Significance
Site-specific modification of epigenetic marks such as DNA methylation to regulate gene expression is a unique approach to enhance economically important crop traits. This approach allows for the maintenance of the introduced modifications in the absence of the initial transgene inducer in subsequent generations and relies largely on methylation of cytosines in the CG-specific sequence context. We have developed a targeted DNA methylation tool based on a bacterial methyltransferase and the CRISPR-Cas9 platform to directly methylate cytosines at CG sites in Arabidopsis. These tools expand the currently available CRISPR-based targeted DNA methylation tools and provide an approach for the establishment of heritable targeted DNA methylation in plants.
Keywords: DNA methylation, Arabidopsis, CRISPR-Cas9, SunTag
Abstract
CRISPR-based targeted modification of epigenetic marks such as DNA cytosine methylation is an important strategy to regulate the expression of genes and their associated phenotypes. Although plants have DNA methylation in all sequence contexts (CG, CHG, CHH, where H = A, T, C), methylation in the symmetric CG context is particularly important for gene silencing and is very efficiently maintained through mitotic and meiotic cell divisions. Tools that can directly add CG methylation to specific loci are therefore highly desirable but are currently lacking in plants. Here we have developed two CRISPR-based CG-specific targeted DNA methylation systems for plants using a variant of the bacterial CG-specific DNA methyltransferase MQ1 with reduced activity but high specificity. We demonstrate that the methylation added by MQ1 is highly target specific and can be heritably maintained in the absence of the effector. These tools should be valuable both in crop engineering and in plant genetic research.
DNA methylation is a modification of the DNA base cytosine and is often associated with transcriptional repression. In plants, the RNA-directed DNA methylation (RdDM) pathway is required for establishment of de novo DNA methylation (1, 2), catalyzed by DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2). Once established, DNA methylation is maintained via three main mechanisms, depending on genomic location and sequence context: CG methylation is maintained by DNA METHYLTRANSFERASE 1 (MET1) (1), non-CG methylation at small transposons and repetitive elements in open chromatin is maintained by RdDM, and non-CG methylation in dense heterochromatin is maintained by CHROMOMETHYLASE 2 (CMT2) and CHROMOMETHYLASE 3 (CMT3) (3, 4). Maintenance of methylation by RdDM in euchromatic regions is complex and depends on small interferring RNAs (siRNA) biogenesis, whereas methylation maintenance in pericentromeric regions by CMT2 and CMT3 depends on a feedback loop with heterochromatic histone modifications (1, 3). In contrast, symmetric CG methylation maintenance by MET1 during DNA replication depends on few additional factors and results in highly efficient copying of the methylation state from parent to daughter strand during cell division and meiosis (1, 4–6). Targeting of methylation directly to cytosines in the CG context should therefore be an effective strategy for establishing de novo DNA methylation that can be stably transmitted both mitotically and meiotically.
CRISPR-based systems have been used to target DNA methylation and trigger silencing of target loci in both plants and animals (7, 8). To date, the CRISPR-based tools developed in plants have used the catalytic domain of the RdDM methyltransferase DRM2, which adds DNA methylation in all sequence contexts but has a preference for the CHH context (8, 9). While these tools are able to cause gene silencing, they work at a relatively low efficiency, and the heritability of the targeted DNA methylation in the absence of the effector transgene is often incomplete (8). In a study utilizing an artificial zinc finger fused to RdDM components to target DNA methylation, Gallego-Bartolome et al. (10) demonstrated that the heritability of targeted DNA methylation was highly dependent on the establishment of high levels of CG methylation. To test the hypothesis that targeting methylation directly to cytosines in the CG context would increase silencing efficiency and heritability relative to the currently available tools, we sought to develop a CRISPR-based CG-specific targeted DNA methylation system for plants. A CG-specific bacterial methyltransferase, MQ1 (SssI), from the bacterium Mollicutes spiroplasma, fused to inactive Cas9 (dCAS9), was recently used to trigger de novo DNA methylation in the CG context at specific target loci in mammalian cell lines and mice (7). However, these lines also exhibited nonspecific gain of CG methylation throughout the genome (7). To reduce nonspecific methylation, Lei et al. (7) used a variant of MQ1 (MQ1(Q147L)) with reduced DNA binding affinity and activity. Here we show that this MQ1(Q147L) variant can be used for accurate targeted de novo CG DNA methylation in Arabidopsis.
Results and Discussion
MQ1v Targets DNA Methylation to FWA Leading to FWA Silencing and an Early-Flowering Phenotype.
We cloned dCAS9 fused to MQ1(Q147L) (hereafter called MQ1v) into a plant binary vector (SI Appendix, Fig. S1A) and targeted MQ1v to the promoter of the FLOWERING WAGENINGEN (FWA) gene (11, 12). FWA is normally silent in the vegetative tissues of wild-type plants due to DNA methylation in its promoter region (13). However, in plants that have stably lost DNA methylation at the FWA promoter (fwa epimutants), FWA is expressed, causing a late-lowering phenotype. Thus, targeting the FWA promoter in fwa epimutants produces a simple phenotypic readout (flowering time) that is linked to the methylation status of the FWA promoter (8, 10, 14, 15). An MQ1v construct containing three previously characterized guide RNAs (gRNAs) (guide 4, guide 10, and guide 18) targeting three different regions of the FWA promoter (SI Appendix, Fig. S1B) (8) was transformed into an fwa epimutant (Col background) and T1 transformants were monitored for an early-flowering phenotype. As a negative control, we transformed fwa epimutant plants with a catalytically inactive mutant of MQ1 (dMQ1) (7) fused to dCAS9, along with the same three guide RNAs (SI Appendix, Fig. S1A). We did not observe any difference in flowering time between the first-generation transformed (T1) MQ1v and control dMQ1 plants, indicating that MQ1v could not trigger sufficient de novo DNA methylation to silence FWA in T1 plants. We examined DNA methylation levels in these T1 plants at the FWA promoter using a restriction-enzyme–based DNA methylation assay (McrBC–quantitative real-time PCR [McrBC-qPCR]). Out of eight MQ1v T1 plants tested, two (T1-3, T1-7) had gained DNA methylation in comparison to matched dMQ1 negative control plants (T1-1) (SI Appendix, Fig. S2A). To confirm this finding, we performed bisulfite PCR sequencing (BS-PCR) on both lines. We found that the two MQ1v T1 plants (T1-3, T1-7) had gained DNA methylation in both the CG and CHH contexts in the FWA promoter, compared to a control dMQ1 T1 plant (T1-1) (Fig. 1A). Thus, MQ1v was able to trigger de novo DNA methylation in these plants.
Fig. 1.
MQ1v targets DNA methylation to FWA and causes an early-flowering phenotype. (A) Bisulfite PCR sequencing of two T1 lines (T1-3, T1-7) transformed with MQ1v and a negative control (T1-1) transformed with dMQ1, over three regions of the FWA promoter region: region 1 (Chr4:13038143–13038272), region 2 (Chr4:13038356–13038499), and region 3 (Chr4:13038568–13038695). Each T1 plant is a result of an independent transgenic event. (B) Stacked bar plot showing the percentage of T2 early- and late-flowering plants in the progeny of two different T1 lines (T1-3 and T1-7), together with wild-type Col, and T2 progenies of a matched dMQ1 (T1-1) control. Percentage of early-flowering plants is denoted in the bar plots. The total numbers (N) of plants assayed for each line were 3, 13, 17, and 32. (C) DNA methylation profile at the FWA promoter region in early-flowering progeny of two T1 plants (T2-3-a, T2-3-b are progeny of T2-3; T2-7-a, T2-7-b are progeny of T2-7), along with Col and a matched dMQ1 (T2-1-a) plant as controls. Only cytosines with at least five overlapping reads are shown. Small, negative values (black) indicate cytosines with five or more overlapping reads but no DNA methylation. Vertical yellow bars indicate the locations of the guide 4, guide 10, and guide 18 binding sites.
Several studies in plants have shown that minor gains of de novo DNA methylation can become amplified after multiple generations of inbreeding (16, 17). To test whether the two MQ1v T1 plants (T1-3, T1-7) with modest gains in DNA methylation exhibited further gains in the following generation, we analyzed the progeny (T2) of both T1 lines. We found several early-flowering T2 plants (Fig. 1B and SI Appendix, Fig. S2B), indicating successful FWA gene silencing. Using whole-genome bisulfite sequencing (WGBS), we examined DNA methylation at the FWA promoter in four of these early-flowering T2 plants (T2-3-a, T2-3-b, T2-7-a, T2-7-b) and their matched T2 dMQ1 control (T2-1-a) (see naming schema in SI Appendix, Fig. S1C). DNA methylation in these plants (Fig. 1C) was more pronounced than in their T1 parents (Fig. 1 A and C). To confirm that the gain of DNA methylation in these plants did not also occur genome-wide, we also examined DNA methylation levels by WGBS. We observed little change in genome-wide DNA methylation levels (SI Appendix, Fig. S3), indicating that MQ1v is able to specifically target DNA methylation at the FWA promoter with minimal off-target effects.
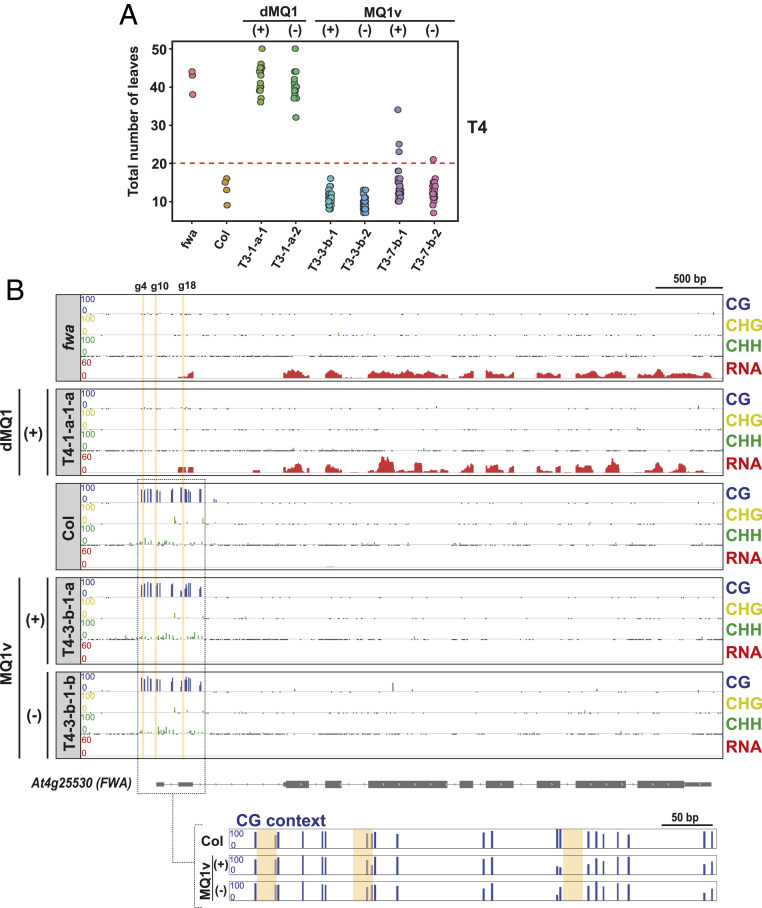
Epigenetic marks such as DNA methylation can often be stably inherited in plants in the absence of the initial effector (8, 10, 15). We therefore examined whether MQ1v-targeted DNA methylation and the associated silencing of FWA were heritable in the absence of MQ1v. We first examined the progeny (T3) of the four early-flowering T2 plants (T2-3-a, T2-3-b, T2-7-a, T2-7-b) (SI Appendix, Fig. S4 A and B). The percentage of early flowering plants among the progeny of these T2 lines ranged from ∼14 to 100% (SI Appendix, Fig. S4B). We selected early-flowering T3 plants (SI Appendix, Fig. S4A) and used PCR-based genotyping to select two plants in which the MQ1v transgene was still present (T3-3-b-1, T3-7-b-1) and two plants in which the transgene had been segregated away (null segregants; T3-3-b-2, T3-7-b-2). These were propagated to the next generation (T4) and again monitored for the flowering-time phenotype (Fig. 2A and SI Appendix, Fig. S4B). Nearly all of the T4 plants were early flowering even after the MQ1v construct had been segregated away, indicating efficient maintenance of silencing of FWA expression in the absence of the original effector (Fig. 2A and SI Appendix, Fig. S4B). We also analyzed the DNA methylation profile at the FWA promoter in a T4 plant maintaining the transgene and a T4 null segregant [T4-3-b-1-a(+), T4-3-b-1-b(−), respectively] by WGBS. Methylation in these T4 plants was comparable to that in Col wild-type plants at the FWA promoter in both T4 plants tested (Fig. 2B). As expected, FWA expression was also strongly repressed in these two plants [T4-3-b-1-a(+), T4-3-b-1-b(−)] relative to the original fwa epimutant and dMQ1 control plants (Fig. 2B and SI Appendix, Fig. S4C). Thus, the DNA methylation and gene silencing targeted by MQ1v can be efficiently maintained in the absence of the transgene.
Fig. 2.
CRISPR-based targeted DNA methylation by MQ1v silences FWA expression and is heritable in the absence of the MQ1v effector. (A) Dot plot of leaf count at flowering time for four T4 MQ1v plants and two control dMQ1 plants. T3 parents are labeled at the bottom. (+), MQ1v transgene positive; (−), MQ1v transgene negative (null segregants). (B) DNA methylation profile at the FWA promoter region in T4 transgene positive (+) and T4 transgene negative (−) early-flowering plants, relative to Col wild-type control, the unmethylated fwa epiallele, and a late-flowering T4 dMQ1 control plant in the fwa background. FWA expression by RNA-seq is also shown (average of three replicates). (Bottom) Zoomed-in view shows strong DNA methylation at most of the CG sites in the targeted region. Vertical yellow bars indicate the locations of the guide 4, guide 10, and guide 18 binding sites.
MQ1v Can Cause Heritable DNA Methylation and Silencing in a drm1 drm2 Mutant Background.
Although MQ1 is a CG-specific DNA methyltransferase, MQ1v-transformed plants also gained non-CG methylation (Figs. 1C and 2B). This is likely due to the RdDM pathway, which is recruited to sites of existing DNA methylation and can add additional DNA methylation at both CG and non-CG sites, acting as a self-reinforcing feedback loop (1). Thus, initial establishment of CG methylation at the FWA promoter likely triggered additional de novo non-CG methylation via RdDM (Figs. 1C and 2B). However, silencing of FWA is known to be primarily dependent on CG methylation (18). To test whether MQ1v-mediated gain of CG methylation alone is sufficient to cause silencing of FWA, we tested MQ1v targeting in fwa epimutants in which drm1 and drm2 had been introgressed (fwa drm1 drm2). In this background, RdDM is abolished, and plants can no longer gain de novo non-CG methylation at FWA (10). We found that several T1 plants in the fwa drm1 drm2 background were early flowering (Fig. 3A). We analyzed DNA methylation patterns at the FWA promoter by BS-PCR for four of these early-flowering plants (T1-R2, T1-R3, T1-R4, T1-R5) and a dMQ1 control plant (T1-R1). The FWA promoter was found to be methylated only in the CG context in these plants, as expected (Fig. 3B). Next, we examined FWA expression in the four early-flowering plants relative to the dMQ1 control plant and confirmed that FWA messenger RNA (mRNA) levels were suppressed in these plants (Fig. 3C). To determine whether loss of DRM1 and DRM2 affected the ability of FWA silencing to persist in the absence of the MQ1v transgene, we also monitored the T2 and T3 progeny plants of the four early-flowering T1 plants (T1-R2, T1-R3, T1-R4, T1-R5) for the early-flowering phenotype. Many T2 plants retained the early-flowering phenotype (Fig. 3D). Using PCR-based genotyping, we identified two T2 early-flowering plants that had segregated away the MQ1v transgene (T2-R5-a, T2-R5-b) and propagated them to the T3 generation. All of these T3 MQ1v plants retained the early-flowering phenotype (Fig. 3E), indicating that CG methylation alone was sufficient to maintain FWA silencing in an RdDM mutant background in the absence of the inducer.
Fig. 3.
MQ1v can target DNA methylation in a drm1 drm2 mutant background. (A) Dot plot of leaf count at flowering time of T1 plants transformed with dCAS9-MQ1v in fwa drm1 drm2, compared to fwa and Col controls. Each T1 plant is the result of an independent transgenic event. (B) Bisulfite PCR sequencing of four early-flowering T1 lines in fwa drm1 drm2 (T1-R2, T1-R3, T1-R4, and T1-R5) over three regions of the FWA promoter: region 1 (Chr4:13038143–13038272), region 2 (Chr4:13038356–13038499), and region 3 (Chr4:13038568–13038695). A dMQ1 T1 plant was used as a negative control (T1-R1). Vertical yellow bars indicate the locations of the guide 4, guide 10, and guide 18 binding sites. (C) FWA expression analysis in four early-flowering MQ1v fwa drm1 drm2 T1 plants relative to dMQ1 negative controls by reverse transcriptase–qPCR analysis. Error bars indicate SDs (n = 3 technical replicates). (D and E) Dot plot of leaf count at flowering time of T2 plants (D) and T3 plants (E) in the fwa drm1 drm2 mutant background compared to fwa and Col controls. The T1 (D) or T2 (E) parent is listed at the bottom. In E, transgene positive (+) and transgene negative (−) lines are also indicated at the top.
It was somewhat paradoxical that FWA gene silencing was more efficient when MQ1v was transformed into the RdDM mutant background (fwa drm1 drm2) compared to the fwa background that contained a wild-type DNA methylation machinery, as indicated by the fact that early flowering plants could be seen in the T1 generation in fwa drm1 drm2 plants (Fig. 3A), whereas MQ1v transformed into the fwa background caused FWA silencing only in the T2 generation (Fig. 1B and SI Appendix, Fig. S2B). This is likely due to increased accumulation of targeting components (MQ1v and/or guide RNAs) in the silencing-defective drm1 drm2 mutant background. Transgenes are often targeted for repression by both transcriptional silencing machinery (TGS) and posttranscriptional silencing machinery (PTGS), namely the RdDM and RNA interference pathways (19, 20). For example, it was reported that PTGS mutants can increase both Cas9/gRNA expression and CRISPR-Cas9 editing efficiency (19). To test whether plants compromised in PTGS also show increased efficiency of methylation targeting by MQ1v, we transformed MQ1v into fwa plants that also carried rdr6, a PTGS pathway mutant (10). Similar to the fwa drm1 drm2 background, we observed early-flowering phenotype in T1 plants (SI Appendix, Fig. S5A), associated with gain of DNA methylation at the FWA promoter (SI Appendix, Fig. S5B) as well as silencing of FWA expression (SI Appendix, Fig. S5C). T2 and T3 progenies of these lines also maintained the early-flowering phenotype in null segregants (SI Appendix, Fig. S5 D and E).
Combining the SunTag System with MQ1(Q147L) Robustly Targets DNA Methylation to FWA to Induce Early Flowering in T1 Plants.
We next examined ways to further increase the efficiency of MQ1-mediated targeted DNA methylation. The CRISPR-based SunTag system enables the recruitment of multiple effectors to the same target site, potentially increasing targeting efficiency compared to straight fusions with dCAS9 (21). We therefore tested whether using the SunTag system to target MQ1(Q147L) to the FWA promoter led to more efficient gain of DNA methylation and FWA silencing. We cloned MQ1(Q147L) into the SunTag construct (SI Appendix, Fig. S1D), hereafter called SunTag-MQ1v. We transformed fwa epimutants with SunTag-MQ1v and monitored the resulting T1 plants for the early-flowering phenotype. We observed many early-flowering plants in the T1 generation (Fig. 4A), indicating that SunTag-MQ1v is more efficient than the MQ1v straight fusion that showed only early-flowering plants in the T2 generation (Fig. 1B and SI Appendix, Fig. S2B). Four early-flowering T1 plants (T1-S1, T1-S2, T1-S3, T1-S4) and four late-flowering T1 plants (T1-S5, T1-S6, T1-S7, T1-S8) were selected for DNA methylation and FWA expression analysis. The four early-flowering T1 plants showed strong DNA methylation at the FWA promoter (Fig. 4B), along with a dramatic reduction in FWA mRNA levels relative to those in fwa (Fig. 4C). In contrast, the late-flowering plants showed little gain of DNA methylation and only partial FWA repression (Fig. 4 B and C). We also performed WGBS on two of the early-flowering T1 plants (T1-S1, T1-S2) and confirmed that these plants gained substantial DNA methylation at the FWA promoter (Fig. 4D). Both of these lines showed minimal changes in genome-wide DNA methylation levels (SI Appendix, Fig. S6). Together, these data demonstrate that MQ1(Q147L) can be efficiently used with the SunTag system to target DNA methylation in Arabidopsis and is highly specific.
Fig. 4.
The CRISPR-based SunTag system is compatible with MQ1v and can target DNA methylation to FWA to induce early flowering. (A) Dot plot of leaf counts at flowering time of T1 plants transformed with SunTag-MQ1v. Each T1 plant is the result of an independent transgenic event. Leaf counts of Col and fwa plants are included as controls. (B) DNA methylation at FWA assayed by McrBC-qPCR in four T1 (T1-S1, T1-S2, T1-S3, T1-S4) early-flowering plants and four T1 (T1-S5, T1-S6, T1-S7, T1-S8) late-flowering plants by reverse transcriptase–qPCR analysis. Error bars indicate SDs (n = 3 technical replicates). (C) FWA expression analysis by qPCR for the same eight T1 plants analyzed in B. Error bars indicate SDs (n = 3 technical replicates). (D) DNA methylation profile at the FWA promoter region in two SunTag-MQ1v T1 plants (T1-S1, T1-S2) as determined by whole-genome bisulfite sequencing. Col and fwa are included in all the panels as controls. Vertical yellow bars indicate the locations of the guide 4, guide 10, and guide 18 binding sites.
Next, we examined whether the targeted methylation at FWA by SunTag-MQ1v was heritable in the absence of the (SunTag-MQ1v) effector transgene. We selected three T1 lines (T1-S1, T1-S2, T1-S3) and analyzed the progeny (T2 plants) of these plants for the early-flowering phenotype (Fig. 5A). Between 98 and 100% of the T2 plants from each line flowered early (SI Appendix, Fig. S7A). We selected two progeny plants for each line that still contained the transgene and two plants that had segregated away the transgene and then analyzed FWA expression and DNA methylation (Fig. 5 A–C). In accordance with the early-flowering phenotype, we observed repression of FWA expression and high levels of DNA methylation at the FWA promoter in these lines compared to the unmethylated fwa epimutants (Fig. 5 B and C). We also examined genome-wide DNA methylation levels in the T2 SunTag-MQ1v lines (SI Appendix, Fig. S8). All 12 T2 plants tested showed no or little change in genome-wide DNA methylation level in comparison to control fwa epimutants (SI Appendix, Fig. S8), indicating that SunTag-MQ1v is highly specific. To further test whether the early-flowering phenotype was maintained in the T3 generation, we examined the progeny of these 12 T2 plants. Nearly all progeny of these 12 plants flowered early (SI Appendix, Fig. S7B), indicating that DNA methylation and FWA silencing are efficiently maintained in the T3 generation even if the transgene was segregated away in an earlier generation. Together these results show that SunTag-MQ1v–targeted DNA methylation at FWA was both specific to the FWA locus and efficiently inherited in the absence of the effector transgene.
Fig. 5.
Heritability of targeted DNA methylation in SunTag-MQ1v T2 plants. (A) Dot plot of leaf counts at flowering time for T2 SunTag-MQ1v plants. T1 parents are labeled at the bottom. fwa and Col plants are included as controls. (B) FWA expression in fwa, Col, and early-flowering T2 SunTag-MQ1v plants both with (+) and without (−) the MQ1v transgene. (C) DNA methylation profile at the FWA promoter region in T2 transgene positive (+) and transgene negative (−) early-flowering plants, relative to a Col wild-type control and the unmethylated fwa epiallele. Vertical yellow bars indicate the locations of the guide 4, guide 10, and guide 18 binding sites.
Previous studies have reported that DNA methylation can be amplified in subsequent generations (16, 17); in line with this, we observed that MQ1v-triggered and SunTag-MQ1v–triggered DNA methylation of FWA and early flowering were both more pronounced in later generations than in the T1 plants (Figs. 1, 2, 4, and 5). We also found that early-flowering SunTag-MQ1v T2 plants were common among the progeny of late-flowering T1s: Among the T2 progeny of seven SunTag-MQ1v T1 late-flowering plants, all lines tested gave rise to some early-flowering T3 plants, although the percentage varied widely (4 to 98%) (SI Appendix, Fig. S7C). This result further suggests that SunTag-MQ1v targeted methylation tends to amplify over successive generations.
Cas9-MQ1v and SunTag-MQ1v Target a Similar Footprint of Methylation to That of FWA.
The SunTag system leads to the recruitment of multiple effector MQ1v proteins attached to an epitope tail while MQ1v recruits only a single MQ1 protein, which could potentially cause the “footprint” or region affected by SunTag to be larger than that of MQ1v. To compare the DNA methylation footprints, we selected lines that exhibited strong DNA methylation at the FWA promoter in progenies of MQ1v and SunTag-MQ1v plants. We observed almost identical DNA methylation footprints at FWA with both tools, with nearly all CG sites in the region showing methylation (SI Appendix, Fig. S9). This is likely due to a combination of factors. Notably, we found two regions flanking the target site at FWA that fully lack CG sites (SI Appendix, Fig. S9), which likely limit the range of both tools. Additionally, we observed CHH methylation over the target locus (Figs. 2B and 5C), indicating that the endogenous DNA methylation machinery, likely RdDM, is also being recruited to the target locus. Initial CG methylation seeded by MQ1v likely helps recruit these pathways, at which point local chromatin context and interactions with other factors would determine the full extent of targeted methylation. Thus, factors such as the genomic context and interactions with other pathways can influence the size of the region affected by these tools.
Conclusion
We have developed two CRISPR-based systems, MQ1v and SunTag-MQ1v, that can directly install CG methylation at a specific locus. Our data suggest SunTag-MQ1v is more potent than MQ1v, and the differential strength of these two tools may provide the flexibility to induce different levels of de novo DNA methylation at different loci. These CRISPR-based tools are distinct from previously developed tools because they specifically target cytosines in a CG context, which likely improves heritability of the added DNA methylation at FWA due to the robustness of CG-methylation maintenance pathways. The CRISPR-based MQ1v systems described here were also highly specific, likely owing to the Q147L mutation that reduces the catalytic activity of the enzyme (7). Indeed, in a separate study, we found that an artificial zinc finger fused to the wild-type SssI enzyme caused broad off-target methylation throughout the genome (22).
Several factors may influence targeted DNA methylation. For example, we observed that the CG methylation added by MQ1v at FWA can eventually establish non-CG methylation, indicating the recruitment of other DNA methylation pathways. In plants, these pathways, including RdDM, are self-reinforcing (1) and likely help strengthen FWA silencing. However, when we transformed MQ1v into a null RdDM mutant background (fwa drm1 drm2), we found that methylation at CG sites alone was able to silence FWA, as previously shown (12). In fact, silencing was more efficient in the drm1 drm2 background. This result was likely due to increased transgene expression rather than a direct consequence of loss of non-CG methylation at FWA, since a similar effect was observed in a background deficient in PTGS (rdr6). These results suggest that efficient transgene expression remains an important limiting factor in CRISPR-based epigenetic editing efforts, as has also been shown for CRISPR-based genome editing (19). This also demonstrates that for some loci, CG methylation is sufficient to silence the expression of a gene. For these loci, the tools described here are likely to be more efficient than the previously developed DRM2-based SunTag system (8). Overall, epigenetic targeting tools such as these may be useful in altering gene expression both for research applications and for crop improvement efforts.
Materials and Methods
Plant Materials and Growth Conditions.
Arabidopsis thaliana [ecotype Columbia-0 (col)] plants were grown under long-day conditions (16 h light/8 h dark) at 23 °C in controlled growth chambers. T1 transgenic plants were selected on 1/2 Murashige and Skoog (MS) medium + 35 μg/mL Hygromycin B (Invitrogen). Flowering time was measured by counting the total number of rosette and cauline leaves at the time of flowering. Red dashed lines in each dot plot of flowering time indicate the cutoff for considering a plant as early flowering, based on the flowering time of Columbia-0 plants that were grown side by side with the tested plants for each individual experiment (usually 20 or 25 leaves).
Cloning dCAS9-MQ1v and SunTag-MQ1v Construct into Plant Binary Vector.
The MQ1v binary vector was generated by amplifying the dCAS9-MQ1(Q147L) region from the pcDNA3.1-dCas9_MQ1(Q147L)EGFP construct (7). The amplified dCAS9-MQ1-147L was inserted between sites HpaI and AscI in the plant binary vector pEG302 that already contained UBQ10 promoter upstream of HpaI and OCS terminator downstream of AscI by using Infusion (Clontech). Guide RNAs were cloned in the KpnI site of the pEG302 vector by using Infusion (Clontech) (8). To clone MQ1v into the SunTag system, MQ1(Q147L) was amplified from the pcDNA3.1-dCas9_MQ1(Q147L)EGFP construct and was inserted into the BsiwI site of the pEG302-SunTag vector that was previously used in another study (8) and contained the three guide RNAs and the SunTag system by using Infusion (Clontech) following the manufacturer’s protocol.
qPCR.
qPCR was performed as previously described (23). Rosette leaves were used for total RNA extraction by using a Direct-zol RNA Miniprep kit (Zymo) and using in-column DNase digestion. SuperScript III First-Strand Synthesis SuperMix (Invitrogen) was used for cDNA synthesis for reverse transcriptase qPCR. Oligos JP7911 (5′-ttagatccaaaggagtatcaaag-3′) and JP7912 (5′- ctttggtaccagcggaga-3′) were used to detect FWA transcripts. The housekeeping gene ISOPENTENYL PYROPHOSPHATE:DIMETHYLALLYL PYROPHOSPHATE ISOMERASE 2 (IPP2) was used to normalize the ct values. IPP2 transcripts were detected using oligos JP11859 (5′-gtatgagttgcttctccagcaaag-3′) and JP11860 (5′-gaggatggctgcaacaagtgt-3′). The delta ct values were calculated relative to the control plants.
McrBC-qPCR.
McrBC-qPCR was performed as previously described (23). Briefly, the cetyl trimethylammonium chloride (CTAB) method was used to extract genomic DNA. McrBC restriction digestion was performed on 500-ng to 1-μg amounts of DNA for 4 h at 37 °C, followed by 20 min at 65 °C. As a negative control, equal amounts of DNA were incubated in buffer without the enzyme for 4 h at 37 °C. Oligos JP15049 (5′-ttgggtttagtgtttacttg-3′) and JP15050 (5′-gaatgttgaatgggataaggta-3′) were used for qPCR of the FWA promoter. The ratio between the digested and the undigested DNA in the samples was calculated using the qPCR data and was expressed relative to the control lines.
WGBS Analysis.
The CTAB-based method was used to extract DNA. A total of 75 to 150 ng of DNA was used to prepare WGBS libraries by using the Ovation Ultralow Methyl-seq kit (NuGEN). Bisulfite treatment was performed by using the Qiagen EpiTect bisulfite kit per manufacturer’s instructions. Libraries were sequenced on Illumina HiSeq 4000 and NovaSeq 6000. Raw sequencing reads were aligned to the Arabidopsis genome (TAIR10) using Bismark (24), which was also used to generate per-position DNA methylation tracks. Reads with three or more consecutive methylated CHH sites were discarded since they are likely to be unconverted reads, as described previously (25). Browser images were obtained from Integrative Genomics Viewer (IGV) (26). Cytosines with a five or more read coverage and zero methylation were represented by a negative black bar, while cytosines with a less than five read coverage are not shown (no bar).
Bisulfite PCR Sequencing.
Bisulfite PCR sequencing was performed as previously described (10, 23). Genomic DNA was extracted using the CTAB-based method followed by sodium bisulfite treatment of the DNA by the Qiagen EpiTect bisulfite kit per manufacturer’s instructions. Libraries were generated from purified PCR products amplified from the bisulfite-treated DNA by using previously described primers (10), using an Ovation Ultralow V2 kit (NuGEN) or a Kapa Kit (Roche) in combination with TrueSeq LT barcodes or IDT UD barcodes (Illumina). Illumina HiSeq 2500 was used to sequence the libraries. Bismark was used to map and process the bisulfite PCR reads (24). As with the WGBS analysis, reads with three or more consecutive methylated CHH sites were discarded since they are likely to be unconverted reads.
Total RNA Sequencing.
Total RNA extraction was performed by using the Direct-zol RNA Miniprep kit (Zymo), using in-column DNase digestion. A total of 1 μg of RNA was used to prepare libraries by using the Illumina TruSeq Stranded Total RNA kit with Ribo-Zero Plant per manufacturer’s protocol and they were run on an Illumina HiSeq 4000 sequencer, with a single-end 50-bp protocol. Reads were first aligned to TAIR10 using the Spliced Transcripts Alignment to a Reference (STAR) package (27). Coverage tracks were generated using bamCoverage [deepTools, options–binSize 1–normalizeUsing CPM (28)], and replicates were then pooled using bigWigMerge from the University of California Santa Cruz (UCSC) toolkit (29). The number of reads mapping to each gene was also calculated by htseq-count (30) using default parameters. Differential expression analysis was performed using the DESeq2 software package (31). Predicted log2(fold change) values and SEs were extracted from the DESeq2 output for FWA (SI Appendix, Fig. S4C). However, due to low counts and high variability caused by failure of the depletion of the rRNA, further analysis of the DE results was not attempted.
Supplementary Material
Acknowledgments
We thank Dr. Margaret Goodell for the gift of the pcDNA3.1-dCas9_MQ1(Q147L)EGFP and pcDNA3.1-dCas9_dMQ1-EGFP vectors and Pranay Hegde, Emily Dembinski, Stephanie Zou, and Breanna Camille Cayabyab for technical help. We thank Mahnaz Akhavan for her help with DNA sequencing and Dr. Jake Harris for bioinformatics help during the preliminary phase of the study. High-throughput sequencing was performed at the University of California, Los Angeles Broad Stem Cell Research Center (BSCRC) BioSequencing Core Facility. C.L.P. was supported by NIH Grant F32 GM136115. This research was supported by grants from the Bill & Melinda Gates Foundation (OPP1210659) and NIH (R35 GM130272). S.E.J. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2125016118/-/DCSupplemental.
Data Availability
Next-generation sequencing raw data have been deposited in Gene Expression Omnibus (GEO) database, http://www.ncbi.nlm.nih.gov/geo (accession no. GSE149840). All study data are included in this article and/or SI Appendix.
References
- 1.Du J., Johnson L. M., Jacobsen S. E., Patel D. J., DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 16, 519–532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matzke M. A., Mosher R. A., RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15, 394–408 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Stroud H., et al., Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21, 64–72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stroud H., Greenberg M. V., Feng S., Bernatavichute Y. V., Jacobsen S. E., Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152, 352–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bostick M., et al., UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Picard C. L., Gehring M., Proximal methylation features associated with nonrandom changes in gene body methylation. Genome Biol. 18, 73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lei Y., et al., Targeted DNA methylation in vivo using an engineered dCas9-MQ1 fusion protein. Nat. Commun. 8, 16026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papikian A., Liu W., Gallego-Bartolomé J., Jacobsen S. E., Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat. Commun. 10, 729 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wada Y., Ohya H., Yamaguchi Y., Koizumi N., Sano H., Preferential de novo methylation of cytosine residues in non-CpG sequences by a domains rearranged DNA methyltransferase from tobacco plants. J. Biol. Chem. 278, 42386–42393 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Gallego-Bartolomé J., et al., Co-targeting RNA polymerases IV and V promotes efficient de novo DNA methylation in Arabidopsis. Cell 176, 1068–1082.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soppe W. J., et al., The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol. Cell 6, 791–802 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Kinoshita Y., et al., Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J. 49, 38–45 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Kinoshita T., et al., One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303, 521–523 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Johnson L. M., et al., SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature 507, 124–128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallego-Bartolomé J., et al., Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc. Natl. Acad. Sci. U.S.A. 115, E2125–E2134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bond D. M., Baulcombe D. C., Epigenetic transitions leading to heritable, RNA-mediated de novo silencing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 112, 917–922 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teixeira F. K., et al., A role for RNAi in the selective correction of DNA methylation defects. Science 323, 1600–1604 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Chan S. W., Zhang X., Bernatavichute Y. V., Jacobsen S. E., Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol. 4, e363 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao Y., et al., Manipulating plant RNA-silencing pathways to improve the gene editing efficiency of CRISPR/Cas9 systems. Genome Biol. 19, 149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan S. W., et al., RNA silencing genes control de novo DNA methylation. Science 303, 1336 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Tanenbaum M. E., Gilbert L. A., Qi L. S., Weissman J. S., Vale R. D., A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Wet al., Ectopic targeting of CG DNA methylation in Arabidopsis with the bacterial SssI methyltransferase. Nat. Commun., 10.1038/s41467-021-23346-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghoshal B., et al., A viral guide RNA delivery system for CRISPR-based transcriptional activation and heritable targeted DNA demethylation in Arabidopsis thaliana. PLoS Genet. 16, e1008983 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krueger F., Andrews S. R., Bismark: A flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cokus S. J., et al., Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorvaldsdóttir H., Robinson J. T., Mesirov J. P., Integrative genomics viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobin A.et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramírez F., Dündar F., Diehl S., Grüning B. A., Manke T., deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent W. J., Zweig A. S., Barber G., Hinrichs A. S., Karolchik D., BigWig and BigBed: Enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anders S., Pyl P. T., Huber W., HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Next-generation sequencing raw data have been deposited in Gene Expression Omnibus (GEO) database, http://www.ncbi.nlm.nih.gov/geo (accession no. GSE149840). All study data are included in this article and/or SI Appendix.