Abstract
Plant disease outbreaks are increasing and threaten food security for the vulnerable in many areas of the world. Now a global human pandemic is threatening the health of millions on our planet. A stable, nutritious food supply will be needed to lift people out of poverty and improve health outcomes. Plant diseases, both endemic and recently emerging, are spreading and exacerbated by climate change, transmission with global food trade networks, pathogen spillover, and evolution of new pathogen lineages. In order to tackle these grand challenges, a new set of tools that include disease surveillance and improved detection technologies including pathogen sensors and predictive modeling and data analytics are needed to prevent future outbreaks. Herein, we describe an integrated research agenda that could help mitigate future plant disease pandemics.
Keywords: emerging plant disease, plant pathology, food security
The United Nations declared 2020 the International Year of Plant Health. It is estimated that food production will need to increase by 60% by 2050 to feed the estimated 10 billion people expected on Earth (1, 2). An increase in production along with a reduction in food loss due to pests and pathogens and food waste will be needed to meet demand (3, 4). Global yield losses due to crop pests and diseases on food crops are large, with mean losses ranging from 21.5% (10.1 to 28.1%) in wheat, 30.3% (24.6 to 40.9%) in rice, 22.6% (19.5 to 41.4%) in maize, 17.2% (8.1 to 21%) in potato, and 21.4% (11 to 32.4%) in soybean (3). In some regions of the world, plant diseases also cause significant preharvest losses for smallholder farmers, with nearly 50% of beans and maize farmers surveyed in Central America and nearly 50% of potato farmers surveyed in South America experiencing loss (4) (SI Appendix, Fig. S1). Plant diseases cause significant losses in food crop production that lead not only to lower yields but also to loss of species diversity, mitigation costs due to control measures, and downstream impacts on human heath (3).
The National Academy of Sciences recently published an ambitious agricultural research agenda that emphasized the need for breakthrough technology for the early and rapid detection and prevention of plant diseases (5). Emerging plant diseases are diseases that 1) have increased in either incidence, geographical, or host range; 2) have changed pathogenesis; 3) have newly evolved; or 4) have been discovered or newly recognized (6). Emerging plant disease and pest outbreaks affect food security, national security, and human health, with serious economic implications for agriculture (6). Emerging plant diseases have already become more frequent, and in coming decades it is expected that shifts in the geographic distributions of pests and pathogens in response to climate change and increased global commerce will make them both more frequent and severe (7–9). Plant diseases influence many components of food security, and effective management will lead to both improved crop production and human health (10). Herein, we discuss the impacts of emerging plant diseases on food production and food security, discuss why plant pathogens emerge, and recommend an integrated research agenda that can be implemented by global science and development agencies to help prevent emerging plant diseases and improve mitigation strategies when plant disease outbreaks do occur.
Plant Diseases Impact Food Security
During the global food crisis of 2007/2008 food price spikes occurred, pushing millions into hunger (11, 12). These price spikes were subsequently associated with political unrest and food riots in the Middle East and Africa (2, 12). This prompted the former US Secretary of State, John Kerry, to remark that “access to food and agriculture needs to be the cornerstone of development strategy” (13). The climate crisis is expected to disrupt food supply chains and increase plant diseases, leading to both lower crop yields and reductions in food stockpiles within countries (12).
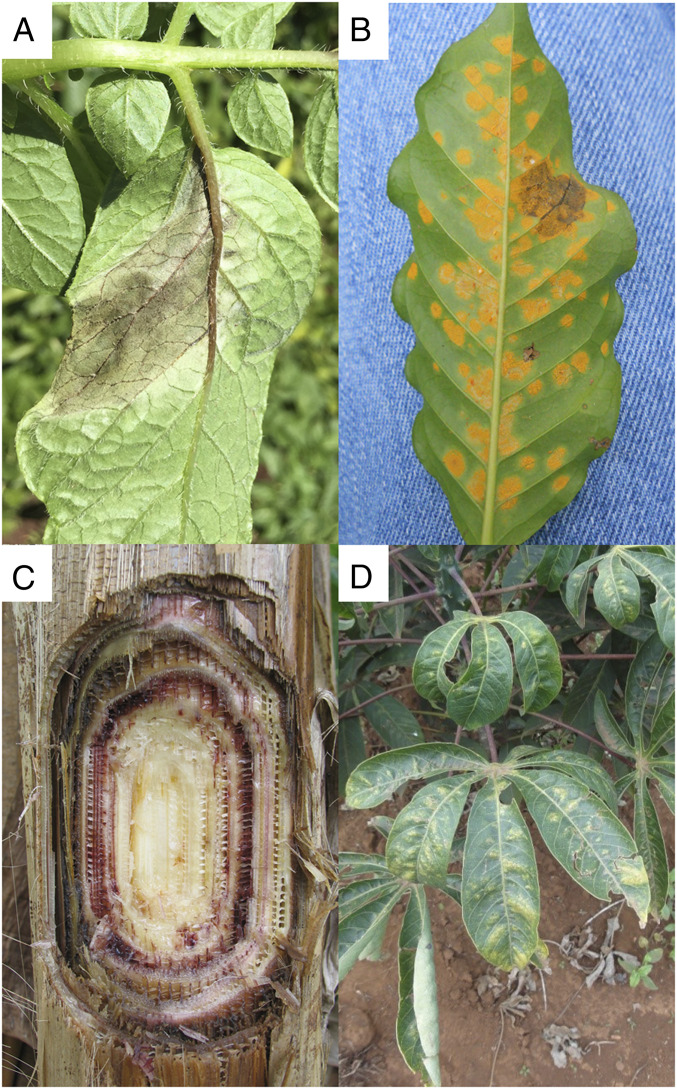
Plant diseases are of global concern and exact a heavy toll on food crop production and the social and political stability of nations. A classic example is the 19th century Irish potato famine. In 1845, Phytophthora infestans destroyed the potato crop and in subsequent years led to the Irish famine with over 2 million deaths and mass migration of people from Ireland (14) (Fig. 1A). The pathogen first emerged in the United States in 1843 with less-severe consequences than in Ireland where dependence on a single food crop, the lack of social and political will, and the delay by the British government to address the hunger problem led to dire food insecurity for the Irish people (14). Conflict, poverty, and the British rule during World War II made the Bengal famine worse as Cochliobolus miyabeanus, the cause of brown spot on rice, spread, resulting in the death of over 2 million Bengalese people. Rice production dropped by 25% and the country’s rice supplies were shifted to feed troops (15). The recent coffee rust outbreaks caused by Hemileia vastatrix in Central America provide yet another example of the displacement of people due to an emerging endemic plant disease and climate change (Fig. 1B) (16). Yield losses in coffee greater than 50% occurred in some regions of Central America and over 400,000 coffee workers lost their livelihoods in the coffee sector in Honduras, El Salvador, and Guatemala, leading to hunger, poverty, and increased migration (17). Monoculture of susceptible coffee varieties, low coffee prices, and climate change in Central America led to the spread of coffee rust to higher elevations where growers were ill-prepared and lacked access to fungicides to control the disease (16).
Fig. 1.
Several important emerging plant diseases that threaten food security, including (A) late blight of potato caused by P. infestans, (B) coffee rust caused by H. vastatrix, (C) Panama disease caused by F. oxysporum f. sp. cubense (TR4) on banana, and (D) cassava mosaic disease caused by East African CMV.
Unlike endemic disease that is usually managed, emerging plant diseases can pose shocks to agricultural productivity, and thus we risk undercutting other input investments in agriculture that alleviate hunger unless the deleterious impacts of plant diseases on crop production are considered in policy discussions (1, 4, 18). Food security exists when all people, at all times, have physical, social, and economic access to sufficient, safe, and nutritious food to meet their dietary needs and food preferences for an active and healthy life (1). The four components of food security include access, availability, utilization, and stability (3), and plant diseases affect these components (Table 1). Fusarium oxysporum f. sp. cubense tropical race 4 (TR4) that causes Panama disease of banana threatens to reduce the availability of banana in some areas of the world (19) (Fig. 1C) (Table 1). The TR4 strain has moved from Asia into Mozambique, Jordan, and, in 2019, Colombia (20). The susceptible cultivar Cavendish is grown in monoculture in Central America and has no resistance to TR4. The disease threatens farms of both subsistence farmers in Asia and Africa and major banana plantations. Thus, the availability of banana may be greatly reduced (19). Cassava is a major staple in the diet of millions of East Africans (Table 1). Cassava viruses (21) threaten access to a stable supply of cassava. The onset of cassava mosaic disease in Uganda in the 1980s led to precipitous drops in cassava production, thus limiting access to this important food source (21). Cassava mosaic virus (CMV) causes over $1 billion in losses in East Africa, and cassava yields have been reduced by 13 million tons annually (Fig. 1D) (21). Conflict, civil war, and drought also exacerbated the problem. The utilization of corn and groundnuts (peanut) as sources of nutrition in Africa has been severely compromised by aflatoxins produced primarily by Aspergillus flavus and other fungi (Table 1) (22). When contaminated corn is consumed, stunting of growth in children and impairment of health, including cancer, can occur. The levels of aflatoxins in corn in East Africa are high, and the lack of uniform standards in Africa for detection of aflatoxins and risk assessment hinders food safety regulations (22). The stability of the premium and fair-trade coffee supply in Central America was severely disrupted and reduced by coffee rust (16, 17) (Fig. 1B and Table 1). Over 120,000 smallholder farmers in Guatemala were impacted by increased production costs of fungicides to control coffee rust, and many were left in poverty due to lack of income from coffee and were forced to emigrate (17).
Table 1.
Impact of emerging plant diseases on four components of food security on key food and subsistence crops
| Component | Definition | Example of a plant disease | Consequence |
| Availability | The existence of food in a particular place and time | F. oxysporum f. sp. Cubense tropical race 4 (TR4) | Cavendish banana, a key food source for many smallholder farmers, is threatened by TR4 race of F. oxysporum f. sp. cubense that migrated from southeast Asia to Mozambique (19, 20). The disease could eliminate production of the crop in some areas of the world. |
| Access | The ability of a person or group to obtain food | Cassava mosaic disease caused by East African CMV (EACMV-!UG2) | A strain of a CMV caused huge losses and cassava fields were abandoned in sub-Saharan Africa. Food shortages and famine-related deaths occurred in Uganda due to dependence on cassava (21). |
| Utilization | The ability to use and obtain nourishment from food (includes food nutritional value assimilation of nutrients) | Mycotoxins on corn caused by A. flavus and Fusarium species | Eighty-seven percent of East Kenyan corn mills had over the legal limit of fumonisins in corn (22). Consuming fumonosin-affected corn affects nutritional value of corn and is carcinogenic. |
| Stability | The absence of significant fluctuation in availability, access and utilization | Coffee rust | Coffee yields reduced by 16 to 31% in Central and South America. Low price of coffee and lack of inputs such as new varieties and fungicides exacerbated disease (16, 17). Smallholder income was lost for food purchases and stability of the commodity in the region was threatened. |
| H. vastatrix |
Why Do Plant Pathogens Emerge?
Plant diseases emerge by a range of means (6). An increase in incidence, geography, or host range of a pathogen can occur by movement of pathogens in infected plant material (9), as was the case in the recent introductions of wheat blast into Bangladesh in infected seed (23). Extreme weather events, such as hurricanes, can transport pathogen spores over continents, as illustrated by soybean rust movement from Brazil to the United States via Hurricane Ivan (24). Years after the introduction of citrus tristeza virus in South America, an insect vector was introduced that led to wider geographic spread of the disease (6). Plant pathogens may shift hosts and gain the ability to infect new hosts when introduced into new regions. For example, CMV does not naturally occur on cassava in South America but moved into cassava in Africa from an unknown host following uptake of cassava as a major food crop there (21). A change in pathogenesis or virulence of an endemic strain can occur, as exemplified by the recent alarm over a new race of the wheat stem rust pathogen in Ethiopia (race TTKTT) with a very high combination of virulence genes and a new race in Sicily that could overcome resistance in a widely grown European cultivars of wheat (25, 26). Newly evolved plant pathogenic species can also occur through interspecific hybridization or mutations within existing pathogen lineages, as illustrated by the emergence of Phytophthora andina in South America and Phytophthora alni in the United Kingdom (27, 28).
A Convergence Research Agenda to Manage Emerging Plant Diseases
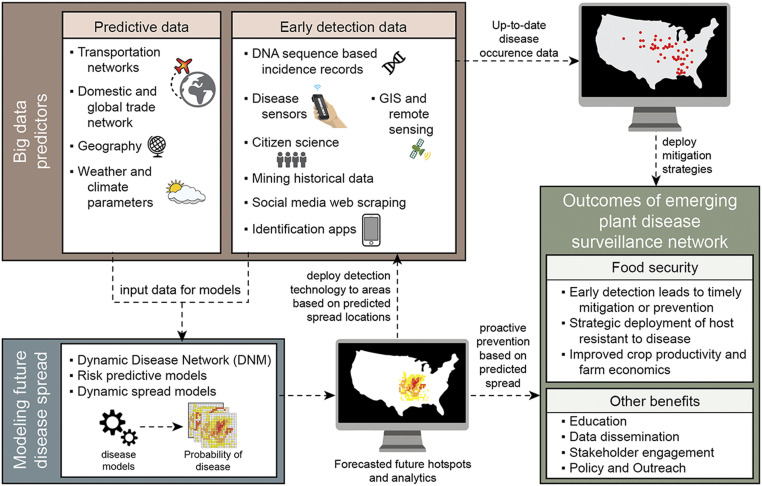
Convergence science is the integration of knowledge, methods, and expertise from different disciplines to form novel frameworks to catalyze scientific discovery and innovation. Science innovation is needed to tackle emerging plant diseases and will require interdisciplinary teams of researchers that work at multiple scales from the molecular to the landscape level (Fig. 2). Fundamental research on pathogen biology is needed to dissect the complexities of disease transmission and emergence of novel pathogen lineages. The integration of such knowledge can be mobilized and accelerated by convergence science. Predictive data such as transportation and trade networks, geography, weather and climate parameters, and early pathogen detection data from DNA sequences, incidence records, disease sensors, citizen science, data mining, and web scraping tools and disease identification applications can be input into models to develop dynamic disease surveillance networks. These data can be used to model and predict future outbreak hotspots on the landscape level (Fig. 2). Breakthroughs in novel technology development such as rapid pathogen detection sensors and artificial intelligence to analyze multiple data streams will enable disease surveillance to be scaled, thus greatly expanding the input datasets that inform disease prediction models. Thus, more rapid responses by stakeholders to contain disease outbreaks and improved farm economies can occur (Fig. 2).
Fig. 2.
Critical components and data analytics needed for an emerging plant disease surveillance network. Data include predictive data such as transportation and trade networks, geography, weather and climate parameters, and early detection data such as DNA sequence data, pathogen detection from sensors, text mining of historical and social media data, citizen science data, and identification applications. These data can be used to model spread of plant diseases and predict future spread. Enhanced monitoring and proactive mitigation strategies can be deployed in forecasted future hotspots. Early detection of plant diseases leads to more timely deployment of mitigation strategies.
The Need for Emerging Plant Disease Surveillance and Monitoring Systems
Microbial threats to human health and food safety are in the daily view of the public and policymakers, as illustrated by the recent pandemic caused by COVID-19. The risks posed by emerging plant disease outbreaks are equally important yet often underreported (3, 6, 29). Coordinated global monitoring and surveillance of emerging human diseases was begun in 1994 by the Program for Monitoring Emerging Diseases (ProMED) (6). ProMED rapidly disseminates written reports of global outbreak information on infectious diseases in humans and animals, and since 1995 in plants, via email and reaches 70,000 subscribers in 185 countries. A team of science experts moderates and reviews ProMed reports before posting to the network. Information is dispersed in email reports. Notably, ProMED monitoring first revealed the Wuhan outbreak of COVID-19 in December 2019 (https://promedmail.org/promed-post/?id=20191230.6864153).
Although there are many national and regional plant protection agencies that monitor disease, a coordinated global reporting mechanism is still lacking (30). The challenges with monitoring and surveillance for emerging plant diseases are that plant diseases are underreported, and reports are overly influenced by the sociology of science (science as a social activity), in addition to the politics and economics of nations and international agriculture (6, 30). There are political considerations around “who should legitimately report” disease outbreaks and who has access to the information (6, 30). Countries are often reluctant to report plant disease outbreaks because of the potential economic consequences, including barriers for export markets. Oftentimes, maps of emerging plant disease are as much an artifact of where projects and scientists are active as where disease is actually emerging (29–31). The areas of highest detected plant disease emergence are often in the developed world, simply due to more funding to conduct and publish work in these areas (8, 31). Policymakers need to be made aware that global plant disease surveillance systems are important, as agricultural food production is inextricably linked to both human and animal health. There is a need to increase data sharing and real-time surveillance of emerging plant diseases to improve the timeliness and accuracy of reporting of plant disease outbreaks globally and improve the food security of all reporting nations.
Geospatial Analytics to Monitor and Understand Outbreaks
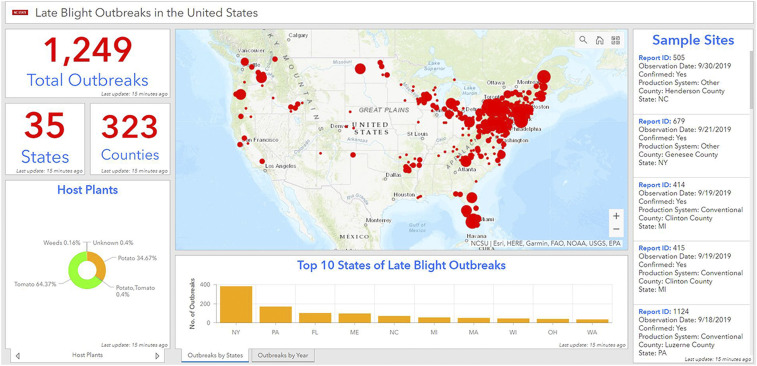
Geographic information systems (GIS) are an essential tool for mapping and analyzing data about healthy and diseased hosts in order to respond to potential disease threats in landscapes and agricultural systems (32, 33). Many forecasts of disease in food production regions are based on routine pest and pathogen monitoring via sentinel plots and weather monitoring and forecasting (24). Real-time web-based GIS applications allow users to see reported crop disease outbreaks within the regions of interest. Unfortunately, only a few plant pathogens are actively monitored globally using disease surveillance systems (30). For example, the disease alert and forecasting systems for late blight in the United States (https://usablight.org/) and in Europe (https://agro.au.dk/forskning/internationale-platforme/euroblight/) uses real-time monitoring to map outbreaks, genotype lineages, and detect fungicide sensitivity of pathogen lineages (32, 33). USABlight.org sends alerts to growers nearby when the pathogen is detected via text or email (32, 33) (Fig. 3). Use of a late blight decision support tool to target fungicide sprays saved growers from $3,706 to $4,201 per hectare depending on the variety planted (34). Another system, the Global Rust Reference Center, tracks the yearly outbreak of yellow rust and stem rust on wheat and provides genotyping and race identification to guide breeding efforts and deployment of resistant varieties (35). There is a need for global plant disease reporting on geographic scales so comparisons of disease can be made across continents and countries. Pathogen detection by citizen scientists using mobile applications and geotagged cell phone images could be used in addition to predictive models to map outbreak risks for many plant diseases (36).
Fig. 3.
Reports of late blight caused by P. infestans on potato and tomato in the United States mapped using geospatial analytics. Data are analyzed from USABlight (2011 to 2019). The total number of outbreaks and the number of states and counties reporting over 9 y are shown. Most of the reports were from tomato and the top 10 states that were hot spots of disease in the eastern United States are indicated.
Disease surveillance systems are used in developed countries, but often the information is not shared 1) because it can confer a competitive economic advantage or 2) because what happens in one region is not viewed as important by growers in another region. More funding by larger intergovernmental organizations such as the Consultative Group for International Agriculture Research (CGIAR), Food and Agriculture Organization of the United Nations, or the World Bank would enable scaling plant disease surveillance (30). This information could be used both by regulators within countries and decision-makers on the ground so that appropriate mitigation strategies can be deployed to prevent pathogen spread once an outbreak is identified. This will require long-term investment by intergovernmental agencies since many surveillance systems are now fragmented and firewalled (30). Greater interagency and international collaboration with the goal of shared access to data for research and disease mitigation could expand disease surveillance. Geospatial surveillance systems funded by intergovernmental agencies that provide shared data on disease outbreaks and that are active in real time are needed by stakeholders to mitigate threats.
Pathogen Risk Assessment and Modeling to Predict Outbreak Spread
Epidemiological tools can be used to predict risk of outbreaks. Models can vary from simple deterministic mathematical models to spatially explicit stochastic simulations and decision support systems (37). Meteorologically driven dispersal models can identify the risks of long-distance movement of spores to show which countries are at risk. For example, five major zones for exchange of rust spores of wheat stem rust throughout Africa, the Middle East, and South Asia were identified. Predominant donor countries, notably Yemen, acted as a stepping stone for wheat rust into and out of Africa, thus identifying sources and countries that were at risk (38). Near-real-time early-warning systems in Ethiopia for within-country spread of wheat stem and stripe rust have been developed that enable growers to apply fungicide effectively to reduce crop losses (39). Critical parameters for dispersal and transmission within the landscapes must be estimated under prevailing environmental conditions for many pathogens, allowing for discontinuities in landscape connectedness (40).
Bayesian statistical methods allow any prior knowledge to inform parameter estimates based on emerging patterns of spread, as well as estimates that become progressively more resolved over time as an outbreak progresses (41). Probability distributions of estimated values may be used to parameterize stochastic, spatiotemporal epidemic models to predict the likely spread of disease across the landscape. This can highlight uncertainties in current knowledge, as well as the variability of epidemic spread driven by both climate as well as the inherent unpredictability of epidemics. Models are also used to inform surveillance programs (42), to optimize disease control (37), and for web-based decision support via “serious games” to help inform stakeholders and policymakers about options for disease control (43). Linking epidemiological with economic models allows additional insights into how to deploy control when resources are limited (44). Open-source code and software that is scalable to multiple projects and shared knowledge across animal, plant, and human systems could help build capacity more rapidly for resource-poor regions (37). Modeling to accurately predict both the potential for spread of plant diseases and translation to stakeholders’ communities is needed to inform policy and control strategies.
Emerging Pathogen Risks through Trade
International trade has a growing role in providing national food supplies across the globe (45, 46). Just seven countries form the backbone of the global agricultural trade network, each trading with greater than 77% of countries worldwide (47). Thus, pathogen movement risk is concentrated where trade is prevalent (48). For example, the United States is a dominant player in global agricultural trade, and its total agricultural trade activity (mass and trade value of imports + exports) surpassed that of any other country in recent years (49). Major trade partners include China, Europe, Canada, and Mexico. Thus, monitoring pest and disease outbreaks from the major trading export country partners is likely to result in greater likelihood of detecting pests or diseases (49).
New epidemiological tools that utilize dynamic network analysis to study the key drivers of pathogen movement with trade are needed (Fig. 2). Dynamic network models (DNMs) can represent a landscape as a system of interconnected suitable patches, defined as nodes, and consider links between nodes based on the probability of movement of pathogens or pests (50). The integration of risk components based on socioeconomic, weather, or trade factors can offer a more complete system perspective. Several studies have begun to use DNMs to examine the structure of global crop breeding and seed systems networks to examine the spread of crop resistance genes at the landscape level (51–53). Networks of postharvest transportation were used to determine risk of pathogen and mycotoxin movement in stored grains (54). By tracing the transport of stored wheat by rail through states using network analysis, hub locations were identified that provide higher risk of transporting pests and pathogens and priority control points for inspection and controls (54). Social network models were used to analyze patterns of maize trade among nations and to determine where vulnerabilities in food security might arise if maize availability were decreased from shortages, trade tariffs, or diversion to nonfood uses (55). Dynamic and social network models and analysis that include trade data to predict plant disease spread are needed and can be used to identify the critical control points where potential introduction risks of emerging pathogens could be stopped or mitigated more quickly to reduce the costs of mitigation.
Earth Observations to Assess Climate-Driven Disease Spread
Along with increased trade connectivity among countries, warming climates may facilitate the spread of new pathogens, since increased climate suitability may expand the range of plant pathogens and their vectors and hosts into uncolonized areas (7, 8). Weather and climate also play an integral role in determining if conditions are suitable for disease at a particular place and time. However, risk analysis requires pest and pathogen occurrence data along with weather modeling, thus emphasizing the important need for surveillance and detection networks (56) (Fig. 2). Decision support systems are under development that use Earth observation data. One such example is the Pest Risk Information Services (PRIZE) that uses environmental data and land-surface temperature and rainfall combined with models of pest life cycles to make pest risk assessments and was recently used to predict and reduce impact of fall armyworm in corn in east Africa (57).
Weather conditions are routinely monitored across the globe, with reliable forecasts days in advance. The wheat rust early warning system in Ethiopia uses 1- to 7-d weather forecast data from the UK Meteorological Office in combination with spore dispersal models and ground surveillance data (35). A shift in the distribution of plant pests and pathogens, and a movement poleward, has been documented using structured archival published datasets from CABI and climate data (7). Latitudinal trends in the data suggest climate impacts on disease.
However, data are limited on measuring actual disease occurrence on whole plants using satellite imagery because it is difficult to resolve specific diseases. Innovation in hyperspectral imaging is now overcoming this barrier (58). The ability to track the movement of plant pathogens in the atmosphere is also important for establishing effective quarantine measures and forecasting disease spread (59). Drones (unmanned aerial vehicles, UAVs) have been used to study the transport of plant pathogens tens to hundreds of meters above crop fields. Sporangia of P. infestans have been detected 90 m and higher above potato fields using UAVs, showing the potential of drones for atmospheric sampling of plant pathogens (59). Images from drones combined with remote sensing are being used to detect plant water stress and monitor disease spread (58). As resolution from remote sensing and satellite imagery improves, disease at the individual plant or field scale will be possible with both UAV and imagery data. Information can be used in conjunction with models to make appropriate plant disease management decisions based on anticipated patterns of spread of pathogens (42, 59). The expanded use of high-resolution earth observation and weather data including hyperspectral and satellite imagery and drones can help identify and discern impacts of specific plant diseases on crops.
Sensors for Detection of Plant Diseases
Research is rapidly expanding in the development of in-field sensors for detection of plant pathogens on food crops (60–62) (SI Appendix, Fig. S2). For example, a microneedle molecular diagnostic platform was developed that couples a quick microneedle nucleic acid extraction with a loop-mediated isothermal amplification–based sensor to detect coinfections in tomato by P. infestans and tomato spotted wilt virus. A smartphone-based plant volatile organic compound (VOC) sensing platform has also recently been developed that uses multiplexed nanoplasmonic and organic dye sensor arrays prepared on low-cost paper substrates for early detection of late blight (P. infestans), early blight, and Septoria blight on tomato (SI Appendix, Fig. S2) (61). The VOC sensors detect disease before symptoms occur. Wearable, wireless sensors for detection of plant disease are on the horizon.
Development and deployment of new information and communication technologies through smartphone applications that incorporate pathogen-specific sensors could help facilitate risk mitigation strategies. Timely and accurate plant disease diagnosis by field clinicians using sensors, particularly in resource-poor areas that lack extension and plant disease clinics, will help expand collection of data on regional and country levels that can be fed into surveillance and prediction models and improve detection and reporting of newly emerged pathogens. Expanded development of electronic and wearable plant disease sensors for rapid detection of plant pathogens will enable more rapid tracking of emerging pathogens.
Data Mining and Big Data Analytics for Geographic Monitoring
Data mining technologies can be used to examine potential pathogen threats to major food crops. Archival data and recently available “big data” streams from multiple structured (published papers, archival printed documents, PubMed and CAB Abstracts, and Google Ngram) and unstructured data sources (social media, image posts, and Twitter feeds) can be used with natural-language processing tools to monitor and map infectious plant diseases and pathogens (63–65). Data mining tools may be used that periodically scan the internet and social media for plant diseases. New information critical to decision-makers can be obtained from “big data” for efficient management of crop diseases and to examine potential disruptions that could make food supplies more resilient to outbreaks. By identifying the control points we may be able to predict the resilience of cropping systems in specific geographic areas and identify hot spots of disease outbreaks (31).
Few large-scale studies using data mining to study the spread of emerging plant disease have been done (65–67). Natural-language processing tools and geoparsing were used to track the 19th-century spread of late blight in the United States around the ports of New York and Philadelphia and revealed methods used to control the disease and speculations on the source of outbreaks (66). In another example, over 12,500 disease reports were mined and collated to examine the genus-wide distribution and invasiveness of Phytophthora species (67). Countries at risk for Phytophthora outbreaks and the invasive potentials of dominant species were predicted. Data mining techniques using natural-language processing and geoparsing, text mining, and crowd sourcing of social media data should be harnessed as a resource for monitoring past and predicting future plant pathogen outbreaks. This will improve global food security vigilance, policy planning, research, and implementation of phytosanitary plant health services.
Population Genomic Surveillance for Monitoring Emerging Plant Pathogens
Currently, with few exceptions, most plant disease surveillance tracking systems do not incorporate molecular genotyping due to expense and lack of infrastructure to process large numbers of samples (33, 35, 68, 69). Plant disease alert and surveillance systems that track pathogen genotypes and races and use bioinformatic predictive tools to identify novel strains of pathogen lineages can help reveal the source of the outbreak strains. The next-generation sequencing revolution introduces exciting opportunities for using low-cost, evolutionary genetic discovery to track movement of plant pathogens. Genomic and bioinformatic approaches can be applied to elucidate the origins, ancestry, and functional evolution of important plant pathogens and lineages, especially when historical collections are available (70). The use of natural history collections including mycological collections has been especially useful for understanding historical outbreaks, identifying pathogen lineages and sources of new outbreaks, and determining whether an outbreak lineage is new or reproducing sexually (28, 33, 68–70). Enhanced digitization of herbarium specimens and data mining can be used to produce baseline reports of outbreaks for major food crops (70). This is particularly exciting in the context of parameterizing pathogen spread models, for which methods have emerged to infer dispersal from successive spatial snapshots (71). Genomic data can be used to constrain chains of transmission more tightly, thereby allowing better model parameterization.
Genomic mining of both archival and present data outbreak strains promises to help trace and time the presence of a particular plant disease and can help identify migration routes of past outbreaks strains (28, 70, 72). Genomic mining can trace the evolution and geographic origins of pathogen species, identify where highest diversity and greatest threat to crop populations may exist, and reveal the particular dynamics of virulence of past outbreaks. New methods that exploit low-depth sequencing (e.g., “genome skimming”) hold power to monitor both extinct and extant plant diseases. Low-depth sequencing of museum specimens, grouped in time and space to form “populations,” could be used for population-level tracking of past plant pathogen outbreaks and push back the dates of first reports in countries (72, 73). Rapid in-field sequencing technologies using Minion sequencers have been used to reduce time and cost to identify strains of rust fungi (74). Nextstrain is currently being used to track COVID-19 outbreak strains and provide real-time phylogenies and open sequence data. These open phylogenetic tools could be used to track plant disease outbreak strains and reduce time of discovery of novel lineages (75, 76). Additional reference genome assemblies for emerging plant pathogens, diverse sequenced panels of globally diverse plant pathogens, and open sequence reporting platforms are needed to fully exploit rapidly advancing population genomics technologies for tracking outbreak strains.
Extension and Digital Advisory Services for Smallholders to Identify Plant Diseases
Lack of access to timely, appropriate, and actionable extension advice is a major cause of poor productivity and crop loss for smallholders in the developing world. Pest and disease outbreaks can flare up unpredictably and cross borders quickly, and often there is no mechanism to rapidly identify the problem and suggest effective, pragmatic solutions. Work in the developing world by organizations such as CABI’s Plantwise clinics has helped support establishment of local “plant clinics” staffed by trained “plant doctors” where smallholder farmers get practical advice (77). Plant disease clinics and digital disease surveillance needs to be expanded in the developing world (30). The African Crop Epidemiology System (ACES) initiative started by the Bill & Melinda Gates Foundation and a set of UK international development partners aims to develop an early-warning plant health system for East Africa on a set of targeted pathogens and crops. Reductions in crop losses can occur by timely identification of problems and implementation of appropriate management practices (30). Plant disease clinics need to be expanded in developing countries so plant health information can be used more efficiently by regional regulatory agencies. Information can then be disseminated to stakeholders in the form of factsheets, distribution maps, diagnostic tools, and pest management information. Expanded plant disease clinics in the developing world networked with electronic data collection devices to record/report pathogen data in a timely fashion to authorized stakeholders will provide opportunities for early disease warning for mitigation response.
Prospects
Plant diseases do not recognize political borders, yet country borders are often used to stop the sharing of information on plant disease outbreaks (31). The migration of plant pathogens with trade is likely to increase with the emergence of new trade relationships, compounding the impacts of climate change on agricultural productivity in some regions of the world (29). More comprehensive surveillance strategies for plant diseases that include strategic partnerships among research universities, development agencies, nongovernmental organizations, the private sector, and CGIAR are needed. Research programs focused on disease surveillance and epidemiology for the high-impact plant pathogens on major food crops such as wheat, potato, cassava, banana, corn, and rice are needed to prevent plant pathogen spread and predict the global burden of plant disease.
Currently, disease surveillance is woefully underfunded and is only done on a global scale for wheat rusts and potato late blight (30, 33, 35). Modeling, surveillance, and bioinformatics tools in many cases are ready to be deployed on larger landscape-level scales by research, phytosanitary, regulatory, and development agencies, but an organized strategy is needed (30). The open sharing of datasets by researchers and policymakers is needed as well as threat scenario training so coordinated responses can be made to control plant diseases that affect crop yields. New outbreaks can be predicted with disease surveillance systems and data can be used to determine the origin of outbreak strains and calculate rates of spread. Global hot spots of new emerging diseases can be identified (Fig. 2). This information will be useful to deploy resistant germplasm, fungicides, or other eradication methods at the landscape level. The time for strategic thinking is now as the food security of resource poor countries is threatened by both plant diseases and the COVID-19 pandemic that has impacted food production, access, supply chains, and the health of agricultural workers.
Supplementary Material
Acknowledgments
J.B.R. thanks Dr. Fred Gould, member of the National Academy of Sciences, for his useful comments on this manuscript. The National Academies Keck Futures Initiative Grant (NAKFIES9) and the Rockefeller Foundation provided funding for a Bellagio Conference Grant (88220) on “Emerging Infectious Diseases of Africa in the Context of Ecosystem Services.” Appreciation is also expressed to the North Carolina State University Provost’s Office; The Nusbaum Foundation; The Kenan Institute; The College of Agriculture and Life Sciences, the College of Engineering, and the College of Sciences at North Carolina State University; and Research Triangle International and Novozymes for funding of a conference on “Emerging Plant Disease and Global Food Security,” where ideas were generated for this work (https://globalfoodsecurity.ncsu.edu/). We thank Shannon Jones for preparing Fig. 2, Xingli Ma for preparing Fig. 3, and Amanda Saville for formatting all figures. The views and opinions expressed in this paper are those of the authors and not necessarily the views and opinions of the United States Agency for International Development or any of the authors’ institutions.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022239118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
Change History
October 1, 2021: Table 1 has been updated; please see accompanying Correction for details.
References
- 1.Food and Agriculture Organization of the United Nations , The State of Food and Agriculture 2019: Moving Forward on Food Loss and Waste Reduction (Food and Agricultural Organization, Rome, Italy, 2019). [Google Scholar]
- 2.Fedoroff N. V., Food in a future of 10 billion. Agric. Food Secur. 4, 11 (2015). [Google Scholar]
- 3.Savary S., et al., The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Delgado L., Schuster M., Torero M., “The reality of food losses: A new measurement methodology” (IFPRI Discussion Paper 1686, International Food Policy Research Institute, Washington, DC, 2017). http://ebrary.ifpri.org/cdm/ref/collection/p15738coll2/id/131530.
- 5.National Academies of Sciences, Engineering, and Medicine , Science Breakthroughs to Advance Food and Agricultural Research by 2030 (National Academies Press, Washington, DC, 2019). [Google Scholar]
- 6.Anderson P. K., et al., Emerging infectious diseases of plants: Pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 19, 535–544 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Bebber D. P., Ramotowski M. A. T., Gurr S. J., Crop pests and pathogens move polewards in a warming world. Nat. Clim. Chang. 3, 985–988 (2013). [Google Scholar]
- 8.Bebber D. P., Range-expanding pests and pathogens in a warming world. Annu. Rev. Phytopathol. 53, 335–356 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Brasier C. M., The biosecurity threat to the UK and global environment from international trade in plants. Plant Pathol. 57, 792–808 (2008). [Google Scholar]
- 10.Larkin B. J., Emerging Technologies to Benefit Farmers in Sub-Saharan Africa and South Asia (National Academies Press, Washington, DC, 2009). [Google Scholar]
- 11.Food and Agriculture Organization of the United Nations , “The state of food insecurity in the World: The multiple dimensions of food security” (Food and Agriculture Organization, Rome, Italy, 2013).
- 12.International Food Policy Research Institute , “Global food policy report: 2016” (International Food Policy Research Institute, Washington, D.C., 2016).
- 13.Kerry J., “Alleviating global hunger: Challenges and opportunities for US leadership, United States Foreign Relations Committee” (United States Government Printing Office, Washington DC, 2009).
- 14.Bourke P. M. A., Emergence of potato blight, 1943-46. Nature 203, 805–808 (1964). [Google Scholar]
- 15.Padmanabhan S. Y., The Great Bengal famine. Annu. Rev. Phytopathol. 11, 11–24 (1973). [Google Scholar]
- 16.Avelino J., et al., The coffee rust crises in Colombia and Central America (2008–2013): Impacts, plausible causes and proposed solutions. Food Secur. 7, 303–321 (2015). [Google Scholar]
- 17.Sieff K., The migration problem is a coffee problem. Washington Post, 11 June 2019. https://www.washingtonpost.com/world/2019/06/11/falling-coffee-prices-drive-guatemalan-migration-united-states/. Accessed 7 October 2019.
- 18.Foley J. A., et al., Solutions for a cultivated planet. Nature 478, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Stokstad E., Devastating banana disease may have reached Latin America, could drive up global prices. Science 365, 207–208 (2019). [DOI] [PubMed] [Google Scholar]
- 20.García-Bastidas F. A., et al., First report of Fusarium wilt Tropical Race 4 in Cavendish bananas caused by Fusarium odoratissimum in Colombia. Plant Dis. 104 (2019). [Google Scholar]
- 21.Legg J. P., et al., Cassava virus diseases: Biology, epidemiology, and management. Adv. Virus Res. 91, 85–142 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Mutiga S. K., Hoffmann V., Harvey J. W., Milgroom M. G., Nelson R. J., Assessment of aflatoxin and fumonisin contamination of maize in western Kenya. Phytopathology 105, 1250–1261 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Inoue Y., et al., Evolution of the wheat blast fungus through functional losses in a host specificity determinant. Science 357, 80–83 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Isard S. A., Russo J. M., Ariatti A., The integrated aeriobiology modeling system applied to the spread of soybean rust into the Ohio Valley during September 2006. Aerobiologia 23, 271–282 (2007). [Google Scholar]
- 25.Battacharya S., Wheat rust back in Europe. Nature 542, 145–146 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Hei N. B., et al., First report of Puccinia graminis f. sp. tritici race TTKTT in Ethiopia. Plant Dis. 103, 982 (2019). [Google Scholar]
- 27.Gladieux P., et al., The population biology of fungal invasions. Mol. Ecol. 24, 1969–1986 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Martin M. D., et al., Genomic characterization of a South American Phytophthora hybrid mandates reassessment of the geographic origins of Phytophthora infestans. Mol. Biol. Evol. 33, 478–491 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bebber D. P., Holmes T., Smith D., Gurr S. J., Economic and physical determinants of the global distributions of crop pests and pathogens. New Phytol. 202, 901–910 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carvajal-Yepes M., et al., A Global surveillance system for crop diseases: Global preparedness minimizes the risk to food supplies. Science 364, 137–1239 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Scherm H., Thomas C. S., Garrett K. A., Olsen J. M., Meta-analysis and other approaches for synthesizing structured and unstructured data in plant pathology. Annu. Rev. Phytopathol. 52, 453–476 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Fry W. E., et al., The 2009 late blight pandemic in the eastern United States - Causes and results. Plant Dis. 97, 296–306 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Ristaino J. B., Cooke D. E. L., Acuña I., Muñoz M., “The threat of late blight to global food security” in Emerging Plant Disease and Global Food Security, Ristaino J. B., Records A., Eds. (American Phytopathological Society Press, 2020), pp. 403–420. [Google Scholar]
- 34.Liu Y., et al., A risk analysis of precision agriculture technology to manage tomato late blight. Sustainability 10, 3108 (2018). [Google Scholar]
- 35.Park R., et al., International surveillance of wheat rust pathogens: Progress and challenges. Euphytica 179, 109–117 (2011). [Google Scholar]
- 36.Meentemeyer R. K., Dorning M. A., Vogler J. B., Schmidt D., Garbelotto M., Citizen science helps predict risk of emerging infectious disease. Front. Ecol. Environ. 13, 189–194 (2015). [Google Scholar]
- 37.Thompson R. N., Brooks-Pollock E., Detection, forecasting and control of infectious disease epidemics: Modelling outbreaks in humans, animals and plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20190038 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer M., et al., Quantifying airborne dispersal routes of pathogens over continents to safeguard global wheat supply. Nat. Plants 3, 780–786 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Allen-Sader C., et al., An early warning system to predict and mitigate wheat rust diseases in Ethiopia. Environ. Res. Lett. 14, 115004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meentemeyer R. K., et al., Application of stochastic epidemiological models to realistic landscapes: Spread of the sudden oak death pathogen in California. Ecosphere 2, 1–24 (2011). [Google Scholar]
- 41.Thompson R. N., Gilligan C. A., Cunniffe N. J., Control fast or control smart: When should invading pathogens be controlled? PLOS Comput. Biol. 14, e1006014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parnell S., van den Bosch F., Gottwald T., Gilligan C. A., Surveillance to inform control of emerging plant diseases: An epidemiological perspective. Annu. Rev. Phytopathol. 55, 591–610 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Cunniffe N. J., Stutt R. O. J. H., DeSimone R. E., Gottwald T. R., Gilligan C. A., Optimizing and communicating options for the control of invasive plant disease when there is epidemiological uncertainty. PLoS Comput. Biol. 11, e1004211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forster G. A., Gilligan C. A., Optimizing the control of disease infestations at the landscape scale. Proc. Natl. Acad. Sci. U.S.A. 104, 4984–4989 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porkka M., Kummu M., Siebert S., Varis O., From food insufficiency towards trade dependency: A historical analysis of global food availability. PLoS One 8, e82714 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mehrabi Z., Ramankutty N., Synchronized failure of global crop production. Nat. Ecol. Evol. 3, 780–786 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Ercsey-Ravasz M., Toroczkai Z., Lakner Z., Baranyi J., Complexity of the international agro-food trade network and its impact on food safety. PLoS One 7, e37810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sikes B. A., et al., Import volumes and biosecurity interventions shape the arrival rate of fungal pathogens. PLoS Biol. 16, e2006025 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacDonald G. K., et al., Rethinking agricultural trade relationships in an era of globalization. Bioscience 65, 275–289 (2015). [Google Scholar]
- 50.Shaw M. W., Pautasso M., Networks and plant disease management: Concepts and applications. Annu. Rev. Phytopathol. 52, 477–493 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Garrett K. A., et al., Resistance genes in global crop breeding networks. Phytopathology 107, 1268–1278 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Buddenhagen C. E., et al., Epidemic network analysis for mitigation of invasive pathogens in seed systems: Potato in Ecuador. Phytopathology 107, 1209–1218 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Pautasso M., et al., Seed exchange networks for agrobiodiversity conservation. Agron. Sustain. Dev. 33, 151–175 (2013). [Google Scholar]
- 54.Hernandez Nopsa J. F., et al., Ecological networks in stored grain: Key postharvest nodes for emerging pests, pathogens, and mycotoxins. Bioscience 65, 985–1002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu F., Guclu H., Global maize trade and food security: Implications from a social network model. Risk Anal. 33, 2168–2178 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosenzweig C., et al., Assessing agricultural risks of climate change in the 21st century in a global gridded crop model intercomparison. Proc. Natl. Acad. Sci. U.S.A. 111, 3268–3273 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mbugua F., Bundi M., Day C., Beale T., Williams F., “PRISE-PAD fall armyworm SMS alert pilot results” (CABI Study Brief No. 35, CABI, Wallingford, UK, 2020).
- 58.Mahlein A. K., Plant disease detection by imaging sensors–Parallels and specific demands for precision agriculture and plant phenotyping. Plant Dis. 100, 241–251 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Schmale D. G. III, Ross S. D., Highways in the sky: Scales of atmospheric transport of plant pathogens. Annu. Rev. Phytopathol. 53, 591–611 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Paul R., et al., Extraction of plant DNA by microneedle patch for rapid detection of plant diseases. ACS Nano 13, 6540–6549 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Li Z., et al., Non-invasive plant disease diagnostics enabled by smartphone-based fingerprinting of leaf volatiles. Nat. Plants 5, 856–866 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Jackson R., et al., How remote sensing is offering complementing and diverging opportunities for precision agriculture users and researchers. Adv. Anim. Biosci. 8, 383–387 (2017). [Google Scholar]
- 63.Collier N., Uncovering text mining: A survey of current work on web-based epidemic intelligence. Glob. Public Health 7, 731–749 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomas C. S., Nelson N. P., Jahn G. C., Niu T., Hartley D. M., Use of media and public-domain Internet sources for detection and assessment of plant health threats. Emerg. Health Threats J. 4, 7157 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Shea J., Digital disease detection: A systematic review of event-based internet biosurveillance systems. Int. J. Med. Inform. 101, 15–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tateosian L., Guenter R., Yang Y., Ristaino J., “Tracking 19th century late blight from archival documents using text analytics and geoparsing” in Free and Open Source Software for Geospatial (FOSS4G) Conference Proceedings, Schwelk C. M., Zia M., Anderson A., Eds. (University of Massachusetts, Amherst, MA, 2017), pp. 146–155. [Google Scholar]
- 67.Scott P., Bader M., Burgess T., Hardy G., Williams N., Global biogeography and invasion risk of the plant pathogen genus Phytophthora. Environ. Sci. Policy 101, 175–182 (2019). [Google Scholar]
- 68.Saville A. C., Martin M. D., Ristaino J. B., Historic late blight outbreaks caused by a widespread dominant lineage of Phytophthora infestans (Mont.) de Bary. PLoS One 11, e0168381 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Islam M. T., et al., Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae. BMC Biol. 14, 84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ristaino J. B., The importance of mycological and plant herbaria in tracking a plant killer. Front. Ecol. Evol. 7, 521 (2020). [Google Scholar]
- 71.Jombart T., et al., Bayesian reconstruction of disease outbreaks by combining epidemiologic and genomic data. PLoS Comput. Biol. 10, e1003457 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bieker V. C., Martin M. D., Implications and future prospects for evolutionary analyses of DNA in historical herbarium collections. Bot. Lett. 165, 409–418 (2018). [Google Scholar]
- 73.Saville A. C., Ristaino J. B., Global historic pandemics caused by the FAM-1 genotype of Phytophthora infestans. BioRxiv [Preprint] (2021). 10.1101/2020.08.25.266759 (Accessed 5 May 2021). [DOI] [PMC free article] [PubMed]
- 74.Radhakrishnan G. V., et al., MARPLE, a point-of-care, strain-level disease diagnostics and surveillance tool for complex fungal pathogens. BMC Biol. 17, 65 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gardy J. L., Loman N. J., Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet. 19, 9–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hadfield J., et al., Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 34, 4121–4123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Romney D., et al., Plantwise: Putting innovation systems principles into practice. Agric. Dev. 18, 27–31 (2013). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.