Significance
The lymphoid tissue inducer (LTi)-like cell is a specialized innate lymphocyte that produces cytokines that promote intestinal epithelial barrier integrity and protective immunity against pathogenic bacteria. Here, we report that the spatial positioning of LTi-like cells within intestinal mucosa enables them to receive signals that control their activity. Deletion of the LTi-like cell chemokine receptor CXCR5 displaced LTi-like cells from their natural niche in gut mucosa and enhanced their capacity to produce cytokines upon activation. LTi-like cell hyperresponsiveness in Cxcr5-deficient mice strongly improved gut barrier integrity and protected mice during infection with the human opportunistic pathogen Clostridium difficile. Our findings demonstrate that the specialized positioning of LTi-like cells within the gut mucosa controls their activity and impacts protective immunity.
Keywords: mucosal immunity, innate lymphoid cells, lymphoid tissue, intestine, CXCR5
Abstract
Lymphoid tissue inducer (LTi)-like cells are tissue resident innate lymphocytes that rapidly secrete cytokines that promote gut epithelial integrity and protect against extracellular bacterial infections.Here, we report that the retention of LTi-like cells in conventional solitary intestinal lymphoid tissue (SILT) is essential for controlling LTi-like cell function and is maintained by expression of the chemokine receptor CXCR5. Deletion of Cxcr5 functionally unleashed LTi-like cells in a cell intrinsic manner, leading to uncontrolled IL-17 and IL-22 production. The elevated production of IL-22 in Cxcr5-deficient mice improved gut barrier integrity and protected mice during infection with the opportunistic pathogen Clostridium difficile. Interestingly, Cxcr5−/− mice developed LTi-like cell aggregates that were displaced from their typical niche at the intestinal crypt, and LTi-like cell hyperresponsiveness was associated with the local formation of this unconventional SILT. Thus, LTi-like cell positioning within mucosa controls their activity via niche-specific signals that temper cytokine production during homeostasis.
Lymphoid tissue inducer (LTi)-like cells belong to a family of tissue resident innate lymphocytes that lack rearranged antigen-specific receptors and act as a first line of defense at barrier tissues. LTi-like cells, along with other group 3 innate lymphoid cells (ILC3), maintain intestinal homeostasis by producing the cytokines IL-22 and IL-17A, which promote gut epithelial cell proliferation, anti-microbial peptide production, and tight junction protein abundance (1, 2). The conditioning of epithelial cells by these cytokines contributes to balanced interactions between the host and commensal microbiota under steady-state conditions, and LTi-like cell-derived IL-22 promotes barrier integrity and protective immunity during infection with the enteric pathogenic bacteria (3).
In addition to providing effector functions, LTi-like cells and their fetal LTi counterparts are required for early steps in lymphoid tissue development. Fetal LTi induce lymph node and Peyer’s patch development during gestation by activating lymphoid tissue organizer cells at primordial lymphoid organs with lymphotoxin (LT)-α1β2 (4–6). Similarly, LTi-like cells are required for the postnatal development of cryptopatches, small lymphoid aggregates in the intestine that have the potential to mature into isolated lymphoid follicles (ILF) in response to signals from microbes (7, 8). In line with their roles in lymphoid tissue organogenesis and maturation, LTi-like cells in adult mouse intestines preferentially localize in solitary intestinal lymphoid tissue (SILT). The microenvironments of these highly specialized niches are expected to support and regulate LTi-like cells; however, their impact on LTi-like cell behavior has not been fully explored.
LTi-like cells express multiple G protein–coupled receptors that facilitate their migration in tissue (9–12). Among these, CXCR5 has a predominant role in the migration of LTi to developing lymphoid structures, with Cxcr5−/− mice exhibiting defects in lymph node and Peyer’s patch development (13). Mice deficient in CXCR5 or its ligand CXCL13 also have delayed cryptopatch development and fail to convert cryptopatches to mature ILF because of impaired recruitment of B cells to these structures (14–16). Dendritic cells (DCs) have been shown to be a local source of CXCL13 in SILT (16) and thus likely retain B cells and LTi-like cells at these structures under homeostatic conditions via the CXCL13–CXCR5 signaling axis. The retention of LTi-like cells in SILT is expected to bring these cells in close proximity to activating and inhibitory signals provided by specialized myeloid cells, neurons that express the vasoactive intestinal peptide (VIP), and lymphocyte populations localized at these sites (17–20). However, the impact of CXCR5 on functions of LTi-like cells beyond those associated with lymphoid tissue maintenance and development remains unknown.
In the current study, we show that CXCR5 expression regulates LTi-like cell function. Deletion of Cxcr5 led to increased numbers of LTi-like cells in the small intestine (SI) and enhanced their ability to produce IL-17A and IL-22. Cxcr5 regulated LTi-like cells via a cell-intrinsic mechanism that did not involve direct suppression by CXCL13. Heightened LTi-like cell activity in Cxcr5-deficient mice was associated with the development of abnormal LTi-like cell aggregates in the SI that were localized in villus lamina propria instead of at the intestinal crypt base. Importantly, augmented production of IL-22 in Cxcr5−/− mice was protective during acute infection with the opportunistic pathogen Clostridium difficile. These data reveal that CXCR5-dependent migration can control innate type 3 immunity by altering the niche of LTi-like cells in intestinal lamina propria.
Results
LTi-Like Cells Are Numerically Dysregulated in Cxcr5−/− Mice.
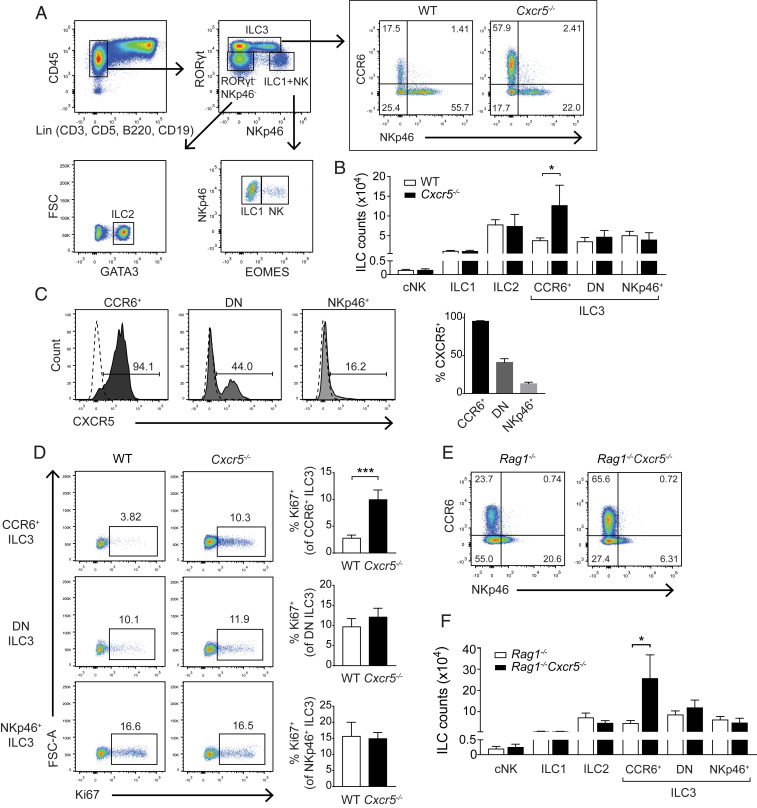
CXCR5 facilitates the migration of fetal and adult LTi-like cells to primordial lymphoid tissues; the mobilization of ILC3 to these sites is necessary for lymphoid tissue maturation during embryonic and postnatal development. To further understand the roles of CXCR5 in ILC3, we examined the innate lymphocyte compartment in adult Cxcr5−/− mice. While the absolute numbers of conventional natural killer cells (cNK), ILC1, and ILC2 were comparable in the SIs of Cxcr5−/− and wild-type (WT) mice, significantly more CCR6+ LTi-like cells were present in Cxcr5−/− mice (Fig. 1 A and B). Numbers of other ILC3 subtypes (NKp46+ and CCR6−NKp46− double negative [DN] ILC3) were unaltered by Cxcr5 deletion (Fig. 1B). Consistent with this differential representation of ILC3 subsets in its absence, CXCR5 was expressed by almost all CCR6+ LTi-like cells, about half of DN ILC3 and less than 20% of NKp46+ ILC3 in WT mice (Fig. 1C).
Fig. 1.
LTi-like cells are numerically dysregulated in Cxcr5−/− mice. (A) Gating strategy used to identify cNK, ILC1, ILC2, and ILC3 populations in SIs of 8- to 12-wk-old mice. (B) Total counts of cNK, ILC1, ILC2, and ILC3 subsets in WT and Cxcr5−/− SIs (n = 4). (C) Frequencies of LTi-like (CCR6+), DN, and NKp46+ ILC3 that express CXCR5 (n = 3). Solid-filled histograms indicate staining of WT cells, and dashed lines indicate staining of Cxcr5−/− cells. (D) Frequencies of Ki67+ LTi-like (CCR6+), DN, and NKp46+ ILC3 populations in SIs of WT and Cxcr5−/− mice (n = 4). (E) Representative plots showing LTi-like (CCR6+), DN, and NKp46+ ILC3 in Cxcr5−/−Rag1−/− and Rag1−/− SIs. Plots were pregated on CD45+Lin−RORγt+ lymphocytes. (F) Total counts of cNK, ILC1, ILC2, and ILC3 in Cxcr5−/−Rag1−/− and Rag1−/− SIs (n = 4). Bar graphs show mean ± SEM. *P < 0.05 and ***P < 0.001 (unpaired Student’s t test). Data shown are representative of three independent experiments (A and B) or two independent experiments (D–F).
LTi-like cells are typically sustained by a low rate of cell division in tissue. We next tested whether LTi-like cells proliferated more in Cxcr5−/− mice than in WT mice. Higher percentages of LTi-like cells expressed the proliferation marker Ki67 in Cxcr5−/− intestines than in those of WT controls at steady state (Fig. 1D), suggesting that the increased abundance of LTi-like cells in Cxcr5−/− mice was caused by enhanced cell proliferation within the gut. DN and NKp46+ ILC3 expressed Ki67 at comparable frequencies in WT and Cxcr5−/− mice.
In addition to being expressed by ILC3, CXCR5 is expressed by T follicular helper (Tfh) cells and B cells. To test whether LTi-like cells in Cxcr5−/− mice were influenced by altered localization of adaptive lymphocytes, Rag1-deficient mice were examined. Similar to Cxcr5−/− mice that contain B cells and T cells, Cxcr5−/− mice lacking Rag1 had higher frequencies of LTi-like cells than did Cxcr5-sufficient Rag1−/− controls (Fig. 1 E and F). Thus, LTi-like cell dysregulation in Cxcr5-deficient mice is independent of adaptive lymphocytes, indicating that CXCR5 expression in the innate compartment regulates the abundance of LTi-like cells in the intestine.
CXCR5 Regulates Cytokine Production in ILC3.
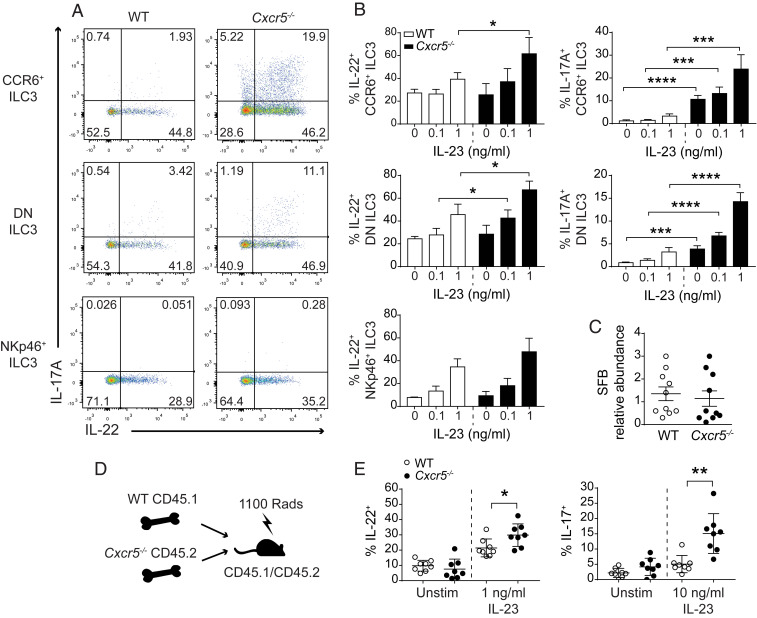
We next tested whether Cxcr5 deficiency affected the effector functions of LTi-like cells. ILC3 produce IL-22 and IL-17A, cytokines that have potent effects on intestinal epithelial cell function. Upon stimulation with IL-23, a greater proportion of Cxcr5−/− LTi-like and DN ILC3 produced IL-22 and IL-17A than did their WT counterparts (Fig. 2 A and B and SI Appendix, Fig. S1 A and B). Cxcr5-deficient and WT NKp46+ ILC3 (relatively few of which express CXCR5) made equivalent amounts of IL-22. As previously reported, NKp46+ ILC3 did not contribute to the IL-17A response (21, 22). The use of cohoused animals in these experiments suggested that LTi-like cell hyperresponsiveness in Cxcr5−/− mice was not caused by major shifts in microbiota composition. Indeed, the relative abundance of segmented filamentous bacteria (SFB), which are associated with heightened ILC3 activation (23), was comparable in feces collected from WT and Cxcr5−/− mice (Fig. 2C). To formally test whether CXCR5 expression was required by LTi-like cells in a cell-intrinsic or -extrinsic manner, mixed bone marrow chimeras were generated. A greater percentage of Cxcr5−/− LTi-like cells produced IL-22 and IL-17A in response to IL-23 than did WT LTi-like cells within the same host (Fig. 2 D and E), indicating that CXCR5 expression regulates IL-17A and IL-22 production in LTi-like cells in a cell-intrinsic manner.
Fig. 2.
CXCR5 regulates cytokine production in ILC3. Representative flow cytometry plots (A) and bar graphs (B) of IL-17 and IL-22 production in CCR6+, DN, and NKp46+ ILC3 stimulated ex vivo for 3 h, with increasing doses of IL-23 as indicated (n = 4). Flow cytometry plots show ILC3 stimulated with 1 ng/mL IL-23. (C) qPCR analysis showing the relative abundance of SFB in feces collected from WT and Cxcr5−/− mice (n = 10). (D) Schematic representation of mixed bone marrow chimera construction. (E) Frequency of IL-17– and IL-22–producing LTi-like cells isolated from the SIs of mixed bone marrow chimeras and stimulated in vitro (n = 8). Bar graphs show mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 (unpaired Student’s t test). Data shown are representative of three independent experiments (A and B) or are pooled from two independent experiments (C and E).
Gene Expression Profiling of Cxcr5−/− LTi-Like Cells.
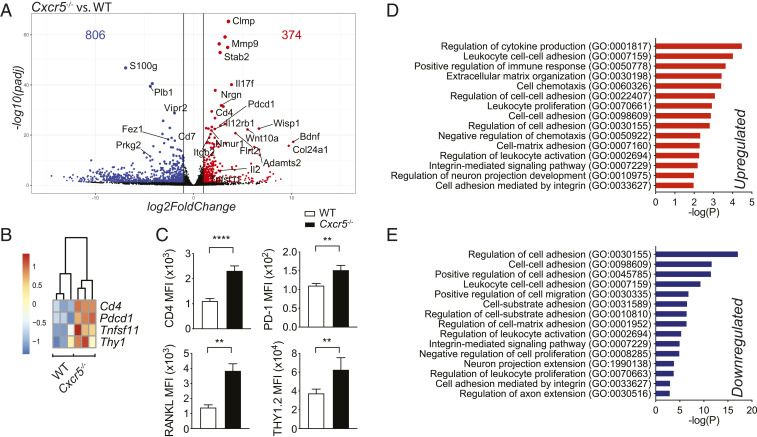
To further examine how Cxcr5 deficiency impacts ILC3, RNA sequencing (RNA-seq) analysis was performed on CCR6+ LTi-like cells sort purified from Cxcr5−/− and WT intestines. A total of 374 genes were up-regulated, and 806 were down-regulated in Cxcr5−/− cells, compared to WT cells (Fig. 3A). Notably, the cytokine-encoding genes Il17f and Il2 and the cytokine receptor subunit Il12rb1 were more highly expressed in Cxcr5−/− LTi-like cells than in WT cells (Fig. 3A). IL12R-β1 is a cytokine receptor chain shared by IL-23R and IL-12R (24), suggesting that LTi-like cell hyperresponsivity to IL-23 in Cxcr5−/− mice may be partially caused by enhanced expression of its receptor. LTi-like cells have been shown to interact with neurons in cryptopatches, and a set of genes associated with neuronal signaling (Nrgn, Bdnf, and the neuropeptide receptors Nmur1 and Vipr2) were differentially expressed in Cxcr5−/− and WT LTi-like cells, suggesting that cross-talk between ILC and enteric neurons may be altered in Cxcr5−/− mice (Fig. 3A). Cd4 and Thy1, genes associated with LTi-like cell identity, and Tnfsf11 and Pdcd1, genes expected to regulate cytokine production, were enriched in Cxcr5−/− LTi-like cells (Fig. 3 A and B). Confirming the RNA-seq data, CD4, THY1.2, PD-1, and RANKL proteins were more abundant in Cxcr5−/− cells than in control cells (Fig. 3C).
Fig. 3.
Gene expression profiling of Cxcr5−/− LTi-like cells. (A) Volcano plot showing differentially expressed genes (P value adjusted < 0.01 and more than twofold change in Cxcr5−/− versus WT LTi-like cells). (B) Heat maps showing Cd4, Pdcd1, Tnfsf11, and Thy1 expression in WT and Cxcr5−/− mice (n = 3). (C) Antibody staining for CD4, PD-1, RANKL, and Thy1.2 in WT and Cxcr5−/− mice (n = 3 to 4). (D and E) Gene ontology (GO) analysis of up-regulated and down-regulated differently expressed genes in Cxcr5−/− LTi-like cells compared to WT cells. Bar graphs show mean ± SEM. **P < 0.01 and ****P < 0.0001 (unpaired Student’s t test). Data shown are representative of two independent experiments (C).
Genes associated with cell adhesion (Stab2, Itgb2, Clmp, and Flrt2) and modulation of the extracellular matrix (Mmp9, Adamts2, and Col24a1) were differentially expressed in Cxcr5−/− LTi-like cells and WT controls (Fig. 3A), suggesting that Cxcr5 deficiency alters how LTi-like cells interact with other cell types and components of the extracellular matrix. Indeed, pathway enrichment analysis revealed that genes involved in the regulation of cell adhesion, organization of the extracellular matrix, and chemotaxis were among the most differentially expressed groups of genes (Fig. 3 D and E). Several pathways associated with the regulation of cell adhesion were listed as both significantly up- and down-regulated based on distinct sets of differentially expressed genes, highlighting a drastic shift in cell adhesion properties. Pathways involved in cytokine production and proliferation were also differentially expressed, consistent with the phenotypes observed in Cxcr5−/− LTi-like cells.
Cxcr5−/− Mice Exhibit Disordered LTi-Like Cell Organization Associated with Heightened Functional Output.
We reasoned that altered LTi-like cell responsiveness in Cxcr5−/− mice could be caused by mechanisms that were either dependent or independent of chemotaxis. To test whether CXCR5 signaling could directly suppress LTi-like cell function, WT cells were stimulated in the presence of the CXCR5 ligand CXCL13. CXCL13 treatment did not suppress IL-22 or IL-17A production in LTi-like cells (SI Appendix, Fig. S2 A and B), but rather, a trend toward increased IL-22 production in cells cultured with the highest doses of CXCL13 was observed (SI Appendix, Fig. S2A). These results indicated that CXCR5 signaling does not suppress cytokine production in LTi-like cells directly and instead implicated CXCR5-dependent cell migration in the suppression of LTi-like cell function.
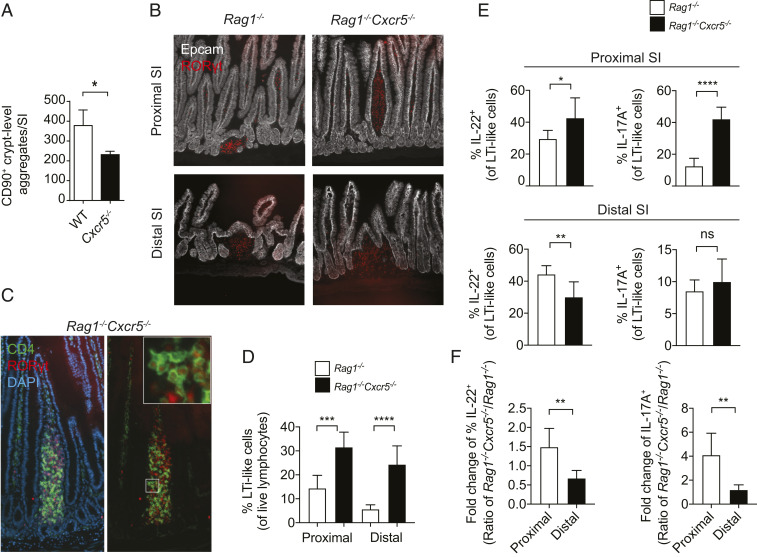
During fetal development, Peyer’s patch organogenesis is dependent on the migration of CXCR5+ fetal LTi. However, Cxcr5−/− neonates and WT controls had comparable numbers of small intestinal LTi at postnatal day 0 (SI Appendix, Fig. S3 A and B), indicating that the expansion of CCR6+ ILC3 occurred during later stages of life. In adult mice, LTi-like cells primarily reside in SILT, a spectrum of lymphoid organs that span immature cryptopatches and mature ILF (25). SILT was previously observed in adult Cxcr5−/− mice, although these structures were delayed in the timing of their development, and they failed to undergo the transition from cryptopatch to ILF (14). Though Cxcr5−/− mice bred in our facility also developed CD90+ (THY1+) aggregates at the crypt base by adulthood, as determined by whole-mount staining, we found fewer of these structures in Cxcr5−/− mice than in WT controls (Fig. 4A). To confirm LTi-like cell identity in these crypt lymphoid aggregates, sections of SI from Cxcr5−/−Rag1−/− mice were stained with CD4, a marker of a subset of LTi-like cells. Larger LTi-like cell-containing SILT was observed in the distal SIs of Cxcr5−/−Rag1−/− mice compared to Rag1−/− controls (SI Appendix, Fig. S4A), consistent with previous findings in mice lacking either Cxcr5 or Cxcl13 (14, 16).
Fig. 4.
Cxcr5−/− mice exhibit disordered LTi-like cell organization. (A) Crypt-level CD90+ (THY1+) aggregates quantified by whole-mount staining of SIs (n = 3). (B) RORγt+ aggregates in proximal and distal sections of Cxcr5−/−Rag1−/− and Rag1−/− SIs. (C) CD4 and RORγt staining in a villus lymphoid aggregate in a Cxcr5−/−Rag1−/− proximal SI. (D) Frequency of LTi-like cells in the proximal and distal SI (n = 6). (E) Frequency of IL-22– and IL-17A–producing, LTi-like cells isolated from the proximal (Top) and distal (Bottom) intestine and stimulated with 1 ng/mL IL-23 (n = 6). (F) Fold change of cytokine production from data shown in E. Bar graphs show mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 (unpaired Student’s t test). ns, not significant. Data are pooled from two independent experiments (D–F).
SILT organization was further examined in Rag1−/− mice to eliminate the effects of B cell localization on the morphology of these structures. In addition to the development of SILT at the level of the crypt base, Cxcr5−/−Rag1−/− mice also developed atypical aggregates of RORγt+ cells that extended up the crypt–villus axis (Fig. 4B and SI Appendix, Fig. S4B). LTi-like cells were contained within these RORγt+ cell aggregates, as determined by the expression of CD4 (Fig. 4C). In addition to containing LTi-like cells, atypical SILT clusters also contained CD11c+ DCs, similar to those seen in conventional clusters (SI Appendix, Fig. S4C) (16). Villus lymphoid structures were frequently found in the proximal region of Cxcr5−/−Rag1−/− SIs but rarely in distal regions (Fig. 4B), indicating that CXCR5 expression is required for restricting SILT development to the crypt base in the proximal SI.
To test whether heightened LTi-like cell activity in Cxcr5-deficient mice was associated with altered localization along the crypt–villus axis, lamina propria cells from the proximal and distal SI were functionally assayed. Frequencies of LTi-like cells were elevated in both intestinal regions in Rag1−/−Cxcr5−/− mice, compared to Rag1−/− control mice (Fig. 4D). However, heightened sensitivity to IL-23 was only observed in Cxcr5-deficient LTi-like cells isolated from the proximal but not distal SI (Fig. 4 E and F). Thus, on a per-cell basis, enhanced LTi-like cell function in Cxcr5−/− mice correlated with intestinal regions where lymphoid aggregates were observed in the villus lamina propria, while the total contribution of LTi-like cell-derived cytokines was enhanced in all areas of the SI because of an increased frequency of LTi-like cells in both the proximal and distal regions (SI Appendix, Fig. S5 A and B).
Although we reasoned that altered localization in the tissue could potentially enhance the ability of LTi-like cells to access microbial metabolites that promote their activation, treatment with broad-spectrum antibiotics did not affect the Cxcr5−/− LTi-like cell phenotype. Cxcr5−/− mice treated with a mixture of VNAM (a mixture of vancomycin, neomycin, amoxicillin, and metranidazole) for 4 wk in drinking water still harbored more LTi-like cells, many with a cytokine hyperresponsive phenotype, than did VNAM-treated WT mice (SI Appendix, Fig. S6 A and B). Moreover, heightened surface expression of CD4, PD-1, and RANKL previously observed in Cxcr5−/− LTi-like cells were still observed after VNAM treatment (SI Appendix, Fig. S6C). Thus, although enhanced LTi-like cell activation in Cxcr5−/− mice correlated with altered cell localization within the tissue, this heightened activity was not due to increased proximity to signals that are dependent on bacteria.
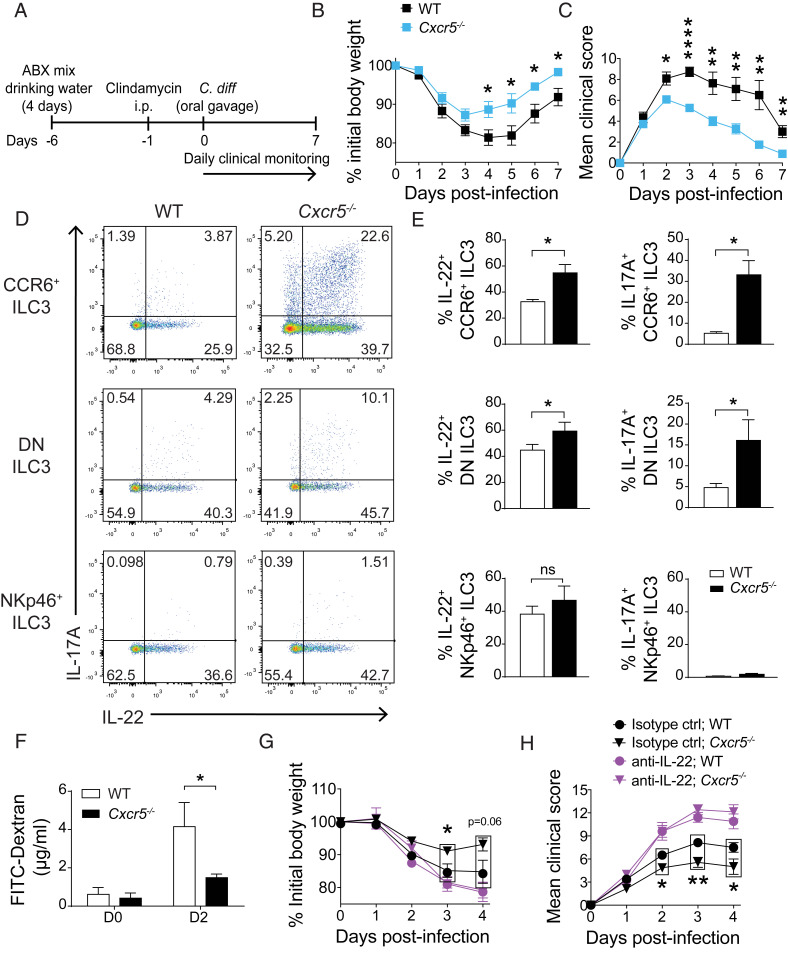
Cxcr5-Deficient Mice Are Protected during Acute Infection with C. difficile.
To test the relevance of CXCR5 deficiency in the setting of bacterial infection, we used an acute model of intestinal infection with the enteric pathogen C. difficile. IL-22 has been shown to be protective during infection with C. difficile (26–29), and CD4+ LTi-like ILC3 are the major source of IL-22 in mesenteric lymph nodes during early infection (26). Cxcr5−/− and WT mice were treated with antibiotics to induce dysbiosis prior to oral inoculation with C. difficile, as previously described (30) (Fig. 5A). Cxcr5−/− mice were protected against infection, with body weight loss blunted (Fig. 5B) and clinical scores less severe (Fig. 5C) than those noted for WT controls. Enhanced protection correlated with greater frequencies of IL-17A and IL-22 producing LTi-like and DN ILC3 in Cxcr5-deficient mice than in WT mice, while comparable proportions of NKp46+ ILC3 from Cxcr5−/− and WT mice produced IL-22 (Fig. 5 D and E).
Fig. 5.
Cxcr5-deficient mice are protected during acute infection with C. difficile. (A) Schematic showing timeline of C. difficile infection protocol. Body weight variation (B) and clinical score (C) of WT and Cxcr5−/− mice infected with C. difficile (n = 11). (D and E) Frequencies of cytokine-producing, LTi-like, DN, and NKp46+ ILC3 that were isolated from SIs of C. difficile-infected WT and Cxcr5−/− mice on day 2 of infection and stimulated in vitro with 1 ng/mL IL-23 (n = 4). (F) FITC-dextran in serum of infected mice at day 2 of infection. Mice were bled 4 h after FITC-dextran administration by oral gavage (n = 4). Percentage of initial body weight (G) and clinical scores (H) of infected WT and Cxcr5−/− mice treated with either anti-IL-22–blocking antibody or isotype control (n = 6 to 8). Bar graphs show mean ± SEM. *P < 0.05, **P < 0.01, and ****P < 0.0001 (unpaired Student’s t test). ns, not significant. Data shown are pooled from two independent experiments (B, C, G, and H) or are representative of two (F) or three independent experiments (D–E).
IL-22 is necessary for the maintenance of intestinal epithelial integrity, and we next measured intestinal permeability at day 2 of infection by administering FITC-dextran by oral gavage. Cxcr5−/− mice had less FITC-dextran in the serum than did WT mice, indicating that barrier integrity was preserved in the absence of CXCR5 (Fig. 5F). To determine whether the protective phenotype was due to heightened amounts of IL-22, an IL-22–blocking antibody was administered to infected mice (Fig. 5 G and H). Neutralization of IL-22 during early stages of infection completely abrogated protection conferred by lack of CXCR5 based on both body weight loss (Fig. 5G) and clinical score (Fig. 5H), indicating that protection against C. difficile observed in Cxcr5−/− mice is mediated by early secretion of IL-22.
Discussion
Here, we show that SI LTi-like cells are functionally regulated by their expression of the chemokine receptor CXCR5. LTi-like cells were increased in number in Cxcr5−/− mice, and a larger proportion produced IL-17 and IL-22 upon stimulation with IL-23 than did their WT counterparts. Tfh cells and B cells, which also express CXCR5, were not responsible for the hyperresponsive LTi-like cell phenotype, as demonstrated by experiments using Rag1-deficient mice. We confirmed that LTi-like cell cytokine production was regulated by CXCR5 expression in a cell-intrinsic manner, which implicated CXCR5-dependent migration in the regulation of LTi-like cell activity.
Among ILC, LTi-like cells are distinct in that they are primarily found in the intestinal SILT in adult mice. SILT structures develop prior to weaning age and are located at the crypt base (31, 32). Consistent with previous findings, we observed that Cxcr5−/− mice developed SILT at the crypt base by adulthood and that ileal SILT in these mice were enlarged. Additionally, we found that CXCR5 deficiency led to the development of atypical LTi-like cell clusters that were localized in villus lamina propria. These villus LTi-like cell clusters were primarily observed in the proximal SI; accordingly, we found hyperfunctional LTi-like cells in the proximal but not distal SI, linking the enhanced effector functions of Cxcr5−/− LTi-like cells with their displacement from the normal crypt niche. In contrast to these regional differences in cytokine secretion, LTi-like cell numbers were increased in both the proximal and distal SI of CXCR5-deficient mice, indicating that distinct mechanisms control LTi-like cell abundance and cytokine secretion.
Previous studies have revealed several ways in which LTi-like cells undergo cross-talk with other cells resident in SILT. LTi-like cell activation is regulated by RANKL-dependent inhibitory cell–cell interactions and by diet-induced secretion of the neural peptide VIP by enteric neurons that innervate SILT (17, 19, 20). IL-23–producing myeloid cells have also been shown to localize to these structures (18). Recently, LTi-like cells were shown to program a specialized DC population in SILT (CIA-DC) to secrete IL-22 binding protein (IL-22BP) via direct cell–contact interactions that activate LT-β receptor (LT-βR) signaling (33). Therefore, the appropriate microlocalization of LTi-like cells within tissue restricts IL-22 availability through at least one additional indirect mechanism, which promotes the sequestration of secreted IL-22. Our data show that CD11c+ DCs were present in the atypical Cxcr5−/− SILT clusters. The nature of the interactions between DCs and LTi-like cells in these villus structures remains to be determined. LTi have also been shown to activate epithelial cells via LT-α1β2 to secrete CCL20 (34, 35), a chemokine that is expressed by SILT and is required for their maturation (7, 36). In line with altered interactions between LTi-like cells and their environment, gene ontology enrichment analysis revealed that several groups of genes associated with cellular adhesion and extracellular matrix organization were among the most differentially expressed in Cxcr5−/− and WT LTi-like cells. Additional studies will be necessary to determine how specific cellular interactions between LTi-like cells and other SILT components are controlled by expression of CXCR5 and other G protein–coupled receptors.
The cytokine IL-22 is required for maintenance of the epithelial barrier integrity and for protective immunity against extracellular bacteria in the intestine. Here, we tested the physiologic relevance of increased total IL-22–producing ILC3 in Cxcr5−/− mice using an acute model of infection with C. difficile. In this model, innate cells and IL-22 mediate protective immunity (26–29). ILC3 are major innate producers of IL-22 and have been shown to be required for protection during C. difficile infection in studies using ILC3-deficient RorcCreAhrfl/fl mice (27). In the present study, we show that Cxcr5−/− mice exhibit improved barrier integrity during C. difficile infection. Accordingly, C. difficile-infected Cxcr5−/− mice were protected from weight loss and had better clinical scores than did WT mice; moreover, this protection was dependent on IL-22. These data suggest that interventions that enhance ILC3 activity may be suitable strategies to prevent acute and recurring C. difficile infections.
Taken together, our data reveal that CXCR5 controls the effector functions of LTi-like cells via cell localization–dependent mechanisms. ILC3 have been reported to exit SILT during colitis induced by anti-CD40 agonistic antibodies (37), and whether this cell behavior enhances tissue inflammation will require further evaluation. Importantly, other ILC3 subtypes do not preferentially localize in SILT and thus are expected to be regulated through unique mechanisms. Indeed, CXCR6-dependent cell localization in tissue has been implicated in the production of IL-22 by NKp46+ ILC3 but not LTi-like cells (38). Defining how the localization of different ILC3 subsets in tissue impacts their effector activities will be crucial for understanding how these cell types collectively regulate gut epithelial function. Future studies will be necessary to understand how the balance of signals from different chemokine receptors expressed by LTi-like cells and other ILCs control their activity under physiologic and pathologic settings.
Materials and Methods
Animals.
C57BL/6J, Cxcr5tm1lipp/J (Cxcr5−/−) (13), B6.sjl-PtprcaPepcb/BoyJ (CD45.1), and Rag1tm1mom/J (Rag1−/−) mice were obtained from The Jackson Laboratory and were maintained in specific pathogen-free facilities at Washington University in Saint Louis. Experiments were conducted with 8- to 14-wk-old mice that were cohoused from birth. All mice were backcrossed to the C57BL/6 background. Experiments were carried out in accordance with the Animal Studies Committee of Washington University in Saint Louis.
VNAM Treatment.
For microbiota depletion, 12-wk-old mice were provided with a mixture of broad-spectrum antibiotics (VNAM: 0.5 mg/mL vanconcomycin, 1 mg/mL neomycin, 1 mg/mL amoxicillin, and 1 mg/mL metronidazole) in drinking water for 4 wk. The VNAM mixture was provided ad libitum and was replaced weekly to ensure proper antimicrobial activity.
Mixed Bone Marrow Chimeras.
CD45.1/CD45.2 recipient mice were irradiated once with 1,100 Rads and within 24 h were injected intravenously with 6 × 106 donor bone marrow cells. Donor cells consisted of Cxcr5−/− and CD45.1 cells mixed at a 1:1 ratio. Mice were analyzed 8 wk after transfer.
Tissue Dissociation to Obtain Single-Cell Suspensions.
SIs were flushed to remove luminal contents, opened lengthwise, and gently agitated for 20 min in a solution of Hank’s balanced salt solution (HBSS) (Gibco) containing 1 M Hepes (Corning), 10% BCS (HyClone), and 0.5 M EDTA (Corning). Intestines were vortexed before subjected to a second round of gentle agitation and vortexing in fresh HBSS/BCS/EDTA buffer to remove remaining epithelial cells. The tissue was then rinsed with HBSS prior to digestion with 100 U/mL Collagenase IV (Sigma) in complete RPMI for 40 min at 37 °C under agitation. Digests were filtered through 100-μm mesh, washed in HBSS, and subjected to density gradient centrifugation using 40 and 70% Percoll (GE Healthcare) solutions. For experiments that involved CXCR5 surface staining, intestines were digested in 1 mg/µL collagenase D (Roche) for 25 min to prevent proteolytic cleavage of cell surface CXCR5. For experiments comparing different segments of the intestine, cells were isolated from 10 cm of the most proximal or most distal SI regions. In experiments with neonatal tissue, SIs were flushed, cut open lengthwise, gently agitated in HBSS/BCS/EDTA solution for 20 min, and then vortexed for 10 s. SIs were then minced and digested with 100 U/mL Collagenase IV in complete RPMI for 40 min at 37 °C.
Cell Culture and Stimulation.
Cells were cultured at 37 °C under 5% CO2 atmosphere in complete RPMI (RPMI 1640 [Sigma] supplemented with sodium pyruvate [Corning], kanamycin sulfate [Gibco], glutamine [Gibco], nonessential amino acids [Corning], β-mercaptoethanol [Sigma], and 10% of BCS [HyClone]). Cells were stimulated immediately after isolation with 10, 1, or 0.1 ng/mL recombinant IL-23 (Biolegend) as indicated. All stimulations were carried out for 3 h in the presence of GolgiPlug (BD Biosciences).
Flow Cytometry.
Single-cell suspensions were incubated with CD16/32 antibodies for 10 min and then stained with antibodies for cells surface markers for 20 min at 4 °C. For detection of cell surface RANKL or CXCR5, cells were cultured in complete RPMI at 37 °C for 2 h prior to staining for 20 min at room temperature. Dead cells were excluded using a Live/Dead Fixable Cell Stain Kit (Thermo Fisher Scientific). Intracellular proteins were stained using the following Fixation/Permeabilization Kits: Cytoperm/Cytofix (BD Biosciences) or FOXP3/Transcription Factor staining buffer set (eBioscience). Cells were run on an FACSCanto II or LSRFortessa (BD Biosciences) and were analyzed using FlowJo (FlowJo LLC). Cells counts were conducted with counting beads (eBioscience). ILCs were identified as live, CD3e−CD5−B220−CD19− lymphocyte-sized cells that were either GATA3hi (ILC2), RORγt+ (ILC3), or RORγt−GATA3intNKp46+ (ILC1+cNK). cNK cells were distinguished from ILC1 based on the expression of Eomes. For intracellular cytokine staining experiments, ILC3 were identified as CD3e−CD5−B220−CD19−CD90.2hiCD45int live lymphocytes.
RNA-seq Analysis.
LTi-like cells were sort purified from SIs of Cxcr5−/− and WT controls using an FACSAria II (BD Biosciences) as live, lymphocyte-sized, and singlet CD90.2hi CD45int CCR6+ cells that were negative for CD3e, CD5, CD19, CD11b, CD11c, NK1.1, Gr-1, B220, KLRG1, and NKp46. Total RNA was extracted from sorted samples using an RNeasy Micro Kit (Qiagen). Total RNA integrity was determined using Agilent Bioanalyzer or 4200 TapeStation. Library preparation was performed with 10 ng total RNA with a Bioanalyzer RNA integrity number score greater than 8. Double standard complementary DNA (cDNA) was prepared using the SMARTer Ultra Low RNA kit for Illumina Sequencing (Takara-Clontech) per manufacturer’s protocol. cDNA was fragmented using a Covaris E220 sonicator using peak incident power 18, duty factor 20%, and cycles per burst 50 for 120 s. cDNA was blunt ended, had an A base added to the 3′ ends, and then had Illumina sequencing adapters ligated to the ends. Ligated fragments were then amplified for 12 to 15 cycles using primers, incorporating unique dual index tags. Fragments were sequenced on an Illumina HiSeq. Basecalls and demultiplexing were performed with Illumina’s bcl2fastq software and a custom Python demultiplexing program with a maximum of one mismatch in the indexing read. RNA-seq reads were then aligned to the Ensembl release 76 primary assembly with Spliced Transcripts Alignment to a Reference version 2.5.1a. Gene counts were derived from the number of uniquely aligned unambiguous reads by Subread:featureCount version 1.4.6-p5 (39). Aligned gene counts were processed using the DESeq2 package with R (version 3.5) (40). Genes with fewer than 10 counts among all of the samples were excluded. Following differential expression analysis, differentially expressed protein-coding genes were filtered by false discovery rate < 0.05 and log2 Fold Change > 1. Gene ontology pathway enrichment analysis of separate lists of statistically significant up- and down-regulated genes was conducted using Metascape (41).
Whole-Mount Staining.
Total numbers of crypt-level SILT in Cxcr5−/− and cohoused WT controls were quantified by whole-mount staining of SIs, as previously described (42).
Immunofluorescence Staining.
SIs were fixed in 4% PFA for 2 h, washed in phosphate-buffered saline (PBS) overnight, treated with 30% sucrose for 24 h prior to being frozen in optimal cutting temperature compound, and sectioned. Then, 8-μm frozen sections of SI were incubated with anti-CD16/32 antibodies, rat serum, and mouse serum before incubation with RORγt, CD4, CD11c, and CD326 antibodies. Sections were stained with DAPI to visualize nuclei. Images were taken using a Zeiss Axio Imager M2.
C. difficile Infection.
The C. difficile VPI 10463 strain was cultivated in brain heart infusion blood agar plates at 37 °C in jars using the AnaeroGen Oxoid system (Thermo Fisher Scientific) to generate an anaerobic atmosphere. Infections were performed by pretreating mice with a mixture of antibiotics (0.4 mg/mL kanamycin, 0.035 mg/mL gentamicin, 0.035 mg/mL colistin, 0.215 mg/mL metronidazole, and 0.045 mg/mL vancomycin [all from Sigma]) in drinking water for 4 d, followed by a single intraperitoneal (i.p.) injection of clindamycin (10 mg/kg, Sigma) to exacerbate dysbiosis. One day after treatment with antibiotics, mice were infected with 108 colony forming units of C. difficile by oral gavage and monitored daily for changes in body weight and clinical score, as previously described (43). To measure intestinal permeability, mice were gavaged with a 250 mg/kg suspension of 70 kDa FITC-dextran (Sigma) on day 2 of infection. Around 4 h after gavage, mice were bled, and the FITC-dextran concentration in serum was measured by fluorescence analysis in a Multimode Microplate Reader (Synergy H1) at 485/528 nm excitation/emission. A standard curve was prepared with serial dilutions of FITC-dextran in PBS.
IL-22 Neutralization In Vivo.
C. difficile–infected mice were injected i.p. with an anti-mouse, IL-22–neutralizing antibody (6 mg/kg; clone 8E11; Genentech) or an equivalent amount of isotype control IgG2a (BioXcell) on days 1 and 3 postinfection. Mice were clinically evaluated until the fourth day of infection.
SFB Quantification.
The relative abundance of SFB was determined by qPCR using SYBR Green Super Mix (Bio-Rad) and the specific primers SFB_736F, 5′-GAC GCT GAG GCA TGA GAG CAT-3′, and SFB_844R, 5′-GAC GGC ACG GAT TGT TAT TCA-3′. DNA used for quantification was extracted from fecal pellets of Cxcr5−/− and WT cohoused controls using a QIAamp fast DNA stool mini kit (Qiagen). For relative quantification, the ΔCT values were compared to the averages of values obtained for the WT group. The mass of bacterial DNA used per reaction was normalized using primers for Eubacteria (EUB_1275: 5′-CCA TTG TAG CAC GTG TGT AGCC-3′ and EUB_1114: 5′-CGG CAA CGA GCG CAA CCC-3′).
Statistical Analysis.
Data were analyzed with PRISM 7 (GraphPad Software) using unpaired Student’s t tests.
Supplementary Material
Acknowledgments
We would like to thank Erica Lantelme, Dorian Brinja, and Pascaline Akitani for assistance using cell sorters. We also thank Simone Brioschi for providing technical expertise in immunofluorescence staining of tissue sections and Genentech for kindly providing the anti-IL-22 antibody used in this study. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by National Cancer Institute (NCI) Cancer Center Support Grant P30 CA91842 to the Siteman Cancer Center and by Institute for Clinical and Translational Sciences (ICTS) Clinical and Translational Sciences Award (CTSA) Grant UL1TR002345 from the National Center for Research Resources (NCRR), a component of the NIH, and NIH Roadmap for Medical Research. This publication is solely our responsibility and does not necessarily represent the official view of NCRR or NIH. This work was funded by NIH Grants U01 AI095542 (to M. Colonna), R01 DE025884 (to M. Colonna), R01 DK124699 (to M. Colonna), K99 DK118110 (to J.K.B.), F30 DK127540-01 (to V.P.), and T32 DK 77653-28 (to V.P.). M. Colonna receives research support from Pfizer.
Footnotes
Competing interest statement: M. Colonna receives research support from Pfizer.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101668118/-/DCSupplemental.
Data Availability
RNA-seq data were deposited in the Gene Expression Omnibus repository under the accession code GSE173144. All other data are included in the manuscript and/or supporting information.
References
- 1.Eyerich K., Dimartino V., Cavani A., IL-17 and IL-22 in immunity: Driving protection and pathology. Eur. J. Immunol. 47, 607–614 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Valeri M., Raffatellu M., Cytokines IL-17 and IL-22 in the host response to infection. Pathog. Dis. 74, ftw111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonnenberg G. F., Monticelli L. A., Elloso M. M., Fouser L. A., Artis D., CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity 34, 122–134 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fütterer A., Mink K., Luz A., Kosco-Vilbois M. H., Pfeffer K., The lymphotoxin beta receptor controls organogenesis and affinity maturation in peripheral lymphoid tissues. Immunity 9, 59–70 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Koni P. A., et al., Distinct roles in lymphoid organogenesis for lymphotoxins alpha and beta revealed in lymphotoxin beta-deficient mice. Immunity 6, 491–500 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Rennert P. D., Browning J. L., Mebius R., Mackay F., Hochman P. S., Surface lymphotoxin alpha/beta complex is required for the development of peripheral lymphoid organs. J. Exp. Med. 184, 1999–2006 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouskra D., et al., Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456, 507–510 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Pabst O., et al., Adaptation of solitary intestinal lymphoid tissue in response to microbiota and chemokine receptor CCR7 signaling. J. Immunol. 177, 6824–6832 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Emgård J., et al., Oxysterol sensing through the receptor GPR183 promotes the lymphoid-tissue-inducing function of innate lymphoid cells and colonic inflammation. Immunity 48, 120–132.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mackley E. C., et al., CCR7-dependent trafficking of RORγ+ ILCs creates a unique microenvironment within mucosal draining lymph nodes. Nat. Commun. 6, 5862 (2015). Correction in: Nat. Commun. 7, 11186 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lügering A., et al., CCR6 identifies lymphoid tissue inducer cells within cryptopatches. Clin. Exp. Immunol. 160, 440–449 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim M. H., Taparowsky E. J., Kim C. H., Retinoic acid differentially regulates the migration of innate lymphoid cell subsets to the gut. Immunity 43, 107–119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Förster R., et al., A putative chemokine receptor, BLR1, directs B cell migration to defined lymphoid organs and specific anatomic compartments of the spleen. Cell 87, 1037–1047 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Velaga S., et al., Chemokine receptor CXCR5 supports solitary intestinal lymphoid tissue formation, B cell homing, and induction of intestinal IgA responses. J. Immunol. 182, 2610–2619 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Ansel K. M., et al., A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature 406, 309–314 (2000). [DOI] [PubMed] [Google Scholar]
- 16.McDonald K. G., McDonough J. S., Dieckgraefe B. K., Newberry R. D., Dendritic cells produce CXCL13 and participate in the development of murine small intestine lymphoid tissues. Am. J. Pathol. 176, 2367–2377 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bando J. K., et al., The tumor necrosis factor superfamily member RANKL suppresses effector cytokine production in group 3 innate lymphoid cells. Immunity 48, 1208–1219.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savage A. K., Liang H.-E., Locksley R. M., The development of steady-state activation hubs between adult LTi ILC3s and primed macrophages in small intestine. J. Immunol. 199, 1912–1922 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seillet C., et al., The neuropeptide VIP confers anticipatory mucosal immunity by regulating ILC3 activity. Nat. Immunol. 21, 168–177 (2020). Correction in: Nat. Immunol. 21, 354 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Talbot J., et al., Feeding-dependent VIP neuron-ILC3 circuit regulates the intestinal barrier. Nature 579, 575–580 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cella M., et al., A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature 457, 722–725 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luci C., et al., Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat. Immunol. 10, 75–82 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Ivanov I. I., et al., Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parham C., et al., A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J. Immunol. 168, 5699–5708 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Eberl G., Inducible lymphoid tissues in the adult gut: Recapitulation of a fetal developmental pathway? Nat. Rev. Immunol. 5, 413–420 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Abt M. C., et al., Innate immune defenses mediated by two ILC subsets are critical for protection against acute Clostridium difficile infection. Cell Host Microbe 18, 27–37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fachi J. L., et al., Acetate coordinates neutrophil and ILC3 responses against C. difficile through FFAR2. J. Exp. Med. 217, jem.20190489 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hasegawa M., et al., Interleukin-22 regulates the complement system to promote resistance against pathobionts after pathogen-induced intestinal damage. Immunity 41, 620–632 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagao-Kitamoto H., et al., Interleukin-22-mediated host glycosylation prevents Clostridioides difficile infection by modulating the metabolic activity of the gut microbiota. Nat. Med. 26, 608–617 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X., et al., A mouse model of Clostridium difficile-associated disease. Gastroenterology 135, 1984–1992 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Finke D., Induction of intestinal lymphoid tissue formation by intrinsic and extrinsic signals. Semin. Immunopathol. 31, 151–169 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Kanamori Y., et al., Identification of novel lymphoid tissues in murine intestinal mucosa where clusters of c-kit+ IL-7R+ Thy1+ lympho-hemopoietic progenitors develop. J. Exp. Med. 184, 1449–1459 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guendel F., et al., Group 3 innate lymphoid cells program a distinct subset of IL-22BP-producing dendritic cells demarcating solitary intestinal lymphoid tissues. Immunity 53, 1015–1032.e8 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Rumbo M., Sierro F., Debard N., Kraehenbuhl J.-P., Finke D., Lymphotoxin beta receptor signaling induces the chemokine CCL20 in intestinal epithelium. Gastroenterology 127, 213–223 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Obata Y., et al., Epithelial-stromal interaction via Notch signaling is essential for the full maturation of gut-associated lymphoid tissues. EMBO Rep. 15, 1297–1304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDonald K. G., et al., CC chemokine receptor 6 expression by B lymphocytes is essential for the development of isolated lymphoid follicles. Am. J. Pathol. 170, 1229–1240 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearson C., et al., ILC3 GM-CSF production and mobilisation orchestrate acute intestinal inflammation. eLife 5, e10066 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satoh-Takayama N., et al., The chemokine receptor CXCR6 controls the functional topography of interleukin-22 producing intestinal innate lymphoid cells. Immunity 41, 776–788 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Dobin A., et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y., et al., Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDonald K. G., Newberry R. D., Whole-mount techniques to evaluate subepithelial cellular populations in the adult mouse intestine. Biotechniques 43, 50, 52, 54 passim (2007). [DOI] [PubMed] [Google Scholar]
- 43.Li Y., et al., Adenosine A2A receptor activation reduces recurrence and mortality from Clostridium difficile infection in mice following vancomycin treatment. BMC Infect. Dis. 12, 342 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data were deposited in the Gene Expression Omnibus repository under the accession code GSE173144. All other data are included in the manuscript and/or supporting information.