Significance
Mechanisms of many human chronic diseases involve abnormal action of the immune system and/or altered metabolism. The microbiome, an important regulator of metabolic and immune-related phenotypes, has been shown to be associated with or participate in the development of a variety of chronic diseases. Viruses of bacteria (i.e., “phages”) are ubiquitous and mysterious, and several studies have shown that phages exert great control over the behavior—and misbehavior—of their host bacteria. This study uses techniques to discover and analyze over 45,000 viruses associated with human bodies. The abundance of over 2,000 specific phages is found to correlate with a variety of common chronic diseases.
Keywords: microbiome, virome, genomics
Abstract
Despite remarkable strides in microbiome research, the viral component of the microbiome has generally presented a more challenging target than the bacteriome. This gap persists, even though many thousands of shotgun sequencing runs from human metagenomic samples exist in public databases, and all of them encompass large amounts of viral sequence data. The lack of a comprehensive database for human-associated viruses has historically stymied efforts to interrogate the impact of the virome on human health. This study probes thousands of datasets to uncover sequences from over 45,000 unique virus taxa, with historically high per-genome completeness. Large publicly available case-control studies are reanalyzed, and over 2,200 strong virus–disease associations are found.
The human virome is the sum total of all viruses that are intimately associated with people. This includes viruses that directly infect human cells (1, 2) but mostly consists of viruses infecting resident bacteria (i.e., phages) (3). While the large majority of microbiome studies have focused on the bacteriome, revealing numerous important functions for bacteria in human physiology (4), information about the human virome has lagged. However, a number of recent studies have begun making inroads into characterizing the virome (5–13).
Just as human-tropic viruses can have dramatic effects on people, phages are able to dramatically alter bacterial physiology and regulate host population size. A variety of evolutionary dynamics can be at play in the phage/bacterium arena, including Red Queen (11), arms-race (14), and piggyback-the-winner (15) relationships, to name just a few. In the gut, many phages enter a lysogenic or latent state and are retained as integrated or episomal prophages within the host bacterium (16). In some instances, the prophage can buttress host fitness (at least temporarily) rather than destroy the host cell. To this effect, prophages often encode genes that can dramatically alter the phenotype of the bacteria, such as toxins (17), virulence factors (18), antibiotic resistance genes (19), photosystem components (20), other auxiliary metabolic genes (21), and CRISPR-Cas systems (22), along with countless genes of unknown function. Experimental evidence has shown that bacteria infected with particular phages (i.e., “virocells”) are physiologically distinct from cognate bacteria that lack those particular phages (21).
There have been a few documented cases in which phages have been shown to be mechanistically involved in human health and disease, sometimes through direct interactions with human cells. This includes roles in increased bacterial virulence (17), response to cancer immunotherapy (23), clearance of bacterial infection (24), and resistance to antibiotics (25). Furthermore, phage therapy, the targeted killing of specific bacteria using live phage particles, has shown increasing promise for treatment of antibiotic-resistant bacterial infections (26). Considering the progress already made, phages represent attractive targets of and tools for microbiome restructuring in the interest of improving health outcomes.
In addition, several studies have conducted massively parallel sequencing on virus-enriched samples of human stool, finding differential abundance of some phages in disease conditions (6, 27–29). A major issue encountered by these studies is that there is not yet a comprehensive database of annotated virus genome sequences, and de novo prediction of virus sequences from metagenomic assemblies remains a daunting challenge (3). Further, though some tools are able to predict virus-derived sequences with high specificity (30, 31), these tools have not been applied to human metagenomes at a large scale [with a possible exception (13)], and, regrettably, most uncovered virus genomes do not end up in central repositories. One study suggests that only 31% of the assembled sequence data in virion-enriched virome surveys could be identified as recognizably viral (32). On the other hand, another study of 12 individuals was able to recruit over 80% of reads from virus-enriched samples to putative virus contigs (11). Still, most of the potential viral contigs from this study were unclassifiable sequences, and a large majority of contigs appeared to represent subgenomic fragments under 10 kb.
The current study sought to overcome the traditional challenges of sparse viral databases and poor detection of highly divergent viral sequences by using Cenote-Taker 2, a new virus discovery and annotation tool (33). The pipeline was applied to sequencing data from nearly 6,000 human metagenome samples. Strict criteria identified over 180,000 viral contigs representing 45,033 specific taxa. In most cases, 70 to 99% of reads from virus-enriched stool datasets could be back-aligned to the Cenote-Taker 2–compiled Human Virome Database. Furthermore, the curated database allowed read-alignment–based abundance profiling of the virome in human metagenomic datasets, enabling the reanalysis of a panel of existing case-control studies. The reanalysis revealed previously undetected associations between chronic diseases and the abundance of 2,265 specific virus taxa.
Results
Characteristics of the Human Virome.
Read data were downloaded from the National Center for Biotechnology Information’s (NCBI’s) Sequence Read Archive (SRA), including data from the Human Microbiome Project (34) and several other studies (25 Bioprojects in total) pursuing massively parallel sequencing of human metagenomic samples (Dataset S1) (11, 35–48). These data spanned multiple body sites, including gut (stool), mouth, nose, skin, and vagina. A subset of the projects performed enrichment for viral sequences (49–52). Almost all of the projects pursued DNA sequencing, but a small number of metatranscriptomic (i.e., ribosome-depleted total RNA) samples were analyzed as well (53). Read data were binned and assembled by Biosample rather than by individual run-in order to combine read sets from the same individual. A total of 5,996 Biosamples were analyzed, encompassing 16,210 sequencing runs.
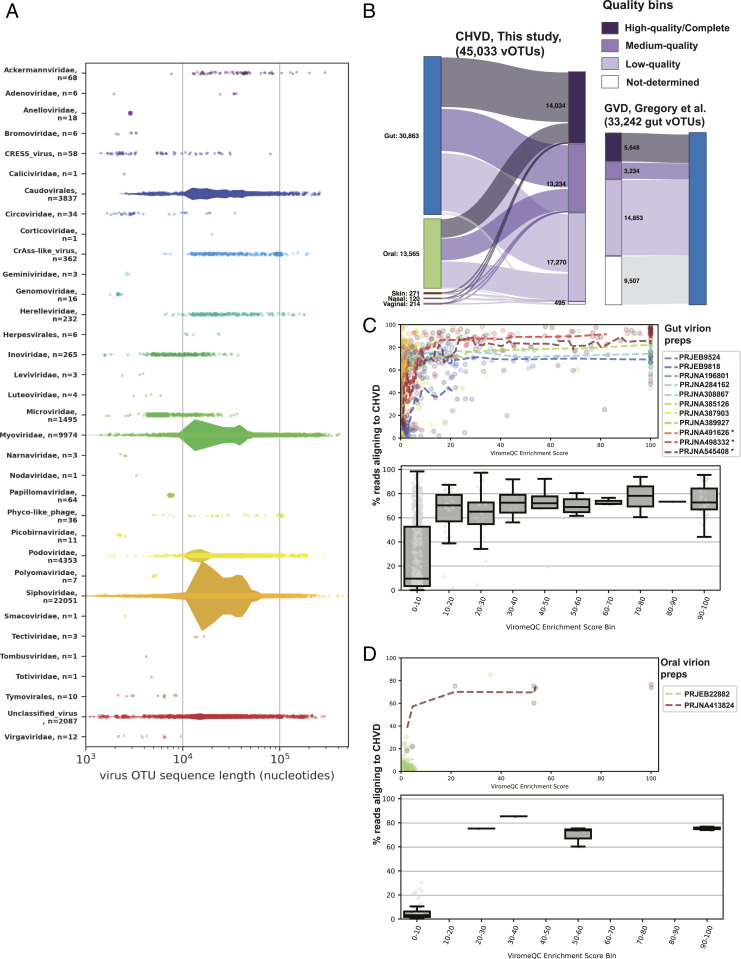
Cenote-Taker 2 (33) was used to check contigs for two common end features of complete viral genomes: direct terminal repeats (DTRs) (suggesting a circular or long terminal repeat–bounded viral genome) or inverted terminal repeats (ITRs). Sequences with DTRs were arbitrarily assumed to represent circular DNA genomes. Sequences were then scanned for the presence of “virus hallmark genes.” Circularized sequences >1,500 nucleotides (nt) with at least one viral hallmark gene and ITR-containing contigs >4,000 nt with at least one viral hallmark gene were designated as putative viruses. Linear (no discernable end features) contigs >12,000 nt with two or more viral hallmark genes were also kept as putative viruses. Since phages are often integrated into bacterial chromosomes, each linear contig was pruned with the Cenote-Taker 2 prophage pruning module to remove flanking host sequences. The analysis resulted in over 180,000 putative viral sequences. The sequences were classified into operational taxonomic units (OTUs) by clustering at 95% average nucleotide identity across 85% of contig length, according to the community-recommended standard (54, 55) (Materials and Methods). A final database of 45,033 sequences representing nonredundant virus OTUs was generated (Fig. 1), and this database will herein be referred to as the Cenote Human Virome Database (CHVD, download available at https://zenodo.org/record/4498884) (56).
Fig. 1.
CHVD metrics. (A) Each classified contig is represented as dot, with the x-axis position representing contig length. Width of violin diagrams represent the density of sequences at a given position and are proportional between categories. Larger (> 4 kb) Circular Rep-Encoding Single Stranded DNA [CRESS] virus OTUs consist of contigs in a previously reported taxon that combines CRESS-like replication genes with inovirus-like virion genes (107, 108). (B) Genome quality bins are derived from CheckV analysis, and body site labels are derived from sample metadata from the exemplar sequence of each virus OTU. (C) Data from virion-enriched stool samples are plotted. To measure the degree to which enrichment for viral sequences was achieved, a ViromeQC Enrichment Score (32) was calculated for each sample (x-axis). The enrichment score is essentially the inverse abundance of known bacterial single-copy marker genes. (Top) Dotted lines of the top panel are moving averages of samples from the same study. Asterisks indicate Bioprojects/samples with data used in the production of the CHVD. (Bottom) Production samples are removed. Data are binned by ViromeQC score, and boxplots represent IQR values, center lines representing median, and whiskers representing 1.5 IQRs. A modified database in which sequences were clustered at 99% identity instead of 95% identity was used for the index to better capture microdiversity and metaviromic islands (109) (e.g., intraspecific structural variations consisting of insertions/deletions of gene cassettes; Materials and Methods). (D) Plots are the same as C but for oral virion preps.
A total of 8,081 virus OTUs consisted of contigs with DTRs, and 112 were ITR bounded. To formally estimate the genome completeness of the virus OTU sequences, CheckV (55) was applied to the entire CHVD dataset (Fig. 1B). A total of 14,034 contigs (31.2%) were estimated to be high-quality (90 to 100% complete), 13,234 contigs (29.4%) were estimated to be medium quality (50 to 90% complete), 17,270 contigs (38.3%) were estimated to be low-quality genome fragments (1 to 50% complete), and 495 contigs (1.1%) were “not-determined.” Not-determined contigs could either be sequences too divergent from the CheckV references to be categorized, too short to categorize, or they could be false positives. Compared to other human-associated metagenome-assembled virus databases, such as the human subset of IMG/VR (versions 1 and 2) (55, 57) (in which ∼75% of contigs were estimated to be low-quality genome fragments or not-determined) or the recently published Gut Virome Database (GVD) (13) (Fig. 1B), CHVD is populated with a higher proportion and number of high- and medium-quality virus genomes, see Fig. 4B.
Fig. 4.
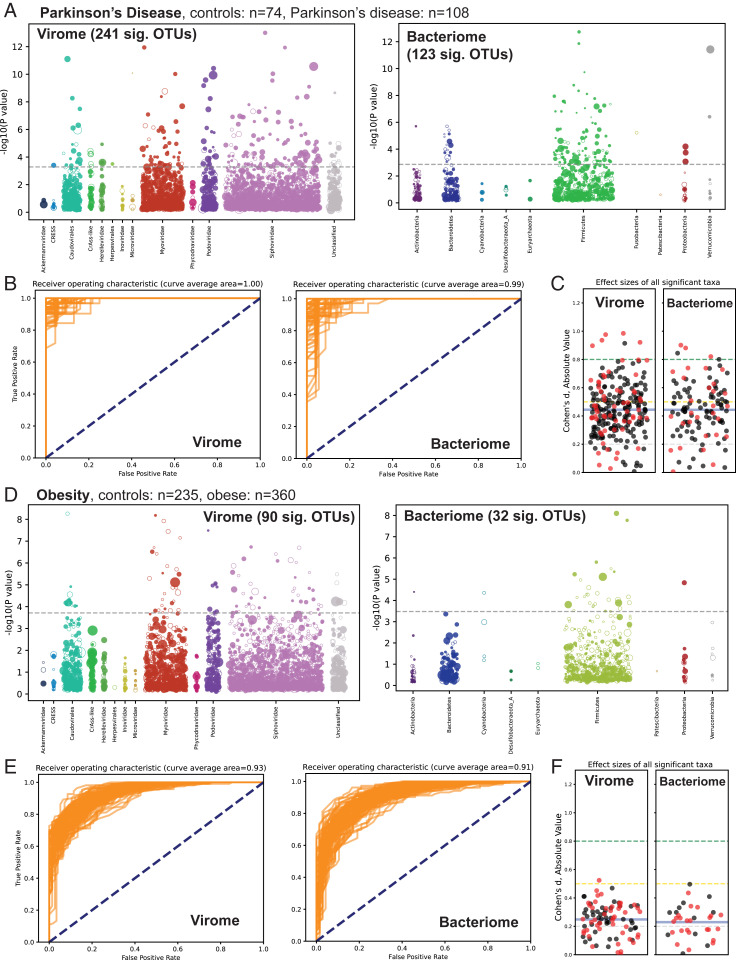
Association of the virome and bacteriome with chronic diseases. (A–C) Analysis of read data from PRJEB17784, a case-control study of stool samples from patients with or without Parkinson’s disease. (A) Virome-wide and bacteriome-wide associations in stool samples from Parkinson’s disease patients (n = 74) and healthy controls (n = 108) represented as Manhattan plots. Each OTU is represented as a dot along the x-axis, with its y-axis value being the inverse log10 P value. The size of each dot corresponds to the median relative abundance of the taxon in the disease cohort. Filled dots represent OTUs found at higher abundance in the diseased state while hollow dots represent decreased abundance in the diseased sate. The dashed gray line represents the false discovery rate < 1% threshold. (B) Receiver operating characteristic plots from 100 differently seeded random forest classifiers trained on the virome (Left) or bacteriome (Right). (C) Swarm plots of Cohen’s d effect sizes (absolute value) of OTUs achieving significant P values. Black dots are positive effect size, and red dots are negative effect size. The mean of all plotted effect sizes is drawn as a blue line. Small effect size = 0.2 to 0.5; medium effect size = 0.5 to 0.8; and large effect size = > 0.8 (84). (D–F) Similar analyses of read data from PRJEB4336, a WGS survey of stool samples from obese and nonobese individuals. Plots D, E, and F are laid out in the same manner as plots A, B, and C, respectively.
Although it is often challenging to obtain very long virus contigs from de novo assemblies, 119 virus OTUs over 200 kilobases (kb) were detected in the survey with the largest being Siphoviridae species ctpHQ1, at 501 kb. A total of 33 family- or order-level taxa were observed, and 2,087 sequences could not be classified by Cenote-Taker 2, representing many unrecognized high-level taxa (SI Appendix, Fig. S1). It is important to note that virus taxonomy, especially taxonomy of double stranded DNA (dsDNA) phages, is currently in flux (54, 58–60), and these taxonomic statistics will likely change as taxonomic groupings are revised. The vast majority of classified sequences represent dsDNA tailed phages in the order Caudovirales (including Siphoviridae, Podoviridae, Myoviridae, Ackermannviridae, Herelleviridae, and Cross-Assembly phage-like viruses [CrAss-like phage]). Relatively small numbers of known human-tropic viruses were uncovered, including members of families Adenoviridae, Anelloviridae, Circoviridae, Herpesviridae, Caliciviridae (Human norovirus), Papillomaviridae, and Polyomaviridae. Most of the human-tropic viruses mapped to previously reported virus species, but 16 previously undiscovered anelloviruses were detected (download available from: https://zenodo.org/record/4498884) (56). Dataset S2 provides spreadsheet information on each virus, including OTU, hallmark genes, CRISPR hits (see Fig. 2), and statistical information. In total, 757/5,996 Biosamples were virus-like particle libraries, resulting in 3,756 virus OTUs in the final database.
Fig. 2.
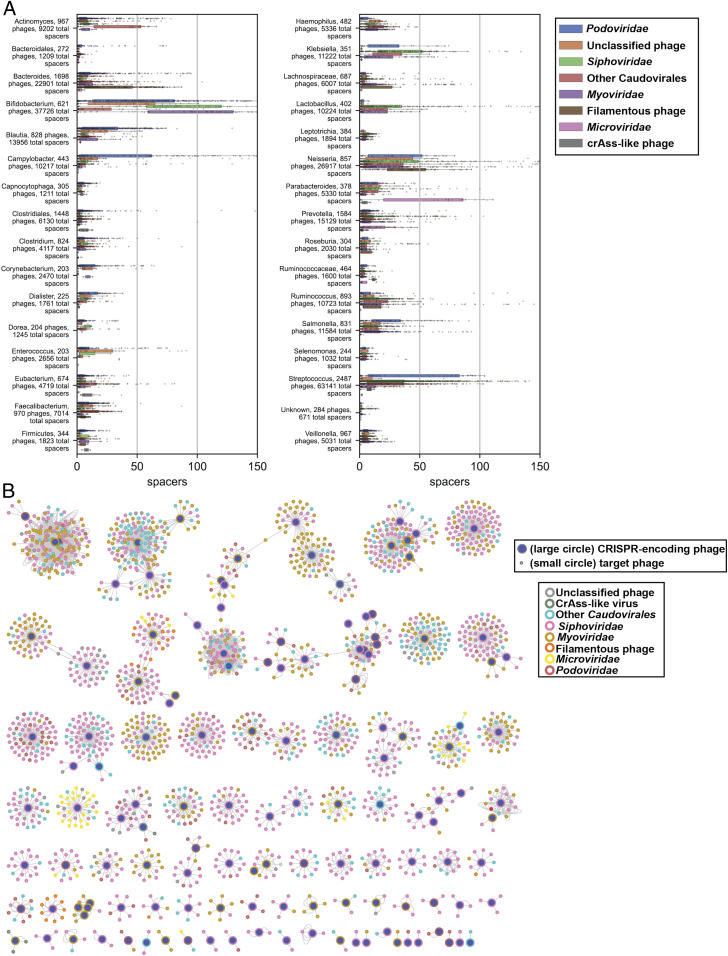
Summary of CRISPR spacer match data. (A) Plots represent matches of bacterially encoded CRISPR spacers to virus contigs. Categories are defined by bacterial genera (or higher taxon when genus is not clearly defined; Materials and Methods). Only genera with 200 or more CRISPR spacer matches to CHVD OTUs are displayed. The x-axis values represent the number of unique bacterial CRISPR spacer hits for each virus OTU. Filamentous phage = Inoviridae and other filamentous phages (e.g., certain CRESS viruses). (B) Network diagram of phage–phage interaction landscape based on CRISPR spacer matches. Each line represents a match of a particular spacer sequence to its target phage.
Fig. 1 presents a graphical summary of observed virus taxa. One taxon, designated “Phyco-like_phage,” is represented by 36 contigs. This is an interesting group of sequences initially binned with Phycodnaviridae due to distant similarity of the terminase/packaging gene of these viruses to a gene encoded by eukaryotic phycodnaviruses (∼30% AA similarity). However, most of the inferred virion structural genes that co-occupy these contigs are distantly similar to those of crAss-like phages, not phycodnaviruses, suggesting that they represent phages. This and the fact that most of the 2,087 “Unclassified” viral sequences have virion hallmark genes corresponding to dsDNA phage models (SI Appendix, Fig. S1) (Cytoscape network file available from https://zenodo.org/record/4498884) (56) supports the idea that substantial phage diversity remains unclassified and undescribed.
To evaluate the degree to which the observed CHVD virus OTUs are already represented in public databases, Mash (61) was used to measure roughly intraspecies-level nucleotide sequence similarity to 23,386 genomes from annotated virus species found in GenBank. With a Mash distance threshold of <0.05 (∼95% Average Nucleotide Identity [ANI]), 334/45,033 (0.7%) CHVD viruses had at least one strict cognate sequence in GenBank (Dataset S3). If the Mash distance threshold is relaxed to <0.1 (∼90% ANI, 2,310/45,033 (5.1%) CHVD viruses have a GenBank cognate (Dataset S3).
Recently, Gregory et al. (13) published a human GVD using different virus discovery methods and some overlapping datasets. The CHVD presented in this manuscript contains sequences from multiple human body sites, so comparisons are not perfect. However, we note that CHVD has 35% more contigs (45,033 versus 33,242) and 143% more sequence information (1.446 gigabases versus 0.596 gigabases) than GVD. The same Mash distance analysis was applied to compare the two datasets. With a Mash distance threshold of <0.05, 5,782 (12.8%) virus sequences from this study had a cognate in the GVD (matching to 5,614 sequences in GVD). Comparing just the subset of CHVD contigs derived from the gut, 5,704/30,863 (18.5%) had a GVD cognate. At a looser threshold (Mash distance <0.1), 18,002 (40.0%) CHVD sequences had cognates to 10,996 GVD sequences (Dataset S4).
Genome maps for all virus genomes (excluding those that were strict cognates to extant entries) were deposited in GenBank and given accession numbers in association with the Bioproject PRJNA573942. Per NCBI guidelines, the files will be released upon publication of this manuscript. Refer to Dataset S2 for accession numbers.
The Large Majority of Reads from Well-Enriched Virome Preparations Are Identifiable.
It is unclear how much of the human virome is cataloged by CHVD. One way to address this question is to look at datasets that are physically enriched for viral sequences and determine what fraction of reads in the dataset are identifiable. Reads from 983 human stool samples (representing 11 different studies) that were physically enriched for virions and subjected to nuclease digestion to remove nonencapsidated nucleic acids were aligned to the CHVD (Fig. 1 C, Upper) (Dataset S5). To quantify how well CHVD recruits reads from previously unanalyzed virion preps, samples used in the production of CHVD were removed, and median and interquartile range were calculated for different bins of ViromeQC score. This analysis shows that the percentage of reads aligning to CHVD scales with the ViromeQC enrichment score. Poorly enriched (<10 ViromeQC score) gut samples aligned about 10% of reads on average, and samples with high enrichment scores (>30) aligned on average 70 to 80% with many samples achieving 99% alignment.
Though well-enriched viromic data were not as available for other body sites, roughly 75% of reads were classifiable in well-enriched oral samples enriched for virus DNA (Fig. 1D) (Dataset S5).
CRISPR Spacer Analysis Reveals Candidate Hosts for Most Phages as well as Phage–Phage Competition Networks.
Many bacteria encode CRISPR-Cas systems, which contain CRISPR spacer arrays of short (∼32-nt) sequences copied from and used against invading mobile genetic elements, especially phages (62). Matching bacterial CRISPR spacer sequences to phage genomes is one way to determine whether a bacterial lineage has previously been exposed to a particular phage. Advances in cataloging of CRISPR spacers from bacterial genomes and optimization of phage/host matching pipelines allowed the association of most of the phages discovered in this project to bacterial hosts (http://crispr.genome.ulaval.ca/) (63). Specifically, 31,259 of the 45,033 virus sequences had at least one CRISPR spacer match from a known bacterium or multiple bacteria, with 369,465 total spacers matched to unique loci in CHVD sequences (Dataset S2). CRISPR spacer density varied dramatically among different bacterial taxa (Fig. 2A). For example, members of genus Bifidobacterium were confirmed to have relatively large and diverse CRISPR spacer libraries (64), while Clostridium, Capnocytophaga, and Leptotrichia typically encoded only one or a handful of spacers per phage.
Phages themselves can encode CRISPR arrays, and some phages have intact and functional CRISPR-Cas systems (22, 65). These CRISPR components can target host defenses as well as other phages competing for the same host (66). Among phage sequences in the CHVD, 1,971 CRISPR spacers were detected in arrays from the genomes of 203 phages. Of these, 799 spacers targeted a total of 2,036 other phages, suggesting complex phage–phage competition networks in human metagenomes (Fig. 2B) (download Cytoscape file from https://zenodo.org/record/4498884) (56). The bacterial host pool of CRISPR-encoding phage and their target phage should be the same. Therefore, bacterial CRISPR spacer matches for phage–phage pairs were documented, and, when a bacterial host could be determined for both the CRISPR-encoding phage and target phage, this bacterial genus was the same for 85.1% of pairs (Dataset S6).
The Most Commonly Abundant Viruses on Several Body Sites.
With this library of viruses and the large sampling effort from the Human Microbiome Project (34, 67), the question of “which viruses are the most commonly abundant” for a given body site can be answered more confidently than was previously possible. It should be noted that the Human Microbiome Project data were collected from healthy Americans between 18 and 40 y of age, and the conclusions here may not be generalizable to other populations.
It is often challenging to precisely determine the border between an integrated prophage and host chromosomal sequences without experimental validation, making virus quantification in whole-genome shotgun (WGS) datasets challenging. Our preliminary analyses revealed that inclusion of even a few hundred nucleotides of flanking host sequence in a viral OTU contig can greatly distort abundance measurements because of inadvertent measurement of uninfected host bacterial sequences. We therefore performed a more stringent analysis in which contigs were trimmed from the first recognized virus hallmark gene through the last virus hallmark gene. While removing some of the virus sequence, this method preserves the most indelible sequences of the virus genome while all but ensuring that no bacterial chromosome will be retained. We refer to these more stringent units as “virus cores” (download from https://zenodo.org/record/4498884) (56).
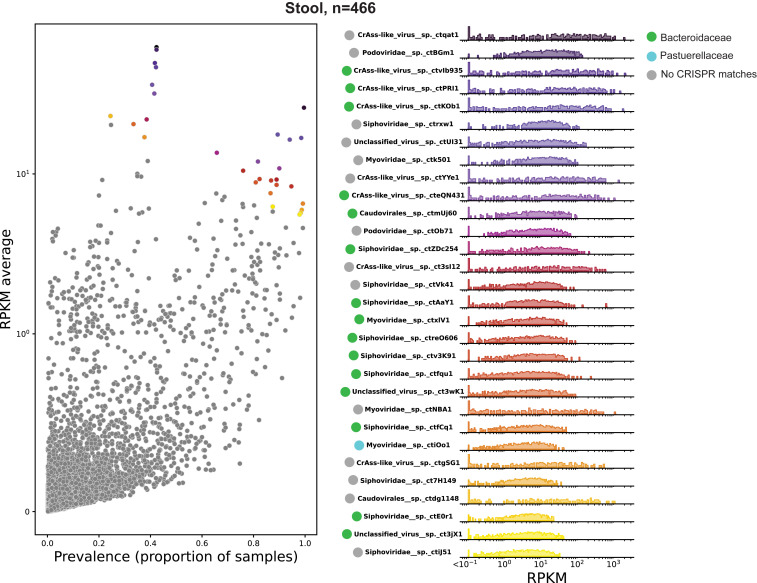
Data were downloaded from SRA and analyzed for hundreds of patients at six body sites (anterior nares, buccal mucosa, posterior fornix, tongue dorsum, supragingival plaque, and gut [stool]). Reads were then aligned to the more stringent virus cores database. As a proxy for the relative abundance of a given virus OTU, the average number of reads per kilobase of virus genome per million reads in the parent dataset (RPKM) was calculated for each sequence (Fig. 3, SI Appendix, Figs. S2 and S3, and Dataset S2). Virus prevalence was determined as proportion of samples with >0.1 RPKM. The most commonly abundant virus OTUs were calculated as (mean RPKM × prevalence). The right panel of Fig. 3 shows the inferred host for each of the top 30 most commonly abundant virus OTUs based on CRISPR spacer target information. A majority of the most commonly abundant viruses appear to infect members of the common bacterial family Bacteroidaceae, which is generally abundant in the human gut. Further, despite a large increase in alignable virus sequences compared to past studies, the observation that crAss-like phages are highly abundant in human gut ecosystems (12) seems to hold remarkably well.
Fig. 3.
Most common viruses, Stool (Gut). (Left) A scatter plot of RPKM (a measure of relative read abundance for a given virus OTU, y-axis) versus prevalence (proportion of samples with >0.1 RPKM, x-axis). For display purposes, the y-axis is a linear scale from 0 to 1 (100) and log10 above 1. The top 30 most commonly abundant virus OTUs (based on the product of coordinates) are colored. (Right) Histogram and rug plot of RPKM values across all samples for the most commonly abundant virus OTUs. Colors of dots in the Left correspond with the colors in the Right. The x- and y-axis are log scale. RPKM values below 0.1 are binned at the left extremity of the plots for display purposes.
The data suggest an interesting bifurcation in prevalence of gut virus OTUs with high abundance (RPKM). Although some virus OTUs, such as Podoviridae sp. ctBGm1 and Siphoviridae sp. ctrxw1, are present in nearly all samples and have an average abundance of >10 RPKM, perhaps representing prophage of ubiquitous bacterial lineages. Others, including all displayed crAss-like viruses and Myoviridae sp. ctNBA1, are absent or low abundance in most samples but highly abundant in a minority of samples. The latter group could either represent viruses that periodically undergo large replicative bursts, or viruses that constitutively dominate the virome in certain individuals but not others.
As expected, most virus OTUs were prevalent at only one body site, but 186 “cosmopolitan” OTUs had a prevalence of >0.2 (i.e., 20% of samples) in at least two body sites (Dataset S7). Bacterial CRISPRs targeted 128/186 sequences, with 36 being targeted by genus Cutibacterium, 17 being targeted by Staphylococcus, 16 being targeted by Strepococcus, and 14 by Bacteroides.
Specific Virus OTUs Are Associated with Human Disease.
A number of prior studies have looked for associations between the virome and human diseases (27–29, 52, 68–72). However, these studies were limited by the lack of a comprehensive virus reference database, and nearly all studies used samples physically enriched for viral sequences (71). Virus enrichment methods can be highly variable, however (Fig. 1C), and can inadvertently remove some viral taxa while failing to significantly select against host sequences (10, 32). Indeed, Gregory et al. (13) report that studies employing different virus enrichment protocols to investigate the same disease state (e.g., inflammatory bowel disease) rarely contain the same virus populations in their data. Instead, studies using similar enrichment protocols (regardless of disease state of patients) shared more virus populations. Furthermore, sequences encapsidated within virions may not be the best reflection of the total viral population, especially in human digestive tracts, where many phages are believed to exist primarily in lysogenic (nonlytic) states (73), and some have been “grounded,” losing their ability to independently excise from the host genome (74). It is possible that the most important phages for human physiology are those that express accessory genes from an integrated provirus state, as opposed to phages that are producing abundant virions. It is thus ideal to examine total DNA (also known as WGS) sequencing, which can detect all DNA virus genomes.
Our study reanalyzed publicly available WGS data from 12 large case-control studies analyzing stool and/or saliva (35, 41, 75–81). These studies examined Parkinson's disease, obesity, colon carcinoma, colon adenoma, liver cirrhosis, type 1 diabetes, ankylosing spondylitis, atherosclerosis, type 2 diabetes, hypertension, and nonalcoholic fatty liver disease. The virus cores database was used to compare the abundance of each virus OTU between case and control cohorts. Fig. 4 shows an analysis of case-control comparisons of Parkinson’s disease (population size, n = 182) (Fig. 4 A–C) and obesity (population size, n = 595) (Fig. 4 D–F). RPKM was used to measure virus OTU abundance in each sample, and Wilcoxon rank-sum tests with 100 bootstraps were conducted for each comparison to calculate the P value (Fig. 4 A and D “Virome,” SI Appendix, Figs. S4 and S5). Statistically significant virus OTUs were determined by a false discovery rate < 1% (Materials and Methods). All analyses compared associations between the virome and the “bacteriome,” measuring the bacteriome in terms of bacterial OTUs (i.e., species-level single-copy bacterial marker gene abundance) using IGGsearch (82) (Fig. 4 A and D “Bacteriome,” SI Appendix, Figs. S4 and S5). A higher number of statistically significant taxa were found for the virome than the bacteriome in eight studies. The four other studies analyzed yielded no significant OTUs for either the virome or the bacteriome (Fig. 4 and SI Appendix, Figs. S4 and S5). P values for each virus OTU detected in each study are documented in Dataset S2. Furthermore, random Forest Classifiers, trained on either all virus OTUs or all bacterial OTUs, were more successful or equally successful, on average, in discriminating healthy and diseased patients using the virome data rather than the bacteriome data in 7/12 case-control populations (Fig. 4 B and E and SI Appendix, Figs. S6 and S7).
The importance of considering effect size when reporting microbiome associations has become apparent in recent years (83, 84). Therefore, for all virus and bacterial OTUs with significant differences between cases and controls, Cohen’s d effect size is reported for each disease state (Fig. 4 C and F and SI Appendix, Figs. S6 and S7).
It is not possible to make one-to-one comparisons of virus OTUs and bacterial OTUs because many phages are capable of infecting and replicating in multiple bacterial species (85), sometimes even in multiple bacterial genera (86), while, at the same time, lineages within a single bacterial species have different abilities to resist or acquire immunity to specific phage (87). Therefore, it is beyond the scope of this study to investigate how specific virus OTUs could be bolstering or depressing the fitness of particular bacterial OTUs in individual gut ecosystems. Nevertheless, it can be informative to qualitatively compare statistically significant virus OTUs and their putative host bacterial genera (per CRISPR spacer match) (SI Appendix, Fig. S8). At a glance, significant virus OTUs targeted by CRISPR spacers from the same bacterial genus seem to all trend the same direction and the same direction of most possible bacterial hosts that have significant differences between cases and controls. This is consistent with lysogenic prophages as well as increase in available hosts supporting larger virus populations (15).
Discussion
This study shows that, by leveraging virus-specific hallmark genes, it is possible to mine human metagenomic data at a large scale to create a database composed largely of previously unknown virus sequences that captures most reads generated from virion-enriched datasets from stool and saliva. This advance, in turn, revealed hidden associations between a variety of chronic disease states and specific virus taxa. It should be stressed that association does not necessarily imply causation, and a variety of associative relationships between viruses and a given disease state are possible. For instance, virus abundance might simply be an epiphenomenon reflecting bacterial host abundance, the human genetics that predispose people to a disease might also provide a more favorable environment for the virus or its bacterial host, the external causes of a disease may create a more favorable environment for the virus, or the virus may contribute to the disease presentation in some way but ultimately does not cause the disease in isolation from other important factors. Verifying the associations we have detected with independent studies of the same diseases in additional populations will be key to understanding the extent to which the findings presented here are generalizable. If the associations are confirmed, it might be possible to experimentally test the causality question by adding or removing phages of interest from gut ecosystems in animal model systems (88).
A limitation of the case-control studies analyzed here is that they only consisted of a single timepoint for each subject. Virome composition can be noisy, and longitudinal data on individual patients might be more effective for discerning stable viral populations (11). This problem may have been partly offset by use of large cohort sizes (mostly over 150 total patients). Furthermore, just as individual bacterial strains host a diversity of nonessential “accessory genes” not shared by all strains within the bacterial species (89), virus strains have unique sets of genes compared to intraspecific relatives (90), reflecting viral pangenomes or “metaviromic islands.” With the current approach, most of the intraspecies accessory gene content is left out due to sequence dereplication, and the importance of these genes was not evaluated. Another limitation is that the analyzed case-control surveys only used DNA WGS methods whereas RNA sequencing of metatranscriptomes might have provided more functional data on expression of specific viral genes, potentially leading to testable hypotheses about possible mechanisms of action. It is also conceivable that correlations for viruses with RNA genomes would be uncovered. Despite these limitations, the current study shows that, using random Forest Classifiers, the virome may well be more diagnostic than the bacteriome for a variety of chronic diseases. The strong associations of specific virus OTUs in chronic diseases, along with medium-to-large–effect sizes for many OTUs, cries out for more mechanistic investigation of possible causal roles for viruses in chronic human disease.
While we maintain that this effort is a significant step forward, it is likely that the CHVD could be improved both in depth and breadth. Metagenomes from more body sites, such as the pulmonary tract (91), could be analyzed, and sequencing runs representing more geographic and lifestyle diversity could be used. Further, analysis of additional metatranscriptomic datasets would likely uncover more RNA viruses.
Even with the relatively inclusive criteria used by Cenote-Taker 2 (discernable amino acid similarity of a viral hallmark gene to a protein the RefSeq virus database), thousands of viruses that live on humans from this dataset could not be taxonomically classified, suggesting that additional families of as-yet-unidentified viruses await formal discovery and categorization.
Materials and Methods
Identification of Viral Contigs in Assemblies.
Human Microbiome Project studies and other arbitrarily selected human metagenome studies (Dataset S1) were downloaded from SRA, and unique Biosamples were delineated. All runs from a given Biosample were downloaded concurrently, preprocessed with Fastp (92), and coassembled with Megahit (93) using default settings. Subsequent contigs were fed to Cenote-Taker 2 (https://github.com/mtisza1/Cenote-Taker2, https://cyverse.org/discovery-environment), which, in short, looks for genome end features (direct terminal repeats and inverted terminal repeats), translates genes into amino acid sequences, compares each amino acid sequence to a hidden Markov model database comprised of virus hallmark gene alignments, keeps contigs with minimum number of genes matching to the hallmark database, then identifies and prunes flanking bacterial chromosome sequences. All remaining gene features are then annotated to create a genome map. Cenote-Taker 2 was used with settings to consider circular or LTR-bearing contigs (minimum 21 nt identical direct repeats at termini of contig) of at least 1500 nt, ITR-containing contigs of at least 4 kb, and linear contigs of at least 12 kb. These contigs were scanned for genes matching viral hallmark models. Terminal repeat–containing contigs with one or more viral hallmark genes were kept, and linear contigs with two or more viral hallmark genes were also kept. For every run, regardless of whether the sample had been physically enriched for virions, the Cenote-Taker 2 prophage pruning module was employed. Cenote-Taker 2 hallmark gene database was the September 15th, 2020 version (https://github.com/mtisza1/Cenote-Taker2). While Cenote-Taker 2 does take steps to earmark potential plasmids and conjugative transposons, extra precautions were taken by removing ∼4,000 putative viral sequences from the nonredundant database that contained replication-associated but not virion- or genome-packaging hallmark genes. For metatranscriptome datasets, all contigs over 1,500 nt with RNA virus hallmark genes were kept as putative viral sequences, regardless of end features.
Clustering Similar Contigs for Dereplication.
A twofold approach was used to cluster genomes. First, contigs were binned with Mash (61), utilizing its ability to handle massive sequence databases, accuracy, and lack of issues arising from genome circularity. All viruses within each higher-level taxon (e.g., Microviridae) were used to create Mash sketches (options -k 16 -s 500), and then these sketches were compared to themselves with Mash’s dist function. Within close genomic distances, the value of the Mash distance score is thought to roughly recapitulate average nucleotide divergence. Virus strain–level distinctions are often defined by <5% average nucleotide divergence (54), so sequence similarity networks were constructed with connections between sequences (nodes) with Mash distance scores ≤0.05 (and P value ≤1 × 10−10). Markov clustering algorithm (MCL clustering) (94) was applied to Mash networks to generate OTU-level clusters. From each cluster of sequences, if circular or ITR-encoding sequences were present, the longest such sequence was used as the representative virus OTU sequence. If only linear sequences were present, the longest linear sequence was used as the representative of the cluster. Singleton contigs (i.e., sequences that were not assigned to any cluster) were also retained for the final database. The same approach was applied for the 99% database (for virus-like particle sequence alignment), but a Mash distance score of ≤0.01 was used.
Following Mash clustering, a basic local alignment search tool (BLAST)–based approach was used for final dereplication. Nucleotide BLAST (BLASTN), anicalc.py, and aniclust.py were used from the CheckV (55) suite of tools as described in the CheckV ReadMe (https://bitbucket.org/berkeleylab/checkv/src/master/), with options “–min_ani 95 –min_qcov 0 –min_tcov 85” used for aniclust to dereplicate sequences into virus OTUs at “95% average nucleotide identity over 85% alignment fraction” per community standards (54). Best representative sequences from aniclust.py were used as virus OTU exemplars comprising the CHVD version 1.1. Future versions will be dereplicated with the “anicalc/aniclust” approach only.
Assessing Genome Completeness of Virus OTUs.
A total of 45,033 dereplicated virus OTU sequences from the CHVD were run through CheckV version 0.7.0 (i.e., checkv end_to_end) (55) with default parameters. Completeness estimates for each sequence were taken from the quality_summary.tsv table, and these values are reported in Dataset S2. This analysis was run in the same manner on the GVD (downloaded from https://datacommons.cyverse.org/browse/iplant/home/shared/iVirus/Gregory_and_Zablocki_GVD_Jul2020/GVD_Viral_Populations). IMG/VR “human-associated” contig metrics were taken from the CheckV manuscript files.
Identifying Cognate Viruses in GenBank and the Human GVD.
Using the NCBI Virus Resource, metadata for all virus genomes listed as complete were downloaded for the following taxa: Adenoviridae, Anelloviridae, Bromoviridae, Caliciviridae, Circoviridae, Cressdnavircota, Herpesviridae, Luteoviridae, Narnaviridae, Nodaviridae, Papillomaviridae, Polyomaviridae, Tombusviridae, Totiviridae, Tymovirales, unclassified viruses, unclassified RNA virus, Virgaviridae, and all bacteriophage (including prophage). The metadata were sorted so that the longest sequence for each unique species name was selected, and these sequences were subsequently downloaded. Additionally, many GenBank virus genomes simply have a family label followed by the indeterminate abbreviation “sp.,” and, as a result, many highly distinct sequences inadvertently share an identical generic label. Therefore, all complete GenBank virus genomes from all nonredundant taxa with an ‘sp.’ designation were downloaded. A Mash sketch was made for the downloaded sequences using options (-k 16 -s 500), and this Mash sketch was compared to the CHVD Mash sketch (see Clustering Similar Contigs for Dereplication). Mash distances of ≤0.05 and P value ≤1 × 10−10 were considered to be strict cognate (intraspecies or intrastrain) sequences. Mash distances of ≤0.1 and a P value of ≤1 × 10−5 were used for “loose” cognate sequences.
The GVD from Gregory et al. (13) was downloaded from https://datacommons.cyverse.org/browse/iplant/home/shared/iVirus/Gregory_and_Zablocki_GVD_Jul2020/GVD_Viral_Populations. The same Mash analyses were applied for comparisons with this dataset as with the GenBank database.
Deposition of Virus Genomes in GenBank.
All sequences from CHVD v1.1 were considered for deposition into GenBank. First, sequences with strict GenBank cognates were discarded. We wanted to minimize any overhanging chromosomal sequences from prophage genomes. Therefore, non–DTR-encoding (i.e., linear) sequences (already trimmed with Cenote-Taker 2 pruning module) were trimmed again with CheckV (v 0.7.0) as we found this approach to be more conservative than Cenote-Taker 2. These double-trimmed sequences were then annotated with Cenote-Taker 2, with full metadata, CRISPR spacer matches, and read coverage information. Then, the corresponding “.sqn” files were sent to GenBank as “TPA assembly” virus genomes. All submitted sequences/genomes are associated with Bioproject PRJNA573942 and will be released upon publication of this manuscript. Accession numbers can be found in Dataset S2.
Gene Sharing Network for Unclassified Viruses.
Vcontact2 (59, 95) was run using all RefSeq v88 bacteriophage genomes with recommended settings and all viruses from the CHVD that were labeled “unclassified” in the taxonomy field. The resulting network was displayed in Cytoscape (96) and colored manually.
Virus Cores.
Using all virus OTUs from CHVD, virus core coordinates were obtained computationally. Cenote-Taker 2 scans contigs for virus hallmark genes and outputs coordinates for each hallmark gene in the context of the contig. The stop and start coordinates for each hallmark gene were compiled, and the lowest and highest coordinates from each contig were taken, and bioawk was used to trim each fasta nucleotide sequence to start and end with these coordinates, discarding peripheral sequences.
Bacteria-Encoded CRISPR Spacer Analysis.
CrisprOpenDB (https://github.com/edzuf/CrisprOpenDB) was used (commit 04e4ffcc55d65cf8e13afe55e081b14773a6bb70) to assign phages to hosts based on CRISPR spacer match using BLASTN (63). Three mismatches were allowed for hits. For hits to bacteria without a currently assigned genus, family-level or order-level taxonomical information was pulled from the output table, when possible.
Phage-Encoded CRISPR Spacer Analysis.
All virus OTU sequences were processed with MinCED (https://github.com/ctSkennerton/minced) to discover CRISPR spacer arrays. As phages can encode CRISPR arrays with spacers as short as 14 nucleotides (97), MinCED was allowed to detect arrays with spacers of 14 or more nucleotides. The CRISPR array regions of phage genomes were masked using Bedtools maskfasta (98), and then all virus OTUs were queried with BLASTN against a database of the CRISPR spacers.
Only hits aligning to the entire length of the spacer and with the following criteria were kept: perfect matches to spacers 16 to 20 nucleotides, matches to spacers 20 to 27 nucleotides in which (mismatches + gaps) is 1 or 0, and matches to spacers ≥ 28 nucleotides in which (mismatches + gaps) are 2 or fewer.
Determining Abundance of Individual Virus OTUs in Metagenomes.
The final database of “virus core” sequences was processed by RepeatMasker to remove low-complexity regions which recruit reads nonspecifically (99). Bowtie2 (100) was used to align reads to the database, and samtools (101) idxstats was used to calculate read coverage and RPKM for each contig.
Comparing OTU Abundance and Discriminatory Ability in Case-Control Studies.
For each Bioproject, case versus control samples were determined, if possible, using categories from Nayfach et al. (82), as patients on confounding medications were removed in this analysis. For other Bioprojects, metadata were taken from SRA (102) run selector (Dataset S8 A–K). For all samples, reads were downloaded from the SRA and trimmed and quality-controlled with Fastp (92). To quantify abundance of bacterial taxa in each sample, IGGsearch was used with default parameters, except the “–all-species” option was employed (82).
Wilcoxon rank-sum test was computed with 100 bootstraps using Python, NumPy, and SciPy (103) for each OTU in a given study in which at least 10% of the total samples had an RPKM of at least 0.05 (bacterial OTUs with “IGGsearch abundance” of at least 0.005 in at least 10% of the samples were kept). False discovery rate (< 1%) was determined with the Benjamini–Hochberg method using SciPy. Cohen’s d effect size was calculated for each OTU above the significance threshold using DaBest Python package (104) with 5,000 bootstraps.
Random Forest Classifiers from scikit-learn were used (105). Training/test set sizes were 70%/30%, number of estimators was 100, and a different seed was used for each of the 100 Random Forest Classifiers trained on each dataset.
Note
Although about 6,000 “Biosamples” encompassing over 16,000 sequencing runs were analyzed in this study, another study (published while this manuscript was under review) was able to mine 28,060 gut metagenome sequencing runs to detect putative bacteriophage sequences (106).
Supplementary Material
Acknowledgments
We acknowledge Gabriel Starrett and Nathan Fons for their helpful discussions on statistical analyses of the data types used in this paper. Also, we are grateful to Linda Frisse and her colleagues at GenBank (National Center for Biotechnology Information, National Library of Medicine, NIH) for assistance with sequence deposits. This work utilized the computational resources of the NIH high performance computing (HPC) Biowulf cluster (https://hpc.nih.gov/). This research was funded by the Intramural Research Program of the NIH and the National Cancer Institute.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023202118/-/DCSupplemental.
Data Availability
Fasta sequence files from CHVD databases can be accessed at (https://zenodo.org/record/4498884) (56). Additionally, all unique virus genomes with metadata and annotated genome maps have been deposited to GenBank under BioProject PRJNA573942. Accession numbers for individual virus OTUs can be found in Dataset S2. Cenote-Taker 2 was accessed at https://github.com/mtisza1/Cenote-Taker2.
References
- 1.Pastrana D. V., et al., Metagenomic discovery of 83 new human papillomavirus types in patients with immunodeficiency. mSphere 3, e00645-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wylie K. M., Weinstock G. M., Storch G. A., Emerging view of the human virome. Transl. Res. 160, 283–290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beller L., Matthijnssens J., What is (not) known about the dynamics of the human gut virome in health and disease. Curr. Opin. Virol. 37, 52–57 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Gilbert J. A., et al., Current understanding of the human microbiome. Nat. Med. 24, 392–400 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manrique P., et al., Healthy human gut phageome. Proc. Natl. Acad. Sci. U.S.A. 113, 10400–10405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reyes A., et al., Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc. Natl. Acad. Sci. U.S.A. 112, 11941–11946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitbart M., et al., Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185, 6220–6223 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minot S., et al., The human gut virome: Inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minot S., et al., Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. U.S.A. 110, 12450–12455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shkoporov A. N., et al., Reproducible protocols for metagenomic analysis of human faecal phageomes. Microbiome 6, 68 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shkoporov A. N., et al., The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe 26, 527–541.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Dutilh B. E., et al., A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 5, 4498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregory A. C., et al., The gut virome database reveals age-dependent patterns of virome diversity in the human gut. Cell Host Microbe 28, 724–740.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gandon S., Buckling A., Decaestecker E., Day T., Host-parasite coevolution and patterns of adaptation across time and space. J. Evol. Biol. 21, 1861–1866 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Silveira C. B., Rohwer F. L., Piggyback-the-Winner in host-associated microbial communities. NPJ Biofilms Microbiomes 2, 16010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirzaei M. K., Maurice C. F., Ménage à trois in the human gut: Interactions between host, bacteria and phages. Nat. Rev. Microbiol. 15, 397–408 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Gamage S. D., Patton A. K., Hanson J. F., Weiss A. A., Diversity and host range of Shiga toxin-encoding phage. Infect. Immun. 72, 7131–7139 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner P. L., Waldor M. K., Bacteriophage control of bacterial virulence. Infect. Immun. 70, 3985–3993 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuch R., Fischetti V. A., Detailed genomic analysis of the Wbeta and gamma phages infecting Bacillus anthracis: Implications for evolution of environmental fitness and antibiotic resistance. J. Bacteriol. 188, 3037–3051 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fridman S., et al., A myovirus encoding both photosystem I and II proteins enhances cyclic electron flow in infected Prochlorococcus cells. Nat. Microbiol. 2, 1350–1357 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Howard-Varona C., et al., Phage-specific metabolic reprogramming of virocells. ISME J. 14, 881–895 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Shayeb B., et al., Clades of huge phages from across Earth’s ecosystems. Nature 578, 425–431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fluckiger A., et al., Cross-reactivity between tumor MHC class I-restricted antigens and an enterococcal bacteriophage. Science 369, 936–942 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Sweere J. M., et al., Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science 363, eaat9691 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarafder A. K., et al., Phage liquid crystalline droplets form occlusive sheaths that encapsulate and protect infectious rod-shaped bacteria. Proc. Natl. Acad. Sci. U.S.A. 117, 4724–4731 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordillo Altamirano F. L., Barr J. J., Phage therapy in the postantibiotic era. Clin. Microbiol. Rev. 32, e00066-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clooney A. G., et al., Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe 26, 764–778.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Norman J. M., et al., Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160, 447–460 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakatsu G., et al., Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology 155, 529–541.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Roux S., Enault F., Hurwitz B. L., Sullivan M. B., VirSorter: Mining viral signal from microbial genomic data. PeerJ 3, e985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kieft K., Zhou Z., Anantharaman K., VIBRANT: Automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome 8, 90 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zolfo M., et al., Detecting contamination in viromes using ViromeQC. Nat. Biotechnol. 37, 1408–1412 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Tisza M. J., Belford A. K., Dominguez-Huerta G., Bolduc B., Buck C. B., Cenote-Taker 2 democratizes virus discovery and sequence annotation. Virus Evol. 7, veaa100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lloyd-Price J., et al., Strains, functions and dynamics in the expanded human microbiome project. Nature 550, 61–66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J., et al., Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 5, 14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karlsson F. H., et al., Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498, 99–103 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Bäckhed F., et al., Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 852 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Horta-Baas G., et al., Intestinal dysbiosis and rheumatoid arthritis: A link between gut microbiota and the pathogenesis of rheumatoid arthritis. J. Immunol. Res. 2017, 4835189 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pehrsson E. C., et al., Interconnected microbiomes and resistomes in low-income human habitats. Nature 533, 212–216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rampelli S., et al., Metagenome sequencing of the Hadza hunter-gatherer gut microbiota. Curr. Biol. 25, 1682–1693 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Heintz-Buschart A., et al., Integrated multi-omics of the human gut microbiome in a case study of familial type 1 diabetes. Nat. Microbiol. 2, 16180 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Karlsson F., Tremaroli V., Nielsen J., Bäckhed F., Assessing the human gut microbiota in metabolic diseases. Diabetes 62, 3341–3349 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olm M. R., et al., Identical bacterial populations colonize premature infant gut, skin, and oral microbiomes and exhibit different in situ growth rates. Genome Res. 27, 601–612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu W., et al., Unique features of ethnic Mongolian gut microbiome revealed by metagenomic analysis. Sci. Rep. 6, 34826 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qin J., et al., A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Oh J.et al.; NISC Comparative Sequencing Program , Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pasolli E., et al., Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176, 649–662.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koenig J. E., et al., Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 108 (suppl. 1), 4578–4585 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maqsood R., et al., Discordant transmission of bacteria and viruses from mothers to babies at birth. Microbiome 7, 156 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yinda C. K., et al., Gut virome analysis of Cameroonians reveals high diversity of enteric viruses, including potential interspecies transmitted viruses. mSphere 4, e00585-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coffey M. J., et al., The intestinal virome in children with cystic fibrosis differs from healthy controls. PLoS One 15, e0233557 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuo T., et al., Gut mucosal virome alterations in ulcerative colitis. Gut 68, 1169–1179 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abu-Ali G. S., et al., Metatranscriptome of human faecal microbial communities in a cohort of adult men. Nat. Microbiol. 3, 356–366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roux S., et al., Minimum information about an uncultivated virus genome (MIUViG). Nat. Biotechnol. 37, 29–37 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nayfach S., et al., CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol., 10.1038/s41587-020-00774-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tisza M. J., Virus sequences and data tables related to the Cenote Human Virome Database v1.1. Zenodo. https://zenodo.org/record/4498884. Deposited 2 February 2021.
- 57.Paez-Espino D., et al., IMG/VR: A database of cultured and uncultured DNA viruses and retroviruses. Nucleic Acids Res. 45, D457–D465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koonin E. V., et al., Global organization and proposed megataxonomy of the virus world. Microbiol. Mol. Biol. Rev. 84, e00061-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bin Jang H., et al., Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 37, 632–639 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Gregory A. C.et al.; Tara Oceans Coordinators , Marine DNA viral macro- and microdiversity from Pole to Pole. Cell 177, 1109–1123.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ondov B. D., et al., Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 17, 132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Makarova K. S., et al., Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9, 467–477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dion M. B., et al., Streamlining CRISPR spacer-based bacterial host predictions to decipher the viral dark matter. Nucleic Acids Res. 49, 3127–3138 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Briner A. E., et al., Occurrence and diversity of CRISPR-Cas systems in the genus Bifidobacterium. PLoS One 10, e0133661 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seed K. D., Lazinski D. W., Calderwood S. B., Camilli A., A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature 494, 489–491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Faure G., et al., CRISPR-Cas in mobile genetic elements: Counter-defence and beyond. Nat. Rev. Microbiol. 17, 513–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turnbaugh P. J., et al., The human microbiome project. Nature 449, 804–810 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao G., et al., Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc. Natl. Acad. Sci. U.S.A. 114, E6166–E6175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fernandes M. A., et al., Enteric virome and bacterial microbiota in children with ulcerative colitis and Crohn disease. J. Pediatr. Gastroenterol. Nutr. 68, 30–36 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kramná L., et al., Gut virome sequencing in children with early islet autoimmunity. Diabetes Care 38, 930–933 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Ma Y., You X., Mai G., Tokuyasu T., Liu C., A human gut phage catalog correlates the gut phageome with type 2 diabetes. Microbiome 6, 24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cornuault J. K., et al., Phages infecting Faecalibacterium prausnitzii belong to novel viral genera that help to decipher intestinal viromes. Microbiome 6, 65 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sutton T. D. S., Hill C., Gut bacteriophage: Current understanding and challenges. Front. Endocrinol. (Lausanne) 10, 784 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramisetty B. C. M., Sudhakari P. A., Bacterial ‘grounded’ prophages: Hotspots for genetic renovation and innovation. Front. Genet. 10, 65 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Le Chatelier E.et al.; MetaHIT consortium , Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546 (2013). [DOI] [PubMed] [Google Scholar]
- 76.Loomba R., et al., Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab. 30, 607 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wen C., et al., Quantitative metagenomics reveals unique gut microbiome biomarkers in ankylosing spondylitis. Genome Biol. 18, 142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bedarf J. R., et al., Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naïve Parkinson’s disease patients. Genome Med. 9, 39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qin N., et al., Alterations of the human gut microbiome in liver cirrhosis. Nature 513, 59–64 (2014). [DOI] [PubMed] [Google Scholar]
- 80.Jie Z., et al., The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 8, 845 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang X., et al., The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 21, 895–905 (2015). [DOI] [PubMed] [Google Scholar]
- 82.Nayfach S., Shi Z. J., Seshadri R., Pollard K. S., Kyrpides N. C., New insights from uncultivated genomes of the global human gut microbiome. Nature 568, 505–510 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Debelius J., et al., Tiny microbes, enormous impacts: What matters in gut microbiome studies? Genome Biol. 17, 217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sawilowsky S. S., New effect size rules of thumb. J. Mod. Appl. Stat. Methods 8, 597–599 (2009). [Google Scholar]
- 85.Kauffman K. M., et al., A major lineage of non-tailed dsDNA viruses as unrecognized killers of marine bacteria. Nature 554, 118–122 (2018). [DOI] [PubMed] [Google Scholar]
- 86.Bradley D. E., Rutherford E. L., Basic characterization of a lipid-containing bacteriophage specific for plasmids of the P, N, and W compatibility groups. Can. J. Microbiol. 21, 152–163 (1975). [DOI] [PubMed] [Google Scholar]
- 87.Nussenzweig P. M., Marraffini L. A., Molecular mechanisms of CRISPR-Cas immunity in bacteria. Annu. Rev. Genet. 54, 93–120 (2020). [DOI] [PubMed] [Google Scholar]
- 88.Arrieta M. C., Walter J., Finlay B. B., Human microbiota-associated mice: A model with challenges. Cell Host Microbe 19, 575–578 (2016). [DOI] [PubMed] [Google Scholar]
- 89.Koonin E. V., Makarova K. S., Wolf Y. I., Evolution of microbial genomics: Conceptual shifts over a quarter century. Trends Microbiol., 10.1016/j.tim.2021.01.005 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bellas C. M., Schroeder D. C., Edwards A., Barker G., Anesio A. M., Flexible genes establish widespread bacteriophage pan-genomes in cryoconite hole ecosystems. Nat. Commun. 11, 4403 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Abbas A. A., et al., Redondoviridae, a family of small, circular DNA viruses of the human oro-respiratory tract associated with periodontitis and critical illness. Cell Host Microbe 26, 297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen S., Zhou Y., Chen Y., Gu J., fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li D., et al., MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 102, 3–11 (2016). [DOI] [PubMed] [Google Scholar]
- 94.Morris J. H., et al., clusterMaker: A multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics 12, 436 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Devisetty U. K., Kennedy K., Sarando P., Merchant N., Lyons E., Bringing your tools to CyVerse discovery environment using docker. F1000 Res. 5, 1442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Su G., Morris J. H., Demchak B., Bader G. D., Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinformatics 47, 8.13.11–8.13.24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pausch P., et al., CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 369, 333–337 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Quinlan A. R., BEDTools: The Swiss-army tool for genome feature analysis. Curr. Protoc. Bioinformatics 47, 11 12 11–11 12 34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen N., Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics Chapter 4, Unit 4 10 (2004). [DOI] [PubMed] [Google Scholar]
- 100.Langdon W. B., Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min. 8, 1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li H.et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kodama Y., Shumway M., Leinonen R.; International Nucleotide Sequence Database Collaboration , The sequence read archive: Explosive growth of sequencing data. Nucleic Acids Res. 40, D54–D56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Virtanen P.et al.; SciPy 1.0 Contributors , SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ho J., Tumkaya T., Aryal S., Choi H., Claridge-Chang A., Moving beyond P values: Data analysis with estimation graphics. Nat. Methods 16, 565–566 (2019). [DOI] [PubMed] [Google Scholar]
- 105.Abraham A., et al., Machine learning for neuroimaging with scikit-learn. Front. Neuroinform. 8, 14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Camarillo-Guerrero L. F., Almeida A., Rangel-Pineros G., Finn R. D., Lawley T. D., Massive expansion of human gut bacteriophage diversity. Cell 184, 1098–1109.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tisza M. J., et al., Discovery of several thousand highly diverse circular DNA viruses. eLife 9, e51971 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roux S., et al., Cryptic inoviruses revealed as pervasive in bacteria and archaea across Earth’s biomes. Nat. Microbiol. 4, 1895–1906 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mizuno C. M., Ghai R., Rodriguez-Valera F., Evidence for metaviromic islands in marine phages. Front. Microbiol. 5, 27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Although about 6,000 “Biosamples” encompassing over 16,000 sequencing runs were analyzed in this study, another study (published while this manuscript was under review) was able to mine 28,060 gut metagenome sequencing runs to detect putative bacteriophage sequences (106).
Fasta sequence files from CHVD databases can be accessed at (https://zenodo.org/record/4498884) (56). Additionally, all unique virus genomes with metadata and annotated genome maps have been deposited to GenBank under BioProject PRJNA573942. Accession numbers for individual virus OTUs can be found in Dataset S2. Cenote-Taker 2 was accessed at https://github.com/mtisza1/Cenote-Taker2.