Significance
Split fluorescent protein (FP) systems have been used to endogenously tag proteins in human cells. However, there have been a limited number of studies to evaluate the potential of multicolor FP11 tags in organisms. Here, we implement the approach for FP11 tagging proteins in Drosophila. We show the use of this approach in creating protein trap lines using MiMIC insertions and enhancing signals through a tandem array of tags. While split GFP has been practical, the availability of a second split FP enables multicolor visualization. Through bioengineering, we develop variants and show two-color FP11 tagging, revealing the differential distribution of proteins in synapses. This approach is advantageous to examine the endogenous localization patterns of multiple proteins in particular cell types.
Keywords: Drosophila, fluorescent protein, fluorescence microscopy, tissue, neuron
Abstract
The impact of the Drosophila experimental system on studies of modern biology cannot be understated. The ability to tag endogenously expressed proteins is essential to maximize the use of this model organism. Here, we describe a method for labeling endogenous proteins with self-complementing split fluorescent proteins (split FPs) in a cell-type–specific manner in Drosophila. A short fragment of an FP coding sequence is inserted into a specific genomic locus while the remainder of the FP is expressed using an available GAL4 driver line. In consequence, complementation fluorescence allows examination of protein localization in particular cells. Besides, when inserting tandem repeats of the short FP fragment at the same genomic locus, we can substantially enhance the fluorescence signal. The enhanced signal is of great value in live-cell imaging at the subcellular level. We can also accomplish a multicolor labeling system with orthogonal split FPs. However, other orthogonal split FPs do not function for in vivo imaging besides split GFP. Through protein engineering and in vivo functional studies, we report a red split FP that we can use for duplexed visualization of endogenous proteins in intricate Drosophila tissues. Using the two orthogonal split FP systems, we have simultaneously imaged proteins that reside in distinct subsynaptic compartments. Our approach allows us to study the proximity between and localization of multiple proteins endogenously expressed in essentially any cell type in Drosophila.
Multicolor fluorescence microscopy provides a powerful means to investigate differential gene expression, protein–protein interactions, or biochemical activities. Engineering a broad color range of fluorescent proteins (FPs) has empowered fluorescence microscopy in the fields of cell and developmental biology (1, 2). The use of FP tags to track and measure the properties of proteins is valuable. By watching proteins’ behavior at high spatiotemporal resolution in living cells, we can gain mechanistic insights into protein trafficking, macromolecular assembly, and other biological events that involve dynamic interactions between proteins (3). When visualizing proteins moving inside cells, we can use traditional transfection and transgenic technologies to express FP-tagged proteins in experimental systems. However, these are subject to limitations due to the significant potential for overexpression artifacts (e.g., protein mislocalization and aggregation and aberrant organelle morphology) (4–6). These artifacts can be averted by using endogenous gene tagging techniques (7), which can be applied to introduce an FP coding sequence into a desired genomic locus. Consequently, the fluorescent fusion proteins are expressed under the control of endogenous regulatory elements. For some genes with low expression levels, however, the fluorescence of FPs is too dim to visualize (8). This limitation has driven researchers to find brighter fluorescent probes.
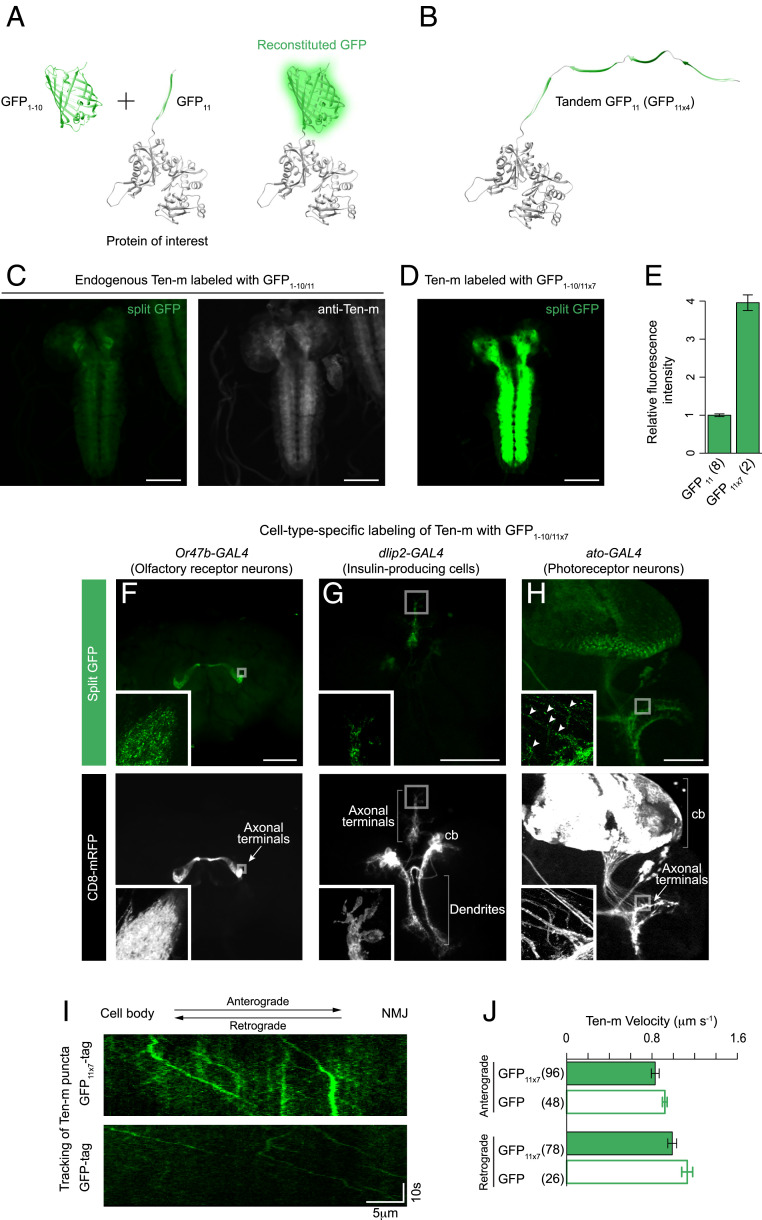
Similar to full-length FPs, split FPs have been widely used for a myriad of applications [e.g., the bimolecular fluorescence complementation assay (9–11), biosensors (12, 13), and optogenetic tools (14)]. In recent years, the self-complementing split GFP1–10/11 system has become an essential tool in live-cell protein imaging (13, 15–17). In this system, the sequence encoding for superfolder (sf) GFP is split between β-strands 10 and 11 (GFP1–10 and GFP11, respectively) (18, 19). The GFP11 fragment is a short 16-residue peptide that constitutes a small protein tag (Fig. 1A). GFP11 tag is fluorescent only when the corresponding GFP1–10 is expressed in the same cell because neither GFP1–10 nor GFP11 by itself produces visible fluorescence. We previously applied the split GFP system to generate a human cell library of fluorescently labeled endogenous proteins via genetic knock-in by CRISPR-mediated homology-directed repair (20). Because the size of GFP11 is small, it improves the knock-in efficiency (21) and simplifies the donor preparation (17, 20). An additional advantage of this system is that the GFP11 fragment can be tandemly linked, providing a multimerization scaffold to recruit multiple GFP1–10 molecules (Fig. 1B) and significantly enhance the fluorescent signal (17, 22). We have shown that tagging Lamin A/C with four repeats of GFP11 (GFP11 × 4) in human cells leads to a fourfold increase in fluorescence intensity compared to GFP11 (20). The enhanced signal can be of great value in imaging molecules difficult to study because of low signal (e.g., less abundant proteins).
Fig. 1.
Labeling endogenously expressed proteins in subsets of neurons. (A and B) Schematic for split GFP labeling of a cellular protein. A protein of interest is labeled with the GFP11 fragment. It becomes fluorescent when the GFP1–10 fragment is expressed in the same cell. In this diagram, the N terminus of β-actin is fused with GFP11 or four copies of GFP11. (C and D) For the visualization of Ten-m, we crossed either the GFP11 or the GFP11 × 7 strain with a panneuronal expression line of GFP1–10. The distribution pattern of Ten-m tagged with GFP11 or GFP11 × 7 is indistinguishable from Ten-m detected with anti–Ten-m. Identical acquisition settings were used for the split GFP images for comparison. (E) Quantification of relative fluorescence intensity of Ten-m labeled with GFP11 or GFP11 × 7, measured by confocal microscopy. n = 2 to 8 larval brains. Error bars are SEM. (F–H) Cell-type–specific labeling of Ten-m proteins. Representative images of adult fly and larval brains showing cell-type–specific fluorescence of Ten-m (Top) and a CD8 membrane marker (Bottom). We crossed the GFP11 × 7 protein trap line of ten-m with different expression lines for GFP1–10. The following GAL4 lines were used to drive UAS-GFP1–10 expression: (F) Or47b-GAL4 (ORN), (G) dlip2-GAL4 (IPCs), and (H) ato-GAL4 (photoreceptor neurons). (H, Inset) Arrowheads mark Ten-m puncta along the axon shafts. (I) Tracking of Ten-m tagged with either GFP11 × 7 (with an expression of GFP1–10 driven by elav-GAL4; Top) or full-length EGFP (Bottom) in larval motor axons (see also Movie S2). Kymograph displays the trajectory of Ten-m puncta. (J) Quantifying the anterograde and retrograde velocity of Ten-m labeled with GFP11 × 7 or full-length EGFP. The number of puncta that we analyzed in each case is indicated in the figure. Error bars are SEM. All images in this and subsequent figures are maximum Z-projections of confocal images. Insets show a higher magnification of boxed regions (single Z-sections). cb, cell bodies. (Scale bars, 100 μm in C through F.)
Drosophila is a valuable and widely used model organism because of the low cost, short generation time, and state-of-the-art genetic tools. Over the past decades, many approaches for endogenous gene tagging in Drosophila have been developed: Minos-mediated integration cassette (MiMIC) insertions and site-specific recombination through cassette exchange (23, 24); TALEN- and CRISPR/Cas9-mediated homologous recombination (25–27); and transposon-based protein traps (28, 29). As a proof-of-concept study, we previously demonstrated genetic knock-in through homologous recombination using a GFP11 donor template (30). We generated a GFP11 knock-in allele of kinesin-1 heavy chain (khc) and expressed GFP1–10 in the dendritic arborization sensory neurons to visualize the localization of Khc. In this report, we set out to establish the GFP11 tag as a general means for labeling endogenous proteins in Drosophila in cell-type–specific and developmental stage–specific patterns. Despite the usefulness of split GFP, there lacks a second split FP1–10/11 for multicolor imaging in Drosophila. Using a screening strategy to engineer self-associating split FPs, we have generated two additional variants. Here, we explore the properties of a red-colored split FP system in cultured cells and flies and demonstrate its utility for duplexed labeling and multicolor imaging of endogenous proteins in Drosophila in vivo.
Results
Cell-Type–Specific Labeling of Cellular Proteins with GFP11 Tag in Drosophila.
To investigate whether the GFP11 fragment can be used as a protein tag to a wide range of cellular proteins in Drosophila cells, we prepared Drosophila expression plasmids encoding five different proteins tagged with GFP11: β-actin (cytoskeleton), α-tubulin (cytoskeleton), histone H3.3A (nucleus), Centrosomin (centrosome), and β2 adrenergic receptors (plasma membrane; GFP11 was fused to β2-AR at the cytoplasmic tail). Each fusion protein was constructed for GFP11 to target its N or C terminus with an appropriate-length linker. The lengths of linkers vary from 0 to 18 amino acids (aa). We could record fluorescence images under confocal microscopy, coexpressing GFP11-containing fusions with cytoplasmic GFP1–10 in cultured Drosophila S2 cells. In contrast, no specific signal was detected with GFP1–10 or GFP11 alone. For all fusions we tested, we obtained bright fluorescence signals reconstituted from split GFP at the predicted subcellular location (SI Appendix, Fig. S1A). For example, S2 cells transfected with split GFP-tagged β-actin exhibit the reconstituted signal in the lamellipodia (SI Appendix, Fig. S1A). Consistent with our observation, Rogers et al. have previously shown a similar subcellular localization of actin fused to full-length GFP (31). Altogether, these results demonstrate that GFP11 efficiently performs as a fusion protein when expressed in S2 cells.
To exploit GFP11 tag for labeling a protein that resides in its target organelle or a compartment inside the organelle, we should carefully consider whether the spatial constraint of a GFP11-tagged protein causes inaccessibility to cytoplasmic GFP1–10, which can result in a lack of complemented GFP signal. Indeed, we have demonstrated that GFP11 targeted to the mitochondrial matrix fails to reconstitute a fluorescence signal when coexpressed with cytoplasmic GFP1–10 (SI Appendix, Fig. S1B). In contrast, mitochondrial matrix-targeted GFP1–10 (GFP1–10mitochondorial matrix) gives rise to complementation with the same GFP11 construct, and the reconstituted signal exhibits distinct localization to the matrix of mitochondria (SI Appendix, Fig. S1C). We expect that as long as both fragments are targeted to the same cellular compartments, complementation fluorescence would be detected in any particular organelle of Drosophila cells. Of note, the split GFP system has previously been adapted to visualize protein localization within the Golgi (32), the endoplasmic reticulum (33), the intermembrane space of mitochondria (34), the inner nuclear membrane (35), and peroxisomes (36) in plants and mammalian cells.
To further demonstrate applications and the potential advantage of split GFP in vivo, we created transgenic flies that express β2AR-GFP11 or GFP1–10 under the control of UAS. SI Appendix, Fig. S2 shows negligible background fluorescence in larval tissues possessing either UAS-β2AR-GFP11 or UAS-GFP1–10 alone. When the GFP1–10 and GFP11 fragments were introduced into the same flies, complementation fluorescence was detected. For instance, when coexpressing UAS-β2AR-GFP11 with UAS-GFP1–10 using engrailed-GAL4 (the engrailed enhancer drives GAL4), we observed a prominent fluorescence signal in the posterior portion of wing imaginal discs (SI Appendix, Fig. S2). The fluorescence pattern reflects where the engrailed gene is expressed (37). We also employed three different GAL4 lines to express split GFP-tagged β2-AR in other larval tissues (such as the central nervous system, the salivary gland, and the fat body). Strong fluorescence signals were observed within these tissues (SI Appendix, Fig. S2). These observations suggest that by overexpressing the two split GFP fragments together, the split GFP system is effective in a broad range of Drosophila tissues.
In Drosophila tissues, the complemented signal from split GFP fusion proteins can be used to localize a variety of proteins. Movie S1 shows actin as an example of proteins tagged in cytoplasmic compartments. Expression of split GFP-tagged β-actin under the control of a panneuronal driver elav-GAL4 produced live images that revealed the retraction and extension of filopodia in an embryo’s growth cones, and these filopodia were filled with the tagged actin (Movie S1). Additionally, by adding an N-terminal endoplasmic reticulum (ER)-import signal to GFP1–10 (GFP1–10sec), we fluorescently labeled the ER luminal Calreticulin protein (Crc-GFP11) in the larval fat body, indicating that split GFP can work within the secretory pathway as well (SI Appendix, Fig. S3A). Using GFP1–10sec, we further aimed to label the extracellular domain of a transmembrane protein. To this end, we made a fusion protein of GFP11, targeting the extracellular domain of the Down syndrome adhesion molecule (Dscam1). GFP11 tag was inserted immediately downstream of the Dscam1 signal sequence. When the UAS-GFP11-dscam1 [exon 17.1] transgene was coexpressed with UAS-GFP1–10sec under the control of 201Y-GAL4 specific for mushroom body (MB) neuron, split GFP formed a complementation signal in the neurons. Moreover, the signal was highly enriched in the cell bodies and dendrites, while the minimal signal was detected on axons (SI Appendix, Fig. S3B). This distribution is consistent with previous results from the full-length GFP fusion (i.e., Dscam1-EGFP [exon 17.1]) (38, 39).
We generated transgenic lines, UAS-GFP1–10 and UAS-GFP1–10sec, to illustrate the utility of the GFP11-tagging approach for investigating the subcellular localization of proteins in tissues. Using these two lines, proteins exposed to the cytoplasm, nucleoplasm, secretory pathway lumen, and extracellular space can be investigated. However, researchers interested in imaging GFP11-tagged proteins in other compartments would need to generate compartment-specific UAS-GFP1–10 lines.
Labeling Endogenous Proteins with GFP11 Tag.
A motivation to adopt the split GFP system for use in Drosophila is the potential to label endogenous proteins fluorescently in cells of specific types. Cell-type–specific labeling is particularly desirable in complex tissues such as the brain, where adjacent and nontargeted cells are unlabeled. Therefore, it enables the measurement of protein distribution at high contrast. By introducing cell-type–specific GFP1–10 expression into a GFP11 knock-in animal, only cells that express both GFP1–10 and GFP11 can be selectively visualized. To demonstrate the feasibility of this approach, we modified some genomic loci to tag with GFP11 by using MiMIC insertions. One of these protein trap lines is a teneurin-m (ten-m) allele. ten-m encodes a type II transmembrane protein. Ten-m is required for the proper establishment and maintenance of synapse formation in the olfactory circuit (40, 41) and at neuromuscular junctions (NMJ) (42). The MiMIC line MI07828 has been converted to a GFP11 protein trap line using recombination-mediated cassette exchange (RMCE). The GFP11 line is homozygous viable. GFP11 is inserted into the intron separating the fourth and fifth coding exons, flanked by splice acceptor and donor site. The GFP11 fragment is located at the cytoplasmic N terminus of Ten-m. GFP1–10 expression under the control of a panneuronal driver elav-GAL4 leads to complementation fluorescence of Ten-m (Fig. 1C), while no complementation fluorescence in the GFP11 allele is observed in the absence of GFP1–10 expression. The tagged Ten-m expression pattern in the central nervous system (CNS) overlaps with the pattern in wild-type larvae revealed with the anti–Ten-m antibody (Fig. 1C), although we notice that split GFP renders a relatively low fluorescence signal.
We have previously demonstrated that inserting a seven tandem GFP11 (GFP11 × 7) tag can substantially enhance the complemented signal of tagged proteins by sevenfold in S2 cells (17). To validate this approach in Drosophila in vivo, we generated a GFP11 × 7 protein trap line of ten-m from the same MiMIC line as above. Like the GFP11 protein trap line, the resulting GFP11 × 7 line is fully viable. For labeling of Ten-m in the nervous system, we crossed the GFP11 × 7 strain with a panneuronal expression line of GFP1–10. We found that the fluorescence distribution resembles that of the single GFP11 fusion (Fig. 1 C and D). Quantifying the overall fluorescent brightness for single-copy and multicopy GFP11, we also confirmed that the GFP11 × 7 fluorescence was ∼4 times brighter than the single copy of GFP11 (Fig. 1E). For the GFP11 × 7 line to be useful, the presence of the tandem tag must not interfere with the function of Ten-m. We addressed this by performing a genetic analysis of the GFP11 × 7 line. It has been previously reported that ten-m mutants alter the number of synaptic boutons and the innervation of a specific skeletal muscle by motor axons (41). Either in the split state or the reconstituted state of GFP11 × 7, however, there has been no observable phenotype in larvae homozygous for the GFP11 × 7 insertion (SI Appendix, Fig. S4). Next, we assessed the distribution of Ten-m in different types of neurons (olfactory neurons, insulin-producing cells [IPCs], and photoreceptor neurons) during larval development and at the adult stage. We used different GAL4 drivers to express GFP1–10 and label Ten-m fluorescently in targeted neurons. As shown in Fig. 1 F–H, the labeled Ten-m proteins localize to the axonal terminals. Regardless of the types of neurons, the proteins tended to form clusters and appeared as fluorescent puncta. Occasionally, a small number of Ten-m puncta was also found along the axon shafts (arrowheads in Fig. 1H). These observations implicate that Ten-m might undergo axonal transport. To assess whether Ten-m is trafficked down axons, we visualized motor neurons in real time. Larval motor axons offer an accessible means to study axonal trafficking (43). With GFP1–10 expressed under elav-GAL4 control, the Ten-m proteins were fluorescently labeled in live larvae. Live imaging revealed that Ten-m puncta rapidly moved in motor axons (Movie S2). After making kymographs (Fig. 1I), we calculated the velocity for Ten-m puncta (0.83 ± 0.03 μm s−1 in the anterograde direction and 0.99 ± 0.03 μm s−1 in the retrograde direction; Fig. 1J). The velocity range (SI Appendix, Fig. S5) coincides with that of other receptors trafficked via microtubule-based axonal transport (44). Importantly, ∼60% of the total particles we observed revealed anterograde bias, supporting the notion that Ten-m puncta are enriched in the motor axon terminals (42). We note that Ten-m labeled with full-length EGFP (i.e., a GFP protein trap) showed similar Ten-m transport kinetics, albeit the GFP signal was much dimmer than that of GFP11 × 7 (Fig. 1 I and J and Movie S2). Altogether, these studies demonstrate that the localization pattern of split GFP-tagged proteins accurately reflects the distribution of endogenous proteins in specific cell types.
Protein Labeling Using the Split sfCherry System in Drosophila.
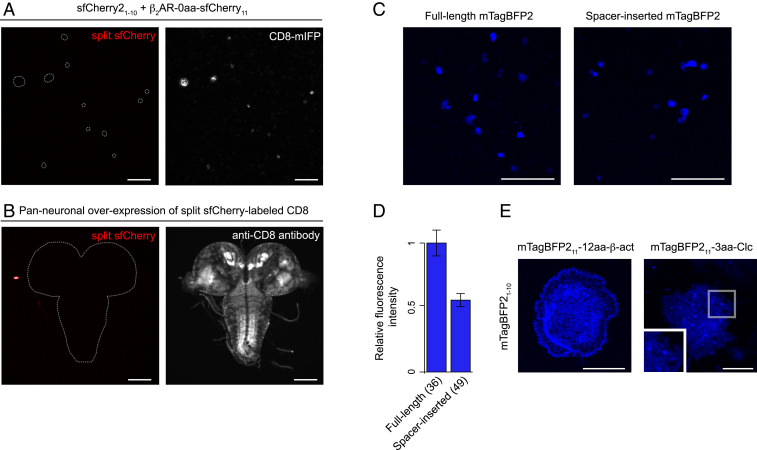
For multicolor imaging experiments, we and others have previously developed the split sfCherry system (17, 45, 46). Although the system has been successfully used in mammalian cells, there is no literature about the performance in Drosophila cells. To examine the utility of the sfCherry11 tag, we designed four fusion proteins: sfCherry211-β-actin, sfCherry211-α-tubulin, β2AR-sfCherry11, and CD8-sfCherry11. With coexpression of sfCherry21–10 in S2 cells, we did not observe fluorescence from any of these fusions (Fig. 2A). Similar results were obtained with in vivo expression analysis (in which a transgenic line of UAS-CD8-sfCherry11 was crossed to a panneuronal sfCherry21–10 line), confirming that the overall fluorescence signal was extremely weak or undetectable in third-instar larvae (Fig. 2B). We also evaluated the performance of sfCherry3V1–10 [sfCherry3V1–10 is one of the most improved sfCherry1–10 variants and is fully capable of binding to sfCherry211 (46)] and found that it produces essentially no fluorescence when coexpressed with sfCherry211 in S2 cells (SI Appendix, Fig. S6). The reason for the reduced overall signal of split sfCherry is currently unknown, but we speculate that it is probably due to the pH of Drosophila cells.
Fig. 2.
Development of other split FPs. (A) Fluorescence images of S2 cells coexpressing β2AR-sfCherry11, sfCherry21–10, and CD8-mIFP (an infrared FP-tagged membrane marker). The dashed lines in the Left indicate where IFP-positive cells are. (B) A transgenic line of UAS-CD8-sfCherry11 was crossed to a panneuronal sfCherry21–10 line. Early third-instar larval brains were dissected and stained with anti-CD8. (C) Fluorescence images of S2 cells expressing either full-length or spacer-inserted mTagBFP2. For direct comparison, we kept all acquisition settings identical for both constructs. (D) Quantification of whole-cell fluorescence intensity of S2 cells expressing full-length or spacer-inserted mTagBFP2. n = 36 to 49 cells. Error bars are SEM. (E) Representative images of mTagBFP211 labeling of cellular proteins in S2 cells. mTagBFP21–10 was coexpressed with mTagBFP211-β-actin or mTagBFP211- Clc. (Scale bars, 10 μm in E and 100 μm in A through C.)
Although not surprisingly, its brightness is partially recovered using the tandem repeat approach. When cotransfecting S2 cells with sfCherry21–10 and either sfCherry211 × 7-β-actin or sfCherry211 × 7-α-tubulin, we observed poor but nonnegligible signal in fluorescence images of actin stress fibers and microtubules, respectively.
Engineering Split FPs in Drosophila S2 Cells.
The weak or undetectable fluorescence signal of complemented split sfCherry in Drosophila makes it challenging to perform multicolor imaging. Thus, we aimed to generate split FP variants in spectral regions other than the green region that retain brightness in Drosophila cells. For this purpose, we chose a set of candidate FPs. Previous studies (1), together with our bioinformatic alignment of full-length FPs, predicted that some FP variants are structurally similar to sfGFP (the split GFP system was derived from sfGFP; Introduction). We hypothesized that, like sfGFP, these FPs could be divided into two fragments between β-strands 10 and 11.
We generated a series of constructs in which we split the candidate FPs with one of the following two ways: for mNeonGreen2, mApple, mKate2, and E2-Crimson, each of the FP11 fragments was fused to one of the well-folded carrier proteins (either Hem oxygenase or sfGFP) and coexpressed with its corresponding FP1–10; and for mCardinal and mTagBFP2, we inserted a 30 amino acid spacer between the 10th and 11th β-strand of the two FPs. In many cases, the long spacer insertion prevents a fluorescent protein from folding correctly, resulting in the drastic reduction of its fluorescence level (45). However, if the complementation of a split FP is good enough (e.g., the well-optimized split GFP system), its fluorescence signal can be kept or slightly reduced with the spacer insertion (45). Among the split FPs we tested in S2 cells, only spacer-inserted mTagBFP2 gave a bright fluorescence signal (Fig. 2C and SI Appendix, Fig. S6). To quantitatively measure the brightness of spacer-inserted mTagBFP2, we performed an image-based assay [previously used for quantitative assessment of split FPs (17, 45)] and compared the brightness between the spacer-inserted and full-length mTagBFP2. To allow direct comparison, we coexpressed either the spacer-inserted or full-length mTagBFP2 with monomeric infrared FP (we used the mIFP signal to normalize the differences of gene expression level). In this assay, we observed that spacer-inserted mTagBFP2 retained ∼53% brightness of full-length mTagBFP2 (Fig. 2D), suggesting the potential for the split mTagBFP2 system to label cellular proteins.
To test whether split mTagBFP2 functions as a fluorescent probe in S2 cells, we constructed Drosophila expression plasmids of mTagBFP211 fusions. We coexpressed mTagBFP21–10 with any one of four mTagBFP211 fusions, including β-actin, α-tubulin, H3.3A, and clathrin light chain (Clc). For the fusion proteins we tested, we obtained blue fluorescence images of actin filaments and clathrin-coated pits (Fig. 2E); however, we did not detect complementation fluorescence with the fusion constructs of α-tubulin and H3.3A (of note, a split GFP-tagged version of the α-tubulin and H3.3A constructs fluoresces in S2 cells, indicating that the performance of split mTagBFP2 is suboptimal). These observations confirm that split mTagBFP2, although it is important to consider optimization of it in the future, enables protein visualization in S2 cells.
Characterizing the Binding Specificities of FP1–10/11 Pairs.
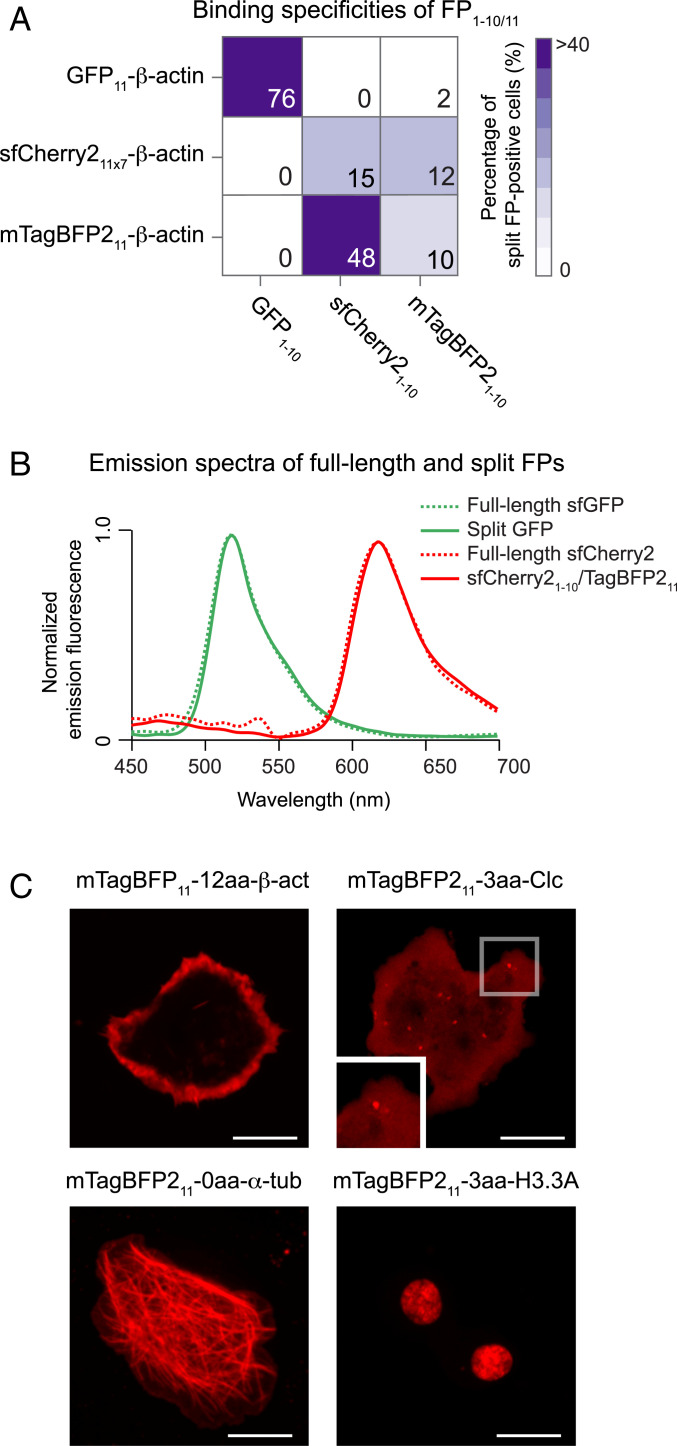
A specific FP1–10/11 pairs ability to bind each other is central to differentially label multiple proteins within the same cell (45). With the three FP1–10/11 pairs (i.e., GFP1–10/11, sfCherry21–10/11 × 7, and mTagBFP21–10/11), we systematically examined their binding specificities. We tested each FP1–10 fragment for binding to any one of FP11 fused to β-actin in S2 cells, in which they were also coexpressed with a reporter protein (i.e., mIFP). Binding between FP1–10 and FP11 was determined as the percentage of split FP-positive cells in the mIFP-positive population. We carried out experiments in a grid format (i.e., 3 × 3 for all FP1–10/11 combinations; Fig. 3A and SI Appendix, Fig. S7). As tested along the grid diagonal, all three FP1–10 fragments reconstituted with their complementary FP11 partners. Also, the pair of sfCherry21–10 and mTagBFP211 rendered a complementation signal. Its reciprocal binding pair (i.e., mTagBFP21–10/sfCherry211 × 7) also gave a similar result. Of note, sfCherry21–10/mTagBFP211 has a similar emission spectrum to full-length sfCherry2, as shown in Fig. 3B. Remarkably, sfCherry21–10/mTagBFP211 exhibited ∼3 to 4 times better performance than sfCherry21–10/11 × 7. This optimal performance of sfCherry21–10/mTagBFP211 might be due to either high molecular brightness or much-enhanced complementation efficiency between the fragments. When comparing the molecular brightness of these red-colored split FPs, we found that sfCherry21–10/mTagBFP211 is similar to sfCherry21–10/11 (SI Appendix, Table S1). Therefore, these data support the notion that sfCherry21–10 has greater complementation efficiency with mTagBFP211 than sfCherry211.
Fig. 3.
Discovery of a bright, red-colored split FP1–10/11 system. (A) Characterizing the binding specificities of three FP1–10/11 pairs (i.e., GFP1–10/11, sfCherry21–10/11 × 7, and mTagBFP21–10/11). We tested each of the FP11 fragments to complement all of the FP1–10 fragments (SI Appendix, Fig. S7). Complementation is indicated as the percentage of fluorescent cells by a color scale and each block’s number. (B) Normalized emission spectra of full-length and split FP variants measured in S2 cells. (C) Fluorescence images of different mTagBFP211 fusions coexpressed with sfCherry21–10 in S2 cells. (Scale bars, 10 μm.)
The pronounced performance of the sfCherry21–10/mTagBFP211 pair holds promises for protein labeling. To illustrate its use to visualize the subcellular localization of proteins, we coexpressed sfCherry21–10 with any of the mTagBFP211 fusions in S2 cells. Contrary to the mTagBFP21–10/11 system (this system fails to yield a fluorescent signal in two out of four targets; Fig. 2E), sfCherry21–10/mTagBFP211 allows us to visualize all the targets we have tested (Fig. 3C). Presumably, the improvement from this pair enables tagging of many targets that we cannot accomplish with mTagBFP21–10/11. Therefore, these results demonstrate that sfCherry21–10/mTagBFP211 is a highly efficient, red-colored split FP system in S2 cells.
Labeling of Endogenous Proteins with sfCherry21–10/mTagBFP211 in Drosophila.
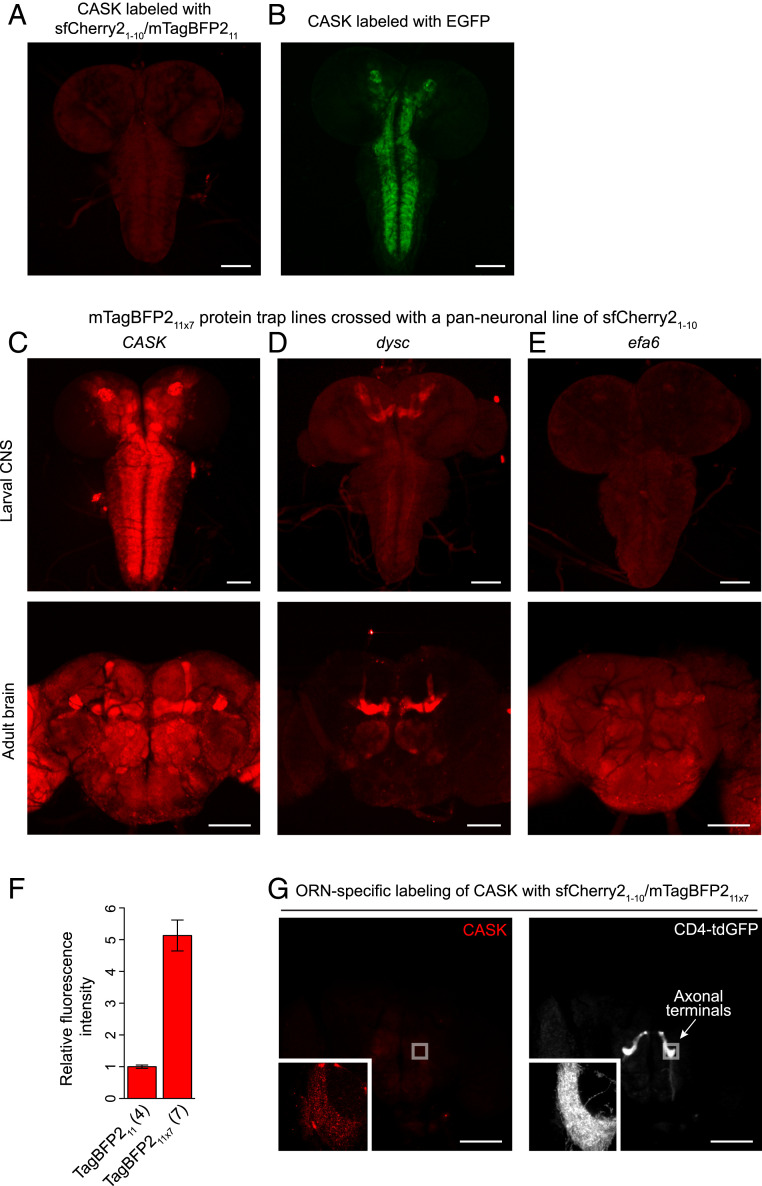
To demonstrate that sfCherry21–10/mTagBFP211 has high enough performance to detect proteins expressed at endogenous levels in Drosophila tissues, we took advantage of the RMCE-based strategy to convert a MiMIC insertion to an mTagBFP211 protein trap line. We tested the CASK gene locus. The CASK gene, which encodes a member of the MAGUK (membrane-associated guanylate kinase) protein family, is moderately expressed in the larval brain (47). When crossed with a panneuronal expression line of sfCherry21–10, the mTagBFP211 line exhibited a weak but significant fluorescence signal in the larval brain (Fig. 4A). On the other hand, we did not observe background fluorescence in larval or adult fly brains from either sfCherry21–10 or mTagBFP211 (SI Appendix, Fig. S8). The tagged CASK protein is ubiquitously expressed in the brain, as previously reported (47). The distribution was also compared to the result assessed by the GFP protein trap line (Nagarkar-Jaiswal et al. have converted the same MiMIC line to label CASK with full-length EGFP) (24). The GFP-tagged CASK has brighter signals in defined regions than the split FP counterpart (Fig. 4 A and B).
Fig. 4.
Labeling of endogenous proteins with sfCherry21–10/mTagBFP211. (A) A fluorescence image of the split FP complementation from an mTagBFP211-tagged CASK line crossed with a panneuronal expression line of sfCherry21–10. (B) A fluorescence image for the localization of EGFP-tagged CASK. (C–E) Representative images of different mTagBFP211 × 7 protein trap lines. We crossed these mTagBFP211 × 7 lines with a panneuronal line of sfCherry21–10. (A, C, and E) The images were taken with the same acquisition settings. (F) Quantification of relative fluorescence intensity of CASK labeled with mTagBFP211 × 1 or mTagBFP211 × 7. n = 4 to 7 larval brains. Error bars are SEM. (G) ORN-specific labeling of CASK in the adult fly brain. The mTagBFP211 × 7 line of CASK was crossed with an ORN-specific expression line of sfCherry21–10 and CD4-tandem GFP. (Scale bars, 100 μm.)
Like other FP1–10/11 systems, tandem mTagBFP211 × 7 tagging enhances the fluorescent signal, thereby promoting the visualization for endogenously expressed proteins. To show the utility of this approach, we created three protein trap lines with mTagBFP211 × 7 tag. In addition to CASK, we tagged two other genes: dyschromic (dysc) and exchange factor for Arf6 (efa6). RNA sequencing–based expression profiles available from the modENCODE consortium (48) have identified that dysc and efa6 are expressed at low levels throughout development. MiMIC insertions for dysc and efa6 have been isolated and converted into protein trap lines with EGFP before (24); we adopted the same MiMIC insertions for mTagBFP211 × 7 into these genes. For its complementation with sfCherry21–10, we crossed the mTagBFP211 × 7 lines with a panneuronal line of sfCherry21–10. We found strong fluorescence signals in both third-instar larval and adult fly brains of all protein traps (Fig. 4 C–E). These tagged protein expression patterns are similar to the patterns in the corresponding GFP protein trap lines (SI Appendix, Fig. S9) and their protein expression annotations (47, 49, 50). By quantifying complementation signals for CASK, we confirmed that the brightness of mTagBFP211 × 7 was ∼5 times greater than that of mTagBFP211 in larval brains (Fig. 4F). We also found the homozygous CASK-mTagBFP211 × 7 line with the panneuronal expression of sfCherry21–10 generated no obvious phenotype, suggesting that this complementary pair is not mutagenic (SI Appendix, Fig. S10).
As an example of cell-type–specific labeling using sfCherry21–10/mTagBFP211, we show the distribution of mTagBFP211 × 7-tagged CASK in a specific class of adult olfactory receptor neurons (ORN), using Or47bGAL4-driven sfCherry21–10. The CASK proteins distribute primarily in axon terminals of Or47b-expressing ORNs (Fig. 4G). For a detailed characterization of CASK localization, we also immunolabeled the axon terminals with the active zone (AZ) maker anti-Bruchpilot (BRP). We found that CASK preferentially formed puncta, and the majority of CASK puncta colocalized with endogenous BRP staining (SI Appendix, Fig. S11). These observations confirm a previous finding that presynaptic CASK functions as a scaffolding protein that tethers multiple signaling molecules at nascent AZs (51).
Dual-Color Endogenous Protein Tagging in the Drosophila Nervous System.
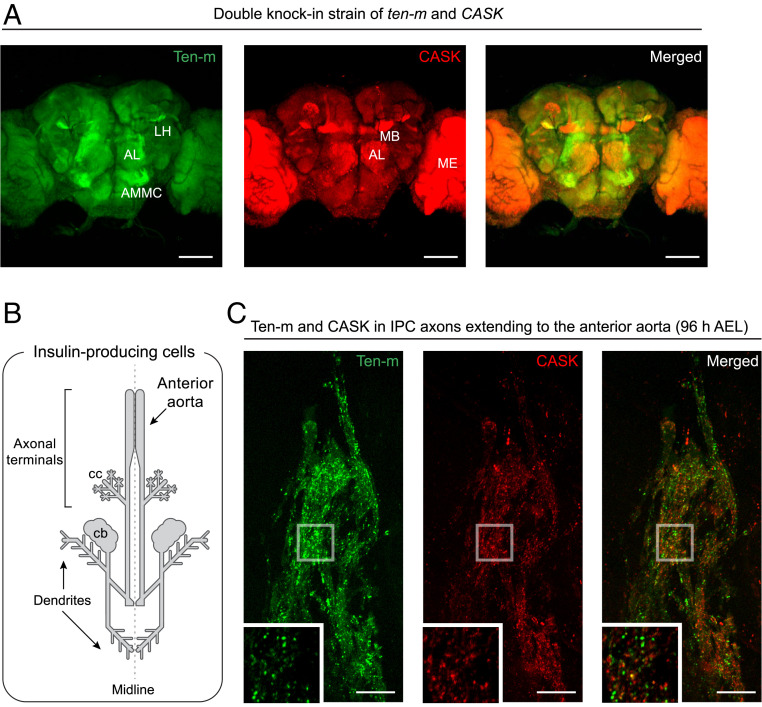
In the grid experiments in Fig. 3A, GFP1–10/11 and sfCherry21–10/mTagBFP211 display minimal cross talk in cultured cells (i.e., GFP1–10 only binds to GFP11 but does not bind to sfCherry211). Thus, these split FP systems can perform duplexed labeling of endogenous proteins in vivo. We tested a double knock-in strain of Ten-m and CASK (tagged with GFP11 × 7 and mTagBFP211 × 7, respectively). To fluorescently label the two different targets in the same animal, we panneuronally expressed both GFP1–10 and sfCherry21–10. The adult flies were dissected and imaged to characterize Ten-m and CASK expressions. We then noticed that their distribution patterns were different. Ten-m is highly concentrated in the antenna-mechanosensory center, the antennal lobe, and the lateral horn, whereas CASK is enriched in some other brain regions (such as the MB and the medulla) (Fig. 5A). These results demonstrate that the orthogonal split FP pairs enable us to visualize the differential spatial distribution of two proteins in a Drosophila tissue.
Fig. 5.
Dual-color endogenous protein tagging in the Drosophila brain. Dual-color fluorescence images of GFP11 × 7-tagged Ten-m and mTagBFP211 × 7-tagged CASK (A) in an adult fly brain, and (C) IPC axonal terminals in a third-instar larval brain. Both GFP1–10 and sfCherry21–10 were (A) panneuronally or (C) IPC-specifically expressed in the double knock-in strain. (B) Schematic drawing of larval IPCs. Their neuronal processes extend to the anterior aorta and the corpora cardiaca (cc) of the ring gland. AL, antennal lobe; AMMC, antenna-mechanosensory and motor center; LH, lateral horn; MB, mushroom body; ME, medulla; cb, cell bodies; and AEL, after egg laying. (Scale bars, 10 μm in C and 100 μm in A.)
A significant advantage of this approach is its ability to compare directly two protein distributions in a given cell type. Here, we took advantage of cell-type–specific labeling of the two synaptic proteins (i.e., Ten-m and CASK) and analyzed presynapse formation in IPCs. Although the morphological development of IPCs has been well characterized (52), the molecular details of synapse formation are largely unknown. Drosophila IPCs are primarily characterized as neuroendocrine cells (53). However, it has been recently reported that IPCs express genes involved in presynapse formation (52, 54), so we hypothesized that these synaptic proteins are trafficked to the axon terminals and assembled into presynaptic structures.
There is a cluster of seven IPCs in each brain hemisphere. IPC axons innervate the anterior aorta and the ring gland and release insulin-like peptides to the hemolymph (52) (Fig. 5B). As observed in nerve cells that release neurotransmitters, we found BRP puncta accumulation in IPC axonal terminals during the third-instar larval stage, suggesting AZ formation or maturation in these areas (SI Appendix, Fig. S12). In larval brains, the dilp2-GAL4 driver is expressed at high levels only in IPCs. Using dilp2-GAL4–driven split FP1–10, we can colabel the FP11 × 7-tagged Ten-m and CASK proteins specifically in IPCs. We found that Ten-m predominantly localizes to IPC axons, while we observed minor fluorescence in IPC cell bodies or dendrites (Fig. 5C and Fig. 1G). In particular, Ten-m displays punctate patterns in the axonal terminals throughout different stages of larval development (SI Appendix, Fig. S13). Strikingly, Ten-m puncta overlap only partially with the AZ protein CASK (Fig. 5C). We carefully quantified the spatial colocalization between Ten-m and CASK. We used the open-source image analysis software Icy (icy.bioimageanalysis.org) to automatically identify puncta in either the Ten-m or CASK channel and isolate them and calculated their spatial distribution with Ripley’s functions. This quantitative analysis confirms that the percentage of colocalization is deficient (13 ± 6%, n = 4 IPC axonal terminals); in contrast, the percentage between CASK and BRP is high (57 ± 4%, n = 4 IPC axonal terminals). Consistent with our observation in the IPC samples, Ten-m and CASK puncta only slightly colocalize in axon terminals of Or47b ORNs (12 ± 3%, n = 4 ORN axonal terminals) (SI Appendix, Fig. S14). From these data, we conclude that Ten-m is trafficked to the presynaptic nerve terminal and resides in subsynaptic compartments outside of AZs (Discussion).
Discussion
This study describes two orthogonal split FP systems for cell-type–specific, duplexed labeling of endogenous proteins in Drosophila. First, we show that split GFP provides bright signals that allow for intensive imaging experiments, including in vivo analysis of axonal transport. Second, by using a screening approach to engineering split FPs, we developed a red-colored split FP system usable for protein tagging in Drosophila. Third, we show that we can use these split FP systems to study synapse organization in specific cell types within the complex CNS.
An ideal approach for protein labeling should be both endogenous and cell-type specific. We have currently performed such labeling by modifying an endogenous locus to express a GFP-tagged protein in the presence of FLP recombinase (55–58). For example, in the absence of FLP, a transcriptional and translational stop sequence blocks the expression of the fusion protein. In contrast, cell-type–specific expression of FLP induces recombination of FRT sites to excise this stop sequence, allowing expression of the fusion protein only in cells of interest. Using GFP-based labeling approaches, tagging of proteins with low expression levels remains a particular challenge: fluorescence of a GFP knock-in strain could be too dim to distinguish from background fluorescence. Approximately 50% of GFP-tagged proteins expressed in Drosophila are detectable, while the other 50% need a stronger fluorescent signal to separate from tissue autofluorescence (59). Using split GFP1–10/11, we demonstrate substantial signal amplification by tagging endogenous proteins with seven copies of the GFP11 fragment in tandem (as seen in Movie S2, the GFP11 × 7 fluorescence is much brighter than the single copy of full-length GFP). Such a bright signal can facilitate imaging applications with low-abundance proteins (20) or high background tissue samples (22, 60). However, we advise using the tandem tag with additional caution because it may disrupt the function of proteins of interest due to its large size. If the functional disruption happens, one may choose to use the single copy of GFP11, accepting that a weak fluorescence signal is possible.
Previously, we applied GFP11 tag to generate a human cell library of fluorescently labeled endogenous proteins via genetic knock-in by CRISPR-mediated homology-directed DNA repair (20). Because GFP11 is only 60 nucleotides long, the required 200-nt single-strand DNA donor is directly synthesized, making this a cloning-free approach. We synthesized donors and knocked GFP11 into ∼50 individual loci encoding various cellular proteins in human cells. Electrophoresis of Cas9/single guide RNA ribonucleoprotein (RNP) complexes led to a knock-in efficiency in the range of 10 to 50%. Because the previous introduction of an RNP into Drosophila resulted in genome editing efficiency comparable to that in human cells (61), the strategy might provide a high FP11 knock-in efficiency in Drosophila. This scalable knock-in strategy would be particularly suited to a project to create a Drosophila library of a specific gene group, for example, the generation of a library with fluorescently labeled endogenous adhesion molecules. The library enables a systematic study of how adhesion molecules specify morphological features (i.e., axon fasciculation sites, dendrite branching points, synapses) in various neural circuits. In this instance, an FP11 coding sequence would be inserted into an array of adhesion molecule genes where GFP1–10 can be expressed in cells of particular types (e.g., motor neuron, olfactory, mechanosensory) using available GAL4 lines. The library would examine their localization relative to morphological features in specific neuronal cells.
As a general rule, red fluorescence provides deep tissue penetration and great spectral separation from tissue autofluorescence (1). Therefore, the discovery of the red-colored split FP should advance the FP11-tagging approach for deep imaging in Drosophila tissues. Combined with split GFP, it can also be applied to dual-color imaging of protein colocalization. Dual-color imaging allowed us to visualize and quantify the spatial relationship between CASK and Ten-m within IPC axonal terminals (Fig. 5C). Although we describe the red-colored split FP as bright, it is ∼40% as bright as a full-length sfCherry in Drosophila cells. As opposed to the highly optimized split GFP, whose overall brightness is comparable to that of its full-length counterpart (17, 46), the red-colored split FP leaves some room for improvement. Recently, we have exploited an evolution strategy to enhance the physical and chemical properties of split FPs via random and targeted mutagenesis (62). Through this strategy, the red-colored split FP may be evolved toward brighter variants. Even slight increases in its fluorescence signal level can reduce the numbers of tandem tags (i.e., mTagBFP211 × 7) required for strong visualization.
One prominent application of these orthogonal split FPs is the study of synaptic development in the Drosophila brain. In Drosophila, the presynaptic AZ contains T-shaped electron-dense structures (i.e., T-bars). T-bars are thought to serve as platforms in recruiting synaptic vesicles to AZs (63). T-bars comprise clusters of BRP protein. These clusters appear as fluorescent puncta when stained with anti-BRP, and each punctum corresponds to a single AZ (64). Strikingly, these BRP puncta overlap with CASK puncta in Or47b ORNs (SI Appendix, Fig. S11) as well as IPC axonal terminals. We, therefore, infer that the tagged CASK proteins also mark AZs in Drosophila. This notion would be consistent with previous studies in vertebrates focused on CASK’s potential roles as a scaffolding protein. CASK directly or indirectly interacts with various cell-surface and cell-junction proteins, organizing a protein complex that recruits synaptic vesicles and Ca2+ channels to AZs (51).
Contrary to CASK, the endogenous localization pattern of Ten-m, which acts as a synaptic cell-surface molecule, is distinct: Ten-m proteins are distributed in a punctate pattern, while Ten-m puncta have only minimal overlap with CASK puncta in IPC axonal terminals (Fig. 5C). These results indicate that Ten-m proteins might be present at subcellular structures outside of the AZs. In a previous study (42), Mosca et al. have demonstrated that presynaptic knock down of Ten-m results in a mild decrease in the number of synaptic boutons at NMJs. They argued that this is due to a requirement for Ten-m to regulate microtubule organization. Therefore, it is conceivable that Ten-m puncta might overlap with microtubules at IPC axonal terminals. Further analysis of the subcellular localization of presynaptic Ten-m may clarify its functional roles in forming IPC axonal terminals.
Materials and Methods
Fusion Protein Construction.
The amino acid sequences of mTagBFP2 (65), mNeonGreen2 (45), mApple (66), mKate2 (67), E2-Crimson (68), and mCardinal (69) were obtained from published reports. We split each of these FPs at a loop between β-strands 10 and 11. The sequences that encode mNeonGreen21–10, mApple1–10, sfCherry21–10 (45), sfCherry31–10 (46), mKate21–10, and E2-Crimson1–10 were ordered as gBlocks (Integrated DNA Technologies). These DNA fragments were ligated into the BglII/XbaI sites of the Drosophila expression vector pACUH (plasmid 58374, Addgene) using an In-Fusion HD cloning system (Takara Bio UAS, Inc.). The DNA fragments of spacer-inserted mTagBFP2 and spacer-inserted mCardinal were directly synthesized by IDT and cloned into the BglII/XbaI sites of pACUH. Each of the spacer-inserted constructs contains a 30-aa spacer (GGGGSEGGGSGGPGSGGEGSAGGGSAGGGS) between β-strands 10 and 11. The sequences of mTagBFP21–10/11, GFP1–10/11, mNeonGreen21–10/11, mApple1–10/11, sfCherry21–10/11, mKate21–10/11, E2-Crimson1–10/11, and spacer-inserted mCardinal are listed in SI Appendix, Tables S2 and S3.
pACUH-GFP11-β-actin, pACUH-GFP11-α-tubulin, pACUH-GFP1–10, and pACUH-GFP11-calreticulin (human calreticulin, NM_004343) were previously described (17). pACUH-SP-GFP1–10 was originally derived from pACUH-SP-GFP1–10-SEHDEL (17). We constructed a version of pACUH-GFP1–10sec that lacks the SEHDEL sequence for this study. To generate the plasmid pACUH-GFP1–10mitochondrial matrix, GFP1–10 with the sequence encoding the N-terminal 29 aa of human COXVIII (NM_004074) was cloned into pACUH vectors. For the other plasmids of FP11 × 1 or FP11 × 7 fusions, DNA fragments encoding various proteins were amplified by PCR with multiple sets of primers using Q5 High-Fidelity Polymerase (New England Biolabs, Inc.) or fully synthesized and then cloned into available restriction enzyme sites of pACUH. Cellular proteins for split FP fusions were derived from the following sources: human β-actin, NM_001101; human α-tubulin, NM_006082; human β2-adrenergic receptor, NM_000024; Drosophila histone H3.3A, NM_078755; Drosophila centrosomin, NM_001169670; Drosophila CG7630, NM_144156; mouse CD8, NM_009857; human clc B, NM_001834; and Drosophila Dscam1, NM_001259252. The oligonucleotides and gene fragments used to make the constructs are listed in SI Appendix, Table S4.
Transgenic Flies.
After being sequenced, all plasmids were injected into either w1118 or attP fly embryos (Rainbow Transgenic Flies, Inc.). We introduced each of seven transgenes into the fly genome. UAS-GFP1–10sec, UAS-GFP11-Calreticulin, UAS-GFP11-β-actin, UAS-β2AR-GFP11, UAS-CD8-sfCherry11, UAS-sfCherry21–10, and UAS-GFP11-Dscam1 were generated by P element-mediated transformation. UAS-GFP1–10 was generated by ΦC31-mediated transformation.
To generate FP11 and FP11 × 7 protein trap lines, we modified the existing protein-trap plasmids for RMCE (plasmids 1298, 1306, and 1314; Drosophila Genomics Resource Center [DGRC]) (23). pBS-KS-attB1-2-PT-SA-SD-FP11 and pBS-KS-attB1-2-PT-SA-SD-FP11 × 7 plasmids were first constructed. In short, the original pBS-KS-attB1-2-PT-SA-SD-EGFP-FIAsH-StrepII-TEV-3xFlag plasmid was digested with BamHI (New England Biolabs, Inc.). The DNA sequence of FP11 or FP11 × 7 flanked by peptide linkers (5′-GGATCC-FP11 × 1 or ×7-GGCGGATCC-3′) was directly synthesized and ligated with the digested plasmid. Protein-trap FP11 × 7 plasmids were made in all three reading frames. We then injected any one of the plasmids into MiMIC insertion lines expressing ΦC31 integrase driven by vasa promoter elements. Microinjections were performed using the following lines: y[1] w[*]; Mi{y[+mDint2]=MIC}Ten-m[MI07828] (Bloomington Drosophila Stock Center [BDSC] stock no. 44887), y[1] w[*]; Mi{y[+mDint2]=MIC}CASK[MI01748]/TM3, Sb[1] Ser[1] (BDSC stock no. 32778), y[1] w[*]; Mi{y[+mDint2]=MIC}dysc[MI01684]/TM3, Sb[1] Ser[1] (BDSC stock no. 34235), y[1] w[*]; Mi{y[+mDint2]=MIC}Efa6[MI00261]/TM6B, Tb[1] (BDSC stock no. 30684), and ΦC31 integrase (BDSC stock no. 34771). Finally, RMCE events were identified by PCR analysis described elsewhere (23).
All Ten-m and CASK protein trap lines are homozygous viable, indicating that insertion of a split FP tag (i.e., either GFP11, GFP11 × 7, mTagBFP211, or mTagBFP211 × 7) does not significantly disrupt gene function. On the other hand, the mTagBFP211 × 7 lines of dysc and efa6 are associated with a lethal phenotype. We have not assessed whether lethality is caused by the tag insertion itself or secondary mutations on the chromosome, which are very common in MiMIC transposon insertions (23). Importantly, note that these lethal lines are viable over deficiencies. Also, most of the fusion proteins maintain restricted localization patterns, as shown in Fig. 4 D and E, suggesting that these proteins are most likely to be folded properly and can localize normally.
Drosophila Stock and Genetics.
The following lines were obtained from the BDSC: engrailed-GAL4 (BDSC stock no. 30564), elav-GAL4 (BDSC stock no. 8760 and 8765), dlip2-GAL4 (BDSC stock no. 37516), CG-GAL4 (BDSC stock no. 7011), 201Y-GAL4 (BDSC stock no. 4440), Or47b-GAL4 (BDSC stock no. 9983), ato-GAL4 (BDSC stock no. 9494), UAS-CD8-GFP (BDSC stock no. 32185), UAS-CD8-mRFP (BDSC stock no. 32218), EGFP-tagged Ten-m (BDSC stock no. 59798), EGFP-tagged CASK (BDSC stock no. 59768), EGFP-tagged dysc (BDSC stock no. 59797), and EGFP-tagged efa6 (BDSC stock no. 60147). UAS-CD4-tdGFP was provided by Yuh Nung Jan, University of California, San Francisco, CA. Stocks were maintained at 25 °C on standard fly food. Specific genotypes in each experiment are listed in SI Appendix, Table S6.
Confocal Microscopy.
Confocal images of cells expressing green and red FPs were captured using an inverted fluorescence microscope (Ti-E, Nikon) with a 100× 1.45 NA oil immersion objective (Plan Apo, Nikon). The microscope was attached to the Dragonfly Spinning disk confocal unit (CR-DFLY-501, Andor). Three excitation lasers (40 mW 488 nm, 50 mW 561 nm, and 110 mW 642 nm lasers) were coupled to a multimode fiber passing through the Andor Borealis unit. A dichroic mirror (Dragonfly laser dichroic for 405-488-561-640) and three bandpass filters (525/50 nm, 600/50 nm, and 725/40 nm bandpass emission wheel filters) were placed in the imaging path. Images were recorded with an electron multiplying charge-coupled device camera (iXon, Andor).
Confocal images of cells expressing mTagBFP2 were acquired on an upright microscope (Axio imager Z2, Zeiss) with a 63× 1.4 NA oil immersion objective (Plan Apo, Zeiss). The upright microscope was attached to an LSM 880 Scan-head (Zeiss) with 32-channel GaAsP spectral photomultiplier tube detector. It was equipped with six laser lines (Diode 405 nm; Argon 458, 488, 514 nm; HeNe 543, 633 nm). The 405-nm diode laser with a 409 to 491 nm barrier filter was used in the blue channel. The other lasers were also used for multichannel fluorescence detection.
Image Quantifications.
To compare the brightness between full-length and spacer-inserted mTagBFP2, we imaged S2 cells coexpressing either of full-length or spacer-inserted mTagBFP2 with mIFP using a 10× objective lens (Plan Apochromat, Zeiss). We took cell images as single optical sections at the center of the cells. Images were processed using ImageJ to calculate the blue-to-infrared signal ratio. The mean ratio within a 10-μm diameter ROI (region of interest) was computed for each cell. We randomly chose ×40 cells per condition. These values were graphed in a bar plot in Fig. 2D.
A brightness comparison between FP11 × 1 and FP11 × 7 was performed by imaging fluorescently tagged Ten-m and CASK in third-instar larvae. Confocal Z-series of larval brains were obtained at 1-μm step size. For each brain, image stacks were projected into a Z-stack (maximum intensity) and then the mean fluorescence intensity within a single ROI (an 18.5 × 18.5 μm square ROI) drawn around neuropils was measured. Per genotype, ∼4 ROIs were quantified and averaged. These average values were graphed in bar plots in Figs. 1E and 4F.
We measured Ten-m motility by imaging fluorescently tagged Ten-m for ∼1 min (2 frames/s) in 1 to 2 motor axons per larva. We chose ∼4 larvae per genotype. We generated kymographs using the Multi Kymograph plugin (ImageJ). Kymographs were further analyzed to quantify the number and velocity of anterograde and retrograde events. Puncta that move only in one direction were counted as anterograde or retrograde transport for statistical analysis. A detailed tracking protocol was described elsewhere (30).
For spectral characterization in vivo, FP-tagged actin and split FP-tagged actin were transfected into S2 cells. Each emission spectrum from 450 to 695 nm was measured on the Zeiss LSM 880 using the software Zen (Zeiss).
In Vitro Characterization of Red-Colored Split FPs.
For in vitro characterization shown in SI Appendix, Table S1, we designed pET plasmids to produce Hisx6-tagged proteins. The amino acid sequences of individual constructs were listed in SI Appendix, Table S5. We lysed bacterial pellets expressing Hisx6-tagged proteins with a French press. We purified recombinant proteins with HisPur Cobalt Resin (Pierce). Proteins were further desalted into phosphate-buffered saline (PBS) pH 7.4 using a GE Healthcare illustra NAP column (GE Healthcare). We then calculated extinction coefficients using Beer–Lambert law. To determine quantum yields, we applied Rhodamine B (Wako) as a standard and measured integrated fluorescence intensities with a fluorescence spectrophotometer (Hitachi F-7100). A detailed measurement protocol was described elsewhere (62).
Colocalization Analysis.
We assessed the spatial overlap between CASK and either Ten-m or BRP puncta at IPC axonal terminals (n = 4 axonal terminals for each case). IPC images were taken as single optical sections. An 18.5 × 18.5 μm square ROI was drawn around the terminals in each image. Within the ROIs, we detected individual puncta in both channels using the spot detector plugin in the software tool Icy (70). To detect individual puncta, we set the size of puncta at 390 nm and the sensitivity at the default value of 100. After automatic detection of puncta, we performed colocalization analysis using the Colocalization Studio plugin (71, 72) in the software. The colocalization percentage was determined using a parametric fit to Ripley’s K function described elsewhere (71).
Immunofluorescence.
Tissues were dissected in PBS, fixed in 4% paraformaldehyde (Electron Microscopy Sciences) for 30 min, washed with TBS (0.1% Triton X-100 in PBS), and incubated in the blocking buffer for 1 h. Dissected tissues were incubated in primary antibodies at 4 °C overnight, washed, and then incubated in secondary antibodies for 2 h at room temperature. After extensive washes with TBS, samples were mounted in 50% glycerol for slide preparation. The following antibodies and dilutions were used: anti-CD8 (53-6.7, 1:1,000, eBioscience), anti–Ten-m (mAb20, 1:500, the Developmental Studies Hybridoma Bank [DSHB]), anti-BRP (mABnc82, 1:100, DSHB), anti-HRP Alexa 647 (1:500, Jackson ImmnoResearch Laboratories, Inc.), anti-rat Alexa 647 (1:500, Invitrogen), anti-mouse Alexa 647 (1:500, Invitrogen), and anti-mouse Atto 488 (1:500, Sigma-Aldrich).
Cell Culture and Transfection.
Drosophila S2 cells (obtained from DGRC) were grown at 25 °C in an SFX-INSECT cell culture medium (HyClone). An actin 5C-Gal4 driver and pACUH expression vectors (100 to 300 ng of each vector per well) were cotransfected by using Effectene (2.5 μL, QIAGEN) into cultures of S2 cells grown on a 24-well plate (Corning). Cells were transferred to Concanavalin A-coated coverslips and imaged 48 h after transfection.
Statistics.
For split FP brightness, Ten-m velocity, and protein colocalization results, data were represented as mean ± SEM. The Student’s t test (two-tailed, equal variance) was used to compare multiple samples. OriginPro (OriginLab) was used for statistical analysis.
Supplementary Material
Acknowledgments
We thank Shannon Walker, Ave Fouriezos, Ece Inal, and Muthugapatti Kandasamy for technical assistance; Kimberly Klonowski for anti-CD8 antibodies; and the Bloomington Drosophila Stock Center for providing fly strains. For a critical reading of the manuscript, we also thank Michael Kim, Edward Kipreos, and all members of the D.K. laboratory. This work was supported by the University of Georgia Faculty Seed Grant (to F.J., J.X., and D.K.), an NIH R01 NS107558 (to R.K., K.B., R.T., M.A.F., and D.K.), and an NSF 1350654 (to A.M.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2024690118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Rodriguez E. A., et al., The growing and glowing toolbox of fluorescent and photoactive proteins. Trends Biochem. Sci. 42, 111–129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chudakov D. M., Matz M. V., Lukyanov S., Lukyanov K. A., Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 90, 1103–1163 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Miyawaki A., Proteins on the move: Insights gained from fluorescent protein technologies. Nat. Rev. Mol. Cell Biol. 12, 656–668 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Cranfill P. J., et al., Quantitative assessment of fluorescent proteins. Nat. Methods 13, 557–562 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J. H., Wang X., Coolon R., Ye B., Dscam expression levels determine presynaptic arbor sizes in Drosophila sensory neurons. Neuron 78, 827–838 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibson T. J., Seiler M., Veitia R. A., The transience of transient overexpression. Nat. Methods 10, 715–721 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Bukhari H., Müller T., Endogenous fluorescence tagging by CRISPR. Trends Cell Biol. 29, 912–928 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Huh W. K., et al., Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Hu C. D., Chinenov Y., Kerppola T. K., Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789–798 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Kerppola T. K., Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37, 465–487 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S., Ma C., Chalfie M., Combinatorial marking of cells and organelles with reconstituted fluorescent proteins. Cell 119, 137–144 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Shu X., Imaging dynamic cell signaling in vivo with new classes of fluorescent reporters. Curr. Opin. Chem. Biol. 54, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feinberg E. H., et al., GFP Reconstitution across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron 57, 353–363 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Deng A., Boxer S. G., Structural insight into the photochemistry of split green fluorescent proteins: A unique role for a his-tag. J. Am. Chem. Soc. 140, 375–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Engelenburg S. B., Palmer A. E., Imaging type-III secretion reveals dynamics and spatial segregation of Salmonella effectors. Nat. Methods 7, 325–330 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinaud F., Dahan M., Targeting and imaging single biomolecules in living cells by complementation-activated light microscopy with split-fluorescent proteins. Proc. Natl. Acad. Sci. U.S.A. 108, E201–E210 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamiyama D., et al., Versatile protein tagging in cells with split fluorescent protein. Nat. Commun. 7, 11046 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cabantous S., Terwilliger T. C., Waldo G. S., Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 23, 102–107 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Romei M. G., Boxer S. G., Split green fluorescent proteins: Scope, limitations, and outlook. Annu. Rev. Biophys. 48, 19–44 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leonetti M. D., Sekine S., Kamiyama D., Weissman J. S., Huang B., A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proc. Natl. Acad. Sci. U.S.A. 113, E3501–E3508 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paix A., et al., Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc. Natl. Acad. Sci. U.S.A. 114, E10745–E10754 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He S., Cuentas-Condori A., Miller D. M. III, NATF (native and tissue-specific fluorescence): A strategy for bright, tissue-specific GFP labeling of native proteins in Caenorhabditis elegans. Genetics 212, 387–395 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venken K. J., et al., MiMIC: A highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8, 737–743 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagarkar-Jaiswal S., et al., A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. eLife 4, e05338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Z., et al., Various applications of TALEN- and CRISPR/Cas9-mediated homologous recombination to modify the Drosophila genome. Biol. Open 3, 271–280 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gratz S. J., et al., Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194, 1029–1035 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Port F., Chen H. M., Lee T., Bullock S. L., Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 111, E2967–E2976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelso R. J., et al., Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster. Nucleic Acids Res. 32, D418–D420 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buszczak M., et al., The carnegie protein trap library: A versatile tool for Drosophila developmental studies. Genetics 175, 1505–1531 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelliher M. T., et al., Autoinhibition of kinesin-1 is essential to the dendrite-specific localization of Golgi outposts. J. Cell Biol. 217, 2531–2547 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers S. L., Wiedemann U., Stuurman N., Vale R. D., Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162, 1079–1088 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park E., Lee H. Y., Woo J., Choi D., Dinesh-Kumar S. P., Spatiotemporal monitoring of Pseudomonas syringae effectors via type III secretion using split fluorescent protein fragments. Plant Cell 29, 1571–1584 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inglis A. J., Page K. R., Guna A., Voorhees R. M., Differential modes of orphan subunit recognition for the WRB/CAML complex. Cell Rep. 30, 3691–3698.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vicario M., et al., A split-GFP tool reveals differences in the sub-mitochondrial distribution of wt and mutant alpha-synuclein. Cell Death Dis. 10, 857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai P. L., Zhao C., Schlieker C., Methodologies to monitor protein turnover at the inner nuclear membrane. Methods Enzymol. 619, 47–69 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belostotsky R., et al., Translation inhibition corrects aberrant localization of mutant alanine-glyoxylate aminotransferase: Possible therapeutic approach for hyperoxaluria. J. Mol. Med. (Berl.) 96, 621–630 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Brower D. L., Engrailed gene expression in Drosophila imaginal discs. EMBO J. 5, 2649–2656 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhan X. L., et al., Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron 43, 673–686 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Wang J., et al., Transmembrane/juxtamembrane domain-dependent Dscam distribution and function during mushroom body neuronal morphogenesis. Neuron 43, 663–672 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Hong W., Mosca T. J., Luo L., Teneurins instruct synaptic partner matching in an olfactory map. Nature 484, 201–207 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mosca T. J., Luo L., Synaptic organization of the Drosophila antennal lobe and its regulation by the Teneurins. eLife 3, e03726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mosca T. J., Hong W., Dani V. S., Favaloro V., Luo L., Trans-synaptic Teneurin signalling in neuromuscular synapse organization and target choice. Nature 484, 237–241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vukoja A., et al., Presynaptic biogenesis requires axonal transport of lysosome-related vesicles. Neuron 99, 1216–1232.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Smith R. B., Machamer J. B., Kim N. C., Hays T. S., Marqués G., Relay of retrograde synaptogenic signals through axonal transport of BMP receptors. J. Cell Sci. 125, 3752–3764 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng S., et al., Improved split fluorescent proteins for endogenous protein labeling. Nat. Commun. 8, 370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng S., et al., Bright split red fluorescent proteins for the visualization of endogenous proteins and synapses. Commun. Biol. 2, 344 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun M., et al., Genetic interaction between Neurexin and CAKI/CMG is important for synaptic function in Drosophila neuromuscular junction. Neurosci. Res. 64, 362–371 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Brown J. B., et al., Diversity and dynamics of the Drosophila transcriptome. Nature 512, 393–399 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jepson J. E., et al., dyschronic, a Drosophila homolog of a deaf-blindness gene, regulates circadian output and Slowpoke channels. PLoS Genet. 8, e1002671 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qu Y., et al., Efa6 protects axons and regulates their growth and branching by inhibiting microtubule polymerisation at the cortex. eLife 8, e50319 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Südhof T. C., The presynaptic active zone. Neuron 75, 11–25 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao J., et al., Insight into insulin secretion from transcriptome and genetic analysis of insulin-producing cells of Drosophila. Genetics 197, 175–192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rulifson E. J., Kim S. K., Nusse R., Ablation of insulin-producing neurons in flies: Growth and diabetic phenotypes. Science 296, 1118–1120 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Meschi E., Léopold P., Delanoue R., An EGF-responsive neural circuit couples insulin secretion with nutrition in Drosophila. Dev. Cell 48, 76–86.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Baena-Lopez L. A., Alexandre C., Mitchell A., Pasakarnis L., Vincent J. P., Accelerated homologous recombination and subsequent genome modification in Drosophila. Development 140, 4818–4825 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y., et al., Cell-type-specific labeling of synapses in vivo through synaptic tagging with recombination. Neuron 81, 280–293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu H., et al., isoTarget: A genetic method for analyzing the functional diversity of splicing isoforms in vivo. Cell Rep. 33, 108361 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fendl S., Vieira R. M., Borst A., Conditional protein tagging methods reveal highly specific subcellular distribution of ion channels in motion-sensing neurons. eLife 9, e62953 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sarov M., et al., A genome-wide resource for the analysis of protein localisation in Drosophila. eLife 5, e12068 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hefel A., Smolikove S., Tissue-specific split sfGFP system for streamlined expression of GFP tagged proteins in the Caenorhabditis elegans germline. G3 (Bethesda) 9, 1933–1943 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee J. S., et al., RNA-guided genome editing in Drosophila with the purified Cas9 protein. G3 (Bethesda) 4, 1291–1295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tamura R., Jiang F., Xie J., Kamiyama D., Multiplexed labeling of cellular proteins with split fluorescent protein tags. Commun. Biol. 4, 257 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harris K. P., Littleton J. T., Transmission, development, and plasticity of synapses. Genetics 201, 345–375 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kittel R. J., et al., Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science 312, 1051–1054 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Subach O. M., Cranfill P. J., Davidson M. W., Verkhusha V. V., An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS One 6, e28674 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shaner N. C., et al., Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 5, 545–551 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shcherbo D., et al., Far-red fluorescent tags for protein imaging in living tissues. Biochem. J. 418, 567–574 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strack R. L., et al., A rapidly maturing far-red derivative of DsRed-Express2 for whole-cell labeling. Biochemistry 48, 8279–8281 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chu J., et al., Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein. Nat. Methods 11, 572–578 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Chaumont F., et al., Icy: An open bioimage informatics platform for extended reproducible research. Nat. Methods 9, 690–696 (2012). [DOI] [PubMed] [Google Scholar]
- 71.Lagache T., Sauvonnet N., Danglot L., Olivo-Marin J. C., Statistical analysis of molecule colocalization in bioimaging. Cytometry A 87, 568–579 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Lagache T., et al., Mapping molecular assemblies with fluorescence microscopy and object-based spatial statistics. Nat. Commun. 9, 698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.