Significance
Fosfomycin is an antibiotic used for the treatment of cystitis. Its activity against both gram-positive and gram-negative pathogens has received strong recent interest. The compound is biosynthesized by both pseudomonads and streptomycetes via two different pathways that converge in the last step to form the natural product. This study investigated the biosynthesis of fosfomycin by Pseudomonas and revealed an oxidative decarboxylation activity within this pathway. Structural and sequence analysis of the enzyme identified key residues important for its activity and demonstrated its role in the biosynthesis of other phosphonate natural products. This newly characterized protein family expands the chemical space of catalysis by metallohydrolase superfamily enzymes.
Keywords: fosfomycin, phosphonate, nonheme iron, decarboxylase, metallohydrolase
Abstract
The epoxide-containing phosphonate natural product fosfomycin is a broad-spectrum antibiotic used in the treatment of cystitis. Fosfomycin is produced by both the plant pathogen Pseudomonas syringae and soil-dwelling streptomycetes. While the streptomycete pathway has recently been fully elucidated, the pseudomonad pathway is still mostly elusive. Through a systematic evaluation of heterologous expression of putative biosynthetic enzymes, we identified the central enzyme responsible for completing the biosynthetic pathway in pseudomonads. The missing transformation involves the oxidative decarboxylation of the intermediate 2-phosphonomethylmalate to a new intermediate, 3-oxo-4-phosphonobutanoate, by PsfC. Crystallographic studies reveal that PsfC unexpectedly belongs to a new class of diiron metalloenzymes that are part of the polymerase and histidinol phosphatase superfamily.
The emergence of bacterial strains resistant to at least one currently available antibiotic continues to be a growing global health concern. In the United States alone, reports of cases of antibiotic resistance exceed 2.5 million, resulting in over 35,000 deaths every year (1). Despite concerted efforts from both the public and private sector, major challenges remain in the treatment of infections from resistant pathogens. Recent advances in genomics, bioinformatics, and analytical methods have resulted in a resurgence of interest in bioactive natural products. The antibiotic fosfomycin (trade name Monurol; Fig. 1) is a clinically approved broad-spectrum antibiotic that has gained recent attention because of its activity against multidrug and pandrug-resistant bacterial infections (2–5). Fosfomycin exerts broad antibacterial activity against both gram-positive and gram-negative pathogens (5). The compound belongs to the phosphonic acid class of secondary metabolites, which are defined by the presence of a carbon–phosphorus linkage that confers stability to hydrolytic cleavage. Fosfomycin was originally discovered in 1969 in a collaborative effort between Merck and Compañia Española de Penicilinas y Antibióticos through screening of cultures of the soil bacterium Streptomyces fradiae (6) and then subsequently reisolated in 1986 from Pseudomonas syringae (7).
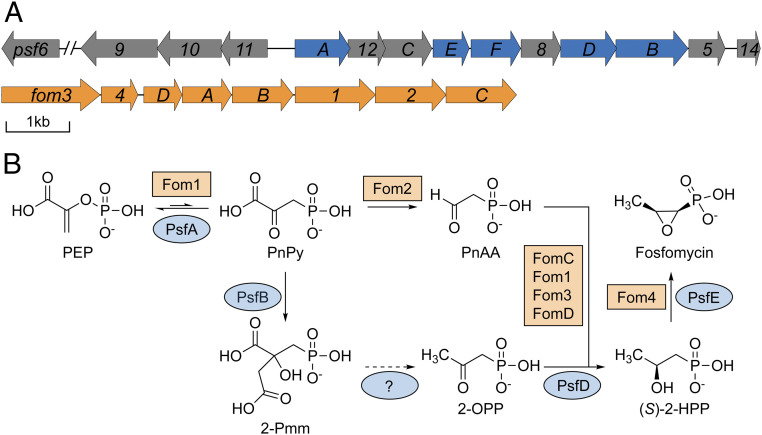
Fig. 1.
Proposed biosynthetic pathways to fosfomycin. (A) BGC responsible for fosfomycin production in P. syringae PB-5123 (Top) and S. fradiae (Bottom, orange). The decarboxylase, Fom2, is not encoded within the Pseudomonas BGC or genome. (B) Previously characterized open reading frames (ORFs) are highlighted in blue in A, and the reactions they catalyze are illustrated in the pathway. ORFs have been renamed from numbers to letters in the apparent order they act in the pathway, based on our experimental data (i.e., psf13 has been renamed psfC to reflect its role after psfAB).
Genome sequencing of the fosfomycin biosynthetic gene clusters (BGCs) revealed that pseudomonads and streptomycetes make fosfomycin through divergent biosynthetic pathways (8–15). The only common steps in the two pathways are the first and last transformations (Fig. 1B). As for all phosphonates for which the biosynthesis has been deciphered, the formation of fosfomycin initiates with the conversion of the central metabolite phosphoenolpyruvate (PEP) to phosphonopyruvate (PnPy) catalyzed by PEP mutase (encoded by pepM) (16). In the streptomycetes pathway, this thermodynamically unfavorable step is driven by the irreversible decarboxylation of PnPy to phosphonoacetaldehyde (17), a common intermediate in many phosphonate biosynthetic pathways. While the genome of the fosfomycin producer P. syringae PB-5123 also contains a pepM, a gene encoding a PnPy decarboxylase is not present. Expression of the 14-gene BGC from P. syringae PB-5123 (Fig. 1A) was sufficient for fosfomycin production in a heterologous host, suggesting that PnPy formation is driven by an alternative strategy (12).
We previously proposed that a homocitrate synthase-like protein (PsfB) catalyzes the condensation of acetate from acetyl coenzyme A with PnPy to form 2-phosphonomethylmalate (2-Pmm; Fig. 1B) (12), a strategy previously observed in the biosynthesis of FR-900098 and pantaphos (18, 19). However, subsequent steps in the fosfomycin biosynthetic strategy in pseudomonads remain elusive. In principle, hydroxylation of 2-Pmm at C-α would facilitate elimination/decarboxylation, followed by tautomerization of the enolate (SI Appendix, Fig. S1). The resulting β-keto acid could decarboxylate to afford 2-oxopropylphosphonate (2-OPP), an intermediate that is on the pathway for fosfomycin and was previously isolated from spent media of P. syringae PB-5123 (20). However, the BGC lacks any identifiable genes encoding enzymes that could catalyze these transformations.
Here, we used a reductionist approach to investigate the fosfomycin biosynthetic route in pseudomonads. Because of the low titer of fosfomycin production in P. syringae PB-5123, studies in this organism are challenging (7). Instead, we engineered the production of the 2-Pmm intermediate in Escherichia coli. Using this platform, we systematically and combinatorially expressed the remaining genes in the Pseudomonas fosfomycin BGC and monitored consumption of 2-Pmm. Thus, we identified a single gene (psfC) that is responsible for performing the oxidative decarboxylation necessary to complete the fosfomycin biosynthetic pathway. Structural characterization reveals PsfC to be a member of the metallohydrolase superfamily, and our studies elaborate an unexpected biochemical function within this superfamily. Sequence- and structure-based network analysis identified diverse phosphonate BGCs that likely involve the intermediacy of 2-Pmm. We further characterized the activity of PsfC through in vitro assays and determined that the enzyme likely contains a diiron active site and requires molecular oxygen for catalysis.
Results
Coexpression of Biosynthetic Genes in E. coli.
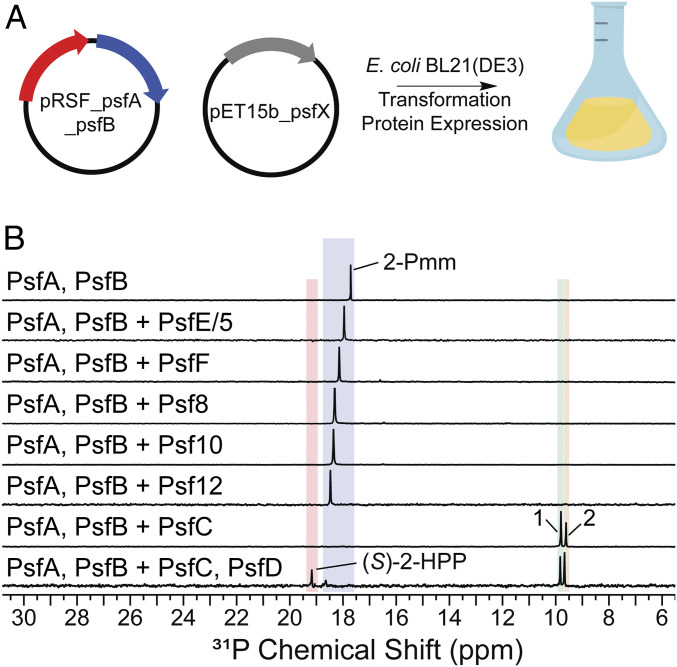
The psfA and psfB genes from P. syringae PB-5123 were cloned into a pRSFDuet vector for coexpression in E. coli (Fig. 2A). Analysis of the cell lysates using 31P NMR spectroscopy demonstrated the formation of 2-Pmm (Fig. 2B). With this platform, we explored the function of the remaining genes of unknown function in the psf BGC (Fig. 1A). Genes were cloned and expressed together with psfAB, and the resultant cell lysates were analyzed by 31P NMR spectroscopy for turnover of 2-Pmm (Fig. 2A). Only coexpression of psfC with psfA and psfB yielded additional resonances in the 31P NMR spectrum that are likely phosphonate derived, given their characteristic chemical shifts in the +6 to +24 ppm range (Fig. 2B) (21).
Fig. 2.
Heterologous expression to screen pathway enzymes. (A) Plasmid design for screening enzymes consisted of a pRSF backbone containing psfA and psfB. A pET15b-based plasmid was used to coexpress the other open reading frames (ORFs) in the BGC. (B) Combinatorial expression screen of ORFs in the BGC showed no new product formation except when psfC was included, which resulted in production of compounds 1 and 2. Additional expression of psfD resulted in the production of the penultimate pathway intermediate, (S)-2-HPP. The chemical shifts of phosphonates in 31P NMR spectroscopy are very sensitive to pH near their pKa values, accounting for the small changes in chemical shifts between experiments.
Characterization of the PsfC Reaction Products Generated in E. coli.
We next carried out two-dimensional NMR (2D-NMR) spectroscopy experiments on cell lysates in order to determine the molecular structure of the two products formed upon coexpression of psfABC. Analysis by 1H–31P heteronuclear multiple bond correlation (HMBC) spectroscopy revealed correlated chemical shifts at ∼3 ppm in the 1H dimension for both intermediates, each present as a doublet because of coupling to the 31P nucleus (SI Appendix, Fig. S2) (22). In a 1H–13C gradient-enhanced HMBC adiabatic (gHMBCAD) experiment, these protons were coupled to carbonyl signals at ∼210 ppm in the 13C dimension (SI Appendix, Fig. S3). An authentic synthetic standard of 2-OPP, previously shown to be an on-pathway intermediate (20), was spiked into the lysates, which confirmed the identity of the product with the downfield 31P signal, product 1, as 2-OPP. The product with the upfield signal, product 2, was unstable and decomposed to form product 1 over time. As we observed similar NMR characteristics for product 2 and 2-OPP, we assigned to compound 2 the preliminary structure of 3-oxo-4-phosphonobutanoic acid (3-OBPn) (SI Appendix, Fig. S3).
To confirm the identity of product 2, we generated a semisynthetic standard of 3-OBPn, following the precedent for the oxidative transformation of citrate to acetonedicarboxylic acid (23–25). 2-Pmm was generated in vitro with PepM and PsfB (see Materials and Methods) and subjected to Fe(II)-catalyzed oxidative decarboxylation to form 3-OBPn, as demonstrated by 2D-NMR analyses. 13C-containing isotopologues of 2-Pmm, and the subsequent labeled analogs of the product 3-OBPn, were also generated for confirmation of the structural assignment (SI Appendix, Figs. S4 and S5). The semisynthetic standard was used to spike cell lysates from psfABC-expressing cells, resulting in an increase in the intensity of the peak assigned to product 2, thus confirming its identity as 3-OBPn (SI Appendix, Fig. S6). During workup of the lysate, we observed that the amounts of 3-OBPn decreased concomitant with increases in the amounts of 2-OPP. However, once partially purified from the cellular components, 3-OBPn proved stable in the solution for >18 h, as determined by NMR spectroscopy. Hence, the conversion of 3-OBPn to 2-OPP is facilitated inside cells, presumably by adventitious activity from E. coli proteins.
To verify that 3-OBPn and 2-OPP are both formed in cells, and that 2-OPP was not solely a byproduct of workup, we sought to trap the initial products by chemical reduction, thus stabilizing 3-OBPn from spontaneous decarboxylation. We first confirmed that treatment with the reductant NaBH4 did not change the ratio of 3-OBPn and 2-OPP and their corresponding reduced forms, 3-hydroxy-phosphonobutanoic acid (3-HBPn) and 2-hydroxypropylphosphonic acid (2-HPP). Next, we treated cell lysates with this reducing agent during different points of the workup and observed both 3-HBPn and 2-HPP products in consistent ratios (SI Appendix, Fig. S7). Thus, our data suggest both 2-OPP and 3-OBPn are formed in vivo, and we conclude that PsfC is sufficient for effecting the transformation of 2-Pmm to 2-OPP, thereby completing the fosfomycin biosynthetic pathway in pseudomonads. The addition of PsfD, a previously characterized oxidoreductase capable of transforming 2-OPP into (S)-2-HPP (20), to the in vivo platform resulted in the formation of (S)-2-HPP in E. coli (Fig. 2B).
Crystal Structure of PsfC.
The primary sequence of PsfC shows sequence similarity with members of the polymerase and histidinol phosphatase (PHP) superfamily of proteins that carry out hydrolysis reactions. However, a sequence alignment of PsfC with characterized members of the family demonstrates that PsfC lacks many of the conserved residues implicated in the canonical trinuclear metal active site architecture associated with the PHP family (SI Appendix, Fig. S8) (26). Moreover, the canonical hydrolase activity observed in members of the PHP protein family (Pfam) does not correspond with the oxidative decarboxylation activity seen in vivo for PsfC (27).
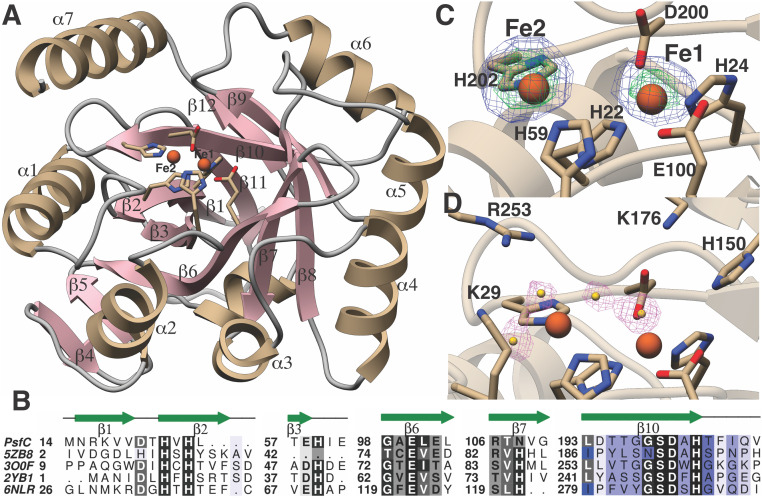
To facilitate understanding of how PsfC catalyzes the transformation of 2-Pmm to 3-OBPn, we pursued crystallographic studies. PsfC crystallized as a dimer in the asymmetric unit and exhibits a distorted (α/β)7 fold, as observed in other members of the PHP superfamily (27–29). Notably, electron density near the presumptive active site was consistent with the presence of a single metal ion, irrespective of attempts at crystallization under aerobic or anaerobic conditions. This form of the enzyme did not display in vitro activity (vide infra). Reasoning that the high concentrations of a dicarboxylate dianion (malonate) in the crystallization media might strip weakly bound metals, alternative crystallization conditions were explored. Anaerobic crystallization of PsfC in malonate-free conditions yielded crystals that could be soaked in precipitant solution supplemented with Fe2+. Structure elucidation yielded a 1.96 Å resolution dataset, showing nearly full occupancy of a bound diiron center (Fig. 3A). The identity of the metal was confirmed by collection of anomalous diffraction data at a wavelength near the Fe K-edge energy. Difference maps calculated with Bijvoet pairs showed strong peaks at the location of the bound Fe/Fe pair.
Fig. 3.
Crystal structure of PsfC, with highlighted residues investigated by site-directed mutagenesis. (A) Crystal structure of the monomeric unit of PsfC. Residues that coordinate the metals are show as stick figures, and the Fe atoms are shown as red spheres. (B) Structure-based multiple sequence alignment between PsfC and other members of the PHP-fold family, showing the identity and location of metal ligands. (C) Anomalous difference Fourier maps calculated using data collected near the Fe absorption edge (1.739 Å) and phases from the final model. The map is contoured at 3 (blue mesh) and 8 σ (green mesh). The coordinates of the final structure are superimposed. (D) Difference Fourier (Fobs-Fcalc) electron density maps calculated with coordinates in which all solvent atoms within 6 Å of the metals were removed and subject to one round of refinement before map calculation.
At present, only a few members of the PHP superfamily have been characterized structurally or functionally (26). A distance‐matrix alignment search against the Protein Data Bank (PDB) shows that the closest characterized structural homologs are a lesion-specific endonuclease from Pyrococcus furiosus (PDB Code 5ZB8, z-score = 18.1, 15% sequence identity, and 201 C-α atoms aligned with an rmsd of 2.9 Å), a metal-dependent phosphoesterase from Bifidobacterium adolescentis (PDB Code 3O0F, z-score = 15, 19% sequence identity, and 170 C-α atoms aligned with an rmsd of 2.8 Å), an amidohydrolase from Chromobacterium violaceum (PDB Code 2YB1, z-score = 12.7, 19% sequence identity, and 157 C-α atoms aligned with an rmsd of 2.9 Å), and histidinol phosphatase from Listeria monocytogenes (PDB Code 6NLR, z-score = 9.8, 16% sequence identity, and 149 C-α atoms aligned with an rmsd of 3.1 Å) (30). The number of bound metal ions in the active sites of these structures varies and is dictated by differences in the composition of the metal-binding residues among these enzymes.
The structure of diiron PsfC shows that residues His22 (strand β2), His24 (strand β2), Glu100 (strand β6), and Asp200 (strand β10) interact with Fe1, and His59 (end of strand β3) and His202 (end of strand β10) interact with Fe2 (Fig. 3 B and C). In the structures of other characterized PHP enzymes, a His residue (His75 in 2YB1) engages a metal ion to form a trinuclear center (31). The equivalent residue in PsfC is Asn108 (strand β7), which is too distant to interact with any of the other residues that could make up the third metal-binding site. Lastly, in some PHP members, an additional Asp (Asp14 in 2YB1) also coordinates Fe2. The equivalent region in PsfC (after strand β2) is deflected away from the active site by a large deletion between strand β2 and helix α1. This deflection creates a cavity that may provide a plausible binding site of the substrate, 2-Pmm. A number of solvent features, which have been modeled as water molecules, complete the coordination of the two Fe atoms (Fig. 3D).
The distance between the two Fe atoms is 4.9 Å, which is considerably longer than the 2.5 to 3.4 Å distances observed in canonical nonheme diiron enzymes (SI Appendix, Table S1) (32). However, similar long Fe–Fe separation distances have been reported in the structures of diiron-reconstituted UndA (33) and SznF (34). In the case of UndA, binding of the substrate triggers the formation of a µ-peroxo-Fe(III/III) with an Fe–Fe separation of 3.21 Å, as determined by extended X-ray absorption fine structure studies. Similar contraction of the diiron separation distance may also occur upon the binding of 2-Pmm, but structural studies could not be carried out because of instability of the metal composition in the presence of the substrate (vide infra). Both His59 and His202 that coordinate Fe2 are located at the end of β strands, allowing for any necessary flexibility during the formation of a productive Fe–Fe complex.
Site-Directed Mutagenesis of PsfC.
The crystallographic studies and sequence alignment suggest several residues that may be involved in catalysis by PsfC. These residues, highlighted in Fig. 3D (expanded in SI Appendix, Fig. S9), were mutated individually to alanine, and the resulting PsfC variants were analyzed by 31P NMR spectroscopy for activity in E. coli (SI Appendix, Fig. S10). Mutation of any of the four residues that bind Fe1 to alanine — His22, His24, Glu100, and Asp200—resulted in complete loss of activity. Mutation of the two residues that bind Fe2—His59 and His202—to alanine similarly abolished activity. A further set of mutations were performed on other conserved residues that are suspected to play a role in either metal binding or performing a catalytic function (SI Appendix, Figs. S9 and S10)—Ser28, Lys29, Lys30, Gln31, Asp35, Glu58, Gln91, His149, His150, Glu172, and Lys176. Mutation of some of these residues (Glu58, His149, His150, and Glu172) to Ala abrogated or strongly decreased activity in vivo and suggests that these residues play a role in the decarboxylation activity (SI Appendix, Figs. S9 and S10). These residues could assist in substrate binding, metal binding, or play a role in the catalytic mechanism (vide infra).
In Vitro Activity of PsfC.
The observed conversion of 2-Pmm to 2-OPP and 3-OBPn in E. coli upon PsfC expression and the observation that select variants abolish this activity provide strong evidence that the enzyme is responsible for this reaction in vivo. To provide further evidence, in vitro studies were explored. PsfC containing one equivalent of iron was inactive with 2-Pmm. Reconstitution of as-isolated PsfC with two equivalents of (NH4)2Fe(SO4)2 did result in conversion of 2-Pmm (1.2 mM) to 2-OPP and 3-OBPn, but the reaction was incomplete (SI Appendix, Fig. S11). 2-Pmm is a close structural analog of citrate, which has a high affinity for iron (Kf 2 × 106 to 7 × 1011) (35). Thus, we suspected that the incomplete in vitro turnover of 2-Pmm might be the result of removal of the weakly bound Fe2 from the enzyme by the substrate. Indeed, incubation of 100-µM PsfC containing 1.58 ± 0.04 equivalents of iron with 2-Pmm (0.8 mM), followed by dialysis and iron analysis, showed that the recovered PsfC contained 1.00 ± 0.03 equivalents of iron and lost its activity. A control in which PsfC was not treated with 2-Pmm did not result in the loss of activity following the same procedure. We then turned to single-turnover experiments using 100-µM enzyme and substrate, which resulted in full turnover to 3-OBPn (SI Appendix, Fig. S12). The transformation required O2, was more efficient in the presence of ascorbate, was dependent on enzyme concentration, and was not due to adventitious iron in solution, as shown with control assays (SI Appendix, Fig. S11). Thus, these data confirm in vitro that PsfC converts 2-Pmm to 3-OBPn. We did not see any evidence that PsfC catalyzes the decarboxylation of 3-OBPn to 2-OPP.
Increasing the 2-Pmm concentration from 100 to 200 µM was sufficient to inhibit the reaction (SI Appendix, Fig. S12). The 3-OBPn product is a much weaker metal ligand than the substrate 2-Pmm. Indeed, when PsfC (100 µM) was reacted with 100-μM 2-Pmm and after a delay of 0.5 min, a second amount of 2-Pmm (100 µM) was added, considerably more product was observed than when 200-µM 2-Pmm was added initially (SI Appendix, Fig. S12). This iterative procedure of substrate addition, enzyme turnover, and new substrate addition resulted in multiple enzyme turnovers (SI Appendix, Fig. S12).
Proposed Mechanism for PsfC.
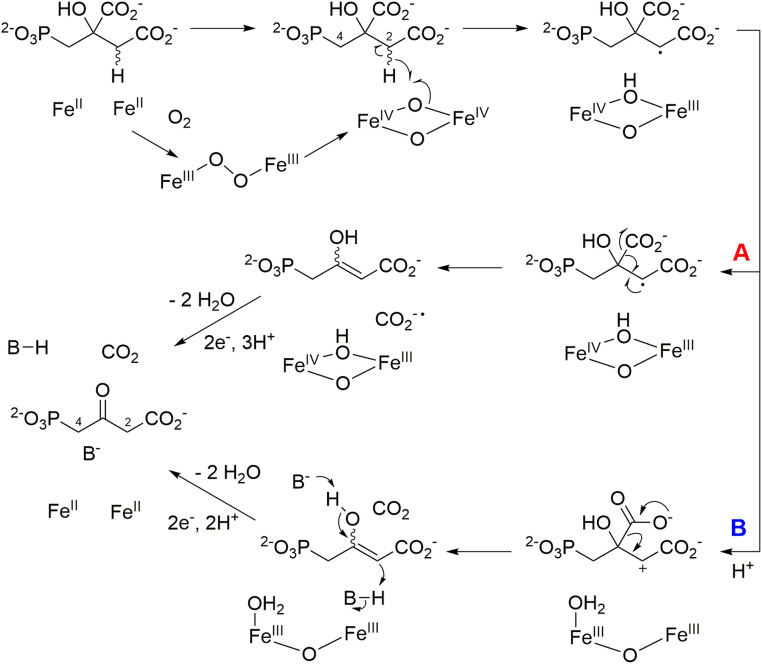
The chemistry catalyzed by PsfC only upon reconstitution of the enzyme, such that it contains more than one equivalent of iron, suggests that the active enzyme likely uses a dinuclear iron center. The findings reported here resemble studies on the iron-dependent decarboxylase UndA that was initially reported to contain only a single iron, based on a crystallographic study (36), but recent mechanistic studies demonstrated the enzyme is a diiron enzyme (33, 37). Scheme 1 shows a putative mechanism for PsfC similar to that proposed for UndA with a diiron center (33). Upon reaction with molecular oxygen, a bridged hydroperoxo-Fe(III)–Fe(III) complex could form, which can initiate hydrogen atom transfer (HAT) from either of the two methylene moieties of 2-Pmm (Scheme 1). As discussed below, we tentatively propose that HAT occurs from C2. After hydrogen atom abstraction, multiple pathways can be envisioned to generate 3-OBPn. Two possibilities are 1) radical-mediated decarboxylation (pathway A, Scheme 1) or 2) generation of a carbocation (pathway B, Scheme 1) (33, 37). Both pathways result in an enol that can tautomerize to form the observed product. In both pathways, the Fe cluster would require regeneration by reduction to its diferrous state, consistent with the requirement of ascorbate for full activity. We note that most likely the substrate 2-Pmm will coordinate to one or both irons, but at present, the substrate-binding geometry is not known, as attempts to obtain cocrystal structures with substrate resulted in removal of the second iron. We also recognize that the set of residues shown above to be important for catalysis (Glu58, His149, His150, and Glu172) could be indicative of an alternative metal coordination geometry in the presence of 2-Pmm substrate.
Scheme 1.
Proposed mechanisms for the PsfC-catalyzed oxidative decarboxylation of 2-Pmm. Pathway A results from homolytic decarboxylation, whereas pathway B involves electron transfer from the radical intermediate to Fe(IV) and subsequent heterolytic decarboxylation.
Regardless of whether HAT occurs from C2 or C4, a proton from the solvent would replace the abstracted hydrogen during enol tautomerization in the mechanism of Scheme 1. Hence, conducting the enzymatic reaction in buffer made in D2O could provide insights into the potential site of HAT. A complication is the possibility of exchange of the protons at both methylene carbons of the 3-OBPn product with the solvent. When we analyzed this exchange, the protons at C2 were observed to exchange faster than the time needed to carry out the enzymatic reaction and analyze the product by NMR spectroscopy (SI Appendix, Fig. S13). Conversely, the protons at C4 did not exchange noticeably over several hours. Thus, 2-Pmm was prepared with a 13C label at the position that becomes C4 in 3-OBPn (SI Appendix). When this substrate was used for a repeated single-turnover reaction (SI Appendix, Fig. S12) with PsfC in buffer made in D2O, the 3-OBPn product was analyzed by 31P- and 1H-decoupled 13C NMR spectroscopy (SI Appendix, Fig. S14). The 1H-decoupled 13C NMR spectrum shows C4 as a doublet because of coupling to the 31P nucleus with no coupling observed because of a possible 2H nucleus (SI Appendix, Fig. S14 B and C). In the 31P NMR spectrum, the phosphorus peak is split into a doublet by the 13C nucleus, and the doublet is split into triplets, indicating the presence of two protons at C4 (SI Appendix, Fig. S14D). Hence, during the PsfC-catalyzed conversion of 2-Pmm to 3-OBPn in D2O, no transfer of deuterium to C4 occurs. We therefore tentatively conclude that HAT must take place at C2 (Scheme 1). An alternative mechanism that would not require HAT from either C2 or C4 would also be consistent with the labeling results (e.g., SI Appendix, Scheme S1).
PsfC Is an Unusual Member of the PHP Domain–Containing Family.
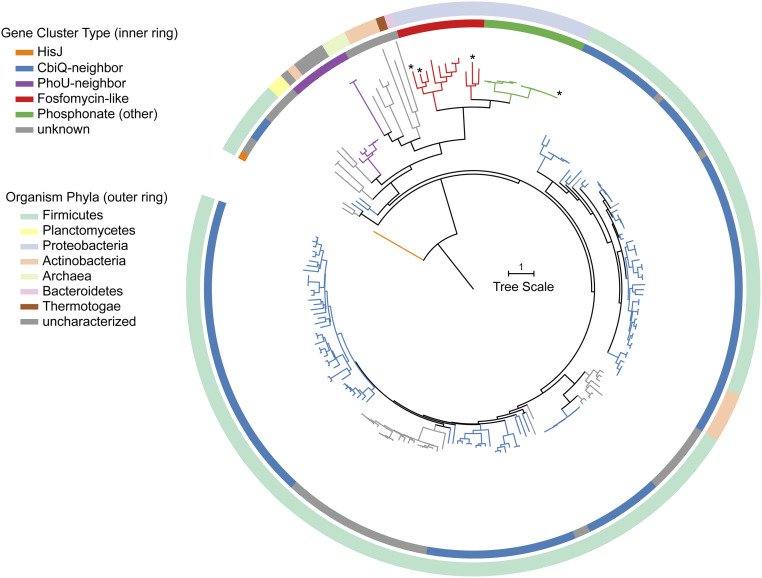
A phylogenetic tree illustrates that the PsfC sequence is quite distant from characterized members of the PHP family (Fig. 4). A position-specific iterative basic local alignment search tool return for PsfC was aligned by multiple alignment using fast Fourier transform and a simulated maximum-likelihood phylogenetic tree was generated in FastTree (38–40). As it appeared that none of the returned sequences belonged to functionally characterized proteins, an outgroup for the Bacillus halodurans C-125 histidinol phosphatase was chosen as the root node (41). Sequences corresponding to the proteins in this tree are included in the SI Appendix, Table S2 and Fig. S15). The tree was organized by the genomic contexts of the associated proteins and colored accordingly (Fig. 4 and SI Appendix, Fig. S15). The local genomic context could be classified into four archetypes. One group of homologs are encoded near phosphonate biosynthetic genes, represented in red and green; a second group of homologs are encoded near a gene from Pfam 02361 (CbiQ like, vide infra), represented in blue; and a third group of homologs are encoded near a gene from Pfam 01895 (PhoU like, vide infra), represented in purple. BGCs whose local genomic context could not be discerned or was not conserved are shown in gray.
Fig. 4.
Phylogenetic analysis of PsfC demonstrates the relationship to characterized members of the PHP superfamily. (Top) Position-specific iterative basic local alignment search tool hits from a PsfC bait sequence and a characterized member of the PHP family (orange branch) were aligned, and a phylogenetic tree was assembled in FastTree and edited in Interactive Tree of Life (40, 56). Branches were colored based on local genetic context. Green and red branches indicate the presence of other phosphonate-associated, PsfC-like proteins. Characterized members are indicated with an asterisk (*).
Branches shown in red and green have a pepM within 10 open reading frames of the PHP-encoding gene, suggesting that these BGCs likely produce a phosphonate. The red branches have similar genetic context to that of the fosfomycin pathway. We sought to determine if 2-Pmm is a substrate for these PsfC homologs. Three representative sequences from the phosphonate clade were coexpressed with PsfAB in E. coli. Sequences from Pseudomonas species p106 (Ps106PsfC, WP_125924462.1), Burkholderia stagnalis (BsPsfC, WP_059908919.1), and an unclassified gammaproteobacterium (GbPsfC, OGT51126.1) were chosen because of their relative distance from PsfC in the phylogenetic analysis and are shown with an asterisk (*) in Fig. 4 (SI Appendix, Fig. S15, bolded in SI Appendix, Table S2). All three experiments produced both 3-OBPn and 2-OPP as major products (SI Appendix, Fig. S16). Thus, despite varying gene architecture in their respective BGCs, the presence of these psfC-like genes confirms 3-OBPn (and possibly 2-OPP) as intermediates in these respective pathways.
The role played by the PsfC-like sequences in non-phosphonate BGCs is still elusive. Branches in blue have a nearby gene from Pfam 02361. This Pfam denotes a family of cobalt transporters, with CbiQ as representative member. CbiQ is implicated in cobalamin biosynthesis in anaerobic organisms, forming a CbiMNQO complex (42–44). The CbiQ homologs encoded near genes for PsfC-like proteins share low homology with canonical CbiQ proteins, suggesting they have an additional or alternative, unknown function.
PhoU, a representative member of Pfam 01895, is implicated as part of the Pho regulon of phosphate uptake in a diverse set of organisms (45). Like CbiQ, its gene is typically found within a larger, conserved operon responsible for high-affinity phosphate uptake. The phoU genes near a psfC homolog, shown as purple branches, did not have the canonical colocalized elements to form such a transport system (46). It is unknown what role these proteins serve with respect to PsfC. The remaining gene cluster types arise from a diverse phylogenetic background in Actinobacteria, Proteobacteria, and a few Archaea. The diversity of sequences and phylogenetic backgrounds implies that PHP domain–containing proteins may support multiple functions outside of the classical hydrolase activity.
Discussion
We identified the enzyme responsible for completing the biosynthesis of fosfomycin in pseudomonads. In the biosynthesis of FR-900098 and pantaphos, the conserved intermediate 2-Pmm undergoes an aconitase-like reaction; however, no such enzyme was encoded in the pseudomonad cluster, questioning the fate of this intermediate (12, 16, 47). By screening the potential function of the uncharacterized psf genes in E. coli, we identified an alternative fate for 2-Pmm. PsfC, previously annotated as a metallohydrolase, catalyzes the oxidative decarboxylation of 2-Pmm in E. coli to 3-OBPn, and three orthologs from diverse genera displayed the same activity. In cells, this intermediate can decarboxylate to 2-OPP, a known intermediate in fosfomycin biosynthesis in P. syringae PB-5123. Thus, PsfC is sufficient to complete the biosynthesis of fosfomycin; although, at present, we anticipate that the decarboxylation step may be facilitated by one or more of the remaining genes of unknown function in the BGC; decarboxylation of β-keto acids is ubiquitous in nature and can be catalyzed by a myriad of proteins (48). The observed substrate inhibition in vitro by removal of the weakly bound Fe2 is likely not important in P. syringae, given the very low production levels (∼0.1 mg/L) and the associated anticipated low concentrations of 2-Pmm. Our findings resemble the discovery that the HD domain family of enzymes, which were presumed to be metalloenzymes with phosphohydrolase activity, encompass members with iron-dependent oxygenase activity (49–52). The discovery of an activity of a PHP family member that is very different from previously reported activities, as well as the phylogenetic analysis, suggests that this enzyme superfamily may catalyze a much larger set of reactions than previously appreciated and adds to the known folds that support dinuclear iron chemistry (32, 34, 53, 54).
Materials and Methods
Coexpression of Fosfomycin Biosynthetic Genes.
An overnight culture of E. coli BL21 (DE3) cells harboring both the PsfA- and PsfB-encoding plasmid and pET15b-based plasmids containing genes of unknown function in the psf BGC, was grown in Luria–Bertani (LB) medium. The cells were used to inoculate a 1-L culture at 1/100 dilution in LB medium in a 2.8 L baffled Fernbach flask supplemented with 100 µg/mL ampicillin and 50 µg/mL kanamycin. The culture was grown at 37 °C with 210 rpm shaking until OD600 reached 0.6 to 0.8. The culture was subsequently cooled to 18 °C on ice, and expression of genes was induced by the addition of isopropyl-1-thio-β-D-galactoside to 0.1 mM. Cultures were grown for an additional 16 h at 18 °C. Cells were harvested by centrifugation, washed in phosphate-buffered saline, centrifuged again, and resuspended in 1.5 equivalents of cold methanol (−20 °C, volume/volume). Cells were suspended by vortexing to homogenize the mixture and submerged in liquid nitrogen. Samples were stored at −80 °C or thawed immediately at 4 °C. Once thawed, equivalent volumes of cold (4 °C) water and chloroform were added to the cell suspension and vortexed vigorously. Samples were centrifuged at 1,000 g for 15 min to clarify the mixture. The aqueous layer was collected and placed on a Savant SPD11V SpeedVac until ∼1 mL solution remained. Concentrated samples were diluted to 20% D2O, EDTA was added to 15 mM, and the samples were analyzed via NMR spectroscopy (SI Appendix, Materials and Methods).
Single-Turnover Assays with PsfC.
Anaerobically purified PsfC-M2 (see SI Appendix, Materials and Methods) was reconstituted with 2.4 equivalents of (NH4)2Fe(SO4)2∙(H2O)6 on ice (300 μM PsfC). The protein was then dialyzed against 1 L storage buffer (vide supra) on ice twice for 3 h. Meanwhile, enzymatically synthesized 2-Pmm solutions were aerated via sparging with O2 for 1 h. After dialysis, PsfC-M2 (100 μM final concentration) was removed from the anaerobic chamber, and the reaction was initiated by the addition of 2-Pmm (100 μM) and ascorbate (5 mM), in a total volume of 400 μL oxygenated buffer; in parallel, multiple-turnover and Fe-stripping experiments were performed with 2-Pmm at 200 and 800 μM (final concentration), respectively, with 100 μM PsfC. The reaction was quenched after 30 s with the addition of EDTA and D2O to 15 mM and 20%, respectively. Where indicated, an additional bolus of 2-Pmm (stock 4 to 8 mM) was added after 30 s, and the activity assay was repeated. Protein was removed via filtration through a 10-kDa Amicon spin filter, and filtrate was analyzed by NMR spectroscopy (SI Appendix, Fig. S12). For Fe-stripping experiments, PsfC-M2 (200 μM) treated with 2-Pmm (800 μM) was dialyzed against an additional 1 L volume storage buffer. Similarly, PsfC-M2 diluted with the same volume of oxygenated buffer was dialyzed as well. Both samples were removed after 3 h and analyzed for Fe content by a Ferene-S–based spectrophotometric assay (55).
For 1D, 13C, and 31P (with 1H coupling) experiments (SI Appendix, Fig. S14), four parallel enzyme assays with three successive additions of 13C-2-Pmm were combined after quenching, lyophilized, and resuspended in 20% D2O for NMR acquisitions. A total of 3 to 4 mg sodium dithionite was added to samples prior to 31P (with 1H coupling) experiments (21).
Supplementary Material
Acknowledgments
We thank Dr. J. Whitteck for the synthesis of 2-OPP and Dr. E. Ulrich for the construction of pET15b_psfB. S.K.N. and C.O. thank the staff at Life Sciences Collaborative Access Team Sector 21 for help with data collection. This work was supported by the NIH (Grant P01 GM077596 to W.A.v.d.D. and S.K.N.) and the HHMI. NMR data were collected in the Carl R. Woese Institute for Genomic Biology Core Facility on a 600-MHz NMR, funded by NIH Grant S10-RR028833, and on a Bruker 500 MHz equipped with a CryoProbe, funded in part by the Roy J. Carver Charitable Trust (Muscatine, Iowa; Grant #15-4521) to the School of Chemical Sciences NMR Laboratory. We thank Dr. L. Zhu and Dr. X. Guan for assistance in NMR data collection.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019863118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Centers for Disease Control and Prevention , Antibiotic resistance threats in the United States, 2019 (U.S. Department of Health and Human Services, CDC, Atlanta, GA, 2019). [Google Scholar]
- 2.Michalopoulos A. S., Livaditis I. G., Gougoutas V., The revival of fosfomycin. Int. J. Infect. Dis. 15, e732–e739 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Sastry S., Doi Y., Fosfomycin: Resurgence of an old companion. J. Infect. Chemother. 22, 273–280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassetti M., Graziano E., Berruti M., Giacobbe D. R., The role of fosfomycin for multidrug-resistant gram-negative infections. Curr. Opin. Infect. Dis. 32, 617–625 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Falagas M. E., Vouloumanou E. K., Samonis G., Vardakas K. Z., Fosfomycin. Clin. Microbiol. Rev. 29, 321–347 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendlin D., et al., Phosphonomycin, a new antibiotic produced by strains of streptomyces. Science 166, 122–123 (1969). [DOI] [PubMed] [Google Scholar]
- 7.Shoji J., et al., Production of fosfomycin (phosphonomycin) by Pseudomonas syringae. J. Antibiot. (Tokyo) 39, 1011–1012 (1986). [DOI] [PubMed] [Google Scholar]
- 8.Seto H., Kuzuyama T., Bioactive natural products with carbon-phosphorus bonds and their biosynthesis. Nat. Prod. Rep. 16, 589–596 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Liu P., et al., Protein purification and function assignment of the epoxidase catalyzing the formation of fosfomycin. J. Am. Chem. Soc. 123, 4619–4620 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Woodyer R. D., et al., Heterologous production of fosfomycin and identification of the minimal biosynthetic gene cluster. Chem. Biol. 13, 1171–1182 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Woodyer R. D., Li G., Zhao H., van der Donk W. A., New insight into the mechanism of methyl transfer during the biosynthesis of fosfomycin. Chem. Commun. (Camb.) 43, 359–361 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Kim S. Y., et al., Different biosynthetic pathways to fosfomycin in Pseudomonas syringae and Streptomyces species. Antimicrob. Agents Chemother. 56, 4175–4183 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato S., Kudo F., Kim S.-Y., Kuzuyama T., Eguchi T., Methylcobalamin-dependent radical SAM C-methyltransferase Fom3 recognizes cytidylyl-2-hydroxyethylphosphonate and catalyzes the nonstereoselective C-methylation in fosfomycin biosynthesis. Biochemistry 56, 3519–3522 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Cho S.-H., et al., Fosfomycin biosynthesis via transient cytidylylation of 2-hydroxyethylphosphonate by the bifunctional Fom1 enzyme. ACS Chem. Biol. 12, 2209–2215 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Sato S., et al., Biochemical and structural analysis of FomD that catalyzes the hydrolysis of cytidylyl (S)-2-hydroxypropylphosphonate in fosfomycin biosynthesis. Biochemistry 57, 4858–4866 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Horsman G. P., Zechel D. L., Phosphonate biochemistry. Chem. Rev. 117, 5704–5783 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Kuzuyama T., Hidaka T., Imai S., Seto H., Studies on the biosynthesis of fosfomycin. V. Cloning of genes for fosfomycin biosynthesis. J. Antibiot. (Tokyo) 46, 1478–1480 (1993). [DOI] [PubMed] [Google Scholar]
- 18.Eliot A. C., et al., Cloning, expression, and biochemical characterization of Streptomyces rubellomurinus genes required for biosynthesis of antimalarial compound FR900098. Chem. Biol. 15, 765–770 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polidore A. L. A., Furiassi L., Hergenrother P. J., Metcalf W. W., A phosphonate natural product made by Pantoea ananatis is necessary and sufficient for the hallmark lesions of onion center rot. mBio 12, e03402–e03420 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olivares P., Ulrich E. C., Chekan J. R., van der Donk W. A., Nair S. K., Characterization of two late-stage enzymes involved in fosfomycin biosynthesis in pseudomonads. ACS Chem. Biol. 12, 456–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peck S. C., Gao J., van der Donk W. A., Discovery and biosynthesis of phosphonate and phosphinate natural products. Methods Enzymol. 516, 101–123 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Metcalf W. W., et al., Synthesis of methylphosphonic acid by marine microbes: A source for methane in the aerobic ocean. Science 337, 1104–1107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abrahamson H. B., Rezvani A. B., Brushmiller J. G., Photochemical and spectroscopic studies of complexes, of iron(III) with citric acid and other carboxylic acids. Inorg. Chim. Acta 226, 117–127 (1994). [Google Scholar]
- 24.Butler A., Theisen R. M., Iron(III)-siderophore coordination chemistry: Reactivity of marine siderophores. Coord. Chem. Rev. 254, 288–296 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grau F. H., Halliday W. J., Oxidation and decarboxylation of citrate in the presence of ferrous iron. Nature 179, 733–734 (1957). [DOI] [PubMed] [Google Scholar]
- 26.Ghodge S. V., Raushel F. M., “Structure, mechanism, and substrate profiles of the trinuclear metallophosphatases from the amidohydrolase superfamily” in Meth. Enzymol. (Elsevier, 2018), 607, pp. 187–216. [DOI] [PubMed] [Google Scholar]
- 27.Ghodge S. V., et al., Structural and mechanistic characterization of L-histidinol phosphate phosphatase from the polymerase and histidinol phosphatase family of proteins. Biochemistry 52, 1101–1112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey S., Wing R. A., Steitz T. A., The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell 126, 893–904 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Omi R., et al., Crystal structure of monofunctional histidinol phosphate phosphatase from Thermus thermophilus HB8. Biochemistry 46, 12618–12627 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Holm L., DALI and the persistence of protein shape. Protein Sci. 29, 128–140 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cummings J. A., et al., Prospecting for unannotated enzymes: Discovery of a 3′,5′-nucleotide bisphosphate phosphatase within the amidohydrolase superfamily. Biochemistry 53, 591–600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jasniewski A. J., Que L. Jr, Dioxygen activation by nonheme diiron enzymes: Diverse dioxygen adducts, high-valent intermediates, and related model complexes. Chem. Rev. 118, 2554–2592 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang B., et al., Substrate-triggered formation of a peroxo-Fe2(III/III) intermediate during fatty acid decarboxylation by UndA. J. Am. Chem. Soc. 141, 14510–14514 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McBride M. J., et al., Structure and assembly of the diiron cofactor in the heme-oxygenase-like domain of the N-nitrosourea-producing enzyme SznF. Proc. Natl. Acad. Sci. U.S.A. 118, e2015931118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamm R. E., Shull C. M. Jr, Grant D. M., Citrate complexes with iron(II) and iron(III). J. Am. Chem. Soc. 76, 2111–2114 (1954). [Google Scholar]
- 36.Rui Z., et al., Microbial biosynthesis of medium-chain 1-alkenes by a nonheme iron oxidase. Proc. Natl. Acad. Sci. U.S.A. 111, 18237–18242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manley O. M., Fan R., Guo Y., Makris T. M., Oxidative decarboxylase UndA utilizes a dinuclear iron cofactor. J. Am. Chem. Soc. 141, 8684–8688 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Altschul S. F., et al., Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price M. N., Dehal P. S., Arkin A. P., FastTree 2–Approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.le Coq D., Fillinger S., Aymerich S., Histidinol phosphate phosphatase, catalyzing the penultimate step of the histidine biosynthesis pathway, is encoded by ytvP (hisJ) in Bacillus subtilis. J. Bacteriol. 181, 3277–3280 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bao Z., et al., Structure and mechanism of a group-I cobalt energy coupling factor transporter. Cell Res. 27, 675–687 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodionov D. A., Hebbeln P., Gelfand M. S., Eitinger T., Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: Evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 188, 317–327 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roth J. R., Lawrence J. G., Rubenfield M., Kieffer-Higgins S., Church G. M., Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J. Bacteriol. 175, 3303–3316 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shinagawa H., Makino K., Nakata A., Regulation of the pho regulon in Escherichia coli K-12. Genetic and physiological regulation of the positive regulatory gene phoB. J. Mol. Biol. 168, 477–488 (1983). [DOI] [PubMed] [Google Scholar]
- 46.Devine K. M., Activation of the PhoPR-mediated response to phosphate limitation is regulated by wall teichoic acid metabolism in Bacillus subtilis. Front. Microbiol. 9, 2678 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao Y., Peng Q., Li S., Deng Z., Gao J., The intriguing biology and chemistry of fosfomycin: The only marketed phosphonate antibiotic. RSC Adv. 9, 42204–42218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walsh C. T., Biologically generated carbon dioxide: Nature’s versatile chemical strategies for carboxy lyases. Nat. Prod. Rep. 37, 100–135 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Bollinger J. M. Jr., Diao Y., Matthews M. L., Xing G., Krebs C., myo-Inositol oxygenase: A radical new pathway for O(2) and C-H activation at a nonheme diiron cluster. Dalton Trans. 38, 905–914 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McSorley F. R., et al., PhnY and PhnZ comprise a new oxidative pathway for enzymatic cleavage of a carbon-phosphorus bond. J. Am. Chem. Soc. 134, 8364–8367 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Wörsdörfer B., et al., Organophosphonate-degrading PhnZ reveals an emerging family of HD domain mixed-valent diiron oxygenases. Proc. Natl. Acad. Sci. U.S.A. 110, 18874–18879 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rajakovich L. J., et al., A new microbial pathway for organophosphonate degradation catalyzed by two previously misannotated non-heme-iron oxygenases. Biochemistry 58, 1627–1647 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Komor A. J., Jasniewski A. J., Que L., Lipscomb J. D., Diiron monooxygenases in natural product biosynthesis. Nat. Prod. Rep. 35, 646–659 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rajakovich L. J., et al., “Emerging structural and functional diversity in proteins with dioxygen-reactive dinuclear transition metal cofactors” in Comprehensive Natural Products III: Chemistry and Biology, Liu H.-W., Begley T. P., Eds. (Elsevier, Amsterdam, 2020), pp. 215–250. [Google Scholar]
- 55.Hennessy D. J., Reid G. R., Smith F. E., Thompson S. L., Ferene — a new spectrophotometric reagent for iron. Can. J. Chem. 62, 721–724 (1984). [Google Scholar]
- 56.Letunic I., Bork P., Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.