Significance
We custom synthesize a fluorescent ligand and use highly sensitive microscopy methods to show that receptors may be targeted to distinct sites of a cell. We provide microscopic images of the localization of β1‐adrenergic receptors (β1-ARs) and β2‐adrenergic receptors (β2‐ARs), main drivers of the contractility of the heart, in heart muscle cells. We show that β2‐ARs are exclusively targeted to specific structures in these cells, the T-tubules, while β1-ARs are found at the entire cell surface and in T-tubules. Such differential localization may explain their different physiological functions, even though they trigger the same biochemical signal, i.e. an increase in cyclic adenosine monophosphate (cAMP). Thus, receptor function may be determined not only by their biochemical signals, but also by their specific cellular localization.
Keywords: GPCR, β-adrenergic receptors, cardiomyocyte, fluorescence microscopy, fluorescence correlation spectroscopy
Abstract
A key question in receptor signaling is how specificity is realized, particularly when different receptors trigger the same biochemical pathway(s). A notable case is the two β‐adrenergic receptor (β‐AR) subtypes, β1 and β2, in cardiomyocytes. They are both coupled to stimulatory Gs proteins, mediate an increase in cyclic adenosine monophosphate (cAMP), and stimulate cardiac contractility; however, other effects, such as changes in gene transcription leading to cardiac hypertrophy, are prominent only for β1‐AR but not for β2-AR. Here, we employ highly sensitive fluorescence spectroscopy approaches, in combination with a fluorescent β‐AR antagonist, to determine the presence and dynamics of the endogenous receptors on the outer plasma membrane as well as on the T-tubular network of intact adult cardiomyocytes. These techniques allow us to visualize that the β2‐AR is confined to and diffuses within the T-tubular network, as opposed to the β1‐AR, which is found to diffuse both on the outer plasma membrane as well as on the T-tubules. Upon overexpression of the β2‐AR, this compartmentalization is lost, and the receptors are also seen on the cell surface. Such receptor segregation depends on the development of the T-tubular network in adult cardiomyocytes since both the cardiomyoblast cell line H9c2 and the cardiomyocyte-differentiated human-induced pluripotent stem cells express the β2‐AR on the outer plasma membrane. These data support the notion that specific cell surface targeting of receptor subtypes can be the basis for distinct signaling and functional effects.
G protein-coupled receptors (GPCRs) represent the largest group of cell surface receptors and regulate almost any physiological function. The several hundred receptors couple to only a limited number of downstream G proteins and second messenger pathways, and thus, often different receptors trigger apparently identical signaling cascades. However, often distinct GPCRs, although coupled to the same second messenger system, generate rather diverse physiological responses (1).
A prominent example for such diverse signaling is the β1-adrenergic receptors (β1-ARs) and β2-adrenergic receptors (β2-ARs) in cardiomyocytes (CMs), which are both activated by the endogenous catecholamines—adrenaline and noradrenaline—and are the key triggers to increase the myocytes’ contractility. Both receptor subtypes couple to Gs proteins and stimulate the production of the second messenger cyclic adenosine monophosphate (cAMP) (2). However, in addition to the stimulation of contractility, the two subtypes lead to distinct physiological and pathophysiological responses: β1-ARs, but not β2-ARs, stimulate protein kinase A (PKA)-mediated phosphorylation of phospholamban and cardiac contractile proteins (3), change gene expression to induce hypertrophy (4, 5), and promote CM apoptosis (6, 7). Prolonged stimulation of β1-ARs, but much less so of β2-ARs, leads to cardiac remodeling and ultimately, development of heart failure, which appears to be the basis for the beneficial effects of β1-AR antagonists in heart failure (7). In part, these distinct effects may be mediated by the ability of β2-ARs to couple to inhibitory Gi proteins following PKA-mediated phosphorylation of the receptors (8, 9).
However, there is also evidence that spatial differences in signaling may be critically involved in such specific downstream effects. Compartmentalization of cAMP and of receptor-mediated cAMP signals has in fact been suggested for decades and by several lines of evidence, including our own previous work (10–14). This effect was directly visualized in adult murine CMs by means of local cAMP readouts using fluorescence biosensors and local stimulation of the receptors using a scanning probe delivery method (15). These results demonstrated that β2-ARs elicited cAMP signals only when the stimulus was delivered directly to the CM T-tubules (TTs), while β1-AR–stimulated cAMP signals could be elicited over the entire cell surface including the TTs. Further evidence for distinct compartments of β1-AR– vs. β2-AR–triggered cAMP was recently provided by Yang et al. (16) who observed that β2-AR stimulation reduced the β1-AR–stimulated cAMP compartments to nanometer sizes.
So far, it has remained unclear how such a specific targeting of β2-AR signaling to TTs may be brought about. In principle, a downstream compartmentalization of the second messenger may be dependent on other biophysical processes (17) or occur due to compartmentalization of or spatial modulation at any of the upstream signaling steps; these include enzymes degrading cAMP such as phosphodiesterases (18), the effectors producing the cAMP (i.e., adenylyl cyclases), receptor to G-protein coupling (19), and ultimately, receptor localization.
Although mounting evidence has shown that GPCR signaling can be spatially and temporally encoded (20), reports of direct observation of receptor or G-protein partitioning at the single-cell level are scarce (21–23). Addressing this important question has been ultimately hampered by the lack of effective microscopic visualization tools; while effective cAMP biosensors were developed over a decade ago (24) and despite enormous progress in the field of high-resolution microscopy over the last two decades (25–27), visualization of endogenous proteins involved in the GPCR signaling cascade, including receptors themselves, remains challenging. At low expression levels of the order of a few receptors per micrometer squared of cell membrane (28), the intensity contrast that can be expected by conventional imaging (wide field, confocal) of primary cell types is very limited. While total internal reflection fluorescence (TIRF) microscopy has allowed some progress in visualizing endogenous individual receptors labeled by fluorescent ligands on the basolateral membrane of primary cell types (29, 30), adult CMs and their intracellular network of T-tubular membranes are not amenable to TIRF imaging.
We, therefore, exploited the possibility of measuring both receptor presence and their dynamics in portions of the CM membranes illuminated by the microscope excitation volume (point spread function [PSF]), in a confocal geometry. Here, small signal fluctuations due to fluorescent molecules entering and exiting the microscope PSF can be used to determine their dynamic fingerprint (i.e., their diffusion coefficient) using fluorescence correlation spectroscopy (FCS) approaches (31–33). This technique is sensitive to very low concentrations (in the nanomolar range) (34), allowing it to work at concentrations well below saturation labeling and to capture the minute concentrations of receptors expected at endogenous expression levels.
A nontrivial requirement is that the target of interest can be labeled specifically with a fluorophore. Since the questions of targeting and compartmentalization concern (primarily) endogenous receptors, this precludes the use of labeling by fusions or tags, which have been very successfully used in transfected cells. Antibodies against GPCRs are notoriously difficult to raise and have mostly insufficient specificity and sensitivity. Labeling with receptor ligands requires highly specific binding with low background, which has rarely been achieved (35). A few fluorescent ligands have been developed to date for β‐ARs (36–39), including commercial analogs from Cisbio and Abcam, the latter line being discontinued. The majority of these ligands has not found widespread application. An ideal ligand should combine high photostability with high affinity and low degree of nonspecific binding. Moreover, given the high intrinsic autofluorescence of adult CMs, fluorophores in the red and far-red regions of the spectrum are advantageous.
We, therefore, set out to develop a ligand (termed JE1319), based on the high-affinity inverse agonist carazolol conjugated to Alexa Fluor 647 (Alexa647), together with a confocal-based line scan–FCS approach (32, 40) in order to visualize the presence and localization of endogenous β1‐ and β2‐ARs in adult murine CMs.
Results
To obtain a high-affinity, nonsubtype selective and antagonistic ligand for β1- and β2-ARs, we based it on the high-affinity inverse agonist carazolol (41), which was fused via an extended linker to the photostable red fluorophore, Alexa647. The resulting compound, abbreviated as JE1319 (Fig. 1A), was synthesized and verified as described in detail in Materials and Methods, and SI Appendix, Supplementary Materials and Methods and Fig. S1 A–C show the absorption, excitation, and emission spectra, which match those of Alexa647.
Fig. 1.
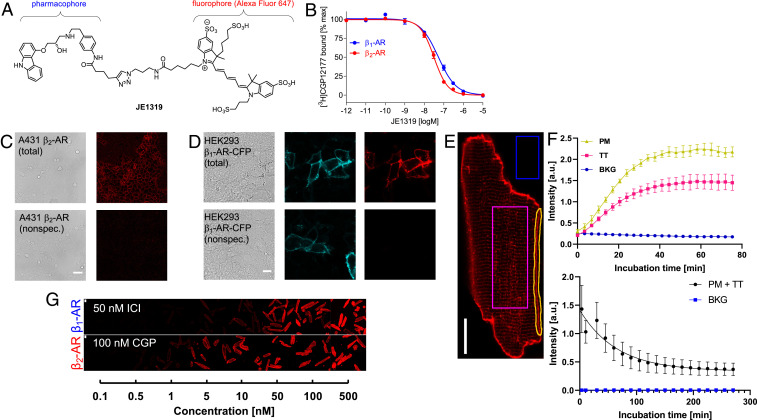
Labeling of β-ARs in live cells with the ligand JE1319. (A) Chemical structure of the fluorescent ligand JE1319, a carazolol–Alexa647 conjugate. (B) Radioligand binding displacement experiments performed on intact HEK293T cells overexpressing either the β1-AR (blue dots) or the β2-AR (red dots), yielding Ki values of 42 ± 8 nM (n = 2 transfections) and 22 ± 3 nM (n = 8 transfections) for β1- and β2-AR, respectively. Error is SEM. (C) Endogenous β2-ARs were stained with 5 nM JE1319 in A431 cells (Upper); nonspecific binding was determined upon displacement with 50 nM ICI 118,551 (Lower). (Scale bar: 50 µm [n = 2 imaging sessions].) (D) HEK293AD cells were transfected with β1-AR-CFP and stained with 5 nM JE1319 (Upper); nonspecific binding was determined in the presence of 100 nM CGP 20712 (Lower). (Scale bar: 10 µm [n = 3 transfections].) (E) Image of a CM stained with 5 nM JE1319. Three regions of interest (ROIs) each were drawn on the membrane (PM), the inside of the cell (TT), and in the background (BKG). (Scale bar: 20 µm [representative of n = 3 CM preparations].) (F, Upper) Time course of ligand binding from the three ROIs, such as those highlighted in E and (F, Lower) time course of ligand dissociation in β2-AR-TG32 CMs. Errors are SD. a.u., arbitrary units. (G) Ligand binding series with different concentrations of JE1319 on adult CMs isolated from β1-AR-TG4 and β2-AR-TG32 mice and cultured for 1 d with either 50 nM ICI118,551 or 100 nM CGP 20712. Image contrast used in G is lower with respect to the one used in E. (Scale bar: 20 µm.)
Competition experiments to determine the affinity of JE1319 for β1- and β2-AR were done with intact human embryonic kidney (HEK) 293T cells transiently transfected with the respective receptor complementary DNA and using the radioligand [3H]CGP 12′177 (Fig. 1B). These experiments yielded affinities (Ki) of 42 ± 8 and 22 ± 3 nM, respectively, indicating that the affinity remained high after coupling to Alexa647, even though there was a substantial loss from the subnanomolar affinity of the parent compound. Comparable values (62 ± 7 nM for β1-AR and 54 ± 9 nM for β2-AR) were obtained by recording fluorescence intensities of HEK293AD cells labeled with increasing concentrations of JE1319, as displayed in SI Appendix, Fig. S1D.
We tested the suitability of JE1319 to visualize β-ARs on human carcinoma A431 cells, a frequently used model for β2-ARs (42), and on transiently transfected HEK293AD cells expressing a cyan fluorescent protein (CFP)-labeled β1-AR. Incubating A431 cells with 5 nM JE1319 for 30 min revealed clear labeling of endogenously expressed cell surface β2-ARs, which were abolished upon displacement by the β2-AR antagonist ICI 118,551 (Fig. 1C). Similarly, HEK293AD cells expressing a CFP-labeled β1-AR showed clear cell surface labeling with JE1319, which was not seen in the presence of the β1-AR antagonist CGP 20712 nor in untransfected HEK293AD cells, indicating very low nonspecific binding of the ligand (Fig. 1D and SI Appendix, Fig. S1E).
We then moved to image ventricular CMs isolated from mice with a cardiac transgene for either the β1-AR or the β2-AR. We used two mouse lines β1-AR-TG4 and β2-AR-TG32, which overexpress the respective β-AR subtype at comparable levels (∼2.5 pmol/mg membrane protein at 3 mo of age) under the control of the CM-specific α-myosin heavy-chain promoter (5). When applied to adult CMs prepared from these mice, the ligand displayed specific staining of both outer plasma membrane (PM) and of T-tubular compartment (Fig. 1E). Kinetic binding experiments analyzing both the cell surface and the T-tubular region showed the expected asymptotic behavior and essentially complete labeling within ∼45 min, while background fluorescence remained very low (Fig. 1F). Dissociation of the ligand took place with a half-life of 30 to 40 min (Fig. 1F). Finally, we studied binding at various ligand concentrations and observed a saturable behavior with optimal binding at 50 nM JE1319, but we clearly recognizable labeling already at 5 nM (Fig. 1G).
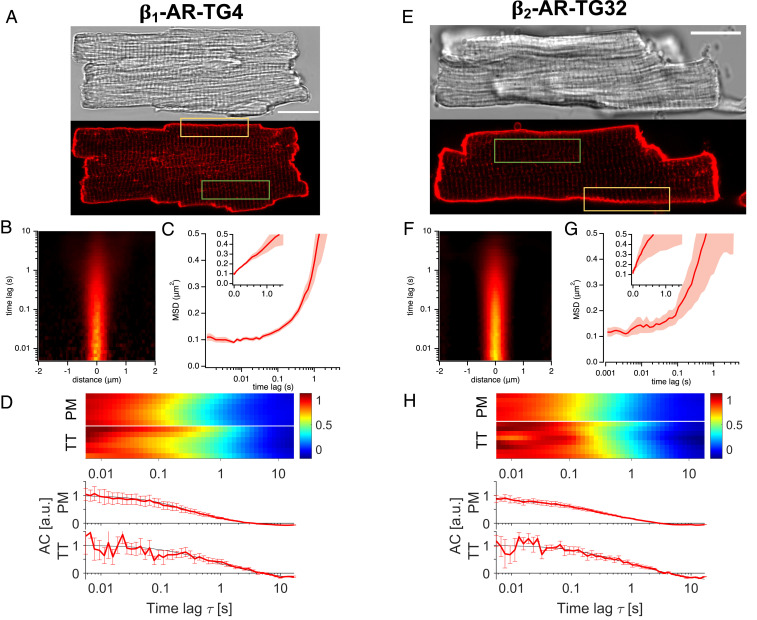
Having determined the ability of our ligand to specifically stain β1- and β2-ARs in live cells, we set out to use it to characterize the localization and dynamics of both β-ARs in adult ventricular CMs, concentrating on the outer PM and the T-tubular membranes, in analogy to the Nikolaev et al. (15) study. Both overexpressed β-AR subtypes were clearly imaged with 5 nM JE1319 in CMs isolated from the transgenic mice overexpressing either the β1- or the β2-ARs (Fig. 2 A and E), both at the outer PM and on the TTs. Given the overexpression of the receptors, working at this ligand concentration yields an experimentally adequate signal to noise ratio both in direct imaging as well as in our fluctuation spectroscopy experiments, as the amplitude of the signal in these methods scales as 1/N, where N is the average number of labeled molecules present in the confocal excitation volume (43).
Fig. 2.
Diffusion of β-ARs in overexpressing CMs along the outer PM and TTs. (A) Microscopic images of an adult CM from a β1-AR-TG4 mouse. (Upper) differential interference contrast (DIC) image. (Lower) Confocal fluorescence image after labeling with 5 nM JE1319 in the presence of 50 nM ICI 118,551. (B) Representative STICS function for β1-AR diffusion along the PM of the CM (yellow rectangle in A). (C) Average MSD curve between 1 ms and 5 s extracted from n = 5 β1-AR overexpressing CMs (two mice). Shading indicates SEM; Inset displays MSD in linear scale. (D) Combined normalized autocorrelation curves (each row represents the normalized autocorrelation curve of one cell) from the PM (yellow rectangle in A) and the TTs (green rectangle in A) of β1-AR overexpressing CMs (n = 6, 2 mice for both compartments). (Lower) In the chart, the corresponding autocorrelation curves (ACs), in arbitrary units (a.u.) together with their fit to two dimensional (2D) (PM) and one dimensional (1D) diffusion (TT) are shown. Error bars indicate SEM. (E) Microscopic images of an adult CM from a β2-AR-TG32 mouse. (Upper) DIC image. (Lower) Confocal fluorescence image after labeling with 5 nM JE1319 in the presence of 100 nM CGP 20712 and (F) corresponding representative STICS function. (G) Average MSD curve between 1 ms and 5 s extracted from n = 5 β2-AR overexpressing CMs (two mice). Shading indicates SEM; Inset displays MSD in linear scale. (H) Combined normalized autocorrelation curve (each row represents the normalized autocorrelation curve of one cell) from the PM (yellow rectangle in E) and the TTs (green rectangle in E) of β2-AR overexpressing CMs (n = 6 for PM, n = 8 for TT, two mice). (Lower) In the chart, the corresponding autocorrelation curves together with their fit to 2D (PM) and 1D diffusion (TT) are shown. Error bars indicate SEM. For TT measurements in D and H, each row represents the normalized average autocorrelation curve of six to seven tubules of one cell. (Scale bars: A and E, 20 µm.)
To measure the abundance and dynamics of these receptors, we used a variant of the general FCS approach, based on performing confocal line scans. This approach is favored by the linear morphology of adult CMs, where the PM has just minor oscillations over length scales of a few micrometers. Conceptually, this is equivalent to conducting in parallel many (of the order of hundreds) single-point FCS experiments (32). The data were visualized in a spatial–temporal representation, known as spatial–temporal image correlation spectroscopy (STICS) function, which provides an intuitive view of the timescales (y axis) and spatial ranges (x axis) of the diffusion process at hand (31, 33, 44) (Fig. 2 B and F), derived from the regions highlighted by the yellow squares in Fig. 2 A and E. The characteristic “plume-like” patterns display the probability of molecular diffusion as a function of space and time and illustrate the characteristic diffusion process for both receptor subtypes.
The representative graph of Fig. 2B can be quantitatively analyzed to extract the molecular mean squared displacement (MSD), a physical quantity that reflects how rapidly a molecule diffuses. The average MSD displayed in Fig. 2C is linear (note the semilogarithmic scale), indicating largely free diffusion of β1-AR along the outer PM of CMs isolated from β1-AR-TG4 mice. Diffusion values and PSF waists for the different conditions are summarized in Table 1.
Table 1.
Diffusion values and time constants (with SEM)
| Location | D STICSPM (µm2/s) | Waist STICSPM (µm) | D FCSPM (µm2/s) | D FCSTT (µm2/s) | τD FCS TT (s) | τD STICS TT (s) |
| β1-AR-TG | 0.081 ± 0.007 (5) | 0.30 ± 0.01 (6) | 0.06 ± 0.01 (6) | 0.08 ± 0.05 (6) | 0.3 ± 0.2 (6) | — |
| β2-AR-TG | 0.14 ± 0.07 (5) | 0.34 ± 0.02 (6) | 0.062 ± 0.003 (6) | 0.05 ± 0.01 (8) | 0.6 ± 0.2 (8) | — |
| β1-AR wt | 0.07 ± 0.01 (17) | 0.29 ± 0.01 (6) | 0.05 ± 0.02 (8) | — | 0.13 ± 0.08 (7)* | 0.13 ± 0.02 (8) |
| β2-AR wt | — | 0.33† ± 0.01 | — | — | 0.09 ± 0.03 (7)* | 0.12 ± 0.01 (8) |
One scan was identified as an outlier by the Chauvenet criterion and excluded from the mean.
From the diameter of fluorescent microspheres.
This type of analysis is particularly advantageous for the PM because a continuous stretch of receptors can be imaged but less for the periodic, interrupted pattern of the TTs. Another way to look at the data—and to visually compare differences between distinct experiments—is to plot the characteristic diffusion time calculated on a pixel-by-pixel basis (SI Appendix, Fig. S2). These can then be combined in a single normalized average fluorescence autocorrelation curve, one for each cell; the color scale reflects the characteristic correlation time τD, from high correlation (i.e., the receptors are still in the PSF at this timescale [red]) to low correlation (i.e., the receptors have diffused out of the PSF by this time [blue]) (SI Appendix, Fig. S2). This approach is convenient, as it allows us to manually pick only those pixels corresponding to the TTs, unlike the spectrogram in Fig. 2 B and F, which is a global analysis over the entire scanned line.
We then performed this analysis both on sections of the PM (yellow box in Fig. 2 A and E) and of the TT network (green box). We imaged those TTs, spaced ∼2 µm apart, having their major axis in the imaging plane. Fig. 2D shows that we observe a consistent dynamic behavior of the β1-AR along the outer PM of the CM, with a characteristic correlation time τD across the PSF of the order of 0.4 s. Assuming 2D diffusion, this value can be converted to a diffusion coefficient D upon using the equation D = ω02/(4τD), where ω0 is the lateral size of the excitation volume. Diffusion coefficient values are indicated in Table 1. Moreover, we can immediately visualize that the dynamic behavior of the receptor in the TTs is analogous to that measured along the PM. In Fig. 2 F–H, we followed the same approach in the analysis of β2-AR overexpressing CMs, except preincubating with CGP 20712 instead of ICI 118,551 to avoid residual binding to endogenous β1-ARs: we observe comparable dynamics (τD ∼ 0.5 s for PM, 0.6 s for TTs, D values are indicated in Table 1) to the β1-AR. Again, for all of the cells examined, we observed a homogeneous behavior of receptor dynamics.
These determinations are further supported by simulations of particle diffusion performed on cylindrical geometries mimicking the TTs. SI Appendix, Fig. S3A displays the fluorescence kymographs arising from simulated line scans across cylinders of a size comparable with the TTs. SI Appendix, Fig. S3B displays the autocorrelation functions (i.e., the profile plots corresponding to the data displayed in Figs. 2 D and H and 3 D and H) for freely diffusing particles on the surface of the TTs. Notably, we obtain a diffusion value (D = 0.107 ± 0.003 µm2/s) for simulated particles diffusing axially as well as radially on the TT (SI Appendix, Fig. S3B), which is in agreement with the experimentally measured one in the overexpressing cells (Table 1). Interestingly, the dynamics of the receptor on the TTs are characterized by two timescales, which appear prominently in reason of the peculiar geometry of this system. A slow diffusion timescale associated with longitudinal motion (along the major axis of the tubule) (SI Appendix, Fig. S3C) and an apparently faster timescale arising from the periodic rotation around the circumference of the tubule (SI Appendix, Fig. S3D).
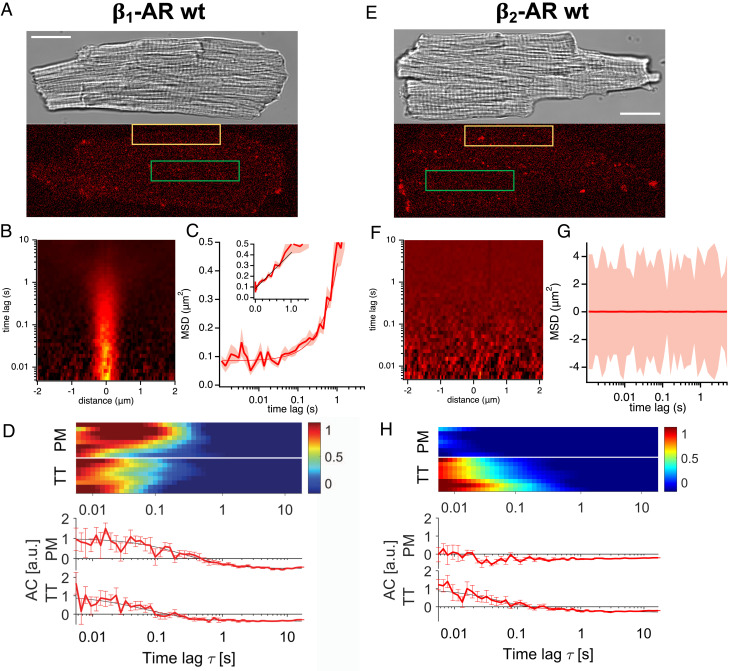
Fig. 3.
Diffusion of β-ARs in wt CM along the outer PM and TTs labeled with 50 nM JE1319. (A) Microscopic images (as in Fig. 2) of an adult CM staining the β1-AR. Receptors are labeled with 50 nM JE1319 after pretreatment with 50 nM ICI 118,551. (B) Example of STICS function for β1-AR on the PM (yellow rectangle in A). (C) Average MSD curve between 1 ms and 5 s extracted from n = 17 CMs (seven mice). Shading is SEM; Inset displays MSD in linear scale. (D) Normalized autocorrelation curves (ACs), in arbitrary units (a.u.), for β1-AR along the PM (eight cells; yellow rectangle in A) and TTs (eight cells; green rectangle in A) of six mice are collected together in one plot separated by a white line. Each row represents the normalized autocorrelation curve of one cell. (Lower) In the chart, corresponding normalized autocorrelation functions with the fits to 2D (PM) and 1D diffusion (TT) are shown. Error bars indicate SEM. (E) Microscopic images of an adult CM staining the β2-AR. Receptors are labeled with 50 nM JE1319 after pretreatment with 100 nM CGP 20712. (F) Example of STICS function for β2-AR on the PM, indicating no detectable diffusion. (G) Average MSD curve between 1 ms and 5 s extracted from n = 23 CMs (five mice). Shading is SEM. (H) Normalized autocorrelation curves for β2-AR along the PM (seven cells; yellow rectangle in E) and TTs (eight cells; green rectangle in E) of six mice are collected together in one plot separated by a white line. Each row represents the normalized autocorrelation curve of one cell. (Lower) In the chart, corresponding normalized autocorrelation functions with the fits to 2D (PM) and 1D diffusion (TT) are shown. Error bars indicate SEM. Contrast in A and E is set to zero. For TT measurements in D and H, each row represents the normalized average autocorrelation curve of six to seven tubules of one cell. (Scale bars: A and E, 20 µm.)
In contrast, when β1-AR/β2-AR–knockout control CMs were labeled with 5 nM JE1319, no visible staining was observed (SI Appendix, Fig. S4A), as well as no diffusion fingerprint (SI Appendix, Fig. S4 B and C).
Having demonstrated our spectroscopy approach on overexpressing systems, we then moved to address the question of the localization and dynamics of the β1- and β2-ARs endogenously expressed in adult wild-type (wt) CMs. As the main question associated with this investigation is whether a differential localization of the β1- and β2-ARs between outer PM and TTs exists, we combined in Fig. 3 the results from our analysis at the PM and in the TTs for each of the two receptors (Fig. 3 A and E). Selective labeling of either the β1- or β2-ARs in wt CMs was again achieved based upon using the selective antagonists ICI 118,551 and CGP 20712, respectively. Here, considering the lower endogenous expressions of the β-ARs, we stained with 50 nM JE1319 and increased the laser power at the TTs by fivefold.
Notably, direct inspection of confocal images of the CM PM acquired even at low speed and high laser power displayed a negligible signal and hardly detectable differences between the two receptors (SI Appendix, Fig. S5). On the other hand, the spectra from the line scans performed along the outer PM are visibly different between the β1-AR, where a profile indicative of diffusion is visible, in 19 of the 30 cells we overall analyzed (Fig. 3B and SI Appendix, Fig. S6A) and the β2-AR (Fig. 3F and SI Appendix, Fig. S6C), where no such profile is visible in any of the cells we measured. The latter observation is similar to the data we observe for β1-AR/β2-AR–knock out (k.o.) CMs (SI Appendix, Fig. S4). For the β1-AR, we recover, from the MSD plot and the autocorrelation curves (Fig. 3D), diffusion rates of 0.07 ± 0.01 µm2/s (D STICS) and 0.05 ± 0.02 µm2/s (D FCS) (Table 1) in agreement with each other and with what was observed in the corresponding overexpressing system.
We then compared the single-cell average correlation profiles for outer PM and TT network in Fig. 3D, which highlights that a correlation profile is visible for both the PM and the TTs. Together with the data in Fig. 3 B and C, these data unambiguously point to our ability to detect the presence and diffusion of β1-AR at the PM and TTs in wt CMs. The correlation profile in Fig. 3 D, Upper shows that we have a characteristic correlation time of the order of 0.4 s, once again indicative of receptor diffusion. The data collected across the TT network remarkably also show a correlation profile with comparable, albeit faster, timescales.
We further collected STICS functions in the TT network of wt CMs (SI Appendix, Fig. S6B) and extracted the corresponding correlation times. Correlation times from FCS and STICS, in agreement within each other, are reported in Table 1.
Strikingly, the β2-AR (Fig. 3E and SI Appendix, Fig. S6C) did not display any diffusion fingerprint on the outer PM of the cells. The STICS function (Fig. 3F) was comparable with that observed in the knockout cells (SI Appendix, Fig. S4), and no molecular MSD can be calculated (Fig. 3G). This is also reflected by the single-cell correlation functions displayed in Fig. 3H. However, the scenario for the β2-AR is sizably different in the TTs (Fig. 3H and SI Appendix, Fig. S6D); here, β2-AR is present, and a diffusional fingerprint can be detected with a τD ∼ 0.1 s, which is consistent to what was observed for the β1-AR in the TTs (Table 1). These data point unambiguously to a differential localization of the β2-AR on the outer PM vs. the TT network in wt adult CMs. Comparable results, although at lower signal to noise ratio, were obtained for the PM when staining the wt CMs with 5 nM JE1319 and are displayed in SI Appendix, Figs. S7 and S8.
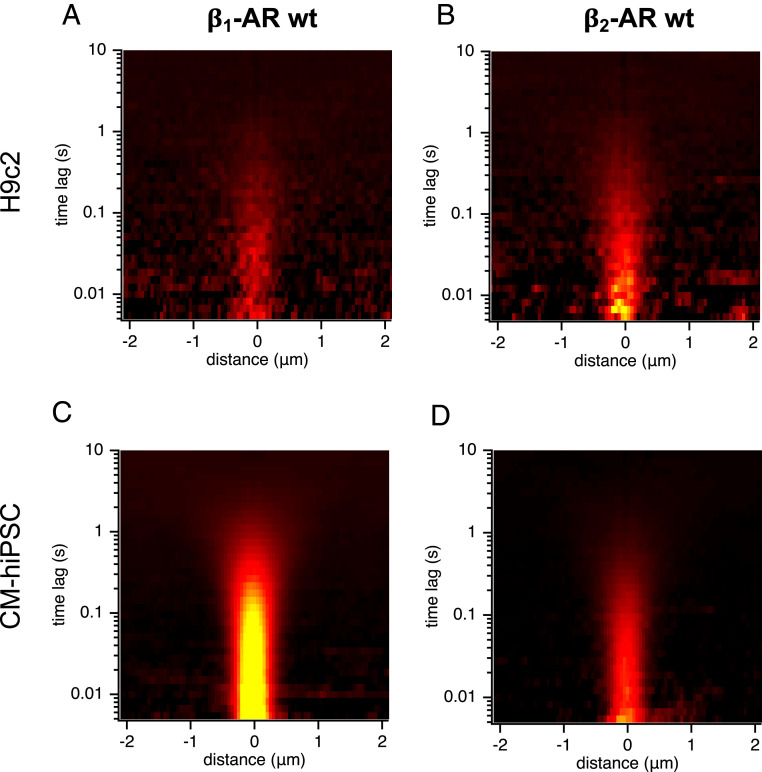
In the light of this stark differential localization, we compared the results obtained in the adult CMs with those obtained in two other cell systems, which have CM-like characteristics but do not possess a T-tubular network. Interestingly enough, in rat cardiomyoblast cells (H9c2), a cell line previously described to mimic the hypertrophic response of primary neonatal CMs (45), diffusion of both β1- and β2-AR was observed along the outer PM of the cells (Fig. 4 A and B and SI Appendix, Fig. S9 A and B). Diffusion coefficients of 0.13 ± 0.01 µm2/s for the β1-AR and 0.05 ± 0.01 µm2/s for the β2-AR are overall in agreement with the expected diffusion rates for these receptors (46).
Fig. 4.
Expression and dynamics of endogenous β-ARs in H9c2 and hiPSC-derived CMs. Average STICS function (n = 4) from line scans collected along the basolateral PM of single H9c2 cells labeled with 50 nM JE1319 and pretreated with (A) 50 nM ICI 118,551 or (B) 100 nM CGP 20712. The same measurements were repeated in matured hiPSC-derived CMs that were cultured for 107 d and again pretreated with (C) 50 nM ICI 118,551 or (D) 100 nM CGP 20712.
The experiment was repeated in human induced pluripotent stem cells (hiPSCs), which were differentiated into CMs followed by a prolonged culture over ∼100 d for further maturation (Materials and Methods). This led to regularly and strongly contracting cardiomyocyte–human induced pluripotent stem cells (CM-hiPSCs) stained positive for typical cardiac markers, such as cardiac troponin T and α-actinin. Even though they are comparable with adult CM in size, they do not possess TTs. Notably, the β2-AR was observed to be present and diffuse at the outer PM (D = 0.12 ± 0.01 µm2/s), together with a robust diffusion fingerprint from the β1-AR (D = 0.1 ± 0.1 µm2/s) (Fig. 4 C and D and SI Appendix, Fig. S9 C and D). These results seem to indicate that the receptor relocates and segregates only upon the formation of the T-tubular network and the specific intracellular environment associated with this specialized compartment of adult CMs.
Discussion
In this work, we addressed the question of the localization and dynamics of endogenous β1-AR and β2-AR in adult CMs. The question of a spatially compartmentalized activity of these receptors has been suggested since the observation, a decade ago, of localized β2-AR–induced cAMP response around the TT of the CM (15). While β-ARs' distribution in adult CM has been explored before by means of viral transduction of the cells with receptor genes coding for an antibody tag (47), the visualization of endogenously expressed β-ARs in wt CMs has to date not been achieved. In this work, we specifically synthesized a fluorescent ligand, displaying high affinity for both the β1-AR and β2-AR and negligible nonspecific binding (Fig. 1 and SI Appendix, Fig. S1), to target endogenous receptors in living cells. We adapted a confocal-based FCS method based on the extraction of spatial–temporal correlations from repeated line scans (40). This allowed us to reveal the presence and dynamics of the labeled receptors, even at the very low endogenous expression levels.
This approach allowed us to make two key biological observations. First, in overexpression systems, β1-ARs and β2-ARs distribute homogeneously across the outer PM and the T-tubular system of the CM. Here, the diffusion rates of the receptors in both compartments are comparable and in line with the expected diffusion rates for a seven-transmembrane receptor, namely ∼0.1 µm2/s (34, 48).
Second, when imaging in wt CM, therefore at endogenous expression level, the β1-AR also displays the localization and dynamic fingerprint observed in the overexpression system (i.e., diffuses both on the TTs and on the PM). On the other hand, strikingly, the endogenous β2-AR appears to be confined to the T-tubular network. While a certain degree of cell to cell variability can be observed in the expression of the β1-AR (only ∼60% of the cells imaged displayed a diffusional fingerprint for this receptor on the outer PM), none of the cells where β2-AR was selectively labeled displayed any trace of diffusion for the receptor on the outer PM.
The most important question that follows from these observations relates to the molecular mechanisms that may allow the β2-AR to segregate differentially from the β1-AR. The β2-AR is known to possess specific C-terminal interactions with cytoskeletal scaffolds (49), which have been shown to localize GPCRs along cytoskeletal stress fibers and the cortical actin network (46, 50). Another possibility might be represented by the dynamic partitioning of the receptors in the region of higher curvature represented by the TTs since recent observations support the notion that GPCRs can dynamically partition in response to varying membrane curvature (22). Our data cannot exclude that in a minority of cells, the β1-AR may also be predominantly localized to the T-tubular network, but a follow-up study with larger statistics would be required to address this specific question.
The observation that receptor compartmentalization is lost in the overexpression phenotypes appears to support a mechanism whereby intracellular protein scaffolds modulate receptor localization. The scaffolds responsible for interacting with the β2-ARs appear therefore to be mainly localized at the level of the T-tubular membranes of the CM and become saturated when the expression level exceeds a certain threshold. The more rapid apparent correlation time observed in those simulated T-tubular membranes when receptors can diffuse only radially (SI Appendix, Fig. S3D), combined with the observation of similar behavior in the experimental datasets from wt CMs, may suggest that receptor confinement may arise due to a suppression of axial diffusion in the TTs, but further experiments will be necessary to address this question. Interestingly, when no T-tubular membranes are present, such as in H9c2 cells and in hiPSCs differentiated to CMs, β2-ARs were observed to diffuse along the cell surface PM of both cell types, suggesting that the progressive formation of the TTs in the adult CM phenotype allows the specific targeting of the diffusing receptors.
Another observation that can be made based on our data is that specific CMs do not appear to express the β1-AR at all (neither on TT nor on PM). In this respect, the notion that GPCRs expression may experience cell to cell variability is not new, including in a cardiac setting; the expression of the muscarinic M2 receptor has been reported to be clearly heterogeneous in isolated CMs (29), and more generally, five different ventricular CM populations, which have distinct transcriptomic profiles, were recently identified (51). In smooth muscle cells and endothelial cells, a substantial expression heterogeneity of GPCRs is observed depending on the anatomical localization of the cells (52). Our results are in general agreement with these earlier observations.
In summary, our results provide a method for the detection of the localization of endogenous β-ARs in live CMs and explain the longstanding observation of compartmentalized cAMP signaling in the TTs of adult CMs by the heterogeneous localization of the receptors themselves. This illustrates that receptor function may be determined not only by the biochemical signals that they elicit but also by their specific cellular localization. Furthermore, not all cells appear to express the same amounts of these receptors. Future studies making use of selective knockouts of PDZ binding scaffolds such as NHERF1, domain swapping of the two β-AR subtypes, or using hiPSCs further differentiated toward adult CMs allowing the formation of a T-tubular network (53) will help address the underlying mechanisms.
Materials and Methods
Fluorescent Ligand Synthesis and Characterization.
The fluorescent ligand JE1319 is based on the pharmacophore carazolol, fused with an Alexa647 derivative as a fluorophore. Detailed chemical synthesis is described in SI Appendix, Supplementary Materials and Methods.
Animal Models and Procedures.
All animal experiments were carried out according to the German Animal Welfare Act considering the guidelines of the NIH and the 2010/63/EU Directive of the European Parliament on the protection of animals used for scientific purposes. Animal experiments were done with approval by the Landesamt für Gesundheit und Soziales (Berlin) under the approval number G 0165/19. The animals had free access to food and water and were kept in individually ventilated cages under a 12-:12-h light/dark regime (light from 6:30 AM to 6:30 PM), a constant 22 °C ± 2 °C temperature, and 55 ± 10% humidity.
The transgenic lines β1-AR-TG4 and β2-AR-TG32 were generated in house and express human β1- or β2-ARs, respectively, under the control of a murine α-myosin heavy-chain promoter (5). β1/β2–AR k.o. mice originally generated and provided by the Kobilka laboratory were obtained from The Jackson Laboratory (stock no. 003810). They are homozygous null for the adrb1 and adrb2 genes (54). For studies on endogenous β-ARs expression levels, wt littermates were used that were generated during heterozygous breeding of the β2-AR-TG32 mouse strain. Further details can be found in SI Appendix, Supplementary Materials and Methods.
CM Isolation.
For the isolation of murine adult ventricular cardiac myocytes, hearts of male and female mice at the age of 8 to 12 wk were used. CM isolations were performed via enzymatic collagen digestion and retrograde perfusion through the aorta using a Langendorff perfusion apparatus as described previously (55). In short, hearts were quickly removed from mice after cervical dislocation and mounted onto a cannula of a custom-built perfusion system. Hearts were initially perfused for 4 min at 3 mL/min with perfusion buffer. In order to destruct the extracellular matrix, perfusion was continued for 8 min using the myocyte (MC) digestion buffer, which corresponds roughly to 5 mg liberase dispase high DH (Roche) enzyme per mouse. As soon as the hearts became limp, they were removed from the perfusion system, and the ventricles were cut into small pieces using scalpels. Digestion was halted by taking up the heart tissue pieces in serum-containing MC stop 1 buffer (SI Appendix, Supplementary Materials and Methods). Further dissociation was achieved by gently pipetting the cell suspension several times using serological plastic pipettes with large openings. After sedimentation, removal of supernatant, and resuspension in stop 2 buffer, cells were passed through a nylon mesh cell strainer (100-µm pore size; Falcon) to remove tissue leftovers. Cells were then stepwise introduced to physiological Ca2+ concentrations (∼1 mM), resuspended in MC plating medium, and seeded on Matrigel-coated µ-slides, which were prepared freshly for each experiment. Further details on the buffers and materials used can be found in SI Appendix, Supplementary Materials and Methods.
Cell Culture.
HEK293AD (BioCat; AD-100-GVO-CB), A431 (ATCC; CRL-1555), and H9c2 (2, 1) (ATCC; CRL-1446) cells were maintained in Dulbecco’s modified Eagle’s medium (PAN-Biotech) supplemented with 10% (vol/vol) fetal bovine serum (FBS) (Sigma-Aldrich), 2 mM l-glutamine (PAN-Biotech), penicillin (100 U/mL; Gibco), and streptomycin (100 μg/mL; Gibco) at 37 °C and 5% CO2. For passaging, HEK293AD and H9c2 (2, 1) cells were detached using trypsin 0.05%/(ethylenedinitrilo)tetraacetic acid (EDTA) 0.02% in phosphate buffered saline (PBS) (PAN-Biotech) or 0.25% trypsin-EDTA (Gibco) in case of A431 cells.
hiPSCs (line BIHi005-A) were received from and validated by the stem cell core facility of the Max Delbrück Center for Molecular Medicine. While pluripotent cultures were grown at 37 °C with 5% CO2 and 5% O2, differentiated cultures were maintained at 5% CO2 and atmospheric (21%) O2. Monolayers of hiPSC were differentiated into CM-hiPSC by modulating Wnt signaling following a small molecule-based cardiac differentiation strategy (56) and enriched for cardiac myocytes by metabolic lactate selection as previously described (57). Further method details for cell culture and materials used can be found in SI Appendix, Supplementary Materials and Methods.
Cell Preparation for Confocal Imaging.
For imaging, cells were seeded in glass-bottom eight-well µ-slides (Ibidi). CMs were plated on eight-well glass-bottom µ-slides (Ibidi) coated with a 1:30 dilution of growth factor reduced Matrigel (Corning) in serum-free MC plating medium. CMs were given time to settle and attach on µ-slides for at least 2 h at 37 °C and 5% CO2. Then, MC plating medium was changed to imaging buffer (pH 7.4, 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [Hepes], 137 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 0.5% bovine serum albumin [BSA]) Transfection of HEK293AD was done using Effectene (Qiagen) according to the manufacturer’s instructions and imaged 2 d after transfection.
Prior to imaging, transfected HEK293ADs, CMs, CM-hiPSCs, and H9c2 cells were preincubated for 40 min to 1 h with either 100 nM CGP 20712 (to image β2-AR) or 50 nM ICI 118,551 (to image β1-AR) diluted in the appropriate imaging buffer. Then, 5 or 50 nM JE1319 as indicated was diluted in imaging buffer and was directly added to the cells (with CGP 20712 or ICI 118,551 as described) for 40 min to 1 h. During the last 15 min of ligand incubation, 0.25× CellMask Green Plasma Membrane Stain (Invitrogen) was added to the cells in case of H9c2 cells and CM-hiPSCs. Untransfected HEK293ADs and A431 cells did not undergo the antagonist preincubation step.
After all incubations, cells were washed three times (except CMs, which were washed only once) using imaging buffer and also imaged in this buffer. CMs were imaged in imaging buffer containing the respective β-AR antagonist as well as 50 µM para-aminoblebbistatin (Optopharma) to inhibit spontaneous CM contractions. Finally, cells were imaged on an SP8 confocal laser scanning microscope (Leica) under physiological conditions (37 °C, 5% CO2, 85% humidity) using a sample incubator (Stage Top Chamber; OKOlab).
Microscopy.
Line scans were acquired on a confocal laser scanning microscope, Leica SP8, with a white-light laser at an acquisition speed of 1,800 Hz with a line size of 256 pixels. All measurements were conducted with an HC PLAP CS2 40× 1.3 numerical aperture (NA) oil immersion objective (Leica). The pixel size was 50 nm, and the number of acquired lines was between 3 × 105 and 6 × 105. The infrared laser-based autofocus of the microscope (Adaptive Focus Control; Leica) was enabled during the acquisition to stabilize the focal position, as described in ref. 40. The ligand was excited at 633 nm with a laser power of 1 or 5%, which corresponded to 0.5 or 3 µW at the sample, respectively. To determine the laser power, a PM100A power meter (Thorlabs) with an S120VC (Thorlabs) photodiode power sensor head was used. Lines were placed either along the PM over the full length or crossing the TTs. Here, the confocal beam is repeatedly scanned at high speed over the same portion of the sample to extract diffusion data from the raw line scans by calculating the autocorrelation function.
Images of cells were acquired on the Leica SP8 with the same objective using either 633 nm (going up to 10% laser power, which corresponds to 6 µW) or 405 nm. Emission was detected on hybrid detectors in photon-counting mode detecting in the range of either 460 to 540 nm (CFP) or 650 to 750 nm (JE1319). Beam waists were determined either by extraction from mean square displacement curves (along the PM) or based on the observation of the profiles of fluorescent microspheres (Tetraspeck; Thermofisher Scientific) as in ref. 40, resulting in a lateral waist of ω0(633 nm) = 0.33 µm and an axial waist of ωz(633 nm) = 1.12 µm (only for β2-AR in wt cells PM and TT).
Acquisition and Analysis of Line Scans.
Two methods were applied and compared to extract the behavior of the diffusing species, namely FCS and STICS.
In the first approach, temporal autocorrelation in each pixel of the line scan was calculated. From the recorded fluorescent intensity time trace arising from the detection volume, a temporal autocorrelation G(τ), which reflects the timescale of the fluorescence intensity fluctuations, can be calculated according to the following formula:
The pointed brackets represent an averaging over all time values t.
By including photophysical processes, the following equations were derived to fit the calculated autocorrelation functions (58):
in which γ is a constant shape factor for the excitation volume, ω0 is the distance in the image plane where the excitation volume intensity decays to the value I(R,Z)/I0 = e−2, D is the diffusion constant, N is the number of particles, τ is the temporal time lag, τT is the lifetime of the photophysical process, T is the fraction of molecules in this process, and G∞ is the limiting value of G(τ) for τ → ∞ (33).
The STICS analysis performs a global spatial and temporal analysis on all pixels and times of the line-scan kymograph, yielding a spatiotemporal correlation function. Such two-dimensional plots, or STICS functions, have two axes and display the process of diffusion in a shape of a “plume,” which broadens in space as a function of time. In STICS, the spatiotemporal image correlations of these line scans are calculated, circumventing the influence of photobleaching (32). The kymographs are corrected for drifts and slow fluctuations using a random number addition detrending within a moving window of ∼250 lines (about 20 s) similar to ref. 40. The spatiotemporal correlation curve is calculated:
where indicates the average over all positions x and scans i (corresponding to time ti) and . ξ represents the spatial lag variable, ti = iT is the time, and τi = iT is the discrete time lag as an integer multiple of the scanning period. Alternatively, by calculating the fast Fourier transform and its complex conjugate, the autocorrelation or STICS function was determined by performing the inverse fast Fourier transform of the product of the two transforms (31).
The spatiotemporal correlation curve for pure diffusion in a one-dimensional line scan can then be fitted to the following fitting model (31, 32):
From horizontal cross-sections of the STICS function, the different time lags of the average MSD can be extracted. From these plots at time τ = 0, the waists of the PSF can be extracted. Further details on the data analysis strategy to extract both the STICS plots as well as the single-point autocorrelation curves can be found in SI Appendix, Supplementary Materials and Methods.
Simulations of Diffusion in T-tubules.
The main purpose of these simulations was the generation of artificial line-scan imaging data with known underlying ground truth. To this end, we integrated the equations of motion and then, at stroboscopic time steps corresponding to the experimental scanning frequency 1,800 Hz = 1/0.556 ms, computed the contribution of every particle to the intensity measured at a specific pixel (with a pixel size of 50 nm) by convolution with a Gaussian PSF (lateral beam waist ω0 = 0.33 µm and axial beam waist ωz = 1.12 µm) (SI Appendix, Fig. S3A). This way, we generate series of pixel graphics (one line per time step, 1,000 lines per image) (SI Appendix, Fig. S3A) suitable for the same analysis as the experimental data with full control about the dynamics of the particles and the geometry they are moving on (composed of prototypical structures such as tubules and planar membranes) as well as the relative orientation and location of the imaging apparatus (SI Appendix, Fig. S3A).
This software has been written in C++ (using Libtiff as well as the gnu scientific library) and is available on GitLab (see Data Availability). Further details on the implementation of these simulations can be found in SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We acknowledge funding from Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) collaborative research center 1423 Project 421152132, Subproject C03 (to M.J.L. and P.A.) and DFG Cluster of Excellence EXC2046 Math+ (to M.J.L. and P.A.). We acknowledge Narasimha Telugu from the Max Delbrück Center stem cell core facility for guidance and advice with the culture and differentiation of hiPSCs and Marlies Grieben for technical assistance.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101119118/-/DCSupplemental.
Data Availability
Code data for the simulation of microscopy datasets have been deposited in GitLab, https://gitlab.com/receptor-signaling-group/movie-generator2. All study data are included in the article and/or SI Appendix.
References
- 1.Maudsley S., Martin B., Luttrell L. M., The origins of diversity and specificity in G protein-coupled receptor signaling. J. Pharmacol. Exp. Ther. 314, 485–494 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiang Y., Kobilka B. K., Myocyte adrenoceptor signaling pathways. Science 300, 1530–1532 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Xiao R. P., Beta-adrenergic signaling in the heart: Dual coupling of the beta2-adrenergic receptor to G(s) and G(i) proteins. Sci. STKE 2001, re15 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Milano C. A., et al., Enhanced myocardial function in transgenic mice overexpressing the β 2-adrenergic receptor. Science 264, 582–586 (1994). [DOI] [PubMed] [Google Scholar]
- 5.Engelhardt S., Hein L., Wiesmann F., Lohse M. J., Progressive hypertrophy and heart failure in β1-adrenergic receptor transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 96, 7059–7064 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Communal C., Singh K., Sawyer D. B., Colucci W. S., Opposing effects of β(1)- and β(2)-adrenergic receptors on cardiac myocyte apoptosis: Role of a pertussis toxin-sensitive G protein. Circulation 100, 2210–2212 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Lohse M. J., Engelhardt S., Eschenhagen T., What is the role of β-adrenergic signaling in heart failure? Circ. Res. 93, 896–906 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Daaka Y., Luttrell L. M., Lefkowitz R. J., Switching of the coupling of the β2-adrenergic receptor to different G proteins by protein kinase A. Nature 390, 88–91 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Xiao R. P., et al., Coupling of β2-adrenoceptor to Gi proteins and its physiological relevance in murine cardiac myocytes. Circ. Res. 84, 43–52 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Buxton I. L., Brunton L. L., Compartments of cyclic AMP and protein kinase in mammalian cardiomyocytes. J. Biol. Chem. 258, 10233–10239 (1983). [PubMed] [Google Scholar]
- 11.Guellich A., Mehel H., Fischmeister R., Cyclic AMP synthesis and hydrolysis in the normal and failing heart. Pflugers Arch. 466, 1163–1175 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Judina A., Gorelik J., Wright P. T., Studying signal compartmentation in adult cardiomyocytes. Biochem. Soc. Trans. 48, 61–70 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaccolo M., Zerio A., Lobo M. J., Subcellular organization of the cAMP signaling pathway. Pharmacol. Rev. 73, 278–309 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikolaev V. O., Bünemann M., Schmitteckert E., Lohse M. J., Engelhardt S., Cyclic AMP imaging in adult cardiac myocytes reveals far-reaching beta1-adrenergic but locally confined beta2-adrenergic receptor-mediated signaling. Circ. Res. 99, 1084–1091 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Nikolaev V. O., et al., Beta2-adrenergic receptor redistribution in heart failure changes cAMP compartmentation. Science 327, 1653–1657 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Yang H. Q., et al., β2-adrenergic stimulation compartmentalizes β1 signaling into nanoscale local domains by targeting the C-terminus of β1-adrenoceptors. Circ. Res. 124, 1350–1359 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bock A., et al., Optical mapping of cAMP signaling at the nanometer scale. Cell 182, 1519–1530.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stangherlin A., Zaccolo M., Phosphodiesterases and subcellular compartmentalized cAMP signaling in the cardiovascular system. Am. J. Physiol. Heart Circ. Physiol. 302, H379–H390 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Sungkaworn T., et al., Single-molecule imaging reveals receptor-G protein interactions at cell surface hot spots. Nature 550, 543–547 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Plouffe B., Thomsen A. R. B., Irannejad R., Emerging role of compartmentalized G protein-coupled receptor signaling in the cardiovascular field. ACS Pharmacol. Transl. Sci. 3, 221–236 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laviv T., et al., Compartmentalization of the GABAB receptor signaling complex is required for presynaptic inhibition at hippocampal synapses. J. Neurosci. 31, 12523–12532 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosholm K. R., et al., Membrane curvature regulates ligand-specific membrane sorting of GPCRs in living cells. Nat. Chem. Biol. 13, 724–729 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Schou K. B., Pedersen L. B., Christensen S. T., Ins and outs of GPCR signaling in primary cilia. EMBO Rep. 16, 1099–1113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikolaev V. O., Bünemann M., Hein L., Hannawacker A., Lohse M. J., Novel single chain cAMP sensors for receptor-induced signal propagation. J. Biol. Chem. 279, 37215–37218 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Bates M., Huang B., Dempsey G. T., Zhuang X., Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science 317, 1749–1753 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rust M. J., Bates M., Zhuang X., Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hell S. W., Wichmann J., Breaking the diffraction resolution limit by stimulated emission: Stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19, 780–782 (1994). [DOI] [PubMed] [Google Scholar]
- 28.Ostrom R. S., Post S. R., Insel P. A., Stoichiometry and compartmentation in G protein-coupled receptor signaling: Implications for therapeutic interventions involving G(s). J. Pharmacol. Exp. Ther. 294, 407–412 (2000). [PubMed] [Google Scholar]
- 29.Nenasheva T. A., et al., Abundance, distribution, mobility and oligomeric state of M2 muscarinic acetylcholine receptors in live cardiac muscle. J. Mol. Cell. Cardiol. 57, 129–136 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoddart L. A., Kilpatrick L. E., Briddon S. J., Hill S. J., Probing the pharmacology of G protein-coupled receptors with fluorescent ligands. Neuropharmacology 98, 48–57 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Di Rienzo C., Gratton E., Beltram F., Cardarelli F., Fast spatiotemporal correlation spectroscopy to determine protein lateral diffusion laws in live cell membranes. Proc. Natl. Acad. Sci. U.S.A. 110, 12307–12312 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ries J., Chiantia S., Schwille P., Accurate determination of membrane dynamics with line-scan FCS. Biophys. J. 96, 1999–2008 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hebert B., Costantino S., Wiseman P. W., Spatiotemporal image correlation spectroscopy (STICS) theory, verification, and application to protein velocity mapping in living CHO cells. Biophys. J. 88, 3601–3614 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scarselli M., et al., Revealing G-protein-coupled receptor oligomerization at the single-molecule level through a nanoscopic lens: Methods, dynamics and biological function. FEBS J. 283, 1197–1217 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Hern J. A., et al., Formation and dissociation of M1 muscarinic receptor dimers seen by total internal reflection fluorescence imaging of single molecules. Proc. Natl. Acad. Sci. U.S.A. 107, 2693–2698 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitronova G. Y., et al., High-affinity functional fluorescent ligands for human β-adrenoceptors. Sci. Rep. 7, 12319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker J. G., Hall I. P., Hill S. J., Pharmacology and direct visualisation of BODIPY-TMR-CGP: A long-acting fluorescent β2-adrenoceptor agonist. Br. J. Pharmacol. 139, 232–242 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daly C. J., et al., Fluorescent ligand binding reveals heterogeneous distribution of adrenoceptors and ‘cannabinoid-like’ receptors in small arteries. Br. J. Pharmacol. 159, 787–796 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hegener O., et al., Dynamics of β2-adrenergic receptor-ligand complexes on living cells. Biochemistry 43, 6190–6199 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Bathe-Peters M., Gmach P., Annibale P., Lohse M. J., Linescan microscopy data to extract diffusion coefficient of a fluorescent species using a commercial confocal microscope. Data Brief 29, 105063 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartsch W., Dietmann K., Leinert H., Sponer G., Cardiac action of carazolol and methypranol in comparison with other β-receptor blockers. Arzneimittelforschung 27, 1022–1026 (1977). [PubMed] [Google Scholar]
- 42.Pippig S., Andexinger S., Lohse M. J., Sequestration and recycling of β2-adrenergic receptors permit receptor resensitization. Mol. Pharmacol. 47, 666–676 (1995). [PubMed] [Google Scholar]
- 43.Elson E. L., Magde D., Fluorescence correlation spectroscopy. I. Conceptual basis and theory. Biopolymers 13, 27 (1974). [DOI] [PubMed] [Google Scholar]
- 44.Zhou C., et al., Motility and segregation of Hsp104-associated protein aggregates in budding yeast. Cell 147, 1186–1196 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watkins S. J., Borthwick G. M., Arthur H. M., The H9C2 cell line and primary neonatal cardiomyocyte cells show similar hypertrophic responses in vitro. In Vitro Cell. Dev. Biol. Anim. 47, 125–131 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Calebiro D., et al., Single-molecule analysis of fluorescently labeled G-protein-coupled receptors reveals complexes with distinct dynamics and organization. Proc. Natl. Acad. Sci. U.S.A. 110, 743–748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nooh M. M., Mancarella S., Bahouth S. W., Novel paradigms governing β1-adrenergic receptor trafficking in primary adult rat cardiac myocytes. Mol. Pharmacol. 94, 862–875 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Işbilir A., et al., Advanced fluorescence microscopy reveals disruption of dynamic CXCR4 dimerization by subpocket-specific inverse agonists. Proc. Natl. Acad. Sci. U.S.A. 117, 29144–29154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hall R. A., β-adrenergic receptors and their interacting proteins. Semin. Cell Dev. Biol. 15, 281–288 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Wheeler D., et al., Regulation of parathyroid hormone type 1 receptor dynamics, traffic, and signaling by the Na+/H+ exchanger regulatory factor-1 in rat osteosarcoma ROS 17/2.8 cells. Mol. Endocrinol. 22, 1163–1170 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Litviňuková M., et al., Cells of the adult human heart. Nature 588, 466–472 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaur H., et al., Single-cell profiling reveals heterogeneity and functional patterning of GPCR expression in the vascular system. Nat. Commun. 8, 15700 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ronaldson-Bouchard K., et al., Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556, 239–243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohrer D. K., Chruscinski A., Schauble E. H., Bernstein D., Kobilka B. K., Cardiovascular and metabolic alterations in mice lacking both beta1- and beta2-adrenergic receptors. J. Biol. Chem. 274, 16701–16708 (1999). [DOI] [PubMed] [Google Scholar]
- 55.O’Connell T., Ni Y., “Isolation of Adult Mouse Cardiac Myocytes from One Heart” (AfCS Procedure Protocol PP00000125, 2002).
- 56.Lian X., et al., Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. U.S.A. 109, E1848–E1857 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tohyama S., et al., Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell 12, 127–137 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Haustein E., Schwille P., Fluorescence correlation spectroscopy: Novel variations of an established technique. Annu. Rev. Biophys. Biomol. Struct. 36, 151–169 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code data for the simulation of microscopy datasets have been deposited in GitLab, https://gitlab.com/receptor-signaling-group/movie-generator2. All study data are included in the article and/or SI Appendix.