of DEGs identified from the analysis of the samples in A (the data from each biological replicate are shown). Selected signaling pathways enriched in DEGs down- and up-regulated in Rhox10-null germ cells are shown in Upper and Lower, respectively. (C and D) STRING analysis of DEGs defined in B.
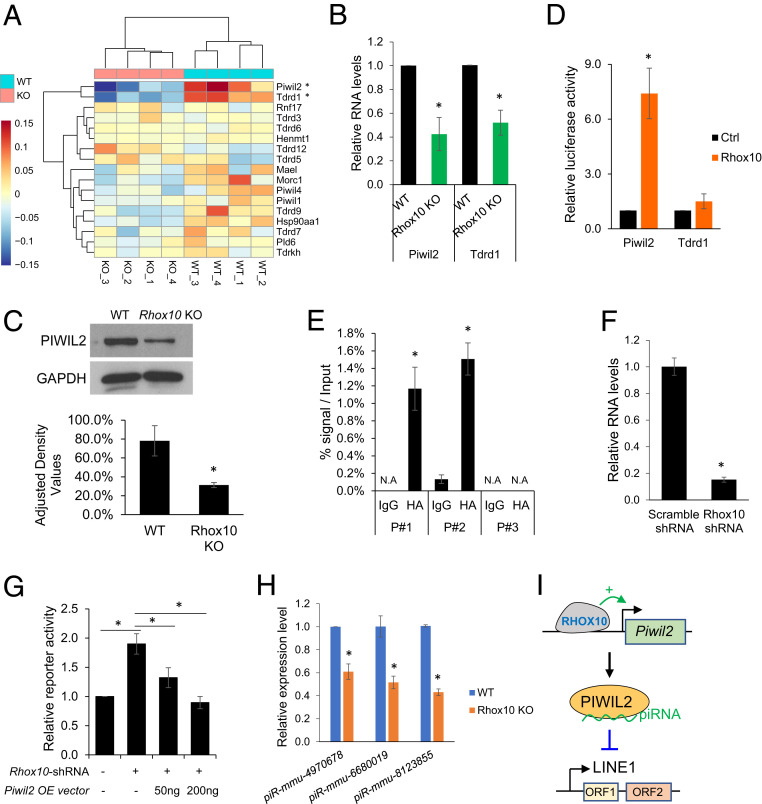
Among the functional categories associated with genes regulated by Rhox10 loss (Fig. 2 C and D) was “piRNA biogenesis.” The piRNA pathway is known to silence LINE1 elements (49, 50), which led us to postulate that Rhox10 acts through the piRNA pathway to suppress LINE1 elements. Among piRNA biogenesis genes, Piwil2 and Tdrd1 were significantly down-regulated in Rhox10-null germ cells (Fig. 3A), which we validated by qPCR analysis (Fig. 3B) and Western blot analysis for PIWIL2 (Fig. 3C). To determine whether RHOX10 might directly regulate their expression, we first employed reporter analysis. The Piwil2 and Tdrd1 promoter regions were separately cloned into a firefly luciferase reporter vector and cotransfected with a Rhox10-expression vector into HEK-293T cells. Rhox10 dramatically increased expression from the Piwil2 promoter (by ∼seven-fold), demonstrating that RHOX10 acts through the Piwil2 promoter to increase Piwil2 expression (Fig. 3D). In contrast, RHOX10 likely acts by a different mechanism to up-regulate Tdrd1, as forced Rhox10 expression did not significantly increase reporter expression from the Tdrd1 promoter.
Fig. 3.
RHOX10 suppresses LINE1 expression via regulating Piwil2 expression. (A) Heatmap of piRNA pathway gene expression determined by the RNA-seq analysis shown in Fig. 2. *, q < 0.05, Basemean > 10, |Log2FC| > 0.25. (B) qPCR validation of Rhox10-mediated regulation of Piwil2 and Tdrd1 expression in germ cells from E16.5 Rhox10-null; Oct4-eGFP+/+ (KO) and Oct4-eGFP+/+ (WT) mice (n = 3). *P < 0.05. (C, Top) Western blot analysis of PIWIL2 protein expression in testes lysates from E16.5Rhox10-null (KO) and littermate WT mice. (Bottom) PIWIL2 quantification when normalized to GAPDH (n= 3). *P< 0.05.(D) Luciferase analysis of Piwil2 or Tdrd1 promoter activity in HEK-293T cells cotransfected with a luciferase reporter vector harboring the Piwil2 or Tdrd1 promoter and a Rhox10-expression vector or an empty expression vector as a control (Ctrl) (n = 4). *P < 0.05. (E) ChIP-qPCR analysis of GS cells transfected with an expression vector encoding HA-tagged RHOX10 and analyzed using an HA antibody. Endogenous RHOX10 was knocked down using a Rhox10-shRNA lentivirus (>80% efficiency) (51). IgG antibody was used as a negative control. The values shown are the DNA signal in the immunoprecipitation samples relative to the input. (n = 3). *P < 0.05. (F) qPCR analysis of Rhox10 expression in GS cells transduced with a Rhox10-shRNA lentivirus or a scramble-shRNA lentivirus as a control (n = 2). *P < 0.05. (G) Luciferase analysis of LINE1 transposition activity in GS cells cotransfected with the ORFeus mouse LINE1 reporter, a Piwil2 expression vector, and a Rhox10-shRNA vector, as shown (n = 3). *P < 0.05. (H) TaqMan‐qPCR analysis of the indicated piRNAs in FACS-purified germ cells from E16.5 Rhox10-null; Oct4-eGFP+/+ (Rhox10 KO) and littermate Oct4-eGFP+/+ (WT) mice (n = 3). *P < 0.05. U6 small nuclear RNA levels were used for normalization. (I) Model: the RHOX10 transcription factor activates Piwil2 transcription to suppresses LINE1 elements.
The finding that the RHOX10 transcription factor strongly up-regulates Piwil2 expression in the male germline in vivo raised the possibility that RHOX10 acts in a circuit with PIWIL2 to suppress LINE1 transposition. In support of this model, we found that RHOX10 occupies the Piwil2 promoter, as shown by chromatin immunoprecipitation (ChIP) analysis (Fig. 3E). This finding, coupled with 1) the coexpression of Rhox10 and Piwil2 in fetal germ cells (16, 51, 36), 2) the ability of RHOX10 to drive expression of the Piwil2 promoter in vitro (Fig. 3D), and 3) the ability of RHOX10 to positively regulate Piwil2 in germ cells in vivo (Fig. 3 B and C), strongly suggests that RHOX10 directly drives transcription of the Piwil2 gene in fetal germ cells. To assess whether RHOX10 suppresses LINE1 expression via Piwil2, we performed a rescue experiment. In this experiment, we asked whether reduced LINE1 defense caused by RHOX10 knockdown could be rescued by adding back the depleted PIWIL2. Indeed, we found that forced expression of PIWIL2 in germline stem (GS) cells reversed the defect in LINE1 transposition caused by RHOX10 knockdown (Fig. 3F), as judged by a cotransfected dual luciferase LINE1 reporter plasmid (ORFeus reporter) (48) (Fig. 3G). Together, these data support a model in which RHOX10 directly transcriptionally activates the Piwil2 gene to provide defense against LINE1 TEs in the developing male germline (Fig. 3I).
PIWIL2 is an endonuclease that produces piRNA intermediates, leading to the formation of mature piRNAs (52). This raises the possibility that by stimulating PIWIL2 expression, RHOX10 also up-regulates the expression of piRNAs. To test this, we selected three piRNAs known to be bound to PIWIL2 in ProSG, each in a different subfamily (L1Md type A, D, and F2) (53). TaqMan analysis showed that all of these piRNAs are significantly down-regulated in FACS-purified germ cells from Rhox10-null E16.5 Oct4-eGFP+/+ mice compared to germ cells from littermate Oct4-eGFP+/+controls (Fig. 3H). These data indicate that Rhox10 promotes piRNA expression in fetal germ cells, which may contribute to Rhox10’s role in LINE1 defense.
Rhox10 Drives LINE1 Promoter DNA Methylation.
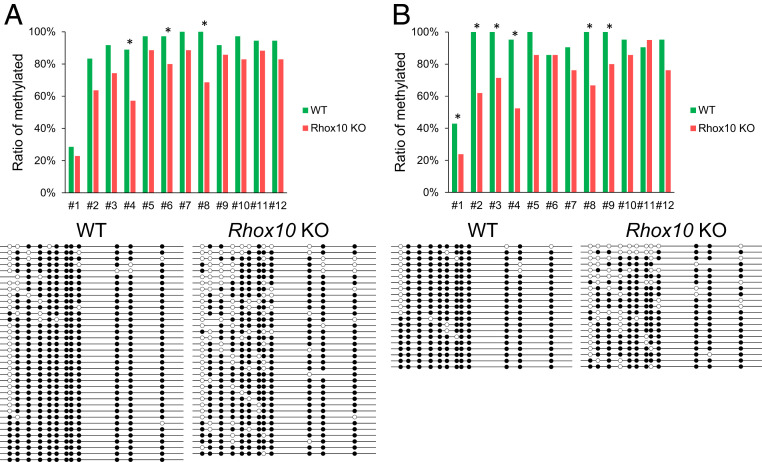
Our evidence that Rhox10 acts, at least in part, through Piwil2 to suppress LINE1 transposition predicts that Rhox10-null mice testes will have hypomethylated LINE1 promoters. This prediction follows from the considerable evidence that the piRNA pathway suppresses LINE1 expression by methylating LINE1 promoters (16, 17). To test this possibility, we performed bisulfite analysis of the SN1 transgene promoter in testes from fetal (E16.5) Rhox10-null and control mice. Among 12 CpG sites, three showed significantly decreased methylation levels in Rhox10-null testes when compared with control testes (Fig. 4A). The total DNA methylation level of all CpG sites was also significantly decreased in Rhox10-null compared to control mice (Fig. 4A). To test whether the methylation of the LINE1 promoter is regulated by Rhox10 specifically in germ cells, we purified germ cells from E16.5 testes by magnetic-activated cell sorting using epithelial cell adhesion molecule, an early germ cell marker (54, 55). Bisulfite analysis showed that 5 of 12 CpG sites in the SN1 transgene promoter exhibited significantly decreased methylation in Rhox10-null germ cells compared to control germ cells (Fig. 4B). Total methylation level was also significantly decreased (Fig. 4B). Together, these results indicated that Rhox10 drives the methylation of LINE1 promoters in germ cells.
Fig. 4.
Rhox10 drives LINE1 promoter methylation. (A, Top) The degree of methylation at each CpG site in the SN1 transgene promoter in testes from E16.5 Rhox10-null; SN1+/− (KO) and SN1+/− (WT) mice. (Bottom) Bisulfite analysis data of individual samples. *P < 0.05. (B) Bisulfite analysis, performed as in A, of purified germ cells from E16.5 Rhox10-null; SN1+/− (KO) and SN1+/− (WT) mice testes. *P < 0.05.
Many Members of the Mouse Rhox Gene Cluster Repress LINE1 Transposition.
The evidence described above indicated that mouse Rhox10 suppresses LINE1 transposition in fetal germ cells, including ProSG. Other members of the mouse Rhox cluster are expressed in other contexts, including at later stages of male germ cell development, in female germ cells, and in somatic cells in the testes and ovary (24, 56). To determine whether other Rhox cluster genes suppress LINE1 transposition, we used the ORFeus dual luciferase reporter assay (48). We cotransfected this LINE1 reporter with expression vectors encoding individual Rhox gene in NIH 3T3 cells, revealing that several of the mouse Rhox genes repress LINE1 transposition (Fig. 5A). As described in Discussion, this finding raises the possibility that individual members of the Rhox cluster coordinate LINE1 suppression across male germline development, as well as acting to suppress LINE1 elements in different cell types in the reproductive tract.
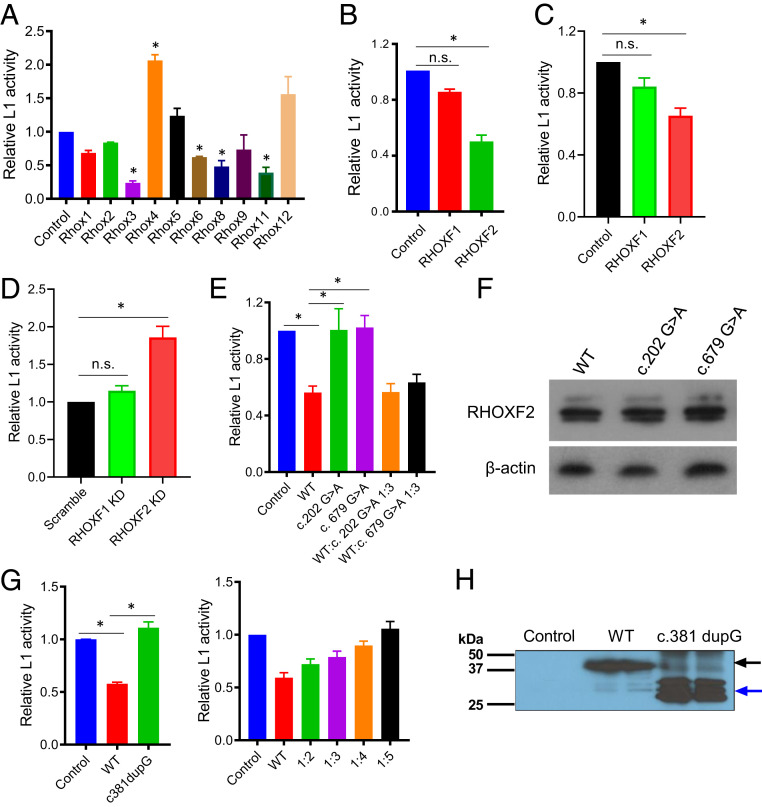
Fig. 5.
Many Rhox family members represses LINE1 transposition. (A) Luciferase analysis of LINE1 transposition activity in NIH/3T3 cells cotransfected with the mouse ORFeus LINE1 reporter and vectors expressing the indicated Rhox genes or an empty expression vector as a control (n = 3). *P < 0.05. (B) Luciferase analysis of LINE1 transposition activity in HEK-293T cells cotransfected with the human L1RP LINE1 reporter and vectors expressing the indicated human RHOX genes or an empty expression vector as a control (n = 4). *P < 0.05. (C) Luciferase analysis of LINE1 transposition activity in TCam-2 cells cotransfected with the human L1RP LINE1 reporter and vectors expressing the indicated human RHOX genes or an empty expression vector (n = 3). *P < 0.05. (D) Luciferase analysis of LINE1 transposition activity in K562 cells cotransfected with the human L1RP LINE1 reporter and the RHOXF1 or RHOXF2 shRNA vector or a randomly scrambled shRNA vector (n = 3). *P < 0.05. (E) Luciferase analysis of LINE1 transposition activity in HEK-293T cells cotransfected with the human L1RP LINE1 reporter and vectors expressing WT or mutated RHOXF2 or an empty expression vector (n = 3). *P < 0.05. (F) Western blot analysis of HEK-293T cells transiently transfected with vectors expressing WT RHOXF2 or the RHOXF2 mutants. β-actin was used for the internal control (n = 2). (G, Left) Luciferase analysis of LINE1 transposition activity in HEK-293T cells cotransfected with the human L1RP LINE1 reporter and vectors expressing WT or truncated nonfunctional RHOXF2 (c.381dupG). (Right) Luciferase analysis of LINE1 transposition activity in HEK-293T cells cotransfected with the human LINE1 (L1RP) reporter, a constant amount of WT RHOXF2, and different amount of c.381dupG RHOXF2. Cells cotransfected with the human LINE1 (L1RP) reporter and an empty expression vector serve as the control (n = 3). *P < 0.05. (H) Western blot analysis of HEK-293T cells transiently transfected with a vector expressing WT RHOXF2 or the RHOXF2 mutants shown. Cells that were transiently transfected with an empty expression vector is the control (n = 2).
Human RHOXF2 Represses LINE1 Transposition.
Rhox homeobox genes are also presents in other mammals, including humans (19, 57), raising the possibility that they encode transcription factors that suppress LINE1 elements in the male germline in humans. Humans harbor only three RHOX genes: RHOXF1, RHOXF2A, and RHOXF2B (also called hPEPP1 and hPEPP2) (58). The latter two are likely to be functionally equivalent, as they are 99.8% identical in sequence in their exons and introns, and only differ by two amino acids in their deduced protein sequence. Both the RHOXF1 and RHOXF2A/B genes are most highly expressed in testes; their encoded proteins are specifically expressed in germ, not somatic, cells in the human testes and ovary (29). To assess whether human RHOXF1 and/or RHOXF2 have LINE1 silencing activity, we cotransfected a human LINE1 reporter plasmid (L1RP reporter) (48) with individual expression vectors encoding RHOXF1 and RHOXF2. This revealed that RHOXF2, but not RHOXF1, has the ability to suppress LINE1 transposition in human HEK-293T cells (Fig. 5B) and the human germ cell line TCam-2 (Fig. 5C). To supplement these gain-of-function studies, we took a loss-of-function approach in K562 cells, which highly express RHOXF2 (58). We generated a RHOXF2-small hairpin (sh)RNA lentivirus to knockdown RHOXF2 in a K562 L1-GFP reporter line that harbors an EGFP transposition cassette inserted into the 3′ untranslated region of a LINE1 element to assay LINE1 transposition (59). Transient knockdown of RHOXF2 with this RHOXF2-shRNA lentivirus increased LINE1 transposition, as judged using this reporter (Fig. 5D). We conclude that human RHOXF2 suppresses LINE1 transposition.
RHOX Mutants from Infertility Patients Lack the Ability to Repress LINE1 Transposition.
Mutations in RHOX genes have been implicated in causing male human infertility (60). Coupled with our findings described above, this raised the possibility that RHOX mutations disrupt the ability of RHOX proteins to suppress LINE1 elements. To test this possibility, we first examined two missense RHOXF2 mutants found in patients with severe oligozoospermia (60). We cotransfected these c.202G > A and c.679G > A mutants with the L1RP reporter (48) into HEK-293T cells and found that neither repressed LINE1 transposition (Fig. 5E). These mutations did not destabilize RHOXF2 (Fig. 5F); thus, we conclude that instead they disrupt the ability of this protein to suppress LINE1 elements. We also tested a frameshift mutant that encodes a truncated RHOXF2 protein—c.381dupG (61)—and found it also lacked the ability to repress LINE1 element transposition (Fig. 5G). Because this mutant encodes a truncated protein completely lacking the homeodomain, we considered the possibility that it is a dominant-negative mutant. In support, cotransfection of a constant amount of WT RHOXF2 with increasing amounts of the c.381dupG RHOXF2 mutant lead to a progressive loss in LINE1-reporter transposition inhibition (Fig. 5G).
Discussion
The Rhox homeobox genes and their encoded proteins have been widely used as cell type–specific and stage-specific germ cell markers in both humans and mice (27, 51, 29, 62, 63). Rhox genes also are widely used to measure androgen signaling (25). Despite their usefulness as markers, little is known about the function of most Rhox genes. With only one exception (Rhox10), KO or knockdown of Rhox genes leads to either relatively subtle defects in spermatogenesis or no detectable defects at all (19, 27, 28, 64). Our discovery that many Rhox genes function in LINE1 defense provides an explanation for such weak phenotypes. Reduced LINE1 germline defense would not be expected to necessarily cause overt defects in the first generation. Instead, the selection pressure to evolve defense against TEs would likely come from the progressive accumulation of mutations in the germline over subsequent generations.
We previously reported that loss of the Rhox10 homeobox gene causes greatly impaired differentiation and migration of ProSG, leading to the formation of very few SSCs (26). Our findings reported herein indicate that Rhox10 also has a second function—LINE1 defense—which raises the possibility that two distinct Rhox10-associated regulatory circuits evolved: one to support germ cell progression and another to defend against TEs. In support of the notion that Rhox10 evolved to regulate ProSG differentiation and migration, we previously showed that Rhox10 regulates numerous differentiation and migration genes (26). However, we cannot rule out that the ProSG differentiation and migration defects observed in Rhox10-null mice are a secondary consequence of elevated LINE1 expression in ProSG. This is an interesting possibility, but if true, it is unlikely to be due to LINE1-induced mutations, as random mutations would presumably not consistently cause the stage-specific ProSG progression defect found in Rhox10-null mice (26). A more likely scenario is that the elevated LINE1 ORF1 expression we observed in Rhox10-null germ cells is responsible, as this and elevated ORF2p expression have both been found to lead to cell toxicity in other contexts (7–9). Another possibility is that LINE1 activation in Rhox10-null ProSG perturbs the epigenetic chromatin landscape in these cells, leading to impaired differentiation. This putative mechanism is supported by studies showing that aberrant TE activation perturbs the chromatin landscape (65).
Our finding that RHOX10 positively regulates Piwil2 expression is intriguing given that Piwil2 encodes a key endonuclease that silences TEs and greatly curbs LINE1 mobilization and thus is essential for germline integrity and normal spermatogenesis (14). Our findings support a model in which RHOX10 transcriptionally activates the Piwil2 gene during the PGC–ProSG transition, leading to production of PIWIL2 protein, which then acts to drive piRNA pathway-mediated suppression of LINE1 promoters and consequent reduced transposition (Fig. 3I). Several findings support this model: 1) Piwil2 is down-regulated in Rhox10-null fetal germ cells (as shown by RNA-seq analysis and verified by qPCR and Western analysis); 2) Rhox10 dramatically increases expression from the Piwil2 promoter (as demonstrated by reporter analysis); 3) RHOX10 occupies the Piwil2 promoter (as shown by ChIP analysis); 4) forced expression of PIWIL2 in a germ cell line reverses the defect in LINE1 transposition caused by RHOX10 knockdown; 5) Piwil2 exhibits enriched expression in T1-ProSG (66); 6) Rhox10 and Piwil2 have both been reported to be first detectably expressed in male PGCs on precisely the same day of fetal development: E12.5 (16, 36); and 7) piRNAs known to be bound to PIWIL2 (53) exhibit reduced expression in Rhox10-null ProSG.
Rhox10 is expressed not only in ProSG but also in spermatogonia and early spermatocytes (51), raising the possibility that it functions in LINE1 defense at these germ cell stages as well. Our results suggest that other Rhox genes might also function in LINE1 defense. For example, our in vivo LINE1 reporter analysis of Rhox10-null versus whole Rhox cluster–null mice suggested that Rhox10 is only responsible for ∼1/2 of LINE1 suppression mediated by the entire Rhox cluster in fetal germ cells. Candidates to also play a role are Rhox1, Rhox2, Rhox4, Rhox5, and Rhox7, all of which are expressed in male fetal germ cells between E12.5 and 15.5 (36, 37). We also demonstrated that mouse Rhox family members expressed in other biological contexts suppress LINE1 transposition. Interestingly, these Rhox genes—Rhox3, Rhox6, Rhox8, and Rhox11—each have a different pattern of expression in the reproductive tract (24). Rhox3 messenger (m)RNA and RHOX3 protein have a postnatal expression pattern consistent with selective expression in spermatids (67), as does Rhox11 mRNA and RHOX11 protein. This raises the possibility that these two RHOX transcription factors function in LINE1 defense in postmeiotic germ cells. Rhox6 is primarily expressed in female PGCs (36), suggesting that it might suppress LINE1 elements in the female germline. Indeed, many Rhox genes are expressed in the female germline (19). Rhox8 is expressed in Sertoli cells and is necessary for normal spermatogenesis (68), raising the possibility that Rhox8 suppresses LINE1 elements in these testicular nurse cells. Finally, we suggest the placenta is another site where Rhox genes may provide LINE1 defense. The placenta has an intimate relationship with LINE1 elements, as this organ has widely co-opted these parasitic elements for its own purposes (69, 70) and has a hypomethylated genome potentially permissive to LINE1 element mobilization (71). Given that all known Rhox genes are expressed in the placenta (19), we suggest they may function to reduce LINE1 mobilization to a manageable level to avoid overt toxicity. Consistent with this, loss of the entire Rhox cluster causes embryonic lethality (26) and mice lacking the orphan Rhox gene, Esx1 (24), exhibit an embryonic growth defect (72). Surprisingly, we obtained in vitro evidence that unlike other Rhox family members, Rhox4 promotes, rather than inhibits, LINE1 transposition. This raises the possibility that individual RHOX transcription factors have heterogenous roles in LINE1 defense, which requires future testing in vivo.
Interestingly, humans have only three RHOX genes—RHOXF1 and two almost identical RHOXF2 genes—all of which are expressed only in the male and female germline (29). Our finding that RHOXF2 represses LINE1 transposition in several different cell lines raises the possibility it also has this activity in humans in vivo at the sites where it is normally expressed: prespermatogonia in human fetal testes, spermatogonia and leptotene spermatocytes in adult male human testes, and oocytes in adult human female ovaries (29).
While several lines of evidence indicated that a PIWIL2-dependent mechanism is responsible, at least in part, for RHOX10-mediated suppression of LINE1 transposition in vivo, RHOX transcription factors may also use other mechanisms to suppress LINE1 transposition. Our in vitro reporter experiments demonstrated that several RHOX transcription factors suppress LINE1 transposition in NIH 3T3, HEK-293T, and K562 cells, despite the fact that these somatic cell lines have trace or undetectable Piwil2/PIWIL2 mRNA expression (SI Appendix, Fig. S2). This suggests that RHOX transcription factors also suppress LINE1 transposition by a PIWIL2-independent mechanism, which may work in conjunction with the PIWIL2-dependent mechanism we uncovered.
The ability of RHOX transcription factors to suppress LINE1 elements provides an explanation for the paradoxical finding that Rhox gene copy number varies greatly (by more than 10-fold) in different mammalian species, including expansion in mice compared to rats, and differential copy number in different primate species (19, 24, 57). In particular, we postulate that the Rhox gene cluster is in an evolutionary arms race with LINE1 elements, leading to Rhox cluster expansion as a means to better repress the deleterious effects of LINE1 elements, including mutagenetic effects to the germline. We suggest that the differential copy number of Rhox genes and their different expression patterns in different species reflect unique selective forces acting in different species to suppress LINE1 elements.
In conclusion, we report that a large set of DNA-binding factors encoded by the X chromosome have the potential to provide defense against TEs in both the germline and soma of mammals. It remains for future studies to determine the full extent of their repertoire, both in terms of the types of TEs targeted and the cellular contexts in which they act.
Methods
Mice.
This study was carried out in strict accordance with the guidelines of the Institutional Animal Care and Use Committee at the University of California San Diego (protocol S09160). All mice were housed under a 12-h light:12-h dark cycle and provided with food and water ad libitum. All mouse strains used for analysis were backcrossed to C57BL/6J for at least eight passages.
Mammalian Cell Culture, Plasmids, Transfections, and Luciferase Analysis.
NIH/3T3, GC1, HEK-293T, and GS cells were used for transfection. For transient transfection experiments, the coding sequences of mouse Rhox and human RHOX genes were cloned into the plasmid cloning (pc)DNA 5/FRT expression vector (Invitrogen). The Piwil2 coding sequence was cloned into pcDNA 3.1 (−) expression vector. RHOXF1 and RHOXF2 shRNA plasmids were generated by inserting small interfering RNA sequences into the pLLU2G backbone. Mouse (ORFeus) and human (L1RP) LINE1 reporter constructs were generated as previously described (48). The mutant RHOXF2 constructs were obtained from two previous studies (60, 61) and cloned into the pIRES-hrGFP expression vector. The Piwil2 and Tdrd1 promoters were separately cloned into the pGL3 vector (Promega). For a detailed description, reference SI Appendix, Materials and Methods.
ChIP Analysis.
GS cells (5 million) were cross-linked, lysed, and homogenized for chromatin preparation. Detailed protocols are described in SI Appendix, Materials and Methods.
qRT-PCR and TaqMan Assay.
Total cellular RNA was isolated as previously described (73). Reverse transcription and qPCR analysis were performed following the manufacturers’ protocol. The primers used are listed in SI Appendix, Table S1. Results were from at least three independent replicates. Statistical significance was determined using the paired Student's t test. For detailed description, reference SI Appendix, Materials and Methods.
Western Blotting Analysis.
Western blot analysis was performed as previously described (74). Quantification of the blots was performed using NIH ImageJ (1.8.0). Statistical significance was determined using the paired Student's t test. For detailed description, reference SI Appendix, Materials and Methods.
RNA-Seq Analysis.
Single testicular cells were isolated from fetal testes using a two-step enzymatic digestion protocol described previously (75). For each sample analyzed, testes from three to four fetuses were pooled. RNA-seq was performed as described previously (76). The average number of reads per sample ranged from ∼38 to 46 million. Detailed RNA-seq analyses are described in SI Appendix, Materials and Methods.
ddPCR.
ddPCR reactions were performed as previously described (38). For detailed description, reference SI Appendix, Materials and Methods.
Bisulfite Sequencing Analysis.
Cells were processed for methylation analysis using the EpiTect Plus LyseAll Bisulfite Kit (Qiagen). Bisulfite treatment of fetal DNA was conducted with the EpiTect Plus DNA Bisulfite Kit (Qiagen). PCR amplification, gel extraction, thymine and adenine cloning, and sequence analysis were performed as previously described (38). For detailed description, reference SI Appendix, Materials and Methods.
Immunofluorescence Analysis.
Immunofluorescence analysis was performed as previously described (66, 77). For a detailed description, reference SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This project would not have been initiated without the inspiration of Dr. Marcy Richardson (formally in the Obstetrics, Gynecology, and Reproductive Sciences Department at the University of California San Diego [UCSD]). We are grateful to Dr. Wenfeng An (South Dakota State University) for providing SN1 transgenic mice and Drs. Alysson Muotri and Angela Macia for providing LINE1 reporter constructs as well as intellectual input. We thank Drs. Anindita Sarkar and Nasun Hah in the F.H.G. laboratory (Salk Institute) for technical support with ddPCR analysis. We also thank Dr. Joanna Wysocka (Stanford University) for providing the K562 L1-GFP cells. Finally, we thank the UCSD Institute for Genomic Medicine for technical support and the San Diego Supercomputer Center for providing data analysis resources. This work was supported by NIH Grant R01 GM119128 (M.F.W.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2024785118/-/DCSupplemental.
Data Availability
RNA-seq data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus database (GSE160600) (78). All other study data are included in the article and/or supporting information.
References
- 1.Platt R. N. II, Vandewege M. W., Ray D. A., Mammalian transposable elements and their impacts on genome evolution. Chromosome Res. 26, 25–43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander E. S.et al. ; International Human Genome Sequencing Consortium , Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). Corrected in: Nature 412, 565 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Esnault C., Maestre J., Heidmann T., Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24, 363–367 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Perez J. L., Doucet A. J., Bucheton A., Moran J. V., Gilbert N., Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 17, 602–611 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tristan-Ramos P., et al., sRNA/L1 retrotransposition: Using siRNAs and miRNAs to expand the applications of the cell culture-based LINE-1 retrotransposition assay. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190346 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodier J. L., Kazazian H. H. Jr, Retrotransposons revisited: The restraint and rehabilitation of parasites. Cell 135, 23–35 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Tiwari B., et al., p53 directly represses human LINE1 transposons. Genes Dev. 34, 1439–1451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallace N. A., Belancio V. P., Deininger P. L., L1 mobile element expression causes multiple types of toxicity. Gene 419, 75–81 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gasior S. L., Wakeman T. P., Xu B., Deininger P. L., The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 357, 1383–1393 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deniz Ö., Frost J. M., Branco M. R., Regulation of transposable elements by DNA modifications. Nat. Rev. Genet. 20, 417–431 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Smallwood S. A., Kelsey G., De novo DNA methylation: A germ cell perspective. Trends Genet. 28, 33–42 (2012). [DOI] [PubMed] [Google Scholar]
- 12.McCarrey J. R., Toward a more precise and informative nomenclature describing fetal and neonatal male germ cells in rodents. Biol. Reprod. 89, 47 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S., et al., PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos. Mol. Cell 56, 564–579 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwasaki Y. W., Siomi M. C., Siomi H., PIWI-interacting RNA: Its biogenesis and functions. Annu. Rev. Biochem. 84, 405–433 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Kuramochi-Miyagawa S., et al., Two mouse Piwi-related genes: Miwi and Mili. Mech. Dev. 108, 121–133 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Kuramochi-Miyagawa S., et al., DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22, 908–917 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kojima-Kita K., et al., MIWI2 as an effector of DNA methylation and gene silencing in embryonic male germ cells. Cell Rep. 16, 2819–2828 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Wilkinson M. F., Kleeman J., Richards J., MacLeod C. L., A novel oncofetal gene is expressed in a stage-specific manner in murine embryonic development. Dev. Biol. 141, 451–455 (1990). [DOI] [PubMed] [Google Scholar]
- 19.Maclean J. A. II, et al., Rhox: A new homeobox gene cluster. Cell 120, 369–382 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Jackson M., et al., A murine specific expansion of the Rhox cluster involved in embryonic stem cell biology is under natural selection. BMC Genomics 7, 212 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris L., Gordon J., Blackburn C. C., Identification of a tandem duplicated array in the Rhox alpha locus on mouse chromosome X. Mamm. Genome 17, 178–187 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Wang X., Zhang J., Remarkable expansions of an X-linked reproductive homeobox gene cluster in rodent evolution. Genomics 88, 34–43 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Geyer C. B., Eddy E. M., Identification and characterization of Rhox13, a novel X-linked mouse homeobox gene. Gene 423, 194–200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacLean J. A. II, Wilkinson M. F., The Rhox genes. Reproduction 140, 195–213 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Z., et al., Androgen-induced Rhox homeobox genes modulate the expression of AR-regulated genes. Mol. Endocrinol. 24, 60–75 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song H. W., et al., The homeobox transcription factor RHOX10 drives mouse spermatogonial stem cell establishment. Cell Rep. 17, 149–164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welborn J. P., et al., Rhox8 ablation in the Sertoli cells using a tissue-specific RNAi approach results in impaired male fertility in mice. Biol. Reprod. 93, 8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Busada J. T., et al., Rhox13 is required for a quantitatively normal first wave of spermatogenesis in mice. Reproduction 152, 379–388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song H. W., et al., The RHOX homeobox gene cluster is selectively expressed in human oocytes and male germ cells. Hum. Reprod. 28, 1635–1646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasaki A. W., et al., The oncofetal gene Pem encodes a homeodomain and is regulated in primordial and pre-muscle stem cells. Mech. Dev. 34, 155–164 (1991). [DOI] [PubMed] [Google Scholar]
- 31.Oda M., et al., DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner. Genes Dev. 20, 3382–3394 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Q., Bartlett D. L., Gorry M. C., O’Malley M. E., Guo Z. S., Three epigenetic drugs up-regulate homeobox gene Rhox5 in cancer cells through overlapping and distinct molecular mechanisms. Mol. Pharmacol. 76, 1072–1081 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Maclean J. A., et al., The rhox homeobox gene cluster is imprinted and selectively targeted for regulation by histone h1 and DNA methylation. Mol. Cell. Biol. 31, 1275–1287 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richardson M. E., Bleiziffer A., Tüttelmann F., Gromoll J., Wilkinson M. F., Epigenetic regulation of the RHOX homeobox gene cluster and its association with human male infertility. Hum. Mol. Genet. 23, 12–23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maeder M. L., et al., Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat. Biotechnol. 31, 1137–1142 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daggag H., et al., The rhox homeobox gene family shows sexually dimorphic and dynamic expression during mouse embryonic gonad development. Biol. Reprod. 79, 468–474 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Pitman J. L., Lin T. P., Kleeman J. E., Erickson G. F., MacLeod C. L., Normal reproductive and macrophage function in Pem homeobox gene-deficient mice. Dev. Biol. 202, 196–214 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Newkirk S. J., et al., Intact piRNA pathway prevents L1 mobilization in male meiosis. Proc. Natl. Acad. Sci. U.S.A. 114, E5635–E5644 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ostertag E. M., et al., A mouse model of human L1 retrotransposition. Nat. Genet. 32, 655–660 (2002). [DOI] [PubMed] [Google Scholar]
- 40.An W., et al., Conditional activation of a single-copy L1 transgene in mice by Cre. Genesis 46, 373–383 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.An W., et al., Active retrotransposition by a synthetic L1 element in mice. Proc. Natl. Acad. Sci. U.S.A. 103, 18662–18667 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muotri A. R., et al., Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435, 903–910 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Babushok D. V., Ostertag E. M., Courtney C. E., Choi J. M., Kazazian H. H. Jr, L1 integration in a transgenic mouse model. Genome Res. 16, 240–250 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kano H., et al., L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 23, 1303–1312 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grandi F. C., Rosser J. M., An W., LINE-1-derived poly(A) microsatellites undergo rapid shortening and create somatic and germline mosaicism in mice. Mol. Biol. Evol. 30, 503–512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coufal N. G., et al., L1 retrotransposition in human neural progenitor cells. Nature 460, 1127–1131 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baillie J. K., et al., Somatic retrotransposition alters the genetic landscape of the human brain. Nature 479, 534–537 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie Y., Rosser J. M., Thompson T. L., Boeke J. D., An W., Characterization of L1 retrotransposition with high-throughput dual-luciferase assays. Nucleic Acids Res. 39, e16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Fazio S., et al., The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480, 259–263 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Reuter M., et al., Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480, 264–267 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Song H. W., et al., Dynamic expression pattern and subcellular localization of the Rhox10 homeobox transcription factor during early germ cell development. Reproduction 143, 611–624 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Wenda J. M., et al., Distinct roles of RNA helicases MVH and TDRD9 in PIWI slicing-triggered mammalian piRNA biogenesis and function. Dev. Cell 41, 623–637.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aravin A. A., et al., A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inoue K., Ichiyanagi K., Fukuda K., Glinka M., Sasaki H., Switching of dominant retrotransposon silencing strategies from posttranscriptional to transcriptional mechanisms during male germ-cell development in mice. PLoS Genet. 13, e1006926 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe T., Cui X., Yuan Z., Qi H., Lin H., MIWI2 targets RNAs transcribed from piRNA-dependent regions to drive DNA methylation in mouse prospermatogonia. EMBO J. 37, e95329 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee S. E., Lee S. Y., Lee K. A., Rhox in mammalian reproduction and development. Clin. Exp. Reprod. Med. 40, 107–114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niu A. L., et al., Rapid evolution and copy number variation of primate RHOXF2, an X-linked homeobox gene involved in male reproduction and possibly brain function. BMC Evol. Biol. 11, 298 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wayne C. M., MacLean J. A., Cornwall G., Wilkinson M. F., Two novel human X-linked homeobox genes, hPEPP1 and hPEPP2, selectively expressed in the testis. Gene 301, 1–11 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Ostertag E. M., Prak E. T., DeBerardinis R. J., Moran J. V., Kazazian H. H. Jr, Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 28, 1418–1423 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Borgmann J., et al., The human RHOX gene cluster: Target genes and functional analysis of gene variants in infertile men. Hum. Mol. Genet. 25, 4898–4910 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frainais C., et al., RHOXF2 gene, a new candidate gene for spermatogenesis failure. Basic Clin. Androl. 24, 3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hwang Y. S., et al., Reconstitution of prospermatogonial specification in vitro from human induced pluripotent stem cells. Nat. Commun. 11, 5656 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rao M. K., Wayne C. M., Meistrich M. L., Wilkinson M. F., Pem homeobox gene promoter sequences that direct transcription in a Sertoli cell-specific, stage-specific, and androgen-dependent manner in the testis in vivo. Mol. Endocrinol. 17, 223–233 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Takasaki N., Rankin T., Dean J., Normal gonadal development in mice lacking GPBOX, a homeobox protein expressed in germ cells at the onset of sexual dimorphism. Mol. Cell. Biol. 21, 8197–8202 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zamudio N., et al., DNA methylation restrains transposons from adopting a chromatin signature permissive for meiotic recombination. Genes Dev. 29, 1256–1270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan K., Song H. W., Wilkinson M. F., Single-cell RNAseq analysis of testicular germ and somatic cell development during the perinatal period. Development 147, dev183251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Song H. W., et al., shRNA off-target effects in vivo: Impaired endogenous siRNA expression and spermatogenic defects. PLoS One 10, e0118549 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown R. M., Davis M. G., Hayashi K., MacLean J. A., Regulated expression of Rhox8 in the mouse ovary: Evidence for the role of progesterone and RHOX5 in granulosa cells. Biol. Reprod. 88, 126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haig D., Going retro: Transposable elements, embryonic stem cells, and the mammalian placenta (retrospective on DOI 10.1002/bies.201300059). BioEssays 37, 1154 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Jangam D., Feschotte C., Betrán E., Transposable element domestication as an adaptation to evolutionary conflicts. Trends Genet. 33, 817–831 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schroeder D. I., et al., Early developmental and evolutionary origins of gene body DNA methylation patterns in mammalian placentas. PLoS Genet. 11, e1005442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Y., Behringer R. R., Esx1 is an X-chromosome-imprinted regulator of placental development and fetal growth. Nat. Genet. 20, 309–311 (1998). [DOI] [PubMed] [Google Scholar]
- 73.Ramaiah M., et al., A microRNA cluster in the Fragile-X region expressed during spermatogenesis targets FMR1. EMBO Rep. 20, e46566 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tan K., et al., Impaired imprinted X chromosome inactivation is responsible for the skewed sex ratio following in vitro fertilization. Proc. Natl. Acad. Sci. U.S.A. 113, 3197–3202 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sohni A., et al., The neonatal and adult human testis defined at the single-cell level. Cell Rep. 26, 1501–1517.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tan K., et al., Transcriptome profiling reveals signaling conditions dictating human spermatogonia fate in vitro. Proc. Natl. Acad. Sci. U.S.A. 117, 17832–17841 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tan K., et al., The role of the NMD factor UPF3B in olfactory sensory neurons. eLife 9, e57525 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tan K., Wilkinson M. F., The Rhox gene cluster suppresses germline LINE1 transposition. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE160600. Deposited 1 November 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Tan K., Wilkinson M. F., The Rhox gene cluster suppresses germline LINE1 transposition. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE160600. Deposited 1 November 2020. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus database (GSE160600) (78). All other study data are included in the article and/or supporting information.