Significance
Designed protein assemblies have many applications in medicine and technology. A long-standing design paradigm generates assemblies through genetic fusion of homo-oligomers via α-helical linkers. Here, we introduce a rigid fusion strategy that finds a greater number of design solutions and reduces formation of unintended assembly states, and a method for doubly anchoring target binding domains in the resulting assemblies that could contribute to cryogenic electron microscopy structure determination of small proteins.
Keywords: protein design, cryo-EM, DARPin, protein fusion, nanomaterials
Abstract
Protein nanomaterial design is an emerging discipline with applications in medicine and beyond. A long-standing design approach uses genetic fusion to join protein homo-oligomer subunits via α-helical linkers to form more complex symmetric assemblies, but this method is hampered by linker flexibility and a dearth of geometric solutions. Here, we describe a general computational method for rigidly fusing homo-oligomer and spacer building blocks to generate user-defined architectures that generates far more geometric solutions than previous approaches. The fusion junctions are then optimized using Rosetta to minimize flexibility. We apply this method to design and test 92 dihedral symmetric protein assemblies using a set of designed homodimers and repeat protein building blocks. Experimental validation by native mass spectrometry, small-angle X-ray scattering, and negative-stain single-particle electron microscopy confirms the assembly states for 11 designs. Most of these assemblies are constructed from designed ankyrin repeat proteins (DARPins), held in place on one end by α-helical fusion and on the other by a designed homodimer interface, and we explored their use for cryogenic electron microscopy (cryo-EM) structure determination by incorporating DARPin variants selected to bind targets of interest. Although the target resolution was limited by preferred orientation effects and small scaffold size, we found that the dual anchoring strategy reduced the flexibility of the target-DARPIN complex with respect to the overall assembly, suggesting that multipoint anchoring of binding domains could contribute to cryo-EM structure determination of small proteins.
There is considerable interest in the design of novel protein assemblies, for example, to develop cryogenic electron microscopy (cryo-EM) scaffolds to aid in structure determination (1, 2) and protein nanoparticle vaccines (3, 4). Many studies have utilized 1) genetic fusion of α-helices at N and C termini or 2) protein interface design to drive assembly of symmetric protein homo-oligomers into a larger symmetric nanoparticle or lattice. The genetic fusion route was originally demonstrated with the creation of a tetrahedral protein nanocage and fiber (5) and has since been used to generate two-dimensional (2D) layers (6), a porous cube (7), additional tetrahedra (8, 9), octahedra (10), and icosahedra (11, 12). Strengths of the fusion approach are that it is relatively straightforward, not inherently destabilizing, and guarantees the intended stoichiometry and specificity between fusion partners. Despite its simplicity and success, several aspects of genetic fusion complicate its use. Genetic fusion is best applied with α-helical termini, as this imparts some rigidity between the fused domains, but termini are not always α-helical or physically accessible. Furthermore, the number of possible sequence alignments between any given pair of building blocks is finite and this, in turn, limits the number of geometric solutions for a desired symmetric architecture. This can complicate or preclude the use of fusion methods in assembly design, especially when a particular protein building block is required for a downstream application. Finally, flexibility is often introduced at the point of fusion, even with α-helical linkers and certainly with disordered linkers. In the best cases, model deviations are subtle (8, 13); however, varying levels of unintended assembly products are also commonly observed. In contrast, the noncovalent protein interface design approach bypasses considerations of termini and their alignment. Interface design is compatible with the free rotation and translation of building blocks along their axes (14), so the set of valid geometric solutions is technically unlimited. The drawback to interface design is that it requires the creation of a new soluble and specific interface without destabilizing either protein. Like fusion, the interface design route to the protein nanomaterial production has achieved considerable success (4, 15–20).
Among applications for genetic fusion is the creation of cryo-EM scaffolds (1, 2, 21–23): If a small target protein can be immobilized and rigidly bound onto a larger symmetric assembly, EM particle images can be more readily aligned and classified, and structures too small to analyze alone could then become amenable to structure determination. Yeates et al. demonstrated the potential of this approach by fusing a designed ankyrin repeat protein (DARPin) (24, 25) to the outside of a previously designed protein nanocage (1); the bound green fluorescent protein (GFP) target was resolved at 3.8 Å resolution with only a single α-helical fusion anchoring the DARPin.
A Computational Method for Rigid Three-Body Symmetric Fusion
We set out to develop a computational method for generating symmetric assemblies by gene fusion that explores many more combinations than previous methods and enforces rigid connections between the building blocks. Previous genetic fusion studies have focused on 1) fusing symmetric oligomeric building blocks together at their N and C termini. We reasoned that a much larger set of possible configurations could be generated by 2) incorporating a variable length rigid monomeric protein spacer between the two oligomers, and 3) allowing fusions at internal residues (not just the termini). The number of accessible configurations in a set of building blocks is on the order of
-
1.
N(oligomer1) × N(oligomer2) for standard fusion at building block helical termini.
-
2.
N(oligomer1) × N(oligomer2) × N(spacer), with the addition of spacer domains.
-
3.
N(oligomer1) × N(oligomer2) × N(spacer) × N(fusion sites per oligomer1) × N(fusion sites per oligomer2) × N(fusion sites per spacer)2 when both spacers and internal fusion sites are allowed. The quadratic term is because the spacer is fused twice (i.e., in both N- and C-terminal directions).
We explored the third option and found that it generally produces thousands of geometric solutions from our small starting set of de novo building blocks, while the first option (pursued in previous studies) produces few or no solutions with the same error tolerances. To ensure rigid junctions between the building blocks, we only allow fusion via alignment and superposition of shared helices from both building blocks being fused and disallow α-helical extension, as DARPin-fusion studies have shown that shared helix fusion (i.e., where the connecting helix is integral to both domains being connected) improves junction rigidity (26, 27). Our use of internal fusion (to access more geometric combinations) causes truncation that would destabilize most globular proteins; to avoid this, we use idealized repeat protein building block spacers (and oligomers, when possible) where every repeat unit is nearly identical. Such proteins are amenable to truncation or fragmentation without undermining folding and stability (28–31).
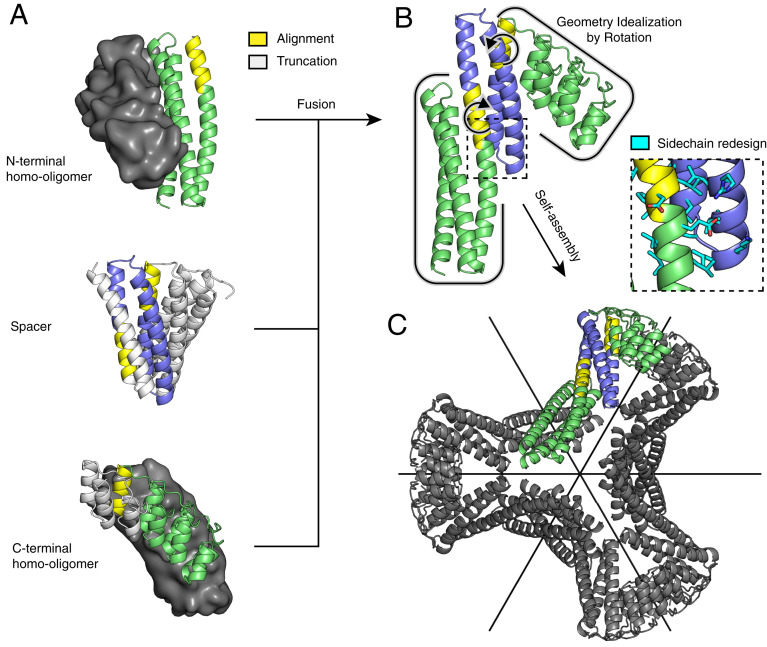
Geometric matches to the desired symmetry are identified for each (oligomer1, spacer, and oligomer2) tuple by the following procedure, which is illustrated schematically in Fig. 1: First, all rigid body transforms (T1) on oligomer1 are enumerated that superimpose one of its helical segments onto one from the spacers. Rigid body transforms T2 (applied to oligomer2) are calculated identically to identify possible fusions between oligomer2 and the spacer. Second, for each (T1 and T2) combination, the transforms are applied to both oligomers, and the arrangement of the axes of the two oligomers is tested for compatibility with the target architecture. For D2 symmetry, axes must intersect at 90°, and for D3, 60°. Since the actual angles between the axes will deviate from these ideal values, each homo-oligomer is then rotated about its corresponding shared helix segment center-of-mass so that the axes meet perfectly at the required angle. In cases where this last rotation (a measure of nonideality) is less than the configured error tolerance, three-dimensional (3D) models of the resulting assemblies are built by superposition of the repositioned structures. Models are discarded if homo-oligomer interface positions are modified, or there are backbone clashes; sidechain clashes are acceptable since redesign can eliminate them.
Fig. 1.
Illustration of the tripartite design strategy for the D3 architecture. (A) The final structure is composed of two homo-oligomers (dimers, top and bottom; the partner subunit is shown as a surface in gray) and a DHR protein (middle). (B) All possible nonclashing backbone alignments are geometrically analyzed and filtered to generate a three-component fusion, which is idealized to the target geometry by small rigid body rotations and redesigned to improve core packing and remove exposed hydrophobics. (C) The result is a D3 assembly with symmetric C2 axes (black) that correspond to those of the original homodimers and a new C3 axis orthogonal and through the center.
In selecting helical segment pairs for fusion in the first step of the above procedure, we use several criteria to try to ensure fusion rigidity. First, we require superpositions with a low backbone rmsd (default 0.5 Å) over multiple residues (default 8, minimum) to ensure the shared helix geometry is consistent with both building blocks. The stringency of backbone rmsd and overlap length is configurable. Unlike traditional symmetric fusion with α-helical linkers, overlapping fusion preserves proximity and sidechain packing of the fusion region with remaining secondary structures from the original building blocks, which reinforces the structure. For efficiency, the rmsd and overlap length thresholding is carried out early in the procedure during geometric-match identification, when alignments are initially computed. Second, we rank fusions based on the number of likely sidechain contacts between secondary structures of previously separate building blocks. This ensures that significant cross-building-block contacts can be made to buttress the interactions along the shared helix. This step also occurs prior to sidechain redesign, so Cα-Cβ vectors are used in place of contact counts as a measure of designability. Third, we redesign with Rosetta the regions adjacent to the new building block junctions to eliminate steric clashes and improve packing between newly joined regions. Fusion-related truncation often exposes hydrophobic residues that were previously buried, so these regions must be redesigned as well.
Design and Characterization of D2 and D3 Symmetric Oligomers
As a proof of concept, we applied the three-body fusion method to design dihedral symmetric assemblies from de novo designed repeat protein monomers and oligomers. Two different designed C2 dimers were fused as described above with a repeat protein monomeric spacer such that the C2 axes intersect at 90° for D2 structures or 60° for D3 structures (Fig. 2). Two different design rounds were performed targeting D2 and D3 symmetries. Even in the second design round, where fewer building blocks and additional selection criteria were imposed, millions of tripartite alignment combinations were scanned per symmetry, nearly 10,000 fusions matched the target geometry within the angular error tolerance (5°), and several hundred passed junction rigidity metrics. The sequences at the junction regions in these assemblies were then optimized using Rosetta as described in SI Appendix, Methods S1.
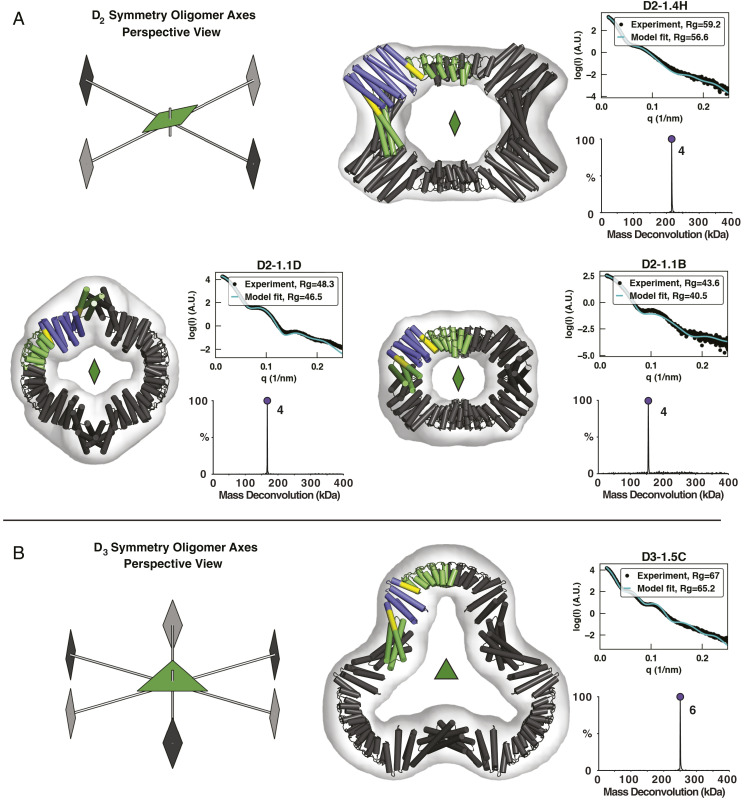
Fig. 2.
Characterization of first-round designs. EM, native-MS, and SAXS experiments are consistent with the formation of intended architectures for four designs: three D2 designs (A) and one D3 design (B). Negative-stain 3D reconstructions are overlaid with the design models, whose asymmetric units are colored according to the constituent building blocks (N- and C-terminal oligomers, green; DHR, blue; shared alignment, yellow). Native-MS deconvolutions show the relative abundance of the determined masses, and the peaks are labeled with their assigned oligomeric states. SAXS plots compare the theoretical (cyan) and experimental (black) scattering intensities (log scale) as a function of q as well as radius of gyration (Rg).
In a first design round, 28 D2 and 9 D3 assemblies that matched the geometric selection criteria and had low energy junctions were selected for experimental characterization and recombinantly produced in Escherichia coli. Of these 37 designs, 7 (3 D2 and 4 D3) were both soluble and eluted as single monodisperse peaks by size exclusion chromatography (SEC) (SI Appendix, Fig. S1). Scattering profiles and radius of gyration determined from small-angle X-ray scattering (SAXS) data were consistent with design models for the three D2 designs and for two of the four D3 designs (Fig. 2 and SI Appendix, Fig. S2). Of these designs, native mass spectrometry (native-MS) verified the expected oligomeric states for all of the assemblies. Despite the limited dataset, clear trends emerged; for example, all five designs corroborated by SAXS data incorporated a C-terminal ankyrin homodimer, which was present in only 60% of the tested designs. In addition, four of five SAXS- and native-MS–corroborated designs incorporated the same N-terminal three-helix homodimer “rop20” despite it being present in only ∼40% of tested designs. The set of 12 designs that combined both the N-terminal rop20 and C-terminal ankyrin dimer contained four of the five successes. Meanwhile, no designs using hairpin helical bundle dimers were successful.
A second round of design was performed with the same procedure, this time using primarily the rop20 helical bundle at the N terminus and any of the three similar ankyrin dimers at the C terminus that had been successful in the first round. We also introduced two additional design constraints with the aim of improving the designs’ suitability as cryo-EM DARPin scaffolds. First, structures with fewer total secondary structures (helix count) were selected, anticipating that they would be more rigid. Although the design method reinforces the point of fusion, the de novo helical repeat (DHR) spacers, and especially the ankyrin dimer building blocks, have small cross-sectional areas, so each additional repeat likely adds flexibility. Second, we selected designs with the ankyrin binding groove facing away from the assembly center (SI Appendix, Fig. S3) to reduce the chance of steric hindrance in multivalent target binding. A total of 31 D2 and 24 D3 designs were ordered, of which 15 and 12, respectively, had high levels of soluble expression and single major peaks by SEC. SAXS and native-MS agreed with the design models of four D2 designs and four D3 designs (Fig. 3). Designs D2-21.29 and D2-21.30 have 85% sequence identity and were not considered independent successes; only D2-21.29 was fully characterized.
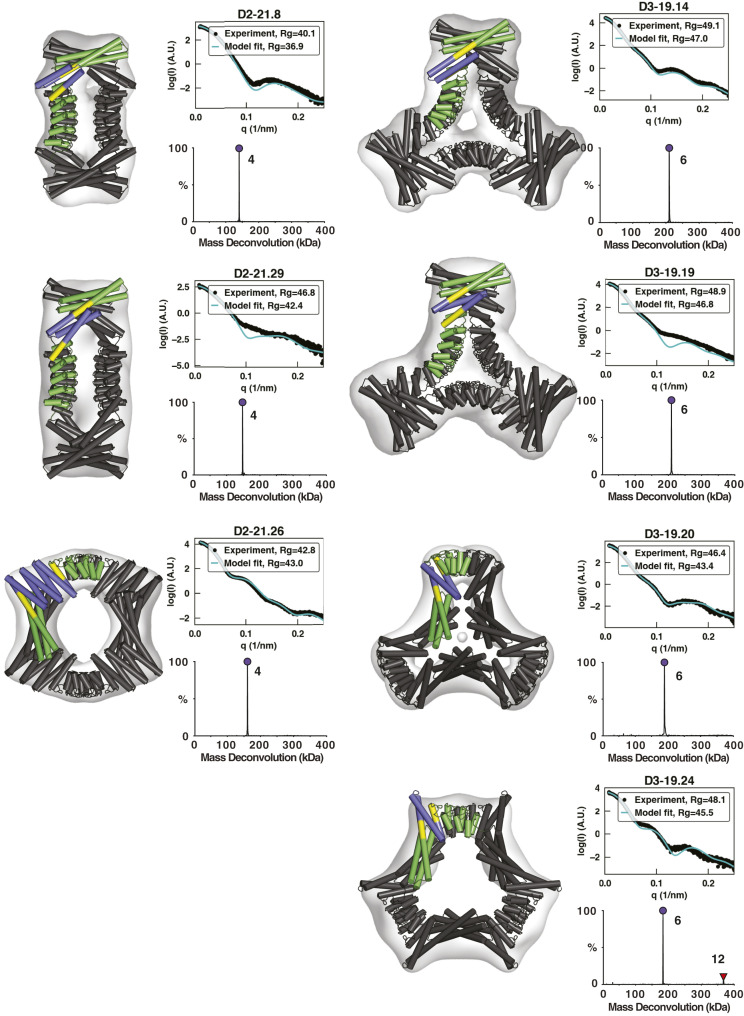
Fig. 3.
Characterization of second-round designs. EM, native-MS, and SAXS experiments are consistent with the formation of intended architectures for six designs. Negative-stain 3D reconstructions are overlaid by design models, whose asymmetric units are colored according to the constituent building blocks (N- and C-terminal oligomers, green; DHR, blue; shared alignment, yellow). Native-MS deconvolutions show the relative abundance of the determined masses, and the peaks are labeled with their assigned oligomeric states. SAXS plots compare the theoretical (cyan) and experimental (black) scattering intensities (log scale) as a function of q as well as Rg. Native-MS for D3-19.24 shows a small amount of 12-mer, likely formed through the association of two designed hexamers.
With the exception of designs D2-21.22 and D3-1.5A2, which may exhibit lower stability, negative-stain EM 2D class averages and 3D reconstructions recapitulated the expected shape for all designs that passed both native-MS and SAXS screening. Most designs have a pronounced central cavity that makes their top views readily identifiable in micrographs. An unusual structural aspect of our D3 designs is their subunit connectivity; in natural D3 architectures, subunits related through C3 rotations normally make direct contact, but in ours, they typically do not. It is noteworthy that the successful assemblies were built in large part from building blocks without crystal structures. Of the 10 successful and sequence-independent designs, 2 do not use a single crystal-verified building block (D2-1.4H and D3-19.24) and another 7 use only one crystal-verified building block out of three (SI Appendix, Table S2).
Cryo-EM of Coassembled DARPin and GFP
In prior studies, imaging scaffolds have been constructed through fusion of a DARPin onto existing symmetric protein assemblies via a shared helix or α-helical extension (1, 2). Our constructs differ in that they incorporate a designed ankyrin homodimer, so the ankyrin (or DARPin postinterface grafting) is anchored to the scaffold through both helical fusion and back-to-back dimerization. To evaluate the designs as cryo-EM scaffolds, we grafted the surface binding residues from a GFP-binding DARPin (25) into six constructs: D2-1.1D, D2-1.4H, D3-1.5C, D2-21.8, D2-21.29, and D3-19.20, taking care not to alter the ankyrin homodimerization interface in the base construct (SI Appendix, Fig. S4). As ankyrins are repeat proteins, the DARPin alignment can be shifted up or down by one or more consensus repeats, which we did to create variants. After SEC purification and cryo-EM of GFP–scaffold cocomplexes, a majority of constructs exhibited high levels of preferred orientation or outright aggregation (SI Appendix, Table S3), but D2-1.4H and D2-21.8 derivatives were promising. Second-round design derivative D2-21.8.GFP.v2 (SI Appendix, Fig. S9) was resolved at a low resolution, while D2-1.4H.GFP.v1 was derived from the first-round design scaffold D2-1.4H and was reconstructed at 4.78 Å resolution (Fig. 4), as measured by the standard Fourier shell correlation (FSC) = 0.143 criterion. The core scaffold is more ordered than the GFP, with local resolutions of ∼4.5 and 5.0 Å, respectively, based on CryoSPARC (32, 33) local resolution estimates.
Fig. 4.
Cryo-EM characterization of DARPin-grafted scaffold with GFP. (A) Design model of four GFP (two in front, two behind) bound to the scaffold D2-1.4H.GFP.v1. GFP-binding DARPin “3G124nc” (24) residues are grafted onto the assembly subunit of D2-1.4H while preserving core residues and homo-oligomer interfaces to form the hybrid structure capable of binding GFP. (B) View of the entire assembly colored by local resolution as calculated by CryoSPARC. Resmap calculates the mean resolution at 5.25 Å and median at 5.0 Å. (C) Surface and clipped (through the central chromophore) views of the GFP β-barrel and DARPin region of the scaffold, colored by local resolution. (D) FSC curves calculated by cryoSPARC. Resolution is 4.78 Å based on the 0.143 FSC level.
Cryo-EM of Coassembled DARPin Scaffold with Human Serum Albumin
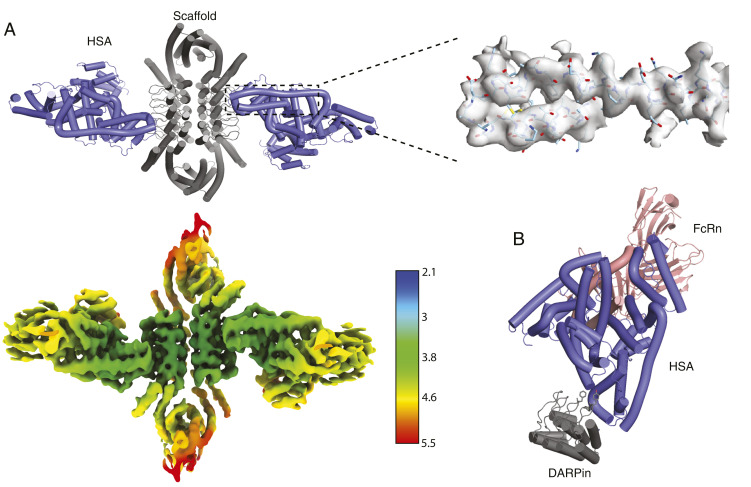
All cryo-EM scaffolding studies based on the GFP-binding DARPin had the benefit of an available cocomplex crystal structure, but in the general use case, cocomplex structural information will be absent, and we sought to assess the feasibility of integrating DARPins in such a scenario. Toward this aim, we incorporated a previously unpublished DARPin sequence targeting human serum albumin (anti-HSA DARPin “C9”; see SI Appendix, Table S3) into second-round scaffolds D2-21.8 and D2-21.29. As before, the DARPin sequence was aligned and grafted onto the assembly scaffold, taking care to only graft surface residues away from the ankyrin homodimer interface so as not to disrupt it. Four sequence-grafted designs were expressed (two scaffolds × two variants where the grafted surface residues are shifted by a repeating unit of the ankyrin). One design based on the D2-21.8 base scaffold proved more soluble than the others (D2-21.8.HSA-C9.v2), and SEC with sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) confirmed binding to HSA, which appeared to be substoichiometric based on SDS-PAGE of column fractions.
The HSA complex was reconstructed by cryo-EM to 4.0 Å resolution with local refinement and 4.53 Å without (Fig. 5), as determined by the FSC = 0.143 cutoff. Consistent with the substoichiometric binding suggested by the SEC and SDS-PAGE results, the HSA binding mode sterically precludes full binding site occupancy, and each face of the dihedral ring has one HSA instead of two. Occupied binding sites occur either directly across or along the diagonal—image classification and refinement focused on the former species. The volumetric map elucidates the binding mode: aromatic residues on the DARPin overlap a hydrophobic patch in domain II of HSA, away from the site where HSA binds to neonatal Fc receptor (FcRn). FcRn binding enables HSA escape from endosomal degradation and long serum half-life, so the noninterference indicates that the HSA DARPin “C9” may serve as a fusion domain for half-life extension. These results demonstrate the feasibility of integrating DARPins and retaining binding, even with the added complexities of maintaining the scaffold homodimer ankyrin interface and lack of prior structural information.
Fig. 5.
Characterization of anti-HSA DARPin assembly in complex with HSA. (A) A cartoon model of scaffold D2-21.8.HSA-C9.v2 and HSA cocomplex docked into a locally refined cryo-EM map obtained from the map below, which is colored by resolution (Å). The density map surrounding the indicated two helices is shown as a transparent gray surface with the model docked. (B) A model of the relative HSA-binding positions of the DARPin and FcRn is built by superposition of this cryo-EM structure (DARPin scaffold + HSA) and an existing crystal structure of HSA with FcRn (Protein Data Bank 4K71). Several hydrophobic DARPin residues involved in binding are shown as sticks.
Discussion
The multifusion assembly design method introduced here remedies two of the long-standing drawbacks to fusion-based assembly construction: the low number of geometric solutions and linker flexibility. Our results demonstrate the feasibility of performing multiple fusions, ranking, and redesign in a single workflow to produce intended architectures with low levels of off-pathway assembly. It is notable that all successful designs in the present work incorporated at least one, usually two, and occasionally even all three design models lacking high-resolution structural validation (SAXS validation only); it is possible that using X-ray crystal structures of the building blocks rather than computational design models might lead to greater success. On the other hand, the success using design models bodes well for future applications of this approach as far more plausible building block structures can be designed than can be solved by X-ray crystallography.
We had hypothesized that DARPins would be more rigidly embedded in our designed assemblies than in the previous designs by Liu et al. (1, 23), as in the present work they form structural components of the assembly and are held in place by both a designed interface and fusion. Indeed, the secondary stabilization provided by the homodimer interface appears to be effective since the DARPin is well-resolved in the overall complex even without multibody refinement. The previous study resolved the GFP target at higher resolution (3.8 Å), but this was after multibody refinement on account of DARPin flexibility relative to the core scaffold. The benefits of our secondary stabilization strategy were likely counteracted by a combination of inherent flexibility of the core scaffold and preferred orientation in cryo-EM; consistent with this, the HSA-scaffold resolution improved appreciably with the use of a tilt series for cryo-EM micrograph collection, so preferred orientation appears to have an impact. The use of tetrahedral or higher-order scaffolds could favor more viewing angles and alleviate the preferred orientation effects. The poorer results with the second-round designs suggest that while shorter closure paths may increase rigidity, they also bias toward smaller designs that are likely more sensitive to destabilization when grafting in the hydrophobic DARPin interface (indeed, D2-1.4H.GFP.v1, which yielded the best GFP density, contains the largest DHR building blocks). Our determination of the DARPin–HSA binding mode using the double tethering method in the absence of prior structure information illustrates the potential of the method for cryo-EM structure determination; using bulkier building blocks and higher-order symmetry nanoparticles should make this approach even more powerful.
Beyond scaffolds for cryo-EM structure determination, the rigid three-body fusion method introduced here is an effective general strategy for constructing protein assemblies. This has been further demonstrated recently by the use of this method to create cyclic, tetrahedral, octahedral, and icosahedral assemblies as described elsewhere (34, 35).
Methods
All experimental methods applied in this article have been published previously. Detailed procedures are described in the SI Appendix, as are further details of the computational design method put forth in the main text. The SI Appendix, Methods describe 1) protein expression with Studier’s autoinduction (36) and purification, 2) oligomeric state determination by online buffer exchange mass spectrometry (37–41), 3) SAXS sample preparation and analytical tools for data collected at the Structurally Integrated Biology for Life Sciences (SIBYLS) Beamline (42–44), 4) negative-stain EM procedures and tools (32, 33, 45, 46), and 5) cryo-EM sample preparation and processing schemes (32, 33, 46–57).
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences (NIGMS) under the NIH under Award Number T32GM008268 to I.V., and the Open Philanthropy Project, HHMI, and NSF grant CHE-1629214 to D.B. NIH grant under award AI150464 provided support to G.J.J. Cryo-EM work under G.J.J. was performed in the Caltech Beckman Institute Resource Center for Transmission Electron Microscopy. We also thank Dr. Songye Chen and Dr. Andrey Malyutin at Caltech for technical assistance. This work was also supported by the National Institute of Allergy and Infectious Diseases (NIAID) grant DP1AI158186, NIH grant HHSN272201700059C, NIGMS grant R01GM120553, a Pew Biomedical Scholars Award, and a Burroughs Wellcome Investigators in the Pathogenesis of Infectious Diseases award to D.V. This work was also supported by NIH grant P41GM128577 to V.H.W. and Swiss National Science Foundation grant 310030_192689 to A.P. In addition, we thank Kathryn Burnett and Greg Hura for SAXS data collection through the SIBYLS mail-in SAXS program at the Advanced Light Source (ALS), a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the Department of Energy, Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies program, supported by the Department of Energy (DOE) Office of Biological and Environmental Research. Additional support comes from the NIH project ALS-ENABLE (grant P30 GM124169) and High-End Instrumentation Grant S10OD018483. A.C. is a recipient of the Human Frontiers Science Program Long Term Fellowship. A.C. and D.D.S. received Washington Research Foundation fellowships. We thank Albumedix for providing high-quality Veltis-grade HSA. I.V. thanks Shane Caldwell (University of Washington) for discussion on SAXS analysis and Vikram Mulligan (Flatiron Institute) for answering questions on RosettaScripts.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. S.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015037118/-/DCSupplemental.
Data Availability
Cryo-EM maps have been deposited in the Electron Microscopy Data Bank (see SI Appendix for details), and the fusion method implementation is available on GitHub (archived with Zenodo at https://zenodo.org/record/4771121) (58). Additional supporting data is deposited with Zenodo (https://zenodo.org/record/4771103) (59) and all other data is included in the article or supporting information.
References
- 1.Liu Y., Huynh D. T., Yeates T. O., A 3.8 Å resolution cryo-EM structure of a small protein bound to an imaging scaffold. Nat. Commun. 10, 1864 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao Q., Weaver S. J., Mock J. Y., Jensen G. J., Fusion of DARPin to aldolase enables visualization of small protein by cryo-EM. Structure 27, 1148–1155.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcandalli J., et al., Induction of potent neutralizing antibody responses by a designed protein nanoparticle vaccine for respiratory syncytial virus. Cell 176, 1420–1431.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ueda G., et al., Tailored design of protein nanoparticle scaffolds for multivalent presentation of viral glycoprotein antigens. eLife 9, e57659 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Padilla J. E., Colovos C., Yeates T. O., Nanohedra: Using symmetry to design self-assembling protein cages, layers, crystals, and filaments. Proc. Natl. Acad. Sci. U.S.A. 98, 2217–2221 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinclair J. C., Davies K. M., Vénien-Bryan C., Noble M. E., Generation of protein lattices by fusing proteins with matching rotational symmetry. Nat. Nanotechnol. 6, 558–562 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Lai Y. T., et al., Structure of a designed protein cage that self-assembles into a highly porous cube. Nat. Chem. 6, 1065–1071 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai Y. T., Cascio D., Yeates T. O., Structure of a 16-nm cage designed by using protein oligomers. Science 336, 1129 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Badieyan S., et al., Symmetry-directed self-assembly of a tetrahedral protein cage mediated by de Novo-designed coiled coils. ChemBioChem 18, 1888–1892 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Sciore A., et al., Flexible, symmetry-directed approach to assembling protein cages. Proc. Natl. Acad. Sci. U.S.A. 113, 8681–8686 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cristie-David A. S., et al., Coiled-coil-mediated assembly of an icosahedral protein cage with extremely high thermal and chemical stability. J. Am. Chem. Soc. 141, 9207–9216 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Cannon K. A., Nguyen V. N., Morgan C., Yeates T. O., Design and characterization of an icosahedral protein cage formed by a double-fusion protein containing three distinct symmetry elements. ACS Synth. Biol. 9, 517–524 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Lai Y. T., Tsai K. L., Sawaya M. R., Asturias F. J., Yeates T. O., Structure and flexibility of nanoscale protein cages designed by symmetric self-assembly. J. Am. Chem. Soc. 135, 7738–7743 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fallas J. A., et al., Computational design of self-assembling cyclic protein homo-oligomers. Nat. Chem. 9, 353–360 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King N. P., et al., Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science 336, 1171–1174 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King N. P., et al., Accurate design of co-assembling multi-component protein nanomaterials. Nature 510, 103–108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonen S., DiMaio F., Gonen T., Baker D., Design of ordered two-dimensional arrays mediated by noncovalent protein-protein interfaces. Science 348, 1365–1368 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Bale J. B., et al., Accurate design of megadalton-scale two-component icosahedral protein complexes. Science 353, 389–394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsia Y., et al., Design of a hyperstable 60-subunit protein dodecahedron. [corrected]. Nature 535, 136–139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salgado E. N., et al., Metal templated design of protein interfaces. Proc. Natl. Acad. Sci. U.S.A. 107, 1827–1832 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kratz P. A., Böttcher B., Nassal M., Native display of complete foreign protein domains on the surface of hepatitis B virus capsids. Proc. Natl. Acad. Sci. U.S.A. 96, 1915–1920 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coscia F., et al., Fusion to a homo-oligomeric scaffold allows cryo-EM analysis of a small protein. Sci. Rep. 6, 30909 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y., Gonen S., Gonen T., Yeates T. O., Near-atomic cryo-EM imaging of a small protein displayed on a designed scaffolding system. Proc. Natl. Acad. Sci. U.S.A. 115, 3362–3367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plückthun A., Designed ankyrin repeat proteins (DARPins): Binding proteins for research, diagnostics, and therapy. Annu. Rev. Pharmacol. Toxicol. 55, 489–511 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Hansen S., et al., Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity. Sci. Rep. 7, 16292 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Batyuk A., Wu Y., Honegger A., Heberling M. M., Plückthun A., DARPin-based crystallization chaperones exploit molecular geometry as a screening dimension in protein crystallography. J. Mol. Biol. 428, 1574–1588 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Wu Y., et al., Rigidly connected multispecific artificial binders with adjustable geometries. Sci. Rep. 7, 11217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watson R. P., et al., Spontaneous self-assembly of engineered armadillo repeat protein fragments into a folded structure. Structure 22, 985–995 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Doyle L., et al., Rational design of α-helical tandem repeat proteins with closed architectures. Nature 528, 585–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michel E., Plückthun A., Zerbe O., Peptide-guided assembly of repeat protein fragments. Angew. Chem. Int. Ed. Engl. 57, 4576–4579 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Correnti C. E., et al., Engineering and functionalization of large circular tandem repeat protein nanoparticles. Nat. Struct. Mol. Biol. 27, 342–350 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Punjani A., Zhang H., Fleet D. J., Non-uniform refinement: Adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Hsia Y., et al., Design of multi-scale protein complexes by hierarchical building block fusion. Nat. Commun. 12, 2294 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Divine R., et al., Designed proteins assemble antibodies into modular nanocages. Science 372, 6537 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Studier F. W., Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005). [DOI] [PubMed] [Google Scholar]
- 37.VanAernum Z. L., et al., Rapid online buffer exchange for screening of proteins, protein complexes and cell lysates by native mass spectrometry. Nat. Protoc. 15, 1132–1157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van de Waterbeemd M., et al., High-fidelity mass analysis unveils heterogeneity in intact ribosomal particles. Nat. Methods 14, 283–286 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Fort K. L., et al., Expanding the structural analysis capabilities on an Orbitrap-based mass spectrometer for large macromolecular complexes. Analyst (Lond.) 143, 100–105 (2017). [DOI] [PubMed] [Google Scholar]
- 40.VanAernum Z. L., et al., Surface-induced dissociation of noncovalent protein complexes in an extended mass range orbitrap mass spectrometer. Anal. Chem. 91, 3611–3618 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marty M. T., et al., Bayesian deconvolution of mass and ion mobility spectra: From binary interactions to polydisperse ensembles. Anal. Chem. 87, 4370–4376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rambo R. P., Tainer J. A., Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneidman-Duhovny D., Hammel M., Tainer J. A., Sali A., Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys. J. 105, 962–974 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneidman-Duhovny D., Hammel M., Tainer J. A., Sali A., FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 44, W424–W429 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grant T., Rohou A., Grigorieff N., cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suloway C., et al., Automated molecular microscopy: The new leginon system. J. Struct. Biol. 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Zheng S. Q., et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grant T., Grigorieff N., Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife 4, e06980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zivanov J., et al., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asarnow D., Palovcak E., Cheng Y., UCSF pyem v0.5. Zenodo (2019). 10.5281/zenodo.3576630. Accessed 1 June 2020. [DOI]
- 53.Terwilliger T. C., Sobolev O. V., Afonine P. V., Adams P. D., Automated map sharpening by maximization of detail and connectivity. Acta Crystallogr. D Struct. Biol. 74, 545–559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pettersen E. F., et al., UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tegunov D., Cramer P., Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vulovic I., et al., Generation of ordered protein assemblies using rigid three-body fusion. Zenodo. https://zenodo.org/record/4771121. Deposited 18 May 2021. [DOI] [PMC free article] [PubMed]
- 59.Vulovic I., et al., Generation of ordered protein assemblies using rigid three-body fusion. Zenodo. https://zenodo.org/record/4771103. Deposited 18 May 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps have been deposited in the Electron Microscopy Data Bank (see SI Appendix for details), and the fusion method implementation is available on GitHub (archived with Zenodo at https://zenodo.org/record/4771121) (58). Additional supporting data is deposited with Zenodo (https://zenodo.org/record/4771103) (59) and all other data is included in the article or supporting information.