Significance
To determine if there is a general base in the DNA synthesis reaction that deprotonates the nucleophile, we systematically removed potential hydrogen-bond acceptors of the nucleophile, the 3′-OH of the primer strand. We then characterized the activity of human DNA Pol η by kinetic, structural, and molecular dynamics simulation analyses. We found that no single or combined perturbations eliminate catalysis. Moreover, removal of two potential proton acceptors of the 3′-OH by mutating the conserved S113 to Ala and addition of 2′-F to the primer end rescued the defects of the S113A mutation alone. Our results support that there is no specific general base and the proton is prone to leave the O3′ upon activation by the three Mg2+ ions.
Keywords: polymerase, reactant alignment, electrostatic, three-Mg2+-ion, catalysis
Abstract
DNA synthesis by polymerases is essential for life. Deprotonation of the nucleophile 3′-OH is thought to be the obligatory first step in the DNA synthesis reaction. We have examined each entity surrounding the nucleophile 3′-OH in the reaction catalyzed by human DNA polymerase (Pol) η and delineated the deprotonation process by combining mutagenesis with steady-state kinetics, high-resolution structures of in crystallo reactions, and molecular dynamics simulations. The conserved S113 residue, which forms a hydrogen bond with the primer 3′-OH in the ground state, stabilizes the primer end in the active site. Mutation of S113 to alanine destabilizes primer binding and reduces the catalytic efficiency. Displacement of a water molecule that is hydrogen bonded to the 3′-OH using the 2′-OH of a ribonucleotide or 2′-F has little effect on catalysis. Moreover, combining the S113A mutation with 2′-F replacement, which removes two potential hydrogen acceptors of the 3′-OH, does not reduce the catalytic efficiency. We conclude that the proton can leave the O3′ via alternative paths, supporting the hypothesis that binding of the third Mg2+ initiates the reaction by breaking the α–β phosphodiester bond of an incoming deoxyribonucleoside triphosphate (dNTP).
DNA polymerases catalyze the incorporation of deoxyribonucleoside monophosphate (dNMP) into an existing primer after binding a deoxyribonucleoside triphosphate (dNTP) complementary to a templating base. The catalytic process is thought to begin with deprotonation of the primer 3′-OH by a general base, continue by nucleophilic attack of the α-phosphate of the dNTP leading to a new phosphodiester bond, and finish with release of pyrophosphate after its protonation by a general acid (Fig. 1A) (1). This mechanism has long been established to require two Mg2+ ions (A and B) (2). However, visualizing the reaction intermediates using time-resolved X-ray crystallography reveals little movement of the protein but a third and transiently bound Mg2+ ion necessary for the DNA synthesis reaction (3, 4). Whether the reaction is driven by the third Mg2+ ion or the nucleophile 3′-OH, the identity of a general base responsible for deprotonating the primer 3′-OH remains unclear.
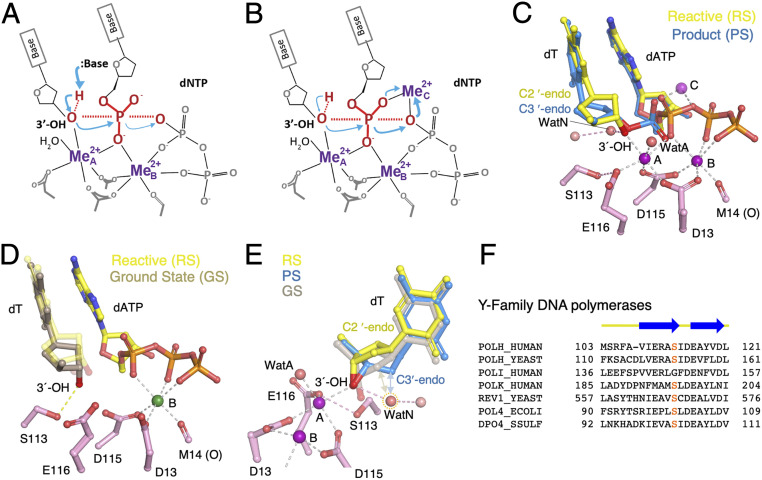
Fig. 1.
DNA synthesis reaction. (A) Deprotonation and activation of the primer-end 3′-OH for the nucleophilic attack is hypothesized to be the first step. (B) In the three–Mg2+-ion catalysis, the first step appears to break the phosphodiester bond between the α- and β-phosphates of the dNTP. (C) Structural superposition of the reaction-ready (RS) and product state of the WT enzyme in extending the dT primer. The two Mg2+ ions (A and B sites) are bound and the 3′-OH is hydrogen-bonded to the transient WatN, which in turn is linked to the bulk solvent via another water molecule. The third Mg2+ (C site) is observed only with the products. (D) In the ground state with one Ca2+ (green sphere), the 3′-OH is hydrogen-bonded with S113 and not aligned for the inline nucleophilic attack in the absence of the A-site Mg2+. The aligned 3′-OH in the RS is shown as semitransparent sticks as a reference. (E) The transient WatN (circled in orange dashes) appears only in the RS and would clash with the C2′ in the GS (wheat) and PS (blue) as indicated by double arrowheads. (F) Sequence alignment of active-site residues around S113 (highlighted in red) of human Pol η, Saccharomyces cerevisiae Pol η (yeast), human Pol ι, human Pol κ, S. cerevisiae Rev1, Escherichia coli Pol IV, and Sulfolobus solfataricus Dpo4. The conserved secondary structures are shown above the sequences.
Crystal structures of various DNA polymerases bound to DNA and dNTP, known as ternary complexes, reveal a conserved catalytic center with two Mg2+ ions coordinated by the conserved catalytic carboxylates, three phosphates of an incoming dNTP, and the primer 3′-OH (Fig. 1C) (5, 6). DNA polymerase has been likened to a right hand with the palm domain containing the catalytic residues, the finger domain closing on top of the nascent or replicating base pair, and the thumb domain binding the upstream DNA (product) duplex (7, 8). Many DNA polymerases undergo a large conformational change involving closing of the finger domain upon binding a correct dNTP (8), which was proposed to be a rate-limiting step (9). However, the rate of finger domain closing is much faster than the rate of chemical reaction (10–13). Moreover, the finger domain of the Y-family DNA polymerases is already closed even in the absence of an incoming nucleotide (14). Although intradomain motions have been implicated in one example (15), the rate-limiting step is probably the chemical reaction (3, 4, 16, 17).
If nucleophilic attack is the first step, deprotonation of the primer 3′-OH by a general base would be essential to initiate the reaction (Fig. 1A). In the more than three decades since the first DNA polymerase structure was determined (7), no residues other than the conserved carboxylates that coordinate the two Mg2+ ions have been found to eliminate the catalytic activity when mutated. Computationally, the conserved carboxylates (1, 18) and the incoming nucleotide together with the water molecule bound to the A-site Mg2+ (WatA) have each been suggested to deprotonate the 3′-OH (12, 19–21). As the carboxylates and dNTPs are necessary for Mg2+ binding and the synthesis reaction, their role in deprotonation is nearly impossible to be experimentally tested.
A third Mg2+ ion (occupying the C site) has been observed to transiently bind to dNTP in the reactions catalyzed by different DNA polymerases (3, 22–24). Unlike the A- and B-site Mg2+ ions, which are coordinated by the catalytic carboxylates, the C-site Mg2+ does not contact the enzyme at all. Its low affinity (kd) matches the minimal Mg2+ concentration required for catalysis (3, 4). When two canonical Mg2+ ions are bound and reactants are aligned for the inline nucleophilic attack, the third Mg2+ ion does not bind readily (3, 4). Only when products are formed is the third Mg2+ ion observed to bind between the product DNA and pyrophosphate with four additional water ligands. Yet without the third Mg2+, no product can form (4). Its binding is thermal energy (temperature)-dependent, and concurrent with the product formation. The third Mg2+ ion may drive dNMP incorporation by breaking the α–β phosphodiester bond of dNTP and pushing the α-phosphate toward the 3′-OH for the new bond formation (4) (Fig. 1B). With the C-site Mg2+, deprotonating the 3′-OH is likely favored and does not need a strong general base.
The in crystallo analysis of the DNA synthesis reaction catalyzed by human polymerase (Pol) η reveals three reaction states and two potential candidates to deprotonate the nucleophile. Initial dNTP binding with one divalent cation only (B site) leads to the ground state (GS) (Fig. 1D). Binding of the second Mg2+ converts the GS to the reactive state (RS), in which the 3′-OH is aligned for the inline nucleophilic attack, and appearance of the third Mg2+ (C site) is coupled with the product formation (product state; PS) (3, 4) (Fig. 1 C and E). In the GS, the 3′-OH is within 2.7 Å and hydrogen-bonded to the hydroxyl group of S113 (3). Although S113 is highly conserved among the Y-family polymerases (Fig. 1F) and may facilitate deprotonation of the 3′-OH, removal of the hydroxyl group by mutating S113 to Ala in human Pol η reduces the catalytic efficiency but does not eliminate catalysis (3). In the RS, the 3′-OH is 2.7 Å from a water molecule, which is termed WatN (N for nucleophile) and hypothesized to shuttle the proton off the 3′-OH to the bulk solvent (3). However, preliminary kinetic analysis showed that displacement of the water molecule by the 2′-OH of a ribonucleotide at the primer end leaves catalysis of DNA synthesis unaltered (3).
Here we investigate in detail how the 3′-OH is deprotonated in the reaction catalyzed by human Pol η. Combining S113A mutant Pol η (S113A) and modified nucleotides at the primer 3′ end, we measured how the altered environment of the 3′-OH perturbed the steady-state kinetics and reaction process in crystallo. In addition, static structures and dynamics simulation are employed to explain observed kinetic parameters. In contrast to the hypothesis of a specific general base, we find that the proton of the 3′-OH can depart via multiple paths.
Results
S113A Exhibits Reduced kcat and Increased KM.
In the Pol η active site, S113 is hydrogen-bonded with the 3′-OH at the primer end in the GS and, when the 3′-OH shifts toward the incoming dNTP in the RS, S113 is hydrogen-bonded with the catalytic carboxylate E116 that chelates the A-site Mg2+ ion (Fig. 1 C and D). The serine-to-alanine mutation was chosen to remove the functional group (OH), while keeping the protein structure minimally changed (3). An equivalent of S113 exists in the A- and B-family polymerases as His and Thr, respectively (SI Appendix, Fig. S1). As S113, the conserved Thr in the B-family polymerases can hydrogen-bond with the primer 3′-OH (25, 26), and the conserved His seems to insulate the active site and thereby stabilizes the primer 3′ end (27–29). Although not essential for the catalysis, these equivalent residues improve catalytic efficiencies (30, 31).
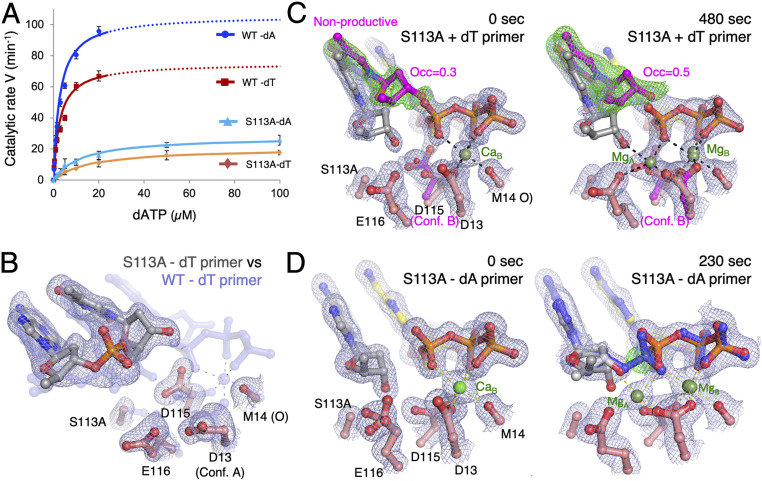
The steady-state kinetic parameters of S113A were measured for nucleotide incorporation in comparison with wild-type (WT) Pol η. Different nucleotides at the primer 3′ end have been shown to affect primer alignment and nucleotide incorporation (32). To include DNA sequence variations, both dA and dT at the primer 3′ end were examined. Previously only a dA-terminated primer (dA-primer) had been studied (3). On average, S113A lost ∼20-fold catalytic efficiency (kcat/KM) compared with WT (Fig. 2A and Table 1), with a 3- to 4-fold reduction in kcat and 4- to 6-fold increase in KM. The catalytic efficiency of the primer ending with dT was a little lower than with dA even with WT Pol η (1.4-fold), and the disparity became more pronounced with S113A (1.8-fold). Interestingly, WT Pol η had a similar KM for different primer ends but, with S113A, KM for dATP was higher with the dT- than the dA-primer. The kinetic data indicate that S113A may have a reduced ability to bind an incoming nucleotide when dT is at the primer end.
Fig. 2.
CS113 anchors the primer end. (A) Steady-state kinetic profiles of WT and S113A mutant Pol η extending primers terminating in dA or dT. Kinetic parameters for each profile are listed in Table 1. Error bars represent standard deviation of triplicate measurements. (B) Superimposed ternary-complex structures of S113A (solid multicolor) and WT (semitransparent blue) Pol η both with 0.1 mM dAMPNPP. In the S113A structure, carbon atoms of the DNA are shown in gray and of Pol η in salmon pink. 2Fo−Fc map of the S113A structure (1σ; gray) is shown as meshes. (C) 2Fo−Fc (1σ; gray) and Fo−Fc maps (3σ; green; calculated by omitting the nonproductive primer conformation) are superimposed onto the 0- (Left) and 480-s (Right) structures of the S113A Pol η–dT-primer complex. The nonproductive conformations of the primer end and D115 are shown in magenta. (D) 2Fo−Fc maps (contoured at 1σ) are superimposed on the 0-s S113A (Left) and 230-s S113A Pol η–dA-primer structure (Right). By 230 s, formation of the product (shown as blue sticks) and the new bond between the primer and dATP are evident in the green Fo−Fc map, which is calculated without the product and contoured at 3σ.
Table 1.
Kinetic data of WT and S113A Pol η
| Experiment | kcat, min−1 | KM, μM | Catalytic efficiency, μM−1⋅min−1 | Relative efficiency | |
| WT | dA | 106 ± 5.0 | 2.7 ± 0.4 | 39.3 | 1 |
| rA | 72.8 ± 2.3 | 2.0 ± 0.3 | 36.4 | 0.93 | |
| 2′F-dA | 95.1 ± 1.0 | 5.2 ± 0.2 | 18.3 | 0.47 | |
| S113A | dA | 28 ± 1.0 | 12.3 ± 1.7 | 2.3 | 1 |
| rA | 79.5 ± 3.1 | 8.4 ± 1.2 | 9.5 | 4.16 | |
| 2′F-dA | 109.2 ± 1.8 | 21.4 ± 0.9 | 5.1 | 2.24 | |
| WT | dT | 75.0 ± 6.0 | 2.7 ± 0.7 | 27.8 | 1 |
| rU | 38.1 ± 1.0 | 4.3 ± 0.4 | 8.9 | 0.32 | |
| S113A | dT | 20.7 ± 0.7 | 16.3 ± 1.5 | 1.3 | 1 |
| rU | 43.0 ± 1.6 | 7.7 ± 0.9 | 5.6 | 4.40 | |
S113A Fails to Dock the 3′ dT in the Active Site.
To understand how S113 assists dNTP binding and facilitates catalysis, we first compared the ternary-complex structures of WT and S113A Pol η with a dT-terminated primer (dT-primer). To stop the polymerization reaction, the incoming nucleotide deoxyadenosine triphosphate (dATP) was replaced by a nonreactive analog, 2′-deoxyadenosine-5′-[(α,β)-imido]triphosphate (dAMPNPP). The structure of S113A with dT at the primer end was determined at 2.27-Å resolution (SI Appendix, Methods and Table S1). WT and S113A Pol η complexed with the same DNA substrate and 1 mM dAMPNPP were indistinguishable, and the rmsd of over 431 pairs of Cα atoms was only 0.22 Å (SI Appendix, Fig. S2A). Nearly superimposable onto the RS of WT Pol η in catalysis, the static structures obtained with dAMPNPP have a slightly tighter packing around the 3′-OH (SI Appendix, Fig. S2B).
Because the KM for incoming nucleotides was higher with S113A, we determined crystal structures of WT and S113A Pol η with 0.1 mM instead of 1 mM dAMPNPP, as 0.1 mM is similar to physiological dNTP concentrations. The ternary complex of WT Pol η formed readily with 0.1 mM dAMPNPP, but S113A failed to make a ternary complex and remained as a binary Pol η–DNA complex (Fig. 2B). Without the hydrogen bond between S113 and the 3′-OH at the primer end, the last nucleotide of the primer shifted toward the dNTP-binding site. Although hydrogen-bonded with the templating base dA, the 3′ terminal dT was no longer base-stacked with the rest of the primer strand (Fig. 2B).
We further compared in crystallo catalytic processes of S113A with WT Pol η. The in crystallo reactions were carried out as previously reported (3). Briefly, crystals were grown with a normal template/primer DNA and 1 mM dATP, but reaction was prevented by substituting Mg2+ with Ca2+. After fully grown, the crystals were “washed” in a stabilization buffer containing no dATP or Ca2+ first, and the reaction was initiated by transferring the crystals to a buffer containing 1 mM Mg2+. By freezing a series of crystals at various reaction time points in liquid nitrogen, the catalytic process was visualized by X-ray diffraction of these crystals (33).
In the GS (before soaking crystals in the Mg2+ buffer), the active site of WT Pol η contains a fully occupied dATP and one Ca2+ occupying the B site coordinated by all three phosphates of dATP, while the 3′-OH at the primer end is misaligned for nucleophilic attack (Fig. 1D) (3). In contrast, before soaking in the Mg2+ buffer, only 70% of the S113A active site contained an incoming dATP (Fig. 2C), and the 3′-OH appeared to assume an RS-like conformation instead of the GS in the absence of the A-site Mg2+ (Fig. 2C). The remaining 30% of Pol η–DNA complexes were devoid of dATP because the primer ends shifted into the incoming nucleotide-binding site (Fig. 2C), as observed in the S113A–DNA binary complex formed with 0.1 mM AMPNPP (Fig. 2B). After exposure to Mg2+ for 60 s, WT Pol η starts to form products and peaks at 230 s (3). With S113A, despite 70% RS-like ternary complexes existing prior to Mg2+ exposure, no product was observed by 480 s, and instead 50% of the primer end became misaligned as dATP was partially diffused out (Fig. 2C). Removal of the hydroxyl group of S113 in Pol η resulted in defective docking of the primer 3′ end in the GS and an inactive RS-like state upon Mg2+ binding, which corresponded to the increased KM and reduced catalytic efficiency (Table 1).
Defective Primer Docking Is Partially Rescued by the 3′-End dA.
The defective docking of the primer 3′ end in the active site and absence of product formation of S113A are partially rescued by dA at the primer 3′ end. The large adenine base favors base stacking (34), and dAMP is often incorporated by DNA polymerases without a template, a phenomenon known as the “A rule” (35). The ternary-complex structure of S113A, DNA with dA at the primer end, and dATP was identical to the GS structures of WT Pol η with the dT-primer (Fig. 2D). The nonproductive primer conformation was eliminated by stable base stacking of dA. S113 may be more important for stabilizing a pyrimidine than purine due to the different intrinsic propensities of base stacking. By forming the hydrogen bond with the 3′-OH, S113 allows WT Pol η similar KM for correct incoming dNTPs regardless of whether a purine or pyrimidine is at the primer end (Table 1).
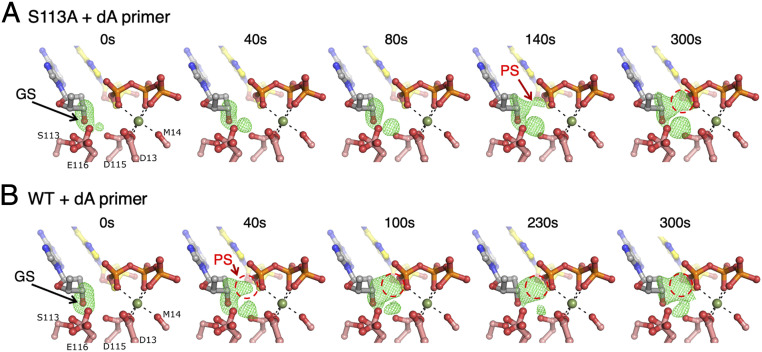
S113A was able to extend a dA-primer in crystallo (Fig. 2D), but more slowly than WT protein (Fig. 3). The Fo−Fc maps of the reaction intermediates calculated by omitting the 3′-OH and A-site Mg2+ revealed a persistent GS (50% of the ternary complexes up to 300 s) and an absence of the C2′-endo RS (Figs. 2D and 3A). Because previous in crystallo reactions were carried out with dT-primers (Fig. 1 C–E), we analyzed in crystallo extension of the dA-primers by WT Pol η (Fig. 3B). After soaking in the Mg2+ reaction buffer, products began to form as early as 40 s and accumulated to over 65% by 230 s, which is similar to the catalytic extent of dT-primers (3, 4). During the reaction course the 3′-end deoxyribose was transformed from C2′-endo in the substrate state to C3′-endo in the product state (Fig. 1E). But the distinct RS with C2′-endo and the 3′-OH aligned and hydrogen-bonded to WatN (Fig. 1 C and E) was absent, and the substrate remained in the GS with the O3′ hydrogen-bonded to S113 (Fig. 3B). As a result, the occupancy of the A-site Mg2+ was nearly constant (0.5) instead of reducing from 0.9 at 40 s to 0.5 at 300 s with the dT-primer (3, 4). We suspect that the dA-primer favors the GS and the RS is too transient to be observed.
Fig. 3.
GS-to-PS transition of a dA-primer. (A) Time course of the S113A mutant Pol η-catalyzed extension reaction of a dA-primer. (B) Time course of the WT Pol η-catalyzed extension of a dA-primer. Fo(S113A or WT)−Fc(WT, GS) maps (3.5σ) calculated with C3′ and O3′ omitted are superimposed onto the GS structure of the WT Pol η–dA–dATP complex (t = 0 s). GS and PS positions are indicated with arrows. Product density is circled in red.
While the GS persisted in the S113A–dA ternary complex after soaking in the Mg2+ reaction buffer, 30 (40 s) to 50% (300 s) of primer ends underwent conformational change from C2′-endo (GS) to C3′-endo, but only half of them were converted to product. In the C3′-endo conformation, the primer end was 3.1 to 3.2 Å from the α-phosphorus of dATP and well-aligned for the nucleophilic attack (Fig. 2D). However, the new phosphodiester bond was inexplicably slow to form (Figs. 2D and 3A), analogous to the inactive RS-like S113A–dT ternary complexes (Fig. 2C). The occupancy of the A-site Mg2+ was slightly higher with S113A than WT Pol η throughout the reaction, and the occupancy of the C-site Mg2+ was similar to the product amount as usual (4). In solution, measurement of Mg2+-concentration dependence also showed that S113A and WT are similar (SI Appendix, Fig. S3). The in crystallo analyses of S113A indicate that the absence of this conserved hydroxyl group that anchors the 3′-OH not only changes the geometry but also the reactivity of the primer end, even when seemingly well-aligned.
Slow Catalysis of S113A Is Rescued by Ribonucleotide at the Primer 3′ End.
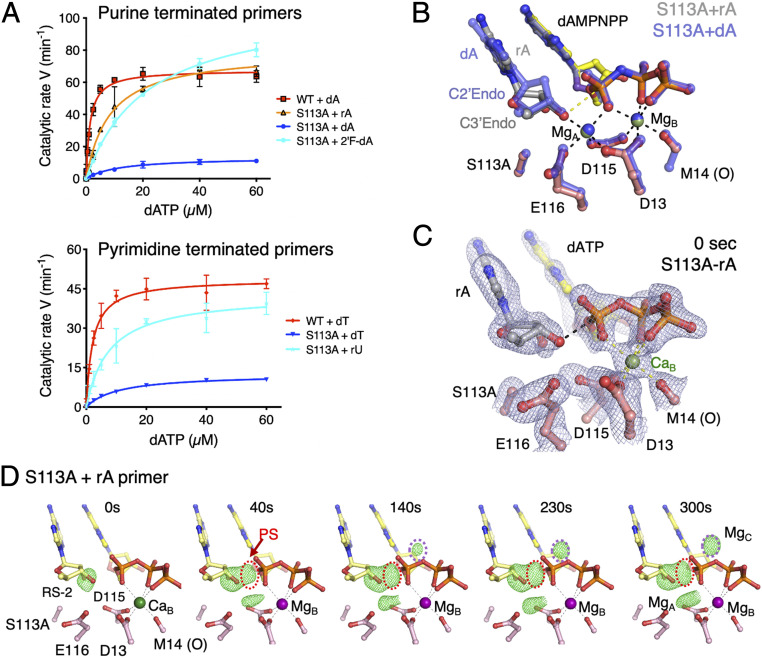
To differentiate conformation and reactivity, we examined Pol η catalysis on RNA-terminated primers. Deoxyribonucleotides predominantly adopt a C2′-endo pucker, while ribonucleotides exist in the C3′-endo conformation (36). We replaced dA with rA (adenylate) and dT with rU (uridylate) at the primer end and repeated kinetic measurements. With WT Pol η these RNA-terminated primers were extended at a slightly reduced kcat (Table 1). But with S113A, RNA-terminated primers restored the catalytic rate to the level of WT enzyme on both rU and rA primers (Fig. 4A).
Fig. 4.
RNA-terminated primer rescues the defects of S113A Pol η. (A) Steady-state kinetic profiles of WT and S113A mutant Pol η extending primers terminating in dA, rA, or 2′-fluoro-dA (Top) and dT or rU (Bottom). Kinetic parameters for each profile are listed in Table 1. Error bars represent standard deviation of triplicate measurements. (B) Superimposed structures of S113A Pol η bound to the dA- or rA-primer and dAMPNPP. The dA-primer structure is shown in semitransparent blue sticks and the rA-primer structure is shown in colored sticks (yellow dAMPNPP, salmon protein residues, and gray primer). The sugar pucker of the terminal primer base is labeled. (C) The 0-s structure of S113A Pol η bound to the rA-primer with the 2Fo−Fc map (1σ) superimposed. The rA adopts the C3′-endo conformation and is reaction-ready (RS-2) prior to Mg2+ binding in the A site. (D) The reaction time course of S113A extending the rA-primer. Reaction time course of the rA-primer extension by S113A. Fo(S113A-rA, 0–300 s)−Fc(WT, GS) maps (3.5σ) with C3′ and O3′ omitted are superimposed onto the 0-s S113A Pol η–rA-primer complex at 0 s. The electron densities for the product and Mg2+C are circled in red and purple, respectively.
To visualize the effects of ribonucleotides on the active-site configuration, we determined the crystal structure of S113A bound to an rA-terminated primer (rA-primer) with Mg2+ and dAMPNPP (SI Appendix, Table S1). The rA at the primer end adopted a C3′-endo pucker as expected, and the rest of the active site remained in the active RS conformation including the O3′ location (Fig. 4B). The reaction intermediates of S113A extending the rA-primer were also examined. Because of the C3′-endo conformation with rA at the primer end, there was no GS and the O3′ was aligned for nucleophilic attack prior to soaking in the Mg2+ buffer (Fig. 4 C and D). In addition, the phosphate of the rA adopted a singular conformation unlike those of dA and dT, which exhibited two alternate conformations (SI Appendix, Fig. S4). The in crystallo reaction took place at a rate matching the WT enzyme with the dA-primer. The occupancy of the incoming dATP was reduced at 0.85, and pyrophosphate product (PPi) was partially released, which was not observed with WT Pol η. Concurrently, both A- and C-site Mg2+ occupancies were lower than with fully occupied dATP and PPi. Nevertheless, the catalytic activity of S113A on the rA-primers approached the level of WT Pol η on DNA (Fig. 4D and Table 1). The similar catalytic efficiency of WT Pol η with a dA- or rA-primer indicates that both C3′-endo and C2′-endo sugar puckers can participate in the nucleophilic attack efficiently. However, C3′-endo at the primer end renders the A-site Mg2+ unnecessary for the 3′-OH to be in the reaction-ready state. In the absence of hydroxyl groups of S113 and WatN surrounding the O3′, the good catalytic efficiency of S113A with the rA-primer suggests that alternative proton-transfer pathways exist.
Molecular Dynamics Analysis of WT and S113A Pol η.
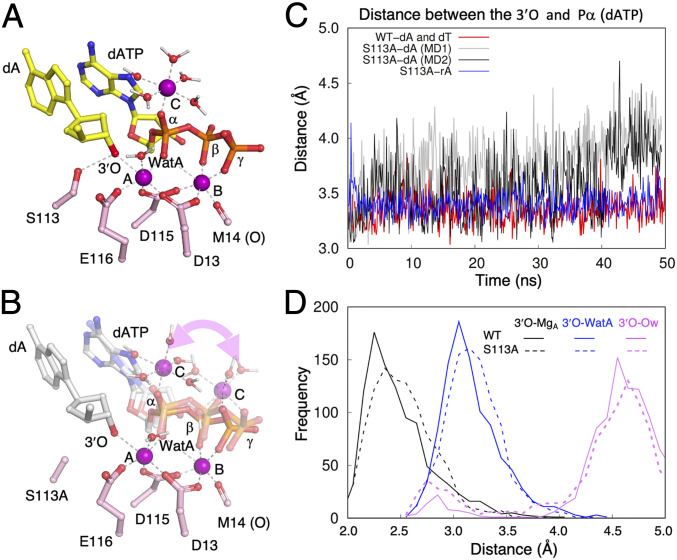
To find out why the well-aligned 3′-OH of dA- and dT-primers is slow to react with S113A, we carried out classical molecular dynamics (MD) simulations of either the C2′- or C3′-endo RS form of WT or S113A Pol η on the scale of 50 to 100 ns (SI Appendix, Methods). As the C-site Mg2+ is necessary for catalysis (4), it is placed near the α-phosphate (Pα) of dATP initially. In a large number of independent MD simulations of S113A, the C-site Mg2+ appeared easily perturbed and moved between the α- and γ-phosphates of dNTP complexed with S113A, in contrast to the stable binding to the α-phosphate (Pα) with WT Pol η prior to the product formation (Fig. 5 A and B). Only one out of seven simulations of S113A with the dA-primer led to the same binding mode as the WT enzyme. Coupled with the changes of the C-site Mg2+ binding mode, positions of the primer end and the incoming nucleotide were also perturbed, leading to a longer O3′-to-Pα distance (Fig. 5C). The observations are qualitatively consistent with the perturbed substrate binding by S113A. These results suggest that the C-site Mg2+ is important for not only product formation but also the favorable nucleophilic attack configuration, thus playing an explicit role in catalysis.
Fig. 5.
MD simulation of the WT and S113A active sites. (A) The C-site Mg2+ is stably bound to the α-phosphate of dATP in the WT active site. Three Mg2+ ions (A, B, and C) are shown as purple spheres. The primer-end dA, dATP, catalytic residues, and inner-shell water molecules of Mg2+ ions are shown as sticks, and Mg2+ coordinations are shown as dashed lines. (B) In multiple independent MD simulations of the S113A mutant, the C-site Mg2+ binds either the α- or β-γ phosphates of dATP. The alternative binding mode is indicated by the curved double arrowhead. The incoming dATP also wobbles during simulations. Two representative intermediate states are shown. (C) During dynamics simulation, the distance between the primer O3′ and Pα of dATP is stable in the WT Pol η active site (red trace) but is varied and longer in the S113A active site. Two traces of the S113A simulation are shown in gray and black. In the S113A-rA simulation, the two reactants (O3′ and Pα of dATP) are as stable (blue trace) as with WT Pol η. (D) Comparison of distances between the O3′ and Mg2+A, WatA, and surrounding water molecules in the WT (solid lines) and S113A mutant Pol η (dashed lines).
With the C-site Mg2+ bound to the α-phosphate, the active-site properties of S113A with the dA-primer were generally similar to the WT enzyme. However, subtle differences are observed in the distributions of O3′-Mg2+A and O3′-WatA, which exhibit peak distributions at slightly longer distances in S113A than in the WT enzyme (Fig. 5D). Meanwhile, removal of the hydroxyl group of S113 exposes the active site and increases the frequency of water molecules (Ow) surrounding the 3′-OH (Fig. 5D). The S113A mutation seems to perturb interactions between the primer end and both A- and C-site Mg2+, leading to either less favorable nucleophilic attack geometries or weaker polarization of the 3′-OH by the A-site Mg2+, and thus a slightly lower catalytic activity.
To understand the observed rescuing effect of the 2′-OH at the primer end, we also carried out MD simulations for S113A with the rA-primer. In contrast to the unstable C-site Mg2+ and increased distance between O3′ and Pα in the simulations of the S113A–dA complex, S113A-rA exhibited characteristics similar to WT Pol η (Fig. 5C). The results from classic MD analyses have established foundations for future in-depth analysis of catalysis of different enzyme and substrate complexes by quantum mechanics/molecular mechanics (QM/MM) studies.
Deprotonation Path Is Flexible.
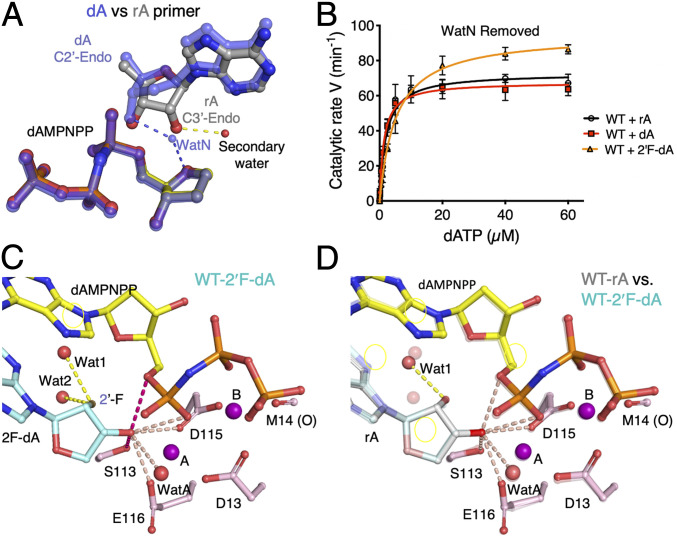
In addition to the altered sugar-pucker conformation, the 2′-OH of the rA at the primer 3′ end also displaced the transient WatN, which is hydrogen-bonded to the 3′-OH and potentially facilitates deprotonation of the nucleophile. The O2′ is competent to accept a proton from the O3′ and release it to solvent. Consistent with such a proton-transfer relay, a water molecule was observed to hydrogen-bond with the O2′ in the ternary complex with an RNA-terminated primer and dAMPNPP (Fig. 6A). The near-normal primer extension on the RNA-terminated primers by WT and S113A Pol η indicates that any role WatN may have in the nucleophile deprotonation can be replaced by the O2′.
Fig. 6.
Deprotonation route is flexible. (A) Structural superposition of the dAMPNPP-bound WT Pol η with dA- (semitransparent blue sticks) and rA- (gray and yellow sticks) primers. The transient water associated with the 3′-OH of dA is shown. The secondary water of the rA structure is shown as a red sphere. Hydrogen bonds between the terminal primer sugar hydroxyl groups and the water molecules are shown as dashes. (B) Steady-state kinetic profiles of WT Pol η extending primers terminating in dA, rA, or 2′F-dA. Kinetic parameters for each profile are listed in Table 1. Error bars represent standard deviation of triplicate measurements. (C) Possible 3′-OH deprotonation routes. The structure of WT Pol η bound to the 2′F-dA–terminated primer is shown. The six atoms within 3.1 Å of the 3′-OH of 2′F-dA are marked by pink dashed lines. Two (the hydroxyl of S113 and O5′ of dAMPNPP) are highlighted in hot pink because these distances are 0.1 to 0.15 Å shorter in the 2′F-dA complex than those in the rA complex. (D) The structure of the rA-primer is superimposed on 2′F-dA (semitransparent). The two are nearly identical. Two water molecules (Wat1 and Wat2) are within hydrogen-bond distance of the 2′-F. Only Wat1 is bound to the 2′-OH of rA.
To test if the 2′-OH of a ribonucleotide is required for deprotonation of the 3′-OH, we replaced rA with 2′-fluoro-dATP (2′F-dA) at the primer 3′ end. Unlike a hydroxyl group, fluorine attached to C2′ would displace the transient water but is unlikely to serve as a proton acceptor. To our surprise, the 2′F-dA–terminated primer was extended by WT enzyme as efficiently as normal DNA. The 2′F-dA even rescued the defects of S113A and improved its kcat and keff to near-WT levels (Fig. 4A and Table 1). In the absence of the two most obvious suspects, S113 and the transient WatN (displaced by 2′-F), another path of 3′-OH deprotonation is needed to facilitate the DNA synthesis reaction.
To identify an alternative proton acceptor or relay path, the structure of WT Pol η bound to the 2′F-dA–terminated primer with Mg2+ and dAMPNPP in the RS-like state was determined at 1.31-Å resolution (SI Appendix, Table S1). At this resolution, weakly bound groups and solvent were observable. Six potential proton acceptors of the O3′ were found, all of which were within hydrogen-bond distance (3.1 Å). They are S113 (OH), D115 (Oδ), E116 (Oε), αO-proRp, and O5′ of dAMPNPP, and a water molecule that chelates the A-site Mg2+ (WatA) (Figs. 1 C–E and 6C). As the αO-proRp of dAMPNPP coordinates the A-site Mg2+ and has an extremely low intrinsic pKa value, it is an unlikely proton acceptor (37). The carboxylates D115 and E116 also directly coordinate the A-site Mg2+ and thus also have lowered pKa values. Although the residue equivalent to Pol η’s E116 in DNA Pol β has been suggested to be the proton acceptor during polymerase catalysis (18, 22), the loss of catalytic activity upon mutating the carboxylate can be due to defective A-site Mg2+ binding instead.
Aside from the three Mg2+ ligands and S113, only two candidates remain. Firstly, the O5′ of an incoming nucleotide may take the proton and pass it to the bulk solvent. Lastly, WatA that coordinates the A-site Mg2+ ion is proximal to the O3′ and the only nonmacromolecule chelators of the A- and B-site Mg2+ ions. WatA has been previously proposed to directly accept the O3′ proton in the Dpo4 and Pol β catalysis using QM/MM simulations (19–21). WatA is usually more than 3.3 Å away from the 3′-OH in all in crystallo reaction time courses but is 3.05 Å away in the static 2′F-dA and rA structures (Fig. 6 C and D) and ∼3.1 Å in MD simulations (Fig. 5D). In these two comparable structures, only S113 (OH) and the O5′ of dAMPNPP became closer to the O3′ with 2′F-dA than with rA, while the other four maintained the same distance (Fig. 6D). The last possible candidates are two water molecules hydrogen-bonded to the 2′-F (Fig. 6C). Although fluorine is not known to be a proton acceptor, it may be a stepping stone to pass the proton from the 3′-OH to the bulk solvent.
Conclusion
Our combined structural, kinetic, and computational analyses suggest that human Pol η does not rely on a specific proton pathway for catalysis. The conserved S113 plays an important role in the primer 3′-end docking and influencing the A- and C-site Mg2+ and incoming dNTP binding. The hydroxyl group of S113, WatN transiently bound to the 3′-OH, WatA of the A-site Mg2+, and O5′ of the incoming dNTP each may be a proton acceptor and shuttle it out to the bulk solvent. In agreement with previous experimental and computational studies, neither the proton acceptor nor the departure path of the nucleophile deprotonation is fixed. When a convenient or preferred path is eliminated, multiple alternatives, including water molecules (38), a protein side chain, or incoming dNTP (12), may aid the proton to depart from the O3′. Such a flexible deprotonation mechanism is consistent with the three–metal-ion catalysis, by which the C-site Mg2+ alters the electrostatic environment around the α-phosphate and 3′-OH and effectively lowers the energy barrier to product formation (39) (Fig. 1B). Rather than requiring a specific general base to deprotonate the 3′-OH and initiate the nucleophilic attack, the Mg2+ likely drives the reaction forward and promotes dissociation of the proton.
Materials and Methods
Pol η 1 to 431 amino acids was cloned, expressed, and purified as described previously (40). Oligos were obtained from Integrated DNA Technologies or the Food and Drug Administration Facility for Biotechnology Resources. The oligo sequences are listed in SI Appendix, Table S2. Kinetic measurements, X-ray crystallographic analyses, and MD simulations were carried out according to established protocols. Please see details in SI Appendix.
Supplementary Material
Acknowledgments
This research was supported by National Institute of Diabetes and Digestive and Kidney Diseases Intramural Grant DK036146 (to W.Y.), Welch Foundation Grant C-2033-20200401 (to Y.G.), and NIH Grant 1R01GM106443 (to Q.C.). Y.G. is a Cancer Prevention and Research Institute of Texas Scholar in Cancer Research. The computational work was performed on the Shared Computing Cluster administered by Boston University’s Research Computing Services (http://www.bu.edu/tech/support/research/).
Footnotes
Competing interest statement: W.Y. and M.-D.T. are coauthors on an article [W.-J. Wu, W. Yang, M.-D. Tsai. Nat. Rev. Chem. 10.1038/s41570-017-0068 (2017)].
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103990118/-/DCSupplemental.
Data Availability
The structure coordinates and structure factors reported in this article have been deposited in the Protein Data Bank (ID codes 7M7L–7M7U, 7M7Y, 7M7Z, 7M8A–7M8D, 7M80–7M89; SI Appendix, Table S1).
References
- 1.Castro C., et al., Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 104, 4267–4272 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steitz T. A., DNA polymerases: Structural diversity and common mechanisms. J. Biol. Chem. 274, 17395–17398 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Nakamura T., Zhao Y., Yamagata Y., Hua Y.-J., Yang W., Watching DNA polymerase η make a phosphodiester bond. Nature 487, 196–201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao Y., Yang W., Capture of a third Mg2+ is essential for catalyzing DNA synthesis. Science 352, 1334–1337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang W., Lee J. Y., Nowotny M., Making and breaking nucleic acids: Two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Yang W., Gao Y., Translesion and repair DNA polymerases: Diverse structure and mechanism. Annu. Rev. Biochem. 87, 239–261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A., Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature 313, 762–766 (1985). [DOI] [PubMed] [Google Scholar]
- 8.Doublié S., Sawaya M. R., Ellenberger T., An open and closed case for all polymerases. Structure 7, R31–R35 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Wong I., Patel S. S., Johnson K. A., An induced-fit kinetic mechanism for DNA replication fidelity: Direct measurement by single-turnover kinetics. Biochemistry 30, 526–537 (1991). [DOI] [PubMed] [Google Scholar]
- 10.Arndt J. W., et al., Insight into the catalytic mechanism of DNA polymerase beta: Structures of intermediate complexes. Biochemistry 40, 5368–5375 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Rothwell P. J., Mitaksov V., Waksman G., Motions of the fingers subdomain of klentaq1 are fast and not rate limiting: Implications for the molecular basis of fidelity in DNA polymerases. Mol. Cell 19, 345–355 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Wang L., Yu X., Hu P., Broyde S., Zhang Y., A water-mediated and substrate-assisted catalytic mechanism for Sulfolobus solfataricus DNA polymerase IV. J. Am. Chem. Soc. 129, 4731–4737 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu C., Maxwell B. A., Brown J. A., Zhang L., Suo Z., Global conformational dynamics of a Y-family DNA polymerase during catalysis. PLoS Biol. 7, e1000225 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang W., Woodgate R., What a difference a decade makes: Insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 15591–15598 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raper A. T., Suo Z., Investigation of intradomain motions of a Y-family DNA polymerase during substrate binding and catalysis. Biochemistry 55, 5832–5844 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah A. M., Li S. X., Anderson K. S., Sweasy J. B., Y265H mutator mutant of DNA polymerase beta. Proper teometric alignment is critical for fidelity. J. Biol. Chem. 276, 10824–10831 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Showalter A. K., Tsai M. D., A reexamination of the nucleotide incorporation fidelity of DNA polymerases. Biochemistry 41, 10571–10576 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Batra V. K., et al., Amino acid substitution in the active site of DNA polymerase β explains the energy barrier of the nucleotidyl transfer reaction. J. Am. Chem. Soc. 135, 8078–8088 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alberts I. L., Wang Y., Schlick T., DNA polymerase beta catalysis: Are different mechanisms possible? J. Am. Chem. Soc. 129, 11100–11110 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Wang Y., Schlick T., Quantum mechanics/molecular mechanics investigation of the chemical reaction in Dpo4 reveals water-dependent pathways and requirements for active site reorganization. J. Am. Chem. Soc. 130, 13240–13250 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roston D., Demapan D., Cui Q., Extensive free-energy simulations identify water as the base in nucleotide addition by DNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 116, 25048–25056 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freudenthal B. D., Beard W. A., Shock D. D., Wilson S. H., Observing a DNA polymerase choose right from wrong. Cell 154, 157–168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vyas R., Reed A. J., Tokarsky E. J., Suo Z., Viewing human DNA polymerase β faithfully and unfaithfully bypass an oxidative lesion by time-dependent crystallography. J. Am. Chem. Soc. 137, 5225–5230 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jamsen J. A., et al., Time-lapse crystallography snapshots of a double-strand break repair polymerase in action. Nat. Commun. 8, 253 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia S., Konigsberg W. H., RB69 DNA polymerase structure, kinetics, and fidelity. Biochemistry 53, 2752–2767 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang F., Yang W., Structural insight into translesion synthesis by DNA Pol II. Cell 139, 1279–1289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doublié S., Tabor S., Long A. M., Richardson C. C., Ellenberger T., Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature 391, 251–258 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Johnson S. J., Taylor J. S., Beese L. S., Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc. Natl. Acad. Sci. U.S.A. 100, 3895–3900 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee Y. S., Gao Y., Yang W., How a homolog of high-fidelity replicases conducts mutagenic DNA synthesis. Nat. Struct. Mol. Biol. 22, 298–303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zakharova E., Wang J., Konigsberg W., The activity of selected RB69 DNA polymerase mutants can be restored by manganese ions: The existence of alternative metal ion ligands used during the polymerization cycle. Biochemistry 43, 6587–6595 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Singh K., Modak M. J., Presence of 18-Å long hydrogen bond track in the active site of Escherichia coli DNA polymerase I (Klenow fragment). Its requirement in the stabilization of enzyme-template-primer complex. J. Biol. Chem. 278, 11289–11302 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y., et al., Mechanism of somatic hypermutation at the WA motif by human DNA polymerase η. Proc. Natl. Acad. Sci. U.S.A. 110, 8146–8151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samara N. L., Gao Y., Wu J., Yang W., Detection of reaction intermediates in Mg2+-dependent DNA synthesis and RNA degradation by time-resolved X-ray crystallography. Methods Enzymol. 592, 283–327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sponer J., Leszczynski J., Hobza P., Nature of nucleic acid-base stacking: Nonempirical ab initio and empirical potential characterization of 10 stacked base dimers. Comparison of stacked and H-bonded base pairs. J. Phys. Chem. 100, 5590–5596 (1996). [Google Scholar]
- 35.Strauss B. S., The ‘A rule’ of mutagen specificity: A consequence of DNA polymerase bypass of non-instructional lesions? BioEssays 13, 79–84 (1991). [DOI] [PubMed] [Google Scholar]
- 36.Dickerson R. E., et al., The anatomy of A-, B-, and Z-DNA. Science 216, 475–485 (1982). [DOI] [PubMed] [Google Scholar]
- 37.McElroy W. D., Glass B., Eds., Phosphorus Metabolism: A Symposium on the Role of Phosphorus in the Metabolism of Plants and Animals (Johns Hopkins Press, Baltimore, MD, 1951). [Google Scholar]
- 38.Qin Y., et al., Direct probing of solvent accessibility and mobility at the binding interface of polymerase (Dpo4)-DNA complex. J. Phys. Chem. A 117, 13926–13934 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang W., Weng P. J., Gao Y., A new paradigm of DNA synthesis: Three-metal-ion catalysis. Cell Biosci. 6, 51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biertümpfel C., et al., Structure and mechanism of human DNA polymerase η. Nature 465, 1044–1048 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structure coordinates and structure factors reported in this article have been deposited in the Protein Data Bank (ID codes 7M7L–7M7U, 7M7Y, 7M7Z, 7M8A–7M8D, 7M80–7M89; SI Appendix, Table S1).