Significance
Fusobacterium nucleatum interacts with many oral microbes and has the ability to spread to the placenta and amniotic fluid, promoting preterm birth. Yet, the molecular mechanisms underlying polymicrobial interactions, termed coaggregation, by Fusobacteria are poorly understood. Here, we revealed that the two-component signal transduction system CarRS regulates expression of genes encoding lysine utilization factors (e.g., KamA) and the coaggregation factor RadD. Extracellular lysine blocks RadD-mediated coaggregation by binding to RadD. Significantly, mutants lacking KamA or CarR (which up-regulates RadD) are attenuated in virulence in a preterm birth model, while mutants devoid of RadD or CarS (which down-regulates RadD) exhibit increased virulence. Our findings unveiled a molecular linkage between coaggregation and lysine metabolism via CarRS-mediated gene regulation that modulates bacterial virulence.
Keywords: Fusobacterium nucleatum, coaggregation, preterm birth, two-component transduction system, virulence
Abstract
A gram-negative colonizer of the oral cavity, Fusobacterium nucleatum not only interacts with many pathogens in the oral microbiome but also has the ability to spread to extraoral sites including placenta and amniotic fluid, promoting preterm birth. To date, however, the molecular mechanism of interspecies interactions—termed coaggregation—by F. nucleatum and how coaggregation affects bacterial virulence remain poorly defined. Here, we employed genome-wide transposon mutagenesis to uncover fusobacterial coaggregation factors, revealing the intertwined function of a two-component signal transduction system (TCS), named CarRS, and a lysine metabolic pathway in regulating the critical coaggregation factor RadD. Transcriptome analysis shows that CarR modulates a large regulon including radD and lysine metabolic genes, such as kamA and kamD, the expression of which are highly up-regulated in the ΔcarR mutant. Significantly, the native culture medium of ΔkamA or ΔkamD mutants builds up abundant amounts of free lysine, which blocks fusobacterial coaggregation with streptococci. Our demonstration that lysine-conjugated beads trap RadD from the membrane lysates suggests that lysine utilizes RadD as its receptor to act as a metabolic inhibitor of coaggregation. Lastly, using a mouse model of preterm birth, we show that fusobacterial virulence is significantly attenuated with the ΔkamA and ΔcarR mutants, in contrast to the enhanced virulence phenotype observed upon diminishing RadD (ΔradD or ΔcarS mutant). Evidently, F. nucleatum employs the TCS CarRS and environmental lysine to modulate RadD-mediated interspecies interaction, virulence, and nutrient acquisition to thrive in the adverse environment of oral biofilms and extraoral sites.
Fusobacterium nucleatum is a gram-negative, obligate anaerobe often found in the human oral cavity. Gene-based imaging of the supragingival plaque revealed that Fusobacteria reside in an annulus between the base and the peripheral layers, in close proximity with Leptotrichia, Capnocytophaga, and Corynebacterium, especially Corynebacterium matruchotii and Corynebacterium durum (1). Remarkably, F. nucleatum has the ability to spread to extraoral sites, such as the placenta, where it has been associated with preterm birth, and the gastrointestinal tract, where it is believed to promote colorectal carcinogenesis (2–5). Classic microbiological experiments have shown that F. nucleatum has the capacity to interact with a wide variety of bacterial species in the oral cavity, including the early colonizers Streptococcus and Actinomyces and numerous late colonizers Veillonella, Actinobacillus, Bacteroides, Capnocytophaga, and Porphyromonas (6, 7) as well as the fungal pathogen Candida albicans (8, 9), thereby ascribed to a “bridging” capability for this organism in the developing oral biofilm (10). Intriguingly, F. nucleatum binds the early oral colonizer Streptococcus sanguis, forming a specific morphological unit termed corncob, that is, a filamentous organism surrounded by the adhering cocci (11). The congregation of F. nucleatum with gram-positive early colonizers, including Streptococcus and Actinomyces, can be inhibited by L-arginine, whereas coaggregation of F. nucleatum to primarily gram-negative late colonizers, such as Actinobacillus and Porphyromonas, is inhibitable by D-galactose (6), indicating the role of specific receptor–ligand interactions in the intermicrobial interactions involving F. nucleatum.
The molecular entities of galactose/arginine-inhibitable adhesins involved in fusobacterial coaggregation began to emerge a decade ago. Pioneering work by Shi and colleagues revealed an outer membrane adhesion protein called RadD, which is required for interspecies interaction and targeted by L-arginine (12). The same group identified another arginine-inhibitable adhesin, Aid1 (Adherence Inducing Determinant 1), which is interestingly dependent on RadD (13). Another outer membrane adhesin named Fap2 (14) was subsequently shown to mediate the galactose-inhibitable binding of F. nucleatum to Porphyromonas gingivalis (15). Importantly, the functions of RadD and Fap2 are not limited to bacterial coaggregation. Mutants devoid of radD and fap2 are severely defective in inducing cell death in immortalized T lymphocytes (14), while the fap2 mutant also exhibits significant reduction in binding to red blood cells, embryonic kidney-derived cells, placental tissues, and colorectal cancer cells (15, 16). These latter findings point to potential physiological roles of these adhesins in fusobacterial virulence, the exact mechanisms of which have yet to be investigated.
Notably, while arginine-inhibitable adhesion by F. nucleatum mostly occurs with many gram-positive early colonizers (6), F. nucleatum adherence to some gram-negative oral bacteria, such as Prevotella intermedia, Prevotella nigrescens, and Capnocytophaga ochracea, is inhibited by L-lysine (17, 18). In this context, it is intriguing that F. nucleatum is capable of utilizing a number of amino acids when grown in a chemically defined medium, which include lysine, glutamine, and asparagine but critically not glycine, phenylalanine, and arginine (19). Consistent with this, F. nucleatum ferments lysine; it might share a similar pathway and lysine metabolic enzymes with the lysine-fermenting clostridia (20). To date, however, little has been disclosed about how lysine inhibits F. nucleatum–mediated coaggregation with other oral bacteria or the identity of these enzymes. A major impediment to molecular analysis of Fusobacteria has been the lack of robust genetic investigations for this organism.
To fill this gap, we recently developed a set of convenient genetic tools for F. nucleatum, including markerless gene deletion technology and Tn5 transposon mutagenesis (21). Here, we performed a genome-wide screen with over threefold coverage for isolating F. nucleatum mutants that are defective in coaggregation with streptococci. From a total of 78 independent coaggregation-defective mutants obtained from this screen, we identified 10 distinct genetic loci, with many of the Tn5 insertion mutations mapped to radD and the genes in the lysine degradation pathway. Remarkably, we discovered that a two-component signal transduction system (TCS) that we named CarRS regulates expression of both radD and the lysine degradation genes, which are members of a large regulon controlled by the CarR response regulator and the CarS sensor kinase. Subsequently, our biochemical analysis indicated that RadD might function as a lysine receptor aiding in scavenging extracellular lysine for bacterial utilization, possibly in the planktonic, prebiofilm phase of fusobacterial growth. Importantly, we show that CarS and RadD are each intimately involved in bacterial virulence in a mouse model of preterm birth. Our study reveals a multitude of genetic determinants that are not only required for fusobacterial interactions with other oral bacteria but also an important aspect of fusobacterial pathogenicity relevant to birth defects.
Results
A Genome-Wide Transposon Mutagenesis Screen Search for Genetic Determinants of F. nucleatum Mediating Polymicrobial Interactions.
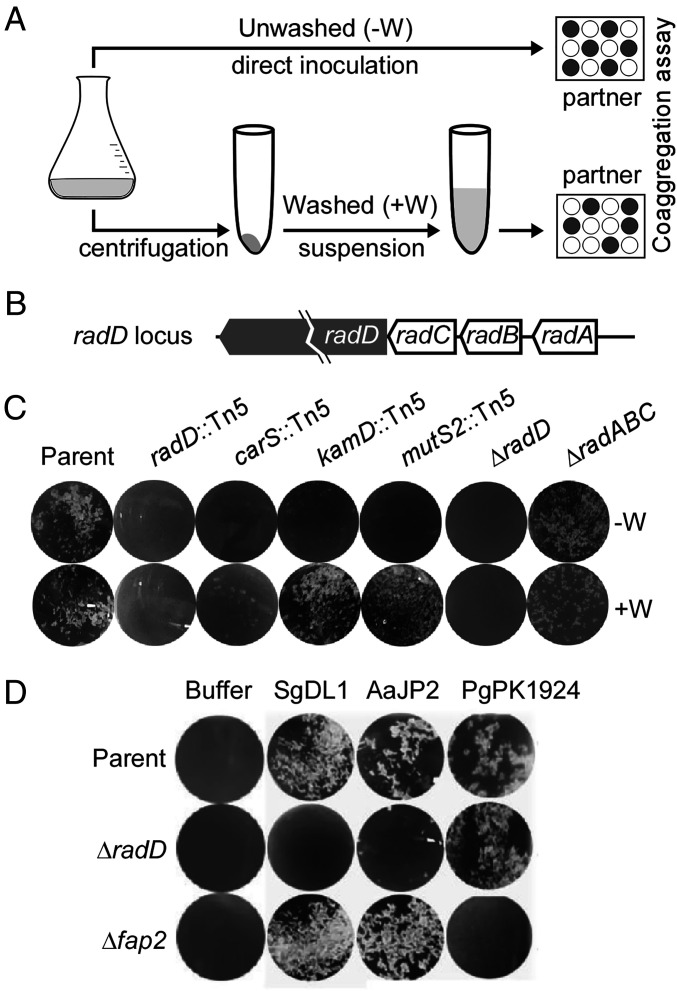
To identify factors that are required for interactions of F. nucleatum with oral bacteria, we performed a high-throughput Tn5 mutagenesis screen for isolating mutants that are defective in coaggregation with the oral bacterium Streptococcus gordonii DL1 (SgDL1). In this screen (Fig. 1 A, Top), S. gordonii cells anaerobically grown in culture were mixed in equal volumes with individual cultures of a library of roughly 7,000 random Tn5 insertion mutants also anaerobically grown in batches of 96-well plates. The resultant coaggregation after a short incubation period was subsequently inspected visually for clump accumulation and clearing of turbidity. This manual screen yielded a set of 78 coaggregation-defective mutants, each of which was validated in follow-up screens to confirm the defect in coaggregation. Next, we mapped the insertion mutations in each of these strains by amplifying the Tn5-targeted loci and DNA sequencing of the amplicons (see Materials and Methods), revealing 27 targeted genes that could be classified into 10 groups based on the nature of encoded proteins and their annotated functions (Table 1). The single most repetitively targeted group represents the radD locus, which is comprised of four genes: radA, radB, radC, and radD in that order (Fig. 1B); note that we hereby named the locus tag HMPREF0397_RS02355 (National Center for Biotechnology Information [NCBI]) as radA. Among our coaggregation-defective mutant library, the radD gene coding for the aforementioned adhesin RadD (12) was independently hit by Tn5 48 times, with insertions in many hot spots, while radB and radC were each targeted by Tn5 just once (Table 1). This clearly indicates the pivotal role of RadD adhesin in coaggregation. Curiously, genes which encode the enzymes for lysine utilization and metabolism, Rnf electron transport complex, vitamin B12 biosynthesis, and coenzyme A biosynthesis were also frequently targeted (Table 1). The remainder of the Tn5-targeted genes code for a histidine kinase, some amino acid transport proteins, outer membrane proteins, protease, and an AMP biosynthesis protein (Table 1).
Fig. 1.
Genome-wide screening of coaggregation-associated factors in F. nucleatum. (A) Presented is a schematic of two coaggregation procedures, one that fusobacterial cells of Tn5 mutant cultures were not washed (−W; Top) and the other washed (+W; Bottom) prior to mixing with coaggregation partners. (B) The Tn5 screen above identified 78 coaggregation-defective mutants with many mutations mapped to the radD locus. (C) Representative coaggregation-defective mutants identified in A were confirmed using the two aforementioned coaggregation procedures. Nonpolar, in-frame deletion mutants devoid of radD or radABC, originated from the parental strain (CW1), were used as reference. (D) Nonpolar, in-frame mutant strains lacking radD or fap2 were examined for coaggregation with S. gordonii DL1 and Aggregatibacter actinomycetemcomitans JP2 and P. gingivalis PK1924.
Table 1.
Coaggregation-defective Tn5 mutants identified by transposon mutagenesis
| Group | Targeted pathway/protein | Targeted gene | Location of Tn5 insertion sites* |
| 1 | radD operon | radD | 10387 (586, 641, 706, 751, 1048, 1383, 1478, 1580, 2062, 2706, 2822, 3167, 3799, 3824, 4082, 4160, 4162, 4925, 6323, 6767, 7172, 7664, 7929, 7957, 7959, 7960, 8655, 8757, 9574, 9595, 9612, 9984) |
| radB | 395 (55) | ||
| radC | 390 (201) | ||
| 2 | Rnf electron transport complex | rnfB | 1158 (222, 806) |
| rnfC | 1326 (220) | ||
| rnfE | 618 (337, 502) | ||
| rnfF | 1020 (619) | ||
| 3 | Histidine kinase | HMPREF0397_RS09120 (carS) | 1338 (143, 586, 761, 784, 793, 969, 1257) |
| 4 | Lysine degradation pathway | kamA | 1279 (791) |
| kamD | 1572 (913, 1528) | ||
| hp | 1017 (143, 376) | ||
| kce | 816 (649) | ||
| kal | 387 (240) | ||
| mutS | 1462 (291, 645) | ||
| 5 | Vitamin B12 biosynthesis | HMPREF0397_RS10070 | 570 (191) |
| cobD | 978 (68, 362) | ||
| cobI | 723 (316) | ||
| cobT | 1065 (120, 615) | ||
| 6 | Coenzyme A biosynthesis | HMPREF0397_RS0349 | 771 (475) |
| HMPREF0397_RS08605 | 510 (458) | ||
| HMPREF0397_RS05915 | 2928 (1402) | ||
| 7 | Amino acid transport | alsT | 1356 (1054) |
| brnQ | 1278 (279) | ||
| 8 | Outer membrane protein | HMPREF0397_RS04065 | 7107 (3267) |
| HMPREF0397_RS04015 | 783 (96) | ||
| 9 | Protease | HMPREF0397_RS01910 | 963 (837) |
| 10 | AMP biosynthesis | purA | 1278 (889) |
The nucleotide length of each gene is shown with the position of Tn5 insertion sites in the targeted gene indicated by numbers in parentheses.
To confirm that the initially identified fusobacterial mutants are indeed defective in coaggregation, we subjected representative candidates to a standard coaggregation assay as previously reported (21), whereby cells of each Tn5 mutant grown anaerobically in overnight cultures were harvested, washed, resuspended in the coaggregation buffer, and mixed together with the oral bacterial partner in equal volumes (Fig. 1 A, Bottom). Strikingly, while the radD::Tn5 and carS::Tn5 mutants were as defective in coaggregation with S. gordonii SgDL1 as a nonpolar deletion mutant devoid of radD (ΔradD), as we previously described (21), the kamD::Tn5 and mutS::Tn5 mutants were coaggregation-proficient comparable to the parental strain, to our surprise (Fig. 1C). In sharp contrast, when we did not wash the same sets of overnight cultures prior to mixing with their partner in the coaggregation assay, each of the above-named mutants exhibited a coaggregation-defective phenotype, exactly as in our initial screen. It is important to note that the triple mutant lacking radA, radB, and radC was able to bind S. gordonii similar to the parental strain, regardless of whether the cultures were washed or not (Fig. 1C). We conclude that the coaggregation defect of the radB::Tn5 and radC::Tn5 mutants is due to the polar effects of the Tn5 transposon insertion element on the transcription of the downstream radD gene, and hence, the observed phenotype is attributed to the loss of expression of radD.
To confirm RadD’s role in fusobacterial interaction with various oral bacterial species, we performed coaggregation assays using the ΔradD mutant with a nonpolar deletion mutant lacking of fap2 as a control. As shown in Fig. 1D, the ΔradD mutant was severely defective in coaggregation with S. gordonii DL1 as well as Aggregatibacter actinomycetemcomitans JP2. By contrast, the ΔradD mutant was able to interact with Porphyromonas gingivalis PK1924, a coaggregation phenomenon that was specifically abolished with the Δfap2 mutant (Fig. 1D), thereby indicating the differential roles of RadD and Fap2 adhesins in interspecies interactions.
Taken together, the above results substantiate that RadD is a major coaggregation factor in F. nucleatum as previously reported (12), and they suggest that the overnight cultures of certain mutants (kamD and mutS) may contain metabolites whose accumulation during bacterial growth inhibits this RadD-mediated coaggregation.
Discovery of the Interconnected Role of a TCS CarRS and RadD Adhesin in Fusobacterial Coaggregation.
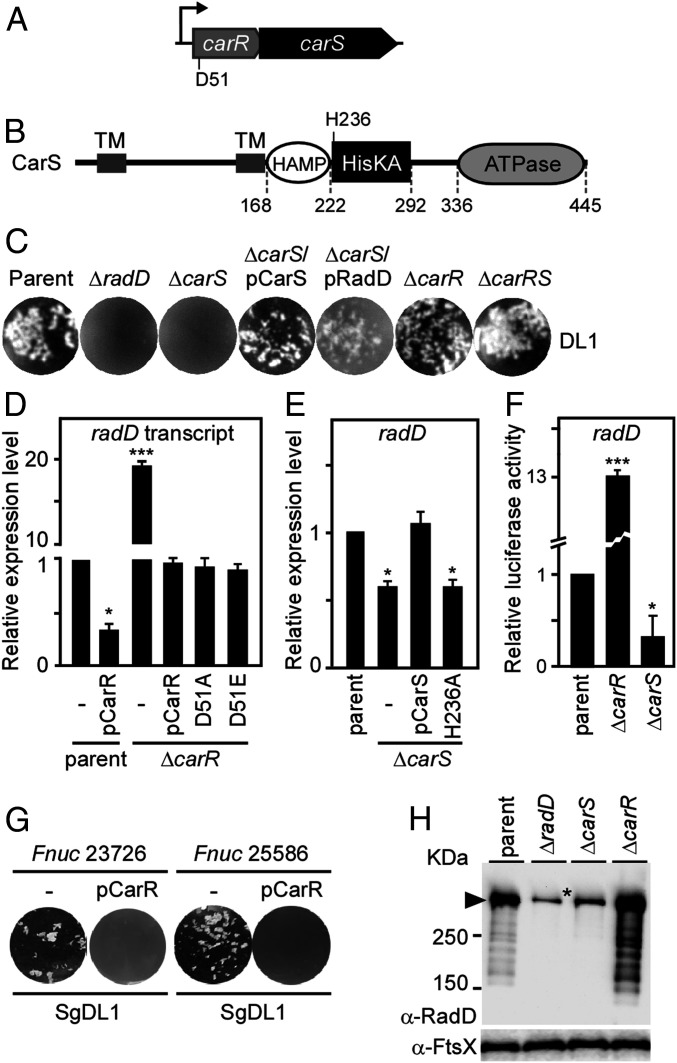
Of the many components uncovered in our screen as candidates required for coaggregation, the role of a sensor kinase was particularly intriguing. Note that HMPREF0397_RS09120, which we named CarS (car for coaggregation regulator), is part of a two-gene locus coding for a putative TCS, designated CarRS, with CarR as the response regulator and CarS the histidine sensor kinase (Fig. 2A). According to protein sequence homology predictions, CarR contains an N-terminal response regulatory domain in which an aspartate (D51) may serve as the site of phosphorylation by CarS, while CarS harbors HAMP (present in histidine kinases, adenylate cyclases, methyl-accepting proteins, and phosphatases and known to be involved in transmission of conformational changes within intradomains), HisKA, and ATPase domains, of which the HisKA domain contains the critical histidine (H236) predicted to participate in a phospho-relay reaction with CarR (Fig. 2B). In addition, the N terminus of CarS is predicted to contain an extracellular receptor domain sandwiched between two obvious transmembrane domains. We next went on to investigate how this kinase may be linked to RadD-mediated coaggregation.
Fig. 2.
Requirement of the two-component system CarRS in coaggregation and radD regulation. (A) Presented is a diagram of the carRS locus in strain ATCC 23726. The response regulator CarR contains a conserved Asp residue (D51). (B) The sensor kinase CarS contains two transmembrane domains (TM) followed by a HAMP domain, histidine kinase domain (HisKA) with the conserved His236 residue, and an ATPase domain. (C) The F. nucleatum parent strain, its isogenic mutants, and rescued strains were examined for coaggregation with S. gordonii DL1. (D and E) Relative expression levels of radD in indicated strains were determined by qRT-PCR with the expression level of radD in the parent strain arbitrarily assigned as 1. Data are presented as the average of three independent experiments performed in triplicate (*P < 0.01; ***P < 0.001; Student’s t test). All real-time RT-PCR values were normalized according to the abundance of 16S rRNA in each sample. (F). A single-copy (chromosomal) luciferase transcriptional fusion to the 3′ end of radD was generated in the parent or its isogenic mutants ΔcarR and ΔcarS. Relative transcript levels of radD were determined by measuring luciferase activity with a Tecan M1000 plate reader. The results are presented as an average of three independent experiments done in triplicate (*P < 0.01; ***P < 0.001; Student’s t test). (G) F. nucleatum ATCC strains 23726 and 25586 and these strains harboring a plasmid expressing carR under the control of the rpsJ promoter were examined for coaggregation with S. gordonii DL1. (H) Whole-cell lysates of fusobacteria grown to the stationary phase were analyzed by immunoblotting using antibodies against RadD (α-RadD), with antibodies against the membrane protein FtsX (α-FtsX) used as control. An asterisk (*) marks a nonspecific band observed in the radD and carS mutants.
Is CarS involved in fusobacterial coaggregation? To address this critical question first, we generated individual nonpolar, in-frame deletion mutants lacking carS, carR, or both and tested the generated mutants in the standard coaggregation assay using the ΔradD mutant as the control. As shown in Fig. 2C, the ΔcarS mutant was clearly defective in coaggregation with SgDL1, and this mutant’s defect was rescued by the ectopic expression of either CarS or RadD, driven by a constitutive promoter. In contrast, the ΔcarR and the ΔcarRS mutants were each coaggregation positive comparable to the parental strain.
Critically, the fact that the ectopic expression of RadD in the ∆carS mutant rescues its coaggregation-negative phenotype leads to the hypothesis that the CarRS system regulates radD expression, specifically that CarS might act as a positive regulator of radD transcription, while CarR might act a repressor since its deletion also rescues the ∆carS defect. To test this hypothesis, we isolated RNA samples from the respective strains and determined the relative level of radD transcripts by RT-PCR with that of the parent strain as the control. Indeed, the deletion of carR led to an appreciable increase in radD transcript level, and the complementation of the mutant by ectopic expression of the wild-type carR (pCarR) or ∆carR mutants with D51 mutated to alanine (A) or glutamate (E) resulted in radD expression at levels similar to that of the parent strain (Fig. 2D). By contrast, deletion of carS diminished the level of the radD transcript (Fig. 2E), while the ectopic expression of wild-type carS but not the carS mutants with H236A mutation in the ∆carS mutant could restore RadD transcription to normal levels in the ∆carS mutant.
To firmly establish the transcriptional regulatory role of CarRS in radD expression, we employed a luciferase reporter assay system, whereby we monitored luciferase activities in the aerobic environment of a microplate reader (22). We generated a single-copy chromosomal luciferase transcriptional fusion of the luciferase reporter gene to the 3′ end of radD (rad::luc), and we measured the luciferase enzymatic activity as the proxy for radD transcriptional activity. Importantly, this luciferase assay confirmed our conclusion made above on the roles of CarS and CarR as the activator and repressor, respectively (Fig. 2F). Furthermore, note that the overexpression of carR in the parent strain causes a significant reduction of the radD transcript as measured by qRT-PCR (Fig. 2D). To further examine the effect of reduced radD expression in Fusobacteria, we introduced the plasmid pCarR into the clinical isolates F. nucleatum ATCC (American Type Culture Collection) 23726 and ATCC 25586 and compared their coaggregation efficiencies. Remarkably, compared to the parental strains, the transformed strains with pCarR failed to mediate bacterial coaggregation with S. gordonii DL1 (Fig. 2G). Clearly, this shows that the repressive effects of CarR on radD transcription is not strain specific.
Next, to confirm whether carRS deletion affects radD expression at the protein level, we monitored the RadD protein in the ΔcarS and ΔcarR mutants by subjecting whole cell lysates to immunoblotting with a polyclonal antibody, which we raised against the N terminus of RadD. As shown in Fig. 2H, the 3,461-amino acid RadD protein was detected as a major band migrating near the 360-KDa marker and many low molecular mass bands, possibly due to degradation. These bands were absent in the ΔradD mutant, although it should be noted that a nonspecific band of size similar to RadD was detected in this mutant. While the nature of this cross-reactivity remains unknown, it is noteworthy that several autotransporter domain-containing proteins homologous to RadD are present in the organism (https://www.biocyc.org). Consistent with RNA measurements reported in Fig. 2 D–F, the ΔcarS mutant diminished the level of RadD protein as compared to the parent, whereas the ΔcarR mutant overproduced RadD protein to levels that resulted in the detection of many antibody-reactive degradation products similar to the parent strain (Fig. 2H). We infer that the expression of the RadD adhesin is tightly regulated in Fusobacteria and that this regulation is mediated by the two-component signal transduction system CarRS, which modulates fusobacterial coaggregation by controlling radD transcription.
Transcriptome Analysis by RNA Sequencing Unveils a Large Regulon Subject to Control by CarR.
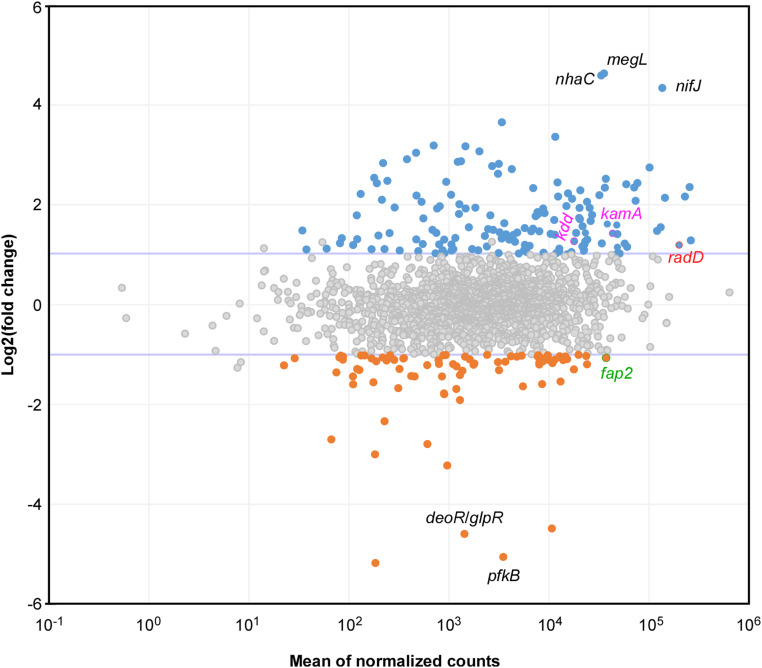
Signal transduction proteins in bacteria often regulate multiple genes and operons. To elucidate whether genes other than radD are modulated by CarR, we performed a comparative genome-wide transcriptome analysis using RNA sequencing (RNA-seq). We isolated total RNA from midlog phase cells of the parental strain and the ΔcarR mutant strain using the TRIzol extraction kit and prepared high-quality, DNA-free RNA suitable for RNA-seq analysis (see Materials and Methods). Roughly 2 μg total RNA from biological triplicates were processed to generate DNA sequence libraries, and six libraries were sequenced using the Illumina platform (see Materials and Methods). The normalized sequence read counts were used to calculate fold changes in specific gene expression and statistical significance, and a comparison was then made between the carR mutant and wild-type transcriptomes using DESeq2 (23) to identify differentially expressed genes.
Strikingly, our data revealed that CarR controls many more genes than RadD and that it acts to not only repress but also activate transcription of distinct sets of genes. When a twofold cutoff (log2 fold change ± 1) was used to compare the RNA-seq reads, 86 genes reached statistical significance for reduced expression in the ΔcarR mutant, while 150 genes reached significance for elevated expression in the absence of CarR (Fig. 3 and SI Appendix, Fig. S1 and Dataset S1). Significant among genes whose expression is diminished in the CarR mutant, that is, CarR-activated genes, are a couple of genes coding hypothetical proteins, pfkB (coding for ribokinase), and a predicted DeoR/GlpR-type transcriptional regulator. That CarR normally up-regulates a transcriptional regulator indicates that some of the genes that are differentially expressed in the ΔcarR mutant are likely not direct targets of CarR. It is noteworthy that fap2, coding for a galactose-inhibitable adhesin shown to be required for coaggregation and placental colonization (15), was down-regulated in the ΔcarR mutant (Fig. 3). As for genes whose expression was elevated in the absence of CarR, that is, CarR-repressed genes, they include megL, nhaC, and nifJ—coding for methionine gamma-lyase, Na+/H+ antiporter family protein, and pyruvate:ferredoxin oxidoreductase, respectively (Fig. 3 and SI Appendix, Fig. S1). Consistent with the qRT-PCR results above, the RNA-seq data showed a greater than twofold up-regulation of radD in the carR mutant; moreover, radB and radC were also up-regulated more than twofold (Dataset S1), consistent with the idea that radB/C/D constitute a single transcriptional unit (Fig. 1B).
Fig. 3.
Transcriptome analysis of the carR mutant by RNA-seq. RNA samples collected from log-phase cultures of the parent strain and its isogenic mutant ∆carR were analyzed by RNA-seq. Differentially transcribed genes in the ∆carR mutant, as compared to the parent strain, are presented in an M/A plot. The purple lines indicate the log2 (fold change) threshold of +1 and −1. Up-regulated and down-regulated genes, based on the threshold, are colored in blue and orange, respectively.
Significantly, the expression of many genes of a predicted lysine metabolic pathway, which was represented in the coaggregation-defective Tn5 mutants described above (Table 1), was also elevated in the ΔcarR mutant. These CarR-repressed genes include kdd, kamA, kamD, hp, kce, atoD, and others (Fig. 3 and Dataset S1). This last piece of data points to a possible link between the lysine metabolic pathway (LMP) and coaggregation in F. nucleatum and the global regulatory role of the two-component system CarRS in this process.
Modulation of Environmental Lysine as a Key Factor in the Genetic and Metabolic Control of Fusobacterial Coaggregation.
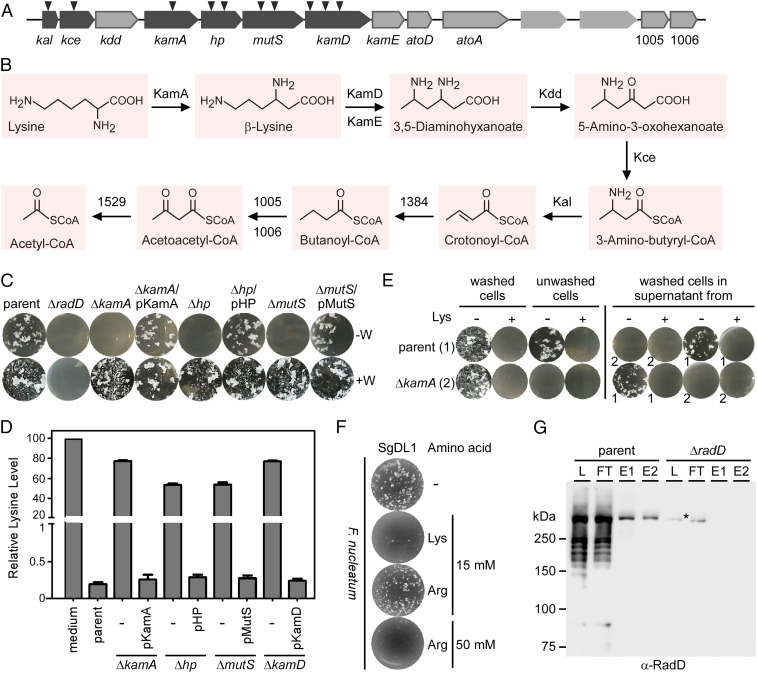
To elucidate the potential connection between lysine metabolism and the coaggregation defect of the Tn5 mutants disrupting the LMP genes (Fig. 4B), we generated nonpolar, in-frame deletion mutants of kamA, hp, mutS, and kamD as they are clustered together in that order (Fig. 4A), and we retested them in the coaggregation assay. When tested as unwashed cell cultures, the ΔkamA, Δhp, and ΔmutS mutants were all defective in coaggregation with SgDL1 similar to that observed with the ΔradD strain (Fig. 4C); this defect of each of these mutants were rescued with plasmids complementing for the respective genes (Fig. 4C). In stark contrast, when the same set of LMP pathway mutant cultures were first washed to free cells from the culture medium prior to mixing with SgDL1, each mutant strain tested proficient in aggregating with SgDL1 at wild-type levels. The results suggest that the culture media of these mutant strains contain one or more substances that mask interspecies interaction.
Fig. 4.
Involvement of an LMP in F. nucleatum coaggregation. (A) Mapping of mutations in the collection of coaggregation-defective mutants reveal many targeted genes in the LMP locus; arrowheads indicate Tn5 insertion sites. (B) A lysine degradation pathway in F. nucleatum strain ATCC 23726 is proposed based on Kyoto Encyclopedia of Genes and Genomes predictions with many factors encoded by the LMP locus in A involved in various steps of lysine degradation. 1384 and 1529 are encoded by HMPREF0397_1384 and HMPREF0397_1529, respectively, which are located elsewhere in the chromosome. (C) Nonpolar, in-frame deletion mutants of representative genes in the LMP locus were generated, and washed (+W) and unwashed (−W) cells of the generated mutants and rescued strains were examined in coaggregation assays with S. gordonii DL1. (D) Culture supernatants of indicated strains were collected, and levels of free lysine in the supernatants were determined by liquid chromatography–mass spectrometry relative to the lysine level in medium, which was arbitrarily assigned a value of 100. The results are presented as the average of three independent experiments. (E, Left) The F. nucleatum parent strain (1) and its isogenic mutant ΔkamA (2) were subjected to coaggregation assays with S. gordonii DL1 using washed and unwashed fusobacterial cells as aforementioned. Lysine was added to the final concentration of 15 mM (+). (Right) Washed cells of the parent and ΔkamA strains were treated with supernatants collected from the parent (1) or ΔkamA (2) cultures prior to mixing with S. gordonii DL1 in the presence or absence of 15 mM lysine. (F) F. nucleatum ATCC 23726 cells were subjected to the coaggregation assay with S. gordonii DL1, as described in Fig. 2C, in the absence or presence of lysine (15 mM) or arginine (15 or 50 mM). (G) RadD was captured from cell-free lysates prepared from the parent strain using L-lysine resin; protein samples obtained from the total lysate (L), flow-through (FT), and eluates (E1 and E2) were analyzed by immunoblotting with antibodies against RadD (α-RadD). The ΔradD mutant strain was used as a control; note that an asterisk marks a nonspecific band in the ΔradD strain.
Considering that lysine is known to inhibit fusobacterial coaggregation with many oral bacterial species (17, 24), we envisioned that it might be the accumulation of lysine in the culture media of the LMP mutant cultures that blocks coaggregation in unwashed cultures. To explore this logical possibility, we first used liquid chromatography–mass spectrometry and directly measured free lysine content in the culture medium from the parent strain and its derivatives harvested after 24 h of cultivation. With the free lysine level in fresh medium arbitrarily assigned to 100%, free lysine was almost completely exhausted in the parental culture spent medium, whereas the ΔkamA, Δhp, ΔmutS, or ΔkamD mutants all retained ∼80% of free lysine in their culture media. Complementing these mutants with plasmids expressing respective genes restored lysine utilization ability to the parental level (Fig. 4D), supporting that the enzymes encoded by each of these genes are required for lysine utilization.
Since lysine blocks RadD-dependent coaggregation in vitro (12, 18, 25, 26), the higher levels of lysine remaining in the culture medium of specific LMP mutants likely contributes to their coaggregation deficiency in situ. Next, we directly verified this hypothesis by adding 15 mM lysine into the coaggregation assay using washed cells of the parent or ΔkamA mutant strain. Indeed, as expected, the added lysine (which did not alter the pH of the medium) abolished coaggregation with each strain (Fig. 4 E, Top and Bottom, circles one and two). By comparison, 50 mM arginine produced a comparable inhibitory affect as that of 15 mM lysine, whereas 15 mM arginine did not have any effect on coaggregation (Fig. 4F). Lastly, the addition of 15 mM lysine also abrogated coaggregation by unwashed parental cells (Fig. 4 E, Top, compare circle three with four from left), and unwashed ΔkamA cells did not aggregate with or without lysine addition, as expected (Fig. 4 E, Bottom, circles three and four from left).
As further evidence that the unused lysine in the culture media of LMP mutant cells interferes with coaggregation in situ, we treated washed cells of the parental or ΔkamA strain with the cell-free culture supernatants (spent medium) obtained from the parental or ΔkamA strain and then performed coaggregation tests with SgDL1 cells. As suspected, no coaggregation was observed with the washed parental cells treated with the ΔkamA supernatant with or without additional lysine (Fig. 4 E, Top; circles five and six from left). In contrast, the washed parental cells were able to coaggregate with SgDL1 cells in the presence of the parental spent medium from the parental culture without added lysine but not in its presence (Fig. 4 E, Top; circles seven and eight from left). The washed ΔkamA cells were able to coaggregate with streptococci when treated with the parental spent medium, whereas no coaggregation was observed under the other environmental conditions (Fig. 4 E, Bottom; compare circle five with circles six through eight).
The above data provided compelling evidence to prompt us to investigate whether lysine interacts with RadD directly to control coaggregation. We therefore prepared whole cell lysates of the parental and ΔradD mutant strains and tested the ability of an L-lysine immobilized resin to trap RadD from the lysates. Remarkably, this pulldown experiment showed that lysine beads captured RadD from the parental lysates efficiently and specifically (Fig. 4G). Since arginine (Arg) at high concentrations can inhibit fusobacterial coaggregation with SgDL1 (Fig. 4F), we also performed a pulldown assay using Arg beads. As shown in SI Appendix, Fig. S2, Arg beads also captured RadD from the parental lysates. Altogether, we posit that fusobacterial RadD is not only the key adhesin for coaggregation but it also serves as a receptor for environmental lysine, whose level may be used as a cue for initiating the development of a multispecies biofilm.
The CarRS TCS Constitutes a Critical Virulence Factor in F. nucleatum.
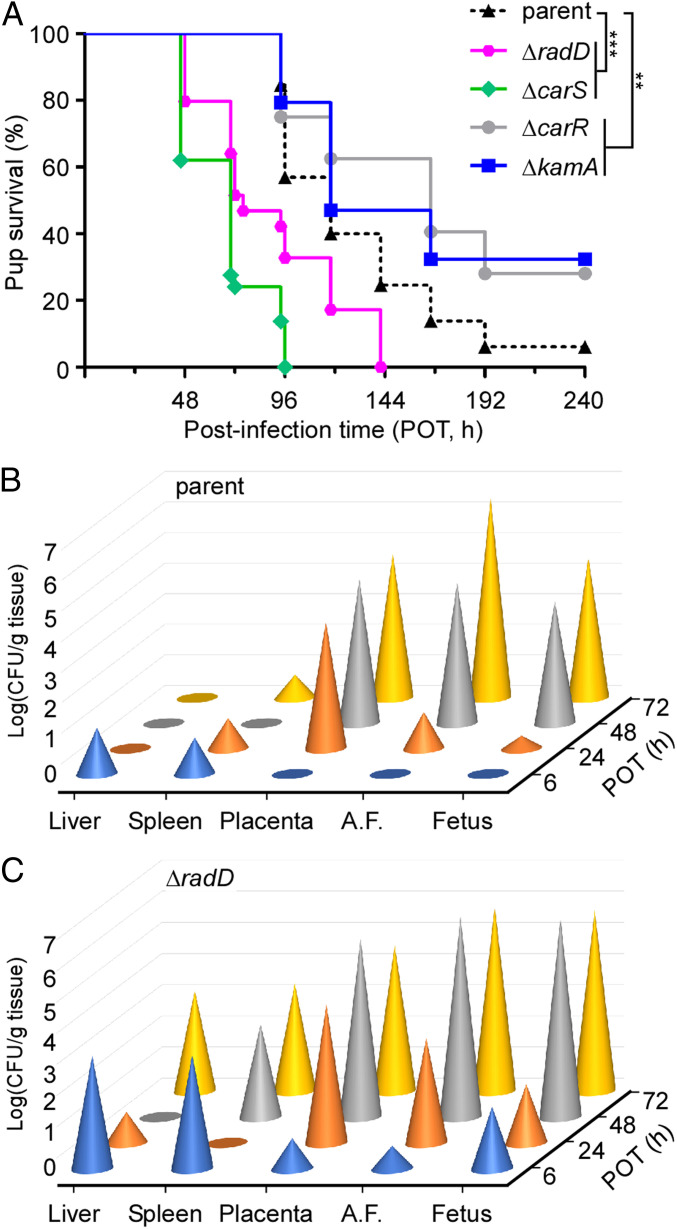
F. nucleatum is one of the most commonly detected microorganisms in the amniotic fluid and placenta from preterm and term pregnancies (2). In pregnant mice as an experimental model, F. nucleatum has been shown to induce premature and term stillbirths (3). Since the deletion of carR/carS affects the expression of radD, lysine metabolic genes, and many other genes, we were motivated to determine if CarR/S are associated with fusobacterial virulence. To do so, we chose to utilize the aforementioned mouse model of preterm birth for F. nucleatum that was utilized in previously reported studies of fusobacterial pathogenesis (3, 15, 27). Accordingly, a group of five pregnant CF-1 mice was injected via the tail vein with ∼5.0 × 107 colony forming units (CFU) of individual bacterial strains on day 16/17 of gestation, and the number of live and stillborn pups was recorded for the next 7 d. Remarkably, the nonpolar, in-frame deletion mutant ΔcarR exhibited a virulence attenuation phenotype as compared to the parental strain, which produced only 6% pup survival at the end point (Fig. 5A; compare the gray line and circles, ΔcarR, with the black dashed line and triangles—parent strain). Intriguingly, this attenuation of virulence is similar to that of the deletion mutant ΔkamA, which is predicted to be defective in lysine utilization in situ (Fig. 5A; blue line and squares). In contrast, the nonpolar, in-frame deletion mutant ΔcarS was significantly increased in virulence, leading to no pup survival at the end point within 96 h postinfection (Fig. 5A; green line and circles). Considering that RadD is a major adhesin and that adhesins typically act as virulence factors, we suspected that the ΔradD mutant would be attenuated in virulence. Contrary to this expectation, the ΔradD mutant mirrored the hyper-virulence phenotype of the ΔcarS mutant in the preterm birth model, with 0% of pups surviving at the end point, albeit a slightly longer time compared to the ΔcarS mutant (Fig. 5A; pink line and circles).
Fig. 5.
Virulence attenuation of the carR mutant. (A) A group of five pregnant CF-1 mice was infected with ∼5.0 × 107 CFU of the parent, ΔcarR, or ΔcarS strain via the tail vein on day 16 or 17 of gestation. The birth outcome was recorded during the next 7 d. The statistical differences were analyzed by Mantel–Cox (**P < 0.01; ***P < 0.001). (B and C) A group of 12 pregnant CF-1 mice was infected with ∼5.0 × 107 CFU of the parent (B) or ΔradD (C) strain via the tail vein on day 16 or 17 of gestation. At indicated time intervals, different organs and AF were harvested for bacterial numeration. Statistical analysis was performed by one-way ANOVA followed by a Newman–Keuls multiple comparison test.
To determine whether the hyper-virulence phenotype of the ΔradD mutant correlates with its ability to colonize distal organs or not, we employed a mouse colonization model as previously described (3). In this experiment, a group of three pregnant CF-1 mice was injected via the tail vein with ∼5.0 × 107 CFU of individual bacterial strains. The liver, spleen, placenta, amniotic fluid (AF), and fetus were harvested 6, 24, 48, and 72 h postinfection for bacterial plating. In animals infected with the parental strain, Fusobacteria were detected in liver and spleen at 6 h postinfection, and the bacterial load was largely diminished at 72 h postinfection, whereas high numbers of Fusobacteria were recovered from the placenta, AF, and fetus at 48 and 72 h postinfection (Fig. 5B). In contrast, in animals infected with the ΔradD mutant, Fusobacteria were already detected in the placenta, AF, and fetus at early time points and continued to increase in numbers at later time points (Fig. 5C).
We conclude that the RadD adhesin and its regulatory system CarRS are not only pivotal for interspecies interaction in vitro but they also contribute significantly to the modulation of fusobacterial virulence in vivo in a mouse model.
Discussion
The oral cavity contains one of the most complex microbial biofilms known to be associated with some common diseases in humans. Yet, the oral cavity constitutes an ever-changing hostile environment, which inflicts significant challenges for the survival of the hundreds of microbial species thus far identified as members of the oral microbiome, including F. nucleatum, the subject of the present study. The constituent microbes live with and communicate with each other as friend or foe while establishing residence and competing for nutrients under additional survival challenges posed by the host immune system. As we stated at the outset, F. nucleatum plays a bridging role for several organisms of which some serve as the seed for fusobacterial colonization, while others are able to colonize using Fusobacteria as the soil. Here, we wished to tackle the central problem of how Fusobacteria achieve this capacity of serving as a “polymicrobial niche.” To foster an unbiased and systematic mechanistic study of this problem, we recently made strides in developing several essential genetic tools for F. nucleatum (21, 27), and we began to dissect some of the key molecular steps in interspecies interactions and biofilm development. Here, we describe the results of a systematic genetic investigation of one of these steps, the fusobacterial coaggregation with streptococci, which uncovered a hitherto unknown genetic network that promotes fusobacterial interactions with other oral bacteria and its metabolic regulation by a physiologically intriguing pathway for fusobacterial communication with the host that may also be connected to the important process of nutrient acquisition by the microbe.
At first, this study sought to identify in a genome-wide scale all the genetic determinants that confer fusobacterial ability to interact and communicate with various other microbes in the oral cavity. To do so, we generated and employed a comprehensive Tn5 transposon mutant library (with greater than or equal to threefold genome coverage) to screen for and isolate any fusobacterial mutant that fails to aggregate with one of its key partners, S. gordonii, an early colonizer of the oral cavity. We succeeded in obtaining a vast collection of such coaggregation-defective mutants (Table 1). Subsequent sequencing of the Tn5-disrupted gene loci determined that the affected genetic loci not only included the well-known coaggregation factor RadD but also disclosed nine additional classes of coaggregation-associated proteins whose function in biofilm development had been unknown so far (Fig. 1 and Table 1). RadD is one of the three outer membrane proteins whose role has been revealed from our coaggregation screen (Table 1). The fact that radD was targeted by Tn5 over 40 times in our library of coaggregation-defective mutants, and that the nonpolar radD deletion mutant displayed the coaggregation defect (Figs. 1 and 2), support that RadD is a major coaggregation factor in F. nucleatum. While future experiments with similar genetic and biochemical approaches will be needed to characterize the other two outer membrane protein candidates (HMPREF0397_RS04065 and HMPREF0397_RS04015), which were targeted only once by Tn5, we suspect that these studies would further validate the critical role of RadD in polymicrobial interactions.
The more significant outcome of the present work is our finding that a large body of previously unknown factors contributes to interspecies interactions in addition to the RadD adhesin. The prime among these newly identified coaggregation gene candidates is defined by the carS::Tn5 mutant (Table 1; group 3). This targeted gene encodes a sensor kinase that we named CarS, which is a part of the hitherto uncharacterized two-component system CarRS (Fig. 2) that is shown to modulate RadD transcription: RadD transcription is reduced in the absence of CarS but greatly elevated with CarR missing. Thus, CarR is a repressor of the radD gene. How CarS acts to stimulate radD transcription in the presence of CarR remains to be dissected. Nevertheless, comparative transcriptome analyses in a genome-wide scale revealed that the response regulator CarR actually modulates hundreds of genes constituting a CarR regulon. Intriguingly, CarR not only represses the expression of a large set of genes, transcripts for some of which are elevated more than 10-fold in the ΔcarR mutant, but CarR also activates the transcription of yet another set of genes, transcripts for some of which are diminished by more than 10-fold in the ΔcarR mutant (Fig. 3). Significantly, CarR is a repressor of many genes that code for enzymes of an LMP in F. nucleatum (20) (Figs. 3 and 4 and Dataset S1). Further characterization of some of these LMP genes, that is, kamA and kamD, provided a deeper insight into the previously described phenomenon that lysine blocks fusobacterial coaggregation with many oral bacteria (17, 18). We showed here that the in-frame deletion mutants kamA and kamD accumulate free lysine in their culture medium, leading to the inhibition of RadD-mediated coaggregation with streptococci (Fig. 4). Considering that kamA and kamD code for lysine 2,3-aminomutase and D-lysine 5,6-aminomutase, respectively, which catalyze early steps of LMP converting L-lysine to the intermediate 3,5-diaminohyxanoate (Fig. 4B), it is conceivable that a block in the first step in lysine utilization results in lysine accumulation in the extracellular milieu, possibly due to reduced lysine uptake. While this logical scenario remains to be tested, it is noteworthy that none of the genes coding for potential lysine transporters (AtoD and AtoA) and enzymes catalyzing the downstream steps of LMP (HMPREF0397_RS01384, HMPREF0397_RS01005, HMPREF0397_RS01006, and HMPREF0397_RS01529) were represented by our genome-wide Tn5 mutagenesis screen (Fig. 4A). It is likely that these are essential genes, as the metabolic intermediate acetyl coenzyme A (acetyl-CoA) generated by the encoded enzymes is involved in many essential bacterial processes including protein, carbohydrate, and lipid metabolism.
The phenotypic convergence of radD and LMP deletion mutants in fusobacterial coaggregation defects led us to ask the critical physiological question of whether lysine acts to bind and inhibit RadD-mediated coaggregation. Indeed, our biochemical investigation demonstrated clearly that lysine-containing agarose beads could capture RadD in the fusobacterial cell lysates (Fig. 4). This simple result gives credence to our speculation that RadD not only serves the role of an interspecies adhesin but it also directly interacts with environmental lysine in situ, raising the intriguing possibility that RadD might serve as a lysine receptor coupled to a lysine acquisition apparatus encoded by the LMP locus. The fact that lysine (Fig. 4) and arginine (12) can each block RadD-mediated coaggregation raises another possibility that these positively charged amino acids may sterically interfere with RadD binding to the ligand present on the streptococcal surface. Future studies should investigate how RadD binds to the ligand(s) present on the streptococcal surface and how these positively charged amino acids are involved in blocking those ligand–receptor interactions.
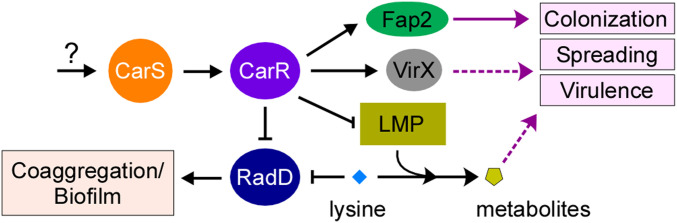
Most significantly, our studies revealed that the phenotypic convergence of RadD and LMP mutants is not to be restricted to just bacterial coaggregation: our investigation of fusobacterial pathogenesis presented here provided the important evidence that the regulated expression of radD and LMP genes through the action of the two-component system CarRS modulates bacterial virulence itself (Fig. 5). We showed that the nonpolar ΔkamA deletion mutant is substantially attenuated in virulence in the mouse model of preterm birth, while the ΔradD mutant exhibited a hyper-virulence phenotype with no pup survival to term. The latter phenotype is entirely consistent with the increased bacterial burden in the placenta, AF, and fetus of pregnant mice infected with the radD mutant (Fig. 5). In this context, it is noteworthy that early studies found that lysine is one of several amino acids that are present in high concentrations in mother’s blood, cord blood, placenta, and AF (28). Considering the role of RadD in polymicrobial interactions (Fig. 1) and biofilm formation (12) and lysine as an important component in fusobacterial metabolism (Fig. 4B) (20), we suggest that when Fusobacteria encounter a sedentary condition or coaggregation/biofilm state, up-regulation of radD and LMP gene expression, via de-repression of the TCS CarRS, may facilitate the acquisition of lysine, which is converted to intermediate metabolites vital to cellular metabolism. Conversely, in response to respective environmental inputs, the CarRS represses expression of radD and LMP genes while modulating expression of many other genes, which diminishes coaggregation and supports bacterial spreading to distal organs, leading to bacterial colonization of new sites and subsequently pathogenicity (Fig. 6). While this model of fusobacterial colonization of pathogenesis remains to be substantiated and dissected, a significant molecular puzzle that needs to be solved is what triggers the activation and inhibition of CarS/CarR and how the phosphorelay reaction affects specific expression of the large sets of genes that are repressed and activated by CarRS. Furthermore, it is conceivable that additional factors, the expression of which may be modulated by CarR, potentially contribute to bacterial spreading and virulence (Fig. 6).
Fig. 6.
A simplified model of bacterial coaggregation and virulence modulated by RadD and LMP via regulation by the TCS CarRS in F. nucleatum. CarRS is proposed to regulate expression of radD, LMP genes, and many others in response to environmental inputs, albeit currently unknown (question mark). The response regulator CarR directly or indirectly represses expression of radD and LMP genes while activating the expression of many other genes, including those encoding Fap2, known to be involved in placental colonization, and possibly unidentified factor(s) (VirX in gray oval), which may contribute to bacterial spreading and virulence (dashed purple arrow). Lysine binds to RadD and inhibits RadD-mediated coaggregation while it can be utilized by Fusobacteria via the LMP system to produce metabolites, which may influence fusobacterial virulence potential.
Lastly, given that most of the remaining groups of coaggregation-associated factors identified here are related to cellular metabolism (Table 1), it is reasonable to speculate that F. nucleatum employs a battery of metabolites to modulate polymicrobial interactions to thrive in the environment of oral biofilms. While this aspect of fusobacterial cell biology also remains to be investigated through further genetic and biochemical characterizations of the remaining factors, the present study represents a significant step forward to inspire the molecular genetic investigations of F. nucleatum pathophysiology in the future.
Materials and Methods
Bacterial Strains, Plasmids, and Media.
Bacterial strains and plasmids used in this study are listed in SI Appendix, Tables S1 and S2. F. nucleatum were grown in tryptic soy broth supplemented with 1% Bacto peptone plus, 0.25% freshly made cysteine (TSPC), or on TSPC agar plates in an anaerobic chamber (10% CO2, 10% H2, and 80% N2). Brain Heart Infusion broth supplemented with 0.5% glucose and Heart Infusion broth were used to grow streptococci and A. oris, respectively. A. actinomycetemcomitans were grown in TSPC in a 5% CO2 incubator. P. gingivalis were grown in TSPC supplemented with 1% vitamin K1-hemin solution. Escherichia coli strains were grown in Luria broth. When needed, kanamycin, chloramphenicol, or thiamphenicol were added into medium at a concentration of 50, 15, or 5 µg ⋅ ml−1, respectively. The growth of Fusobacteria was monitored by optical density at 600 nm. Reagents were purchased from Sigma unless indicated otherwise.
Plasmid Construction.
A detailed list of generated plasmids and primers used in this study can be found in SI Appendix, Table S1. 1) For pCarR, the primer sets PcatP-F1/R1 and com-carR-F/R (SI Appendix, Table S2) were used to amplify the catP promoter from plasmid pHS30 (29) and the carR coding region from the genomic DNA of F. nucleatum ATCC 23726, appending KpnI/NdeI or NdeI/XhoI sites for cloning purposes, respectively. The PCR products were cut by KpnI/NdeI or NdeI/XhoI and cloned into the KpnI and XhoI sites of pCWU6 (21). 2) For pCarRrpsJ, primers rpsJ-F and rpsJ-R (SI Appendix, Table S2) containing SacI and KpnI sites were used to amplify the untranslated region of rpsJ from F. nucleatum ATCC 23726 using genomic DNA as a template. The carR coding sequence was amplified by using primers com-carR-F2/R containing KpnI and XhoI sites. Both fragments were subcloned into pCWU6 at SacI and XhoI sites. 3) For pHP and pMutS, two primer pairs, com-HP-F/R and com-mutS-F/R, were used to amplify the HP (HMPREF0397_0997) and mutS coding sequences while appending KpnI and XhoI sites for cloning purposes. The two PCR-amplified DNA fragments were digested with KpnI and XhoI and ligated into the KpnI/XhoI-cut vector pCarRrpsJ, respectively. 4) For pCarS, the primer sets PcatP-F1/R1and com-carS-F/R (SI Appendix, Table S2) were used to amplify while appending KpnI and NdeI or NdeI and BgIII sites to the amplicons, the catP promoter region, and the carS coding sequence from pHS30 and chromosomal DNA of F. nucleatum ATCC 23726, respectively. The PCR-amplified fragments were digested with KpnI and NdeI and NdeI and BglII, respectively, and ligated into the vector pCWU6 precut with KpnI and BglII. 5) For pGalK-PcatP, the primer set PcatP-F2/R2 was used to amplify the catP promoter from plasmid pHS30. The PCR products were treated with SacI and KpnI and cloned into the SacI and KpnI sites of pCM-GalK (21). 6) For pRadD, due to its large size (10,387 nucleotides), the cloning of radD was performed in two steps. First, the radD region encompassing the nucleotide 1 to the 4,450th was amplified with the primer set RadDhalf1-F/R while appending KpnI and PstI sites. Second, the primer set RadDhalf2 –F/R was used to amplify the remainder of radD while appending PstI and XhoI sites. The generated PCR products were treated with KpnI and PstI or PstI and XhoI restriction enzymes and subsequently ligated into the KpnI and XhoI sites of the vector pGalK-PcatP while appending the catP promoter to the radD coding region. The generated plasmid was electroporated into the ΔcarS strain and integrated at a radD homologous locus into the chromosome, resulting in a radD direct duplication. One radD expression is under the control of its native promoter, and the other one is driven by the catP promoter. 7) For pKamA, the primer pair com-kamA-F/R was used to amplify the coding region of F. nucleatum kamA while adding KpnI and XhoI sites. The resulting PCR product was digested with KpnI and XhoI and cloned into pCarRrpsJ precut with KpnI and XhoI.
Site-Directed Mutagenesis of Recombinant Plasmids.
1) To generate Asp-to-Ala or Glu mutations within CarR, the mutation sites were incorporated into the 5′ end of synthesized primers. The wild-type carR fragment was cut from pCarR by NdeI and XhoI and cloned into pCM-GalK at NdeI and XhoI sites. The resulting plasmid, pGalK-carR, was used as a template for PCR amplification with Pfu DNA polymerase using appropriate primers sets (SI Appendix, Table S2). The PCR products were purified by gel extraction and phosphorylated to facilitate religation of the amplicons into circular plasmids, which were then transformed into E. coli DH5α. Resultant mutations were verified by DNA sequencing, and carR fragments carrying desired mutations were subcloned into pCarR at NdeI and XhoI sites. 2) For CarS-truncated mutants, appropriate primer sets (SI Appendix, Table S2) were used in reverse PCR to selectively amplify the plasmid pGalK-carS, which was obtained by cloning the carS fragment from pCarS into pCM-GalK at NdeI and BglII sites. To generate the His-to-Ala mutation within CarS, the same procedure as the above was employed.
Gene Deletion in F. nucleatum.
The generation of nonpolar, in-frame deletion mutants in F. nucleatum was performed following a previously published protocol (21). Briefly, 1 kb fragments up- and downstream of a targeted gene were cloned with the integrative vector pCM-GalK, which expresses thiamphenicol resistance and galK genes (21). The resulting plasmid was electroporated into F. nucleatum CW1, which lacks a functional galK gene. The integration of the vector into the bacterial chromosome via homologous recombination was selected on TSPC agar plates containing 5 μg ⋅ ml−1 thiamphenicol. The excision of the vector via a second recombination event, resulting in gene deletion or reconstitution of the wild-type genotype, was selected by 0.2% 2-deoxygalactose (2-DG) on plates. 2-DG-resistant and thiamphenicol-sensitive colonies were screened for the expected gene deletion by PCR amplification and/or Western blot analysis. The same procedure was employed for the construction of double, triple deletion mutants.
High-Throughput Screening for Coaggregation-Defective Mutants.
Using our previously generated library containing ∼24,000 Tn5 clones (21), we set up a pilot screen with roughly 7,000 Tn5 mutants for F. nucleatum mutants defective in coaggregation with S. gordonii DL1. Individual Tn5 mutants were inoculated into 96-well plates containing 200 μL of TSPC in an anaerobic chamber at 37 °C. After 48 h growth, 80 μL of each culture was transferred into new 96-well plates. Separately, overnight cultures of S. gordonii DL1 were prepared, and streptococcal cells were harvested, suspended in coaggregation buffer (0.1 mM CaCl2, 0.1 mM MgCl2, 0.02% NaN3, and 1 mM Tris, pH 8.0), and normalized to OD600 of 2.0. A total of 80 μL aliquots of streptococcal suspension was added to each well of F. nucleatum plates prepared above. Each plate was sealed by Adhesive Sealing Films for Microplates (Sigma-Aldrich) and vortexed for 30 s, and coaggregation was visually examined and recorded. Positive (F. nucleatum 23726 and S. gordonii DL1) and negative controls (F. nucleatum ΔradD devoid of RadD and S. gordonii DL1) were included for comparison. A total of 78 coaggregation-defective mutants were identified from this screen, and subsequently, single-primer one-step PCR was employed to map Tn5 insertion sites in these mutants as detailed in our published procedure (21).
Bacterial Coaggregation Assays.
Coaggregation between F. nucleatum and other bacterial species was performed as previously described (6) with some modification. Briefly, stationary-phase cultures of F. nucleatum and other bacterial strains grown with the appropriate media were harvested by centrifugation. Bacterial cells were washed in coaggregation buffer. Cells of F. nucleatum were normalized to an OD600 of 0.8, whereas others were adjusted to an OD600 of 2.0. A total of 0.2 mL aliquots of fusobacterial and streptococcal cell suspension were mixed in 24-well plates for a few minutes on a rotator shaker, and coaggregation was recorded by an Alpha Imager (Alpha Innotech). In the coaggregation experiments with unwashed cells, a similar procedure as the above was employed, except that the fusobacterial cultures were directly used without washing. To determine if coaggregation could be blocked by lysine, fusobacterial cells were suspended in a coaggregation buffer containing 15 mM of lysine.
RNA-Seq Analysis.
F. nucleatum RNA was extracted using RNeasy Mini Kits (Qiagen) according to the manufacturer’s protocol. Fusobacterial cell pellets from 3 mL log-phase culture (OD600 ∼0.6) were harvested and resuspended into 200 μL chilled 10 mM RNA-free Tris-EDTA (TE) buffer [10 mM Tris HCl, pH 8; 1 mM ethylenediaminetetraacetic acid (EDTA)]. Each suspension was transferred to a fast protein tube (Qbiogene), which contained 700 μL RNeasy lysis buffer for lysing cells and tissues (RLT) (RNeasy Mini Kit, Qiagen) and 7 μL β-mercaptoethanol (Thermo Fisher Scientific). Cells were lysed by using Ribolyser (Hybaid), and supernatants were obtained by centrifugation at 13,000 × g for 5 min at 4 °C. RNA was then purified from the supernatant accordingly, and purified RNA was treated with Deoxyribonuclease (DNase; Qiagen) and cleaned by using an RNeasy clean up kit (Qiagen). The quality of RNA samples was determined by RNA integrity number values greater than 8 using an Agilent 2100 Bioanalyzer (Agilent Technologies).
For RNA-seq, complementary DNA (cDNA) libraries were prepared, and sequencing was performed in the paired-end mode on an Illumina HiSeq as previously described (30). The processing and mapping of the paired-end reads and differential gene expression analysis was performed by using DESEq2 as previously reported (30). Genes with a log2 (fold change) above +1.0 or below −1.0, respectively, were considered to be differentially transcribed under the examined conditions. The RNA-seq data were deposited in the NCBI Gene Expression Omnibus (GEO) database with the accession number of GSE168051.
Real-Time PCR.
F. nucleatum cells anaerobically grown in TSPC to an OD600 of 0.8 at 37 °C were collected by centrifugation. Cell pellets were suspended in 1 mL ice-cold TRIzol (Sigma) and disrupted with 0.1 mm Zirconia Beads using a Minibead-Beater (BioSpec). After centrifugation, supernatants were collected and RNA purified using an Ambion RiboPure-Bacteria Kit according to the manufacturer’s protocol. The extracted RNA samples were treated with DNase I (Ambion) to remove traces of chromosomal DNA, and RNA samples were further cleaned with a Qiagen RNeasy MinElute Cleanup kit. Total RNA (300 ng) was used for cDNA synthesis using Stratascipt RT (Stratagene) based on the manufacturer’s instruction. Real-time PCR was performed using the CFX96 Real-Time System (Bio-Rad), and reactions were prepared using the SYBR Green PCR Master Mix with appropriate primers (SI Appendix, Table S2). The changes in gene expression were calculated using the ∆∆CT method as follows, ∆CT = CT (target) − CT (housekeeping gene); ∆∆CT = ∆CT1 − ∆CT2; fold changes were calculated as 2-∆∆Ct. The 16S rRNA gene was used as a reference, and reactions without reverse transcriptase were used as a control to assess genomic DNA contamination.
Luciferase Assay.
To measure gene expression using a luciferase assay, a radD-luc reporter was constructed using the vector pCWU5, which contains a chloramphenicol/thiamphenicol resistance marker (catP) (21). To construct a single-copy luciferase transcription fusion to the rad operon, the 3′ portion of radD was amplified by PCR from the chromosomal DNA of strain ATCC 23726 with primers radD3′F and radD3′R (SI Appendix, Table S2). The luc ORF including its ribosome-binding site was amplified from pFW5-luc (31). The radD3′ and luc PCR product were digested with BamHI/XhoI and XhoI/HindIII, respectively, and ligated into the pCWU5 to form plasmid pCWU5::3′radD-luc. Plasmid pCWU5::3′radD-luc was transformed into the wild-type strain or ΔcarR mutant, and transformants were selected on TSPA plates containing thiamphenicol (5 μg/mL).
For a luciferase assay as reported previously (22), 25 μL 1 mM D-luciferin (Molecular Probe) suspended in 100 mM citrate buffer, pH 6, was added to 100 μL of the cell culture anaerobically grown during the log phase (OD600 ∼0.6), and we gently pipetted the mixture eight times aerobically. Luciferase activity was measured by using a Tecan Infinite M1000 reader.
Detection of Lysine by Liquid Chromatography–Mass Spectrometry.
The cultures of the parental strain and its derivatives were grown to a stationary phase, and bacterial supernatants were obtained from 1 mL of each culture by centrifugation twice at 13,000 g for 2 min. A total of 5 μl internal lysine standards was added to 500 μL bacterial supernatants. The resulting mixture was subjected to filtration using 3-kDa filters (Amicon Ultracel-3K Membrane; Millipore Corporation). A total of 10 μl of samples was analyzed by a 6490 Triple Quadrupole mass spectrometer (Agilent Technologies) coupled to a high-performance liquid chromatography (HPLC) system (Agilent Technologies) at the Metabolomics Core at Baylor College of Medicine. The source parameters were set up as follows: the gas temperature was at 200 °C, the gas flow was 14 l/min, the nebulizer was 20 psi, the sheath gas temperature was at 300 °C, the sheath gas flow 11 l/min, the capillary was 3,000 V positive and 3,000 V negative, and the nozzle voltage was 1,500 V positive and 1,500 V negative. Approximately eight to 11 data points were acquired per detected metabolite. For mass spectrometry, an electrospray ionization positive mode was used. For HPLC, samples were separated by a C18 column (ZORBAX Eclipse XDB; 80 Å, 4.6 × 50 mm, 1.8 µm), using mobile phase A and B of 0.1% formic acid in water and acetonitrile, respectively.
Western Blotting Analysis.
For immunoblotting, polyclonal antibodies against recombinant RadD were generated according to a published protocol (32). Briefly, primers LIC-RadD-5 and LIC-RadD-3 were used to PCR amplify the DNA region encoding an N-terminal domain of RadD (residues 41 to 360). The generated amplicons were cloned into the expression vector pMCSG7. The recombinant plasmid was introduced into E. coli BL21 (DE3). The purification of the recombinant protein H6-RadD41–360 was carried out by affinity chromatography based on a published protocol. The purified proteins were used for antibody production (Cocalico Biologicals, Inc.).
To detect RadD in Fusobacteria, stationary-phase cultures (OD600 of 1.2) of the wild-type strain and its derivatives were harvested by centrifugation. The cell pellets were washed twice with water and suspended in a sample buffer containing sodium dodecyl sulfate (SDS). The samples were boiled 10 min and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) using 8% Tris-glycine gels followed by immunoblotting with antibodies against RadD (1:5,000 dilution for α-RadD). Of note, the radD mutant was used to deplete nonspecific antibodies from the α-RadD antibodies. A polyclonal antibody against FtsX (21) (1:1,000 dilution) was used as a control.
For RadD and lysine/arginine interactions, the wild-type culture (250 mL) grown to OD600 of 1.0 was harvested by centrifugation, and the cell pellets were washed twice with water prior to suspending in 25 mL suspension buffer (150 mM NaCl and 50 mM Tris HCl, pH 7.5) containing 2 mM phenylmethylsulfonyl fluoride. The cells were lysed by French press, and cell debris was removed by centrifugation at 10,000 g at 4 °C for 10 min. The cell membrane was collected by ultracentrifugation at 150,000 g for at 4 °C for 2 h and was suspended in coaggregation buffer containing 100 mM n-octyl-β-glucopyranoside. After overnight incubation at 4 °C, supernatants were obtained by centrifugation at 4,750 g for 10 min. The obtained supernatant was treated with 500 μL L-lysine/L-arginine agarose overnight at 4 °C, and the suspension was passed through a column. RadD-bound agarose was washed twice in coaggregation buffer prior to treatment with 4 mL coaggregation buffer containing 150 mM L-lysine/L-arginine. Eluates were subjected to trichloroacetic acid precipitation, and protein pellets were suspended in sample buffer for SDS-PAGE and immunoblotting with α-RadD.
Induction of Preterm Birth and Colonization of Fusobacteria in Mice.
The experimental procedure was based on a published protocol (3). Briefly, 10-wk-old CF-1 mice purchased from Charles Rivers Laboratories were mated at the female to male ratio of 2:1. The presence of a mating plug during daily checkups was used to mark the start of pregnancy. On day 16 or 17 of gestation, pregnant mice were infected via tail vein injection with ∼5 × 107 CFU of individual fusobacterial strains suspended in Dulbecco's phosphate-buffered saline. The number of live and stillborn pups was recorded for the next 7 d. Five pregnant mice were used in each group, and each experiment was repeated twice. Statistical analyses were carried out relative to the parental strain, and the significance was determined via Mantel–Cox testing using GraphPad.
To determine fusobacterial colonization, a published procedure was followed (3) with some modifications. Breeding was performed as described above; however, animals were killed at 6, 24, 48, or 72 h postinfection. Immediately following killing, the AF and various tissues from different organs, including liver, spleen, and placenta, were harvested, weighted, and homogenized (except AF) for bacterial plating. Three pregnant mice per group were used, and each experiment was repeated twice. Statistical analysis was carried out relative to the parental strain, and the significance was determined via t test. All animal procedures were approved by the University of California, Los Angeles (UCLA) Animal Research Committee.
Supplementary Material
Acknowledgments
We thank Vincent Lee (University of Maryland), Nicholas Ramirez (UCLA), and our laboratory members for a critical review of the manuscript and discussion. This work was also supported by federal funds from the National Institute of Dental and Craniofacial Research/NIH under Award Nos. DE026574 (to C.W.) and DE026758 and DE017382 (to H.T-T).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006482118/-/DCSupplemental.
Data Availability
RNA-seq data have been deposited in NCBI GEO (GSE168051). All other study data are included in the article and/or supporting information.
References
- 1.Mark Welch J. L., Rossetti B. J., Rieken C. W., Dewhirst F. E., Borisy G. G., Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. U.S.A. 113, E791–E800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Payne M. S., Bayatibojakhi S., Exploring preterm birth as a polymicrobial disease: An overview of the uterine microbiome. Front. Immunol. 5, 595 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han Y. W., et al., Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: Implication of oral bacteria in preterm birth. Infect. Immun. 72, 2272–2279 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kostic A. D., et al., Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubinstein M. R., et al., Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 14, 195–206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolenbrander P. E., Andersen R. N., Moore L. V., Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect. Immun. 57, 3194–3203 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosen G., Nisimov I., Helcer M., Sela M. N., Actinobacillus actinomycetemcomitans serotype b lipopolysaccharide mediates coaggregation with Fusobacterium nucleatum. Infect. Immun. 71, 3652–3656 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimaudo N. J., Nesbitt W. E., Coaggregation of Candida albicans with oral Fusobacterium species. Oral Microbiol. Immunol. 12, 168–173 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Wu T., et al., Cellular components mediating coadherence of Candida albicans and Fusobacterium nucleatum. J. Dent. Res. 94, 1432–1438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rickard A. H., Gilbert P., High N. J., Kolenbrander P. E., Handley P. S., Bacterial coaggregation: An integral process in the development of multi-species biofilms. Trends Microbiol. 11, 94–100 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Lancy P. Jr, Dirienzo J. M., Appelbaum B., Rosan B., Holt S. C., Corncob formation between Fusobacterium nucleatum and Streptococcus sanguis. Infect. Immun. 40, 303–309 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplan C. W., Lux R., Haake S. K., Shi W., The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol. Microbiol. 71, 35–47 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplan A., et al., Characterization of aid1, a novel gene involved in Fusobacterium nucleatum interspecies interactions. Microb. Ecol. 68, 379–387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaplan C. W., et al., Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infect. Immun. 78, 4773–4778 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coppenhagen-Glazer S., et al., Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect. Immun. 83, 1104–1113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abed J., et al., Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe 20, 215–225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okuda T., et al., Synergistic effect on biofilm formation between Fusobacterium nucleatum and Capnocytophaga ochracea. Anaerobe 18, 157–161 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Okuda T., et al., Synergy in biofilm formation between Fusobacterium nucleatum and Prevotella species. Anaerobe 18, 110–116 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Dzink J. L., Socransky S. S., Amino acid utilization by Fusobacterium nucleatum grown in a chemically defined medium. Oral Microbiol. Immunol. 5, 172–174 (1990). [DOI] [PubMed] [Google Scholar]
- 20.Barker H. A., Kahn J. M., Hedrick L., Pathway of lysine degradation in Fusobacterium nucleatum. J. Bacteriol. 152, 201–207 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu C., et al., Forward genetic dissection of biofilm development by Fusobacterium nucleatum: Novel functions of cell division proteins FtsX and EnvC. mBio 9, e00360-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He X., et al., The cia operon of Streptococcus mutans encodes a unique component required for calcium-mediated autoregulation. Mol. Microbiol. 70, 112–126 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.George K. S., Falkler W. A. Jr, Coaggregation studies of the Eubacterium species. Oral Microbiol. Immunol. 7, 285–290 (1992). [DOI] [PubMed] [Google Scholar]
- 25.Takemoto T., et al., Characteristics of multimodal co-aggregation between Fusobacterium nucleatum and streptococci. J. Periodontal Res. 30, 252–257 (1995). [DOI] [PubMed] [Google Scholar]
- 26.Signat B., Roques C., Poulet P., Duffaut D., Fusobacterium nucleatum in periodontal health and disease. Curr. Issues Mol. Biol. 13, 25–36 (2011). [PubMed] [Google Scholar]
- 27.Peluso E. A., Scheible M., Ton-That H., Wu C., Genetic manipulation and virulence assessment of Fusobacterium nucleatum. Curr. Protoc. Microbiol. 57, e104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Velázquez A., Rosado A., Bernal A., Noriega L., Arévalo N., Amino acid pools in the feto-maternal system. Biol. Neonate 29, 28–40 (1976). [DOI] [PubMed] [Google Scholar]
- 29.Kinder Haake S., Yoder S., Gerardo S. H., Efficient gene transfer and targeted mutagenesis in Fusobacterium nucleatum. Plasmid 55, 27–38 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wittchen M., et al., Transcriptome sequencing of the human pathogen Corynebacterium diphtheriae NCTC 13129 provides detailed insights into its transcriptional landscape and into DtxR-mediated transcriptional regulation. BMC Genomics 19, 82 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kreth J., Merritt J., Zhu L., Shi W., Qi F., Cell density- and ComE-dependent expression of a group of mutacin and mutacin-like genes in Streptococcus mutans. FEMS Microbiol. Lett. 265, 11–17 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Siegel S. D., et al., Structure and mechanism of LcpA, a phosphotransferase that mediates glycosylation of a Gram-positive bacterial cell wall-anchored protein. mBio 10, e01580-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in NCBI GEO (GSE168051). All other study data are included in the article and/or supporting information.