Significance
Mutations in profilin-1 (PFN1), an actin-binding protein, cause amyotrophic lateral sclerosis (ALS) through an unknown mechanism. Here, we carried out a series of biophysical and cell biological analyses to investigate the effects of ALS-linked mutations on PFN1 function. We found that some moderately misfolded ALS-linked variants bind certain formin proteins with higher affinity and also promote enhanced formin-mediated actin polymerization. Mutation-induced flexibility within actin- and polyproline-binding regions of PFN1 may underlie these phenotypes. However, severe misfolding of PFN1 leads to loss of function with respect to formin-mediated actin polymerization. Our results indicate that ALS-linked PFN1 perturbs actin dynamics, but the mechanism by which this perturbation occurs appears dependent upon the thermodynamic stability of the mutant protein.
Keywords: profilin-1, actin dynamics, amyotrophic lateral sclerosis, protein misfolding, formins
Abstract
Profilin-1 (PFN1) plays important roles in modulating actin dynamics through binding both monomeric actin and proteins enriched with polyproline motifs. Mutations in PFN1 have been linked to the neurodegenerative disease amyotrophic lateral sclerosis (ALS). However, whether ALS-linked mutations affect PFN1 function has remained unclear. To address this question, we employed an unbiased proteomics analysis in mammalian cells to identify proteins that differentially interact with mutant and wild-type (WT) PFN1. These studies uncovered differential binding between two ALS-linked PFN1 variants, G118V and M114T, and select formin proteins. Furthermore, both variants augmented formin-mediated actin assembly relative to PFN1 WT. Molecular dynamics simulations revealed mutation-induced changes in the internal dynamic couplings within an alpha helix of PFN1 that directly contacts both actin and polyproline, as well as structural fluctuations within the actin- and polyproline-binding regions of PFN1. These data indicate that ALS-PFN1 variants have the potential for heightened flexibility in the context of the ternary actin–PFN1–polyproline complex during actin assembly. Conversely, PFN1 C71G was more severely destabilized than the other PFN1 variants, resulting in reduced protein expression in both transfected and ALS patient lymphoblast cell lines. Moreover, this variant exhibited loss-of-function phenotypes in the context of actin assembly. Perturbations in actin dynamics and assembly can therefore result from ALS-linked mutations in PFN1. However, ALS-PFN1 variants may dysregulate actin polymerization through different mechanisms that depend upon the solubility and stability of the mutant protein.
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease that primarily affects motor neurons. Symptom progression is rapid, and the disease is universally fatal. Although most cases of ALS are sporadic, ∼10% of all ALS cases are familial (FALS) due to an inheritable genetic defect (1). Identification of FALS-linked genes has informed on the biological mechanism(s) underlying ALS pathogenesis. The ∼25 known ALS proteins appear to cluster within three functional processes: cytoskeletal dynamics, protein homeostasis, and RNA processing (1, 2). These categorizations are not mutually exclusive, however, in that some ALS-linked proteins can affect more than one process. For example, mutations in the actin-binding protein profilin-1 (PFN1) were recently linked to FALS (3–6), providing further support for disrupted cytoskeletal dynamics as a pathogenic mechanism in ALS (7, 8). In addition, ALS-linked mutations destabilize the PFN1 tertiary structure and induce PFN1 aggregation, which is expected to impact cellular protein homeostasis (3, 9–13). Despite the emerging evidence that mutant PFN1 adopts a misfolded conformation, little is known about how these conformational changes affect the biological functions of PFN1. Furthermore, it has been unclear whether mutation-induced destabilization and/or altered actin dynamics contribute to PFN1-mediated ALS.
Efficient actin assembly requires coordinated interactions between PFN1, actin, and cytoskeletal proteins containing polyproline (poly-Pro) motifs, such as the formin family of proteins (14, 15), Ena/Vasp (16) as well as all nucleation promoting factors (17). Monomeric actin (G-actin) is the building block of filamentous actin (F-actin) (14), and formins are a class of proteins that function in actin nucleation and elongation (18, 19). G-actin–bound PFN1 binds poly-Pro motifs within the formin homology (FH)1 domain, forming a ternary complex that facilitates incorporation of G-actin onto the FH2-bound barbed end of a growing actin filament (14). Given the pivotal role of PFN1 in actin assembly, ALS-linked mutations have been assumed to induce functional deficits related to actin binding and/or formin-mediated actin assembly (20). In support of this notion, co-immunoprecipitation (co-IP) experiments from cultured cells showed impaired binding between mutant PFN1 and actin (3). However, subsequent studies with recombinant proteins showed that PFN1 mutants retain wild-type (WT) binding affinity for actin (9). Furthermore, mutant PFN1 binds a generic poly-L-proline peptide with similar affinity as PFN1 WT, although similar binding experiments using specific poly-Pro sequences derived from biological proteins have not been reported (9). Therefore, the effects of ALS-linked mutations on PFN1 ligand-binding and function have yet to be determined.
Here, we conducted an unbiased immunoprecipitation–mass spectrometry (IP-MS) study with mammalian cells to identify potential differences in the protein interactomes of PFN1 WT and several ALS-linked PFN1 variants. Most of the PFN1 protein interactions were the same across all variants tested and mainly involved cytoskeletal proteins. However, the IP-MS analyses revealed enhanced interactions between two PFN1 variants, M114T and G118V, with a subset of formin proteins. Actin polymerization assays showed that these PFN1 variants retain the ability to facilitate formin-induced actin polymerization and that this function becomes enhanced in the context of a cellular environment. In contrast, the C71G ALS variant did not exhibit enhanced binding to formins and was deficient in promoting formin-mediated actin polymerization in vitro. In fact, PFN1 C71G was the most destabilized protein in all assays performed herein, resulting in PFN1 C71G turnover by the proteasome and reduced steady-state expression.
Altogether, our results demonstrate that ALS-linked mutations do not simply abrogate binding of PFN1 to actin, formins, or other cytoskeletal factors. Rather, the effect of these mutations on the actin cytoskeleton appears to be more complex than previously thought. The outcomes of our various analyses indicate that some degree of mutation-induced destabilization (i.e., caused by M114T and G118V mutations) confers enhanced flexibility within mutant PFN1 in regions that bind actin and poly-Pro sequences. PFN1 flexibility appears to positively correlate with enhanced formin-mediated actin polymerization in vitro and in cells. However, severe mutation-induced destabilization leads to unproductive interactions between PFN1 C71G and cytoskeletal proteins, resulting in loss of PFN1 function. Notably, both classes of mutation alter formin-mediated actin polymerization, which may represent a general dysregulation of actin dynamics that contributes to PFN1-mediated ALS pathogenesis.
Results
ALS-Linked PFN1 M114T and G118V Variants Exhibit Enhanced Binding to Select Formin Proteins.
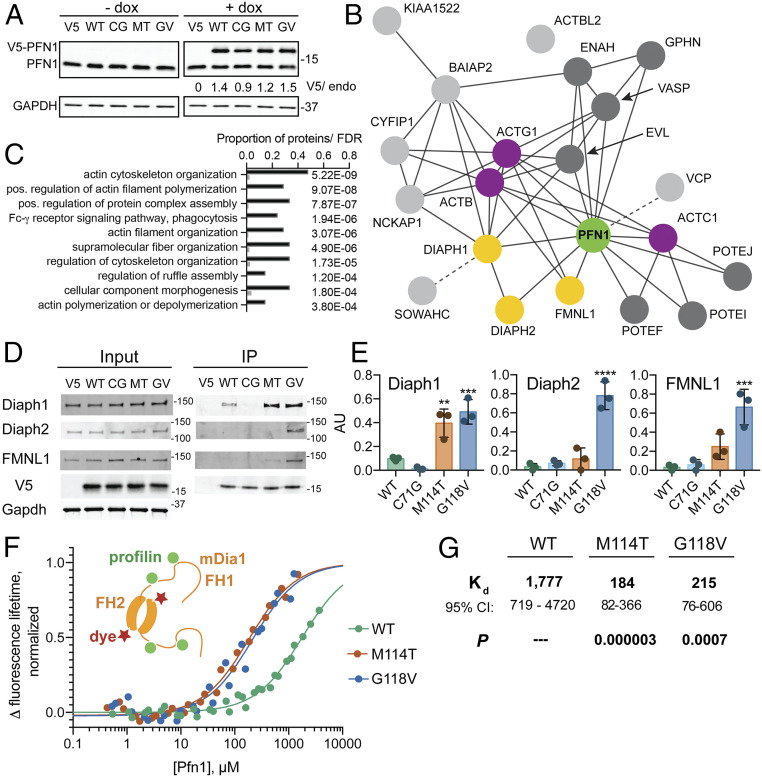
ALS-linked mutations induce conformational changes within PFN1 (9). However, little is known about how these changes affect PFN1 function. We performed an unbiased proteomics analysis to determine whether the functional protein interactions of PFN1 are altered by disease-causing mutations. To this end, stable, doxycycline-inducible HeLa cell lines expressing V5-tagged PFN1 WT and three ALS-associated variants (C71G, M114T, and G118V) were generated for an IP-MS analysis of PFN1 binding proteins. Exogenous PFN1 proteins were expressed at ∼1.2- to 1.5-fold over endogenous PFN1 and at similar levels to each other with the exception of PFN1 C71G, which expressed at lower levels compared with endogenous PFN1 (Fig. 1A). As described in Materials and Methods, lysates were diluted such that all input samples for the IP contained similar levels of V5-PFN1 as well as total protein. IP of V5-PFN1 proteins was achieved with an anti-V5 antibody, and eluates from three independent experiments were processed for MS analysis (Dataset S1) (21).
Fig. 1.
Select formins exhibit enhanced binding to ALS-linked PFN1 variants over PFN1 WT. (A) Western blot of cell lines treated with doxycycline (dox) or vehicle for 48 h, probed with anti-PFN1 and anti-GAPDH antibodies. V5-tagged PFN1 (top band) and endogenous PFN1 (bottom band) are labeled. CG = C71G, MT = M114T, EG = E117G, and GV = G118V. Numbers (V5/endo) represent the ratio of V5-tagged PFN1 to endogenous PFN1 based on densitometry. (B) STRING diagram for IP-MS results. Shown are gene names of PFN1-binding proteins enriched over the V5 negative control, most of which are known interactors of PFN1 (green) and actin (purple), including formin proteins that are validated in this study (yellow). Additional direct and indirect PFN1 interactions are shown in dark and light gray, respectively, with dotted lines connecting interactions not yet documented in STRING. (C) Results of a STRING gene ontology (GO) term analysis for the top 10 “process” terms. The graph includes the GO term, frequency of the term in this IP-MS dataset (black bars) versus approximate frequency in the total proteome (gray bars), and the false discovery rate (FDR). (D) Representative Western blots of IP inputs and eluates. V5-PFN1 variants, formin candidates, and the GAPDH loading control are labeled accordingly. (E) Quantification of D with n = 3 biological replicates. For each IP reaction, the intensity of the formin band was normalized to the V5-PFN1 band. Bars represent the mean ± SD. Statistics were determined using one-way ANOVA with Dunnett’s multiple comparison test compared with PFN1 WT: **P < 0.01, ***P < 0.001, and ****P < 0.0001. (F) Results of TCSPC measurements with a fluorescently labeled recombinant GST-mDia1-FH1-FH2-(His)8 construct upon titration with the indicated untagged PFN1 variants. A cartoon of this complex is shown on the left. Each curve is fitted using a single-site binding model based on measurements from at least two independent, 12-point baseline adjusted titrations. The binding curves are normalized for the total lifetime change upon binding and the concentration of GST-Dia1-FH1-FH2-(His)8. (G) Kd values (micromolar, with CI) were determined from F. P values for Kd results were determined from the extra sum-of-squares F-test for each PFN1 variant versus PFN1 WT.
To identify high-confidence interactions of PFN1, we employed the Significance Analysis of INTeractome (SAINT) algorithm to compare the V5-PFN1 samples with the V5 negative control cell line (expressing the V5 plasmid without PFN1) (22). The SAINT scores show the expected bimodal distribution between 0 and 1 (SI Appendix, Fig. S1A), with a SAINT score of 1 representing a high-confidence PFN1 interaction (22). The mean fold change in spectral counts for a given protein between the experimental and negative control conditions also show a positive correlation with the SAINT score (SI Appendix, Fig. S1B). Furthermore, proteins with intensity-based absolute quantification values that were enriched in PFN1 IPs over the negative control largely overlapped with the SAINT analysis (Dataset S1). Twenty-one proteins were found to be significantly enriched within at least one PFN1 cell line over the negative control line with SAINT scores ≥0.8 (Table 1). A Search Tool for Recurring Instances of Neighbouring Genes (STRING) gene ontology analysis of these proteins revealed a network of cytoskeletal-associated proteins (23), many of which were already known to bind PFN1 (P < 1 × 10−16) (Fig. 1 B and C and Dataset S1) (15). For example, high-confidence interactions with several actin isoforms were detected for all PFN1 variants, which is not surprising given that monomeric actin binds PFN1 with high affinity in cells (14). Conversely, some known PFN1 ligands were only weakly detected for PFN1 WT, such as Formin-like 1 (FMNL1), resulting in a low SAINT score (Table 1). Differences in abundance, binding strength, and stability among PFN1-ligand complexes are expected to influence how much of a particular ligand co-IPs with PFN1 and could explain why some PFN1 ligands were more enriched in the IP-MS study than others. For the high-confidence PFN-WT ligands with SAINT scores ≥0.8, no loss of interaction due to ALS-linked mutations was detected (Table 1).
Table 1.
Proteins enriched in PFN1 lines compared with the V5 negative control line
| WT | C71G | M114T | G118V | |||||
| Gene | FC | SAINT | FC | SAINT | FC | SAINT | FC | SAINT |
| GPHN | 29.74 | 1 | 30.46 | 1 | 32.16 | 1 | 28.25 | 1 |
| ENAH | 22.94 | 1 | 22.89 | 1 | 22.61 | 1 | 23 | 1 |
| VASP | 16.84 | 1 | 16.8 | 1 | 17.69 | 1 | 17.1 | 1 |
| PFN1 | 4.78 | 1 | 4.4 | 1 | 3.8 | 1 | 4.54 | 1 |
| ACTB | 4.58 | 1 | 4.72 | 1 | 4.53 | 1 | 4.52 | 1 |
| ACTG1 | 4.56 | 1 | 4.7 | 1 | 4.53 | 1 | 4.54 | 1 |
| ACTC1 | 4.14 | 1 | 4.2 | 1 | 3.67 | 1 | 3.9 | 1 |
| ACTBL2 | 3.99 | 1 | 3.96 | 1 | 3.44 | 1 | 3.49 | 1 |
| POTEF | 3.58 | 1 | 3.58 | 1 | 3.29 | 1 | 3.3 | 0.96 |
| POTEI | 3.38 | 0.98 | 3.5 | 1 | 3.29 | 1 | 3.33 | 0.94 |
| BAIAP2 | 4.87 | 0.66 | 5.53 | 0.96 | 8.64 | 1 | 10.25 | 1 |
| CYFIP1 | 4.25 | 0.59 | 5.9 | 0.93 | 8.95 | 1 | 8.25 | 1 |
| EVL | 4.73 | 0.64 | 6.54 | 0.89 | 6.8 | 0.98 | 5.97 | 0.99 |
| DIAPH1 | 18.97 | 0.59 | 16.04 | 0.29 | 130.52 | 1 | 145.21 | 1 |
| FMNL1 | 2.17 | 0.01 | 2.22 | 0 | 13.01 | 1 | 24.05 | 1 |
| SOWAHC | 1.36 | 0 | 0 | 0 | 18.61 | 1 | 15.31 | 1 |
| POTEJ | 4.07 | 0.79 | 4.06 | 0.99 | 4.08 | 1 | 4.08 | 0.76 |
| NCKAP1 | 2.84 | 0.38 | 3.85 | 0.66 | 7.92 | 0.98 | 6.35 | 0.99 |
| KIAA1522 | 2.89 | 0.57 | 1.71 | 0.26 | 4.81 | 0.92 | 5.35 | 0.93 |
| DIAPH2 | 2.17 | 0 | 0 | 0 | 21.14 | 0.79 | 48.32 | 1 |
| VCP | 2.21 | 0.73 | 2.16 | 0.68 | 2.2 | 0.89 | 1.98 | 0.66 |
Genes encoding proteins with a SAINT score ≥0.8 versus the V5 negative control line for at least one V5-PFN1 line. The mean fold change (FC) in spectral counts for each protein in the V5-PFN1 line versus the V5 negative control line is also shown. Proteins are ranked by the number of V5-PFN1 cell lines showing a SAINT score ≥0.8, then by maximum SAINT score, then by average FC. SAINT scores below the 0.8 threshold are shown in italics.
To identify proteins that preferentially bind mutant PFN1, the IP-MS results from the PFN1 WT line were compared with those of all three ALS-linked variants using SAINT. No significant differences were detected between PFN1 WT and C71G. However, four proteins were highly enriched in the PFN1 M114T and G118V IPs compared with PFN1 WT, including the formins Diaphanous (Diaph) 1 and 2, FMNL1, and the ankyrin repeat domain family member sosondowah homolog C (SOWAHC) (Table 2). SOWAHC was excluded from further analysis, as this protein is not expressed in the central nervous system and therefore is less relevant to ALS (24). Independent IP and Western blot analyses confirmed that FMNL1, Diaph1, and Diaph2 were significantly enriched in the PFN1 G118V IP compared with PFN1 WT IP (Fig. 1 D and E). Analogous results were observed for M114T, except the enrichment of Diaph2 and FMNL1 did not reach statistical significance in the IP–Western analysis. Diaph2 levels appeared lower in PFN1 C71G versus PFN1 WT eluates, although this difference also did not reach statistical significance.
Table 2.
ALS profilin 1 variants show increased interaction with formin family proteins
| C71G | M114T | G118V | ||||
| Gene | FC | SAINT | FC | SAINT | FC | SAINT |
| DIAPH2 | 0 | 0 | 8.75 | 0.97 | 19.86 | 1 |
| SOWAHC | 0 | 0 | 11.55 | 1 | 9.53 | 0.89 |
| DIAPH1 | 0.85 | 0 | 6.78 | 1 | 7.55 | 0.7 |
| FMNL1 | 1.02 | 0 | 5.43 | 0.95 | 9.94 | 0.92 |
All identified PFN1-interacting proteins in ALS-PFN1 variant cell lines were evaluated for enrichment over the PFN1 WT cell line using SAINT. Several formin family proteins (DIAPH1, DIAPH2, FMNL1) are more abundant. All proteins with a SAINT score ≥0.5 versus WT with any variant are shown. Proteins are ranked by maximum SAINT score versus WT, then by average fold change (FC) as calculated by the SAINT algorithm. SAINT scores below the 0.8 interaction threshold are shown in italics.
We wondered whether the preferential binding between select formins and PFN1 variants resulted from an altered subcellular localization of the proteins. Reliable antibodies for coimmunofluorescence (IF) analysis were identified for V5 (PFN1), Diaph1, and FMNL1. However, there were no obvious differences in cellular expression or localization of these formins among the PFN1 lines (SI Appendix, Fig. S2). Next, we considered the possibility that ALS-linked mutations in PFN1 result in an inherently tighter binding affinity for formin. PFN1 interactions are mediated through multiple poly-Pro motifs within the FH1 domain, whereas FH2 domains promote formin dimerization (Fig. 1F) (18). Diaph1 was most enriched in binding PFN1 proteins in the IP-MS study (Table 1) and was therefore prioritized for in vitro binding studies with recombinant proteins (9, 25). Recombinant mDia1, the mouse homolog of human Diaph1, was used here in line with our previous work (25, 26). GST-mDia1-FH1-FH2-(His)8 was labeled with the fluorescent Red-Tris-NTA dye at the poly-His tag, and the fluorescence lifetime of the fluorophore was measured by time-correlated single-photon counting (TCSPC) as a function of increasing PFN1 concentration (27, 28). Titration of untagged PFN1 increased the fluorescence lifetime of the labeled mDia1 construct by ∼1 ns, a change that was detected at much lower concentrations of M114T and G118V compared with PFN1 WT (Fig. 1F). A global fit of the fluorescence lifetime binding curves resulted in a significantly lower dissociation constant (Kd) for PFN1 M114T and G118V relative to PFN1 WT, indicative of tighter binding between these variants and mDia1 (Dataset S2) (29). As none of the binding curves reached a full plateau, there were large confidence intervals for the Kd values, particularly for PFN1 WT (Fig. 1G). This assay was limited by the concentrations of PFN1 needed to detect an mDia1 binding signal, in which ∼4 mM PFN1 WT was insufficient to saturate the binding curve, and PFN1 C71G could not be included due to its low solubility (Fig. 1F) (9). Nonetheless, the TCSPC assay reports on a binding event between PFN1 and mDia1 that is tighter for the ALS variants relative to PFN1 WT, consistent with the IP-MS results.
We then performed a binding assay with a small peptide (PPPPPFGFGVPAAPVL) derived from the first poly-Pro track in the FH1 domain of mDia1, adjacent to the FH2 domain, and followed changes in intrinsic PFN1 tryptophan fluorescence as a read-out for binding. Binding of all PFN1 variants to this peptide occurred with Kd values between 0.9 and 2.9 µM, with WT PFN1 exhibiting the highest affinity (SI Appendix, Fig. S3). These binding affinities are tighter than what was detected by TCSPC with GST-mDia1-FH1-FH2-(His)8 and in other reports with generic poly-L-proline peptides (9, 30). The amino acids C-terminal to the P5 motif may contribute to a lower Kd between this peptide and PFN1 compared with generic poly-L-proline peptides, as observed for other profilins and proline-rich FH1 domains (31). The poly-Pro track(s) within mDia1 that contribute to the TCSPC results have yet to be identified. While the PPPPPFGFGVPAAPVL sequence probably does not promote enhanced binding to ALS-PFN1 variants, we cannot exclude the possibility that PFN1 binding occurs at multiple poly-Pro tracks during the TCSPC experiment but that the Red-Tris-NTA dye may be sensitive to only one of these binding events. Furthermore, it is possible that the presence of the FH2 domain, which dimerizes, is tethered to FH1, and is present in the TCSPC experiment but not the peptide binding assay, influences the binding between PFN1 and FH1. We also acknowledge that the binding affinities between profilins and poly-Pro–containing ligands can be influenced by assay conditions, the context of the poly-Pro sequence and profilin sequence (30, 31).
Formin-Induced Actin Polymerization Is Altered by ALS-PFN1 Variants In Vitro and in Cells.
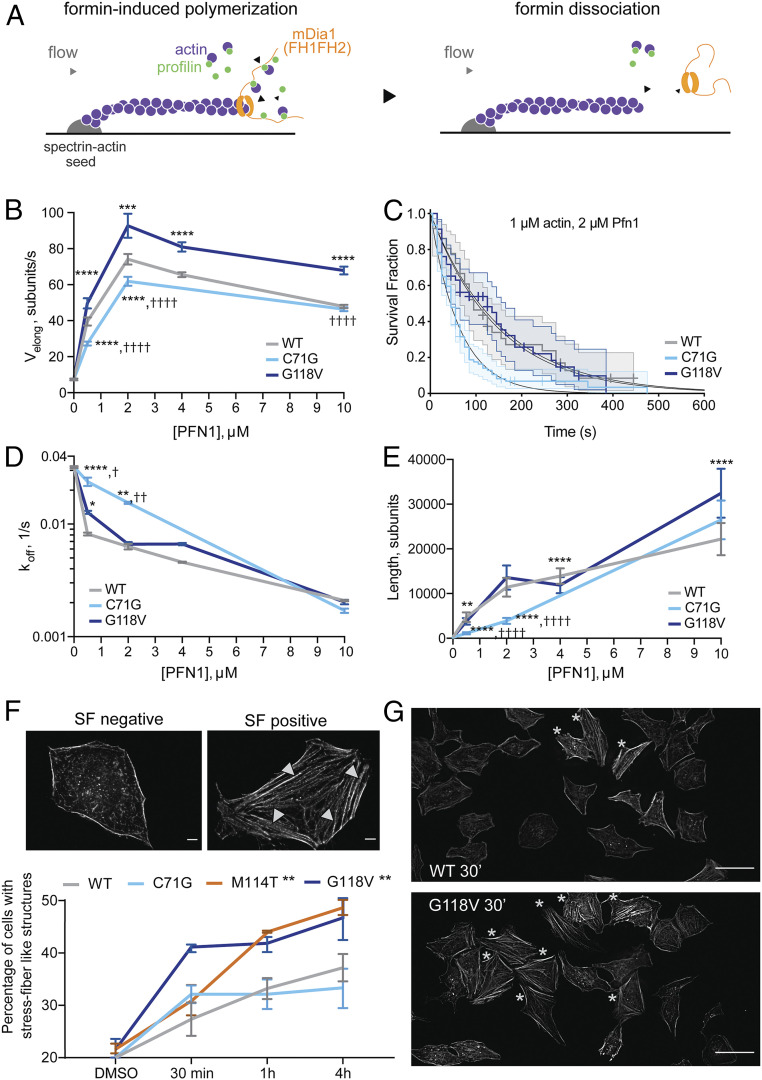
Next, we investigated the functional consequences of altered binding between ALS-linked PFN1 variants and diaphanous formins. First, mDia1-induced elongation of individual actin filaments was measured as a function of untagged, recombinant PFN1 concentration using a microfluidics approach in vitro (Fig. 2A and Movie S1) (25, 32). As expected, the rate of barbed-end actin filament elongation increases as a function of PFN1 WT concentration in the presence of 1 µM actin, peaking at an elongation rate of ∼70 subunits per second with 2 µM PFN1 (Fig. 2B) (25, 33, 34). Two ALS-linked PFN1 proteins were also examined, including the G118V variant that exhibited enhanced binding to diaphanous formins and the C71G variant that is severely destabilized (Fig. 1) (9). Both variants promoted mDia1-induced actin elongation. However, compared with PFN1 WT, the rate of elongation was significantly higher in the presence of PFN1 G118V but significantly lower with PFN1 C71G (Fig. 2B). While formation of F-actin filaments in the presence of PFN1 G118V occurred on a similar timescale as PFN1 WT in a pyrene–actin assembly assay, simulations of this process indicated that faster actin elongation rates for PFN1 G118V can be offset by higher numbers of formin-nucleated filaments in the presence PFN1 WT or C71G in this assay (SI Appendix, Fig. S4) (35). In the presence of PFN1 C71G, the time that mDia1 remains productively associated with the barbed end of elongating actin filaments in the microfluidics assay was reduced (Fig. 2C), resulting in faster rates of mDia1 dissociation (koff) from actin filament barbed ends compared with PFN1 WT and G118V (Fig. 2D). Together, these in vitro measurements indicate that PFN1 C71G is less competent at promoting formin-induced actin elongation, as slower actin elongation rates and faster dissociation of mDia1 from actin filaments in the presence of this variant culminates in low mDia1 processivity and shorter actin filaments being assembled (Fig. 2E). Although the elongation rates were higher overall for G118V than WT (Fig. 2B), mDia1 also tended to dissociate faster from actin barbed ends in the presence of PFN1 G118V (Fig. 2D), resulting in actin filaments that were of similar length as for PFN1 WT, except at high PFN1 concentrations (Fig. 2E).
Fig. 2.
ALS-linked mutations have differential effects on formin-induced actin assembly. (A) Actin filaments (1 µM actin, 10% Alexa488-labeled) elongate from surface-anchored spectrin–actin seeds in a microfluidics platform designed to measure actin elongation with purified proteins. Upon exposure to a solution of recombinant mDia1(FH1-FH2-DAD), the mDia1 FH2 domain associates with the barbed end of actin, and the mDia FH1 domain associates with actin-bound PFN1, resulting in accelerated actin filament elongation (Left). Upon formin dissociation, the barbed end reverts to slower elongation (Right). This assay was used to determine both formin-induced actin filament elongation rates, and formin processivity in the presence of PFN1 WT (gray), PFN1 C71G (cyan), and PFN1 G118V (dark blue). (B) The rate of actin elongation (Velong; subunits per second) at the barbed end is plotted as a function of PFN1 variant and concentration (micromolar). Each data point represents the mean ± SD of n = 20 to 88 filaments, t test with Bonferroni correction. (B, D, E): *difference from WT; †difference from G118V; *,†P < 0.05; **,††P < 0.01; ***,†††P < 0.001; and ****,††††P < 0.0001. (C) The survival fraction of mDia1(FH1-FH2-DAD) that remains bound to the actin barbed end over a 600 s time course was determined as a function of PFN1 variant and concentration (shown is 2 µM PFN1). Decay curves were fitted to a single exponential function to obtain the formin dissociation rate (koff). Each curve shows n = 36 to 94 filaments. Log-rank test: WT versus G118V, P = 0.894; WT versus C71G, P = 0.000765; and G118V versus C71G, P = 0.000503. (D) mDia1 koff (from C) are plotted as a function of PFN1 as in B; n = 24 to 138 filaments, log-rank test, P values adjusted with Bonferroni correction: *difference from WT; †difference from G118V. (E) Mean formin-induced actin filament length (Velong/koff) ± SD; n = 20 to 138 filaments, t test with Bonferroni correction. (F) HeLa cells expressing V5-PFN1 variants were treated with 10 µM IMM-01 and assessed for the presence of actin stress fiber (SF)–like structures with phalloidin. An SF-negative and -positive cell are shown. (Scale bar, 5 µm.) Only four fibers are denoted by arrowheads for clarity. The percentage of SF-positive cells for each line at the indicated time point is shown (n = 3 biological replicates, mean ± SEM, two-way ANOVA, and Dunnet’s multiple comparison test for difference from the WT cell line, **P < 0.01). (G) The indicated cell line is stained with phalloidin after 30 min of IMM-01 treatment. Stars denote SF-positive cells. (Scale bar, 50 µm.)
To complement the above reconstitution assay, we sought to investigate the effects of ALS-linked variants on formin-induced actin polymerization within the complex environment of a cell. To this end, V5-PFN1 HeLa cell lines were treated with intramimic-01 (IMM-01), a small molecule agonist of mammalian diaphanous proteins. IMM-01 binds the diaphanous inhibitory domain with a half maximal inhibitory concentration of ∼140 nM, thereby relieving diaphanous autoinhibition (36). Activation of Diaph1 formins induces an increase in the formation of actin stress fibers (37, 38). Here, addition of 10 µM IMM-01 to V5-PFN1–expressing cells resulted in robust phalloidin-positive F-actin assemblies that resembled stress fibers (Fig. 2F). In the presence of IMM-01, significantly more PFN1 M114T- and G118V-expressing cells exhibited stress fibers compared with PFN1 WT and C71G (Fig. 2F). G118V-expressing cells exhibited the most pronounced effect of IMM-01 after only 30 min and throughout the 4 h time course (Fig. 2F). We note that IMM-01–induced actin assembly occurred rapidly in the V5 negative control line compared with cells expressing V5-PFN1 variants (SI Appendix, Fig. S2). Given that the balance between F-actin networks within cells is complex and dependent on PFN1 levels (39), it is not surprising that exogenous expression of V5-PFN1 has an effect on actin dynamics compared with cells without exogenous PFN1. Taking this into account, we focused on differences among V5-PFN1–expressing cell lines, which express similar levels of PFN1. Indeed, the effects of IMM-01 follow the same trend as the IP-MS study for Diaph1, which was most enriched in PFN1 G118V, followed by M114T > WT ∼ C71G (Fig. 1 D and E). This trend is not influenced by PFN1 levels, as both exogenous and endogenous PFN1 levels are comparable in V5-PFN1 WT and G118V lines (Fig. 1A). Therefore, the effects of PFN1 variants on stress fiber formation are not due to differential expression of PFN1 between lines.
The M114T and G118V Mutations Perturb Internal Dynamic Couplings in PFN1.
To gain insight into how ALS-linked mutations perturb the PFN1/formin interaction, we performed molecular dynamics (MD) simulations on the different PFN1 variants. The X-ray crystal structures of WT and M114T mutant PFN1 served as the starting conformations for the simulation of WT and M114T PFN1 variants (9). For MD simulations of C71G and G118V, the initial conformation was generated from the X-ray structure of WT PFN1 after mutation of Cys71 to Gly and Gly118 to Val, respectively. For each system, five independent trajectories were run. Each of the 20 simulations was equilibrated for 100 ns, and data were collected during a subsequent 220 ns trajectory. While the structures of WT, M114T, and G118V were stable over the entire trajectories, the structure of PFN1 C71G did not reach equilibrium and started to unfold during the simulation, precluding further analysis. This observation is consistent with the destabilized nature of PFN1 C71G (9), and this variant was therefore excluded from further analysis.
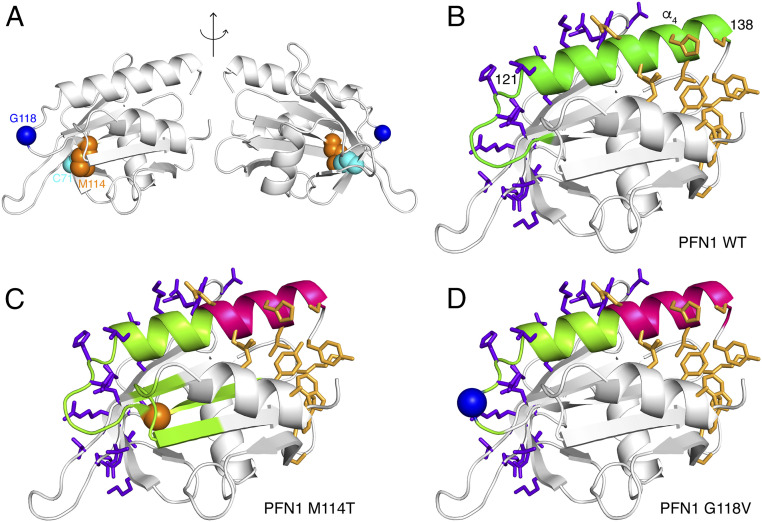
Network theory was used to analyze our MD simulations of PFN1 WT, M114T, and G118V (40). This approach can detect how subtle changes in conformation and flexibility, caused by a mutation, affect dynamical correlation between distant regions of the protein that are important for allosteric communication. For each system, we determined a network describing the internal dynamics of PFN1 that identifies communities of residues whose dynamics are correlated. To further characterize this network, we calculated the betweenness centrality for each residue, a measure of its importance for communication between correlated communities (40, 41). The observed differences in betweenness centrality for the networks of PFN1 variants indicate alterations in dynamical coupling for both variants compared with PFN1 WT (SI Appendix, Fig. S5). We also identified differences in the community structures, or regions within PFN1 that constitute a community of residues with correlated dynamics, among the PFN1 variants (highlighted in different colors in SI Appendix, Fig. S6 A–F). A striking difference involves the C-terminal helix (α4, residues 121 to 138, SI Appendix, Fig. S6 G and H; highlighted in green in Fig. 3B and lime and magenta in Fig. 3 C and D). Intriguingly, this helix is the only secondary structural element that contains residues that make direct contact with both actin and poly-Pro residues in the crystal structure of the actin–PFN1–poly-Pro ternary complex, in which the poly-Pro is derived from VASP (42) (Fig. 3 and SI Appendix, Fig. S6). For PFN1 WT, all the residues within this C-terminal helix are part of the same community (Fig. 3B). However, for both PFN1 M114T and G118V, this helix is split into two communities, each containing either the actin-binding residues or the formin-binding residues (Fig. 3 C and D and SI Appendix, Fig. S6), indicating an uncoupling of the two binding sites.
Fig. 3.
MD simulations show that M114T and G118V mutations perturb internal dynamic couplings in PFN1. (A) Cartoon representation of the structure of PFN1 highlighting the residues C71 (cyan), M114 (orange), and G118 (blue). Difference in correlation networks is observed between (B) WT and ALS-linked variants (C) M114T and (D) G118V PFN1. The side chains of residues that directly interact with actin and poly-Pro in the ternary complex comprised of PFN1-actin and the last poly-Pro motif of human VASP (Protein Data Bank 2PAV) are shown in stick representation in purple and gold, respectively. The C-terminal helix of PFN1 (α4, residues 121 to 138) binds both actin and poly-Pro. Actin and poly-Pro–binding residues located within this C-terminal helix are part of the same community with correlated dynamics (green) in WT (B), whereas this helix is separated into two distinct communities (lime and magenta) in both M114T (C) and G118V (D).
The fragmentation of this C-terminal helix (α4) community in the mutants corresponds with increased structural fluctuations throughout the mutant proteins (SI Appendix, Fig. S7A) and a few localized backbone dihedral fluctuations (SI Appendix, Fig. S7B), both of which affect the actin and poly-Pro interfacial residues. For both mutants, these structural fluctuations correspond to larger fluctuations (i.e., SDs) in the surface areas of PFN1 that bind actin and poly-Pro (SI Appendix, Fig. S7C). This is particularly apparent for the poly-Pro–binding region within PFN1 G118V (SI Appendix, Fig. S7 A and C). Despite these dynamical changes, the mutant proteins do not exhibit disrupted secondary structure relative to WT PFN1 (SI Appendix, Fig. S7D). However, when comparing more subtle structural differences between WT PFN1 and the G118V variant, a structural rearrangement within the ternary complex with actin and poly-Pro is predicted. Superimposing the structure of PFN1 G118V from the MD simulations onto the structure of WT PFN1 in the ternary complex (Protein Data Bank ID 2PAV, SI Appendix, Fig. S8) reveals a steric clash between PFN1 V118 and both T351 and Q354 within actin, suggesting that a rearrangement within PFN1 G118V and/or actin is necessary for binding. Additional details for the MD analyses are located within SI Appendix, Extended Analysis of the MD Trajectories.
PFN1 C71G Is Particularly Susceptible to Proteasomal Degradation in Cells.
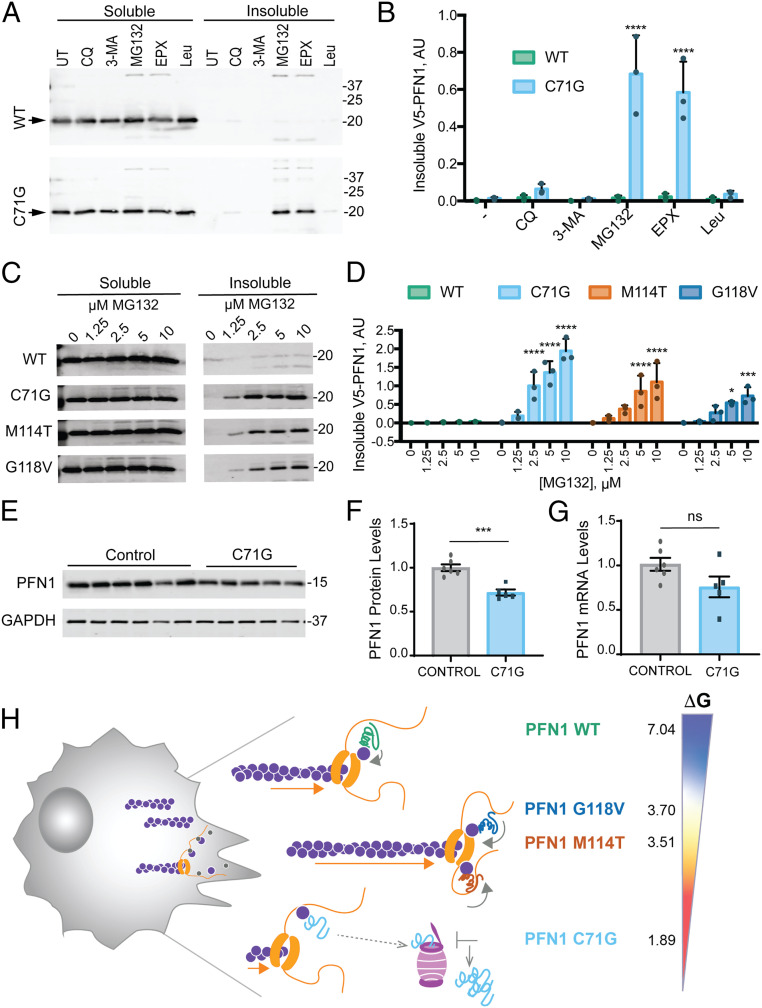
Throughout this study, efforts to examine PFN1 C71G by biochemical and MD approaches were hindered by the inherent instability of this variant. Our previous report demonstrated a reduced half-life of V5-PFN1 C71G in cells (9), in agreement with lower steady-state expression levels of this variant in our stable cell lines (Fig. 1A) and in yeast (13). To elucidate the mechanism for mutant PFN1 turnover, cells were treated with pharmacological inhibitors of the proteasome and autophagy pathways, and cell lysates were assessed for V5-PFN1 by Western blot analysis following separation of the soluble and insoluble fractions (Fig. 4A) (43). PFN1 C71G robustly accumulated within the insoluble fraction of HeLa cells treated with the proteasome inhibitors MG132 and epoxomicin, in contrast to HeLa cells treated with autophagy inhibitors chloroquine or 3-methyladenine, or with the lysosomal protease inhibitor leupeptin (Fig. 4B). Insoluble V5-PFN1 WT was negligible under all conditions (Fig. 4 A and B). To determine whether other ALS-linked PFN1 variants are also targeted to the proteasome, a similar study was carried out with all four V5-PFN1 lines treated with increasing concentrations of MG132. Indeed, insoluble V5-PFN1 M114T and G118V also accumulated within the insoluble fraction but to a lesser extent than PFN1 C71G (Fig. 4 C and D).
Fig. 4.
The PFN1 C71G variant is robustly targeted by the proteasome and exhibits reduced steady-state protein expression levels. (A) Western blot analysis of detergent soluble and insoluble fractions from HeLa cells expressing V5-PFN1 WT or C71G treated with proteasome or autophagy/lysosome inhibitors for 24 h prior to cell lysis. Soluble and insoluble fractions were processed on the same gel. Immunoblots were probed with anti-V5 antibody; arrowheads indicate V5-PFN1 WT and C71G. Abbreviations: UT: untreated, CQ: chloroquine (100 μM), 3-MA: 3-methyladenine (5 mM), MG: MG132 (10 μM), EPX: epoxomicin (10 μM), and Leu: leupeptin (100 μM). (B) Quantification of insoluble fractions from A (n = 3 biological replicates, mean ± SD, two-way ANOVA with Dunnett’s multiple comparison test for difference from untreated, ****P < 0.0001). (C) The indicated V5-PFN1 cell lines were treated with increasing concentrations MG132 for 24 h and processed as described in A. (D) Quantification of insoluble fractions from C (n = 3 biological replicates, mean ± SD, two-way ANOVA with Dunnett’s multiple comparison test for difference from WT, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (E) Western blot analysis of endogenous, untagged PFN1 protein levels and GAPDH loading control from immortalized human lymphoblast cells derived from controls and ALS patients harboring the C71G PFN1 mutation. (F) Quantification of E. For each biological replicate, PFN1 protein levels were normalized to the average level of the control cell lines. Each data point represents the average protein levels of n = 2 biological replicates for individual cell lines. Bar graphs show mean ± SEM among lymphoblast lines. Statistics were determine using student’s t test (***P < 0.001). (G) Relative mRNA levels of PFN1 normalized to the average level of the control cell lines. Each data point represents the average mRNA levels of n = 2 to 3 biological replicates for individual lines. Bar graphs show mean ± SEM among lymphoblast lines. (H) Proposed model for the effects of ALS-linked mutations on PFN1 in the context of formin-induced actin polymerization. Our MD simulations and the lower free energy of folding [ΔG°; determined by Boopathy et al. (9)] indicate regions within PFN1 that contact both actin and poly-Pro are more flexible in ALS-linked variants compared with PFN1 WT. This conformational flexibility may contribute to enhanced formin binding and processivity (depicted by orange arrows), potentially by affecting the ring complex (i.e., PFN1-actin bound to FH1 in contact with the FH2-bound barbed end). PFN1 C71G appears to be the least competent at promoting formin-induced actin polymerization. This variant also exhibits the lowest ΔG° (kcal/mol); 4% of PFN1 C71G exist in the unfolded state, compared with <0.3% for M114T and G118V and <0.0007% for WT. These data are consistent with the C71G variant adopting an unfolded and/or misfolded conformation that is more robustly targeted to the proteasome. Compared with PFN1 WT, all ALS-linked variants dysregulate formin-induced actin polymerization.
While V5-PFN1–inducible cells represent a tractable system for biochemical and functional studies, we also assessed the expression of endogenous, untagged mutant PFN1 in human cells derived from ALS patients. Specifically, PFN1 levels were quantified in immortalized human lymphoblast cells derived from five ALS patients harboring the heterozygous C71G PFN1 mutation and six controls (3). Steady-state PFN1 levels were significantly lower by ∼25% in the mutant line compared with controls (Fig. 4 E and F). Although PFN1 messenger RNA (mRNA) levels tended to be lower in mutant cells, the difference compared with controls did not reach statistical significance (Fig. 4G). Unfortunately, we do not have access to collections of lymphoblasts lines with the other ALS-linked PFN1 mutations for a similar analysis. Nonetheless, results from both the V5-PFN1 HeLa (Fig. 1A) and lymphoblast (Fig. 4 E and F) cell lines indicate that PFN1 C71G is more susceptible to proteasomal degradation and is expressed at lower levels relative to other PFN1 variants.
Discussion
To gain insight into the mechanisms underlying PFN1-mediated ALS, we conducted an unbiased proteomics screen to identify potential differences in PFN1 interactions caused by mutation. Overall, the same proteins were identified here across IP-MS samples, irrespective of PFN1 genotype, and most proteins were previously identified as PFN1 binding partners (15). In contrast to a previous co-IP experiment, we did not observe impaired binding between ALS-PFN1 variants and actin (3). Rather, our collective results demonstrate that all ALS-PFN1 variants tested herein are able to bind actin. For example, multiple actin isoforms were detected by MS in all the IP eluates across all PFN1 variants. Furthermore, our single-filament microfluidics experiments confirmed that PFN1 variants can promote formin-induced actin elongation, which requires formation of a functional G-actin/PFN1 complex (Fig. 2) (14). These observations are consistent with similar concentration-dependent effects of WT-PFN1 and ALS-linked PFN1 variants on suppression of the spontaneous polymerization of pyrene‐labeled actin monomers (9). Although no loss of PFN1 interactions due to ALS mutations were identified in this study, our IP-MS approach may have been limited in detecting weak or transient interactions of PFN1. While interactions that become diminished due to mutations in PFN1 could also be relevant to ALS, a protocol that involves proximity labeling in situ may be required to uncover this phenotype.
Importantly, this IP-MS protocol identified proteins with enhanced binding to ALS-linked PFN1 variants. Specifically, Diaph1 and 2 and FMNL1 appear to bind more tightly with PFN1 M114T and G118V (Fig. 1). Does enhanced binding of mutant PFN1 to these formins accelerate actin polymerization? Indeed, the formin/actin/profilin system is complex, as there are multiple steps and modes of interaction between these proteins during actin polymerization (34). PFN1 plays a critical role in barbed-end elongation by mediating the interaction between poly-Pro tracks within FH1 and actin (35, 44, 45). Binding of PFN1-actin to FH1 poly-Pro tracks appears to be rate limiting for addition of actin onto the FH2-bound barbed end of a growing filament (35, 44). FH1 domains are disordered domains and thus able to form flexible loops that place PFN1/actin/poly-Pro in close proximity to the barbed end in the form of a “ring complex” for effective actin transfer (Fig. 4H) (25, 34). With 14 poly-Pro tracks in mDia1 FH1, it is not clear whether one or multiple of those poly-Pro tracks contribute to the apparent increase in binding affinity for PFN1 M114T and G118V. Our results indicate the poly-Pro track closest to the FH2 domain probably does not account for the enhanced binding observed in our IP and TCSPC studies. In fact, the affinities for all ALS-linked PFN1 variants were weaker for this poly-Pro sequence compared with PFN1 WT. While lower affinity interactions between PFN1–actin and poly-Pro tracks nearest the FH2 domain may favor a faster transfer of actin to the barbed end, higher affinity interactions at distal poly-Pro tracks could stabilize the PFN1/actin/poly-Pro ternary complex and thus compensate for a slower loop closure from distal tracks (44). Accordingly, PFN1 M114T and G118V may bind more tightly to distal poly-Pro tracks than PFN1 WT (i.e., accounting for more Diaph1 in the co-IP) but weaker at tracks proximal to FH2. Both effects on poly-Pro binding could conceivably accelerate actin elongation in vitro and in cells (Fig. 2).
Other factors may also contribute to the effects of PFN1 on actin polymerization. Our MD analyses identified a network of residues within α4, which contacts both actin and poly-Pro, that are dynamically correlated in PFN1 WT but fragmented by ALS mutations (Fig. 3). These observations implicate this C-terminal helix in mediating allosteric communication within the ternary complex during actin elongation (41). Additionally, our MD analyses uncovered heightened structural fluctuations within mutant PFN1 at both the poly-Pro and actin-binding regions. It is uncertain whether heightened flexibility of PFN1 directly impacts actin polymerization, but this flexibility could modify interactions at the barbed end that in turn influence actin polymerization, including formation of the ring complex between FH1 and the barbed end, transfer of actin to the filament, and dissociation of PFN1 from the barbed end (Fig. 4H). Flexibility within the PFN1 structure due to ALS mutations may also confer differential binding with certain formins, potentially by allowing the poly-Pro–binding site to better accommodate certain sequences. While other poly-Pro–containing proteins were detected in our IP-MS study such as ENAH, EVL, NCKAP1, and VASP, they did not exhibit enhanced binding to PFN1 mutants. These results imply some degree of sequence specificity for the enriched PFN1/formin interactions, consistent with observations that ALS-linked PFN1 variants bind generic poly-L-proline peptides with similar affinity as PFN1 WT (9). Structural studies of PFN1 variants in complex with poly-Pro sequences from different formins should inform on the residues that contribute to their differential binding.
In contrast to a gain of function for PFN1 M114T and G118V with respect to actin polymerization, the C71G mutation appears to cause a loss of PFN1 function. The C71G mutation is severely destabilizing, likely due to an internal cavity formed by mutation of Cys to Gly (9). M114 and G118 are closer to the surface of PFN1, and mutation of these residues only partially destabilize PFN1 (Fig. 4H) (9). The instability and insolubility of PFN1 C71G precluded several analyses herein, including the TCSPC binding experiments and MD simulations. Single-filament microfluidics experiments, which require lower concentrations of protein, were possible with PFN1 C71G and revealed significant impairment of mDia1-induced actin elongation. We speculate that the C71G mutation causes a pathological loosening, or misfolding, of the PFN1 tertiary structure. The extent of this misfolding in the context of PFN1 C71G may destabilize the ternary complex and hamper normal actin polymerization. In support of this model, the C71G variant is most robustly targeted by the proteasome in cells, indicative of extensive misfolding (Fig. 4) (43). Furthermore, mDia1 dissociates prematurely from elongating actin filaments in the presence of PFN1 C71G, consistent with an ineffective ternary complex (Fig. 2). That steady-state levels of PFN1 C71G are reduced can also contribute to loss-of-function phenotypes (9, 13), although these phenotypes are often masked by coexpression of endogenous PFN1 WT as in the case of our IMM-01–treated cells (Fig. 2). It is possible that as the efficiency of the quality control pathways decline with age (43), PFN1 C71G and other ALS-PFN1 variants may exhibit a gain of toxicity related to pathological protein aggregation (Fig. 4H) (46, 47).
The finding that ALS-linked mutations affect formin-induced actin polymerization provides insight into the role of PFN1 in neurodegeneration. The actin cytoskeletal network is a tightly regulated, complex system that requires functional PFN1 (14). In fact, PFN1 appears to maintain a homeostatic balance among different actin networks by controlling the accessibility of G-actin and directly influencing the activity of other actin-binding proteins (39, 48). Formin-regulated actin polymerization is important for cell division, motility, and mechanotransduction (19). Dysregulation of these functions, through both loss- and gain-of-function mechanisms, is expected to affect the development and maintenance of motor neurons, which degenerate during ALS disease progression (49, 50). Intriguingly, PFN1 is highly expressed in microglia (51), which require diaphanous proteins for complement receptor-mediated phagocytosis (52). Given that microglia contribute to motor neuron degeneration in ALS (53), altered interactions between mutant PFN1 and diaphanous formins in microglia may also be relevant to PFN1-mediated ALS pathogenesis. It will be interesting to examine the effects of ALS-linked PFN1 variants in vivo and to dissect their loss and gain of functions within the different cell types that drive disease pathogenesis.
Materials and Methods
Generation and Culturing of V5-PFN1 Stable HeLa Lines.
Standard molecular biology techniques were used to clone N-terminal V5, V5-WT-PFN1, or V5-ALS-PFN1 constructs (3) into the pLenti-CMV-TO-Puro Destination vector (Addgene, 17293). V5-PFN1 and Tet-R (Addgene, 17492) lentiviral particles (∼106 titer) were generated using human embryonic kidney 293T cells and delivered to HeLa cells as described previously to create stable doxycycline-inducible V5-PFN1 expressing HeLa cell lines (54). Briefly, HeLa Tet-R cells were generated following lentiviral transduction of Tet-R virus and selection with 1.5 µg/mL blasticidin (Cellgro 30-100-RB). HeLa Tet-R stable lines were then transduced with V5-PFN1 viral particles followed by puromycin (Thermo Fisher Scientific, A1113803) selection (1 µg/mL). Stable cell lines were maintained in Dulbecco’s Modified Eagle Medium (Invitrogen, 10213021) supplemented with 10% (vol/vol) fetal bovine serum (tetracycline free, Sigma-Aldrich, F-6178 or F2442), 1% (wt/vol) penicillin and streptomycin (Invitrogen, 15140122), and selection drugs as indicated above under standard culture conditions (37 °C, 5% CO2/95% air). For induction, fresh doxycycline (1 µg/mL, Sigma-Aldrich D9891-1G) was added to the growth media for 24 to48 h, as indicated. Methods are continued in the SI Appendix.
Supplementary Material
Acknowledgments
We are thankful to Drs. Scott Shaffer, John Leszyk, and Roshanak Aslebagh at the Mass Spectrometry Facility at the University of Massachusetts Medical School (UMMS) for help with the proteomics studies; Dr. Robert Brown and Diane McKenna-Yasek for providing human lymphoblast cell lines; Guillaume Romet-Lemonne (Université de Paris) for help in designing and interpreting data from the microfluidics studies; and Drs. Yen-Chen Lin, Zuoshang Xu and Desiree Baron, and other members of the UMMS community for help with experiments and useful discussions. We are grateful to the following funding sources that supported this research: US NIH/National Institute on Neurological Disorders and Stroke R01 NS090352 (D.A.B.); the European Research Council (Grant StG-679116 to A.J.); and US NIH/National Institute of General Medical Sciences R01 GM139316 (F.M.) and R01 GM137529 (F.M. and D.A.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2024605118/-/DCSupplemental.
Data Availability
MS data have been deposited in the Proteomics Identifications database (https://www.ebi.ac.uk/pride/archive/projects/PXD015602) (21).
References
- 1.Brown R. H., Al-Chalabi A., Amyotrophic lateral sclerosis. N. Engl. J. Med. 377, 162–172 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Bartoletti M., et al., Phenotypic suppression of ALS/FTD-associated neurodegeneration highlights mechanisms of dysfunction. J. Neurosci. 39, 8217–8224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu C. H., et al., Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature 488, 499–503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith B. N., et al., Novel mutations support a role for profilin 1 in the pathogenesis of ALS. Neurobiol. Aging 36, 1602.e17–27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y., et al., PFN1 mutations are rare in Han Chinese populations with amyotrophic lateral sclerosis. Neurobiol. Aging 34, 1922.e1–5 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Ingre C., et al., A novel phosphorylation site mutation in profilin 1 revealed in a large screen of US, Nordic, and German amyotrophic lateral sclerosis/frontotemporal dementia cohorts. Neurobiol Aging 34, 1708.e1–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burk K., Pasterkamp R. J., Disrupted neuronal trafficking in amyotrophic lateral sclerosis. Acta Neuropathol. 137, 859–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morfini G. A., et al., Axonal transport defects in neurodegenerative diseases. J. Neurosci. 29, 12776–12786 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boopathy S., et al., Structural basis for mutation-induced destabilization of profilin 1 in ALS. Proc. Natl. Acad. Sci. U.S.A. 112, 7984–7989 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Poggetto E., Bemporad F., Tatini F., Chiti F., Mutations of profilin-1 associated with amyotrophic lateral sclerosis promote aggregation due to structural changes of its native state. ACS Chem. Biol. 10, 2553–2563 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Del Poggetto E., et al., Stability of an aggregation-prone partially folded state of human profilin-1 correlates with aggregation propensity. J. Biol. Chem. 293, 10303–10313 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nekouei M., et al., Changes in biophysical characteristics of PFN1 due to mutation causing amyotrophic lateral sclerosis. Metab. Brain Dis. 33, 1975–1984 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figley M. D., Bieri G., Kolaitis R. M., Taylor J. P., Gitler A. D., Profilin 1 associates with stress granules and ALS-linked mutations alter stress granule dynamics. J. Neurosci. 34, 8083–8097 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollard T. D., Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 8, a018226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Witke W., The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 14, 461–469 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Reinhard M., et al., The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 14, 1583–1589 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campellone K. G., Welch M. D., A nucleator arms race: Cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11, 237–251 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Courtemanche N., Mechanisms of formin-mediated actin assembly and dynamics. Biophys. Rev. 10, 1553–1569 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimmermann D., Kovar D. R., Feeling the force: Formin’s role in mechanotransduction. Curr. Opin. Cell Biol. 56, 130–140 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Alkam D., Feldman E. Z., Singh A., Kiaei M., Profilin1 biology and its mutation, actin(g) in disease. Cell. Mol. Life Sci. 74, 967–981 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt E. J., Bosco D. A., IP-MS of ALS-linked PFN1 variants. Proteomics Identifications database. https://www.ebi.ac.uk/pride/archive/projects/PXD015602. Deposited 26 September 2019.
- 22.Choi H., et al., SAINT: Probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods 8, 70–73 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szklarczyk D., et al., STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thul P. J., et al., A subcellular map of the human proteome. Science 356, eaal3321 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Cao L., et al., Modulation of formin processivity by profilin and mechanical tension. eLife 7, e34176 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jégou A., Carlier M. F., Romet-Lemonne G., Formin mDia1 senses and generates mechanical forces on actin filaments. Nat. Commun. 4, 1883 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Meyners C., Mertens M., Wessig P., Meyer-Almes F. J., A fluorescence-lifetime-based binding assay for class IIa histone deacetylases. Chemistry 23, 3107–3116 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Berezin M. Y., Achilefu S., Fluorescence lifetime measurements and biological imaging. Chem. Rev. 110, 2641–2684 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prinz H., Hill coefficients, dose-response curves and allosteric mechanisms. J. Chem. Biol. 3, 37–44 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petrella E. C., Machesky L. M., Kaiser D. A., Pollard T. D., Structural requirements and thermodynamics of the interaction of proline peptides with profilin. Biochemistry 35, 16535–16543 (1996). [DOI] [PubMed] [Google Scholar]
- 31.Neidt E. M., Scott B. J., Kovar D. R., Formin differentially utilizes profilin isoforms to rapidly assemble actin filaments. J. Biol. Chem. 284, 673–684 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Jégou A., et al., Individual actin filaments in a microfluidic flow reveal the mechanism of ATP hydrolysis and give insight into the properties of profilin. PLoS Biol. 9, e1001161 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovar D. R., Harris E. S., Mahaffy R., Higgs H. N., Pollard T. D., Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124, 423–435 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Vavylonis D., Kovar D. R., O’Shaughnessy B., Pollard T. D., Model of formin-associated actin filament elongation. Mol. Cell 21, 455–466 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paul A. S., Pollard T. D., The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr. Biol. 18, 9–19 (2008).Corrected in: Curr. Biol. 18, 233 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lash L. L., et al., Small-molecule intramimics of formin autoinhibition: A new strategy to target the cytoskeletal remodeling machinery in cancer cells. Cancer Res. 73, 6793–6803 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Tojkander S., Gateva G., Lappalainen P., Actin stress fibers–Assembly, dynamics and biological roles. J. Cell Sci. 125, 1855–1864 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Watanabe N., Kato T., Fujita A., Ishizaki T., Narumiya S., Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1, 136–143 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Rotty J. D., et al., Profilin-1 serves as a gatekeeper for actin assembly by Arp2/3-dependent and -independent pathways. Dev. Cell 32, 54–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Girvan M., Newman M. E., Community structure in social and biological networks. Proc. Natl. Acad. Sci. U.S.A. 99, 7821–7826 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao X. Q., et al., Dynamic coupling and allosteric networks in the α subunit of heterotrimeric G proteins. J. Biol. Chem. 291, 4742–4753 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferron F., Rebowski G., Lee S. H., Dominguez R., Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J. 26, 4597–4606 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pohl C., Dikic I., Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 366, 818–822 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Courtemanche N., Pollard T. D., Determinants of formin homology 1 (FH1) domain function in actin filament elongation by formins. J. Biol. Chem. 287, 7812–7820 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Funk J., et al., Profilin and formin constitute a pacemaker system for robust actin filament growth. eLife 8, e50963 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fil D., et al., Mutant profilin1 transgenic mice recapitulate cardinal features of motor neuron disease. Hum. Mol. Genet. 26, 686–701 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang C., et al., Mutant PFN1 causes ALS phenotypes and progressive motor neuron degeneration in mice by a gain of toxicity. Proc. Natl. Acad. Sci. U.S.A. 113, E6209–E6218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suarez C., et al., Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Dev. Cell 32, 43–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kweon J. H., Kim S., Lee S. B., The cellular basis of dendrite pathology in neurodegenerative diseases. BMB Rep. 50, 5–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ragagnin A. M. G., Shadfar S., Vidal M., Jamali M. S., Atkin J. D., Motor neuron susceptibility in ALS/FTD. Front. Neurosci. 13, 532 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y., et al., An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Colucci-Guyon E., et al., A role for mammalian diaphanous-related formins in complement receptor (CR3)-mediated phagocytosis in macrophages. Curr. Biol. 15, 2007–2012 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Ilieva H., Polymenidou M., Cleveland D. W., Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J. Cell Biol. 187, 761–772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baron D. M., et al., Quantitative proteomics identifies proteins that resist translational repression and become dysregulated in ALS-FUS. Hum. Mol. Genet. 28, 2143–2160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MS data have been deposited in the Proteomics Identifications database (https://www.ebi.ac.uk/pride/archive/projects/PXD015602) (21).