Significance
It has long been observed that activation of p53 can yield two main cell fates: either apoptotic cell death or cell cycle arrest. However, what determines this cell fate remains to be fully elucidated. Here we show that the state of mitochondrial priming prior to activation of p53 can determine cell fate. Highly primed cells commit to apoptosis, while less-primed cells commit to cell cycle arrest. We show that modulation of priming in either direction can alter cell fate; increasing priming in poorly primed cells yields apoptosis, while decreasing priming in highly primed cells yields arrest. Drugs that increase priming enforce an apoptotic fate, suggesting a p53-independent strategy for augmenting efficacy of cancer chemotherapy drugs that kill via p53.
Keywords: p53, apoptosis, cell cycle arrest, cell fate
Abstract
Reactivation of p53 in established tumors typically results in one of two cell fates, cell cycle arrest or apoptosis, but it remains unclear how this cell fate is determined. We hypothesized that high mitochondrial priming prior to p53 reactivation would lead to apoptosis, while low priming would lead to survival and cell cycle arrest. Using a panel of Kras-driven, p53 restorable cell lines derived from genetically engineered mouse models of lung adenocarcinoma and sarcoma (both of which undergo cell cycle arrest upon p53 restoration), as well as lymphoma (which instead undergo apoptosis), we show that the level of mitochondrial apoptotic priming is a critical determinant of p53 reactivation outcome. Cells with high initial priming (e.g., lymphomas) lacked sufficient reserve antiapoptotic capacity and underwent apoptosis after p53 restoration. Forced BCL-2 or BCL-XL expression reduced priming and resulted in survival and cell cycle arrest. Cells with low initial priming (e.g., lung adenocarcinoma and sarcoma) survived and proceeded to arrest in the cell cycle. When primed by inhibition of their antiapoptotic proteins using genetic (BCL-2 or BCL-XL deletion or BAD overexpression) or pharmacologic (navitoclax) means, apoptosis resulted upon p53 restoration in vitro and in vivo. These data demonstrate that mitochondrial apoptotic priming is a key determining factor of cell fate upon p53 activation. Moreover, it is possible to enforce apoptotic cell fate following p53 activation in less primed cells using p53-independent drugs that increase apoptotic priming, including BH3 mimetic drugs.
The most commonly mutated tumor-suppressor gene is TP53, with ∼50% of human cancers harboring mutations in this locus (1). The remaining 50% of human cancers typically harbor mutations that inactivate the p53 pathway, usually through down-regulation or mutation of its main upstream activator p14Arf (p19Arf in mice), or genomic amplification or overexpression of its main negative regulator Mdm2 (2). As a consequence, great effort has been placed on elucidating p53’s properties in tumor suppression and drug response.
Upon activation by a variety of stress signals, such as DNA damage, oncogenic stress, and hypoxia, p53 transcriptionally activates a large repertoire of genes. This can lead to a variety of outcomes, ranging from cell cycle arrest, senescence, or apoptosis, to changes in metabolism and autophagy, among others (3–5). Due to the prevalence of inactivating mutations in p53, recent efforts have focused on strategies to pharmacologically reactivate mutant p53 as a possible option for treating human cancers that harbor p53 mutations (6). This concept has been supported by experiments in genetically engineered mouse models (GEMMs) of cancer (7–10). We previously employed a GEMM with temporal control of p53 expression in established, autochthonous tumors. This mouse model expresses a Lox-STOP-Lox (LSL)-p53 allele in the germline (9). Homozygous mutant mice (p53LSL/LSL) are phenotypically identical to homozygous knockout mice (p53−/−) (11, 12). However, upon induction of Cre recombinase [which in this case is expressed from the ubiquitously expressed Rosa26 locus (13) as an estrogen receptor fusion protein called Cre-ERT2] and excision of the LSL cassette, p53 transcription is restored. Utilizing this immunocompetent mouse model, our group demonstrated that established tumors that spontaneously arise in the context of p53 deficiency (mostly soft tissue sarcomas and lymphomas) (11, 12) remain exquisitely sensitive to the reactivation of p53 (9). Notably, two contemporary studies by Evan and colleagues (8) and Lowe and colleagues (10) reached the same conclusion using estrogen receptor-regulatable systems and inducible-reversible RNAi technologies, respectively.
In a subsequent study, our group crossed the p53 restorable mouse to the KrasLA2 mouse model, which bears a latent allele of oncogenic KrasG12D that gets spontaneously activated in vivo (14), for the purposes of studying the effects of p53 reactivation in established lung tumors (7). Surprisingly, we found that the tumor-suppressive effects of p53 reactivation in established lung tumors growing in immunocompetent mice are stage-specific, whereby p53 triggers the elimination of highly advanced lesions but spares low-grade lesions. Mechanistically, high-grade lesions harbor hyperactive MAPK signaling, which in turn leads to potent induction of the p19Arf tumor-suppressor gene, leading to stabilization and activation of the p53 tumor suppressor (7). A contemporaneous study by Evan and colleagues (15) using an estrogen receptor-regulatable allele of p53 reached the same conclusion. More recently, Lowe and colleagues (16) demonstrated that p53 reactivation in advanced pancreatic ductal adenocarcinoma tumors leads to a combination of cell cycle arrest, senescence, and differentiation. Collectively, these GEMM-based studies have convincingly demonstrated that p53 can mediate different tumor-suppressive responses in different tumor types and even at different stages of tumor progression, and lend strong support to efforts aimed at pharmacologically reactivating p53 as a possible cancer therapeutic strategy (6).
Despite the fact that p53 has been extensively studied for over 30 y, the molecular mechanisms behind its highly context-specific tumor-suppressive responses remain widely unknown (17). Many models have focused on the quality, quantity, and dynamics of the p53 transcriptional response as determinants of cell fate (18). In these models, increased intensity of p53 activation and transcription induction promotes apoptotic cell fate.
Pretreatment mitochondrial apoptotic priming can determine cell fate in response to a number of drugs, including those that can kill in a p53-dependent fashion (19–21). This prompted us to ask whether the level of mitochondrial apoptotic priming in a cell dictates whether p53 reactivation promotes cell death or cell cycle arrest. To answer this question, we made use of the restorable p53 allele that we have used previously in GEMMs to give a uniform p53 signal in the setting of different levels of mitochondrial apoptotic priming. Our results support a model in which the level of mitochondrial apoptotic priming is a critical cell-autonomous determinant of cell fate upon p53 restoration.
Results
Tumor-Specific Responses to p53 Restoration.
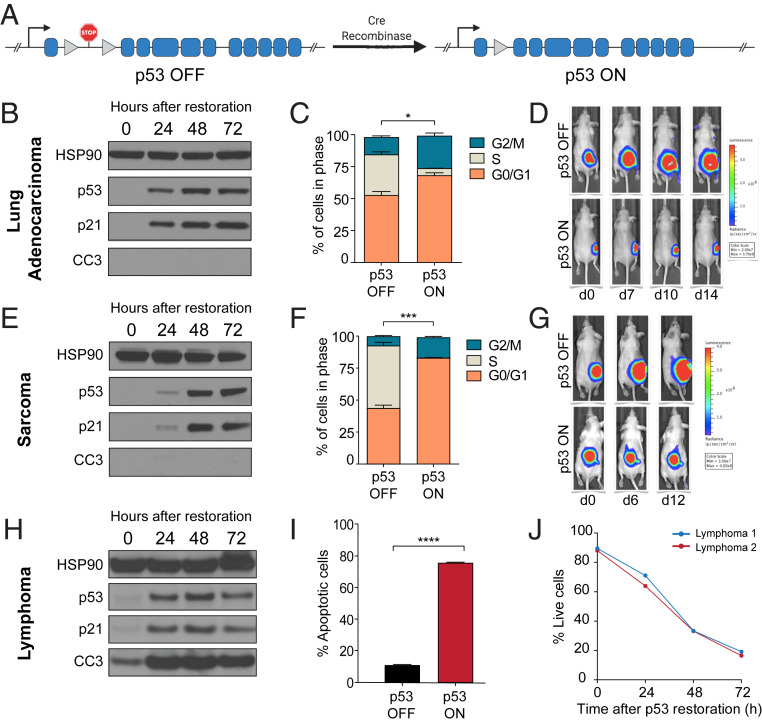
We hypothesized that tumor-specific responses to p53 restoration could be explained at least in part by the level of mitochondrial priming on each specific tumor type. Based on our model, we predicted that highly primed cells would readily undergo apoptosis upon p53 restoration, but poorly primed cells would instead undergo cell cycle arrest. To test this, we derived cell lines from three different tumor types (lung adenocarcinoma, sarcoma, and lymphoma) that arose spontaneously in KrasLA2/+;Trp53LSL/LSL;Rosa26CreERT2/CreERT2 mice (7). These mice harbor a KrasLA2 allele, which leads to spontaneous expression of the KrasG12D oncogene (14), as well as two LSL p53 alleles (9), which prevent expression of endogenous p53 and phenocopy p53-deficient mice. Cell lines derived from these tumors also express CreERT2, which allows for inducible Cre activity upon the addition of 4-hydroxytamoxifen (4-OHT), leading to excision of the STOP cassette upstream of Trp53, thereby restoring wild-type p53 expression (Fig. 1A). Although they all share the same genetic background, these cells experience two nonoverlapping cell fates upon p53 restoration (Fig. 1 B–J). Lung adenocarcinoma and sarcoma cell lines predominantly undergo cell cycle arrest, as shown by: 1) robust induction of p21 expression (Fig. 1 B and E), 2) significant accumulation of p53-restored cells in the G0/G1 stage of the cell cycle (Fig. 1 C and F), 3) pronounced tumor stasis upon restoration of p53 in established tumors in vivo (Fig. 1 D and G), and 4) marked absence of cleaved caspase 3 (CC3), even after prolonged restoration of p53 for 72 h (Fig. 1 B and E). Remarkably, lymphoma cells also exhibit robust and stable up-regulation of p21 upon p53 restoration (Fig. 1H) but fail to undergo cell cycle arrest. Instead, p53 restoration in lymphoma cells leads to robust apoptosis, as indicated by induction of CC3, annexin V staining, and propidium iodide staining over time (Fig. 1 H–J). These data demonstrate that the initiation of cell cycle arrest signaling does not prevent the induction of apoptosis, as all three cell types are fully capable of initiating cell cycle arrest signaling upon p53 restoration, but only lymphoma cells undergo apoptosis. We next asked, what determines whether apoptosis will or will not occur upon p53 restoration?
Fig. 1.
Differences in cell fate upon p53 restoration. (A) Schematic of p53 restorable allele. A stop cassette flanked by loxP sites prevents expression of p53 until Cre excises the cassette and restores wild-type p53. (B, E, and H) Western blots showing restoration of p53 and p21 induction in all tumor types but cleavage of caspase 3 only in lymphoma. Three independent samples were tested for each tumor type. (C and F) Cell cycle analysis showing reduction or loss of S-phase cells after p53 restoration in lung adenocarcinoma and sarcoma cells. (D and G) p53 restoration in lung adenocarcinoma and sarcoma tumors in vivo results in growth arrest but not clearance. (I) Loss of viability in lymphoma cells after p53 restoration at one timepoint and (J) over time. *P < 0.05, ***P < 0.001, ****P < 0.0001.
The Level of Mitochondrial Apoptotic Priming Explains Tumor-Specific Responses.
It is known that both normal and malignant tissues have different propensities toward apoptosis and sensitivity to chemotherapy based on their mitochondrial apoptotic priming (20, 22). Therefore, we asked whether differences in mitochondrial apoptotic priming could account for the differences in cell fate upon p53 restoration. We first measured the degree of mitochondrial priming in the cell lines before p53 restoration. To do this, we used BH3 profiling, a technique that interrogates the mitochondria using synthetic peptides based on proapoptotic BH3-only proteins (23–25). The degree of mitochondrial apoptotic priming is inversely related to the quantity of BH3 peptide required to exceed the threshold for mitochondrial outer membrane permeabilization (MOMP). When the apoptotic threshold is exceeded, proteins BAX and BAK can oligomerize to form pores on the outer mitochondrial membrane, leading to MOMP. BH3 profiling employs surrogate markers, including cytochrome c release or mitochondrial potential, to measure MOMP. The less peptide needed to cause MOMP, the more primed the cells are for apoptosis.
BH3 profiling of multiple cell lines per tissue of origin demonstrated a clear difference between those cell lines fated to cell cycle arrest and those that undergo apoptosis. Sarcoma and lung adenocarcinoma cell lines showed far lower priming across all peptides (Fig. 2A). Using the broadly interacting BIM peptide as a global measure of priming (23), sarcoma and lung adenocarcinoma cell lines were not significantly different from each other, but both were significantly less primed than lymphoma cell lines (Fig. 2B). We hypothesized that while lung adenocarcinoma and sarcoma cell lines survive following p53 induction, they nonetheless were able to initiate apoptotic signaling. To test this, we examined mitochondrial priming in the lung adenocarcinoma and sarcoma cell lines during p53 restoration. Regardless of tissue of origin, priming increased in a time-dependent manner upon p53 restoration (Fig. 2 C and D and SI Appendix, Fig. S1). This suggests that increased priming is a general effect of p53 restoration, but that only cells with high enough initial priming are forced to commit to apoptosis by such signaling. Cells that begin with low priming would still experience p53-dependent increased priming, but because priming levels stay below a critical threshold, cell cycle arrest dominates over apoptosis. If this were true, modification of priming would be expected to alter cell fate upon p53 restoration.
Fig. 2.
BH3 profiling reveals differential priming at baseline and during p53 reactivation. (A) Baseline mitochondrial priming profiles of sarcoma, lung adenocarcinoma, and lymphoma cell lines showing increased priming in lymphoma relative to all other cell lines. Three independent samples were tested for each tumor type. (B) Comparison of priming measured by BIM peptide at 0.75 μM showing the difference between lymphoma priming and all other cell lines. ****P < 0.0001; n.s., not significant. (C and D) BH3 profiling showing increase in priming with time after p53 restoration in lung adenocarcinoma and sarcoma cell lines, respectively.
Increasing Priming Is Sufficient to Switch Lung Adenocarcinoma and Sarcoma Cell Fate to Apoptosis.
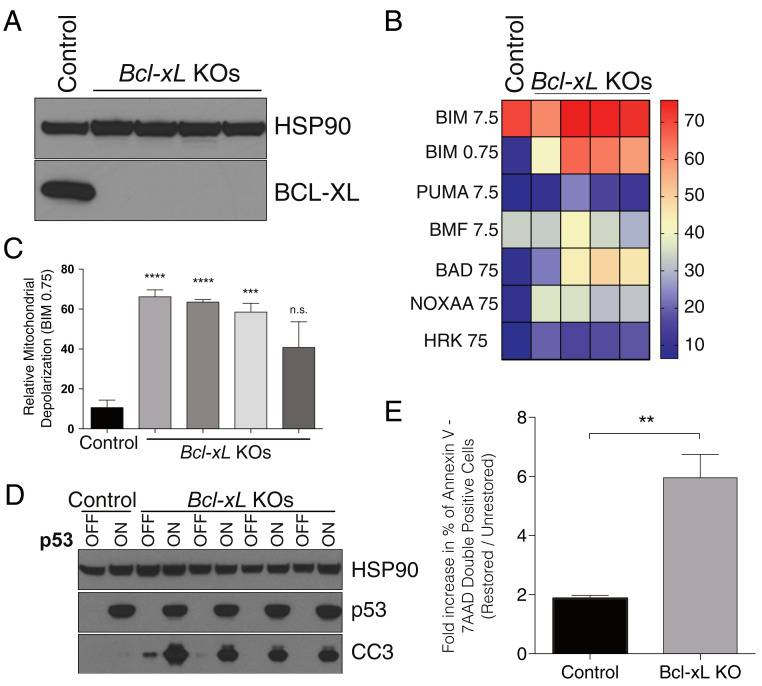
If basal priming levels determine whether cells can survive the increase in priming caused by p53 restoration, then increasing the priming of lung adenocarcinoma and sarcoma cells should change their fate from cell cycle arrest to apoptosis. We used multiple experimental approaches to test this hypothesis. First, we genetically ablated the antiapoptotic protein BCL-XL in lung adenocarcinoma cells using CRISPR-Cas9 (26) to test whether its deletion would increase the basal level of mitochondrial apoptotic priming in these poorly primed cells. Indeed, BH3 profiling of cells lacking BCL-XL (Fig. 3A) demonstrated a clear increase in global priming (Fig. 3 B and C). Restoration of p53 led to a robust increase in CC3 and a decrease in cell viability in cells lacking BCL-XL but not in the parental line (Fig. 3 D and E and SI Appendix, Fig. S2). Similarly, CRISPR-mediated deletion of BCL-XL in sarcoma cells resulted in increased priming levels comparable to those observed in lung adenocarcinoma (SI Appendix, Fig. S3 A–C), and knockdown of either BCL-2 or BCL-XL was sufficient for p53 restored cells to show cleavage of caspase 3, indicating they were undergoing apoptosis (SI Appendix, Fig. S3D). Thus, increasing initial mitochondrial apoptotic priming was sufficient to bias cells toward apoptosis upon additional apoptotic priming due to p53 restoration.
Fig. 3.
Genetic priming of lung adenocarcinoma cell lines is sufficient to switch cell fate of p53-restored cells from cell cycle arrest to apoptosis. (A) CRISPR-mediated deletion of Bcl-xL in lung adenocarcinoma cell lines. (B) BH3 profile of four Bcl-xL knockout lung adenocarcinoma cell lines showing that Bcl-xL deletion increases mitochondrial apoptotic priming. (C) Priming measured by BIM peptide is significantly increased in four Bcl-xL knockout lung adenocarcinoma cell lines. (D) Bcl-xL deletion is sufficient to switch cell fate of p53-restored lung adenocarcinoma cells from cell cycle arrest to apoptosis. Data shown is from cells harvested 72 h after p53 restoration. (E) Fold-change in the percentage of Annexin V–7-AAD double-positive control or Bcl-xL knockout cells 72 h after p53 restoration. Data in C and E represent the mean ± SEM, n = 3 or more. Statistics were calculated with two-sided Student’s t test: **P < 0.01, ***P < 0.001, ****P < 0.0001; n.s., not significant.
Although a useful proof-of-concept, genetic ablation of antiapoptotic proteins cannot be applied to a patient. However, small-molecule BH3-mimetic drugs are now available, including ABT-263 (navitoclax), which is a dual BCL-2/BCL-XL antagonist that binds in the BH3-binding pocket of these antiapoptotic proteins and competes for binding with proapoptotic BCL-2 family members (27). Addition of ABT-263 to lung adenocarcinoma and sarcoma cell lines increased their priming (Fig. 4 A, B, D, and E) in a similar manner to deletion of BCL-XL. While ABT-263 by itself was unable to induce apoptosis, restoring p53 in the presence of ABT-263 resulted in robust induction of apoptosis, as measured by nuclear fragmentation and an increase in the sub-G1 fraction (Fig. 4 C and F). Therefore, the addition of BH3 mimetics to therapeutic p53 reactivation or induction strategies may sensitize tumors that would otherwise undergo cell cycle arrest, raising the possibility that combining these pharmacological agents with drugs like MDM2 inhibitors in cases of wild-type p53 tumors might increase tumor apoptosis.
Fig. 4.
Pharmacological priming of lung adenocarcinoma and sarcoma cell lines is sufficient to switch cell fate of p53-restored cells from cell cycle arrest to apoptosis. (A) BH3 profile of lung adenocarcinoma cells after navitoclax addition showing increased priming. (B) Priming measured by BIM peptide is significantly increased upon ABT263 treatment. (C) Lung adenocarcinoma cells show increased cell death as indicated by nuclear degradation. (D) BH3 profile of sarcoma cells after navitoclax addition showing increased priming. (E) Priming measured by BIM peptide is significantly increased upon ABT263 treatment. (F) Sarcoma cells show increased cell death as indicated by nuclear degradation. *P < 0.05, **P < 0.01, *** P < 0.001; ns, not significant.
To test this hypothesis in vivo, we engineered sarcoma cells to overexpress the BH3-only sensitizer protein Bad (Fig. 5A) (28). We confirmed that while Bad overexpression by itself was insufficient to cause cell death (SI Appendix, Fig. S4), it nonetheless led to a measurable increase in the levels of mitochondrial apoptotic priming prior to p53 restoration (Fig. 5B). We then injected these cells subcutaneously into immunodeficient (nude) mice and after 2 wk of growth restored of p53 function (Fig. 5C). Remarkably, restoration of p53 in established sarcomas resulted in robust cell death as gauged by a substantial increase in the number of cleaved caspase 3+ cells compared to nonrestored cells or cells expressing empty vector (Fig. 5D). Collectively, these data demonstrate that altering the levels of mitochondrial apoptotic priming in tumors is sufficient to alter the fate of tumor cells in vivo upon p53 reactivation.
Fig. 5.
Increased mitochondrial priming by forced expression of BAD is sufficient to switch the fate of sarcoma tumors to apoptosis in vivo. (A) Western blot to verify Bad overexpression in stably transduced sarcoma cells. MIG-Empty = empty vector. MIG-Bad = Bad cDNA expressing vector. (B) BH3 profile of sarcoma cells transduced with empty or BAD-expressing vector showing increased priming by forced expression of BAD. (C) Schematic of experiment for assessing cell fate in vivo. (D) Immunohistochemistry of sarcoma sections stained for cleaved caspase 3 to indicate apoptosis. BAD expressing tumors show much greater apoptosis as indicated by the increased number of cells with cleaved caspase 3 staining. (Magnification: D, 4×; D, Inset, 40×.) *P < 0.05, **P < 0.01; ns, not significant.
Reduction of Priming in Lymphoma Is Sufficient to Bias Cells Away from Apoptosis and toward Cell Cycle Arrest.
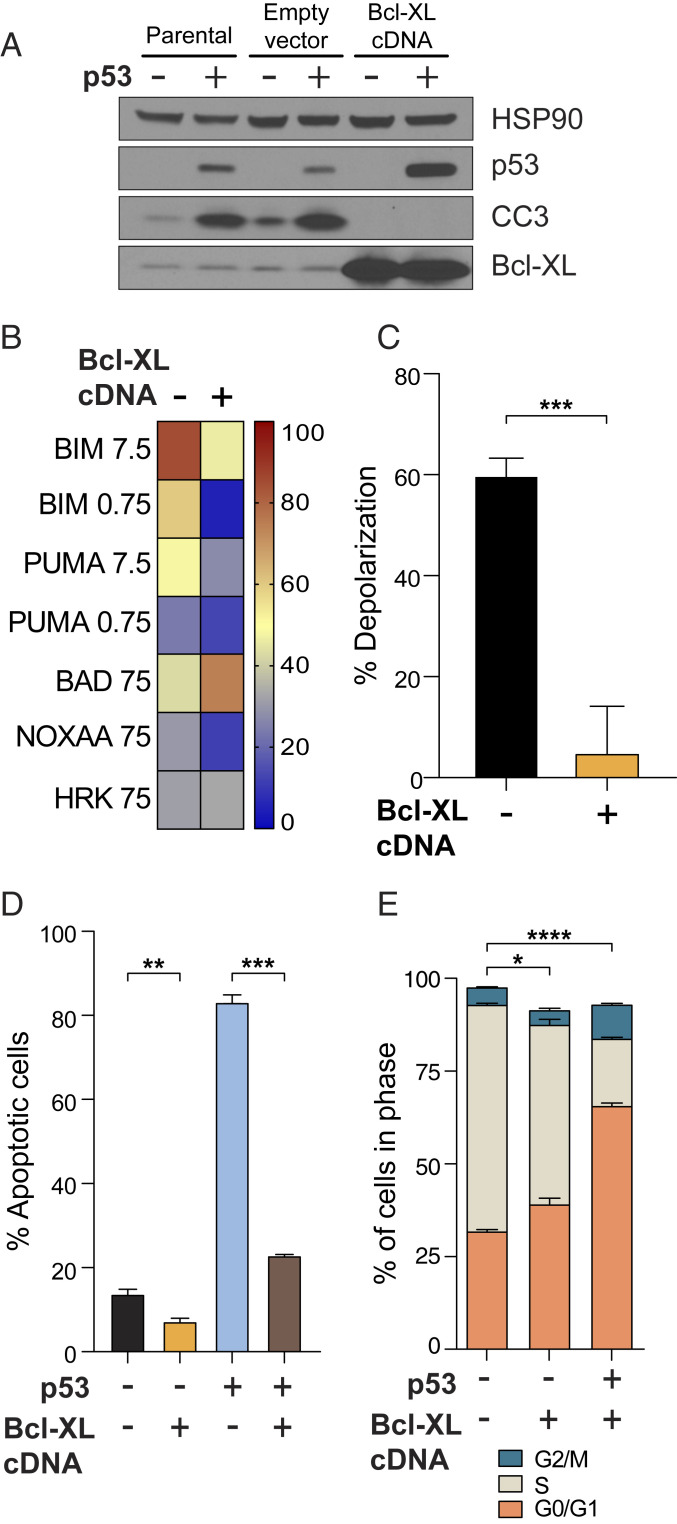
If our hypothesis were correct and the fate of cells upon p53 restoration depends on the initial level of mitochondrial priming, then it might be possible to change the fate of lymphoma cells by reducing their priming so that priming is more like that of adenocarcinoma and sarcoma. To test this, we engineered lymphoma cells to overexpress the antiapoptotic protein BCL-XL (Fig. 6A). Expression of BCL-XL was sufficient to reduce mitochondrial priming in lymphoma cells (Fig. 6 B and C). This decrease in mitochondrial apoptotic priming was sufficient to switch the fate of lymphoma cells upon p53 restoration, as demonstrated by the absence of CC3 (Fig. 6A) and a significant decrease in the percentage of annexin V+ cells to levels comparable to p53-null cells (Fig. 6D). These cells were now able to survive p53 restoration and demonstrated a statistically significant smaller S-phase population and a build-up of cells in G0/G1 (Fig. 6E), suggesting that the cell cycle arrest program is functionally engaged upon p53 restoration and only becomes apparent when apoptosis is no longer eliminating the cells.
Fig. 6.
Decreased mitochondrial priming by forced expression of BCL-XL is sufficient to switch the fate of p53-restored lymphoma cell lines to cell cycle arrest. (A) Forced expression of BCL-XL prevents caspase 3 cleavage after p53 restoration and (B) globally reduces mitochondrial priming in lymphoma cell lines. (C) Reduction of mitochondrial priming by BCL-XL expression as measured with BIM 0.75 μM. (D and E) Reduction of mitochondrial priming by BCL-XL expression significantly blunts p53 restoration-mediated apoptosis (D) and leads to a statistically significant decrease in the accumulation of S-phase p53-restored lymphoma cells and a concomitant accumulation in G1 and G2 (E). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Discussion
Using a set of defined GEMM-derived p53 restorable lung adenocarcinoma, sarcoma, and lymphoma cell lines in combination with BH3 profiling, we have demonstrated that the level of mitochondrial apoptotic priming is a critical determinant of cell fate in the context of p53 restoration in vitro and in vivo. We further show that modulation of mitochondrial apoptotic priming via genetic or pharmacological means is sufficient to change cell fate upon p53 restoration: increasing priming enforces an apoptotic cell fate, whereas decreasing priming enforces cell cycle arrest.
In considering the adoption of cell fates after p53 activation, one can consider whether commitment to cell cycle arrest or apoptosis is first adjudicated. If arrest were adjudicated first, then manipulation of apoptotic signaling would not affect the proportion of cells undergoing cell cycle arrest. However, our data show that we can affect the proportion of cells that undergo cell cycle arrest simply by manipulating apoptotic signaling (Fig. 7A). Specifically, we show that increasing priming can decrease the proportion of cells that undergo cell cycle arrest, whereas decreasing priming can increase this proportion. Therefore, our data suggest that one could in principle order the adjudication of fates, with apoptosis first being adjudicated followed by cell cycle arrest.
Fig. 7.
The fate of p53 restored cells depends on mitochondrial apoptotic priming. (A) Model depicting a scenario in which increasing mitochondrial apoptotic priming decreases the proportion of cells that undergo cell cycle arrest, whereas decreasing priming would increase this proportion. Diagram created with BioRender.com. (B) Pharmacological priming of cancer cells is sufficient to switch cell fate upon p53 restoration: I, Poorly primed sarcoma and lung adenocarcinoma cells increase mitochondrial priming in response to p53 restoration but do not reach the threshold of apoptosis; II, dual inhibition of BCL-XL and BCL-2 by navitoclax increases mitochondrial priming such that additional priming by p53 restoration crosses the apoptotic threshold; III, lymphoma cells have higher basal mitochondrial priming, and additional p53 induced priming is sufficient to cross the apoptotic threshold; IV, reduction of basal mitochondrial priming by expression of an antiapoptotic protein is sufficient to block p53-restored cells from crossing the apoptotic threshold and instead leads to cell cycle arrest and senescence.
The issue of apoptotic priming and cell fate after p53 activation has previously arisen in a different context. Bowen et al. (18) used BH3 profiling to show that two different cell types, human neural crest cells (hNCC) and early smooth muscle cells, exhibited similar mitochondrial apoptotic priming. Nonetheless, when p53 was activated, either with Nutlin 3A or with doxorubicin, hNCCs exhibited a greater propensity to adopt an apoptotic cell fate. They went on to show that this was related to a significantly greater transcriptional induction of p53-driven genes in the case of the apoptosis-driven hNCC cells. Thus, in this case, the different cell fates seemed better attributed to differential magnitude of p53 activation rather than differential priming, which has also been observed in other contexts (29).
In our experiments, we instead compared cells with different priming and very similar postactivation p53 levels and signaling intensity as gauged by protein levels of p21 (encoded by the Cdkn1a gene) and gene-expression levels of several direct and indirect p53 targets (SI Appendix, Figs. S5 and S6). We do not consider our results incompatible with those of Bowen et al. (18). Instead, a parsimonious model would take into account both preactivation apoptotic priming, as well as the intensity of the p53 signal that follows induction. That is, one can imagine two cells with similar priming adopting different cell fates if they are exposed to different magnitudes of p53 proapoptotic transcription induction, with the cell enduring more signaling more likely to adopt an apoptotic cell fate. Similarly, two cells enduring similar magnitudes of p53 proapoptotic transcription induction would adopt different cell fates if their preactivation apoptotic priming were different, with the more primed cell more likely to adopt an apoptotic cell fate. We note, in fact, that in a developmental context, cell fate following the p53-dependent response to DNA-damaging agents has been related to differential apoptotic priming (21). The relation among cell fate (F), priming (P), and p53 activation (A) might hence be considered analogous to the equation F = P + A, where an apoptotic cell fate is chosen when F exceeds a threshold.
The goal of many types of cancer chemotherapy is to induce apoptosis selectively in cancer cells. Often, the apoptotic signaling induced by these cancer cells is p53-dependent. Our results here suggest that we can guide cancer cells toward adopting an apoptotic cell fate by increasing their apoptotic priming prior to inducing p53, either with drugs or genetic manipulation (Fig. 7B). Clinically, drug treatment is far more relevant. We have shown previously that we can use dynamic BH3 profiling to measure drug-induced apoptotic signaling to identify drugs that selectively prime cancer cells (30, 31). Our results here suggest a rational strategy of constructing combinations, at least in those cancer cells with functional p53, by combining agents that prime cancer cells with those known to induce p53. Importantly, we show that drugs acting independently of p53 can be rationally chosen to enhance apoptosis after p53 activation.
Materials and Methods
Generation of Cell Lines and Cell Culture.
Bcl-xL knockout cells were generated by transient transfection using the pX458 vector (Addgene, 48138) essentially as described in the protocol by Ran et al. (32) using sgBcl-xL.2 (5′-G CCCAGCTTCACATAACCCCA-3′), where the G in bold was added for ensuring appropriate U6 transcription. Bcl-2 and Bcl-xL knockdown cell lines were generated using MSCV-LTR-MIR30-SV40-GFP (MLS) retroviral vectors (33) expressing potent validated short-hairpin RNA (34, 35). Bcl-xL and Bad overexpressing cell lines were generated using pMIG-Bcl-XL (Addgene, #8790) and pMIG-Bad (Addgene, #8787) retroviral constructs, respectively (28). The pMIG-Empty retroviral construct (Addgene, #9044) was used as control. Retroviral particles were generated and packaged using Phoenix cell system (G. Nolan, Stanford University, Stanford, CA). Cells were maintained in DMEM (lung adenocarcinoma and sarcoma cell lines) or IMDM (lymphoma cell lines) supplemented with 10% fetal bovine serum and gentamicin. For p53 restoration experiments, 4-hydroxy-tamoxifen (Sigma) was added at a final concentration of 250 to 500 nM for a total of 72 h.
Antibodies.
The following antibodies were used for Western blotting experiments: anti–Bcl-xL (CST, #2764; 1:1,000), anti-p53 (Novocastra, NCL-p53-505; 1:1,000), anti-Hsp90 (BD, 610418; 1:10,000 to 1:15,000), anticleaved caspase 3 (CST, #9661; 1:1,000), anti-Bad (CST, #9292; 1:1,000), and anti-p21 (Santa Cruz, sc-6246).
Western Blotting.
Cells were lysed with RIPA buffer (BP-115, Boston BioProducts) supplemented with 1× protease inhibitor solution (cOmplete EDTA-free, 11873580001, Roche). Protein concentration of cell lysates was determined by Pierce BCA protein assay (23225, Thermo Fisher Scientific). Total protein (50 μg) was separated on 4 to 12% Bis-Tris gradient SDS-PAGE gels (Life Technologies) and then transferred to PVDF membranes (IPVH00010, EMD Millipore) for blotting.
Cell Cycle Analysis.
For cell cycle analysis, we used a FITC BrdU Flow Kit (559619, BD Biosciences). Briefly, 1 million cells were plated in triplicate with or without p53 restoration for a total of 72 h. Cells were then labeled with 10 μM BrdU for 30 to 60 min, and subsequently fixed and stained with anti-BrdU and 7-AAD and analyzed by flow cytometry.
Annexin V–7-AAD Analysis.
Cells were cultured identically as described for cell cycle experiments. Seventy-two hours after p53 restoration, cells were analyzed using the BioLegend assay kit (640919) following the manufacturer’s guidelines.
Standard BH3 Profiling.
BH3 profiling was performed essentially as described in Ryan and Letai (24). Cells were harvested, by trypsinization for lung adenocarcinoma and sarcoma cell lines, and washed once in PBS before being suspended in MEB buffer (150 mM mannitol, 50 mM KCl, 0.02 mM EDTA, 0.02 mM EGTA, 0.1% protease free BSA, 5 mM succinate, 10 mM Hepes, final pH 7.5) at 6.5 × 105 cells/mL. Fifteen microliters of cell suspension was added to each well of a black 384 well plate containing 2× mixtures of each peptide in MEB supplemented with 2 μM JC-1 dye, 0.005% digitonin, 20 μg/mL oligomycin, 10 mM 2-mercaptoethanol. Mitochondrial potential measurements were taken every 5 min for 3 h using a Tecan Safire2 plate reader exciting at 545 nm and reading the red fluorescence of JC-1 at 590 nM with bandwidth of 20 nm on excitations and emission at 30 °C. The area under each curve was normalized to the areas under the DMSO and FCCP curves as follows: % depolarization = 1 − [(Sample – FCCP)/(DMSO – FCCP)].
iBH3 Profiling.
Lymphoma cells were collected after 24 h of p53 restoration and washed once in PBS before being resuspended MEB2P25 buffer (150 mM Mannitol, 150 mM KCl, 5 mM succinate, 1 mM EDTA, 1 mM EGTA, 0.1% BSA, 2.5g/L Polaxamer 188, 10 mM Hepes pH 7.4) and added to assay plates containing MEB2P25 with 0.002% (wt/vol) digitonin and peptides at 2× final concentration. Cells were exposed to peptides and digitonin for 1 h, fixed for 10 min by the addition of formaldehyde to a final concentration of 1%, and neutralized by adding a volume of 3 M Tris base equal to half that of the formaldehyde. Retained cytochrome c was stained by adding clone 6H2.B4 in 10× BD perm buffer to a final dilution of 1:400 overnight. Cells were analyzed on an Intellicyt iQue Screener Plus, gated on cells with intact nuclei (G1-M-G2), and cytochrome c quantified as the median fluorescence intensity (MFI) of the cytochrome c antibody. MFI values were normalized to Buffer (full stain) and no antibody (no stain) controls and expressed as percent release as follows:
Animal Experiments.
All animal studies described in this study were approved by the Massachusetts Institute of Technology Institutional Animal Care and Use Committee. A total of 500,000 sarcoma cells were injected subcutaneously into n = 5 Nude mice from Taconic at the age of 6 to 8 wk. Tamoxifen (Sigma) was dissolved in corn oil (Sigma) and a single injection was administered intraperitoneally at 200 μg per gram of total body weight, as in Feldser et al. (7).
Immunohistochemistry.
All immunohistochemistry procedures were performed essentially as described in Sánchez-Rivera et al. (36). Mice were killed by carbon dioxide asphyxiation. Sarcoma tumors were fixed with 4% paraformaldehyde (PFA) overnight, transferred to 70% ethanol, and subsequently embedded in paraffin. Sections were cut at a thickness of four micrometers and stained with H&E for routine pathological examination. Immunohistochemistry was performed on a Thermo Autostainer 360 machine. Slides were antigen-retrieved using Thermo citrate buffer, pH 6.0 in the pretreatment module. Sections were treated with Biocare rodent block, primary antibody, and anti-Mouse (Biocare) or anti-rabbit (Vector Labs) HRP-polymer. The slides were developed with Thermo Ultra DAB and counterstained with hematoxylin in a Thermo Gemini stainer and coverslipped using the Thermo Consul coverslipper. Cleaved caspase 3 was detected using anticleaved caspase 3 (CST, #9661; 1:1,000). All pictures were obtained using a Nikon 80i microscope with a DS-U3 camera and NIS-elements software.
Statistical Analyses.
All statistical analyses and plots were produced using Prism 8 (GraphPad). Error bars represent SD, unless otherwise noted. We used Student’s t test (unpaired, two-tailed) to assess significance between treatment and control groups, and to calculate P values. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank members of the T.J., A.L., and M.T.H. laboratories, as well as Edward R. Kastenhuber (Weill Cornell Medicine) and José Reyes (Memorial Sloan Kettering Cancer Center) for scientific discussions; K. Cormier and C. Condon from the Hope Babette Tang (1983) Histology Facility for technical support; and G. Paradis, M. Griffin, M. Jennings, and M. Saturno-Condón for flow cytometry and FACS support. This work was supported by the HHMI, by National Cancer Institute (NCI) R35CA242427, and in part by Cancer Center Support (core) Grant P30-CA14051 from the NCI. F.J.S.-R. was supported by a Repligen Corporation KIICR Graduate Fellowship and the Massachusetts Institute of Technology (MIT) Biology NIH Pre-Doctoral Training Grant T32GM007287. Y.M.S.-F. was supported by the NCI of the NIH under Award F31-CA183405 and the MIT Biology NIH Pre-Doctoral Training Grant T32GM007287. M.T.H. is the Eisen and Chang Associate Professor of Biology. T.J. is a HHMI Investigator, the David H. Koch Professor of Biology, and a Daniel K. Ludwig Scholar.
Footnotes
Competing interest statement: A.L. discloses consulting and sponsored research agreements with AbbVie, Novartis, and Astra-Zeneca. He serves on the Scientific Advisory Board of Flash Therapeutics, Dialectic Therapeutics, and Zentalis Pharmaceuticals. The following are US Patents regarding BH3 profiling owned by Dana-Farber: 10,393,733; 9,902,759; 9,856,303; 9,540,674; 8,221,966; and 7,868,133. A.L. and J.R. are inventors on patent applications US20180128813A1 and US20180120297A1 held/submitted by the Dana-Farber Cancer Institute that covers high-throughput BH3 profiling. T.J. is a member of the Board of Directors of Amgen and Thermo Fisher Scientific. He is also a cofounder of Dragonfly Therapeutics and T2 Biosystems. T.J. serves on the Scientific Advisory Board of Dragonfly Therapeutics, SQZ Biotech, and Skyhawk Therapeutics. None of these affiliations represent a conflict of interest with respect to the design or execution of this study or interpretation of data presented in this report. T.J.’s laboratory currently also receives funding from the Johnson & Johnson Lung Cancer Initiative and The Lustgarten Foundation for Pancreatic Cancer Research, but this funding did not support the research described in this report.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019740118/-/DCSupplemental.
Data Availability
All study data are included in the article and SI Appendix.
References
- 1.Polager S., Ginsberg D., p53 and E2f: Partners in life and death. Nat. Rev. Cancer 9, 738–748 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Momand J., Jung D., Wilczynski S., Niland J., The MDM2 gene amplification database. Nucleic Acids Res. 26, 3453–3459 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lane D., Levine A., p53 Research: The past thirty years and the next thirty years. Cold Spring Harb. Perspect. Biol. 2, a000893 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu X., Tied up in loops: Positive and negative autoregulation of p53. Cold Spring Harb. Perspect. Biol. 2, a000984 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kastenhuber E. R., Lowe S. W., Putting p53 in context. Cell 170, 1062–1078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khoo K. H., Verma C. S., Lane D. P., Drugging the p53 pathway: Understanding the route to clinical efficacy. Nat. Rev. Drug Discov. 13, 217–236 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Feldser D. M., et al., Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature 468, 572–575 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martins C. P., Brown-Swigart L., Evan G. I., Modeling the therapeutic efficacy of p53 restoration in tumors. Cell 127, 1323–1334 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Ventura A., et al., Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Xue W., et al., Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–660 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donehower L. A., et al., Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356, 215–221 (1992). [DOI] [PubMed] [Google Scholar]
- 12.Jacks T., et al., Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4, 1–7 (1994). [DOI] [PubMed] [Google Scholar]
- 13.Zambrowicz B. P., et al., Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. U.S.A. 94, 3789–3794 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson L., et al., Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 410, 1111–1116 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Junttila M. R., et al., Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature 468, 567–571 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris J. P. 4th, et al., α-Ketoglutarate links p53 to cell fate during tumour suppression. Nature 573, 595–599 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson J. G., Post S. M., Lozano G., Regulation of tissue- and stimulus-specific cell fate decisions by p53 in vivo. J. Pathol. 223, 127–136 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Bowen M. E., et al., The spatiotemporal pattern and intensity of p53 activation dictates phenotypic diversity in p53-driven developmental syndromes. Dev. Cell 50, 212–228.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Letai A. G., Diagnosing and exploiting cancer’s addiction to blocks in apoptosis. Nat. Rev. Cancer 8, 121–132 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Ni Chonghaile T., et al., Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science 334, 1129–1133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J. C., et al., High mitochondrial priming sensitizes hESCs to DNA-damage-induced apoptosis. Cell Stem Cell 13, 483–491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarosiek K. A., et al., Developmental regulation of mitochondrial apoptosis by c-Myc governs age- and tissue-specific sensitivity to cancer therapeutics. Cancer Cell 31, 142–156 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Certo M., et al., Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell 9, 351–365 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Ryan J., Letai A., BH3 profiling in whole cells by fluorimeter or FACS. Methods 61, 156–164 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryan J., Montero J., Rocco J., Letai A., iBH3: Simple, fixable BH3 profiling to determine apoptotic priming in primary tissue by flow cytometry. Biol. Chem. 397, 671–678 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Sánchez-Rivera F. J., Jacks T., Applications of the CRISPR-Cas9 system in cancer biology. Nat. Rev. Cancer 15, 387–395 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tse C., et al., ABT-263: A potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 68, 3421–3428 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Cheng E. H. Y. A., et al., BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol. Cell 8, 705–711 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Le Pen J., et al., Constitutive p53 heightens mitochondrial apoptotic priming and favors cell death induction by BH3 mimetic inhibitors of BCL-xL. Cell Death Dis. 7, e2083 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhola P. D., et al., High-throughput dynamic BH3 profiling may quickly and accurately predict effective therapies in solid tumors. Sci. Signal. 13, eaay1451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montero J., et al., Drug-induced death signaling strategy rapidly predicts cancer response to chemotherapy. Cell 160, 977–989 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ran F. A., et al., Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dickins R. A., et al., Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat. Genet. 37, 1289–1295 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Jiang H., Pritchard J. R., Williams R. T., Lauffenburger D. A., Hemann M. T., A mammalian functional-genetic approach to characterizing cancer therapeutics. Nat. Chem. Biol. 7, 92–100 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pritchard J. R., et al., Defining principles of combination drug mechanisms of action. Proc. Natl. Acad. Sci. U.S.A. 110, E170–E179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sánchez-Rivera F. J., et al., Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 516, 428–431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.