Significance
Our study makes significant progress understanding asymmetry in icosahedral viruses that would be otherwise masked by forcing homogeneity through icosahedral averaging. Using an asymmetric approach revealed the atomic-resolution structure of a complex between canine parvovirus and a strain-specific neutralizing antibody. Since species jumping is a rare event in DNA viruses, the emergence of an antibody that binds more avidly to the canine-adapted virus (and not ancestral feline equivalent) is of special interest. The Fab-bound and -unbound epitopes were solved on the same virus capsid with an atomic-resolution asymmetric map. Fab 14 stabilizes a capsid loop within the same binding site used by the receptor, suggesting capsid conformational change or steric competition with the receptor contributes to the mechanism of antibody neutralization.
Keywords: virus–fab complex, cryo-EM, parvovirus, epitope, ISECC
Abstract
Canine parvovirus is an important pathogen causing severe diseases in dogs, including acute hemorrhagic enteritis, myocarditis, and cerebellar disease. Overlap on the surface of parvovirus capsids between the antigenic epitope and the receptor binding site has contributed to cross-species transmission, giving rise to closely related variants. It has been shown that Mab 14 strongly binds and neutralizes canine but not feline parvovirus, suggesting this antigenic site also controls species-specific receptor binding. To visualize the conformational epitope at high resolution, we solved the cryogenic electron microscopy (cryo-EM) structure of the Fab–virus complex. We also created custom software, Icosahedral Subparticle Extraction and Correlated Classification, to solve a Fab–virus complex with only a few Fab bound per capsid and visualize local structures of the Fab-bound and -unbound antigenic sites extracted from the same complex map. Our results identified the antigenic epitope that had significant overlap with the receptor binding site, and the structures revealed that binding of Fab induced conformational changes to the virus. We were also able to assign the order and position of attached Fabs to allow assessment of complementarity between the Fabs bound to different positions. This approach therefore provides a method for using cryo-EM to investigate complementarity of antibody binding.
Canine parvovirus (CPV) emerged as a host range variant virus in the mid-1970s, subsequently causing a pandemic of disease in dogs during 1978 (1, 2). Since that time, multiple variants have emerged with additional mutations in the viral capsid (3, 4). Extensive genetic and biochemical studies have shown that specific mutations displayed on or near the capsid surface alter binding to the host receptor, transferrin receptor type-1 (TfR). Since the specific host ranges of canine and feline parvoviruses are primarily controlled by the ability of the virus to bind TfR, changes in the binding site alter the ability of the virus to infect different hosts (3, 5, 6).
The virus capsid is highly antigenic, and an infection elicits many different host antibodies, which recognize specific structures on the surface of the virus that are primarily displayed as conformational epitopes. Most antibodies efficiently neutralize virus as IgGs, whereas they vary in their neutralization abilities when tested as Fabs (7, 8). In a number of cases, selection of antibody escape mutations by antibodies also selects for host range variation in the viruses, and conversely, selection for host range variation alters the antigenic structure recognized by specific antibodies (6, 9, 10). Although some of these changes appear to result from overlap of the receptor and antibody binding sites, it is still not clear how different selections operate in the natural evolution of the viruses. Understanding the mechanisms of host recognition and the dynamics of the binding by antibodies and receptors would provide insight into the connections between antigenic and host range variation, enabling us to predict the ability of a given virus capsid to change hosts or to escape host immunity. The coordinated overlap between antibody and receptor binding has also been seen in other viruses, including SARS-CoV-2 and influenza viruses (11–13).
CPV has a small, 26-nm diameter, T = 1 icosahedral capsid that packages a single-stranded DNA (ssDNA) genome of about 5,000 bases. The capsid shell is composed of VP2 (∼90%) and VP1 (∼10%), which are generated by differential messenger RNA splicing events so that the entire sequence of VP2 is also contained within VP1. Both proteins fold into the same eight-stranded, antiparallel β-barrel structure, where the β-strands are connected by loops that make up the surface features of the capsid. A raised region known as the threefold spike surrounds each icosahedral threefold axis and contains most of the antigenic structures recognized by different antibodies (14, 15). MAb 14 is a mouse monoclonal antibody generated against CPV capsids that has particularly interesting properties. MAb 14 binding, hemagglutination inhibition properties, and neutralization are all virus-strain specific, and it bound with significantly higher affinity to CPV capsids than to the closely related but host range variant–virus that infected cats, feline panleukopenia virus (FPV) (16–18). The virus-specific binding of MAb is controlled by the capsid surface residue 93, which is Lys in FPV and Asn in CPV (19–21). In addition to antibody recognition, residue 93 also controls canine host range, since Asn93 allows binding to canine TfR and infection of canine cells, whereas Lys93 in the equivalent position on the FPV capsid prevents both of these processes (20).
Despite the central role of antibodies in protecting animals against virus infections and allowing recovery from disease, in many cases, we still lack a detailed understanding of epitope characteristics, the dynamics of binding processes, and the viral neutralization mechanisms. Previous X-ray crystallography and cryo-electron microscopy (cryo-EM) structures include the Fab of MAb 14 (Fab 14), CPV and FPV capsids, and a Fab 14–CPV capsid complex at moderate resolution of 12.5 Å (Protein Data Bank [PDB] IDs: 2CAS, 1C8F, 3IY0, and 3GK8) (18, 22, 23). Crystal structures of Fab 14, CPV, and FPV fitted into the cryo-EM map of Fab–virus complex have allowed us to predict protein interactions in the binding interface (18). Although this was the most rigorous approach at the time, the resulting pseudoatomic structure based on the fitting did not explain why variation in residue 93 controlled Fab binding, identify likely mechanisms of antibody neutralization, or explain how the minor changes in the site also affect TfR binding. Recent technological advances in cryo-EM now allow us to solve Fab–virus structures at high enough resolution to build atomic models directly into the density map for identifying interactions unambiguously.
The binding and occupancy of Fabs on the CPV capsid have also been determined previously using charge detection mass spectrometry, which revealed that some of the tested monoclonal antibody–derived Fabs, including Fab 14, could fully occupy all 60 epitopes of the capsid but with some differences in the kinetics of attachment (24). Incubating with excess Fab molecules to occupy all icosahedrally equivalent sites on capsids has long been the preferred cryo-EM structural approach since this allows icosahedral symmetry averaging to be imposed during the reconstruction process for maximizing resolution (18, 25, 26). However, there are few other studies confirming the occupancies of Fabs on viral capsids, and the larger IgG likely does not saturate the entire surface of the capsid so that an undersaturated capsid (with fewer than 60 bound Fab in the case of parvoviruses) would more closely mimic the physiologically relevant complexes. Solving such an asymmetric structure at atomic resolution is now possible due to advances in cryo-EM and the reconstruction approaches.
Here, we define an atomic model of Fab 14 bound to the capsid of CPV based on cryo-EM of the complex and examine the functional mechanisms that affect binding by testing antibody mutants. Of the two data sets used to reconstruct Fab–virus complex maps, one had close to complete occupancy of the 60 capsid epitopes, whereas the other had an average of 10 Fabs bound per capsid. These data were used initially to solve the icosahedrally averaged structures of fully Fab-occupied and partially Fab-occupied complexes to resolutions of 3.2 and 2.3 Å, respectively. An asymmetric, partially Fab-occupied virus map calculated with local reconstruction approaches attained 2.4-Å global resolution and revealed the Fab-occupied and unoccupied sites on the same virus capsid. These structures allowed unambiguous identification of residues and side chains involved in the Fab–virus binding interface and also revealed local conformational changes in the antibody binding site induced by Fab binding. The partial occupancy of the capsids by Fab provided an opportunity to develop an innovative algorithm to test for complementarity of Fab binding to different positions on the capsid. Notably, it was the asymmetric approach and not the traditional icosahedrally averaged reconstruction that revealed the details of antibody binding.
Results
Independent CPV and Fab 14 Incubations Generated Complexes with Complete and Partial Fab Occupancies.
Here, we examine the data and compare the results obtained from two separately prepared samples of Fab 14–CPV complexes. As can be seen in the micrographs (Fig. 1), one of the resulting complexes had near saturation of the Fab binding sites, whereas the other showed only partial Fab occupancy. The complexes showing close to 60 Fab 14 molecules bound per capsid, were termed “full-Fab” and allowed icosahedral symmetry averaging to reveal bound Fab. The complexes with partial Fab occupancy of binding sites were called “low-Fab,” and these data were used for an initial icosahedrally averaged reconstruction that was followed by an asymmetric exploration of the complex and the development of algorithms to assess binding patterns.
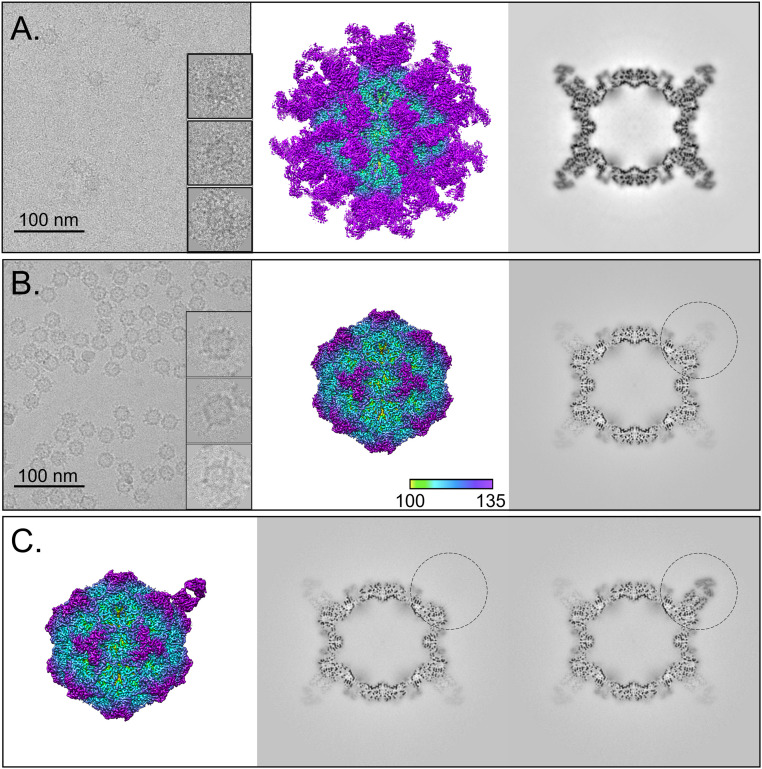
Fig. 1.
Full-Fab and low-Fab data reconstructed with and without icosahedral symmetry averaging. (A and B, Left) In the cryo-EM micrographs, an excess of Fab in the incubation results in Fab attached to most capsid binding sites, which can be seen as full-Fab complexes that have a spidery appearance. In comparison, complexes resulting from lower ratio of Fab:virus incubation have obvious low-fab occupancy (Scale bar, 100 nm). (A and B, Center and Right) The 3.2- and 2.3-Å icosahedrally averaged cryo-EM maps of Fab 14–CPV full-Fab and low-Fab complexes are surface rendered and colored according to radius, with key Inset. The Center section of each corresponding map shows the magnitude of Fab density relative to that of the capsid. (C) Asymmetric reconstruction of low-Fab data using localized classification approach. Although there were many more Fab-unoccupied particles per particle, we intentionally selected only as many Fab-unoccupied subparticles as there were Fab-occupied subparticles from each particle. (Left) A 2.4-Å surface-rendered asymmetric map is colored according to radius (as above) and shown with (Center and Right) the central sections of the fab-unoccupied and -occupied subparticles (black circle) in the context of the capsid. The low-magnitude Fab density seen in other positions in the Center section corresponds to an average of the other 10 Fab molecules averaged over the remaining 59 sites.
Reconstruction of the Full-Fab Data.
Using icosahedral symmetry averaging in cryoSPARC, the full-Fab data were refined to a 3.2-Å–resolution map (Fig. 1 and SI Appendix, Fig. S2) (27). The crystal structures for Fab 14 (PDB ID: 3GK8) (18) and the CPV capsid (PDB ID: 2CAS) (22) were fitted into the corresponding densities to initiate the builds followed by manual adjustment in Coot and simulated annealing in PHENIX. The resulting capsid structure superimposed on 2CAS with an rmsd of 0.503 (with C-alpha), with the only significant difference mapping to loop 228 (His-222–Thr-230) in the virus structure. All other areas that worsened the rmsd were the result of other flexible loop movements during simulated annealing in the weak density of fivefold loop residues 156 to 162 as well as residues 360 to 375 and 510 to 520. The only substantial change between the crystal structures and the cryo-EM density was in the binding interface, where Fab residues 100 and 101 were out of density. This discrepancy corresponds to a polymorphism of Fab 14 residue 100 in the H chain, which was identified as Phe in the original Fab sequence (18); however, a Ser was identified in this position on resequencing of the variable domain (SI Appendix, Table S2). The density in our structure supports the assignment as Ser-100. After changing the identity of the side chain, Ser-100 and His-101 were adjusted to fit into the Fab density within the complex. With these modifications, no other significant density differences were interpreted in the binding interface, and the fitted Fab crystal structure was used without further refinement for interpretations.
Local resolution mapping showed that the capsid shell had somewhat better resolution than the bound Fab, which likely correlates to slight flexibility. Virus-to-Fab contacts were identified as residues having atoms separated by less than 0.4-Å van der Waal’s radius (28). The newly identified Fab 14 footprint is different from what was estimated previously from the 12-Å–resolution capsid–Fab structure (18) (Table 1). The main interface interactions took place between capsid surface loops containing residues 93 and 228 that interacted with complementarity determining regions (CDRs) H2 and 3 and L1 of the antibody.
Table 1.
Footprint of Fab 14 on CPV capsid
| Molecule | Loop | Residue | 2.4-Å asymmetric map | 3.2-Å icosahedral map | 12-Å map (18) |
| CPV chain 1 | 87 to 93 | MET 87 | X | ||
| ASP 88 | X | X | X | ||
| LYS 89 | X | X | X | ||
| ALA 91 | X | ||||
| VAL 92 | X | X | |||
| ASN 93 | X | X | X | ||
| 222 to 229 | HIS 222 | X | X | ||
| THR 223 | X | X | X | ||
| GLY 224 | X | X | X | ||
| THR 225 | X | X | X | ||
| SER 226 | X | X | |||
| GLY 227 | X | X | X | ||
| THR 228 | X | X | |||
| PRO 229 | X | ||||
| CPV chain 2 | 305 to 309 | ASP 305 | X | ||
| ILE 306 | |||||
| GLY 307 | X | ||||
| VAL 308 | X | X | |||
| GLN 309 | X | X | |||
| 422 to 427 | LEU 422 | X | X | ||
| PRO 423 | X | X | |||
| THR 425 | X | X | X | ||
| ASN 426 | X | X | |||
| ASP 427 | X | X | X |
Reconstruction of the Low-Fab Data.
In an initial step, the low-Fab data were reconstructed while imposing full icosahedral symmetry, which resulted in a 2.3-Å–resolution map (Fig. 1). As expected, the low occupancy of Fab resulted in weak density due to the 60-fold averaging of the fewer than 60 Fab molecules per capsid. Consequently, the Fab structure could not be interpreted. The capsid build was initiated with the fitted crystal structure (PDB ID: 2CAS) and further refined in Phenix (17, 29). Unresolved regions included the 226 to 228 loop where the density was noncontinuous, weak, and uninterpretable.
An Asymmetric Reconstruction of the Low-Fab Data Resolved the Bound Fab and Allowed the Fab Structure to Be Built in the Context of the Virus.
Using the icosahedrally averaged reconstruction of low-Fab data as a starting point, an asymmetric reconstruction was done to resolve the Fab density in the context of the whole capsid. By centering on the fab binding site, subparticle extraction and three-dimensional (3D) classification were performed to classify subparticles as either Fab occupied or unoccupied (Fig. 1 and SI Appendix, Fig. S1). This analysis showed an average of 10 Fab were bound per capsid in the low-Fab data. The results of subparticle classification were used to generate an asymmetric map after superimposing all the Fab density in a standard orientation (ISECC_symbreak), resulting in a final asymmetric map at 2.4-Å global resolution (Fig. 1 and SI Appendix, Fig. S2). In this asymmetric map, the virus–Fab interface had stronger density at higher resolution than the icosahedrally averaged full-Fab map (SI Appendix, Fig. S3). The Fab 14 crystal structure was fitted to initiate the build with the identity of residue 100 corrected from Phe to Ser, as above, and additional refinement was done in Coot and PHENIX (29, 30).
Both Fab-Occupied and -Unoccupied Virus Capsid Structures Were Reconstructed from the Low-Fab Data.
To compare the Fab-occupied and -unoccupied structures, equal number of subparticles were selected per capsid to generate equivalent or matched maps from the same particles, orienting them to feature no Fab density at that same location (Fig. 1) (see asymmetric reconstruction section for particle matching in Methods). Direct comparability was maintained between the two asymmetric reconstructions, such that individual particles contributed equally to both maps, differing only in their orientations. For example, if particle A had 11 Fab molecules bound, it was incorporated in only 11 orientations in both maps by discarding the surplus unoccupied sites. This selection criteria ensured that particles contributed equally to both reconstructions, a process we termed “particle matching.” The resulting 2.4-Å–resolution asymmetric maps resolved the unoccupied and occupied Fab-binding sites (SI Appendix, Fig. S4).
The Fab-Occupied and -Unoccupied Capsid Epitopes Differ at the 228 Loop.
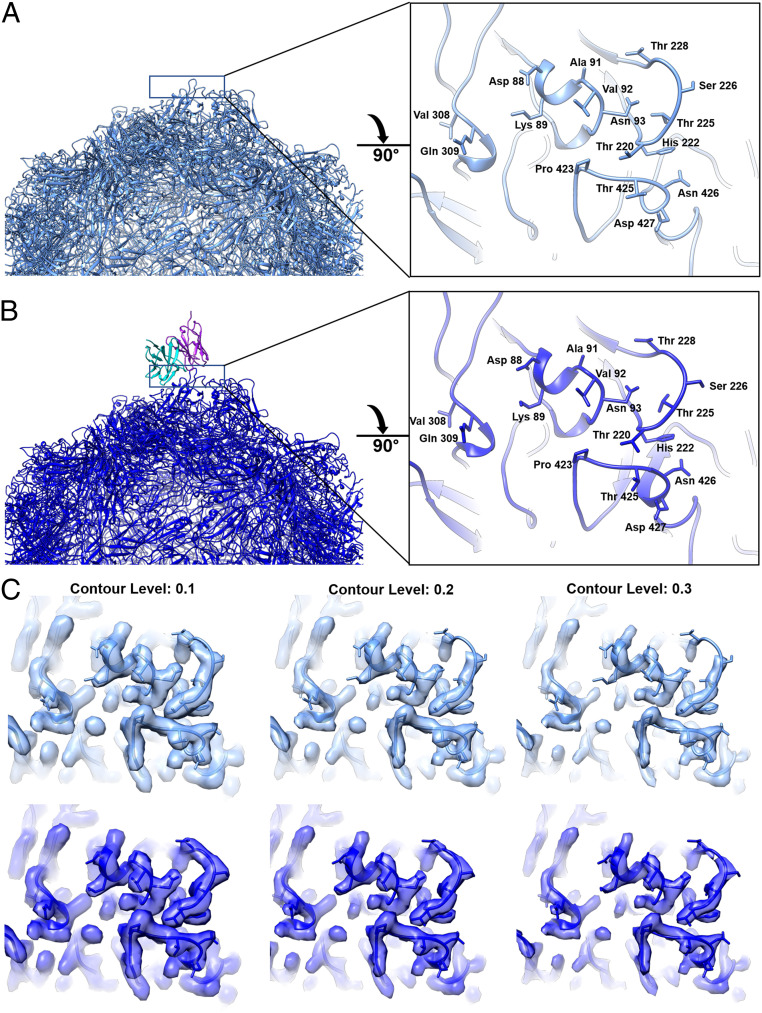
To test for local conformational changes induced by bound Fab, a difference map was calculated between the particle-matched asymmetric reconstructions of Fab-bound and -unbound epitopes (SI Appendix, Fig. S5). In addition to the expected Fab difference density, there was significant capsid difference density corresponding to virus loop 228 in the Fab-binding interface. The density of loop 228 is stronger and ordered in the bound capsid epitope compared to the unbound epitope (Fig. 2). As described above, the fitted crystal structure of the capsid protein (PDB ID: 2CAS) was refined in Coot and PHENIX for the Fab-bound and -unbound maps (SI Appendix, Table S3) (22). Inspection of the bound and unbound models suggests a hinge-like motion of the 228 loop (1.9-Å maximum Cα–Cα displacement), which makes room for Fab heavy chain residue His101 (SI Appendix, Fig. S5 and Movies S1 and S2). The bound and unbound structures superimposed with a C-α rmsd of 0.200 Å, indicating minor variation between the two structures due to reconfiguration of residues 226 to 229 in the Fab interface.
Fig. 2.
Structure of unbound and bound epitopes on the virus surface. (A and B, Left) Half capsid ribbon structures of capsid with unbound (A) (medium blue) and bound (B) (dark blue) Fab 14 heavy (cyan) and light chains (purple). (A and B, Right) Zoomed view of area (black solid line) rotated 90° from the panel A and B, Left to view the Fab binding footprint on the capsid surface with (B) antibody structure removed for visualization. The difference in the 228 loop is also displayed in two supplemental movies (Movies S1 and S2). (C) Same zoomed view of the conformational epitope surface rendered for unbound (medium blue) and bound (dark blue) with increasing contours of the map from left to right. This panel illustrates the weak, discontinuous density in the unbound epitope 228 loop compared to the stabilized continuous density in the Fab-bound epitope.
The Fab Footprint Identified Using the Asymmetric Map Is Nearly the Same as the Footprint Identified Using the Icosahedrally Averaged Map.
After Fab and capsid structural refinement, residues in the interface were defined as contacts using the same method as with the full-Fab icosahedrally averaged map (Table 1). Predicted contacts in the 222 to 229 loop were identical in both the 3.2 and 2.4-Å–resolution maps. For the 87 to 93, 305 to 309, and 422 to 427 loops, the improved resolution of the asymmetric map moved a few residues (Met87, Ala91, Gly307, Leu422, and Pro423) either in or out if the strict contact criteria cutoff (Table 1 and Fig. 3). Virus capsid residues 93, 222, and 224 identified in the footprint have also been shown previously to influence Mab 14 binding and identified as selected escape mutations (16, 17, 20, 21). Local resolution mapping showed small improvements in resolution of the surface epitope upon Fab binding, including residues 423 to 426, 88 to 93, and 309 (Fig. 4). Residues not involved in direct Fab interaction had similar local resolution in the two maps, suggesting that Fab binding stabilizes loops in the epitope by interacting with and burying the capsid surface.
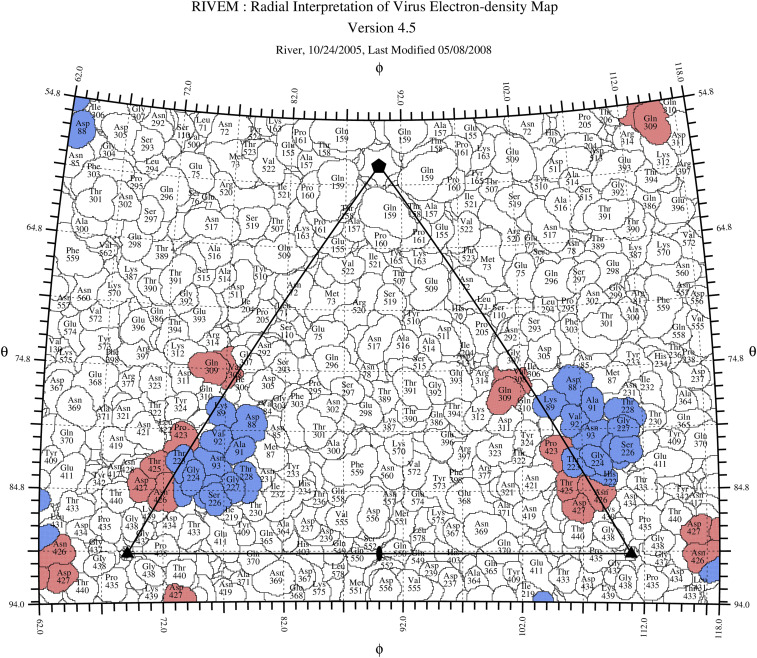
Fig. 3.
Road map of the Fab 14 footprint on the CPV surface. The capsid surface is shown as a stereographic projection where the polar angles phi and theta represent the latitude and longitude of a point on the virus surface (31). The virus surface is represented as a quilt of amino acids (32), and the icosahedral asymmetric unit of the virus is indicated by the triangular boundary. The footprint of Fab 14 has contributions from symmetry-related copies of the capsid protein (red and blue) as in Fig. 2 and SI Appendix, Fig. S4.
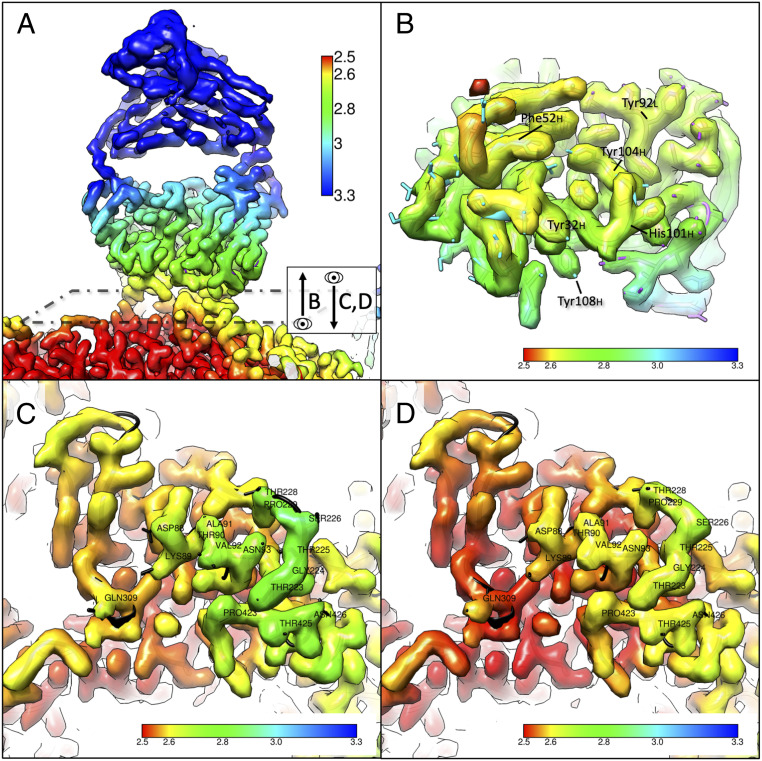
Fig. 4.
Local resolution maps of the Fab–virus interface show stabilization induced by binding. (A) The surface-rendered map for the Fab reconstructed by subparticle classification shows that distal portions of the Fab have poorer resolution, likely due to flexibility of the elbow. The dashed box represents the plane of view for B–D, showing B as a slice looking up at the Fab from the capsid surface and C and D as a slice looking down onto the capsid surface from the Fab. (B) The Fab density was segmented out of the map and rotated 90° to show the region of contact with the virus, with resolution ranging from ∼2.6 to 2.8 Å. (C) Region of contact on the virus capsid for unoccupied Fab binding site. (D) Region of contact on the virus capsid for occupied Fab binding site with obscuring fab density removed by segmentation. Improved local resolution of D versus C suggests that the Fab binding stabilizes the 228 loop. The overall resolution of the antigenic epitope is improved upon Fab binding, suggesting the overall stabilization of the epitope.
Targeted Mutations Inhibit Fab 14 Binding.
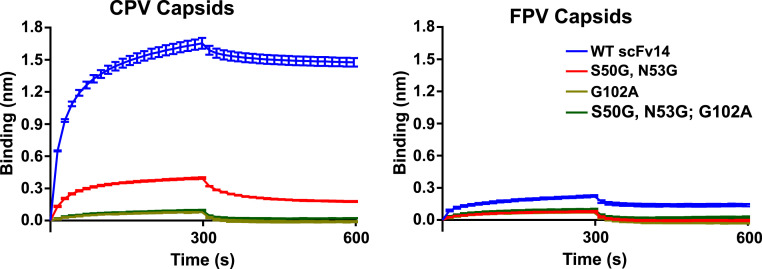
Mutations of the virus that influenced Mab 14 binding were identified in previous studies as residues 93, 222, and 224 (16, 17, 20, 21). Specifically, a naturally occurring change in residue 93 (Asn in CPV) abrogated binding as demonstrated by the diminished ability of FPV (Lys-93) to bind Mab 14. Mutations G224R, G224E, and H222Y also interfere with the Mab 14 binding. For further testing, the Fab 14 was expressed as an scFv-Fc, which allowed us to examine its binding to the capsid and to create mutations in selected CDR loops. The single chain variable fragment-fragment, crystallizable (scFv-Fc) bound to CPV capsids to a higher level than to FPV capsids (Fig. 5), as expected due to the known specificity of the antibody for CPV (16, 21). Mutations introduced in two different loops within the CDRs of the antibody resulted in greatly reduced binding to CPV (and even less binding to FPV), confirming that those were involved in critical contacts in the binding of the antibody to the capsid (Fig. 5).
Fig. 5.
Blitz binding data. Levels of binding to (A) CPV or (B) FPV capsids of wild-type scFv of Fab 14 or scFv14 variants with mutations in the complementarity-determining region. Protein A biolayer interferometry probes were incubated with scFv-Fc to the same level of bound protein, washed, incubated with CPV-2 or FPV empty capsids, and then incubated in buffer. Mutant scFv14 forms were Ser50Gly and Asn53Gly together, Gly316Ala alone, or all three mutations.
To understand the effect of the FPV-encoded Lys at position 93 on the antigenic epitope, the FPV and CPV crystal structures were fitted into the Fab-bound density map for comparison. The longer side chain of Lys-93 in FPV followed the same trajectory as that of Asn-93 of CPV but extended out of cryo-EM density and has been predicted to form two hydrogen bonds to the carbonyl oxygen atoms of residues Thr-225 and Gly-227 (33). The 228 loop is immediately adjacent to residue 93 (SI Appendix, Fig. S6). In the crystal structures of CPV and FPV that were fitted into the unoccupied epitope, the loop containing residue 228 extended out of density. Both CPV and FPV are within density when fitted into the Fab-bound epitope, consistent with the ordering of the 228 loop due to Fab binding. Movement of the 228 loop can be seen when comparing the cryo-EM structures of the unbound and bound epitope (Movies S1 and S2)
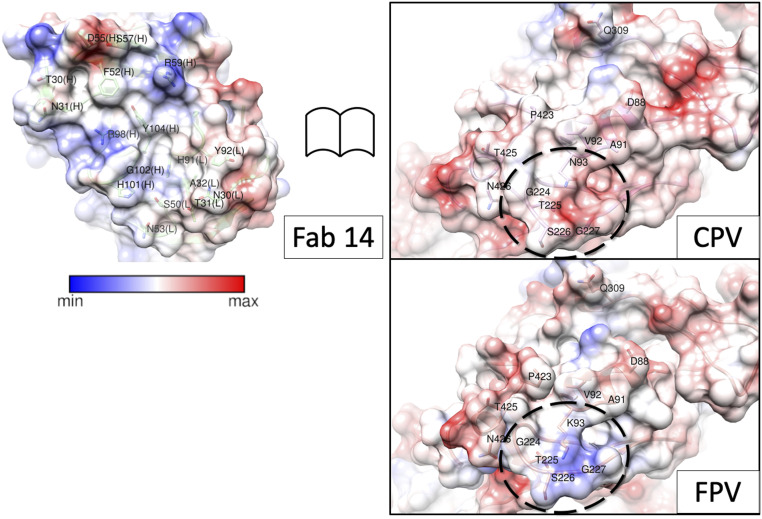
Other differences in the Fab-binding interface of CPV compared to FPV were investigated by mapping the electrostatic surface potentials to understand how the Lys (FPV) or Asn (CPV) at residue 93 might alter the surface charge. There was positive charge at the Fab surface where it interacts with virus residue 93, consistent with a favorable CPV and impaired FPV interaction, due to the positively charged Lys-93 of FPV (Fig. 6). With this change in identification, length, and charge to residue 93, the conformation of the adjacent 228 loop is also altered, resulting in a different topology of the Fab binding site
Fig. 6.
Open book view of the binding interface. Surface-rendered maps depicted with columbic potential (the key indicates blue is positive and red is negative charge) for Fab and CPV illustrate the complementarity of the binding surfaces outlined by dashed lines on virus capsid surface. For comparison, the same region of FPV was rendered, and the change at residue 93 can be seen to alter the surface charge within the region that comprises the epitope.
Correlated Local Classification Provides a Fab Fingerprint of the Complex.
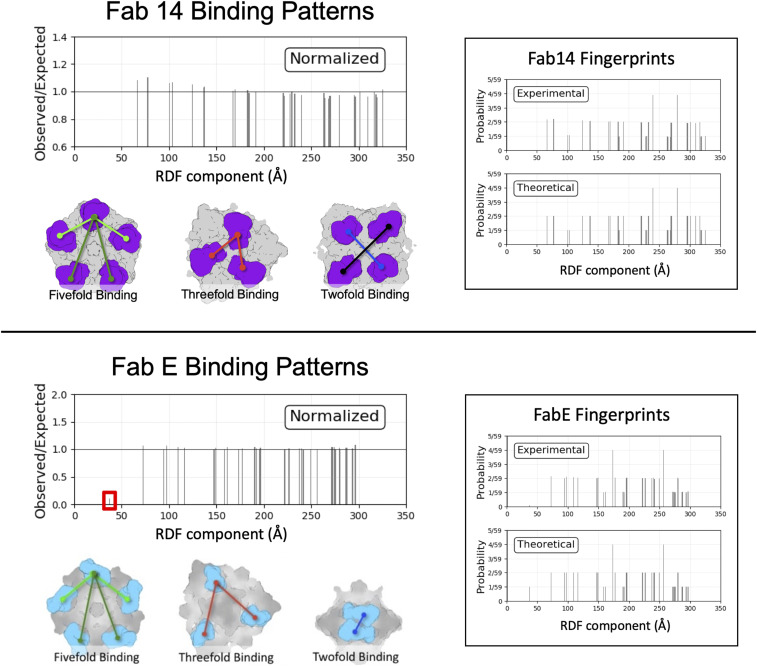
For the low-Fab dataset, each individual complex was analyzed to identify which of the 60 binding sites were occupied by Fab relative to each other on the same virus capsid. After extraction and classification of the originally defined subparticles (see above and Methods), the Fab-occupied particles, distinguished by density (Fig. 1C), were identified per complex. Each complex was defined according to the 3D distribution of Fab molecules relative to each other in the context of the capsid, which we termed “Fab fingerprint” (Fig. 7). This fingerprint is effectively a binarized version of a radial distribution function (RDF), consisting of the center-of-mass distances between all Fab-occupied subparticles. These per-particle Fab fingerprints were pooled and normalized against a hypothetical fully saturated particle. The normalized RDF plots can reveal deviation of the dataset from expected, random-chance binding patterns.
Fig. 7.
ISECC. Binding Patterns for Fab 14 and Fab E are described as RDF evaluated at binding sites. For Fab 14 (Top), a modest excess of proximal RDF components was detected after normalization against a theoretical particle with unbiased binding. For Fab E (Bottom), a severe deficit of the most proximal RDF component was observed, consistent with twofold symmetry clash of Fab E. Example geometry corresponding to RDF components are shown in each case.
Over the entire low-Fab dataset, Fab 14 binding patterns largely matched random-chance predictions but with a slight excess of the proximal RDF components, corresponding to increased binding events among nearby sites (Fig. 7, Upper). Thus, our analysis suggested that there might be binding cooperativity of Fab 14 molecules at the local level since they tend to cluster on the capsid rather than disperse randomly across the 60 symmetry-related epitopes.
For comparison, we used the same correlated local classification method to calculate fingerprints for another Fab–CPV complex. That previously determined cryo-EM structure showed that the Fab of monoclonal antibody E (Fab E) recognized a different epitope on the capsid located close to the twofold axis, with moderate clash between symmetry-related Fabs (34). The virus–Fab E map central section revealed a magnitude of Fab E density less than half that of the capsid itself. As expected, correlated local classification showed a strong deficit of the most proximal RDF component, consistent with steric clash of Fab E molecules across the twofold symmetry axis. All other RDF components matched expected values (Fig. 7, Lower). This negative cooperativity contrasts with the modest positive local cooperativity observed with Fab 14. Residual signal from the twofold Fab E RDF component may be due to low levels of classification error in the modest-resolution dataset.
To further validate RDFs as a tool for correlated classification, two undersaturated enterovirus–Fab complexes were analyzed, one without any steric clashes induced by Fab binding, and the other complex featuring strong steric collisions about each threefold symmetry axis (SI Appendix, Fig. S7). The behavior of CPV–Fab 14 matched that of the clashless enterovirus–Fab complex. Likewise, the behavior of CPV–FabE matched that of the enterovirus–Fab complex with the threefold symmetry clash. These results establish correlated classification as a tool for detecting and quantifying cooperativity of Fab binding.
Discussion
The small and seemingly simple parvovirus capsid performs many different functions in the process of packaging the ssDNA genome, trafficking between cells and animals, and entering into a permissive host. Critical events include effective receptor binding, endocytosis, and transport of the genome to the nucleus, as well as binding and potential evasion of the host antibody response. Interactions with antibodies require recognition and attachment to epitopes displayed on the assembled virus capsid, which may lead to neutralization of infection or selection for antigenic variation of the virus. The relationships between antibody binding sites and evolution that results in antibody escape are complicated by the overlap with receptor binding sites needed for cell infection. The footprint of the Mab 14 examined here almost completely overlaps the binding site of the TfR, which is required for capsid infection and which also controls the viral host range differences between CPV and FPV. The results of this study explained the specificity of binding to CPV compared to FPV and also revealed Fab binding patterns.
The traditional structural approach to reveal a specific interaction on the surface of an icosahedral virus is to saturate all the potential binding sites on the capsid by incubating purified particles with excess protein. This imposed symmetrical approach often results in a virus–protein complex that is coated with protein and allows for icosahedral symmetry averaging during the reconstruction process. The averaging often allows higher resolution to be achieved, but various factors may limit success, including low affinity and steric collisions of symmetry-related bound molecules or bulky proteins. Complexes made with full occupancy of host protein binding sites on icosahedral viruses may also poorly represent in vivo virus–host interactions, which are more often asymmetric or heterogeneous. Thus, we used an asymmetric reconstruction and subparticle classification techniques to solve the structures of Fab-bound and -unbound epitopes. As expected, the footprint revealed by our high-resolution model differs from that estimated previously from a 12-Å–resolution structure (18), and with the improved resolution, there is now an unambiguous definition of the interaction site.
Antibody Binding and Virus-Strain–Specific Recognition.
We identified a local conformational change to the virus capsid induced by Fab binding (Figs. 2 and 4, and SI Appendix, Fig. S5 and Movies S1 and S2) that is likely possible in CPV and not FPV due to the substitution of Asn (CPV) for Lys (FPV) at VP2 residue 93 (SI Appendix, Fig. S6). At the local resolution of this region (∼2.8 Å), movement of the 228 loop by 1.9 Å is not definitive; however, this loop shows higher B factors in the crystal structures of CPV than FPV, supporting flexibility (PDB IDs: 2CAS, 1C8D, and 4DPV) (22, 23, 35). The movement or ordering of the residues comprising loop 228, which may be essential for high-affinity Fab 14 binding, appears to represent a case of induced fit resulting from the binding and has been seen in other antibody binding interactions (36–38).
The structures derived from the Fab-bound and -unbound asymmetric maps also suggest a model for how the side chain of residue 93 controls species-specific binding. Compared to CPV (PDB ID: 2CAS), the FPV loop 228 (PDB ID: 1C8F) is positioned further away from the main part of the conformational epitope and also stabilized due to the Lys-93 side chain and the new bonds that are formed with the carbonyl oxygen atoms of residues Thr-225 and Gly-227, and those effects would clearly reduce Fab 14 recognition of FPV. In addition, local charge differences may play a role in controlling specific antibody binding due to the positively charged patch on Fab 14 that correlates to the interaction with virus residue 93 (Fig. 6). The previously mapped escape mutations such as His-222 also support this model overall, due to interactions with the 426 loop of the neighboring capsid protein subunit, which itself possesses two residues involved in Fab binding (Thr-425 and Asn-426). Mutation of Fab-buried residue Gly-224 likely directly interferes with Fab binding via steric interference. Thus, the identified mutations may directly block Fab binding, or they may alter the configuration of loops within the Fab footprint. We also confirmed key interactions associated with the antibody structure by mutating two CDR loops and expressing the protein as single chain variable fragments (scFvs) linked to the fragment crystallizable (Fc) of human IgG1. Those changes in light chain CDR2 (Ser50Gly and Asn53Gly) and heavy chain CDR3 (Gly316Ala) reduced the binding to both CPV and FPV capsids. These same changes also influence the specific binding of the canine TfR by CPV, suggesting that the host-specific interaction with receptor is controlled by the same changes in the capsid structure that dictate the antibody binding here.
The Local Classification and Correlation Approach Revealed Additional Details Compared to Simple Icosahedral Averaging.
Besides the structural solution, we used local classification to test the low-Fab dataset for positive or negative cooperativity in the binding (Fig. 7). Implementing an RDF-style analysis of the low-Fab complexes allowed interpretation of Fab binding patterns, including steric collision in cases of suspected atomic clash, and suggesting positive cooperativity where symmetry clash is not a factor. For correlated local classification results to be interpreted reliably, the input data should include a typical single-particle dataset to allow averaging of multiple particles, comparison to known binding patterns (Fig. 7 and SI Appendix, Fig. S7), or other corroborating biological data or assays. The amount of data are needed to ensure the accuracy of the classification results. With these controls, negative local cooperativity due to steric clash is seen in the deficit of proximal components in the RDF plots, whereas positive cooperativity is suggested by an excess of proximal components. Comparison to additional complexes will help establish the threshold for distinguishing true biophysical phenomena. This approach also suggests that undersaturated occupancies of host protein molecules on an icosahedral virus capsid can be interpreted and may allow the comparison of occupied and unoccupied binding sites.
Overall, this study confirmed that the binding of Mab 14 with the CPV capsid is a complex process involving multiple loops of the antibody and virus. Capsid changes that have been selected as host range mutations also alter antibody binding, confirming that single-point mutations on either the capsid or antibody are sufficient to prevent attachment (21, 39, 40). We were also able to analyze partially occupied capsids and predict the patterns of Fab binding using a cryo-EM structure.
Methods
Production of Viruses, Antibody, and Fab.
Viral capsids were produced as previously described (41, 42). Briefly, Norden Laboratory Feline Kidney cells were infected with FPV or CPV and incubated for 5 d. The culture supernatants were collected and clarified by centrifugation at 10,000 RPM for 15 min, then capsids were precipitated overnight with polyethylene glycol 6000, resuspended, and banded in a linear 10 to 30% sucrose gradient at 100,000 × g for 3 h, then full and empty capsids were collected separately. The IgGs were purified from hybridoma supernatants by Protein G chromatography and the Fab isolated after digestion with pepsin as described previously (8). Briefly, the Fab fragment was removed by protein G binding and the monomeric Fab isolated by size-exclusion chromatography in an S100 column.
Sample Preparation and Data Collection.
Two preparations of virus capsids and Fab were incubated at room temperature for 1 h to produce fully occupied Fab and partially occupied Fab complexes, designated full-Fab and low-Fab, respectively. A total of 3 μl of each incubation was applied to separate Quantifoil grids (Quantifoil Micro Tools GmbH), blotted to remove excess, and plunge frozen into a liquid ethane–propane mixture or ethane alone using an Mk III Vitrobot (FEI). For the full-Fab data, the sample was applied to R2/1 grids coated with a 2-nm layer of continuous carbon, and low-dose micrographs were recorded using an FEI Polara G2 microscope operating at 300 kV in nanoprobe and a nominal magnification of 78,000× with defocus values ranging from −0.5 to −4.2 μm. Images were collected under the software control of the E Pluribis Unum (EPU) program using an FEI Falcon 3 direct electron detector (DED) with postcolumn magnification of 1.4× yielding a calibrated pixel size at the sample of 1.35 Å (SI Appendix, Table S1). For the low-Fab sample, data were recorded using an FEI Titan Krios microscope operating at 300 kV and a nominal magnification of 59,000× with defocus values ranging from −0.7 to −4.9 μm. Images were also collected under control of EPU on a Falcon 3 DED with a calibrated pixel size at the sample of 1.1 Å. The total dose per exposure was set to 60e-/Å2. Both data sets were recorded in movie mode by recording multiple frames corresponding to one field, allowing for correction of beam-induced movement.
Reconstruction approaches.
Icosahedral reconstruction.
Particles were autopicked using manually selected templates and contrast transfer functions (CTF) were estimated using GCTF (43). RELION was used for motion correction, movie refinement, and particle polishing (44), whereas cryoSPARC was used for particle sorting and high-resolution icosahedral refinement (27). CTF refinement with correction for higher-order aberrations was performed in RELION. Atomic models were built using Coot (29) and Phenix (30), using crystal structures 2CAS (CPV) and 3GK8 (Fab 14) as starting models (18, 22), before validation in MolProbity (45).
Localized Classification.
Subparticle classification was done with localized reconstruction scripts and 3D classification in RELION (44, 46). The subparticle was defined by docking a crystal structure of Fab 14 (PDB ID: 3GK8) into the density map (18) and extracted using icosahedral subparticle extraction and correlated classification (ISECC) (47). These subparticles then underwent 3D classification in RELION without translations or rotations. This allowed for discrimination between Fab-occupied and -unoccupied subparticles. Subparticles were classified into six classes, resulting in strict distinction between occupied and unoccupied epitopes. There was a diversity of unoccupied states, featuring varying amounts of Fab density from the immediately adjacent epitopes (SI Appendix, Fig. S1).
Asymmetric Reconstructions.
After 3D classification into six subparticle classes to distinguish between Fab-occupied and -unoccupied subparticles, a symmetry-break operation was accomplished using ISECC_symbreak. For the two states, Euler angles for each subparticle were reassigned to their corresponding whole-complex image. This produced two separate data files containing particle orientations (.star files), corresponding to an either Fab-occupied or -unoccupied state at a selected site. Even though there were far more Fab-unoccupied sites, in order to make valid comparisons, an equal number of Fab-occupied and Fab-unoccupied subparticles were selected from each particle. This process, which we termed “particle matching,” ensured equivalent scaling of both the occupied and unoccupied maps (SI Appendix, Fig. S1). Both maps contained the same particle images in identical numbers, differing only in the orientations. Orientations were derived from strict symmetry expansion of the original icosahedral refinement without local refinement, ensuring equivalent accuracy angles in the final maps. This matching approach provided equal data sets for a total of 1,657,372 particle orientations per reconstruction. DeepEMhancer was used to improve local sharpening of maps during postprocessing (48). Difference maps were calculated by subtracting the Fab-occupied and -unoccupied maps in EMAN (49).
Correlated Local Classification.
To describe the configuration of individual bound Fab molecules on each Fab–virus complex, we implemented a system analogous to a RDF. Each component of the RDF represents the 3D distance between occupied sites on the capsid, as determined by 3D classification and the vectors used in subparticle extraction. On a per-virus basis, the distance between each pair of subparticles was calculated, yielding a list of (n2 − n) / 2 distances, where n = the number of Fab molecules bound to the given complex. Fab binding patterns were derived by normalizing the observed RDFs to a hypothetical particle with all binding sites occupied. This combined approach was termed correlated local classification. The custom software package, ISECC, is available for download at https://github.com/goetschius/isecc.
Design of scFv-Fcs, Mutagenesis, and Binding Assays.
The heavy and light variable chains of Mab 14 (45) were joined by a linker sequence of 3 × (Gly, Gly, Gly, Ser) to prepare a single-chain variable fragment (scFv) and cloned behind the gp68 signal sequence. This was linked through an additional flexible linker containing a thrombin cleavage site to the Fc portion of human IgG1 and a C-terminal 6-His tag added (10). Residues changed in the antigen binding region of the scFv included those predicted to be directly interacting with the viral capsid or to be interacting with capsid residues that control specific Mab 14 binding, including capsid residue 93 (18). Mutagenesis was conducted through the use of Phusion Site-Directed Mutagenesis Kit (Thermo Fisher Scientific). Bacmids were produced through recombination between the DH10Bac (Invitrogen) vector and the pFastbac donor plasmids. Sf9 insect cells (Invitrogen) were then transfected with the bacmids to produce the initial stock of baculovirus. Proteins were expressed from High Five cells over 5 d incubation at 28 °C. The culture supernatant was centrifuged at 10,000 RPM for 30 min before being dialyzed into 50 mM Tris HCl, 150 mM NaCl, and 0.05% NaN3, (pH = 7.5). The scFv-Fc was isolated with a Protein G column, passed through a Sephacryl S-200 chromatography column in phosphate-buffered saline (PBS) (GE Healthcare), and the protein in the monomeric protein peak was collected.
The BLItz system (ForteBio) was used to measure binding kinetics, using Protein A biosensors which were first blocked with kinetics buffer (PBS with 0.01% bovine serum albumin, 0.02% Tween 20, and 0.005% NaN3). Incubations included 300 s baseline, 300 s loading, 60 s baseline, 300 s association, and 300 s dissociation. Antibodies were tested at various dilutions and used at their optimum loading concentration. Viral capsids were added at 0.3 mg/mL. The BLItz Pro software (ForteBio) was used to analyze the data, and statistical analyses of binding assays were conducted with GraphPad Prism 5 (GraphPad Software, Inc.) Error bars on Fig. 5 represent the mean ± SEM obtained through three independent experiments.
Preparation of enterovirus complexes for RDF controls: Virus was propagated and purified as described previously (33). Briefly, confluent HeLa cells were rinsed with PBS, followed by the addition of stock inoculum for a multiplicity of infection of 1 to 5. After incubation at 37 °C for 1 h, fresh medium was added and infection proceeded for 48 h before cells were harvested, pooled, and stored at −80 °C. Virus was purified by sucrose gradient ultracentrifugation, virus bands collected, and diluted in buffer. Virus and Fab were incubated at room temperature for 30 min at subsaturating molar ratio to form complexes that were subsequently vitrified, data collected, and reconstructed as described above.
Supplementary Material
Acknowledgments
Funding was provided by the Pennsylvania Department of Health Commonwealth Universal Research Enhancement (CURE) funds. Research reported in this publication was supported by the Office of the Director, NIH, under award numbers S10OD019995 (J.F.C.) and S10RR031780 (S.L.H.), as well as NIH grants R01AI107121 (S.L.H.), R01AI092571 (C.R.P.), and T32CA060395 (L.J.O.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025452118/-/DCSupplemental.
Data Availability
Protein Structure and EM Maps data have been deposited in PDB and Electron Microscopy Data Bank (PDB: 7M3L, 7M3M, 7M3N, 7M3O; EMDB: EMD-23656, EMD-23657, EMD-23658, EMD-23659). All other study data are included in the article and/or supporting information.
References
- 1.Parrish C. R., Kawaoka Y., The origins of new pandemic viruses: The acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu. Rev. Microbiol. 59, 553–586 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Truyen U., et al., Evolution of the feline-subgroup parvoviruses and the control of canine host range in vivo. J. Virol. 69, 4702–4710 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stucker K. M., et al., The role of evolutionary intermediates in the host adaptation of canine parvovirus. J. Virol. 86, 1514–1521 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoelzer K., Parrish C. R., The emergence of parvoviruses of carnivores. Vet. Res. 41, 39 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaelber J. T., et al., Evolutionary reconstructions of the transferrin receptor of Caniforms supports canine parvovirus being a re-emerged and not a novel pathogen in dogs. PLoS Pathog. 8, e1002666 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allison A. B., et al., Single mutations in the VP2 300 loop region of the three-fold spike of the carnivore parvovirus capsid can determine host range. J. Virol. 90, 753–767 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.VanBlargan L. A., Goo L., Pierson T. C., Deconstructing the antiviral neutralizing-antibody response: Implications for vaccine development and immunity. Microbiol. Mol. Biol. Rev. 80, 989–1010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson C. D. S., Palermo L. M., Hafenstein S. L., Parrish C. R., Different mechanisms of antibody-mediated neutralization of parvoviruses revealed using the Fab fragments of monoclonal antibodies. Virology 361, 283–293 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voorhees I. E. H., et al., Limited intrahost diversity and background evolution accompany 40 years of canine parvovirus host adaptation and spread. J. Virol. 94, e01162-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callaway H. M., et al., Complex and dynamic interactions between parvovirus capsids, transferrin receptors, and antibodies control cell infection and host range. J. Virol. 92, e00460-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCarthy K. R., Raymond D. D., Do K. T., Schmidt A. G., Harrison S. C., Affinity maturation in a human humoral response to influenza hemagglutinin. Proc. Natl. Acad. Sci. U.S.A. 116, 26745–26751 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu N. C., et al., Major antigenic site B of human influenza H3N2 viruses has an evolving local fitness landscape. Nat. Commun. 11, 1233 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weisblum Y., et al., Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife 9, e61312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mietzsch M., Pénzes J. J., Agbandje-McKenna M., Twenty-five years of structural parvovirology. Viruses 11, 362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsao J., et al., The three-dimensional structure of canine parvovirus and its functional implications. Science 251, 1456–1464 (1991). [DOI] [PubMed] [Google Scholar]
- 16.Parrish C. R., Carmichael L. E., Antigenic structure and variation of canine parvovirus type-2, feline panleukopenia virus, and mink enteritis virus. Virology 129, 401–414 (1983). [DOI] [PubMed] [Google Scholar]
- 17.Strassheim M. L., Gruenberg A., Veijalainen P., Sgro J. Y., Parrish C. R., Two dominant neutralizing antigenic determinants of canine parvovirus are found on the threefold spike of the virus capsid. Virology 198, 175–184 (1994). [DOI] [PubMed] [Google Scholar]
- 18.Hafenstein S., et al., Structural comparison of different antibodies interacting with parvovirus capsids. J. Virol. 83, 5556–5566 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Govindasamy L., Hueffer K., Parrish C. R., Agbandje-McKenna M., Structures of host range-controlling regions of the capsids of canine and feline parvoviruses and mutants. J. Virol. 77, 12211–12221 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hueffer K., Govindasamy L., Agbandje-McKenna M., Parrish C. R., Combinations of two capsid regions controlling canine host range determine canine transferrin receptor binding by canine and feline parvoviruses. J. Virol. 77, 10099–10105 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang S. F., Sgro J. Y., Parrish C. R., Multiple amino acids in the capsid structure of canine parvovirus coordinately determine the canine host range and specific antigenic and hemagglutination properties. J. Virol. 66, 6858–6867 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu H., Rossmann M. G., The canine parvovirus empty capsid structure. J. Mol. Biol. 233, 231–244 (1993). [DOI] [PubMed] [Google Scholar]
- 23.Simpson A. A., et al., Host range and variability of calcium binding by surface loops in the capsids of canine and feline parvoviruses. J. Mol. Biol. 300, 597–610 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Dunbar C. A., Callaway H. M., Parrish C. R., Jarrold M. F., Probing antibody binding to canine parvovirus with charge detection mass spectrometry. J. Am. Chem. Soc. 140, 15701–15711 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Lindner J. M., et al., Human memory B cells harbor diverse cross-neutralizing antibodies against BK and JC polyomaviruses. Immunity 50, 668–676.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Gurda B. L., et al., Capsid antibodies to different adeno-associated virus serotypes bind common regions. J. Virol. 87, 9111–9124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Pettersen E. F., et al., UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Adams P. D., et al., PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao C., Rossmann M. G., Interpretation of electron density with stereographic roadmap projections. J. Struct. Biol. 158, 182–187 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossmann M. G., Palmenberg A. C., Conservation of the putative receptor attachment site in picornaviruses. Virology 164, 373–382 (1988). [DOI] [PubMed] [Google Scholar]
- 33.Organtini L. J., et al., Near-atomic resolution structure of a highly neutralizing fab bound to canine parvovirus. J. Virol. 90, 9733–9742 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapman M. S., Rossmann M. G., Single-stranded DNA-protein interactions in canine parvovirus. Structure 3, 151–162 (1995). [DOI] [PubMed] [Google Scholar]
- 35.Wang W., et al., Conformational selection and induced fit in specific antibody and antigen recognition: SPE7 as a case study. J. Phys. Chem. B 117, 4912–4923 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Sinha N., Smith-Gill S. J., Molecular dynamics simulation of a high-affinity antibody-protein complex: The binding site is a mosaic of locally flexible and preorganized rigid regions. Cell Biochem. Biophys. 43, 253–273 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Rini J. M., Schulze-Gahmen U., Wilson I. A., Structural evidence for induced fit as a mechanism for antibody-antigen recognition. Science 255, 959–965 (1992). [DOI] [PubMed] [Google Scholar]
- 38.Parker J. S., Parrish C. R., Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid. J. Virol. 71, 9214–9222 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Llamas-Saiz A. L., et al., Structural analysis of a mutation in canine parvovirus which controls antigenicity and host range. Virology 225, 65–71 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Agbandje M., McKenna R., Rossmann M. G., Strassheim M. L., Parrish C. R., Structure determination of feline panleukopenia virus empty particles. Proteins 16, 155–171 (1993). [DOI] [PubMed] [Google Scholar]
- 41.Palermo L. M., Hafenstein S. L., Parrish C. R., Purified feline and canine transferrin receptors reveal complex interactions with the capsids of canine and feline parvoviruses that correspond to their host ranges. J. Virol. 80, 8482–8492 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen V. B., et al., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ilca S. L., et al., Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat. Commun. 6, 8843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lauver M. D., et al., Antibody escape by polyomavirus capsid mutation facilitates neurovirulence. eLife 9, e61056 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez-Garcia R., et al., DeepEMhancer: A deep learning solution for cryo-EM volume post-processing 10.1101/2020.06.12.148296 (14 September 2020). [DOI] [PMC free article] [PubMed]
- 48.Ludtke S. J., Baldwin P. R., Chiu W., EMAN: Semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Hafenstein S., et al., Interaction of decay-accelerating factor with coxsackievirus B3. J. Virol. 81, 12927–12935 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Protein Structure and EM Maps data have been deposited in PDB and Electron Microscopy Data Bank (PDB: 7M3L, 7M3M, 7M3N, 7M3O; EMDB: EMD-23656, EMD-23657, EMD-23658, EMD-23659). All other study data are included in the article and/or supporting information.