Significance
The dynein–dynactin nanomotor is required for vastly diverse cellular functions, including cell division and cargo transport. Why dynactin interacts separately with the dynein motor and with microtubules is poorly understood and controversial. Here we directly interrogate the function of native dynein–dynactin teams on single phagosomes extracted from cells after disrupting the above interactions in a targeted manner ex vivo. Perturbing the dynactin–dynein interaction reduces dynein’s on rate to microtubules. However, perturbing the dynactin–microtubule interaction increases dynein’s off rate markedly when dynein generates force against an optical trap. Our results therefore ascribe separable functions to different components within the dynein–dynactin machine and provide mechanistic insight into dynein–dynactin dysfunction.
Keywords: dynein, dynactin, optical trap, supported lipid bilayer, intracellular transport
Abstract
The dynein–dynactin nanomachine transports cargoes along microtubules in cells. Why dynactin interacts separately with the dynein motor and also with microtubules is hotly debated. Here we disrupted these interactions in a targeted manner on phagosomes extracted from cells, followed by optical trapping to interrogate native dynein–dynactin teams on single phagosomes. Perturbing the dynactin–dynein interaction reduced dynein’s on rate to microtubules. In contrast, perturbing the dynactin–microtubule interaction increased dynein’s off rate markedly when dynein was generating force against the optical trap. The dynactin–microtubule link is therefore required for persistence against load, a finding of importance because disease-relevant mutations in dynein–dynactin are known to interfere with “high-load” functions of dynein in cells. Our findings call attention to a less studied property of dynein–dynactin, namely, its detachment against load, in understanding dynein dysfunction.
Transport of organelles inside cells is driven by motors of the kinesin and dynein families that generate force, respectively, toward the plus and minus ends of microtubules (MTs). Cytoplasmic dynein, aided by many regulators such as dynactin (Fig. 1A), executes bewilderingly diverse cellular functions (1). Dynactin’s largest subunit P150 has a coiled-coil (CC) region that contains a cytoskeleton-associated protein glycine-rich (CAPGly) domain along with a stretch of basic residues (2). Dynactin binds dynein through its CC1 domain, and dynactin also binds MTs through its CAPGly and basic domains (Fig. 1A). Additional interactions between dynein heavy chain and dynactin’s Arp domain may stabilize the complex (3, 4). How all these linkages help dynein–dynactin to function is hotly debated (5, 6). Of particular interest is the recruitment of dynein–dynactin to cargoes by different adaptor proteins (1), as also revealed by the cryogenic electron microscopy (cryo-EM) structure of dynein–dynactin–adaptor complexes wherein two dimeric dynein motors were found to be recruited as a pair (3, 4). Dynein undergoes significant conformational changes in this process, allowing both motor domains to align along the MT for effective force generation. Notably, recruitment of dynein in pairs to phagosomes was suggested by us on the basis of force measurements inside cells (7).
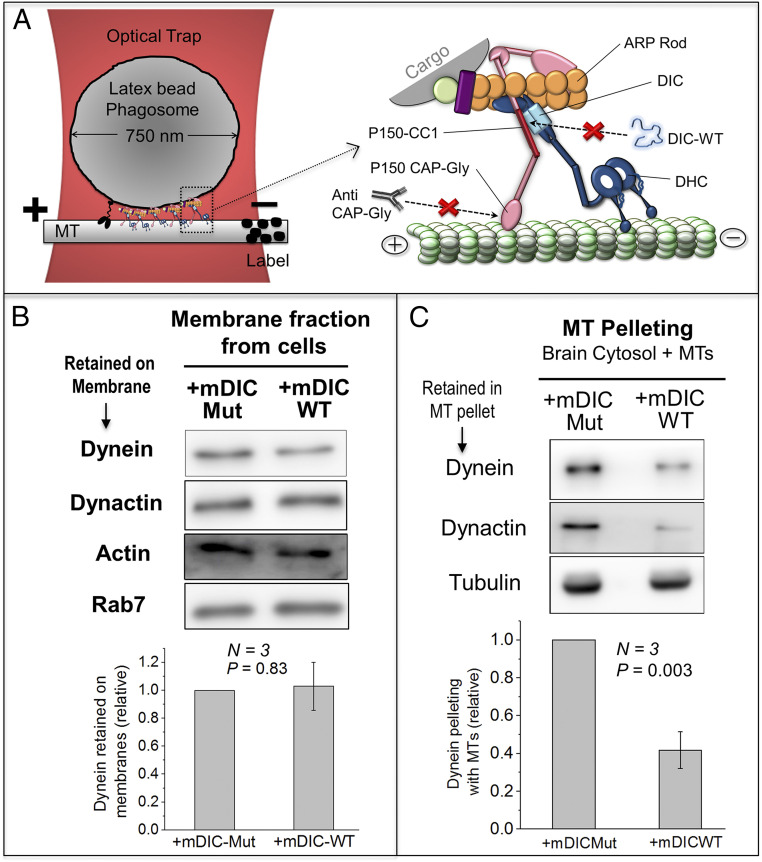
Fig. 1.
Effects of DIC-WT on dynein-membrane and dynein–MT binding. (A) (Left) Schematic of a single phagosome driven by a team of dyneins and a kinesin held in an optical trap over a MT. (Right) Magnified schematic of one dynein–dynactin complex. Treatments to target the DIC–CC1 interaction (with recombinant DIC protein) and CAPGly–MT interaction (with anti-CAPGly antibody) are shown. DIC, dynein intermediate chain; DHC, dynein heavy chain; ARP, actin-related protein. (B) Western blot showing levels of dynein (DIC) and dynactin (P150) retained on organelle membranes after treatment with mDIC-WT or mDIC-Mut. Membranes were prepared from RAW mouse macrophage cells. Rab7 is a marker for late endosomes. Actin is a loading control. Lower panel shows quantification of dynein band intensity across three experiments. The mDIC-Mut band intensity was always taken equal to 1 and mDIC-WT intensity calculated relative to mDIC-Mut. No significant difference is seen between mDIC-WT and mDIC-Mut treatments. Error bars, SEM. P value was calculated using Student’s t test. (C) Goat brain cytosol was treated with mDIC-WT or mDIC-Mut, followed by addition of exogenously polymerized MTs. Western blot shows levels of dynein (DIC) and dynactin (P150) that copelleted with MTs. Tubulin from the resuspended MT pellet is used for normalization. Lower panel shows quantification across three experiments. The mDIC-Mut band intensity was always taken equal to 1 and mDIC-WT intensity calculated relative to mDIC-Mut. Dynein and dynactin binding to MTs is reduced upon addition of mDIC-WT. Error bars, SEM. P values were calculated using Student’s t test.
Two kinds of studies, classified here as in vitro and in vivo, have investigated dynein–dynactin function. In vitro, dynactin was suggested to work as a brake for dynein (8); however, others found dynactin to enhance processive motion of dynein-driven beads (2, 9, 10). Purified dynein and dynactin, when complexed with bicaudal-D (Bic-D) activated dynein for motion (3, 11–14). An elegant demonstration of such activation came from McKenney et al. (12, 15) and Schlager et al. (14). MT binding by dynactin was suggested to initiate processive transport along tyrosinated MTs in vitro by reducing detachment of the dynein–dynactin complex from MTs (12, 15). In vivo studies have relied on overexpression or genetic perturbation of dynactin subunits inside cells. Studies targeting the dynactin–MT binding in neurons suggested that dynactin is not needed once retrograde transport from neurite tips has been initiated (16, 17). Perturbing dynactin–MT binding had no effect on processivity and step size of dynein, but affected MT organization in cells (18). In vitro, however, a CAPGly antibody reduced run length of dynein–dynactin-driven motion (9, 19) and loss of the CAPGly domain increased dynein detachment from MTs (8). The in vitro assays described above are bereft of many (known and unknown) protein regulators of dynein. Further, native-like function requires motors to be assembled on a lipid membrane on the cargo (20), a component missing in most in vitro assays. On the other hand, in vivo studies often cannot extract mechanistic details of function at the single-cargo level and could also suffer from off-target effects arising from genetic perturbations that are introduced in the cells.
To address these controversies, we fed micrometer-sized latex beads into Dictyostelium cells and allowed ingested beads to mature into phagosomes, wherein dynein and kinesin assembled in situ on the phagosome membrane. Phagosomes were then extracted from cells and motion and force generation of single phagosomes against an optical trap were interrogated ex vivo on polarity-labeled MTs (Fig. 1A). Most parameters of phagosome motion inside cells (7) were replicated in this ex vivo assay (21). Recombinant proteins and antibodies were then used to induce targeted perturbation of specific interactions within the dynein–dynactin machinery on phagosomes ex vivo (Fig. 1A). The consequence of these targeted perturbations was assayed by optical trapping and biochemical studies. Lastly, we reconstituted endogenous dynein–dynactin complexes on a lipid membrane that was assembled artificially on a bead (called supported lipid bilayer [SLB]). SLBs exhibit vigorous dynein–dynactin-driven motion, and targeted perturbation of dynein–dynactin on the SLBs replicates the effects seen on phagosomes. These SLB assays establish that the perturbations and their effects are indeed specific to the dynein–dynactin complex.
Perturbing the interaction between dynein intermediate chain (DIC) and the CC1 domain of dynactin reduces the binding rate (KON) of dynein–dynactin to MTs, but has little effect on the detachment rate (KOFF) of dyneins from the MT. In contrast, perturbing the dynactin–MT interaction increases KOFF most appreciably when dynein attempts to move against opposing force (i.e., load) exerted by an optical trap. Therefore, the DIC–CC1 interaction maintains dynein in an initial (unloaded) conformation that binds rapidly to MTs (high KON) to initiate motion. Once motion is initiated, the dynactin–MT interaction ensures tenacious binding (low KOFF) of dynein–dynactin to MTs against opposing force, allowing dynein–dynactin to compete against other motors and/or obstacles. We suggest a mechanism wherein a load-dependent communication operates across the dynein–dynactin scaffold and is necessary for the nanomachine to generate persistent force against opposition.
Results
Recombinant DIC Binds to CC1 without Grossly Disrupting the Dynein–Dynactin Complex.
An N-terminal domain peptide (residues 1 to 100) of the recombinant mammalian DIC protein competes against endogenous dynein for binding dynactin, but its constitutively phosphorylated mutant (Mut) (S84D) cannot bind dynactin (22). The DIC protein pulls down dynactin and disrupts organelle transport inside cells, but DIC-S84D has no such effects (23). Dictyostelium DIC has 56% sequence similarity to rat DIC and interacts with dynactin P150 via conserved residues (24). The N terminus region (1 to 126 amino acids [aa]) of Dictyostelium DIC was cloned with a C-terminal His-tag (hereafter called DIC-wild type [WT]). A pairwise sequence alignment mapped S84 of rat DIC to the S79 residue in Dictyostelium (SI Appendix, Fig. S1A). We therefore generated DIC-S79D (DIC-Mut), expecting it to function as a negative control. Dictyostelium as well as the mammalian counterparts of these proteins, mDIC-WT (=mDIC-S84A) and mDIC-Mut (=mDIC-S84D) were also prepared.
We first verified that Dictyostelium DIC-WT and DIC-Mut show phosphorylation-dependent affinity for dynactin. DIC-WT, but not DIC-Mut bound to CC1 in a dot blot assay (SI Appendix, Fig. S1B). The DIC proteins were next run on polyacrylamide gel electrophoresis (PAGE) under nondenaturing condition, overlaid with Dictyostelium lysate and probed using antibody against Dictyostelium P150 (SI Appendix, Fig. S1C). DIC-WT, but not DIC-Mut bound to P150, suggesting that it may competitively inhibit the DIC–CC1 interaction. We also confirmed that mDIC-WT (but not mDIC-Mut) binds to the mammalian CC1 domain of dynactin (SI Appendix, Fig. S1D). Direct interaction between DIC-WT and purified CC1 was demonstrated by a pull-down (SI Appendix, Fig. S1E). DIC-WT could also pull down endogenous dynactin from Dictyostelium cytosol (SI Appendix, Fig. S1F).
NudE is another regulator of dynein that binds DIC (25). Addition of DIC-WT could potentially sequester NudE away from phagosomes to affect dynein–dynactin function. However, NudE bound with similar affinity to DIC-WT and DIC-Mut (SI Appendix, Fig. S2A). Therefore, any differential effects of DIC-WT versus DIC-Mut (see below) cannot be explained via a NudE pathway. We next added FLAG-BicD to goat brain cytosol to form DDB complexes (12), then pulled down these complexes using FLAG antibody conjugated beads. The pull-down sample was separated into two equal portions that were treated with mDIC-WT or mDIC-Mut. FLAG beads were pelleted, washed, and checked for presence of mDIC-WT or mDIC-Mut using His antibody. Significantly more mDIC-WT was found in the pellet compared to mDIC-Mut (SI Appendix, Fig. S2B), showing that DIC-WT interacts with preformed DDB complexes, and may therefore be able to interfere with the dynein–dynactin interaction within such complexes. FLAG-BicD was next incubated with goat brain cytosol to form DDB complexes. The DDB complexes were pulled down using anti-FLAG beads, treated with mDIC-WT or mDIC-Mut, and then repelleted. There was no difference in dynein or dynactin between the two pellets (SI Appendix, Fig. S2C), showing that DIC-WT does not break apart the DDB complex. Taken together, DIC-WT interacts with CC1 within preformed DDB complexes to potentially perturb the dynein–dynactin interaction without grossly disrupting the complex.
DIC-WT Does Not Remove Dynein/Dynactin from Membranes and Mimics Dynein Distribution on Late Phagosomes.
If we wish to study the effect of DIC-WT, then DIC-WT addition should not remove dynein–dynactin from the cargo. To test this, we prepared a membrane fraction enriched in endosomes from mouse macrophage cells. Our motility assays used 0.25 μM DIC-WT to disrupt the dynein–dynactin interaction on phagosomes. We incubated the membrane fraction with a higher (=0.5 μM) concentration of mDIC-WT or mDIC-Mut, pelleted the membranes, and estimated levels of dynein and dynactin retained in the membrane pellet. No significant difference in dynein or dynactin was observed between the samples (Fig. 1B). Similar results were obtained using a membrane fraction extracted from Dictyostelium (SI Appendix, Fig. S3A). DIC-WT therefore does not remove dynein–dynactin from membranous cargoes, suggesting that its association to the membrane involves other dynein-dynactin subunits independent of DIC–CC1 interaction (23). We have shown (21) that dynein is distributed uniformly on early phagosomes (EPs), but appears punctate on late phagosomes (LPs). EPs and LPs were incubated with DIC-WT, after which DIC-WT was visualized on individual phagosomes. DIC-WT showed a uniform distribution on EPs, but was clearly punctate on LPs (SI Appendix, Fig. S3 C–F), suggesting a specific interaction of DIC-WT with dynein–dynactin within dynein-rich puncta on the LP membrane.
DIC-WT Reduces Binding of Dynein and Dynactin to MTs.
Cryo-EM images suggest that dynactin and BicD reorient dynein for efficient binding to MTs (3, 4, 11). Perturbing the dynein–dynactin interaction by DIC-WT could therefore reduce dynein binding to MTs. This reduction could happen because dynein’s on rate to MTs (= KON) is reduced, or also because dynein detaches rapidly after attachment (i.e., its off rate KOFF from MTs is increased). To test these possibilities, we first used a MT-pelleting assay. Dynein and dynactin can associate with each other in cytosol (26, 27). Clarified goat brain cytosol treated with mDIC-WT or mDIC-Mut was incubated with in vitro polymerized MTs, after which MTs were pelleted. This experiment was done in the absence of adenosine triphosphate (ATP), where dynein’s KOFF from MTs becomes ∼100-fold lower than at physiological ATP (28). The MT-pelleting assay therefore reports largely on the KON of dynein under the assumption that dynein’s detachment is very slow (KOFF is very small) in the absence of ATP, and that this (small) value of KOFF remains unchanged after adding DIC-WT. We will return to an analysis of KOFF below. Significantly less dynein pelleted with MTs in presence of mDIC-WT (Fig. 1C), suggesting that dynein’s KON to MTs is reduced when the DIC–CC1 interaction is perturbed. mDIC-WT also reduced dynactin in the MT pellet, suggesting that dynactin binds to MTs via dynein in this experiment. A reduction in dynein was also observed for Dictyostelium cytosol treated with DIC-WT (SI Appendix, Fig. S4 A and B).
DIC-WT Inhibits Binding of Dynein to MTs and Blocks the Initiation of Transport.
We used DIC-WT to perturb the DIC–CC1 interaction on LPs (Fig. 1A), and then assayed the motion of LPs ex vivo. We expected that DIC-WT would compete against endogenous DIC to bind dynactin, with this equilibrium shifting in favor of DIC-WT when its concentration exceeded the endogenous DIC on LPs. Because mDIC-WT (used at 0.25 μM; Fig. 1C) reduced the amount of dynein pelleting with MTs, we devised an experiment (SI Appendix, Fig. S4C) to estimate that DIC-WT in our motility assay was present in ∼50-fold excess compared to endogenous DIC present on the LPs. Because an N-terminal DIC protein containing 100 aa is equally effective as full-length DIC in disrupting endosome motion inside cells (23), our 126 aa DIC-WT should bind dynactin with similar affinity as endogenous DIC. The probability of finding an “unaffected” dynein–dynactin complex (i.e., without DIC-WT bound to it) is therefore ∼2% [=(1/50)×100]. Because LPs are transported by ∼10 dyneins (21, 29), the probability of finding one or more unaffected dynein–dynactin complexes in this team is extremely low (∼1%; 10 binomial trials with 2% success rate).
We next used an optical trap to place individual LPs, which are primarily driven by dynein (7, 21, 29), on single-polarity-labeled MTs (30, 31). Excursions against the trap toward the MT minus end were observed for 20 s, an arbitrary but experimentally convenient threshold (SI Appendix, Fig. S5). The same LPs were caught in the trap away from MTs, and their SD in position (SDPOS ∼10 nm) was measured using a quadrant photo diode. An excursion was scored when the LP moved >30 nm (= 3×SDPOS) away from the trap center. An LP exhibiting at least one such excursion within a 20-s period was scored as “motile” (SI Appendix, Fig. S5). The % probability of motion PMOTION for each condition was determined across multiple experiments. Only directional displacements toward the MT minus end were used to score PMOTION. Rapid transient events could be detected easily, making it unlikely that missed events caused PMOTION to be underestimated (SI Appendix, Fig. S5). PMOTION represents the ability of a team of dynein–dynactin complexes to bind and move on a MT. Although PMOTION does not directly measure the single-molecule binding rate (KON) of individual dyneins, one expects that a reduction of KON for dyneins within a dynein team will lower PMOTION. We therefore used PMOTION to infer relative changes in KON of dyneins on a cargo in response to perturbations (for off rates, see next section). DIC-WT (used at 0.25 μM) reduced PMOTION by ∼50%, but DIC-Mut had no effect (Fig. 2A). Therefore, in agreement with the MT-pelleting results of Fig. 1C, DIC-WT reduced KON of dynein–dynactin on LPs to reduce the number of active dyneins. In agreement with this, fewer DIC-WT-treated LPs could escape from the trap (Fig. 2B), signaling an overall reduction in force. Because DIC-WT did not remove dynein/dynactin from membranes (Fig. 1B) and did not grossly disrupt the dynein–dynactin complex, these effects must arise from subtle alterations in dynein–dynactin function on phagosomes in response to DIC-WT.
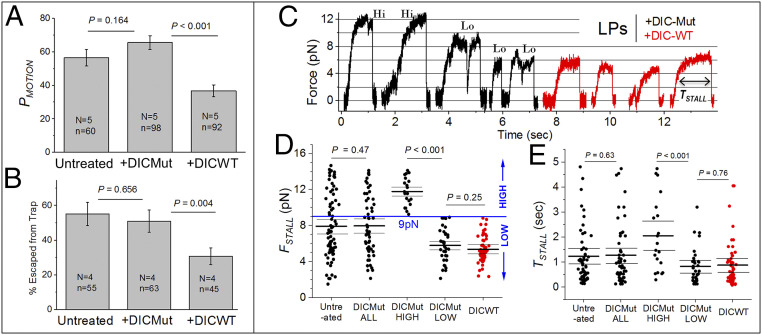
Fig. 2.
The dynein–dynactin interaction is required to initiate, but not to sustain motion. (A) Probability of motion (PMOTION expressed as %) is scored as the fraction of LPs that exhibit at least one excursion within 20 s when held above a MT using an optical trap. LPs were treated with 0.25 µM DIC-WT or 0.25 µM DIC-Mut. DIC-WT reduces PMOTION significantly. Experiments were done at 1 mM ATP in presence of the ATP regenerating system. Each experiment was repeated N times using different phagosome preparations, total of n LPs were used (both mentioned in the figure). Error bars, SEM. P values were calculated using Student’s t test. (B) Percentage of LPs that escape from the trap within 20 s when held above a MT. Error bars, SEM. P values were calculated using Student’s t test. (C) Representative stall force records for dynein on LPs. Experiments were done at 1 mM ATP in presence of the ATP regenerating system. LPs treated with 0.25 µM DIC-Mut displayed stalls of high force (Hi >9 pN; see next panel) and low force (Lo <9 pN). DIC-WT-treated LPs displayed only low-force stalls (<9 pN). (D) Stall forces (FSTALL) for untreated, DIC-Mut- and DIC-WT-treated LPs. DIC-Mut-treated LPs showed stalls from ∼2 pN to ∼14 pN (DICMut-ALL). This force range is divided into DIC-Mut-high (FSTALL >9 pN) and DICMut-low (FSTALL <9 pN) groups. DIC-WT-treated LPs always showed FSTALL <9 pN. FSTALL for DIC-Mut (low) and DIC-WT population is statistically the same. Mean, thick black line. The 95% confidence interval of means (thin black lines) was calculated using a bootstrap algorithm. Comparison of DIC-Mut (low) against DIC-WT: bootstrap P = 0.25; Kolmogorov–Smirnov (KS) test P = 0.14, D = 0.24; Mann–Whitney U test = 1,095, probability > U = 0.21. Phagosomes used: untreated 73; DIC-ALL 55; DIC-Mut high 20; DIC-Mut low 35; DIC-WT 54. (E) The time spent by an LP above half-maximal force (TSTALL) for the corresponding categories in D. TSTALL for DIC-Mut (low) and DIC-WT population is statistically the same. Mean, thick black line. The 95% confidence interval of means (thin black lines) was calculated using a bootstrap algorithm. Comparison of DIC-Mut (low) against DIC-WT: bootstrap P = 0.76; KS test P = 0.95, D = 0.10; Mann–Whitney U test = 982, probability > U = 0.76.
DIC-WT Has No Effect on KOFF of Dynein against Load.
DIC-WT reduces KON of dynein–dynactin to MTs on LPs (Fig. 2A), but once bound, does dynein–dynactin generate force with equal persistence? Fig. 2C shows representative stalls for LPs against an optical trap in the presence of DIC-Mut and DIC-WT (both at 0.25 μM). The plateau (defined as region with slope <20 nm/s) corresponds to the stall force (FSTALL) generated by motors (32). Because single-dynein stalls have a plateau of ∼0.1 s (10), only stalls with a duration >0.1 s were chosen for analysis. We have shown that native dynein–dynactin on phagosomes generates ∼1 pN force (29), although others find higher force (∼2 pN) for full-length dynein in bead assays, depending on trap stiffness (33, 34). The stall force histogram of dynein on LPs inside cells shows ∼2 pN periodicity (7, 21, 29), possibly because two dyneins are recruited by a dynactin (3, 4, 7). We therefore proceeded with a single-dynein force of ∼1 pN on LPs and assumed that forces are additive for multiple dyneins (32, 35–37). FSTALL varied from ∼2 pN to ∼15 pN for DIC-Mut-treated LPs (Fig. 2D, DICMut ALL). Because LPs are driven by multiple dyneins, the force histogram has multiple peaks and is not normally distributed (7, 21, 29). We therefore used bootstrapping (SI Appendix) to estimate 95% confidence interval of the mean in FSTALL for DIC-Mut-treated LPs (Fig. 2D). A comparison of untreated versus DICMut-ALL LPs showed no difference in FSTALL using multiple statistical tests, validating DIC-Mut as a negative control.
Treatment with DIC-WT reduced FSTALL, which now never exceeded 9 pN (in red; Fig. 2 C and D). With this upper limit in mind, the DIC-Mut stalls were segregated into low (FSTALL <9 pN) and high (FSTALL ≥9 pN) categories (Fig. 2D). This segregation ensures that the same average number of dyneins generate force in DIC-Mut-low and DIC-WT categories. The time spent by an LP above half-maximal force is called TSTALL (Fig. 2C). TSTALL measures the tenacity of motors against load (7, 21, 29, 31), and therefore for a motor-team pulling against load, the net KOFF = (1/TSTALL). TSTALL increases linearly with FSTALL for dynein on LPs because more dyneins can generate higher force over longer duration (7). The similar FSTALL of DIC-Mut-low and DIC-WT categories (Fig. 2D) ensures that the same average number of dyneins are engaged in these categories, allowing a fair comparison of the effect of DIC-WT on TSTALL (i.e., on KOFF). In other words, comparison of KOFF becomes insensitive to the KON of dyneins in our analysis (recall that KON is lowered by DIC-WT). TSTALL for dynein on beads is distributed exponentially (10). We therefore used bootstrapping (Materials and Methods) to determine 95% confidence intervals of TSTALL (Fig. 2E). DIC-Mut-low and DIC-WT categories showed no difference in TSTALL. DIC-WT therefore reduced KON, causing fewer bound dyneins, but the dyneins that did bind generated force normally (they had KOFF equal to unperturbed dynein–dynactin).
Antibody that Disrupts the CAPGly–MT Interaction Has No Effect on Dynein’s KON.
Next we addressed the requirement for MT binding by the CAPGly domain of dynactin, an issue that is hotly debated (8, 9, 15, 16, 19, 38). Abrogating the CAPGly–MT interaction could affect initiation of transport because it reduces KON, or also because it increases KOFF. This important mechanistic distinction has never been made, but we addressed it here by high-resolution measurements of force generation (SI Appendix, Fig. S5). Following earlier in vitro work (2, 9, 10, 19), we generated an antibody against the CAPGly domain (amino acids 1 through 78) of Dictyostelium dynactin (hereafter called anti-CAPGly). Anti-CAPGly blocked the pelleting of CAPGly-His in a MT-pelleting assay (SI Appendix, Fig. S6A), suggesting that it inhibits binding of dynactin’s CAPGly domain to MTs. We incubated anti-CAPGly or IgG with a membrane fraction from Dictyostelium, consisting largely of endosomes (31), pelleted the membranes, and probed the pellet to find that anti-CAPGly removed neither dynein nor dynactin from the membranes (SI Appendix, Fig. S6B). The effect of anti-CAPGly was specific to dynactin–MT binding because it did not inhibit the MT-binding domain of dynein from binding to MTs (SI Appendix, Fig. S6C).
We next used the MT-pelleting assay to check the effect of anti-CAPGly on binding of dynein–dynactin complexes to MTs. Dictyostelium cytosol was incubated with anti-CAPGly or IgG control (both used at 0.5 μM), MTs were added to bind motors, followed by MT pelleting in the absence of ATP, where KOFF of dynein becomes very low (see above). Therefore, the pelleting assay should be used only to infer KON. There was no difference in the amount of dynein or dynactin pelleting with MTs between IgG- and anti-CAPGly-treated samples (Fig. 3A). Anti-CAPGly therefore had no effect on KON of the dynein–dynactin–MT interaction. We next tested if anti-CAPGly inhibits dynein-driven transport of LPs at physiological ATP (1 mM). To do this we incubated LPs with 0.5 μM anti-CAPGly (or IgG control) and observed LPs in an optical trap. The number of anti-CAPGly molecules added to the motility mix was ∼35 times higher than the number of dynactins on LPs in the motility assay (estimated similarly to SI Appendix, Fig. S4C). We calculated PMOTION for a dynein–dynactin team in presence of anti-CAPGly exactly as described for DIC-WT. Anti-CAPGly at 0.5 μM had no effect on PMOTION (i.e., on KON of dynein–dynactin to MTs; Fig. 3B). Therefore, taken together with the MT-pelleting data (Fig. 3A), we conclude that the CAPGly domain does not significantly regulate KON for the dynactin–MT interaction.
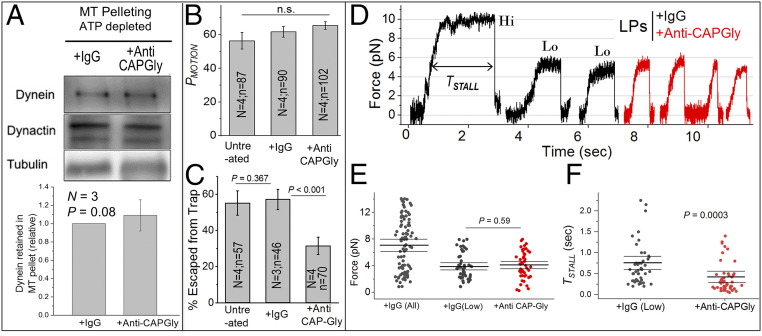
Fig. 3.
Anti-CAPGly has little effect on binding, but increases detachment of dynein from MTs markedly under load. (A) High-speed supernatant from Dictyostelium cytosol was treated with 0.5 µM IgG or anti-CAPGly. This was followed by depleting cytosolic ATP, addition of exogenously polymerized MTs, incubation, and centrifugation to pellet MTs. Western blot shows dynein (DHC) and dynactin (P150) in the MT pellet. Tubulin from the resuspended MT pellet was used for normalization. Lower panel shows quantification across three experiments. IgG band intensity was always taken equal to 1 and anti-CAPGly intensity was calculated relative to IgG. Error bars, SEM. P values were calculated using Student’s t test. (B) Probability of motion (PMOTION) when held above a MT. LPs were treated with 0.5 µM IgG or anti-CAPGly. Each experiment was repeated N times using different LP preparations; the total number of LPs tested (n) is also indicated. Error bars, SEM. P value was calculated using Student’s t test. (C) Percentage of LPs escaped from the trap within 20 s. Each experiment was repeated N times using different LP preparations; the total number of LPs tested (n) is also indicated. Escapees were significantly reduced upon 0.5 µM anti-CAPGly addition. Error bars, SEM. P value was calculated using Student’s t test. (D) Representative minus-directed stalls of LPs. LPs treated with 0.5 µM IgG displayed stalls of both high force (Hi >8 pN) as well as low force (Lo <8 pN). Anti-CAPGly-treated LPs always displayed low (<8 pN) forces. (E) FSTALL for LPs treated with IgG and anti-CAPGly. FSTALL is statistically the same between IgG-low and anti-CAPGly-treated populations. Mean, thick black line. The 95% confidence interval of means (thin black lines) was calculated using a bootstrap algorithm. Comparison of IgG (low) against anti-CAPGly: bootstrap P = 0.59; KS test P = 0.25, D = 0.21; Mann–Whitney U test = 1,034, probability > U = 0.39. Phagosomes used: IgG (all) 90; IgG (low) 48; +anti-CAPGly 48. (F) TSTALL for IgG (low) and anti-CAPGly-treated LPs. TSTALL was reduced by anti-CAPGly treatment. Mean, thick black line. The 95% confidence interval of means (thin black lines) was calculated using a bootstrap algorithm. Comparison of IgG (low) against anti-CAPGly: bootstrap P = 0.0003; KS test P < 0.0001, D = 0.46; Mann–Whitney U test = 1,761, probability > U is less than 0.0001. n.s., not significant.
Very interestingly, anti-CAPGly still caused fewer LPs to escape from the trap (Fig. 3C), suggesting that it reduced the net force from dyneins. If KON is unchanged, then one explanation for reduced force is that anti-CAPGly increases KOFF, causing fewer dyneins to remain engaged against load. This provided the first hint that DIC-WT and anti-CAPGly may inhibit dynein–dynactin function by different mechanisms. The targets of these reagents, respectively, the DIC–CC1 and dynactin–MT interactions, may therefore have distinct functions in transport. Thus, the impaired initiation of transport seen by others after disrupting the CAPGly–MT interaction (15, 16, 38) may actually stem from enhanced KOFF. This possibility was next tested by directly measuring the effect of anti-CAPGly on KOFF.
Anti-CAPGly Causes Premature Detachment of Dynein from Microtubules against Load.
Fig. 3D shows representative stalls for LPs treated with IgG or 0.5 μM anti-CAPGly. FSTALL ranged from ∼2 pN to ∼14 pN for IgG-treated LPs (Fig. 3E), the same as untreated LPs (Fig. 2D). However, anti-CAPGly significantly reduced FSTALL (Fig. 3E), which is expected because it prevented LPs from escaping the trap (Fig. 3C). Because the highest FSTALL on anti-CAPGly-treated LPs was 8 pN (Fig. 3E), we separated the IgG stalls into low [FSTALL <8.0 pN; marked “lo”] and high (FSTALL >8 pN; marked “hi”) categories. The average FSTALL for IgG-low was similar to anti-CAPGly (∼5 pN), suggesting that ∼5 dyneins drive LP motion in both these categories. Fig. 3F plots TSTALL for IgG-low and anti-CAPGly categories. TSTALL was significantly reduced (i.e., KOFF increased) in presence of anti-CAPGly. Dynein on anti-CAPGly-treated LPs therefore detaches frequently from MTs, even when the average number of dyneins driving these LPs is similar (Fig. 3E; compare IgG-low and anti-CAPGly).
Verification of the Effects of Anti-CAPGly on a SLB System.
It is possible that anti-CAPGly interacts with other unknown proteins on the LP membrane, and such interactions indirectly affect dynein–dynactin function. To address this concern, we developed an assay to study function of dynein–dynactin complexes on a bead enclosed by an artificial membrane, SLB (see Insets in Fig. 4A and SI Appendix, Fig. S6D). Dynein–dynactin can be recruited from cytosol to phosphatidic acid-containing membranes (39). We therefore used phosphatidylcholine (PC) and phosphatidic acid (PA) to prepare SLBs. Dictyostelium cytosol was incubated with polymerized MTs in the absence of ATP, motor complexes were released by adding ATP, and this ATP releasate was added to SLBs. Robust long-distance motion was observed for SLBs (86% moved; 82 out of 95) toward the minus end of MTs. As expected, DIC-WT reduced PMOTION of SLBs (SI Appendix, Fig. S6D), confirming that SLBs are driven by dynein–dynactin.
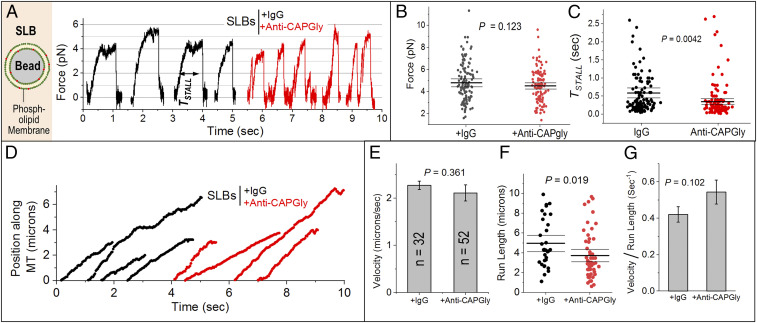
Fig. 4.
Validation of the effects of anti-CAPGly on supported lipid bilayers. (A) Image on Left is a schematic of a single supported lipid bilayer (SLB). Bead is 500 nm in diameter. Lipid membrane consists of phosphatidyl choline (PC) and phosphatidic acid (PA). SLBs were incubated with ATP releasate from Dictyostelium and then treated with 0.5 µM IgG or anti-CAPGly. Representative stalls are shown. (B) FSTALL is statistically the same for IgG- and anti-CAPGly-treated SLBs. Mean, thick black line. The 95% confidence interval of means (thin black lines) was calculated using a bootstrap algorithm. Comparison of IgG against anti-CAPGly: bootstrap P = 0.123; KS test P = 0.225, D = 0.125; Mann–Whitney U test = 8,803, probability > U = 0.30. Phagosomes used: +IgG = 128; +anti-CAPGly = 128. (C) TSTALL for SLBs treated with IgG and anti-CAPGly. Mean, thick black line. The 95% confidence interval of means (thin black lines) was calculated using a bootstrap algorithm. Comparison of IgG against anti-CAPGly: bootstrap P = 0.0042; KS test P < 0.0001, D = 0.273; Mann–Whitney U test = 10,801, probability > U is less than 0.0001. (D) Representative position-time plots of individual SLBs treated with 0.5 µM IgG or anti-CAPGly. All runs ended in detachment of SLB from the MT. (E) Velocity of SLBs treated with IgG or anti-CAPGly. Error bars, SEM. P value was obtained using Student’s t test. n = number of SLBs used. (F) SLB run lengths after IgG and anti-CAPGly treatment. Mean, thick black line. The 95% confidence interval of means (thin black lines) was calculated using a bootstrap algorithm. Comparison of IgG against anti-CAPGly: bootstrap P = 0.019; KS test P = 0.016, D = 0.339; Mann–Whitney U test = 1,114, probability > U = 0.01. (G) (velocity)/(run length), interpreted as KOFF for SLBs treated with IgG or anti-CAPGly. There is no significant change across the samples. Student’s t test was used to obtain P values.
SLBs were next treated with 0.5 μM anti-CAPGly and force generation was assayed on single SLBs (Fig. 4A). Similar to the analysis for LPs, we selected stalls within the same range of FSTALL for control and anti-CAPGly-treated SLBs (Fig. 4B). Assuming that this selection allows us to compare SLBs driven by the same average number of dyneins, anti-CAPGly reduced TSTALL for dynein teams on the SLBs (Fig. 4C), as also seen for LPs (Fig. 3F). We therefore believe that the effects of anti-CAPGly on LPs are specific to the CAPGly–MT interaction on LPs. We next tracked freely moving SLBs (Fig. 4D). For a single motor, KOFF is inverse of the time for which the motor remains bound to MTs (40) and can be estimated using KOFF = (velocity/run length). Anti-CAPGly had no effect on velocity (Fig. 4E), but caused a weak reduction in run length of SLBs (Fig. 4F). The KOFF for freely moving SLBs accordingly showed a small increase after adding anti-CAPGly (Fig. 4G). Anti-CAPGly, however, had no detectable effect on dynein pelleting with MTs in the presence of 1 mM ATP (SI Appendix, Fig. S6E). Therefore, perturbing the CAPGly–MT interaction increases KOFF markedly when dynein is opposed by a load force (Fig. 4C), but has only a weak effect in the absence of an opposing force.
DIC-WT and Anti-CAPGly Do Not Affect Kinesin Activity.
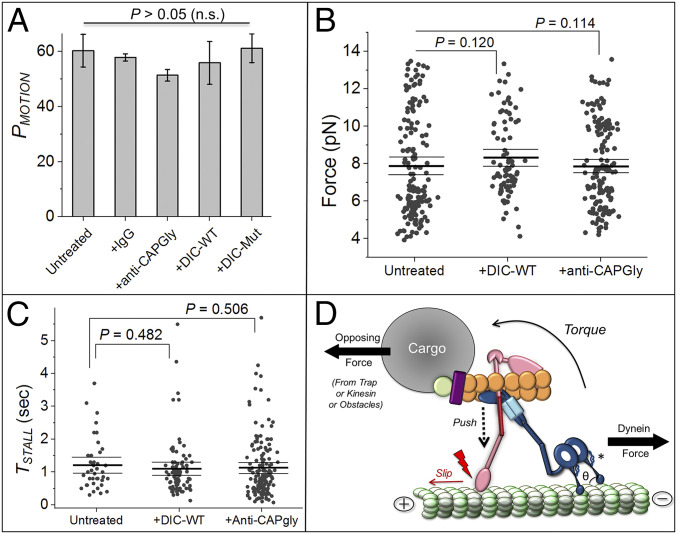
EPs in Dictyostelium are driven by dynein and kinesin-3 (31, 41). EPs exhibit stalls in plus and minus directions, but removing dynein by treatment with the CC1 domain of dynactin elicits only plus-directed kinesin-driven stalls (29). If the effects of DIC-WT and anti-CAPGly are indeed restricted to the dynein–dynactin complex, then neither should affect kinesin-3 activity. To test this, we treated EPs with DIC-WT or DIC-Mut and assayed plus-directed motion on polarity-labeled MTs in an optical trap. We observed no effect of DIC-WT and anti-CAPGly on PMOTION (Fig. 5A), FSTALL (Fig. 5B), and TSTALL (Fig. 5C) of such EPs. Collectively, these data confirm that kinesin-3 and dynein function independently of each other on phagosomes/endosomes as also shown earlier (29), and the effects of DIC-WT and anti-CAPGly are specific to interactions within the dynein–dynactin–MT machinery.
Fig. 5.
Disrupting dynein–dynactin interaction or dynactin–MT interaction does not alter kinesin function. (A) PMOTION of EPs estimated using only excursions toward plus direction of MTs. Error bars, SEM. P values were calculated using Student’s t test. There was no effect on PMOTION across all treatments shown (n.s., not significant). At least 50 EPs were used per condition. (B) FSTALL in plus direction for untreated, 0.25 µM DIC-WT and 0.5 µM anti-CAPGly-treated EPs. There is no change in force generation due to these treatments. Mean, thick black line. The 95% confidence interval of means (thin black lines) and P values were calculated using a bootstrap algorithm. Kolmogorov–Smirnov and Mann–Whitney tests also yielded no difference between the categories shown. SLBs used: untreated 157; +DIC-WT 84; +anti-CAPGly 159. (C) TSTALL in plus direction for categories shown in B. There is no change in TSTALL for plus-directed stalls due to the stated treatments. Mean, thick black line. The 95% confidence interval of means (thin black lines) and mentioned P values were calculated using a bootstrap algorithm. Kolmogorov–Smirnov and Mann–Whitney U tests also yielded no difference. (D) Speculative mechanism by which breaking the CAPGly–MT interaction (red lightning sign) can increase the detachment rate (KOFF) of dynein against an opposing force. The P150 arm is suggested to buttress the complex against slippage induced by a torque (and resultant push) created when dynein’s force is opposed by a load force. Theta (θ) is the angle between dynein’s stalk head and MT. Another buttress connecting dynein’s ring and MT-binding stalk is shown with an *. See main text for details.
Discussion
How dynactin assists dynein is a long-standing controversy (2, 5, 6, 12). Here we quantified the ability of native dynein–dynactin teams to generate force after perturbing the dynein–dynactin and dynactin–MT interactions on phagosomes and SLBs. While dynactin’s role in initiating motion at the MT plus end via plus-tip tracking proteins is well documented (42), the stop-and-go bidirectional motion of cargoes (31, 43) requires frequent reinitiation of dynein-driven motion at locations away from the MT plus end. Such events could be studied in our ex vivo assay because motion of phagosomes/SLBs could be engineered anywhere on a MT by using the optical trap. Our analysis permitted estimation of KON and KOFF for native-like dynein–dynactin teams when they were generating force. We showed using force-generation assays that association of dynein to dynactin via DIC–CC1 keeps dynein in a state where it binds MTs efficiently (high KON), a finding in agreement with structural studies (3, 4, 11, 44). Once dynein has engaged, the DIC–CC1 interaction is not needed to sustain dynein’s force. The dynactin–MT interaction instead appears necessary for this purpose, and because it supports dynein’s force generation even when the DIC–CC1 interaction is perturbed, additional links must permit communication between dynein and dynactin. In agreement with this, dynein and dynactin were still complexed together in presence of DIC-WT.
Several known mutations in dynein–dynactin associated with neurological disorders induce detachment of dynein from MTs (45, 46). When dynein pulls cargo against an opposing force, persistence is likely determined by the load-dependent KOFF. Mutations (e.g., G59S, Q74P, and Q71R) in the CAPGly domain that abrogate dynactin–MT interaction cause motor neuron disease. In addition, deleting the CAPGly domain in yeast dynactin prevents movement of the mitotic spindle and nucleus into the neck of a yeast bud, a process that requires maximal force generation (47). This deletion has minimal effect on other “low-force” functions of dynein, such as sliding of free MTs. These observations are remarkably similar to the finding made here that anti-CAPGly increases KOFF drastically for multiple dynein motions against the trap (Figs. 3F and 4C), but this effect is weaker for free motion (no load; Fig. 4F). Others have suggested that dynactin–MT binding is needed to initiate motion by a single DDB complex in the absence of load (12, 15, 19). We make the counterintuitive suggestion that these problems in transport initiation may be caused by increased KOFF (rather than reduced KON). The increased KOFF has a stronger effect on single-dynein function (12, 15, 19) but appears not to affect multiple dynein-driven motions so significantly in the absence of load (Fig. 4F). A more pronounced requirement of the dynactin–MT interaction may be expected for single dynein when compared to multiple dyneins having independent attachment/detachment kinetics. To elicit an effect of perturbing dynactin–MT interaction on multiple dyneins (perhaps a more physiologically relevant scenario), an external load force is needed to further increase KOFF (Fig. 4 F and C; also see ref. 47). All these observations focus attention on the load-dependent KOFF for dynein–dynactin. Indeed, this parameter emerged as the key controller of bidirectional transport in a computational study (48).
It was recently suggested that the CAPGly–MT attachment is an allosteric activator of dynein (19). Taking this idea further, we suggest that dynein–dynactin function against load requires two-way communication between the CAPGly–MT connection and dynein–MT connection. One may speculate on how the CAPGly–MT attachment prevents premature detachment of dynein against load. The dynein stalk interacts with MTs through hydrogen bonds and salt bridges that are highly sensitive to orientation and distance (49). When dynein is opposed (e.g., by trap/kinesin/obstacles), a torque must develop as shown in Fig. 5D to push downward (dashed arrow) on the P150 arm of dynactin. If the CAPGly–MT attachment is strong enough to prevent slippage against this push, then the P150 arm can function as a buttress that restricts the angle between dynein’s stalk head and the MT (= θ; Fig. 5D) within a permissive range. Disrupting the CAPGly–MT interaction would induce slippage (Fig. 5D), causing θ to attain values where the aforesaid hydrogen bonds and salt bridges also fail and dynein detaches from MTs. In support of this, another structure thought also to act as a buttress emerges from the dynein ring (see * in Fig. 5D) to control force-induced sliding of the helices in dynein’s stalk (50). Buttressing requires mechanical rigidity. The persistence length of coiled-coil helices is ∼150 nm (51), suggesting that the ∼50 nm projecting arm of P150 should have some rigidity. However, the P150 arm may also require flexibility to permit a range of θ values when dyneins in a team strain against load. Such flexibility has indeed been suggested (10). To investigate this mechanical balance, the critical buckling load for P150 would have to be measured under compression, a technical challenge for force microscopy.
Materials and Methods
Antibodies.
Antibodies were purchased or generated in house, as described in SI Appendix.
Cloning and Protein Purification.
Dictyostelium DIC-WT was cloned into pET22b vector with a C terminus His-tag. Site-directed mutagenesis was performed on the DIC-WT clone to generate DIC-Mut. Both DIC-WT and -Mut were purified by affinity chromatography using nickel-nitrilotriacetic acid (Ni-NTA) beads. Further details are in SI Appendix.
MT-Pelleting Assay.
Rat brain or Dictyostelium cytosol was treated with respective DIC proteins, IgG, or Dictyostelium anti-CAPGly antibody on ice for 20 min. Motors from cytosol were then allowed to bind with in vitro polymerized taxol-stabilized MTs. To measure dynein on rate, ATP in cytosol was depleted by adding 3 mM glucose, 15 U/mL hexokinase, 4 mM MgCl2, 1 mM adenylyl imidodiphosphate (AMP-PNP) to the cytosol–MT mix. MTs along with bound motors were pelleted and resuspended in phosphate buffered saline (PBS) containing 0.1% sodium dodecyl sulphate (SDS) followed by Western blotting.
Phagosome Preparation, Motility Assays, and Optical Trapping.
Latex beads were fed to Dictyostelium cells and chased (5-min chase for EPs; 45 min for LPs) at 22 °C. Cell lysate was centrifuged, phagosomes were collected along with the high-speed supernatant, and frozen. Each aliquot of phagosome was either untreated (control) or treated with 0.25 μM DIC (WT/Mut), or with 0.5 μM IgG, or anti-CAPGly for 10 min on ice before flowing in the samples onto polarity-labeled MTs (30) that were stuck to a coverslip using poly-L-lysine. Motility experiments were done at 1 mM ATP concentration in presence of an ATP regenerating system. Motion was observed using differential interference contrast microscopy with a 100×, 1.4-NA oil objective (Nikon), and images were recorded at 30 frames/s. The optical trapping setup has been described [21], also see SI Appendix.
Treatment of Mammalian DDB Complexes with DIC Proteins.
DDB complexes were formed by adding recombinant FLAG-tagged BicD to goat brain cytosol. DDB complexes were then pulled down using beads conjugated to FLAG antibody. The bead-containing pellet was treated with either mDIC-WT or mDIC-Mut for 20 min on ice. Unbound proteins were removed by washing the pellet three times with PBS. The DDB complex with bound proteins was eluted with SDS dye. Levels of dynein, dynactin, and recombinant DIC proteins that copelleted with BicD were assayed by Western blotting.
Supported Lipid Bilayer Preparation.
For SLB preparation, 5 μL of carboxylated latex beads (500-nm diameter), 1 μL of 1 M NaCl, 69 μL of autoclaved double-distilled water, and 20 μL of liposomes (prepared by the freeze–thaw method) were mixed and vortexed for 30 min with intermittent rest, followed by washing and resuspension of SLBs in 200 μL double-distilled water. Further details are in SI Appendix.
ATP Releasate Preparation.
Dictyostelium cytosol was incubated with taxol-stabilized MTs for motor binding. ATP from cytosol was depleted by adding 3 mM glucose, 15 U/mL hexokinase, 4 mM MgCl2, and 4 mM AMP-PNP. Bound motors were copelleted with MTs and active motors were released from MTs by addition of 10 mM (ATP + MgCl2). Further details are in SI Appendix.
Dot Blot Assay.
To detect Dictyostelium DIC–P150 CC1 domain interaction, glutathione S-transferase (GST) or GST-CC1 were spotted on a nitrocellulose membrane, dried, and overlaid with DIC-WT and Mut proteins (1 mg/mL) overnight. The membrane was washed with PBS with 0.05% Tween, blocked with 5% milk, and probed with anti-His antibody to detect His-tagged DIC proteins bound to CC1. For DIC–NudE interaction, mDIC proteins were spotted and the membrane was overlaid with rat brain lysate (1 mg/mL) overnight and probed using NudE antibody. Further details are in SI Appendix.
Blot Overlay Assay.
Dictyostelium DIC or mammalian CC1 proteins were run on a nondenaturing PAGE, transferred on polyvinylidene fluoride membrane and overlaid with 1 mg/mL Dictyostelium lysate (for DIC proteins) or with mDIC proteins (for CC1 interaction) for 2 h. To detect Dictyostelium dynactin P150 binding, the blot was probed with anti-P150 antibody; to detect mDIC proteins, binding the blot was probed with anti-His antibody.
Pull-Down Assays.
To show interaction of His-tagged DIC-WT protein with purified CC1 and P150 dynactin, 20 μg of purified DIC-WT was bound to Ni-NTA beads for 1 h at 4 °C. DIC-WT on beads was further incubated with 20 μg of pure CC1 or 1 mg of freshly prepared Dictyostelium cytosol for 3 h. Beads were washed three times with PBS containing 0.3 M NaCl to remove nonspecific proteins, and the final elution was performed in 1× SDS reducing dye.
Data Representation, Analysis, and Statistical Methods.
Unless otherwise stated, error bars denote the SE of the mean (SEM). Where data were known or assumed to be normally distributed, Student’s t test was used to test significance (two-tailed test, 95% confidence with the null hypothesis that distributions are the same). Statistical inferences for run length, FSTALL, and TSTALL were based on bootstrapping and/or nonparametric tests because these quantities are known not to be normally distributed (see main text). Three different tests were employed for each case, as described in the main text. Further details are in SI Appendix.
Supplementary Material
Acknowledgments
We thank R. Jha and A. D’Souza for help with experiments and comments, K. Vaughan for mDIC constructs, R. McKenney for BicD constructs, T. Schroer for pVEX-CC1 constructs, M. Koonce for dynein MT1 constructs, and A. Akhmanova for comments. R.M. acknowledges funding from a Wellcome Trust International Senior Research Fellowship (Grant WT079214MA). P.S. acknowledges funding from a Department of Biotechnology–Wellcome Trust India Alliance Early Career Fellowship (Grant IA/E/15/1/502298).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103383118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Reck-Peterson S. L., Redwine W. B., Vale R. D., Carter A. P., The cytoplasmic dynein transport machinery and its many cargoes. Nat. Rev. Mol. Cell Biol. 19, 382–398 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Culver-Hanlon T. L., Lex S. A., Stephens A. D., Quintyne N. J., King S. J., A microtubule-binding domain in dynactin increases dynein processivity by skating along microtubules. Nat. Cell Biol. 8, 264–270 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Grotjahn D. A., et al., Author Correction: Cryo-electron tomography reveals that dynactin recruits a team of dyneins for processive motility. Nat. Struct. Mol. Biol. 25, 355 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Urnavicius L., et al., Cryo-EM shows how dynactin recruits two dyneins for faster movement. Nature 554, 202–206 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tirumala N. A., Ananthanarayanan V., Role of dynactin in the intracellular localization and activation of cytoplasmic dynein. Biochemistry 59, 156–162 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Liu J. J., Regulation of dynein-dynactin-driven vesicular transport. Traffic 18, 336–347 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Rai A. K., Rai A., Ramaiya A. J., Jha R., Mallik R., Molecular adaptations allow dynein to generate large collective forces inside cells. Cell 152, 172–182 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Ayloo S., et al., Dynactin functions as both a dynamic tether and brake during dynein-driven motility. Nat. Commun. 5, 4807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King S. J., Schroer T. A., Dynactin increases the processivity of the cytoplasmic dynein motor. Nat. Cell Biol. 2, 20–24 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Tripathy S. K., et al., Autoregulatory mechanism for dynactin control of processive and diffusive dynein transport. Nat. Cell Biol. 16, 1192–1201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang K., et al., Cryo-EM reveals how human cytoplasmic dynein is auto-inhibited and activated. Cell 169, 1303–1314.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenney R. J., Huynh W., Tanenbaum M. E., Bhabha G., Vale R. D., Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 345, 337–341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdhury S., Ketcham S. A., Schroer T. A., Lander G. C., Structural organization of the dynein-dynactin complex bound to microtubules. Nat. Struct. Mol. Biol. 22, 345–347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlager M. A., Hoang H. T., Urnavicius L., Bullock S. L., Carter A. P., In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 33, 1855–1868 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKenney R. J., Huynh W., Vale R. D., Sirajuddin M., Tyrosination of α-tubulin controls the initiation of processive dynein-dynactin motility. EMBO J. 35, 1175–1185 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moughamian A. J., Holzbaur E. L. F., Dynactin is required for transport initiation from the distal axon. Neuron 74, 331–343 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nirschl J. J., Magiera M. M., Lazarus J. E., Janke C., Holzbaur E. L. F., α-Tubulin tyrosination and CLIP-170 phosphorylation regulate the initiation of dynein-driven transport in neurons. Cell Rep. 14, 2637–2652 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H., et al., Microtubule binding by dynactin is required for microtubule organization but not cargo transport. J. Cell Biol. 176, 641–651 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Q., Gicking A. M., Hancock W. O., Dynactin p150 promotes processive motility of DDB complexes by minimizing diffusional behavior of dynein. Mol. Biol. Cell 31, 782–792 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pathak D., Mallik R., Lipid–Motor interactions: Soap opera or symphony? Curr. Opin. Cell Biol. 44, 79–85 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Rai A., et al., Dynein clusters into lipid microdomains on phagosomes to drive rapid transport toward lysosomes. Cell 164, 722–734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaughan P. S., Leszyk J. D., Vaughan K. T., Cytoplasmic dynein intermediate chain phosphorylation regulates binding to dynactin. J. Biol. Chem. 276, 26171–26179 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Towns W. L., Tauhata S. B. F., Vaughan P. S., Vaughan K. T., Transfection-induced defects in dynein-driven transport: Evidence that ICs mediate cargo-binding. Cell Motil. Cytoskeleton 66, 80–89 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma S., Triviños-Lagos L., Gräf R., Chisholm R. L., Dynein intermediate chain mediated dynein-dynactin interaction is required for interphase microtubule organization and centrosome replication and separation in Dictyostelium. J. Cell Biol. 147, 1261–1274 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKenney R. J., Weil S. J., Scherer J., Vallee R. B., Mutually exclusive cytoplasmic dynein regulation by NudE-Lis1 and dynactin. J. Biol. Chem. 286, 39615–39622 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaughan K. T., Vallee R. B., Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J. Cell Biol. 131, 1507–1516 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King S. J., Brown C. L., Maier K. C., Quintyne N. J., Schroer T. A., Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol. Biol. Cell 14, 5089–5097 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imamula K., Kon T., Ohkura R., Sutoh K., The coordination of cyclic microtubule association/dissociation and tail swing of cytoplasmic dynein. Proc. Natl. Acad. Sci. U.S.A. 104, 16134–16139 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanghavi P., et al., Coin tossing explains the activity of opposing microtubule motors on phagosomes. Curr. Biol. 28, 1460–1466.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soppina V., Rai A., Mallik R., Simple non-fluorescent polarity labeling of microtubules for molecular motor assays. Biotechniques 46, 543–549 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Soppina V., Rai A. K., Ramaiya A. J., Barak P., Mallik R., Tug-of-war between dissimilar teams of microtubule motors regulates transport and fission of endosomes. Proc. Natl. Acad. Sci. U.S.A. 106, 19381–19386 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mallik R., Petrov D., Lex S. A., King S. J., Gross S. P., Building complexity: An in vitro study of cytoplasmic dynein with in vivo implications. Curr. Biol. 15, 2075–2085 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Belyy V., et al., The mammalian dynein-dynactin complex is a strong opponent to kinesin in a tug-of-war competition. Nat. Cell Biol. 18, 1018–1024 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brenner S., Berger F., Rao L., Nicholas M. P., Gennerich A., Force production of human cytoplasmic dynein is limited by its processivity. Sci. Adv. 6, eaaz4295 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blehm B. H., Schroer T. A., Trybus K. M., Chemla Y. R., Selvin P. R., In vivo optical trapping indicates kinesin’s stall force is reduced by dynein during intracellular transport. Proc. Natl. Acad. Sci. U.S.A. 110, 3381–3386 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hendricks A. G., Holzbaur E. L. F., Goldman Y. E., Force measurements on cargoes in living cells reveal collective dynamics of microtubule motors. Proc. Natl. Acad. Sci. U.S.A. 109, 18447–18452 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mallik R., Rai A. K., Barak P., Rai A., Kunwar A., Teamwork in microtubule motors. Trends Cell Biol. 23, 575–582 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Lloyd T. E., et al., The p150(Glued) CAP-Gly domain regulates initiation of retrograde transport at synaptic termini. Neuron 74, 344–360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muresan V., et al., Dynactin-dependent, dynein-driven vesicle transport in the absence of membrane proteins: A role for spectrin and acidic phospholipids. Mol. Cell 7, 173–183 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Shastry S., Hancock W. O., Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Curr. Biol. 20, 939–943 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollock N., de Hostos E. L., Turck C. W., Vale R. D., Reconstitution of membrane transport powered by a novel dimeric kinesin motor of the Unc104/KIF1A family purified from Dictyostelium. J. Cell Biol. 147, 493–506 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinmetz M. O., Akhmanova A., Capturing protein tails by CAP-Gly domains. Trends Biochem. Sci. 33, 535–545 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Hancock W. O., Bidirectional cargo transport: Moving beyond tug of war. Nat. Rev. Mol. Cell Biol. 15, 615–628 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urnavicius L., et al., The structure of the dynactin complex and its interaction with dynein. Science 347, 1441–1446 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoang H. T., Schlager M. A., Carter A. P., Bullock S. L., DYNC1H1 mutations associated with neurological diseases compromise processivity of dynein-dynactin-cargo adaptor complexes. Proc. Natl. Acad. Sci. U.S.A. 114, E1597–E1606 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ori-McKenney K. M., Xu J., Gross S. P., Vallee R. B., A cytoplasmic dynein tail mutation impairs motor processivity. Nat. Cell Biol. 12, 1228–1234 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore J. K., Sept D., Cooper J. A., Neurodegeneration mutations in dynactin impair dynein-dependent nuclear migration. Proc. Natl. Acad. Sci. U.S.A. 106, 5147–5152 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohashi K. G., et al., Load-dependent detachment kinetics plays a key role in bidirectional cargo transport by kinesin and dynein. Traffic 20, 284–294 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Redwine W. B., et al., Structural basis for microtubule binding and release by dynein. Science 337, 1532–1536 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rao L., Berger F., Nicholas M. P., Gennerich A., Molecular mechanism of cytoplasmic dynein tension sensing. Nat. Commun. 10, 3332 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolgemuth C. W., Sun S. X., Elasticity of α-helical coiled coils. Phys. Rev. Lett. 97, 248101 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.