Significance
Androgen plays essential roles for sex differentiation of external genitalia through regulating male-biased genes. Even females can differentiate into male-type external genitalia depending on the degree of androgenic exposure. However, the mechanisms underlying androgen responsiveness in females are poorly defined. Here, we demonstrate Sp1 regulates the enrichment of H3K27ac on androgen receptor-mediated regulatory elements in both males and females, indicating transcriptional competency for male-biased genes is conserved by Sp1 in both sexes. Compound-MafB mutant mice lacking a tissue-specific enhancer in the MafB 3′ UTR show incomplete male-type external differentiation similar to female-type differentiation. These findings underscore the idea that transcriptional competency for male-biased genes is preserved in both sexes, thus helping define the potential sexual fate of external genitalia.
Keywords: androgen responsiveness, sex differentiation, external genitalia, Sp1, histone modifications
Abstract
Testicular androgen is a master endocrine factor in the establishment of external genital sex differences. The degree of androgenic exposure during development is well known to determine the fate of external genitalia on a spectrum of female- to male-specific phenotypes. However, the mechanisms of androgenic regulation underlying sex differentiation are poorly defined. Here, we show that the genomic environment for the expression of male-biased genes is conserved to acquire androgen responsiveness in both sexes. Histone H3 at lysine 27 acetylation (H3K27ac) and H3K4 monomethylation (H3K4me1) are enriched at the enhancer of male-biased genes in an androgen-independent manner. Specificity protein 1 (Sp1), acting as a collaborative transcription factor of androgen receptor, regulates H3K27ac enrichment to establish conserved transcriptional competency for male-biased genes in both sexes. Genetic manipulation of MafB, a key regulator of male-specific differentiation, and Sp1 regulatory MafB enhancer elements disrupts male-type urethral differentiation. Altogether, these findings demonstrate conservation of androgen responsiveness in both sexes, providing insights into the regulatory mechanisms underlying sexual fate during external genitalia development.
Testicular androgen is the key endocrine factor driving sex differentiation of hormone-responsive tissues such as the reproductive tract. Each reproductive tract possesses a critical time window for sex differentiation, which is dependent on androgen (1–5). External genitalia are organs highly representative of sex differences between males and females and are an excellent model for understanding male- and female-specific differentiation (6, 7). External genitalia initially arise as a common anlage, the genital tubercle (hereafter “embryonic external genitalia; eExG”), in both sexes, which subsequently differentiates into the penis in males (male-type external genitalia) or the clitoris in females (female-type external genitalia) (8, 9). Morphological sex differences in the external genitalia are manifested as sex-specific urethral structure and organ size, influenced by the presence or absence of androgen (2, 7, 10). Although a size difference appears prominently after birth, male-type urethral formation occurs in the prenatal period in an androgen-dependent manner (7, 10).
The activities of androgen are mediated by androgen receptor (AR), a member of the steroid hormone receptor superfamily. The extent of androgenic exposure is known to affect the development of external genitalia (11–13). Ar knock-out (Ar KO) male mice show feminized external genitalia (14–16). Excess androgen production induces male-type external genitalia differentiation in 46,XX individuals in a condition termed congenital adrenal hyperplasia (17). Exogenous androgen leads to male-type urethral differentiation even in genetic females within a critical time window upon induction of androgen-regulated sex-biased genes (hereafter “male-biased genes”) (1, 10, 13, 16, 18). However, it is unclear how androgen responsiveness, which drives male-biased gene expression, is established during the process of sex differentiation.
AR translocates from the cytoplasm to the nucleus in a ligand-dependent manner. It forms as a homodimer and recognizes a sequence motif known as the androgen response element (ARE), located in cis-regulatory elements, promoters, and/or enhancers of target genes. It has been shown that collaborative transcription factors of hormone receptors such as AR and estrogen receptor (ER) are required to facilitate their transcriptional activities by modulating the genomic environment of their downstream target genes. Histone modifications including acetylation and methylation are key epigenetic marks to regulate gene expression through modulating chromatin structure (19, 20). The histone H3 at lysine 4 monomethylation (H3K4me1) and H3K27 acetylation (H3K27ac) modifications are correlated with activation of cis-regulatory elements (21). Collaborative transcription factors are also required to guide AR to the appropriate genomic locus in several adult tissues, such as FoxA1 in the prostate, Hnf4α in the kidney, and AP-2α in the epididymis (22). However, collaborative transcription factors of AR have not been identified during eExG sex differentiation.
V-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B (MafB) is an essential regulatory gene for male-type urethral differentiation in mice (10). MafB is expressed in male urethral bilateral mesenchyme, which is an essential region targeted by androgens for male-type urethral differentiation (10, 23). AR directly regulates MafB expression through an eExG enhancer located immediately downstream of the MafB coding region (24). Of note, MafB expression is also induced in genomic females after androgen exposure (10, 24), suggesting that transcriptional competency of male-biased genes is maintained in females. In the current study, we identified Specificity protein 1 (Sp1; official name: transacting transcription factor 1) as a collaborative transcription factor of AR in the sex differentiation of eExG. Sp1 belongs to the Sp/Klf transcription factor family, which binds to a consensus GC box motif (GGGGCGGGGC) (25, 26). Whereas it was originally identified as a regulator of ubiquitously expressed housekeeping genes (26), the current study suggests that Sp1 is required for androgen responsiveness for regulating male-biased genes in both sexes. We generated a MafB mutant mouse line in which we deleted Sp1 regulatory H3K27ac elements downstream of the MafB coding region (MafBeΔ/eΔ) using the CRISPR-Cas9 genome editing system. Intriguingly, our analyses on a series of MafB mutant mice utilizing MafBeΔ/eΔ and MafB-GFP knock-in alleles showed a spectrum of male-type urethral differentiation dependent on remnant MafB expression levels. Our data provide insights into the mechanisms of androgen responsiveness for the expression of male-biased genes to regulate the sexual fate of developing external genitalia.
Results
Identification of the Collaborative Transcription Factor of AR for the Expression of Male-Biased Genes.
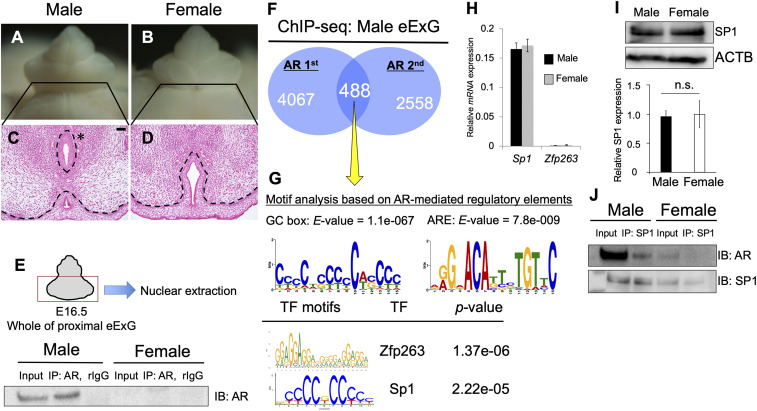
Whereas eExG size is not notably different between males and females at embryonic day (E)16.5, male-type urethral differentiation is clearly evident at that stage (Fig. 1 A–D). Mesenchymal cells migrate to fuse in the midline with elimination of epithelial cells for incorporating the urethra into glans (male-type urethra) (Fig. 1C, asterisk). In contrast, the urethral plate epithelium remains in female eExGs (Fig. 1D).
Fig. 1.
Analysis of the regulatory genes for AR transcriptional activity. (A–D) Histological analysis of male and female eExG at E16.5. The asterisk indicates male-type urethra. The urethral plate epithelium remained in female eExG. (Scale bar, 50 μm.) (E) Purified nuclear fraction of whole proximal eExG was utilized for immunoprecipitation with anti-AR or normal rabbit IgG followed by Western blotting with AR antibody. (F) ChIP-seq derived AR cistrome shown as Venn diagram. (G) Motif enrichment analyses from 488 peaks based on AR ChIP-seq (yellow arrow), followed by TOMTOM and MEME analysis. The E-value was utilized to detect significant motif from these analyses. TF, transcription factor. (H) Quantitative real-time PCR from eExG urethral bilateral region mRNA was performed to detect the expression levels of Sp1 and Zfp263 normalized with levels of Gapdh. (I) The levels of SP1 protein normalized by levels of ACTB was detected by Western blotting. Three independent biological replicates were performed. (J) Purified nuclear fraction of whole proximal male and female eExG was utilized for immunoprecipitation with anti-SP1 followed by Western blotting with AR or SP1 antibody. Dashed lines, epithelial–mesenchymal border. n.s. indicates not significant, P > 0.05.
AR signaling is activated by binding androgen and translocating from the cytoplasm into the nucleus in a ligand-dependent manner. To investigate whether AR signaling was activated during urethral sex differentiation, an immunoprecipitation assay on the eExG nuclear fraction was performed. AR was prominently located in the nuclear fraction in males, indicating that AR was activated in male eExG (Fig. 1E). To identify the collaborative transcription factors of AR, we used motif analyses based on AR chromatin immunoprecipitation sequencing (ChIP-seq) performed on male eExG tissue. Venn diagram analyses showed that 488 peaks were identified as AR-mediated regulatory elements between two independent experimental trials (Fig. 1F). In addition to the 15-base pair (bp) canonical ARE (AGAACAnnnTGTTCT), GC box motifs identified Zinc finger protein 263 (Zfp263) and Specificity protein 1 (Sp1) as the top candidate genes (Fig. 1G). Although Zfp263 was rarely expressed in the eExG, both Sp1 mRNA and SP1 protein were highly expressed in both sexes (Fig. 1 H and I). Furthermore, SP1 physically interacted with AR in male eExG but not in female eExG (Fig. 1J), consistent with the absence of AR protein in the female nuclear fractions (Fig. 1E). Hence, we focused on the roles of Sp1 as a collaborative transcription factor of AR during urethral sex differentiation.
Sp1 Regulates the Expression of Male-Biased Genes during Urethral Sex Differentiation.
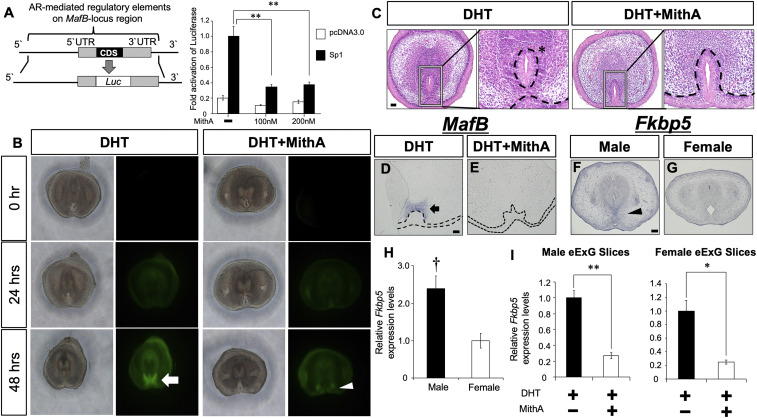
Mithramycin A (MithA) is well characterized as a Sp1 inhibitor, which competitively antagonizes binding of Sp1 at its recognition sites. We performed luciferase reporter assays, which also indicated that MithA significantly inhibited Sp1 transcriptional activity on the MafB regulatory region (MafB-locus region) (Fig. 2A). To investigate the role of Sp1 during urethral sex differentiation, we analyzed the effect of MithA on the expression of male-biased genes and male-type urethral differentiation by utilizing our eExG slice culture system (18). A MafB-GFP knock-in mouse line (MafB+/GFP) was utilized to monitor the inhibitory effects of MithA (10, 27). 5α-dihydrotestosterone (DHT) is a major androgen in male-type external genitalia development (28). DHT-induced MafB-GFP expression and male-type urethral differentiation after a 48-h treatment (Fig. 2 B and C). MithA inhibited DHT-induced male-type urethral differentiation with decreased MafB-GFP expression (Fig. 2 B and C). Urethral plate epithelium remained in the presence of MithA, which was similar to female eExG (Figs. 1D and 2C). We also confirmed the reduction of MafB-mRNA expression in MithA-treated urethral bilateral mesenchyme via in situ hybridization (Fig. 2 D and E). FK506-binding protein 51 (Fkbp5), another male-biased gene, was highly expressed in urethral bilateral mesenchymal cells in male eExG compared to female (Fig. 2 F–H). DHT-induced Fkbp5 expression was also significantly inhibited by MithA treatment (Fig. 2I). These results suggest that Sp1 is required for AR transcriptional activities for the expression of male-biased genes.
Fig. 2.
Inhibition of Sp1 suppresses the androgen-induced urethral sex differentiation. (A) Sp1 transactivation of the AR-mediated regulatory elements of the MafB-locus region was inhibited by 100 nM or 200 nM MithA treatment for 24 h. CDS, coding sequence. (B) 10 nM DHT or 10 nM DHT + 200 nM MithA was applied to female MafB+/GFP eExG slices. White arrow and arrowhead represent MafB (GFP) expression. (C) 10 nM DHT or 10 nM DHT + 200 nM MithA was applied to female eExG slices. While DHT induced male-type urethral differentiation (asterisk), urethral plate incorporation was inhibited in the presence of MithA. (Scale bar in the hematoxylin-eosin staining, 50 μm.) (D and E) Section in situ hybridization for MafB expression was performed for 10 nM DHT or 10 nM DHT + 200 nM MithA-treated male eExG slices. Black arrow indicates MafB expression as dependent on DHT stimulation. (Scale bar, 50 μm.) (F and G) Section in situ hybridization for Fkbp5 expression in E16.5 male and female eExG. Black arrowhead indicates Fkbp5 expression in male eExG mesenchyme. (Scale bar, 100 μm.) (H) Relative Fkbp5 expression normalized to levels of Gapdh was analyzed for eExG urethral bilateral regions at E16.5. † P < 0.05 versus female. (I) Relative Fkbp5 expression normalized to levels of Gapdh was analyzed from 10 nM DHT or 10 nM DHT + 200 nM MithA-treated eExG slices. Three to four independent biological replicates were performed. *P < 0.05, **P < 0.01. Dashed lines, epithelial–mesenchymal border.
Sp1 Regulates AR Transcriptional Activity via Acetylation of H3K27 at Enhancers of Male-Biased Genes.
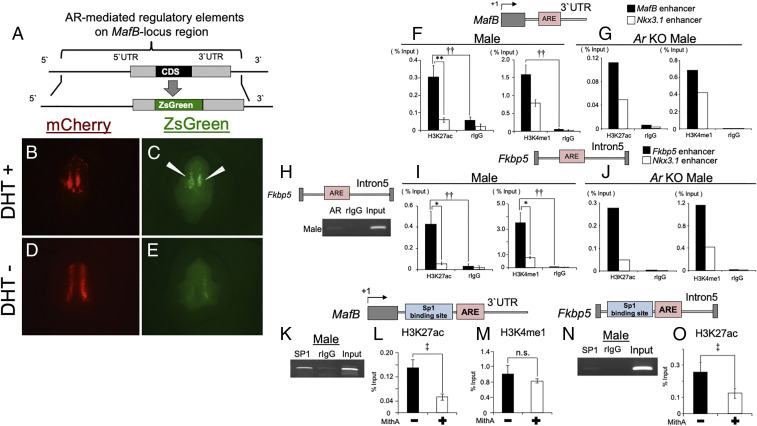
We previously reported that an eExG enhancer of MafB is located at its 3′ untranslated region (UTR), which was identified by in vitro enhancer assays (24). To confirm whether AR actually regulates MafB expression through the eExG enhancer in vivo, we performed an in vivo eExG-derived ZsGreen reporter assay utilizing sonoporation. The sonoporation system is effective for transducing exogenous genes into the mesenchyme (29). We constructed a ZsGreen reporter vector of AR-mediated MafB regulatory regions including the putative eExG enhancer downstream of the coding region (Fig. 3A). mCherry fluorescence, driven by cytomegalovirus immediate early promoter, was utilized as a control of gene transduction (Fig. 3 B and D). While ZsGreen reporter signal was weakly detected in the absence of DHT due to endogenous Wnt/β-catenin–dependent regulation (24), ZsGreen fluorescence was enhanced by DHT stimulation, suggesting that the eExG enhancer regulates MafB expression in an androgen-dependent manner (Fig. 3 C and E).
Fig. 3.
H3K27ac is enriched by Sp1 at enhancers containing an ARE in male eExG. (A) Schematic of ZsGreen fluorescence reporter construct for MafB-locus region. (B–E) Investigation of the effect of DHT stimulation on ZsGreen signal after 24 h by sonoporation. White arrows indicate ZsGreen signal dependent on DHT stimulation. mCherry signal was utilized checking efficiency for the gene transduction in the eExG. (F–J) Whole proximal male eExGs derived from wild-type and Ar KO mice were utilized for ChIP-PCR and qPCR at E16.5. Single ChIP-qPCR analysis (n = 1: 27 pieces of eExG/experiment) was performed for Ar KO males. (F, G, I, and J) ChIP-qPCR analyses for H3K27ac, H3K4me1, and rIgG at MafB, Fkbp5, and Nkx3.1 enhancers containing AREs. Three to five independent biological replicates were performed. (H) ChIP-PCR analysis for AR and rIgG for the Fkbp5 enhancer. The Fkbp5 enhancer is located within its Intron5. (K) ChIP-PCR by male SP1 for the MafB enhancer containing an Sp1 binding site and ARE. (L and M) Inhibition of Sp1 resulted in the reduction of H3K27ac enrichment but not of H3K4me1. Two to three independent biological replicates with two to three technical replicates were performed. (N) SP1 ChIP-PCR for the Fkbp5 enhancer containing an Sp1 binding site and ARE. (O) Effect of Sp1 inhibition on the enrichment of H3K27ac at the Fkbp5 enhancer. Two to three independent biological replicates with two to three technical replicates were performed. †† and **P < 0.01, * and ‡ P < 0.05, n.s. indicates not significant, P > 0.05.
Next, we investigated the requirement of Sp1-mediated regulatory elements for AR transcriptional activity for MafB expression. DHT-induced luciferase activity was significantly inhibited by MithA on the MafB-locus region (SI Appendix, Fig. S1A). Moreover, DHT-induced AR transcriptional activity through the 3′ UTR was reduced by MithA treatment but not through the 5′ upstream region (SI Appendix, Fig. S1 B and C). These results indicate that Sp1 may regulate AR transcriptional activities via the eExG enhancer located 3′ UTR of MafB.
Sp1 is reported to regulate histone acetylation for its downstream gene expression (30). We next investigated the statuses of active histone marks, H3K27ac and H3K4me1, at the regulatory elements of the male-biased genes MafB and Fkbp5. NK3 Homeobox 1 (Nkx3.1) has been reported as an androgen-regulated gene in the prostate, and AREs of its 3′ UTR act as the prostate enhancer (31, 32). In contrast to the prostate, Nkx3.1 expression was undetectable in the eExG (SI Appendix, Fig. S2A). ChIP-seq analyses from eExG samples revealed that active histone marks, AR, and SP1 were not enriched at the Nkx3.1 prostate enhancer (SI Appendix, Fig. S2B). Thus, we utilized the Nkx3.1 prostate enhancer as a control locus to compare with male-biased gene enhancers in this study. AR was detected at an Fkbp5 enhancer containing an ARE during urethral sex differentiation (Fig. 3H) (33). H3K27ac and H3K4me1 were enriched at AREs located in MafB and Fkbp5 enhancers (Fig. 3 F and I). We further analyzed the status of active histone marks in Ar KO males. Both active histone marks were still detected in MafB and Fkbp5 enhancers in Ar KO male mice, suggesting a possible AR-independent mechanism for active histone modifications at the enhancers of male-biased genes (Fig. 3 G and J). To investigate whether SP1 binds to AR-mediated regulatory elements during urethral sex differentiation, we performed SP1 ChIP-PCR using male eExG. SP1 was located adjacent to the ARE at the MafB and Fkbp5 enhancers (Fig. 3 K and N). These results prompted us to investigate whether Sp1 modulates histone statuses at the enhancer of male-biased genes. MithA-treated eExG resulted in significantly reduced enrichment of H3K27ac, but not of H3K4me1, at the MafB eExG enhancer (Fig. 3 L and M). MithA also inhibited the enrichment of H3K27ac at the Fkbp5 enhancer (Fig. 3O), suggesting that the active histone mark H3K27ac is regulated by Sp1. Taken together, these results suggest that Sp1 regulates AR transcriptional activity of male-biased genes.
Sp1 Is Required for the Enrichment of H3K27ac at AR-Mediated Regulatory Elements in Both Sexes.
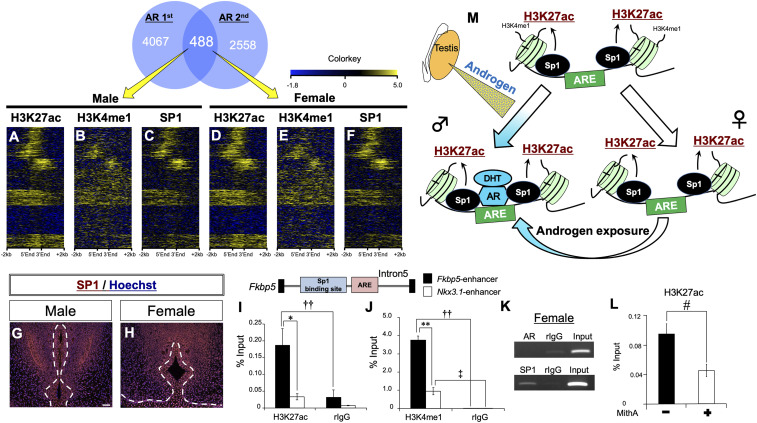
Exogenous androgen induces male-type urethral differentiation in female eExG (1, 10) (SI Appendix, Fig. S3 A–H). However, the mechanisms underlying this androgen responsiveness in female eExG have not been elucidated. To investigate the possibility that the epigenetic status of active histone marks is similar at AR-mediated regulatory elements in both sexes, we performed enrichment analysis of active histone marks and SP1 in female eExG. Heatmaps of both active histone marks and SP1 binding ChIP-seq signals showed a similar pattern at AR-mediated regulatory elements in both sexes (Fig. 4 A–F). Coefficient plots showed that the presence of SP1 was more highly correlated with the presence of H3K27ac as compared to H3K4me1 in both sexes (SI Appendix, Fig. S3I). In addition to genome-wide analyses, immunohistochemical analyses showed that SP1 was expressed in both male and female eExG (Fig. 4 G and H). We further investigated the status of active histone marks and SP1 binding at the enhancers of male-biased genes in females. H3K27ac and H3K4me1 were enriched at the Fkbp5 enhancer (Fig. 4 I and J). These results suggest that epigenetic statuses are conserved at AR-mediated regulatory elements in both males and females. Although AR was not detected at the ARE in the Fkbp5 enhancer, SP1 was detected adjacent to the ARE in females (Fig. 4K). Furthermore, MithA reduced the enrichment of H3K27ac at Fkbp5 enhancer in females (Fig. 4L). These results suggest that Sp1 regulates male-biased genes through the acetylation of H3K27 at AR-mediated regulatory elements in females as well as in males (Fig. 4M).
Fig. 4.
Active histone modifications and SP1 binding status are similar between males and females. (A–F) Heatmaps of H3K27ac, H3K4me1, and SP1 ChIP-seq signal for male and female eExGs from 488 reproducible peaks based on AR ChIP-seq. Heatmaps are represented as centered at gene bodies within ± 2 kb windows. (G and H) Immunofluorescence staining for SP1 on E16.5 eExGs. (Scale bar, 50 μm.) (I and J) H3K27ac, H3K4me1, and rIgG enrichment analyses for E16.5 female eExG by ChIP-qPCR at the Fkbp5 enhancer. Three to five independent biological replicates were performed. (K) ChIP-PCR for AR, SP1, and rIgG at the Fkbp5 enhancer. (L) Inhibition of Sp1 resulted in the reduction of H3K27ac enrichment at the Fkbp5 enhancer. Two independent biological replicates with two to three technical replicates were performed. (M) A model showing the conservation of transcriptional competency for male-biased genes for urethral sex differentiation in both sexes. Active histone marks, H3K27ac and H3K4me1, and Sp1 are enriched at AR-mediated regulatory elements for regulating the expression of male-biased genes in both sexes. Testicular androgen regulates expression of male-biased genes through Sp1-activated AR-mediated regulatory elements during male-type eExG differentiation. A similar gene regulatory mechanism through AR is conserved even in female eExG. Exogenous androgen also regulates expression of male-biased genes during urethral sex differentiation in females. * and ‡ P < 0.05, ** and †† P < 0.01, # P < 0.05 versus DMSO. Dashed lines, epithelial–mesenchymal border.
The Sp1 Regulatory eExG Enhancer Is Essential for Male-Type Urethral Differentiation.
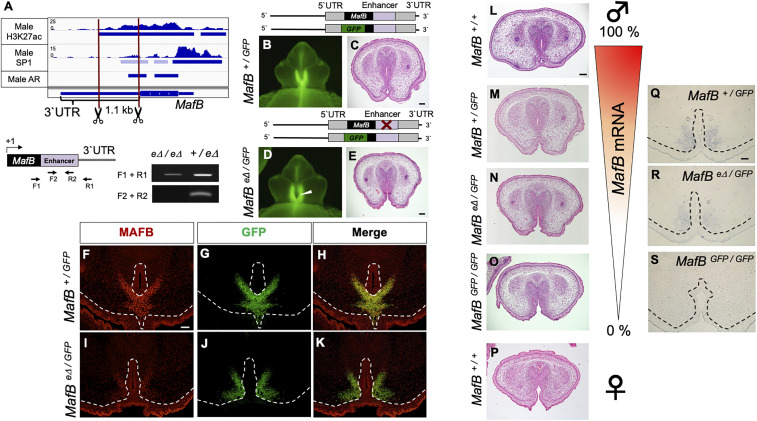
Next, to investigate whether Sp1 regulatory H3K27ac elements possess essential roles for male-type eExG differentiation, we generated a mouse line in which we deleted the H3K27ac element at the MafB eExG enhancer (MafB eΔ/eΔ mice) using the CRISPR-Cas9 genome editing system (Fig. 5A and SI Appendix, Fig. S4). Although MafB+/GFP and MafB eΔ/eΔ mice did not show any significant urethral abnormalities, MafBeΔ/GFP compound-male mutant mice showed incomplete male-type urethral differentiation, which resembled that in females (Fig. 5 B–E, and P). This mutant phenotype was milder compared with that of MafB-null (MafBGFP/GFP) mice (10). Furthermore, native MAFB expression was markedly reduced in these compound-mutants compared with MafB+/GFP mice in the MafB-GFP–expressing mesenchymal region (Fig. 5 F–K). While MafB-null mice die immediately after birth (27, 34), MafBeΔ/GFP mice were alive after birth and survived until at least postnatal day 8 (n = 4/4). These results suggest that the Sp1 regulatory H3K27ac element in the MafB 3′ UTR acts as a male-type enhancer for urethral sex differentiation.
Fig. 5.
Several degrees of male-type urethral differentiation dependent on the androgenic regulation of MafB. (A) The introduction of a deletion into the MafB eExG enhancer (MafB eΔ) mouse line by CRISPR-Cas9 genome editing system is shown below an Integrated Genomics Viewer screenshot of genomic tracks of H3K27ac, SP1, and AR ChIP-seq peak regions detected by MACS2 v2.1.1. on the MafB eExG enhancer in male mice. Schematic of primer pairs and gel images show representative results of PCR genotyping for detecting MafB wild type (+) or MafB eΔ alleles. (B–E) Histological analyses from MafB+/GFP and MafBeΔ/GFP mice at E16.5. White arrowhead indicates incomplete male-type urethral differentiation in MafBeΔ/GFP mice (n = 11/13). (Scale bar in C and E, 100 μm.) (F–K) Immunostaining for MAFB and GFP on MafB+/GFP and MafBeΔ/GFP mice at E16.5. MAFB mesenchymal expression was reduced in the MafBeΔ/GFP as compared to MafB+/GFP. The epithelial staining is due to nonspecific signals. (Scale bar, 50 μm.) (L–O) Histological images of E16.5 male eExG showing a spectrum of male-type urethral differentiation dependent on the androgenic regulation of MafB. (P) Histological image of wild-type female eExG at E16.5. (Scale bar, 100 μm.) (Q–S) Section in situ hybridization for MafB-mRNA expression in MafB+/GFP, MafB eΔ/GFP, and MafBGFP/GFP male eExG at E15.5. (Scale bar, 50 μm.) Dashed lines, epithelial–mesenchymal border.
Discussion
Requirement of Sp1 for the Expression of Male-Biased Genes during Urethral Sex Differentiation.
External genitalia show prominent sex differences between males and females in the presence or absence of androgen activity. Male-type urethral differentiation occurs with dynamic transcriptional changes of male-biased genes such as MafB and Fkbp5 (10, 23). In addition to AR expression, MafB and Fkbp5 are predominantly expressed in male urethral bilateral mesenchyme, which is an essential region for male-type urethral differentiation (8, 10, 23). AR regulates its downstream genes with collaborative transcription factors in a cell-specific manner. FoxA1 has been widely shown to be the collaborative transcription factor of AR in hormone-dependent cancers (35). FoxA1 and FoxA2 double-KO mice exhibit defective male-type urethral differentiation (36). However, FOXA1 was expressed in epithelial cells of eExG but not in the urethral bilateral mesenchyme (SI Appendix, Fig. S5). Epithelial Ar conditional KO mice do not display any defects of eExG sex differentiation (1). Thus, FoxA1 may not be a collaborative transcription factor of AR in eExG. Although only 488 peaks were identified in common by duplicate AR ChIP-seq analyses utilizing the eExG nuclear fraction, we identified SP1 as a candidate collaborative transcription factor for urethral sex differentiation. It may be difficult to get highly reproducible results of ligand-dependent AR ChIP-seq analyses from the specimens derived from carefully and accurately dissected tissues (37). SP1 was expressed in the urethral bilateral mesenchyme and was detected adjacent to the AREs at the enhancers of MafB and Fkbp5. Of note, inhibition of Sp1 resulted in defective male-type urethral differentiation and the reduction of MafB and Fkbp5 expression. Furthermore, we showed that Sp1 physically interacted with AR in male eExG. Taken together, the current study suggests that Sp1 may regulate AR transcriptional activities for male-biased gene expression during urethral sex differentiation.
Sp1-Mediated Active Histone Modification Is Required for the Expression of Male-Biased Genes.
Changes in chromatin structure by posttranslational histone modifications play an essential role to regulate gene expression (38). Nuclear receptors, such as ER-α, regulate epigenetic histone modifications in the case of the reproductive organs. Administration of 17β-estradiol accelerates acetylation of H4K12 at enhancers via ER-α (39). Diethylstilbestrol, which is a synthetic estrogen, induces the acetylation of H3K27 at ER-α–bound enhancers in the neonatal mouse uterus (40). In addition to ER-α–regulated histone modifications, AR also modulates chromatin status through H3T11 phosphorylation and H3K4 methylation through interactions with histone modifiers (41, 42). We showed that active histone marks, H3K27ac and H3K4me1, were enriched at AR-mediated regulatory elements of male-biased genes in both sexes. Collaborative transcription factors are closely implicated in the histone modifications driving transcriptional activities of sex hormone receptors. Gata2 and FoxA1 are well known as collaborative transcription factors for ER-α, AR, and glucocorticoid receptor (35). FoxA1 regulates H3K4 methylation at ER-α–dependent transcriptional regulatory elements by interacting with the methyltransferase MLL3 (43). Gata2 regulates the acetylation of H3K27 at enhancers of AR target genes by recruiting p300 (44). In addition to reproductive organs, Hnf4-α was identified as a collaborative transcription factor of AR in the adult kidney (22). The current study demonstrates that the ubiquitously expressed transcriptional factor Sp1 is a collaborative transcription factor of AR for urethral sex differentiation. The lysine residues of histone tails are modified by reversible reactions, which are performed by acetyltransferases (HATs) and histone deacetylases. Histone acetylation by HATs such as p300 is a major modulator of chromatin condensation and gene transcription (19). Sp1 has been shown to interact with p300 (30), suggesting that the transcriptional regulation of male-biased genes depends on Sp1 interacting with such key histone modifiers. Although further studies are necessary for identifying histone modifiers interacting with Sp1, we propose that Sp1 may regulate AR transcriptional activities through modulating active histone status (e.g., H3K27ac) during urethral sex differentiation (Fig. 4M).
Conserved Transcriptional Competency for Male-Biased Genes in Both Sexes for the Sexual Fate of eExG.
Murine external genitalia develop as a common anlage from E10.5 in both sexes (45). Sex differentiation of external genitalia is subsequently dependent on androgen activity within a critical time window (6, 7, 16). Male-biased genes are expressed from E14.5 in male urethral bilateral mesenchymal cells. Conversely, these genes are not expressed prominently in female eExG (10, 23). Histological analyses revealed that male-type urethral differentiation was rapidly induced, within 24 h, after testosterone propionate (TP) administration, with the induction of MafB expression (SI Appendix, Fig. S3 A–H). What are the mechanisms underlying male-biased gene induction leading to male-type urethral differentiation in females? Our observations suggest that the transcriptional competency of male-biased genes for male-type urethral differentiation is conserved in both sexes independently of AR signaling. Of note, active histone marks, H3K27ac and H3K4me1, were enriched at AR-mediated regulatory elements in females as well as in males at E16.5. Genome-wide analyses revealed that SP1 was enriched at AR-mediated regulatory elements of male-biased genes in both males and females. Furthermore, H3K27ac was enriched at the enhancer of male-biased genes by Sp1. Therefore, Sp1 may recruit AR through regulating the enrichment of active histone modifications, such as H3K27ac, at AR-mediated regulatory elements of male-biased genes in both sexes (Fig. 4M). Our ChIP-seq analyses are reports describing the genome-wide status of transcription factors and active histone modifications during eExG sex differentiation.
MafB is a pivotal AR target gene during male-type urethral differentiation. In the current study, we generated a MafB eExG enhancer KO mouse line (MafB eΔ/eΔ). MafB+/GFP and MafB eΔ/eΔ mice showed no obvious phenotypes in eExG development. However, MafBeΔ/GFP compound-mutant mice exhibited incomplete male-type urethral differentiation, which resembled female eExG differentiation. The current study is suggesting that the sexual fate of external genitalia is dependent on the degrees of androgenic regulation for male-biased gene expression (Figs. 4 A–F and 5 L–P). In fact, MafB expression was reduced in a manner consistent with the severity of the mutant phenotypes (Fig. 5 Q–S). In the current study, we uncovered that the transcriptional machinery of male-biased genes through AR directly to cis-regulatory elements was conserved in both sexes during male-type urethral differentiation.
Several studies report nongenomic AR activity is mediated by membrane-associated AR (mAR), which is independent of canonical AR signaling (46, 47). mAR regulates the activation of kinase signaling pathways such as Src (48). Male-biased genes regulated by mAR may also play a role in androgen-induced eExG sex differentiation. Further analyses will be necessary for understanding mAR signaling associated with eExG sex differentiation. Taken together, our results based on genome-wide ChIP-seq analyses suggest mechanisms of androgen responsiveness for the regulation of male-biased gene expression during eExG sex differentiation.
Materials and Methods
Animals, Whole and Slice Organ Culture.
MafB+/GFP, Ar KO, and CD1 (Jcl:ICR, CLEA Japan, Inc.) mice were utilized in this study. MafB+/GFP and Ar KO mice were previously described (14, 27). Mafb+/GFP and ICR mice were utilized for organ and slice culture treatment experiments. For androgen treatment, each pregnant ICR mouse was injected with TP 100 mg/kg (200-17532, FUJIFILM Wako Pure Chemicals) or DMSO (043-07216, FUJIFILM Wako Pure Chemicals) dissolved in sesame oil via intraperitoneal injection. Noon on the day of the vaginal plug was defined as E0.5. For the culture experiments, MithA (200 nM, 18378-89-7, Cayman Chemical), DHT (10 nM, A8380, Sigma-Aldrich), and DMSO (D8418, Sigma-Aldrich) were utilized. For whole and slice organ culture, male and female eExG were dissected from E14.5 mouse embryos. Whole eExG organ culture was performed for ChIP-qPCR assays to detect active histone modifications as a previously described (24). ChIP-qPCR analyses were performed 24 h after MithA treatment. eExG slice culture was performed as previously described (18). Male and female mice were identified by PCR genotyping, using Zfy1 primers designed as previously described (49). Mice were housed under a 12-h dark–light cycle (light from 08:00 to 20:00) at a constant temperature of 22 ± 1 °C, with free access to food and water. The Animal Care and Use Committee of Wakayama Medical University School of Medicine approved the protocols for animal experiments. Animal care and all methods were performed in accordance with relevant guidelines and regulations.
Reporter Assay for eExG Explants by Sonoporation.
Whole E14.5 eExG organs were utilized for this analysis. The detailed procedures of gene transfer by sonoporation were previously described in detail (29). Briefly, ultrasound was irradiated via a Sonitron 2000N (Richmar) after injecting a microbubble (SV-25, Nepa Gene Co., Ltd.) and reporter vector mixture. DHT was added after ultrasound irradiation for 24 h. pmCherry-N1 (632523, Clontech) and pZsGreen-N1 (632448, Clontech) vectors were utilized in this assay. A ZsGreen reporter construct of AR-mediated regulatory elements within the MafB-locus region was generated as follows: DNA fragments of ZsGreen were amplified utilizing PrimeSTAR Max DNA polymerase (Takara Bio) (5′-GCC TCG CTT TTA GCG ATG GCC CAG TCC AAG CAC GG -3′ and 5′- CAG CTC CGC GGC CAT TCA GGG CAA GGC GGA GCC-3′). The luciferase gene was deleted from the MafB-locus vector by inverse PCR using PrimeSTAR GXL DNA polymerase (Takara Bio) (5′-GTC CTG GCG GGT CCG GCC CCC GCC C-3′ and 5′-CGC TAA AAG CGA GGC TCA GCC GCC G-3′) (24). After deleting the luciferase gene, ZsGreen and MafB coding sequences (CDS) were inserted between the 5′ UTR and 3′ UTR via an In-Fusion HD Cloning Kit. Lastly, MafB CDS was deleted by inverse PCR utilizing PrimeSTAR Max DNA polymerase (Takara Bio) (5′-TCC GCC TTG CCC TGA ATG GCC GCG GAG CTG AGC A-3′ and 5′-CGG ACC CGC CAG GAC TCA CAG AAA GAA CTC AGG AGA GGA G-3′).
Luciferase Reporter Assay.
All reporter vectors were utilized as previously described (24). The mouse AR expression vector cloned into the pCS2+MT vector was utilized as previously reported (24). The human SP1-FLAG tagged expression vector, subcloned into pcDNA3.0, was described previously (50). For luciferase assays, procedures and conditions were based on a previous report (24). HepG2 cells (obtained from HB-8065, ATCC, and JCRB1054, Japanese Collection of Research Bioresources Cell Bank) were utilized for several assays. Briefly, 6 h after transfecting plasmid vectors, MithA was pretreated for 24 h. After pretreatment, cells were incubated with or without DHT and/or MithA for 24 h. Luciferase activity was measured by chemiluminescence using the dual-luciferase reporter assay system (Promega). Values were normalized to Renilla luciferase activity.
Histology and Immunohistochemistry.
Embryos and eExG slices were fixed overnight in 4% (weight[wt]/volume[vol]) paraformaldehyde in phosphate-buffered saline (PBS) and dehydrated using methanol. Serial sections (6 μm) embedded in paraffin were prepared for hematoxylin/eosin staining and immunohistochemistry. Dilutions of primary antibodies were as follows: rabbit anti-SP1 (07-645, Millipore) was used at 1:500 with 1% goat serum in PBS containing 0.05% Tween 20; goat anti-FOXA1 (anti–HNF-3α/β, C-20; Santa Cruz Biotechnology) was 1:200 with 1% horse serum in PBS; and rabbit anti-MAFB (1:1,000, IHC-00351, Bethyl Laboratories, Inc.) and mouse anti-GFP (1:200, 11814460001, Roche) were used with 1% goat serum in PBS. Antigen retrieval for SP1 was performed as follows: dilution of HistoVT One (Nacalai Tesque) ratio 1:10, incubated at 105 °C for 15 min. Antigen retrieval for FOXA1, MAFB, and GFP was performed via citrate buffer, incubated at 121 °C for 1 min. Immunostaining was visualized by fluorescent staining (an Alexa 488-conjugated IgG and/or an Alexa 546-conjugated IgG secondary antibody) (Molecular Probes) and counterstained with Hoechst 33342 (Sigma-Aldrich).
Section mRNA In Situ Hybridization for Gene Expression.
Section mRNA in situ hybridization procedures and protocol were performed as previously described (23). Then, 6- or 12-μm serial sections were prepared after embedding in paraffin. Probes for the following genes were utilized: MafB and Fkbp5 (23, 24).
RNA Isolation and qPCR Analysis.
Total RNA was isolated utilizing the ISOGEN II system (Nippon Gene Co., Ltd.) and reverse transcribed utilizing PrimeScript RT Master Mix (Perfect Real time) (Takara Bio) according to the manufacturer’s instructions. qPCR was performed utilizing the StepOnePlus Real-Time PCR system (Applied Biosystems) with SYBR Premix Ex Taq II (Tli RNaseH Plus) (Takara Bio) according to the manufacturer’s instructions. MafB and Fkbp5 primer pairs were obtained from Takara. Gapdh mRNA was utilized as an internal control. Primer sequences were as follows: Gapdh F: AAC GAC CCC TTC ATT GAC CTC, R: CCT TGA CTG TGC CGT TGA ATT; Sp1 F: GGC ATC AAC GTC ATG CAG GTG ACA, R: ATG CTA CCT TGC ATC CCG GGC TTA; Zfp263 F: TTG AGG TAG CCT GCT CAG GGA TGG, R: CTC AGG AGC CCA AAC TCT CCT GCT; and Nkx3.1 F: TGA TAG AAA GCT GGC GGC TTG CAC, R: GGA CTC CTC CGG CTT TGG TAT GGT.
Immunoprecipitation Assay and Western Blot Analysis.
A total of 20 pieces of the proximal part of eExG from E16.5 ICR embryos were collected from both males and females. Organs were homogenized in hypotonic cellular buffer (10 mM Hepes pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 1% Nonidet P-40, 1 mM DTT, cOmplete EDTA-free protease inhibitor mixture from Roche) by Dounce Tissue Grinder. After homogenization, organs were incubated on ice for 1 h and centrifuged at 4 °C for 20 min at 10,600 rpm. The pellets were reconstituted in RIPA buffer (150 mM NaCl, 1.0% TritonX-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris HCl pH 8.0, 1 mM EDTA, cOmplete EDTA-free protease inhibitor mixture) and were incubated in 4 °C for 30 min. After incubation, the samples were centrifuged at 15,000 rpm for 5 min at 4 °C, and the nuclear fraction of the supernatant was collected. Nuclear fractions (∼150 to 350 μg) were incubated with specific antibodies (Sp1 2.4 μg, A303-944A, Bethyl Laboratories; AR 2.0 μg, N-20, Santa Cruz Biotechnology) in Protein A-Sepharose 4B conjugate (Invitrogen) at 4 °C overnight. Protein–antibody complexes were washed in PBS and reconstituted in RIPA added sample buffer (0.1 M Tris HCl pH 6.8, 4% SDS, 20% Glycerol, Bromophenol Blue) with 2-mercaptoethanol (Nacalai Tesque). Analysis for protein levels of SP1 and ACTB were performed on whole lysates of the proximal part of eExG tissues. Samples were boiled for 5 min just before SDS-polyacrylamide gel electrophoresis (PAGE) analysis. SuperSep Ace 5 to 20% (FUJIFILM Wako Pure Chemicals) was utilized for protein loading. The proteins, separated by SDS-PAGE, were blotted onto an Immobilon-P (PVDF; polyvinylidene difluoride) membrane (Millipore). The PVDF membrane was blocked for 1 h with 1% skim milk (BD Difco) in Tris-buffered saline containing 0.1% Tween 20. The PVDF membrane was incubated with primary antibodies (AR at 1:1,000, N-20 Santa Cruz Biotechnology; SP1 at 1:5,000, A303-944A, Bethyl Laboratories; ACTB 1:3,000, A1978, Sigma-Aldrich) in signal enhancer HIKARI for Western blotting and ELISA (Nacalai Tesque) at 4 °C, overnight. After an overnight incubation, PVDF membranes were then washed and incubated in horseradish peroxidase (HRP) goat-conjugated anti-rabbit IgG (H+L) (Invitrogen), HRP rabbit-conjugated anti-goat IgG (H+L) (Invitrogen), or HRP goat-conjugated anti-mouse IgG (H+L) (Invitrogen) antibody. Signals were detected by utilizing a Chimi-Lumi detection kit (Nacalai Tesque) and ChemiDoc XRS+ system (Bio-Rad Laboratories Inc.).
ChIP and PCR, qPCR Analysis.
ChIP assays were performed as previously described (24). ICR and Ar KO mice were utilized for these assays. ICR mice were utilized as controls. For each antibody reaction, 30 to 50 μg DNA was utilized. The following primary antibodies were used: rabbit anti-AR (3.0 μg, N-20 Santa Cruz Biotechnology); rabbit anti-Sp1 (4.0 μg, 07-645, Millipore); rabbit anti-H3K27ac (2.0 μg, ab4729; abcam); and rabbit anti-H3K4me1 (2.0 μg, ab8895, abcam). Negative control rabbit immunoglobulin fraction (X0936, Dako) was utilized as a mock control. Primer sets were designed as follows: MafB enhancer F: CAG CCT CTT AGA CTT GGG CAG, R: CTG AGT CCT GGG CTG CAA AG; Fkbp5 enhancer F: AAG CCA TTC CAG TAG TCA GGA CC, R: GGA CTC TTG TAA AGC AGC GAG G; and Nkx3.1 enhancer F: TTC TTT CGC AGG GCT GTG TG, R: AGA TCC CTC AGC AGT GCC AG. The PCR thermal cycling program was performed using the following conditions: 5 min at 95 °C and 40 cycles of 95 °C for 10 s, 55 to 63 °C for 30 s, 72 °C for 1 min, and 72 °C for 10 min. Finally, PCR products were loaded onto 1.5% (wt/vol) agarose gel and visualized by gel staining with ethidium bromide. qPCR thermal cycling was performed using the following conditions: a holding stage of 95 °C for 30 s; a cycling stage of 40 cycles of 95 °C for 3 s and 60 °C for 30 s; and a melt curve stage; 95 °C of 15 s, 60 °C for 1 min, and 95 °C for 15 s.
ChIP-Seq.
ChIP procedure was performed as described above. The following primary antibodies were used: rabbit anti-AR (3.0 μg, N-20, Santa Cruz Biotechnology); goat anti-Sp1 (2.4 μg, A303-944A, Bethyl Laboratories, Inc.); rabbit anti-H3K27ac (2 μg, ab4729, abcam); and rabbit anti-H3K4me1 (2 μg, ab8895, abcam). An immunoprecipitated DNA library for Ion Torrent was prepared following the methods in NEBNext Fast DNA Library Prep Set for Ion Torrent (New England BioLabs Inc.). An immunoprecipitated DNA library for NextSeq was prepared following the methods in KAPA LTP Library preparation Kit (KAPA), SeqCap Adaptor Kit A (Roche), and Agencourt AMPure XP Beads (Beckman Coulter). High-throughput sequencing was performed using Ion Proton system (Thermo Fisher Scientific) and NextSeq 500 desktop sequencer (Illumina) in accordance with the manufacturer’s instructions. The qualified reads were aligned onto the mouse reference genome University of California, Santa Cruz mm10 using Bowtie2 v2.3.2 (51) with the option “-k 2.” Peak calling was performed utilizing MACS2 v2.1.1.20160309 with the option “–to-large–nomodel–extsize 300” (52). Motif enrichment analysis was carried out by using the MEME program in the MEME Suite (53) followed by the TOMTOM program (54) to compare the identified motifs with known ones. The motif database was utilized by JASPAR (an open-access database for eukaryotic transcription factor binding profiles) (55). Heatmaps of ChIP-seq data using K-means clustering on AR peak regions were generated with ngs.plot v.2.61 (56). All of the 488 common AR peaks were significantly enriched in comparison with nonoverlapped AR peaks judged by MACS2 score. Two independent biological replicates were performed for ChIP-seq (Sp1 for single analysis). ChIP-seq data have been deposited in Gene Expression Ominibus (GEO) under accession number GSE158279.
Generation of MafB-3′ UTR Mutation Mouse by CRISPR-Cas9 Genome Editing.
Mice lacking the H3K27ac regulatory (enhancer) element in the MafB-3′UTR were generated by CRISPR-Cas9 genome editing tools. Pronuclear injections of guide RNAs (gRNAs) into C57BL/6 × DBA/2 (obtained from Sankyo Lab Service) genetic background mixed embryos were performed to generate enhancer deletion F0 mice. All experimental procedures and protocols for generation of this mutant mouse line were approved by the Animal Care and Use Committee of the National Center for Child Health and Development. Two gRNAs and Cas9 protein were injected into fertilized eggs for deletion of the H3K27ac regulatory region: gRNA-d (5′- TCT GTG AGT CCT GGC GGG TCC GG-3′) and gRNA-c (5′-TGC CGA GAT CCA CAT CGT GCA GG-3′). gRNA information was obtained from CRISPRdirect (57). F0 mice were further crossed with C57BL/6J mice. F1 mice were genotyped by direct sequencing of PCR products using ExoSAP-IT (ThermoFisher Scientific) and BigDye Terminator v1.1 (ThermoFisher Scientific) and subsequently by Applied Biosystems 3500/3500 × L Genetic analyzer. Male mice at the F2 generation or later were utilized for analyses. For PCR genotyping for MafBeΔ/eΔ mice, the following primer pair was used: F 5′-CCC AGT CGT GCA GGT ATA AAC-3′ and R 5′-CAC GTA GCA AGA GGT GGA AG-3`. For genotyping of the MafB wild-type allele, the same primer pair was utilized same as for detection of the MafB eExG enhancer in ChIP analyses (as described above).
Statistics Analysis.
All bar graph data are shown as mean ± SEM. Statistical differences between groups were analyzed using the Student’s t test or Welch’s t test and one-way ANOVA followed by Tukey–Kramer test and Dunnett’s test. P value < 0.05 and P value < 0.01 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Drs. Sadaaki Maeda, Satoru Takahashi, Miho Terao, Takanori Hirano, Gerald Thiel, Yukiko Ogino, Takashi Baba, Yuichi Shima, Makoto Tachibana, Daisuke Matsumaru, Aki Murashima, Hiroko Suzuki, Kenji Toyota, Chikako Yokoyama, Mami Miyado, Yoshihiro Komatsu, Yu Hirano, Kei-ichi Katayama, Izumi Sasaki, Kenji Shimamura, Fan-Yan Wei, Shingo Usuki, Takashi Seki, Kei-ichiro Ishiguro, and Sho Morioka. We would like to express our appreciation to our laboratory colleagues and Benjamin Phillis. We also express our appreciation to Tomiko I. Iba and Yugi Rim for their valuable assistance. This work was supported by the Joint Usage/Research Center for Developmental Medicine, Institute of Molecular Embryology and Genetics, Kumamoto University and Japan Society for the Promotion of Science KAKENHI Grants 19K24051, 17H06432, 18H02474, 18K06837, and 18K06938.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2024067118/-/DCSupplemental.
Data Availability
ChIP-seq data have been deposited in National Center for Biotechnology Information GEO under accession no. GSE158279. All other study data are included in the article and/or SI Appendix.
References
- 1.Miyagawa S., et al., Genetic interactions of the androgen and Wnt/beta-catenin pathways for the masculinization of external genitalia. Mol. Endocrinol. 23, 871–880 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Welsh M., et al., Identification in rats of a programming window for reproductive tract masculinization, disruption of which leads to hypospadias and cryptorchidism. J. Clin. Invest. 118, 1479–1490 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toivanen R., Shen M. M., Prostate organogenesis: Tissue induction, hormonal regulation and cell type specification. Development 144, 1382–1398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murashima A., Xu B., Hinton B. T., Understanding normal and abnormal development of the Wolffian/epididymal duct by using transgenic mice. Asian J. Androl. 17, 749–755 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larkins C. E., Enriquez A. B., Cohn M. J., Spatiotemporal dynamics of androgen signaling underlie sexual differentiation and congenital malformations of the urethra and vagina. Proc. Natl. Acad. Sci. U.S.A. 113, E7510–E7517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsushita S., et al., Regulation of masculinization: Androgen signalling for external genitalia development. Nat. Rev. Urol. 15, 358–368 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Welsh M., MacLeod D. J., Walker M., Smith L. B., Sharpe R. M., Critical androgen-sensitive periods of rat penis and clitoris development. Int. J. Androl. 33, e144–e152 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georgas K. M., et al., An illustrated anatomical ontology of the developing mouse lower urogenital tract. Development 142, 1893–1908 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amândio A. R., Lopez-Delisle L., Bolt C. C., Mascrez B., Duboule D., A complex regulatory landscape involved in the development of mammalian external genitals. eLife 9, e52962 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki K., et al., Sexually dimorphic expression of Mafb regulates masculinization of the embryonic urethral formation. Proc. Natl. Acad. Sci. U.S.A. 111, 16407–16412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quigley C. A., et al., Androgen receptor defects: Historical, clinical, and molecular perspectives. Endocr. Rev. 16, 271–321 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Petiot A., Perriton C. L., Dickson C., Cohn M. J., Development of the mammalian urethra is controlled by Fgfr2-IIIb. Development 132, 2441–2450 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Wang S., Lawless J., Zheng Z., Prenatal low-dose methyltestosterone, but not dihydrotestosterone, treatment induces penile formation in female mice and Guinea pigs†. Biol. Reprod. 102, 1248–1260 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumoto T., Takeyama K., Sato T., Kato S., Androgen receptor functions from reverse genetic models. J. Steroid Biochem. Mol. Biol. 85, 95–99 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Yeh S., et al., Generation and characterization of androgen receptor knockout (ARKO) mice: An in vivo model for the study of androgen functions in selective tissues. Proc. Natl. Acad. Sci. U.S.A. 99, 13498–13503 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Z., Armfield B. A., Cohn M. J., Timing of androgen receptor disruption and estrogen exposure underlies a spectrum of congenital penile anomalies. Proc. Natl. Acad. Sci. U.S.A. 112, E7194–E7203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iezzi M. L., et al., Clitoromegaly in childhood and adolescence: Behind one clinical sign, a clinical sea. Sex Dev. 12, 163–174 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Acebedo A. R., et al., Mesenchymal actomyosin contractility is required for androgen-driven urethral masculinization in mice. Commun. Biol. 2, 95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhaumik S. R., Smith E., Shilatifard A., Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 14, 1008–1016 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Öst A., Pospisilik J. A., Epigenetic modulation of metabolic decisions. Curr. Opin. Cell Biol. 33, 88–94 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Calo E., Wysocka J., Modification of enhancer chromatin: What, how, and why? Mol. Cell 49, 825–837 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pihlajamaa P., et al., Tissue-specific pioneer factors associate with androgen receptor cistromes and transcription programs. EMBO J. 33, 312–326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishida H., et al., Gene expression analyses on embryonic external genitalia: Identification of regulatory genes possibly involved in masculinization processes. Congenit. Anom. (Kyoto) 48, 63–67 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Matsushita S., et al., Androgen regulates Mafb expression through its 3'UTR during mouse urethral masculinization. Endocrinology 157, 844–857 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Letovsky J., Dynan W. S., Measurement of the binding of transcription factor Sp1 to a single GC box recognition sequence. Nucleic Acids Res. 17, 2639–2653 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Connor L., Gilmour J., Bonifer C., The role of the ubiquitously expressed transcription factor Sp1 in tissue-specific transcriptional regulation and in disease. Yale J. Biol. Med. 89, 513–525 (2016). [PMC free article] [PubMed] [Google Scholar]
- 27.Moriguchi T., et al., MafB is essential for renal development and F4/80 expression in macrophages. Mol. Cell. Biol. 26, 5715–5727 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imperato-McGinley J., Zhu Y. S., Androgens and male physiology the syndrome of 5alpha-reductase-2 deficiency. Mol. Cell. Endocrinol. 198, 51–59 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Ohta S., et al., Gene transduction by sonoporation. Dev. Growth Differ. 50, 517–520 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Flodby P., et al., Cell-specific expression of aquaporin-5 (Aqp5) in alveolar epithelium is directed by GATA6/Sp1 via histone acetylation. Sci. Rep. 7, 3473 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prins G. S., Putz O., Molecular signaling pathways that regulate prostate gland development. Differentiation 76, 641–659 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie Q., Wang Z. A., Transcriptional regulation of the Nkx3.1 gene in prostate luminal stem cell specification and cancer initiation via its 3′ genomic region. J. Biol. Chem. 292, 13521–13530 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magee J. A., Chang L. W., Stormo G. D., Milbrandt J., Direct, androgen receptor-mediated regulation of the FKBP5 gene via a distal enhancer element. Endocrinology 147, 590–598 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Rondon-Galeano M., et al., MAFB modulates the maturation of lymphatic vascular networks in mice. Dev. Dyn. 249, 1201–1216 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Pihlajamaa P., Sahu B., Jänne O. A., Determinants of receptor- and tissue-specific actions in androgen signaling. Endocr. Rev. 36, 357–384 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Gredler M. L., Patterson S. E., Seifert A. W., Cohn M. J., Foxa1 and Foxa2 orchestrate development of the urethral tube and division of the embryonic cloaca through an autoregulatory loop with Shh. Dev. Biol. 465, 23–30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jung Y. H., et al., Maintenance of CTCF- and transcription factor-mediated interactions from the gametes to the early mouse embryo. Mol. Cell 75, 154–171.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jambhekar A., Dhall A., Shi Y., Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 20, 625–641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagarajan S., Benito E., Fischer A., Johnsen S. A., H4K12ac is regulated by estrogen receptor-alpha and is associated with BRD4 function and inducible transcription. Oncotarget 6, 7305–7317 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jefferson W. N., et al., Widespread enhancer activation via ERα mediates estrogen response in vivo during uterine development. Nucleic Acids Res. 46, 5487–5503 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J. Y., et al., A role for WDR5 in integrating threonine 11 phosphorylation to lysine 4 methylation on histone H3 during androgen signaling and in prostate cancer. Mol. Cell 54, 613–625 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Metzger E., et al., Phosphorylation of histone H3 at threonine 11 establishes a novel chromatin mark for transcriptional regulation. Nat. Cell Biol. 10, 53–60 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jozwik K. M., Chernukhin I., Serandour A. A., Nagarajan S., Carroll J. S., FOXA1 directs H3K4 monomethylation at enhancers via recruitment of the methyltransferase MLL3. Cell Rep. 17, 2715–2723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu D., et al., Three-tiered role of the pioneer factor GATA2 in promoting androgen-dependent gene expression in prostate cancer. Nucleic Acids Res. 42, 3607–3622 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin C., Yin Y., Long F., Ma L., Tissue-specific requirements of beta-catenin in external genitalia development. Development 135, 2815–2825 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garza-Contreras J., Duong P., Snyder B. D., Schreihofer D. A., Cunningham R. L., Presence of androgen receptor variant in neuronal lipid rafts. eNeuro 4, ENEURO.0109-17.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tenkorang M. A. A., Duong P., Cunningham R. L., NADPH oxidase mediates membrane androgen receptor-induced neurodegeneration. Endocrinology 160, 947–963 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng J., Watkins S. C., Walker W. H., Testosterone activates mitogen-activated protein kinase via Src kinase and the epidermal growth factor receptor in sertoli cells. Endocrinology 148, 2066–2074 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Gubbay J., et al., Inverted repeat structure of the Sry locus in mice. Proc. Natl. Acad. Sci. U.S.A. 89, 7953–7957 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujita N., et al., MCAF mediates MBD1-dependent transcriptional repression. Mol. Cell. Biol. 23, 2834–2843 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng J., Liu T., Qin B., Zhang Y., Liu X. S., Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7, 1728–1740 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bailey T. L., et al., MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gupta S., Stamatoyannopoulos J. A., Bailey T. L., Noble W. S., Quantifying similarity between motifs. Genome Biol. 8, R24 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fornes O., et al., JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen L., Shao N., Liu X., Nestler E., ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15, 284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Naito Y., Hino K., Bono H., Ui-Tei K., CRISPRdirect: Software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq data have been deposited in National Center for Biotechnology Information GEO under accession no. GSE158279. All other study data are included in the article and/or SI Appendix.