Significance
ATR is a central molecule involved in the DNA replication stress response and repair that ensures genome stability. Whether chromatin modifiers or chromatin modifications also regulate ATR activation, however, is unclear. We conclude that SETD2-mediated H3K14me3 recruits the RPA complex to chromatin and thus promotes ATR activation during conditions of replication stress. Using a series of biological and molecular approaches, we carefully delineated this new SETD2-H3K14me3-RPA-ATR axis that promotes ATR activation and improves cancer cell survival in response to DNA replication stress. Our data open up future opportunities to generate cancer therapeutics strategies based on this pathway.
Keywords: replication stress, ATR activation, RPA, SETD2, H3K14 trimethylation
Abstract
Ataxia telangiectasia and Rad3 related (ATR) activation after replication stress involves a cascade of reactions, including replication protein A (RPA) complex loading onto single-stranded DNA and ATR activator loading onto chromatin. The contribution of histone modifications to ATR activation, however, is unclear. Here, we report that H3K14 trimethylation responds to replication stress by enhancing ATR activation. First, we confirmed that H3K14 monomethylation, dimethylation, and trimethylation all exist in mammalian cells, and that both SUV39H1 and SETD2 methyltransferases can catalyze H3K14 trimethylation in vivo and in vitro. Interestingly, SETD2-mediated H3K14 trimethylation markedly increases in response to replication stress induced with hydroxyurea, a replication stress inducer. Under these conditions, SETD2-mediated H3K14me3 recruited the RPA complex to chromatin via a direct interaction with RPA70. The increase in H3K14me3 levels was abolished, and RPA loading was attenuated when SETD2 was depleted or H3K14 was mutated. Rather, the cells were sensitive to replication stress such that the replication forks failed to restart, and cell-cycle progression was delayed. These findings help us understand how H3K14 trimethylation links replication stress with ATR activation.
Precise DNA replication is essential to accurately transmit genetic information and maintain genomic integrity (1). When cells sense replication stress, the DNA replication forks either progress slowly or stall, which results in DNA damage and chromosome breakage, rearrangement, and mis-segregation (2, 3). Replication stress induces the generation of single-stranded DNA (ssDNA) that is required to activate Ataxia telangiectasia and Rad3-related (ATR) checkpoint signaling (4–7). Replication protein A (RPA), a heterotrimeric complex composed of RPA70, RPA32, and RPA14 (8, 9), directly binds ssDNA through its multiple oligonucleotide/oligosaccharide-binding (OB) fold domains (10). The RPA–ssDNA complex then establishes a platform to recruit numerous factors to the replication forks. This process promotes ATR transautophosphorylation to potentiate ATR activation (6, 11). RPA–ssDNA is also essential for the recruitment of TopBP1 and Ewing tumor associated antigen 1 (ETAA1) to further activate the ATR signaling pathway (12–14). The RPA–ssDNA platform is thus critical for ATR activation during replication stress, but the mechanisms underlying its formation require further investigation.
Numerous studies have shown that chromatin-modifying enzymes and the corresponding histone marks have essential roles in the replication stress response and DNA damage repair (15–17). Enhancer of zeste homolog 2 (EZH2), a component of the Polycomb repressive complex 2 (PRC2), promotes stalled replication fork degradation by recruiting the MUS81 endonuclease through histone H3K27 trimethylation (18). SETD1A-mediated H3K4 methylation protects stalled forks via the nucleosome chaperone activity of Fanconi anemia group D2 (FANCD2) (19). In yeast cells, Set1-catalyzed H3K4me3 is also required to ensure progression through synthesis phase (S phase) during replication stress. Consistently, Set1-depleted cells and H3K4R mutant cells are sensitive to replication stress caused by hydroxyurea (HU), a ribonucleotide reductase inhibitor (20). These data imply that chromatin modifications regulate signal transmission and processing pathways during replication stress. The correlation between chromatin modifications and the initiation of ATR activation, however, is still unclear.
A histone mark, H3K14 trimethylation, was identified to be catalyzed by Regulator of methylation A (RomA) in mammalian cells after Legionella pneumophila infection. Interestingly, RomA is a unique SET-domain–containing methyltransferase in L. pneumophila that does not exist in mammals (21). However, the occurrence of H3K14me3 in human cells under physiological conditions was not shown. More recently, Zhao et al. reported that H3K14me3 does exist in mammalian cells without pathogenic infection, and the histone lysine demethylase (KDM4) family catalyzes H3K14me3 demethylation to H3K14me2 (22). The endogenous methyltransferase that catalyzes H3K14 trimethylation and the physiological function of this mark in mammals remains to be explored.
The SETD2 methyltransferase exhibits H3K36-specific activity (23, 24). SETD2 is the sole known methyltransferase that mediates cellular H3K36 trimethylation, using H3K36me2 as a substrate (25–27). SETD2 also methylates nonhistone substrates, such as microtubules (28) and the transcription factor STAT1 (29), implying that SETD2 might have multiple biological functions outside of H3K36 trimethylation. Indeed, several histone modification enzymes exhibit enzymatic activity on multiple histone residues; for example, a previous study showed that G9a-like protein (GLP) also methylated H4K16 in response to DNA damage except H3K9 methylation (30, 31). In addition, ASH1L catalyzes both H3K4 and H3K36 methylation (32, 33). These observations raise the possibility that SETD2 might also mediate histone modification on other lysine site(s) beyond H3K36.
In this study, we report that a histone mark H3K14me3 promotes ATR activation. In response to replication stress, methyltransferase SETD2 catalyzes H3K14 trimethylation and facilitates the RPA complex loading to chromatin. Loss of SETD2 disrupts the RPA recruitment to the chromatin and dismisses the activation of ATR signaling pathway under stress stimulation. In summary, our study identifies SETD2 as a critical regulator of genomic stability at stalled forks by its enzymatic substrate H3K14me3 after replication stress.
Results
H3K14 Is Methylated in Mammalian Cells.
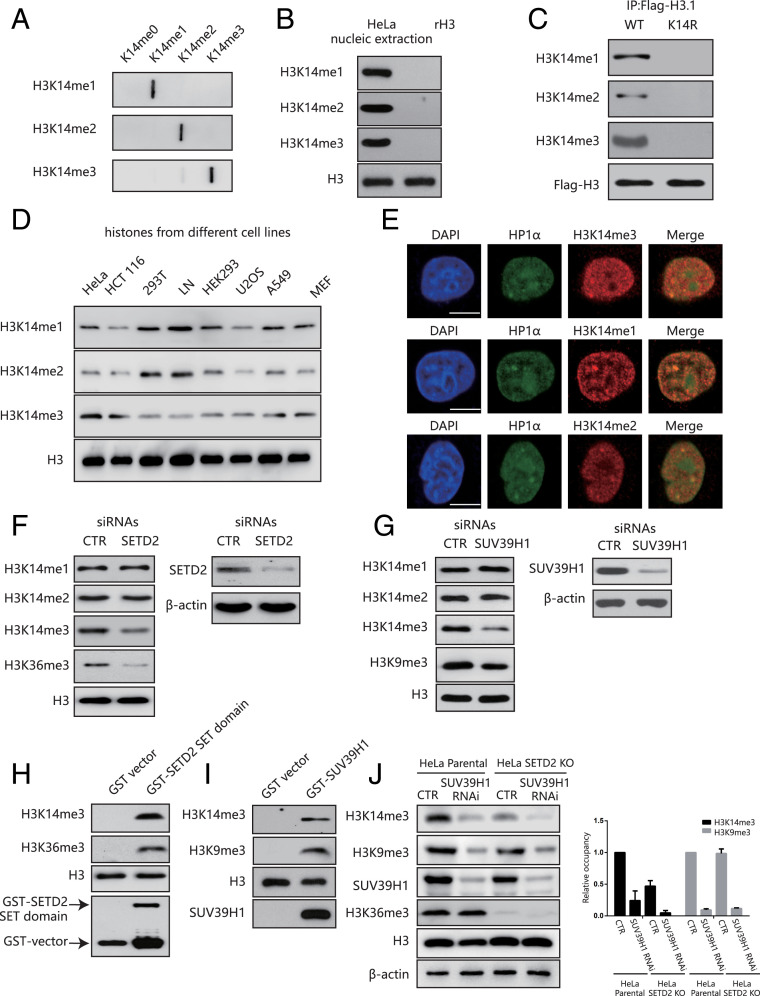
First of all, we generated antibodies that can recognize H3K14 monomethylation, dimethylation, and trimethylation. We validated the specificity of these antibodies by immuno–slot-blot assay and peptide competition assay (Fig. 1A and SI Appendix, Fig. S1 A and B). We detected H3K14 methylation in nuclear extracts of human cervical cancer HeLa cells but not in recombinant unmodified H3 peptides, suggesting that these antibodies can detect endogenous histone H3 in mammalian cells (Fig. 1B). Next, we transfected HeLa cells with a Flag-tagged H3.1 wild type (WT) or a mutant H3.1K14R plasmid, and we analyzed the nuclear immunoprecipitates with anti-Flag or anti-H3K14 methylation antibodies. We detected H3K14 methylation in H3.1 WT transfected cells but not in H3.1K14R transfected cells (Fig. 1C). Importantly, we did not detect any nonspecific bands of these antibodies (SI Appendix, Fig. S1C). Our newly generated antibodies thus recognize H3K14 methylation with high specificity and affinity.
Fig. 1.
H3K14 is methylated in mammals by SETD2 and SUV39H1. (A) A slot-blot assay showing the specificity of the H3K14 methylation antibodies. (B) H3K14 methylation expression in HeLa cells. (C) HeLa cells were transfected with a Flag-H3.1 WT or a Flag-H3.1K14R before anti-Flag immunoprecipitation and Western blotting. (D) The H3K14 methylation expression levels in different cell lines. (E) HeLa cells were labeled with anti-H3K14me1, H3K14me2, H3K14me3 (red), or HP1-α (green) antibodies and analyzed under a confocal microscope. (Scale bar, 10 μm.) (F) HeLa cells were transfected with SETD2 siRNAs or a nonspecific siRNA (CTR). The whole-cell lysates and histones were extracted for Western blotting. (G) HeLa cells were transfected with SUV39H1 siRNAs or a nonspecific siRNA (CTR). The whole-cell lysates and histones were extracted for Western blotting. (H) In vitro methylation assay using a recombinant SETD2 SET domain as the enzyme. Free recombinant histone H3 was used as the substrate. (I) In vitro methylation assay using recombinant SUV39H1 as the enzyme. Free recombinant histone H3 was used as the substrate. (J) HeLa parental cells and SETD2-KO HeLa cells were transfected with or without SUV39H1 siRNAs or a nonspecific siRNA (CTR). The whole-cell lysates were extracted for Western blotting (Left). Quantification of the band density (Right). The band density of HeLa parental cells in “CTR” was normalized to 1. Data are shown as means ± SD (n = 3).
We next detected H3K14 methylation in several human and mouse cell lines and mouse tissues by Western blotting. We confirmed that H3K14 methylation events occurred in all the cell lines and tissues tested, which suggests the universal existence of these H3K14 methylation events in mammals (Fig. 1D and SI Appendix, Fig. S1D). By immunofluorescence analysis, we detected H3K14 methylation signals throughout the nucleus in HeLa cells, with partial overlap with heterochromatin protein 1-α (HP1-α) (Fig. 1E). These data suggest that H3K14 is methylated in mammalian cells and that H3K14 methylation has no preference for euchromatin or heterochromatin.
SETD2 Catalyzes H3K14 Trimethylation In Vivo and In Vitro.
We next explored which histone methyltransferase (HMT) is responsible for H3K14 methylation in vivo. We first transfected HeLa cells with small interfering RNAs (siRNAs) against several well-known HMTs, including SETD2, SUV39H1, SET7/9, MLL, and Dot1l (Fig. 1 F and G and SI Appendix, Fig. S1 J–L). Although we saw no effect on H3K14 monomethylation and dimethylation, H3K14 trimethylation was markedly decreased only in SUV39H1 and SETD2 knockdown cells (Fig. 1 F and G). We obtained similar results in HCT116 colon cancer cells (SI Appendix, Fig. S1 E and H). Next, we transfected HeLa cells with two different siRNAs targeting SETD2, and we obtained consistent results (SI Appendix, Fig. S1F). In addition, we used two different SETD2 knockout cell lines (HeLa SETD2-KO) constructed by CRISPR-Cas9 technology and checked H3K14 methylation levels and got similar results (SI Appendix, Fig. S1G). Considering that SUV39H2 is a paralog of SUV39H1, we also transfected HeLa cells with siRNAs targeting SUV39H2 to see if this molecule regulates H3K14me3. Here, we observed no significant change in H3K14me3 levels after knocking down SUV39H2 compared with HeLa cells transfected with nonspecific siRNAs (SI Appendix, Fig. S1I). In addition, knock down of both SUV39H1 and SUV39H2 did not further decrease H3K14me3 levels when compared to SUV39H1 knock down alone (SI Appendix, Fig. S1I).
We then performed an in vitro methylation assay for HMT activity using a glutathione S-transferase (GST)-purified SET domain fragment of the SETD2 protein or GST-purified SUV39H1, with free recombinant, unmodified histone H3 as the substrate. Consistent with the findings from the siRNA assay, both the SETD2 SET domain and SUV39H1 could catalyze H3K14 trimethylation (Fig. 1 H and I). The H3K14me3 levels correlated with the amount of enzyme in the in vitro assay (SI Appendix, Fig. S1 M and N). Moreover, WT-H3 and K14 mutant H3 (H3K14R) were used as the substrates for in vitro HMTase assay. H3K14 was trimethylated by both the SETD2 SET domain and SUV39H1 in WT-H3 but not in K14R-H3 conditions (SI Appendix, Fig.S1 O and P).
To further delineate the role of SETD2 and SUV39H1 in catalyzing H3K14me3, we transfected HeLa parental cells and SETD2-KO HeLa cells with or without siRNAs against SUV39H1. H3K14me3 was dramatically decreased in both SETD2-KO and SUV39H1 knockdown cells. Interestingly, depletion of both SETD2 and SUV39H1 further blocked H3K14me3 (Fig. 1J). These data suggest that SETD2 and SUV39H1 are the major endogenous methyltransferases that catalyze H3K14 trimethylation.
SETD2-Mediated H3K14me3 Levels Increase in Response to Replication Stress.
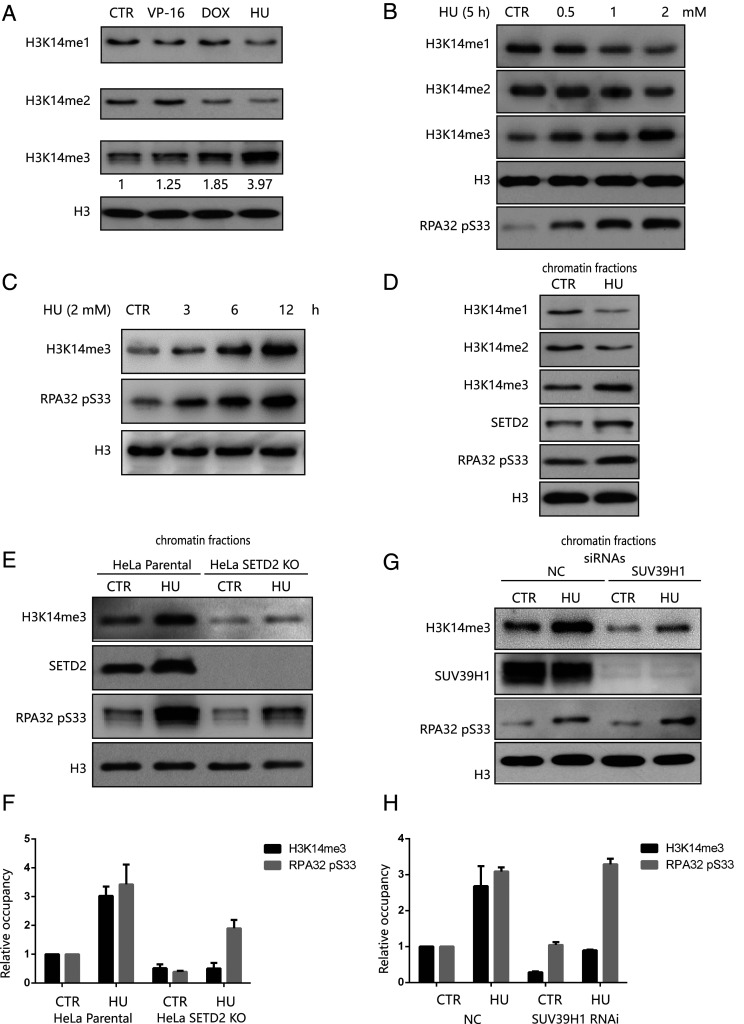
We next wanted to understand the biological functions of H3K14 methylation in human cancer cells. We first treated HeLa cells with different genotoxic (doxorubicin [DOX] or etoposide [VP-16] or HU) agents. We found that the H3K14me3 levels significantly increased following HU but only moderately increase by VP-16 or DOX treatment (Fig. 2A). Correspondingly, H3K14 monomethylation and dimethylation levels decreased after HU treatment (Fig. 2A). To confirm the change of H3K14me3 levels observed with HU, VP-16, and DOX treatment, we treated HeLa cells with ultraviolet (UV) or ionizing radiation (IR) to mimic replication stress and DNA double strand breaks, respectively. We detected increased levels of H3K14me3 after UV irradiation but not IR irradiation (SI Appendix, Fig. S2 A–C). This induction of H3K14 trimethylation and accompanied decrease of H3K14 monomethylation and dimethylation after HU treatment occurred in a time-dependent and dose-dependent manner (Fig. 2 B and C). Finally, immunofluorescent analyses confirmed that H3K14me3 signals were enhanced after HU treatment (SI Appendix, Fig. S2D).
Fig. 2.
SETD2-mediated H3K14me3 levels increase in response to replication stress. (A) Histone extracts from HeLa cells were treated with 40 μM etoposide (VP-16) for 4 h, 1 mM DOX for 8 h, or 6 mM HU for 3 h. “CTR” indicates the cells without any treatment. (B) HeLa cells were treated with the indicated concentrations of HU for 5 h. The chromatin fractions were extracted for Western blotting. “CTR” indicates the cells without any treatment. (C) HeLa cells were treated with 2 mM HU for the indicated times. The chromatin fractions were extracted for Western blotting. “CTR” indicates the cells without any treatment. (D) HeLa cells were treated with 4 mM HU for 5 h. The chromatin fractions were extracted for Western blotting. “CTR” indicates the cells without any treatment. (E) HeLa parental cells and SETD2-KO HeLa cells were treated with or without 4 mM HU treatment for 5 h. The chromatin fractions were extracted for Western blotting. “CTR” indicates the cells without any treatment. (F) A statistical analysis of E was performed by scanning the density of H3K14me3 and RPA32 pS33 band of the Western blots. The band density of HeLa parental cells in “CTR” was normalized to 1. Data are shown as means ± SD (n = 3). (G) HeLa cells transfected with SUV39H1 siRNAs or a nonspecific siRNA (NC) were treated with or without 4 mM HU treatment for 5 h. The chromatin fractions were extracted for Western blotting. “CTR” indicates the cells without any treatment. (H) A statistical analysis of G was performed by scanning the density of H3K14me3 and RPA32 pS33 band of the Western blots. The band density of HeLa cells transfected with “NC” siRNAs in “CTR” was normalized to 1. Data are shown as means ± SD (n = 3).
Interestingly, the levels of chromatin-bound SETD2 increased after HU treatment (Fig. 2D), suggesting a role for SETD2 in responding to replication stress. Conversely, we saw no change in the levels of chromatin-bound SUV39H1 after replication stress (SI Appendix, Fig. S2 E and F). These results suggest that although both SETD2 and SUV39H1 catalyze H3K14 trimethylation, SETD2 seems to mediate the increase of H3K14me3 after HU treatment. Correspondingly, loss of SETD2 disrupted the increase of H3K14 trimethylation levels after HU treatment (Fig. 2 E and F).
We also saw that RPA32-S33 phosphorylation levels were decreased in SETD2-KO HeLa cells compared to the levels in HeLa parental cells (Fig. 2 E and F). By contrast, even though knock down of SUV39H1 decreased endogenous H3K14me3 levels without stress, treating these SUV39H1 knockdown cells with HU robustly induced H3K14me3 compared with parental HeLa cells. RPA32-S33 phosphorylation after HU treatment was also unaffected by SUV39H1 deficiency (Fig. 2 G and H).
To determine whether SUV39H1 and SUV39H2 both contribute to H3K14me3 levels after replication stress, we knocked down either SUV39H1 or both SUV39H1 and SUV39H2 in HeLa cells (SI Appendix, Fig. S2G). ATR activation after HU treatment was unaffected in SUV39H1 deficient or SUV39H1 and SUV39H2 double knockdown cells. The increase in H3K14me3 levels in SUV39H1-deficient cells after HU treatment was also unaffected by further SUV39H2 knock down (SI Appendix, Fig. S2G). These findings imply an important role for SETD2 in activating the ATR signaling pathway during replication stress.
Because DNA replication is a dynamic process in S phase and is challenged by multiple stimuli that can induce replication stress, we checked the distribution of H3K14 trimethylation throughout the cell cycle. HeLa cells were synchronized in G1 phase using double-thymidine treatment and then the cells were released back into the cell cycle. We found that the cells entered S phase at 2 to 4 h, went into G2/M phase at 8 h, and then returned to G1 at 10.5 h upon drug withdrawal (SI Appendix, Fig. S2H). Interestingly, the H3K14me3 levels were high in S phase and comparatively low when the cells went into G2/M phase (SI Appendix, Fig. S2I). The levels of chromatin-bound SETD2 showed a similar pattern as H3K14me3 (SI Appendix, Fig. S2I), while the levels of total SETD2 protein did not vary across the different cell-cycle phases (SI Appendix, Fig. S2J). By contrast, we saw no marked changes in the levels of chromatin-bound SUV39H1 during the cell cycle (SI Appendix, Fig. S2K). These results further support that SETD2, but not SUV39H1, is involved in replication-associated H3K14 trimethylation.
H3K14me3 Interacts with RPA Complex.
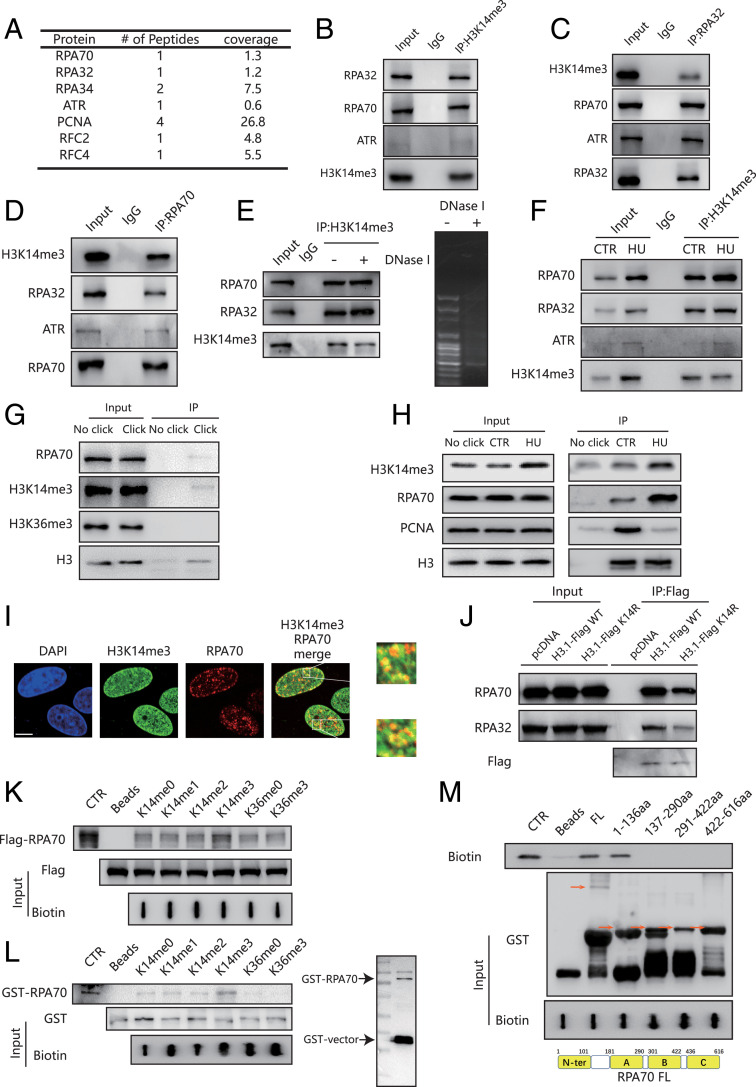
HU treatment affects DNA polymerases and contributes to replication stress (34). Considering that H3K14me3 levels seem to be increased in response to HU treatment, we next investigated the role of H3K14me3 in the replication stress response. To identify the H3K14me3-interacting proteins, we used a histone H3 peptide containing H3K14me3 to precipitate HeLa nuclear extracts treated with or without HU. Potential interacting proteins were identified by mass spectrometry analysis (Dataset S1). Several proteins involved in DNA replication progression or the replication stress response pathways were pulled down after HU treatment (Fig. 3A).
Fig. 3.
H3K14me3 interacts with the RPA complex. (A) HeLa cells were treated with or without 4 mM HU for 5 h, and the nuclear proteins were extracted for pull-down assay with biotin-labeled H3 peptides containing K14me3. The biotin immunoprecipitates were separated by SDS/polyacrylamide gel electrophoresis (SDS/PAGE), and the entire lane was excised from the gel and analyzed by mass spectrometry. The table details some of the proteins identified in HU-treated HeLa cells by mass spectrometry. (B–D) HeLa cells were treated with 4 mM HU for 5 h. The nuclear proteins were extracted and subjected to immunoprecipitation using an anti-H3K14me3 (B), anti-RPA32 (C), or anti-RPA70 (D) antibody. IP, immunoprecipitates. (E) Nuclear proteins were extracted with or without DNase I treatment from HeLa cells treated with 4 mM HU for 5 h and subjected to immunoprecipitation using an anti-H3K14me3 antibody. DNA gel on the right showed the digestion efficiency of DNase I. IP, immunoprecipitates. (F) Nuclear proteins were extracted from HeLa cells treated with or without 4 mM HU for 5 h and subjected to immunoprecipitation using anti-H3K14me3 antibody. “CTR” indicates no HU treatment. IP, immunoprecipitates. (G) HeLa cells were incubated with EdU and HU. Replication fork proteins were isolated by isolation of proteins on nascent DNA (iPOND) and immunoblotted with indicated antibodies. IP, immunoprecipitates. (H) HeLa cells were treated with or without HU and then incubated with EdU. Replication fork proteins were isolated by iPOND and immunoblotted with indicated antibodies. “CTR” indicates no HU treatment. IP, immunoprecipitates. (I) HeLa cells were treated with 4 mM HU for 5 h. The cells were then fixed and stained with an anti-RPA70 antibody (red) and H3K14me3 antibody (green). (Scale bar, 10 μm.) (J) HeLa cells were transfected with pcDNA3.1, Flag-H3.1 WT, and Flag-H3.1K14R for 60 h and then treated with 4 mM HU for 5 h. Mononucleosomes were extracted and subjected to immunoprecipitation using M2 beads. IP, immunoprecipitates. (K) HeLa cells were transfected with Flag-RPA70 for 48 h and then treated with 4 mM HU for 5 h. Nuclear extractions were then pulled down by biotin-labeled H3 peptides containing various forms of histone H3K14 and H3K36, as indicated. “CTR” served as a positive control for Flag-RPA70. (L) Recombinant RPA70 was purified and then pulled down by biotin-labeled H3 peptides containing various forms of histone H3K14 and H3K36, as indicated. “CTR” served as a positive control for GST-RPA70. (M) Recombinant RPA70 fragments were purified, and biotin-labeled H3K14me3 peptides were incubated in vitro in GST pull-down buffer for GST pull-down assay. “CTR” served as a positive control for Biotin-H3K14me3 (Upper). The schematic structure of RPA70. FL, full length. N-terminus, N-ter. A, B, and C are four DNA-binding domains of RPA70 (Lower).
To confirm the possible interactions between these identified proteins and H3K14me3, coimmunoprecipitation (Co-IP) assays were performed and endogenous RPA70, ATR, and RPA32 were immunoprecipitated by H3K14me3 (Fig. 3B). Reciprocally, H3K14me3 was immunoprecipitated by RPA32 and RPA70, separately (Fig. 3 C and D). To exclude the possibility that the RPA complex interacts with H3K14me3 by directly binding with ssDNA, we treated the precipitates with DNase I: we found no difference in the interaction between H3K14me3 and the RPA complex with or without DNase I (Fig. 3E). Although we detected H3K14me3 binding with the RPA complex and ATR under physiological conditions, the interaction was increased after HU treatment (Fig. 3F). Next, we performed an isolation of proteins on nascent DNA assay to investigate whether H3K14me3 is deposited into chromatin immediately surrounding the stressed replicating fork, we found that H3K14me3 was enriched on the stalled replication fork (Fig. 3G). Interestingly, H3K14me3 on the replication fork was increased after HU treatment (Fig. 3H). We confirmed the colocalization of H3K14me3 and RPA70 after HU treatment by immunofluorescence analysis (Fig. 3I).
Next, we overexpressed Flag-H3.1 and H3.1K14R in HeLa cells and performed a mononucleosome Co-IP assay. We found that the H3K14R mutation decreased the interaction between mononucleosome with RPA compared to WT H3 (Fig. 3J). We also overexpressed Flag-RPA70 in HeLa cells and extracted the nuclear fractions after HU treatment for a peptide pull-down assay. RPA70 showed a stronger interaction with peptides containing H3K14me3, compared with the unmodified peptide or peptides containing other modifications (H3K14me1, H3K14me2, or H3K36me3) (Fig. 3K). Moreover, we performed a peptide pull-down assay with purified RPA70 and H3 peptides to see if this interaction is direct. We indeed detected direct binding between RPA70 and H3 peptides (Fig. 3L). Importantly, RPA70 had a higher binding affinity for H3K14me3 peptides than H3 peptides without modifications or with other forms of modifications (Fig. 3L). In addition, we found that the RPA70 N terminus was responsible for this direct interaction (Fig. 3M). These data thus suggest that H3K14me3 may interact with the RPA complex via a direct interaction with RPA70 that occurs in vivo and in vitro.
SEDT2 Deletion Impairs ATR Activation.
Thus far, we have shown that SETD2-mediated H3K14me3 might be associated with the recruitment of RPA complex to chromatin in response to DNA replication stress. We next asked whether SETD2 participates in the ATR signaling pathway. As RPA32, CHK1, and ATR are specific ATR kinase substrates, inhibiting ATR signaling by using ATR inhibitors dramatically decreased RPA32 pS33, CHK1 pS345, and ATR pT1989 levels (SI Appendix, Fig. S3A), as expected. We thus used these markers to monitor ATR activation in our subsequent analyses.
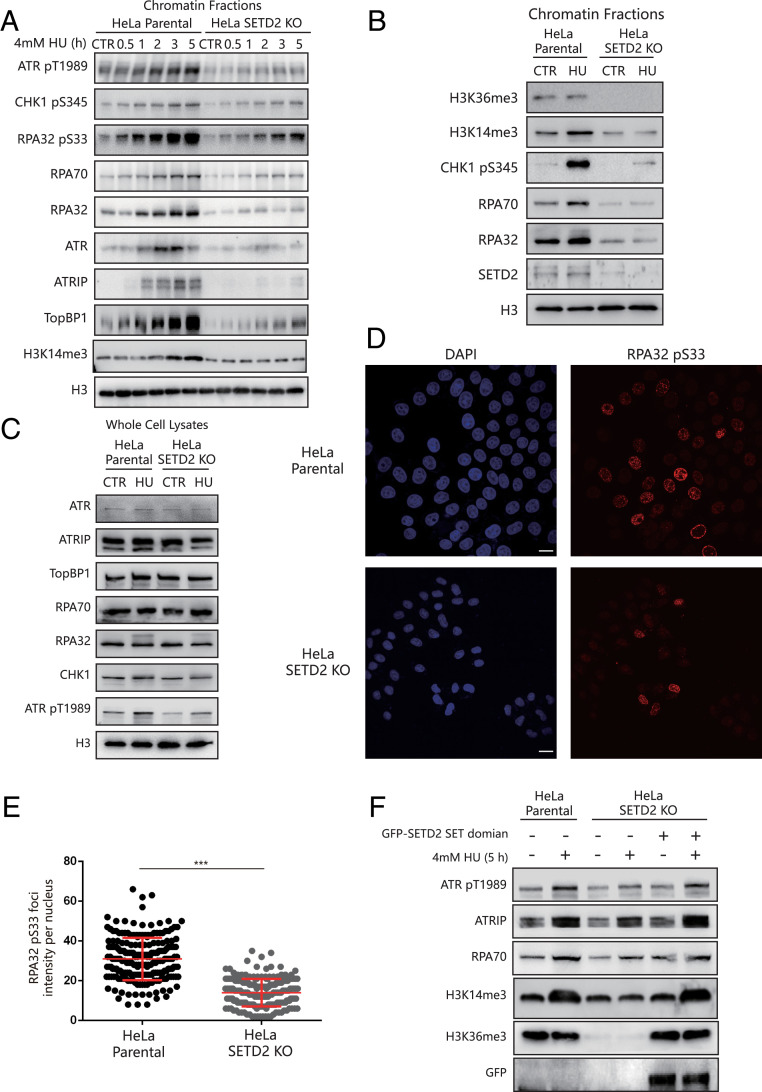
We first treated HeLa parental cells or SETD2-KO HeLa cells with 4 mM HU for 0.5 h up to 5 h and extracted the chromatin fractions for Western blotting. We found that RPA complex recruitment to chromatin was markedly decreased in the SETD2-KO HeLa cells (Fig. 4A). RPA–ssDNA is required to recruit activators, such as TopBP1 and ATRIP, to induce ATR kinase activity (6, 14). Here, we found that the levels of chromatin-bound ATR signaling pathway–related proteins (ATRIP, TopBP1, as well as ATR itself) and the phosphorylation levels of ATR substrates (ATR pT1989, CHK1 pS345, or RPA32 pS33) were notably reduced in SETD2-KO HeLa cells compared to HeLa parental cells (Fig. 4A). We confirmed these phenotypes in two different SETD2-KO HeLa cell lines and siRNA-mediated SETD2 knockdown cells (SI Appendix, Fig. S3 B and C). However, knock down of SUV39H1 alone did not affect ATR signaling (SI Appendix, Fig. S3D). In addition, knock down of SUV39H1 alone or both SUV39H1 and SUV39H2 in SETD2-KO HeLa cells did not show additional effects in ATR signaling, compared with SETD2-KO HeLa cells (SI Appendix, Fig. S3E).
Fig. 4.
SETD2 depletion impairs ATR activation. (A) HeLa parental cells or SETD2-KO HeLa cells were treated with 4 mM HU for the indicated times, and the chromatin fractions were analyzed by Western blotting. “CTR” indicates the cells without HU treatment. (B) HeLa parental cells or SETD2-KO HeLa cells were treated with 4 mM HU for 5 h, and the chromatin fractions were analyzed by Western blotting. “CTR” indicates the cells without HU treatment. (C) HeLa parental cells or SETD2-KO HeLa cells were treated with 4 mM HU for 5 h, and the whole-cell lysates were analyzed by Western blotting. “CTR” indicates the cells without HU treatment. (D) HeLa parental cells or SETD2-KO HeLa cells were treated with 4 mM HU for 5 h. The cells were then fixed and stained with an anti-RPA32 pS33 antibody (red). (Scale bar, 20 μm.) (E) A statistical analysis of D. The RPA32 pS33 foci per nucleus were counted from a minimum of 200 cells. The data represent the means ± SD, ***P < 0.001 (Student’s t test). (F) SETD2-KO HeLa cells were transfected with or without a vector expressing a GFP-tagged SETD2-SET domain for 60 h. Then, HeLa parental cells or SETD2-KO HeLa cells were treated with or without 4 mM HU for an additional 5 h. The chromatin fractions were extracted for Western blotting.
Although SETD2 catalyzes both H3K14me3 and H3K36me3, only H3K14me3 levels were increased in response to HU treatment in HeLa parental cells (Fig. 4B). This finding supports the unique role of SETD2 in mediating H3K14me3 under replication stress. In addition, we did not see any difference in the expression levels of the ATR activators or the RPA proteins between the HeLa parental cells and SETD2-KO HeLa cells (Fig. 4C). These results support that SETD2 promotes ATR activation by facilitating effector recruitment to chromatin.
As RPA32 is a substrate of ATR, and RPA32-S33 phosphorylation is a marker for ATR activation in response to DNA replication stress (35), we performed an immunofluorescence assay to see whether phosphorylated RPA32-S33 foci formation is affected in SETD2-KO HeLa cells. As expected, phosphorylated RPA32-S33 foci formation was reduced in SETD2-KO HeLa cells compared with HeLa parental cells after HU treatment (Fig. 4 D and E). We observed similar results for phosphorylated CHK1-S345 foci formation in SETD2-KO HeLa cells versus HeLa parental cells (SI Appendix, Fig. S4 A and B). Similar results were observed in ATR inhibitor–treated cells, indicating that ATR signaling in SETD2-KO cells was impaired (SI Appendix, Fig. S4 C and D).
Finally, we performed a rescue experiment to investigate whether SETD2 catalytic activity is required for ATR activation. Here, we transfected SETD2-KO HeLa cells with a green fluorescent protein (GFP)-SETD2 SET domain fragment and found that H3K14 trimethylation, ATR phosphorylation, and ATRIP recruitment to the chromatin fraction were restored upon HU treatment (Fig. 4F). Taken together, loss of SETD2 impairs ATR activation in response to replication stress.
H3K14 Trimethylation Promotes ATR Activation.
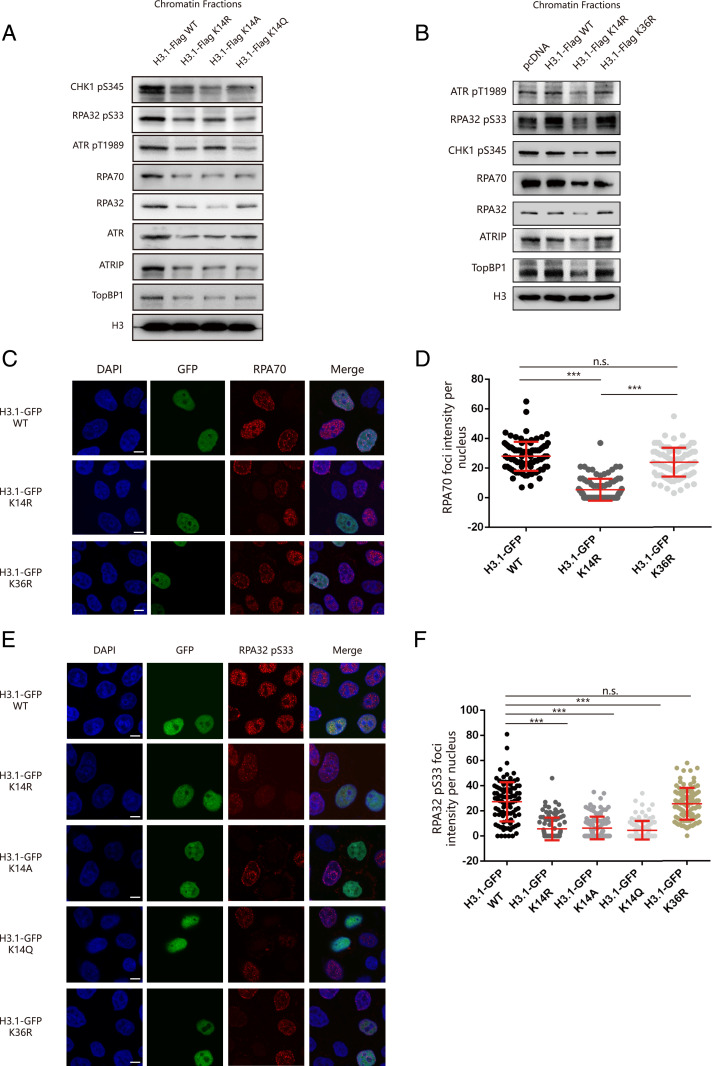
We next investigated whether the role of SETD2 in the ATR signal pathway during replication stress is mediated through H3K14 trimethylation. We constructed a histone H3 mutant by mutating lysine 14 to an alanine (H3K14A) or an arginine (H3K14R) to abolish SETD2-mediated methylation. We also constructed H3K14Q to mimic the acetylation status at this site. We then generated HeLa cell lines stably expressing Flag-tagged histone H3.1 WT or mutant histone H3.1 K14 (K14R, K14A, or K14Q). After HU treatment, we found that the levels of chromatin-bound ATR signaling pathway–related proteins (ATRIP, TopBP1, and RPA complex) and the phosphorylation levels of ATR substrates (ATR pT1989, CHK1 pS345, or RPA32 pS33) were decreased in HeLa cells expressing H3.1-K14 mutant constructs (K14R, K14A, and K14Q) compared to HeLa cells expressing H3.1K14 WT constructs (Fig. 5A).
Fig. 5.
H3K14 trimethylation promotes ATR activation. (A) HeLa cells expressing Flag-H3.1 WT, H3.1K14R, H3.1K14A, or H3.1K14Q were treated with 4 mM HU for 5 h. Chromatin fractions were extracted and analyzed by Western blotting. (B) HeLa cells were transfected with an empty vector or plasmids expressing Flag-H3.1 WT, H3.1K14R, or H3.1K36R. The chromatin fractions were extracted after 4 mM HU treatment for 5 h and analyzed by Western blotting. (C) HeLa cells were transfected with reconstituted GFP-H3.1 WT, H3.1K14R, or H3.1K36R for 48 h and then treated with 4 mM HU for 5 h. The cells were then fixed and stained with an anti-RPA70 antibody (red). (Scale bar, 10 μm.) (D) A statistical analysis of C. The RPA70 foci per nucleus were counted from a minimum of 200 cells. The data represent the means ± SD; n.s., not significant; ***P < 0.001 (Student’s t test). (E) HeLa cells were transfected with reconstituted GFP-H3.1 WT, H3.1K14R, H3.1K14A, H3.1K14Q, or H3.1K36R for 48 h and then treated with 4 mM HU for 5 h. The cells were then fixed and stained with an anti-RPA32 pS33 antibody (red). (Scale bar, 10 μm.) (F) A statistical analysis of E. The RPA32 pS33 foci per nucleus were counted from a minimum of 200 cells. The data represent the means ± SD; n.s., not significant; ***P < 0.001 (Student’s t test).
One of the most important targets of SETD2 is H3K36 trimethylation, which is reportedly crucial for DNA mismatch repair and homologous recombination (36, 37). To determine whether H3K36 methylation affects ATR activation during replication stress, we transfected a Flag-tagged H3.1K36 mutant (K36R) plasmid into HeLa cells to see the effects of H3K36 methylation in ATR activation. Surprisingly, we saw no obvious difference in the levels of chromatin-bound ATR signaling pathway–related proteins or ATR substrate phosphorylation between HeLa cells expressing H3.1K36R compared with cells expressing H3.1 WT or an empty vector (Fig. 5B). We then transfected GFP-tagged histone H3.1 WT and mutant histone H3.1 (K14R or K36R) into HeLa cells and monitored RPA70 foci formation after HU treatment by immunofluorescence. As expected, RPA70 foci formation was distinctly compromised in cells overexpressing GFP-tagged histone H3.1K14R but not in the WT or K36R counterpart cells (Fig. 5 C and D). Strikingly, phosphorylated RPA32 S33 foci formation was markedly decreased in cells expressing either H3.1K14 mutant H3K14R, H3K14A, or H3K14Q compared to H3.1 WT (Fig. 5 E and F). We made similar observations for phosphorylated CHK1 S345 foci formation (SI Appendix, Fig. S4 E and F). These data suggest that defective H3K14me3 restrains RPA70 loading onto chromatin in response to replication stress, which ultimately impairs ATR activation.
SETD2-Induced H3K14me3 Is Not Dispensable for Stalled Forks Restart.
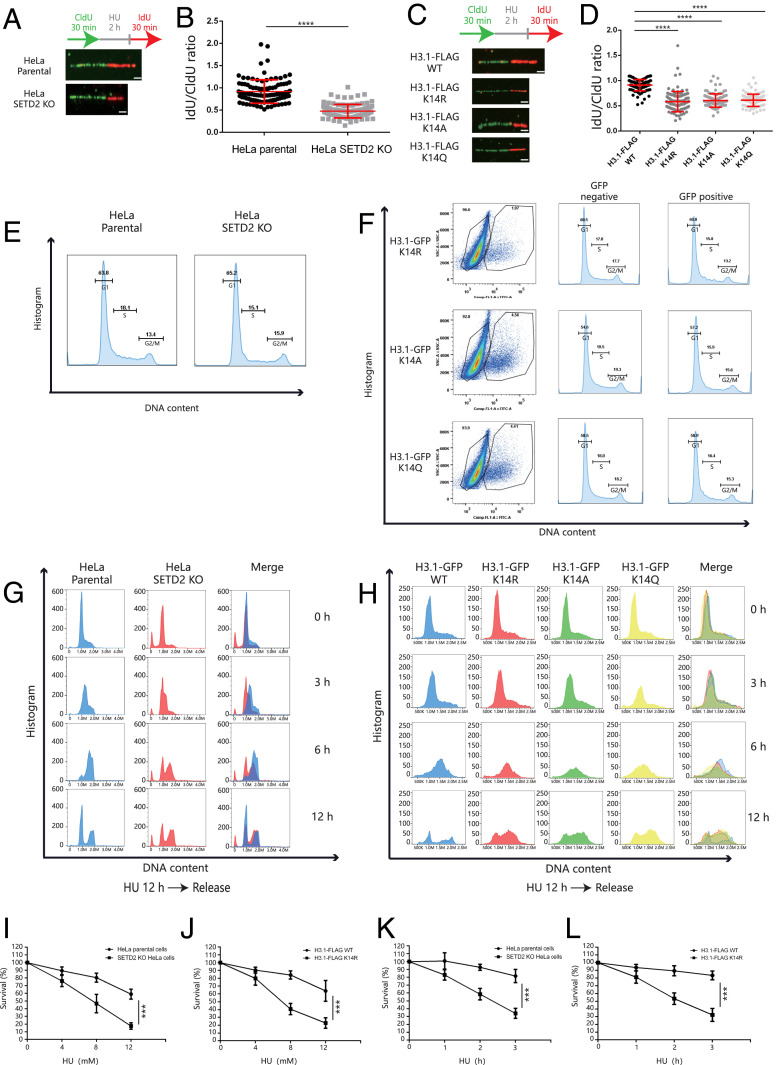
Because ATR activation is required for replication forks to restart (5), we monitored replication fork recovery by single-molecule DNA fiber analysis in HeLa parental cells and SETD2-KO HeLa cells. We found that fork restart was >50% slower in SETD2-KO HeLa cells compared to HeLa parental cells after HU treatment (Fig. 6 A and B). To confirm a role for H3K14me3 in fork restart, we analyzed HeLa cells expressing H3.1 WT or mutant H3.1 (K14R, K14A, or K14Q). Here, we found that cells expressing H3K14 mutants displayed slower recovery rates than cells expressing WT H3 (Fig. 6 C and D). These results support that SETD2-mediated H3K14me3 is important for replication fork restart.
Fig. 6.
SETD2-induced H3K14me3 is required for stalled forks restart. (A) HeLa parental cells or SETD2-KO HeLa cells were pulse labeled with 40 μM CldU for 30 min, treated with 5 mM HU for 2 h, and then released in media containing 100 μM IdU for 30 min. The CldU and IdU track lengths were measured using ImageJ software. (Scale bar, 1 μm.) (B) The IdU/CldU ratio. At least 200 fibers were counted. The means ± SD are shown in the dot plot relative to one representative experiment, and Student’s t test was used for the statistical analysis. ****P < 0.0001. (C) HeLa cells expressing Flag-H3.1 WT, H3.1K14R, H3.1K14A, or H3.1K14Q were pulse labeled with 40 μM CldU for 30 min, treated with 5 mM HU for 2 h, and then released in media containing 100 μM IdU for 30 min. The CldU and IdU track lengths were measured using ImageJ software. (Scale bar, 1 μm.) (D) The IdU/CldU ratio. At least 200 fibers were counted. The means ± SD are shown in the dot plot relative to one representative experiment, and Student’s t test was used for the statistical analysis. ****P < 0.0001. (E) HeLa parental cells and SETD2-KO HeLa cells were harvested for flow cytometry analysis. The histograms show the cell-cycle distribution. (F) HeLa cells were transfected with GFP-H3.1K14R, H3.1K14A, and H3.1K14Q and harvested after 48 h for flow cytometry analysis. The histograms show the cell-cycle distribution of HeLa cells expressing GFP-H3.1K14R, H3.1K14A, and H3.1K14Q (GFP positive) and normal HeLa cells (GFP negative). (G) HeLa parental cells or SETD2-KO HeLa cells were treated with 4 mM HU for 12 h and then released in fresh media for up to 12 h. Cells were harvested at the indicated time for flow cytometry analysis. The histograms show the cell-cycle distributions. (H) HeLa cells expressing GFP-H3.1 WT, H3.1K14R, H3.1K14A, or H3.1K14Q were treated with 4 mM HU for 12 h and then released in fresh media for up to 12 h. Cells were harvested at the indicated time for flow cytometry analysis. The histograms show the cell-cycle distributions. (I) HeLa parental cells or SETD2-KO HeLa cells were treated with indicated doses of HU for 2 h and analyzed by colony formation assay. The cell survival of HeLa parental cells and SETD2-KO HeLa cells in “0” was normalized to 1. The data represent the means ± SD (n = 3). ***P < 0.001 (Student’s t test). “0” indicates the cells without HU treatment. (J) HeLa cells expressing FLAG-H3.1 WT or H3.1K14R were treated with indicated doses of HU for 2 h and analyzed by colony formation assay. The cell survival of Flag-H3.1 WT HeLa cells and H3.1K14R HeLa cells in “0” was normalized to 1. The data represent the means ± SD (n = 3). ***P < 0.001 (Student’s t test). “0” indicates the cells without HU treatment. (K) HeLa parental cells or SETD2-KO HeLa cells were treated with 8 mM HU for indicated time and analyzed by colony formation assay. The cell survival of HeLa parental cells and SETD2-KO HeLa cells in “0” was normalized to 1. The data represent the means ± SD (n = 3). ***P < 0.001 (Student’s t test). “0” indicates the cells without HU treatment. (L) HeLa cells expressing FLAG-H3.1 WT or H3.1K14R were treated with 8 mM HU for indicated time and analyzed by colony formation assay. The cell survival of FLAG-H3.1 WT HeLa cells and H3.1K14R HeLa cells in “0” was normalized to 1. The data represent the means ± SD (n = 3). ***P < 0.001 (Student’s t test). “0” indicates the cells without HU treatment.
Next, we analyzed the cell-cycle distribution of HeLa parental cells and SEDT2-KO HeLa cells; there is a moderate change in cell-cycle progression between HeLa parental cells and SETD2-KO HeLa cells (Fig. 6E). To investigate if H3K14 trimethylation influences the cell cycle, we expressed GFP-tagged K14 mutant H3.1 (K14R, K14A, and K14Q) in HeLa cells and saw no difference in the cell-cycle distribution in HeLa cells expressing GFP-tagged K14R/K14A/K14Q compared with GFP-negative HeLa cells (Fig. 6F). These findings suggest that the H3K14 mutant has minimal effects on cell-cycle progression in the absence of replication stress.
We also synchronized HeLa parental cells and SETD2-KO HeLa cells after HU treatment for 12 h followed by wash out. Here, the HeLa parental cells rapidly resumed DNA synthesis and moved into the S phase of the cell cycle by 6 h and completed cell division by 12 h. By contrast, SETD2-KO HeLa cells showed a much slower recovery, and fewer cells were able to complete the cell division cycle (Fig. 6G). We also analyzed HeLa cells expressing GFP-tagged H3.1 WT or mutant H3.1 (K14R, K14A, and K14Q) to determine the role of H3K14 methylation in cell-cycle progression in the presence of replication stress. By analyzing the GFP-positive cells successfully expressing the corresponding plasmids, we detected impaired cell-cycle progression in mutant H3.1–expressing cells compared with H3.1 WT–expressing cells after HU treatment (Fig. 6H). These data suggest that H3K14 trimethylation is required for cell-cycle progression after replication stress.
SETD2 Is Required for Cells to Tolerate Replication Stress.
In our final set of analyses, we tested whether SETD2-mediated H3K14me3 determines cell survival after HU treatment. We performed a colony formation assay to compare the survival of HeLa parental cells versus SETD2-KO HeLa cells. Under physiological conditions, we found that SETD2-KO HeLa cells grew at a 20% slower rate than HeLa parental cells (SI Appendix, Fig. S5A). Interestingly, after 8 mM HU treatment for 2 h, the SETD2-KO HeLa cells grew at a 60% slower rate than HeLa parental cells (SI Appendix, Fig. S5A). Next, we treated HeLa parental cells and SETD2-KO HeLa cells with either three different doses of HU for the same times or with the same dose of HU for three different exposure times and then performed clonogenic survival analyses. The results of the colony formation assay confirmed that the cell survival rate decreased significantly in SETD2-KO HeLa cells compared with HeLa parental cells (Fig. 6 I and J). HeLa cells stably expressing Flag-H3.1K14R also showed a much lower cell survival rate than cells expressing H3.1 WT after HU treatment (Fig. 6 K and L). In addition, we treated two different SETD2-KO HeLa cell lines with different genotoxic agents to see the sensitivity of SETD2 deficient cells to various genotoxic agents (SI Appendix, Fig. S5B). We also got similar results in SETD2 knockdown HeLa cells by transfection with two different siRNAs (SI Appendix, Fig. S5C). Collectively, these results show that SETD2-mediated H3K14me3 is required for cells to survive during conditions of replication stress.
Discussion
In this study, we report that the methyltransferase SETD2 and SUV39H1 are the primary enzymes responsible for catalyzing the newly discovered histone mark H3K14me3 in mammalian cells. However, SETD2- but not SUV39H1-mediated H3K14me3 serves as a dock to recruit the RPA complex to chromatin and has a critical role in promoting ATR signaling in response to replication stress.
H3K14me3 was recently identified in mammalian cells (21, 22). Consistently, we found that H3K14me3 was endogenously expressed in all the human cancer cell lines and mouse tissues that we tested in this study. A previous report showed that H3K14 methylation occurred only after L. pneumophila infection in THP-1 cells but not in uninfected THP-1 cells (21). This difference with the results of our study might be due to the different cell lines used. For example, we detected low H3K14me3 levels in 293T and LN cells but high levels in HeLa and HCT116 cells (Fig. 1D). In addition, the difference may derive from the different antibodies used. We generated our own H3K14me3 antibody against the H3 peptide containing the first 22 amino acids with K14 trimethylated. Our results are, however, consistent with a previous report by Zhao et al., who also detected H3K14me3 in mammalian cells, and showed that H3K14me3 was associated with active transcription regions in the genome (22). Interestingly, we found that H3K14me3 partially colocalized with HP1-α, a marker of heterochromatin, suggesting a role for H3K14me3 beyond transcriptional regulation.
Prior to our study, the endogenous enzyme for catalyzing H3K14 was unknown. We found that the amino acid sequence before the H3K14 site (TGG-K14) was similar to the sequence preceding the H3K36 site (TGGV-K36). This similarity led us to hypothesize that the methyltransferase that catalyzes H3K36 methylation might also be responsible for catalyzing H3K14 methylation. Using sequence similarities to identify possible methyltransferase seems to be a reasonable approach. For example, a previous study explained that sequence similarities between H3K56 and H3K9 (RK9ST; QK56ST) led to the discovery of H3K56me1 by G9a (38). However, different from H3K36me3 that is merely dependent on SETD2, we found that SETD2 knock down did not abolish H3K14me3 during physiological conditions. As we confirm, other methyltransferases, such as SUV39H1, have a redundant role in maintaining H3K14me3. SUV39H1 and its paralog of SUV39H2 both play essential roles in the catalysis of H3K9me3 at constitutive heterochromatin regions on the genome (39, 40). It is possible, therefore, that these enzymes have redundant roles in maintaining H3K14me3 levels in certain circumstances such as HU treatment. Unfortunately, SUV39H2 knock down did not have additional roles in reducing H3K14me3 levels in SUV39H1 deficient cells, suggesting that SUV39H1 has a unique role in catalyzing H3K14me3.
The premise of an enzymatic reaction is the recruitment of the enzyme to the substrate. For example, after DNA damage, GLP (an enzyme for H3K9me1 and H3K9me2) is recruited to double strand breaks (DSBs) where it catalyzes H4K16 methylation to promote 53BP1 recruitment (31). In this study, we found that SETD2 but not SUV39H1 was recruited to chromatin after HU treatment. Therefore, although both enzymes can trimethylate H3K14, we speculate that SETD2 and SUV39H1 might function in distinct cellular environments or in response to different stimuli. SUV39H1 might contribute to preexisting basal H3K14me3 levels and location overlap with HP1-α. Due to the limited number of enzymes that we tested, we cannot exclude the possibility that other methyltransferases can also catalyze H3K14 methylation.
Notably, SETD2-mediated H3K36me3 is essential for MSH6 recruitment during S phase and for DNA mismatch repair (36); SETD2 thus has a key role in mediating DNA repair pathways. SETD2-mediated H3K36me3 also regulates the expression of RRM2, which is responsible for dNTP synthesis during S phase. As such, SETD2-defficient cells exhibit RRM2 reduction and critical dNTP depletion, replication stress, and S-phase arrest (41). Another study showed that loss of SETD2 function in RCC4 cells induces replication stress through defective nucleosome assembly during S phase, impairing replication fork progression (42). These studies support that SETD2 activity (in terms of H3K36 methylation) is essential for DNA replication.
In this study, however, we show that under conditions of replication stress, SETD2-mediated H3K14me3 has a unique role in directly recruiting the RPA complex to chromatin and promoting ATR activation. SETD2-dependent RPA complex recruitment in response to replication stress is specifically associated with H3K14me3. We provide several lines of evidence to support that H3K14 methylation but not H3K36 methylation has a critical role in response to replication stress: first, H3K14me3 but not H3K36me3 increases after HU treatment; second, in vitro peptide pull-down assay showed no difference in the binding affinities of H3K36me3 or unmethylated H3 peptide for the RPA complex; and third, an H3K36 mutation does not affect the interaction between RPA and histone or RPA32 pS33 foci formation. The underlying mechanism of the different phenotypes between H3K14me3 and H3K36me3 in response to replication stress now needs to be explored. We hypothesize that during HU treatment, SETD2 might be post-translationally modified (by a yet unknown mechanism) to specifically recognize K14 but not K36 on the histone H3 tail.
It is well known that histone methylation provides dock sites for proteins to bind and exert their function (43). Many proteins contain domains that permit lysine methylation recognition, such as the Tudor, PHD, MBT, BRCT2, and Chromo domains (15, 43). We found that H3K14 trimethylation could pull down the RPA70 complex and enhance RPA loading onto chromatin. Interestingly, unmodified H3 or other modified peptides such as H3K14me1, H3K14me2, and H3K36me3 also exhibited a weak interaction with RPA70. This finding is consistent with a previous study that showed that RPA directly binds with free H3 and H4 (44). However, our results here suggest that H3K14me3 promotes the association between histone H3 and the RPA complex. Even though RPA70 has not been reported to contain any known lysine methylation recognition domains, specific binding between H3K14me3 and RPA70 is not surprising. Much evidence shows that lysine methylation recognition domains are not absolutely required for protein interactions. For example, a recent paper suggests that H3K36me3 directly interacts with METTL14, which does not have a methylation recognition domain, and guides m6A RNA modification (45). In addition, H3K56me1 directly associates with PCNA (38) despite PCNA not having any known lysine methylation recognition domains. Furthermore, the PWWP domain of the Lens epithelium-derived growth factor (LEDGF) binds H3K36me3, and the PWWP module reportedly binds DNA without sequence specificity (46). Interestingly, RPA is an ssDNA-binding protein with multiple DNA-binding domains. Whether these DNA-binding domains contribute to the interaction between RPA and H3K14me3 needs to be further explored. Overall, these previous discoveries and our latest data suggest that a lysine methylation recognition domain–independent mechanism exists for protein interactions with methylated histones. Of course, we cannot rule out that RPA70 might contain H3K14me3-specific recognition domains with a yet unknown sequence motif. Although our data supports that the interaction between RPA70 and H3K14me3 is likely direct, we could not exclude the possibility that the role of SETD2 in RPA recruitment may be through other unexplored pathways.
RPA is an essential ssDNA-binding protein that participates in various biological processes including DNA replication and DNA repair (44, 47). We found that only H3K14me3 was increased after HU treatment, suggesting that H3K14me3-dependent RPA recruitment is involved in DNA replication stress but not in DNA repair pathways. The RPA–ssDNA complex forms a platform to help activate ATR kinase. However, several other DNA-binding proteins compete with RPA to bind with ssDNA. For example, BRCA2-mediated Rad51-ssDNA formation dissociates RPA from the DNA at the DSB site to stimulate recombination (48). Besides an interaction with RPA, it is possible that H3K14me3 prevents other proteins from competing for ssDNA to protect the RPA–ssDNA complex. In addition, some proteins, such as SLFN11, disassociate RPA from ssDNA by interacting with the RPA complex (49). H3K14me3 might occupy the RPA70 domains that bind with these proteins and stabilize the RPA complex at the replication forks. It is noteworthy that the RPA complex and ATR activator recruitment to chromatin was not completely abolished in SETD2-deficient or H3K14 mutant cells. This finding implies that SETD2-mediated H3K14me3 is not the only factor required for RPA–ssDNA platform formation and stabilization.
ATR activation in vertebrate cells relies on the TopBP1 and ETAA1 pathways (12–14), both of which require the preexistence of the RPA–ssDNA complex. Even though histone modifications are known to regulate replication fork stability and indirectly affect ATR activation, our study pinpoints that histone methylation has an important role in initiating ATR activation. Interestingly, preclinical data suggest that ATR kinase inhibitors can have promising therapeutic effects against cancer cells. These inhibitors sensitize cancer cells but not normal cells to DNA damage through oncogene-induced replication stress (50). We also found that SETD2-deficient cells are highly sensitive to HU-induced replication stress. SETD2 mutations have been detected in various cancers, including clear cell renal cell carcinoma (51, 52). Cancer cells that harbor SETD2 mutations might thus be particularly sensitive to replication stress, such as HU treatment. Conversely, we need to carefully consider the benefits of ATR inhibitor treatment in patients with SETD2 mutations as these inhibitors might not work as efficiently as they do in patients without SETD2 mutations.
Overall, our findings demonstrate how histone modification regulates RPA–ssDNA platform formation and ATR activation (SI Appendix, Fig. S5D). We reinforce the importance of HMT activity and histone modification in protecting stalled replication forks and maintaining genome stability. Our data expand the role of SETD2 beyond transcription and DNA damage repair. This function for SETD2 in mediating the replication stress response provides us with an angle to exploit its role in chemo-resistance and in the design of cancer therapeutics.
Materials and Methods
H3K14 Methylation Antibody Generation.
The H3K14 methylation antibodies were raised in rabbits against the H3K14 methylation peptides ARTKQTARKSTGG-(monomethyl)K-APRKQLAT, ARTKQTARKSTGG-(di-methyl)K-APRKQLAT, or ARTKQTARKSTGG-(trimethyl)K-APRKQLAT coupled to keyhole limpet hemocyanin. Rabbits were injected with the immunogen three times under a typical boost time schedule. At 1 wk after the third injection, the rabbits were bled for the first time to perform the enzyme linked immunosorbent assay (ELISA) and dot blot assays. The rabbits with high specificity of antisera were boosted again for the last time 1 wk later and exsanguinated 10 d following the final boost. The antisera were captured with protein A resin and purified with antigen peptide conjugated resin. The crossing peptide conjugated resin was used to deplete the cross-reactivity. The purified antibody was tested by dot blot assay and Western blotting.
In Vitro HMTase Assay.
In vitro HMTase assays were performed as previously described (53). In brief, 2 μg substrate was incubated with different enzymes in a methylation reaction buffer (50 mM Tris ⋅ HCl [pH 8.0], 5 mM MgCl2, 4 mM dithiothreitol, 0.5 mM SAM) at 37 °C for 3 h before analysis by Western blotting. Histone H3 and H4 were obtained from New England Biolabs. GST, the GST-SET domain of SETD2, and GST-SUV39H1 were purified from Escherichia coli.
See more detailed materials and methods in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Guo-min Li at Southwestern University, Texas, for kindly sharing the SETD2-KO HeLa cell line with us. We thank Dr. Bing Zhu at the Institute of Biophysics, Chinese Academy of Sciences, for kindly providing the GST-SETD2 (containing the SET domain) plasmid and mono-nucleosome extraction protocol. The manuscript was edited for English language by Dr Jessica Tamanini of ETediting, UK, prior to submission. This work was supported by the National Natural Science Foundation of China (Grants 32090030, 81720108027, and 81530074), the National Key R&D Program of China (2017YFA0503900), the Science and Technology Program of Guangdong Province in China (Grant 2017B030301016), the Shenzhen Municipal Commission of Science and Technology Innovation (Grants JCYJ20170818092450901, JCYJ20200109114214463), and the China Postdoctoral Science Foundation (Grant 2018M643191).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011278118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Masai H., Matsumoto S., You Z., Yoshizawa-Sugata N., Oda M., Eukaryotic chromosome DNA replication: Where, when, and how? Annu. Rev. Biochem. 79, 89–130 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Burrell R. A., et al., Replication stress links structural and numerical cancer chromosomal instability. Nature 494, 492–496 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berti M., Vindigni A., Replication stress: Getting back on track. Nat. Struct. Mol. Biol. 23, 103–109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cimprich K. A., Cortez D., ATR: An essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9, 616–627 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saldivar J. C., Cortez D., Cimprich K. A., The essential kinase ATR: Ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 18, 622–636 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou L., Elledge S. J., Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Menolfi D., Zha S., ATM, DNA-PKcs and ATR: Shaping development through the regulation of the DNA damage responses. Genome Instab. Dis. 1, 47–68 (2020). [Google Scholar]
- 8.Wold M. S., Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Bae S. H., Bae K. H., Kim J. A., Seo Y. S., RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412, 456–461 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Kim C., Paulus B. F., Wold M. S., Interactions of human replication protein A with oligonucleotides. Biochemistry 33, 14197–14206 (1994). [DOI] [PubMed] [Google Scholar]
- 11.Nam E. A., et al., Thr-1989 phosphorylation is a marker of active ataxia telangiectasia-mutated and Rad3-related (ATR) kinase. J. Biol. Chem. 286, 28707–28714 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haahr P., et al., Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat. Cell Biol. 18, 1196–1207 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Bass T. E., et al., ETAA1 acts at stalled replication forks to maintain genome integrity. Nat. Cell Biol. 18, 1185–1195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumagai A., Lee J., Yoo H. Y., Dunphy W. G., TopBP1 activates the ATR-ATRIP complex. Cell 124, 943–955 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Cao L. L., et al., ATM-mediated KDM2A phosphorylation is required for the DNA damage repair. Oncogene 35, 301–313 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Bannister A. J., Kouzarides T., Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sulli G., Di Micco R., d’Adda di Fagagna F., Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nat. Rev. Cancer 12, 709–720 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Rondinelli B., et al., EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat. Cell Biol. 19, 1371–1378 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Higgs M. R., et al., Histone methylation by SETD1A protects nascent DNA through the nucleosome chaperone activity of FANCD2. Mol. Cell 71, 25–41.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faucher D., Wellinger R. J., Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 6, e1001082 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rolando M., et al., Legionella pneumophila effector RomA uniquely modifies host chromatin to repress gene expression and promote intracellular bacterial replication. Cell Host Microbe 13, 395–405 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Zhao B., et al., H3K14me3 genomic distributions and its regulation by KDM4 family demethylases. Cell Res. 28, 1118–1120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao M., et al., Identification of genes expressed in human CD34(+) hematopoietic stem/progenitor cells by expressed sequence tags and efficient full-length cDNA cloning. Proc. Natl. Acad. Sci. U.S.A. 95, 8175–8180 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun X. J., et al., Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J. Biol. Chem. 280, 35261–35271 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Edmunds J. W., Mahadevan L. C., Clayton A. L., Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 27, 406–420 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan W., et al., H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 286, 7983–7989 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner E. J., Carpenter P. B., Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 13, 115–126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park I. Y., et al., Dual chromatin and cytoskeletal remodeling by SETD2. Cell 166, 950–962 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen K., et al., Methyltransferase SETD2-mediated methylation of STAT1 is critical for interferon antiviral activity. Cell 170, 492–506.e14 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Tachibana M., et al., Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 19, 815–826 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu X., et al., GLP-catalyzed H4K16me1 promotes 53BP1 recruitment to permit DNA damage repair and cell survival. Nucleic Acids Res. 47, 10977–10993 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka Y., Katagiri Z., Kawahashi K., Kioussis D., Kitajima S., Trithorax-group protein ASH1 methylates histone H3 lysine 36. Gene 397, 161–168 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Gregory G. D., et al., Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol. Cell. Biol. 27, 8466–8479 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koç A., Wheeler L. J., Mathews C. K., Merrill G. F., Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J. Biol. Chem. 279, 223–230 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Olson E., Nievera C. J., Klimovich V., Fanning E., Wu X., RPA2 is a direct downstream target for ATR to regulate the S-phase checkpoint. J. Biol. Chem. 281, 39517–39533 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Li F., et al., The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell 153, 590–600 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfister S. X., et al., SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 7, 2006–2018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu Y., et al., Histone H3 lysine 56 methylation regulates DNA replication through its interaction with PCNA. Mol. Cell 46, 7–17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peters A. H., et al., Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Lachner M., O’Carroll D., Rea S., Mechtler K., Jenuwein T., Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Pfister S. X., et al., Inhibiting WEE1 selectively kills histone H3K36me3-deficient cancers by dNTP starvation. Cancer Cell 28, 557–568 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanu N., et al., SETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repair. Oncogene 34, 5699–5708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kouzarides T., Chromatin modifications and their function. Cell 128, 693–705 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Liu S., et al., RPA binds histone H3-H4 and functions in DNA replication-coupled nucleosome assembly. Science 355, 415–420 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Huang H., et al., Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 567, 414–419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stec I., Nagl S. B., van Ommen G. J., den Dunnen J. T., The PWWP domain: A potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 473, 1–5 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Maréchal A., Zou L., RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 25, 9–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jensen R. B., Carreira A., Kowalczykowski S. C., Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467, 678–683 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mu Y., et al., SLFN11 inhibits checkpoint maintenance and homologous recombination repair. EMBO Rep. 17, 94–109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fokas E., et al., Targeting ATR in DNA damage response and cancer therapeutics. Cancer Treat. Rev. 40, 109–117 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Dalgliesh G. L., et al., Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duns G., et al., Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 70, 4287–4291 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Yang Q., et al., G9a coordinates with the RPA complex to promote DNA damage repair and cell survival. Proc. Natl. Acad. Sci. U.S.A. 114, E6054–E6063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.