Summary
Smokers are more likely than non-smokers to harbour Porphyromonas gingivalis, they are more susceptible to destructive periodontal disease and smokers may, ultimately, benefit from tobacco-specific preventive and treatment strategies. A Mariner transposon insertion library for P. gingivalis ATCC 33277 was exploited to define 256 genes as essential for P. gingivalis survival in a tobacco-rich environment. Genes whose products play roles in protein transport and catabolism, nicotinamide processing, protection against oxidative stress, drug resistance and transcriptional regulation have all been identified as essential for CSE survival. Many of these tobacco-essential genes are also requisite for epithelial colonization and abscess formation, suggestive of a core stress-related P. gingivalis genome. Single-gene deletions in several of the TnSeq-implicated genes led to significantly reduced P. gingivalis fitness upon competition with the parent strain, under conditions of cigarette smoke extract-induced stress (1000 ng/ml nicotine equivalents). This study identifies, for the first time, a subset of P. gingivalis genes required for surviving the plethora of insults present in cigarette smoke. Such conditionally essential genes may delineate bacterial persistence strategies and represent novel therapeutic foci for the prevention of P. gingivalis infection and related diseases in smokers and in general.
Keywords: cigarette smoke, nicotinamide, oral bacteria, periodontitis, proteases, transposon sequencing library
Graphical Abstract
Porphyromonas gingivalis, an important periodontal pathogen, thrives in the oral cavity of tobacco smokers. Through transposon sequencing and competitive fitness analyses, this study identifies a subset of P. gingivalis genes required for surviving the plethora of insults present in cigarette smoke. Such conditionally essential genes may delineate bacterial persistence strategies and represent novel therapeutic foci for the prevention of P. gingivalis infection and related diseases in smokers and in general.
Introduction
Destructive periodontal diseases are infectious diseases of the tissues surrounding the teeth which lead to significant oral debilitation. In addition, they are associated with multiple serious systemic sequelae and represent a significant economic burden. At the population level, smoking may account for most cases of periodontitis in adults in developed nations, such as Scandinavia, New Zealand and the US (Bergstrom, 2014; Haisman-Welsh & Thomson, 2012; Sanders & Slade, 2013; Tomar & Asma, 2000). Clearly, bacteria are required to induce disease yet the mechanisms by which smoking predisposes to periodontal diseases are not well elucidated.
Contemporary data also show that P. gingivalis, a causative agent of periodontal diseases, and other Bacteroidetes, are more likely to infect and persist in smokers compared to non-smokers (Chigasaki et al., 2018; Feres et al., 2014; Ge, Rodriguez, Trinh, Gunsolley, & Xu, 2013; Guglielmetti et al., 2014; Hanioka et al., 2019; Joshi et al., 2014; Kumar, 2012; Mason et al., 2014). This is consistent with earlier, more traditional, studies showing P. gingivalis to be more prevalent and present in higher numbers in smokers than in non-smokers (Eggert, McLeod, & Flowerdew, 2001; Haffajee & Socransky, 2001; Kamma, Nakou, & Baehni, 1999; Zambon et al., 1996). Indeed, P. gingivalis is resistant to very high doses of cigarette smoke and tobacco constituents (Bagaitkar et al., 2011; Bagaitkar et al., 2009; Cogo et al., 2009).
To induce or exacerbate periodontal diseases in smokers, P. gingivalis must be able to survive cigarette smoke insults, primarily soluble and systemically distributed smoke components. We developed a Mariner transposon insertion library for P. gingivalis ATCC 33277, a model Gram-negative anaerobic periodontopathogen, and employed it to identify a core genome permitting replication of this P. gingivalis strain in rich medium (Hutcherson et al., 2016). We have also assessed the fitness of specific mutants during epithelial cell colonization and survival in a murine abscess model (Miller et al., 2017). We have now utilized this tool to test the hypothesis that multiple P. gingivalis genes are essential for survival upon exposure to physiologically relevant doses of cigarette smoke extract (CSE). We establish that P. gingivalis strains with mutations in a subset of these genes exhibit an absolute reduction in fitness [PGN_1444 (Δcps)], or reduced fitness when competed against the parent strain [PGN_0088 (ΔsinR1), PGN_0287 (Δmfa1), PGN_0388 (Δtpx), PGN_0770 (Δrnz), PGN_1200 (a DNA-dependent ATPase) and PGN_1524 (Δptk1)] under tobacco-induced stress, confirming CSE conditional essentiality.
Methods
Materials
Gifu anaerobic medium (GAM) was obtained from Nissui Pharmaceutical (Tokyo, Japan). Sheep blood was obtained from Lampire Biological Laboratories (Pipersville, PA). Gentamicin, erythromycin, tetracycline, ampicillin, dichloromethane, sodium sulfate and analytical nicotine standards were from Sigma-Aldrich (St. Louis, MO). 3R4F research cigarettes were purchased from the Centre for Tobacco Research Products (University of Kentucky, Lexington, KY). All primers were from Biosynthesis Inc. (Lewiston, TX). Lonza flash gels came from Lonza (Rockland, ME). Wizard SV and PCR clean-up kits were from Promega (Madison, WI). T4 DNA ligase, MmeI; and TAE buffer came from New England Biolabs (Ipswich, MA). HiFi Hotstart Ready mix was from KAPA Biosystems (Wilmington, MA) and PCR SuperMix came from Invitrogen (Carlsbad, CA). Fast digest restriction enzymes and ready to use X-gal and plasmid isolation kits came from Thermo Fisher Scientific (Waltham, MA).
Bacterial culture
P. gingivalis ATCC 33277 and isogenic mutants ΔPGN_0088 (sinR), ΔPGN_0287 (mfa1), ΔPGN_0388 (a putative thiol peroxidase), ΔPGN_0491 (ltp1, a tyrosine protein phosphatase), ΔPGN_0770 (rnz), ΔPGN_1200 (putative ATPase), ΔPGN_1444 (carbamoyl phosphate synthase, cps), ΔPGN_1524 (ptk1) and a non-negatively selected control mutant, ΔPGN_1753 (2-oxoglutarate oxidoreductase-related gene) were cultured anaerobically in GAM, cigarette smoke-conditioned GAM (GAM-CSE) or on GAM agar plates supplemented with defibrinated sheep’s blood at 37°C.
P. gingivalis transposon library
Full details of the 80,000 colony transposon library employed have been previously reported (Hutcherson et al., 2016). The P. gingivalis 33277 library was maintained in GAM containing 50 μg/ml gentamicin and 5 μg/ml erythromycin, the input library aliquoted at 1010 CFU and stored at −80°C.
Cigarette smoke conditioning
GAM was cigarette smoke conditioned, as previously described (Bagaitkar et al., 2010). Essentially, 3R4F standard reference cigarette smoke (GAM-CSE) was drawn through GAM and filtered (0.2 μm). Nicotine content was determined by GLC and GAM-CSE adjusted to 1000 ng/ml nicotine, pH 7.2.
Construction and sequencing of DNA libraries and analysis of sequencing data
The P. gingivalis ATCC 33277 TnSeq mutant library was grown to mid-log phase, as determined by optical density, and passaged twice in GAM (input) and GAM-CSE (output). Double-stranded DNA (dsDNA) adapter molecules were created using primers listed in Supplemental Table 4. The sequencing platform, sequence sorting, read trimming, normalization and alignment to the annotated gene list of P. gingivalis ATCC 33277 were performed as recently reported (Miller et al., 2017). Significance was determined using CLC Bioinformatics Workbench v7.2 software. Genes with ≥ 200 genes in the input pool, ≥ 50 fold change in input and output pools and a Bonferroni-adjusted p-value < 0.05 were considered significant for fitness (Miller et al., 2017).
Mapping and enrichment of CSE-essential genes
Genes on which P. gingivalis is reliant in a tobacco-rich environment were mapped to the 33277 genome, along with P. gingivalis TDC60 and W83 orthologues, using the CGViewer software (http://stothard.afns.ualberta.ca/cgview_server/) [data not shown]. CSE essential genes were distributed throughout the bacterial chromosome. Relationships between essential genes were determined using the KEGG (www.genome.jp/kegg) database. Potential functional annotations for those genes considered hypothetical in the KEGG system were also examined by processing through the NCBI pipeline (https://www.ncbi.nlm.nih.gov/genome/annotation_prok/).
Mutant construction
Mutants employed herein are described in Supplemental Table 3. The P. gingivalis deletion mutants created specifically for the current study were generated as we have previously described (Nguyen, Travis, & Potempa, 2007) [PGN_0388] or (Miller et al., 2017; Simionato et al., 2006) [PGN_0088, PGN_1753], using primers presented in Supplemental Table 4. Following anaerobic selection on erythromycin (10 μg/ml) or tetracycline (1 μg/ml) seeded plates, as appropriate (Supplemental Table 3), colonies arising from transformed clones were screened for the correct mutations by PCR and sequencing
Competitive fitness assays
In competitive growth assays, P. gingivalis ATCC 33277 and individual mutant strains were anaerobically co-cultured 1:1 in GAM-CSE (1000 ng/ml nicotine equivalents). After 2 days, strain growth was quantified by CFU counts on GAM agar plates supplemented with defibrinated sheep’s blood, with and without the mutant-specific resistance antibiotic. All experiments were carried out in triplicate. Differences in percentage survival rates were determined by paired t-test using InStat v3.06 (GraphPad, San Diego, CA, USA).
Results
Identification of essential CSE survival genes
A total of 256 genes conditionally essential for survival in the GAM-CSE model were identified. The genes were distributed throughout the P. gingivalis chromosome. In silico analyses were used to identify the functions of these essential genes, as presented in Supplemental Table 1 (functionally annotated genes) and Supplemental Table 2 (genes encoding hypothetical proteins).
Relationships between essential CSE survival genes
KEGG analysis revealed predicted functional relationships between CSE-essential genes, as delineated in Supplemental Figure 1. Interestingly, 7/13 (54%) of the bacterium’s nicotinate and nicotinamide metabolism genes were essential, as shown in Figure 1. Multiple genes involved in protection against oxidative stress or in protein and / or peptide catabolism were also determined to be essential, as presented in Tables 1 and 2.
Figure 1: Essential nicotinate and nicotinamide metabolism genes in P. gingivalis 33277.
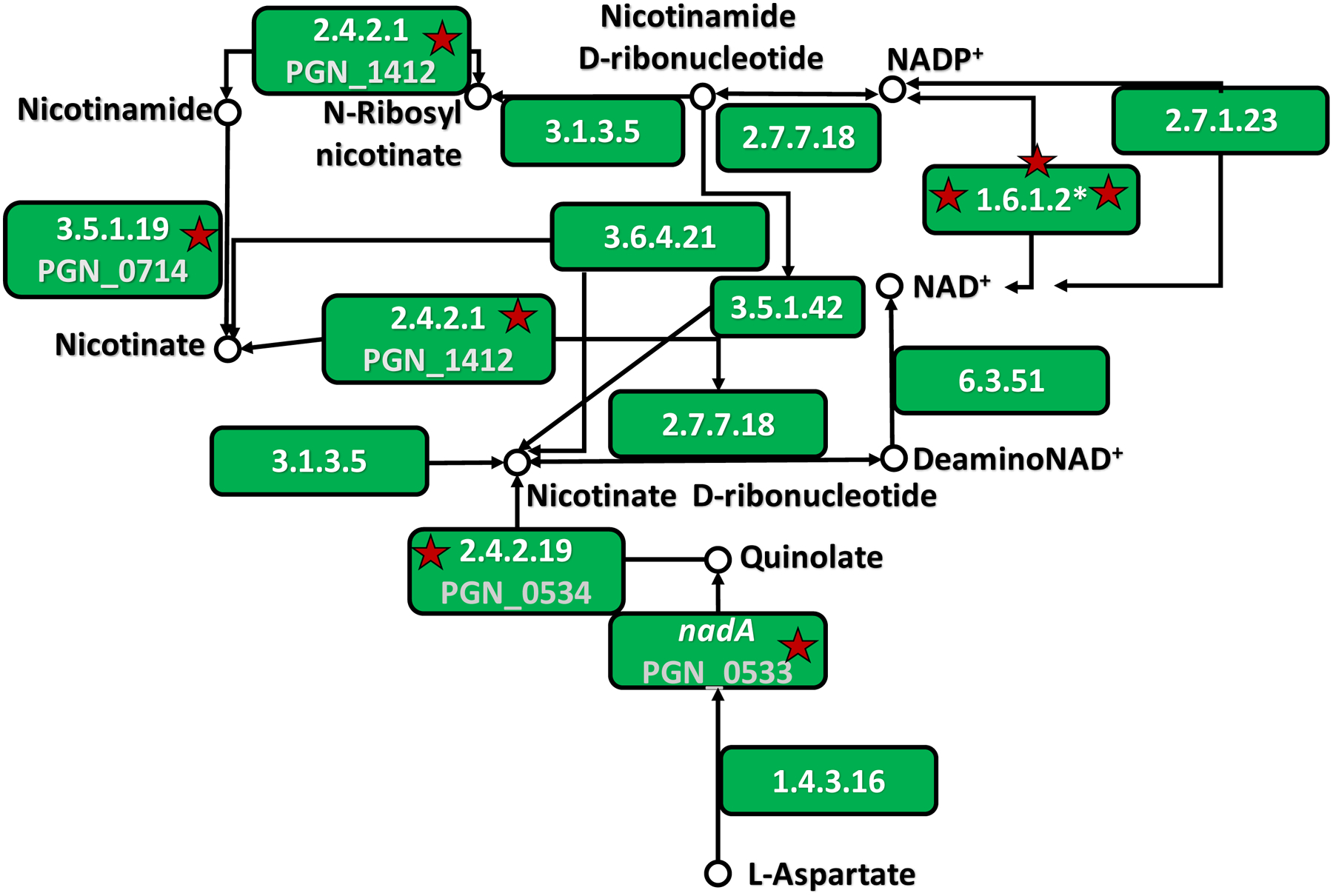
P. gingivalis 33277 homologues of genes in nicotinate and nicotinamide metabolic pathways were enriched by KEGG analysis (www.genome.jp/kegg). Stars denote CSE-essential genes. PGN_0533, PGN_0534, PGN_0714, PGN_1412 are presented as their enzyme commission number and substrates or products. The commission number 1.6.1.2* is represented by three CSE-essential P. gingivalis genes, PGN_1120, PGN_1121 and PGN_1122.
Table 1.
CSE-essential protein catabolism genes of P. gingivalis 33277.
| Protein catabolism genes | |
|---|---|
| Gene number | Annotation |
| PGN_0271 | pepO endopeptidase |
| PGN_0295 | Arg-/Lys-gingipain proteinase C-terminal domainb |
| PGN_0303 | Zinc protease, M16 family |
| PGN_0335 | Zinc carboxypeptidase domain protein |
| PGN_0561 | prtT, cysteine protease |
| PGN_0607 | Dipeptidyl peptidase 11 |
| PGN_0637 | htrA, heat shock related protease |
| PGN_0754 | NlpC/P60 peptidase familya |
| PGN_0771 | NlpC/P60 peptidase familya |
| PGN_0780 | prtQ proteasec |
| PGN_0788 | Peptidyl dipeptidase |
| PGN_0900 | Thiol proteased |
| PGN_1335 | Hypothetical peptidase |
| PGN_1349 | Dipeptidyl aminopeptidase |
| PGN_1466 | rgpB, gingipain B |
| PGN_1479 | S46 family dipeptidyl-peptidasea |
| PGN_1694 | Alanyl dipeptidyl peptidase |
| PGN_1777 | Bleomycin hydrolase |
| PGN_2064 | M48 family peptidase |
Annotated functions of CSE-essential genes (>500 reads on input, >50 fold change, p < 0.05) were annotated using KEGG.
Peptidolytic annotation from the NCBI pipeline.
A Blast search of the MEROPS database (https://www.ebi.ac.uk/merops/) reveals PGN_0295 to be a CTD-secreted protein of unknown function.
No proteolytic motifs were identified upon crystallization (Schacherl, Montada, Brunstein, & Baumann, 2015).
We have previously characterized this protein as periodontain (Nelson, Potempa, Kordula, & Travis, 1999).
Table 2.
CSE-essential oxidative stress genes of P. gingivalis 33277.
| Oxidative stress genes | |
|---|---|
| Gene number | Annotation |
| PGN_0168 | wbpB, LPS biosynthesis protein/oxidoreductase |
| PGN_0302 | Rubrerythrin |
| PGN_0373 | Thioredoxin |
| PGN_0388 | Thiol peroxidase |
| PGN_0533 | nadA, Quinolinate synthetase A |
| PGN_0534 | nicotinate-nucleotide pyrophosphorylase |
| PGN_0564 | Superoxide dismutase Fe-Mn |
| PGN_0604 | Ferritin |
| PGN_0709 | Indolepyruvate ferredoxin oxidoreductase, beta subunit |
| PGN_1268 | oxidoreductase |
| PGN_2073a | oxidoreductase |
| PGN_2077a | 2Fe-2S binding protein |
Annotated functions of CSE-essential genes (>500 reads on input, >50 fold change, p < 0.05) were annotated using KEGG.
Annotation from the NCBI pipeline.
Common and unique essential genes in multiple disease-relevant models.
We have previously determined the core essential genome of P. gingivalis ATCC 33277, that is, genes required for survival in standard rich culture conditions (Hutcherson et al., 2016). More recently, we have determined a subset of genes required for P. gingivalis to invade epithelial cells and to produce a subcutaneous abscess in a mouse model (Miller et al., 2017). We now report that, from a total of 256 genes essential for surviving CSE-induced stress, a subset are specific to this particular microenvironment while most are common between the stress conditions in which this library has been exploited. Genes uniquely required for surviving in cigarette smoke extract conditioned medium include PGN_0065 (traG), PGN_0415 (a restriction endonuclease) and PGN_0584 (topoisomerase). Interestingly, PGN_1440 (vanW) is a CSE-essential gene. PGN_0323, likely outer membrane protein with some homology to PorT protein transporter was also CSE-essential. CSE genes that exhibited the largest differentials between input and output libraries, as well as those whose functional annotations have clear relevance to CSE survival, are discussed further below. Essential genes that are uniquely required for CSE survival as well those commonly requisite for fitness under CSE-induced stress, epithelial invasion and/or abscess formation are presented in Supplemental Tables 1 and 2. The relative distributions of essential genes of P. gingivalis between the three selection pressures are presented in Figure 2A (TIGK > abscess > CSE), with epithelial colonization the environment requiring the largest proportion of the genome. The distributions are compared to those that may happen by chance alone, as determined by the generation of randomized integer sets (Figure 2B).
Figure 2. Distribution of essential genes of P. gingivalis between CSE, TIGK and murine abscess selection pressures.

A. Negatively selected genes between selection pressures are presented. A total of 231 genes were essential for all conditions. The volume of each sphere represents the ratio of essential genes per condition (TIGK > abscess > CSE). B. The Random Integer Set Generator tool (random.org) was employed to generate random n = 541, n = 496 and n = 256 integer arrays from n = 1874, the total number of genes outwith the previously determined 33277 core genome (Hutcherson et al., 2016).
Confirmation of conditionally essential CSE survival genes
To verify the loss of fitness in strains bearing mutations in TnSeq-defined essential genes, we conducted in vitro competition assays between the parental P. gingivalis strain and strains with deletion mutations in 8 different genes. As a control, a strain bearing a mutation in the PGN_1753 locus, which was not shown to be essential in the CSE Tn-seq experiments, was employed. First, we assessed relative growth characteristics to ensure that mutant strains had similar growth rates compared to the parental strain thus preventing false positives or negatives in our competition experiments. Growth of the control mutant, ΔPGN_1753, was not compromised in tobacco-rich environments, either in CSE-conditioned monoculture compared to non-conditioned medium (p > 0.05) or in competition with the parent strain (p > 0.05). ΔPGN_1444 exhibited a reduced fitness phenotype in monoculture, i.e., mutant growth was significantly diminished in CSE-conditioned medium compared to unconditioned GAM (Table 3). In other words, deletion of PGN_1444 alone results in compromised growth of CSE-exposed P. gingivalis. Conditional essentiality was confirmed by competitive fitness assays against parent P. gingivalis in 6/7 (86%) mutant strains tested, as presented Table 3. Growth of the ltp1 mutant, ΔPGN_0491, was not compromised by CSE in monoculture or in competition against wild type P. gingivalis ATCC 33277. Nevertheless, most mutants screened exhibited the library-predicted CSE-related phenotype, suggesting that TnSeq is a powerful tool for the identification of conditionally essential bacterial genes.
Table 3.
CSE- essential genes determined by absolute or competitive fitness assays.
| Mutant | Survival on agar [% (s.d)] |
|---|---|
| PGN_0088 (ΔsinR1) | 22.4 (6.5)* |
| PGN_0287 (ΔfimA) | 0.0 (0.0)*** |
| PGN_0388 (Δtpx) | 2.7 (2.1)*** |
| PGN_0491 (Δltp1) | 40.3 (26.2) |
| PGN_0770 (Δrnz) | 15.8 (5.8)** |
| PGN_1200 (ΔDNA-dependent ATPase) | 17.8 (15.5)* |
| PGN_1524 (Δptk1) | 1.1 (1.8)*** |
P. gingivalis ATCC 33277 and mutant strains were anaerobically co-cultured at a 1:1 in GAM-CSE (1000 ng/ml nicotine equivalents). Growth was quantified by CFU counts on GAM agar plates supplemented with defibrinated sheep’s blood, with and without the mutant-specific resistance antibiotic. Differences in percentage survival rates were determined by paired t-test.
p < 0.05, 0.01 and 0.001, respectively.
Additionally, ΔPGN_1444 exhibited an absolute reduced fitness phenotype in monoculture, i.e., mutant growth was significantly diminished in CSE-conditioned medium (1000 ng/ml nicotine equivalents) compared to unconditioned GAM (0 ng/ml nicotine equivalents).
Discussion
Using a transposon sequencing library, we have previously established a core set of 281 genes, out of a total of 2155 ORFs, that are absolutely required to support the growth of P. gingivalis ATCC 33277 in rich medium (Hutcherson et al., 2016), in common with a second, independently generated and validated P. gingivalis ATCC 33277 library (Klein et al., 2012). More recently we have determined those P. gingivalis genes that are essential for P. gingivalis survival in two disease-related model systems, epithelial cell colonization (n = 541 genes) and subcutaneous abscess formation in mice (n = 496 genes). Interestingly, 482 of these genes were common to each model, suggesting a high degree of overlap in the fitness determinants required for these environments. We now identify 256 P. gingivalis genes that are essential for survival in a tobacco-rich environment. Of these, the majority are common amongst all disease-relevant models tested (CSE exposure, abscess formation and epithelial colonization). Such commonality amongst data sets may be indicative of a core stress-related genome in P. gingivalis. As we have recently discussed the physiological relevance of these common genes, with respect to gingival adhesion, iron acquisition, established stress responses, conjugation and tetratricopeptide repeat (TPR) protein-related virulence (Miller et al., 2017), we shall not revisit this topic in depth herein.
The 5 genes exhibiting the greatest change between input (no CSE) and output (CSE) libraries were PGN_0407, PGN_0025 (spoU), PGN_1494 (coproporphyrinogen III oxidase), PGN_0388 (ptx) and PGN_0604 (ferritin). Unfortunately, efforts to generate mutants in PGN_0025 (spoU) and PGN_1494 (coproporphyrinogen III oxidase) proved unsuccessful. There is no functional annotation available for PGN_0407. P. gingivalis ferritin has been reported to be highly upregulated under oxidative stress, although it is insufficient to fully protect against oxygen exposure (Meuric, Gracieux, Tamanai-Shacoori, Perez-Chaparro, & Bonnaure-Mallet, 2008; Ratnayake et al., 2000). PGN_0604 mutants have not been evaluated, while the CSE-related phenotype of a PGN_0388 mutant is addressed below.
Nicotinamide is a structural analogue of nicotine, long known to be toxic to bacteria (Koser & Kasai, 1947; Murray, 2003), that may be present in cigarette smoke itself (Buyske, Flowers, Hobbs, & Wilder, 1956). Nicotine has also been reported to inhibit the growth of multiple Gram-negative bacteria, including Escherichia coli, Klebsiella pneumoniae, and Borrelia burgdorferi (Gandhi, Athmaram, & Arunkumar, 2016; Pavia, Pierre, & Nowakowski, 2000; Salman et al., 2016). Therefore, it is of particular interest that PGN_0714 was identified as CSE-essential in our TnSeq screen, as well as several genes involved in nicotinate and nicotinamide metabolism. Some nicotinate- and nicotinamide-related gene products exhibit functional overlap with an oxidative stress response. PGN_0714 encodes a nicotinamidase which produces nicotinate from nicotinamide. PGN_0533 (quinolate synthetase A) and PGN_0534 (nicotinate-nucleotide pyrophosphorylase) are involved in generation of the key redox-related molecule, NAD, while PGN_0533 and PGN_1120 (NADPH-NAD transhydrogenase/alanine dehydrogenase) have also be shown to be essential for the black pigmentation associated with heme binding (Klein et al., 2017).
The precise role in protection against tobacco toxins are not immediately obvious for several of the CSE-specific essential genes. PGN_0415, PGN_0584, and PGN_1116 encode a restriction endonuclease, a topoisomerase and an aminotransferase, respectively. PGN_1440 (vanW) is a vancomycin B-type resistance protein that is, as yet, understudied. However, it is possible that a detoxification-related VanW function may be relevant to protection against cigarette smoke components. The hypothetical protein, PGN_0323, exhibits some homology to PorT, while our TnSeq screen also identifies another PorT family member, PGN_0156, as CSE-essential. Indeed PGN_0156 is in the top 10 of input and output library differentials. As we have previously reported (Nguyen et al., 2009), PorT is important in the maturation and secretion of gingipains, which are critical to P. gingivalis colonization and to several other key aspects of P. gingivalis virulence. Multiple other CSE-essential genes may provide insight into P. gingivalis robustness in tobacco-rich environments. Histidine kinases, for example, are key to bacterial signal transduction and, subsequently, virulence and may represent novel antibacterial targets (Bem et al., 2015). PGN_0082 is an AraC-family transcriptional regulator known to be involved in quorum sensing and stress response in other bacterial species (Romero-Lastra et al., 2017). Indeed, in the related bacterium, Bacteroides thetaiotaomicron, greater than 80% of a reported 44 hybrid histidine kinases contain AraC-like DNA-binding domains (Galperin, 2006). PGN_0302 encodes rubrerythrin. This is notable, as P. gingivalis, which lacks catalase, uses rubrerythrin to protect against oxygen- and nitrogen-related stresses (Mydel et al., 2006). PGN_0447 is an ABC transporter permease whose most closely related orthologues are multidrug and other efflux transporters in Tannerella (BFO_0463 and BCB71_07875) and Parabacteroides (CI960_00995) spp. PGN_0675 is a DJ-1 family protein. DJ-1 is a multifunctional protein with declycase activity in mammals whose bacterial homologues have recently been shown to be important mediators of genomic stability in bacteria through their nucleotide glycation repair activity (Richarme et al., 2017). This is likely relevant to the multiple mutagenic compounds known to be present in cigarette smoke. PGN_0754 and PGN_0771 belong to the NlpC/P60 family of proteins, typically encompassing papain-like domain endopeptidases with specificity for peptidoglycan-embedded amino acids (Xu et al., 2015). While such P. gingivalis genes are poorly characterized, they may be essential for modification of the P. gingivalis cell-envelope during stress induced by CSE components. PGN_0200 is now thought to encode TraB and joins traA, traG and traI as stress-related essential conjugation genes.
The dependence of P. gingivalis on proteolysis for survival and subsequent virulence in the oral cavity is a well-established phenomenon. Those genes involved in protein catabolism that are essential for CSE survival contain a combination of established proteolytic virulence factors and a larger set of seemingly more mundane conditionally essential proteolytic genes. The former include pepO, an endopeptidase whose deletion lowers epithelial invasion efficacy (Ansai, Yu, Urnowey, Barik, & Takehara, 2003), at least in P. gingivalis 381, periodontain and the archetypal virulence factor, rgpA. However, many of the remaining CSE-essential protease-related genes are not well characterized. As with other genes encoding essential but understudied proteins, such proteolytic genes may represent novel therapeutic targets for the control of P. gingivalis infection.
Several CSE-essential genes were previously recognized as key to subcutaneous abscess formation and/or epithelial colonization that, at the time of discovery, were without functional annotation. Such stress genome constituents include PGN_1928 (cmr6, type III CRISPR module RAMP protein), PGN_1930 (cmr4, type III CRISPR module RAMP protein) and PGN_1932 (cmr2, type III CRISPR-associated protein). While these genes are considered to be involved in bacterial innate defense (Wang & Li, 2012), their essential role in protecting against environmental stress is currently elusive. Of particular interest is the α2-macroglobulin-encoding PGN_2070. α2-macroglobulin, as we have also shown, is the only efficient endogenous proteinaceous inhibitor of Rgp gingipains described to date (Gron et al., 1997). Gram-negative prokaryotic α2-macroglobulins are thought to locate to the periplasm (Wong & Dessen, 2014). It is a possibility, then, that the PGN_2070 product may help control translocating gingipain activity in a manner that is essential in three different P. gingivalis environments. Certainly, bacterial α2-macroglobulins have been shown to sequester exogenous proteases as part of a rudimentary defense system (Wong & Dessen, 2014). The type IX secretion system (T9SS) is restricted to a subset of bacteria of the Bacteroidetes phylum that, as we have recently summarized, is critical for the transport of over 30 P. gingivalis proteins bearing a conserved C-terminal domain (CTD), including primary virulence factors, such as the gingipains (Lasica, Ksiazek, Madej, & Potempa, 2017). It is interesting that multiple CTD-proteins (PGN_0291, PGN_0295 (MEROPS search), PGN_0335, PGN_0561, PGN_0852, PGN_0898, PGN_0900, PGN_1116 and PGN_1416) (Lasica et al., 2017), are CSE-essential, but RgpA and Kgp are not among them.
Klein et al (Klein et al., 2017) have recently identified a set of P. gingivalis ATCC 33277 genes that are required for heme acquisition that exhibits only limited overlap with genes that are both functionally annotated and requisite for CSE survival (mfa1, nadA, htrA, PGN_0862 [type III restriction enzyme], PGN_1116 [aminotransferase], PGN_1120 [NADPH-NAD transhydrogenase], PGN_1618 [wbpB], PGN_1722 [uridine kinase]. PGN_1777 [bleomycin hydrolase], and PGN_2017 [YjeF-family], as well as the hypothetical protein-encoding genes, PGN_1313 and PGN_1591). On the other hand, common CSE-, TIGK- and abscess-essential genes show considerably more overlap than would be expected by chance alone, as determined by the generation of random integers, strengthening the core stress genome hypothesis.
Finally, and critically, we established the tobacco-related phenotype of a cross-section of CSE-related genes in competitive growth assays. Interestingly, although compromised growth under CSE-induced stress was not observed for PGN_0491 (Δltp1), the other essential mutants tested [PGN_0088 (ΔsinR1), PGN_0287 (Δmfa1), PGN_0388 (Δtpx), PGN_0770 (Δrnz), PGN_1200 (ΔATPase) and PGN_1524 (Δptk1)] were outcompeted by the parent strain of P. gingivalis. PGN_1444 (Δcps) exhibited suppressed growth under CSE-induced stress, an absolute reduction in fitness. PGN_0088 (ΔsinR1) encodes a transcriptional regulator. This protein has been reported to inhibit exopolysaccharide synthesis (Yamamoto et al., 2013), in keeping with our own finding that CSE-exposure leads to P. gingivalis capsule diminishment (Bagaitkar et al., 2011; Bagaitkar et al., 2009). PGN_1524 encodes the bacterial tyrosine kinase, Ptk1, which is a key component of a signaling pathway that controls community development with Streptococcus gordonii (Wright et al., 2014). We have previously shown that Ptk1 is also necessary for exopolyscaccharide production (Wright et al., 2014). Several CSE essential minor fimbrial genes were identified. PGN_0287 (Δmfa1) encodes the minor fimbrial antigen, PGN_0288 encodes FimB/Mfa2, while PGN_0291 encodes Mfa5. It is unclear how minor fimbrial proteins may abet survival in CSE-rich batch culture. However, we have previously shown that smoking augments P. gingivalis monospecies and P. gingivalis-S. gordonii biofilms in an Mfa1-related manner (Bagaitkar et al., 2011; Bagaitkar et al., 2010). Since then, there have been multiple reports of tobacco-enhanced biofilm formation in several microbes, as we have recently summarized (Hutcherson, Scott, & Bagaitkar, 2015). PGN_0388 is an anti-oxidant thiol peroxidase, thus the conditional essentiality of an oxidative stress gene is established. The PGN_0770 product is RNase Z, a 6S RNA-/σ70-related transcriptional regulator, at least in Escherichia coli (Chen, Dutta, & Deutscher, 2016). The gene targets of PGN_0770 in P. gingivalis, and their relevance to cigarette smoke, have yet to be ascertained. PGN_1200 is functionally annotated as a DNA-dependent ATPase (RarA-related replication-associated recombination protein MgsA/RarA) and has been reported to contribute to genome stability in E. coli (Shibata et al., 2005). PGN_1444, as we recently hypothesized, may play a key role in P. gingivalis virulence by generating the metabolic cue, arginine (Miller et al., 2017).
In summary, we have determined a subset of genes that delineate, for the first time, essential bacterial strategies relevant to persistence in an environment representing the primary risk factor for periodontitis – tobacco smoking. Genes whose products play roles in protein transport and catabolism, nicotinamide processing, protection against oxidative stress, drug resistance and transcriptional regulation have all been identified. Conditional essentiality has been confirmed using a broad range of bacterial mutants [PGN_0088 (transcriptional regulator, ΔsinR1), PGN_0287 (minor fimbrial antigen, Δmfa1), PGN_0388 (thiol peroxidase, Δtpx), PGN_0770 (ribonuclease z, Δrnz), PGN_1200 (DNA-dependent ATPase) and PGN_1524 (tyrosine kinase, Δptk1)]. Such essential genes may represent appropriate therapeutic targets for the treatment of P. gingivalis infections specifically in tobacco users. Perhaps more importantly, however, we have determined a subset of >200 genes that are commonly required for surviving three different disease relevant conditions, CSE exposure, epithelial colonization and murine abscess formation. The CSE model exposes P. gingivalis to a host of environmental toxins. The epithelial model requires attachment to and / or passage through a eukaryotic biological membrane and resistance to the important, but limited, epithelial defense mechanisms. To facilitate abscess formation, P. gingivalis strains must withstand a robust mammalian immune response. This common stress-related gene set contains both established and novel potential virulence factors. Interestingly, epithelial colonization appears to exert the highest selective pressure on this important periodontal pathogen, while CSE survival required the least number of genes. However, genes with overlapping conditional essentiality may represent particularly attractive preventative targets for P. gingivalis-induced disease in general.
Supplementary Material
Acknowledgments:
These studies were supported by grants from NIDCR and NIGMS (DE026963, DE011111, DE012505, DE017921, DE023193, DE022597 and GM125504). The authors declare that they have no conflicted interests.
References
- Ansai T, Yu W, Urnowey S, Barik S, & Takehara T (2003). Construction of a pepO gene-deficient mutant of Porphyromonas gingivalis: potential role of endopeptidase O in the invasion of host cells. Oral Microbiol Immunol, 18(6), 398–400. [DOI] [PubMed] [Google Scholar]
- Bagaitkar J, Daep CA, Patel CK, Renaud DE, Demuth DR, & Scott DA (2011). Tobacco smoke augments Porphyromonas gingivalis - Streptococcus gordonii biofilm formation. PLoS One, 6(11), e27386. doi:doi: 10.1371/journal.pone.0027386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagaitkar J, Demuth DR, Daep CA, Renaud DE, Pierce DL, & Scott DA (2010). Tobacco upregulates P. gingivalis fimbrial proteins which induce TLR2 hyposensitivity. PLoS One, 5(5), e9323. doi: 10.1371/journal.pone.0009323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagaitkar J, Scott DA, Williams LR, Renaud DE, Daep C, Martin M, & Demuth DR (2009). Tobacco-induced alterations to Porphyromonas gingivalis-host interactions. Environmental Microbiology, 11(5), 1242–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bem AE, Velikova N, Pellicer MT, Baarlen Pv, Marina A, & Wells JM (2015). Bacterial histidine kinases as novel antibacterial drug targets. ACS Chem Biol, 10(1), 213–224. doi: 10.1021/cb5007135 [DOI] [PubMed] [Google Scholar]
- Bergstrom J (2014). Smoking rate and periodontal disease prevalence: 40-year trends in Sweden 1970–2010. J Clin Periodontol, 41(10), 952–957. doi: 10.1111/jcpe.12293 [DOI] [PubMed] [Google Scholar]
- Buyske DA, Flowers JM Jr., Hobbs ME, & Wilder P Jr. (1956). Nicotinic and glutamic acids, nicotinamide, and glutamine in cigarette tobacco smoke. Science, 124(3231), 1080–1081. doi: 10.1126/science.124.3231.1080 [DOI] [PubMed] [Google Scholar]
- Chen H, Dutta T, & Deutscher MP (2016). Growth Phase-dependent Variation of RNase BN/Z Affects Small RNAs: REGULATION OF 6S RNA. J Biol Chem, 291(51), 26435–26442. doi: 10.1074/jbc.M116.757450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigasaki O, Takeuchi Y, Aoki A, Sasaki Y, Mizutani K, Aoyama N, … Izumi Y (2018). A cross-sectional study on the periodontal status and prevalence of red complex periodontal pathogens in a Japanese population. J Oral Sci, 60(2), 293–303. doi: 10.2334/josnusd.17-0223 [DOI] [PubMed] [Google Scholar]
- Cogo K, Calvi BM, Mariano FS, Franco GC, Goncalves RB, & Groppo FC (2009). The effects of nicotine and cotinine on Porphyromonas gingivalis colonisation of epithelial cells. Arch Oral Biol, 54(11), 1061–1067. [DOI] [PubMed] [Google Scholar]
- Eggert FM, McLeod MH, & Flowerdew G (2001). Effects of smoking and treatment status on periodontal bacteria: evidence that smoking influences control of periodontal bacteria at the mucosal surface of the gingival crevice. J Periodontol, 72(9), 1210–1220. [DOI] [PubMed] [Google Scholar]
- Feres M, Teacher H, Bernal MA, Matarazzo F, Faveri M, Duarte PM, & Figueiredo LC (2014). Subgingival bacterial re-colonization after scaling and root planing in smokers with chronic periodontitis. Aust Dent J. doi: 10.1111/adj.12225 [DOI] [PubMed] [Google Scholar]
- Galperin MY (2006). Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol, 188(12), 4169–4182. doi: 10.1128/JB.01887-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi PT, Athmaram TN, & Arunkumar GR (2016). Novel nicotine analogues with potential anti-mycobacterial activity. Bioorg Med Chem, 24(8), 1637–1647. doi: 10.1016/j.bmc.2016.02.035 [DOI] [PubMed] [Google Scholar]
- Ge X, Rodriguez R, Trinh M, Gunsolley J, & Xu P (2013). Oral microbiome of deep and shallow dental pockets in chronic periodontitis. PLoS One, 8(6), e65520. doi: 10.1371/journal.pone.0065520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gron H, Pike R, Potempa J, Travis J, Thogersen IB, Enghild JJ, & Pizzo SV (1997). The potential role of alpha 2-macroglobulin in the control of cysteine proteinases (gingipains) from Porphyromonas gingivalis. J Periodontal Res, 32(1 Pt 1), 61–68. [DOI] [PubMed] [Google Scholar]
- Guglielmetti MR, Rosa EF, Lourencao DS, Inoue G, Gomes EF, De Micheli G, … Pannuti CM (2014). Detection and Quantification of Periodontal Pathogens in Smokers and Never-Smokers With Chronic Periodontitis by Real-Time Polymerase Chain Reaction. J Periodontol. doi: 10.1902/jop.2014.140048 [DOI] [PubMed] [Google Scholar]
- Haffajee AD, & Socransky SS (2001). Relationship of cigarette smoking to the subgingival microbiota. J Clin Periodontol, 28(5), 377–388. [DOI] [PubMed] [Google Scholar]
- Haisman-Welsh RJ, & Thomson WM (2012). Changes in periodontitis prevalence over two decades in New Zealand: evidence from the 1988 and 2009 national surveys. N Z Dent J, 108(4), 134–138. [PubMed] [Google Scholar]
- Hanioka T, Morita M, Yamamoto T, Inagaki K, Wang PL, Ito H, … Ogawa H (2019). Smoking and periodontal microorganisms. Jpn Dent Sci Rev, 55(1), 88–94. doi: 10.1016/j.jdsr.2019.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutcherson JA, Gogeneni H, Yoder-Himes D, Hendrickson EL, Hackett M, Whiteley M, … Scott DA (2016). Comparison of inherently essential genes of Porphyromonas gingivalis identified in two transposon sequencing libraries. Mol Oral Microbiol, 31, 354–364. doi: 10.1111/omi.12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutcherson JA, Scott DA, & Bagaitkar J (2015). Scratching the surface - tobacco-induced bacterial biofilms. Tob Induc Dis, 13(1), 1. doi: 10.1186/s12971-014-0026-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi V, Matthews C, Aspiras M, de Jager M, Ward M, & Kumar P (2014). Smoking decreases structural and functional resilience in the subgingival ecosystem. J Clin Periodontol. doi: 10.1111/jcpe.12300 [DOI] [PubMed] [Google Scholar]
- Kamma JJ, Nakou M, & Baehni PC (1999). Clinical and microbiological characteristics of smokers with early onset periodontitis. J Periodontal Res, 34(1), 25–33. [DOI] [PubMed] [Google Scholar]
- Klein BA, Cornacchione LP, Collins M, Malamy MH, Duncan MJ, & Hu LT (2017). Using Tn-seq To Identify Pigmentation-Related Genes of Porphyromonas gingivalis: Characterization of the Role of a Putative Glycosyltransferase. J Bacteriol, 199(14). doi: 10.1128/JB.00832-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BA, Tenorio EL, Lazinski DW, Camilli A, Duncan MJ, & Hu LT (2012). Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics, 13, 578. 10.1186/1471-2164-13-578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koser SA, & Kasai GJ (1947). The effect of large amounts of nicotinic acid and nicotinamide on bacterial growth. J Bacteriol, 54(1), 20. [PubMed] [Google Scholar]
- Kumar PS (2012). Smoking and the subgingival ecosystem: a pathogen-enriched community. Future Microbiol, 7(8), 917–919. doi: 10.2217/fmb.12.71 [DOI] [PubMed] [Google Scholar]
- Lamont RJ, El-Sabaeny A, Park Y, Cook GS, Costerton JW, & Demuth DR (2002). Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology, 148(Pt 6), 1627–1636. [DOI] [PubMed] [Google Scholar]
- Lasica AM, Ksiazek M, Madej M, & Potempa J (2017). The Type IX Secretion System (T9SS): Highlights and Recent Insights into Its Structure and Function. Front Cell Infect Microbiol, 7, 215. doi: 10.3389/fcimb.2017.00215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MR, Preshaw PM, Nagaraja HN, Dabdoub SM, Rahman A, & Kumar PS (2014). The subgingival microbiome of clinically healthy current and never smokers. ISME J. doi: 10.1038/ismej.2014.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuric V, Gracieux P, Tamanai-Shacoori Z, Perez-Chaparro J, & Bonnaure-Mallet M (2008). Expression patterns of genes induced by oxidative stress in Porphyromonas gingivalis. Oral Microbiol Immunol, 23(4), 308–314. doi: 10.1111/j.1399-302X.2007.00429.x [DOI] [PubMed] [Google Scholar]
- Miller DP, Hutcherson JA, Wang Y, Nowakowska ZM, Potempa J, Yoder-Himes DR, … Lamont RJ (2017). Genes Contributing to Porphyromonas gingivalis Fitness in Abscess and Epithelial Cell Colonization Environments. Front Cell Infect Microbiol, 7, 378. doi: 10.3389/fcimb.2017.00378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MF (2003). Nicotinamide: an oral antimicrobial agent with activity against both Mycobacterium tuberculosis and human immunodeficiency virus. Clin Infect Dis, 36(4), 453–460. doi: 10.1086/367544 [DOI] [PubMed] [Google Scholar]
- Mydel P, Takahashi Y, Yumoto H, Sztukowska M, Kubica M, Gibson FC 3rd, … Potempa J (2006). Roles of the host oxidative immune response and bacterial antioxidant rubrerythrin during Porphyromonas gingivalis infection. PLoS Pathog, 2(7), e76. doi:05-PLPA-RA-0249R4 [pii] 10.1371/journal.ppat.0020076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D, Potempa J, Kordula T, & Travis J (1999). Purification and characterization of a novel cysteine proteinase (periodontain) from Porphyromonas gingivalis. Evidence for a role in the inactivation of human alpha1-proteinase inhibitor. J Biol Chem, 274(18), 12245–12251. [DOI] [PubMed] [Google Scholar]
- Nguyen KA, Travis J, & Potempa J (2007). Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of Gram-Negative bacteria? J Bacteriol, 189(3), 833–843. doi:JB.01530–06 [pii] 10.1128/JB.01530-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen KA, Zylicz J, Szczesny P, Sroka A, Hunter N, & Potempa J (2009). Verification of a topology model of PorT as an integral outer-membrane protein in Porphyromonas gingivalis. Microbiology, 155(Pt 2), 328–337. doi:155/2/328 [pii] 10.1099/mic.0.024323-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavia CS, Pierre A, & Nowakowski J (2000). Antimicrobial activity of nicotine against a spectrum of bacterial and fungal pathogens. J Med Microbiol, 49(7), 675–676. doi: 10.1099/0022-1317-49-7-675 [DOI] [PubMed] [Google Scholar]
- Ratnayake DB, Wai SN, Shi Y, Amako K, Nakayama H, & Nakayama K (2000). Ferritin from the obligate anaerobe Porphyromonas gingivalis: purification, gene cloning and mutant studies. Microbiology, 146 (Pt 5), 1119–1127. [DOI] [PubMed] [Google Scholar]
- Richarme G, Liu C, Mihoub M, Abdallah J, Leger T, Joly N, … Lamouri A (2017). Guanine glycation repair by DJ-1/Park7 and its bacterial homologs. Science, 357(6347), 208–211. doi: 10.1126/science.aag1095 [DOI] [PubMed] [Google Scholar]
- Romero-Lastra P, Sanchez MC, Ribeiro-Vidal H, Llama-Palacios A, Figuero E, Herrera D, & Sanz M (2017). Comparative gene expression analysis of Porphyromonas gingivalis ATCC 33277 in planktonic and biofilms states. PLoS One, 12(4), e0174669. doi: 10.1371/journal.pone.0174669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman S, Idrees F, Pervaiz S, Shah FH, Badshah S, Abdullah, … Idrees J (2016). Short Communication: Evaluation of antimicrobial activities of Harmine, Harmaline, Nicotine and their complexes. Pak J Pharm Sci, 29(4), 1317–1320. [PubMed] [Google Scholar]
- Sanders A, & Slade G (2013). State cigarette excise tax, secondhand smoke exposure, and periodontitis in US nonsmokers. Am J Public Health, 103(4), 740–746. doi: 10.2105/AJPH.2011.300579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacherl M, Montada AA, Brunstein E, & Baumann U (2015). The first crystal structure of the peptidase domain of the U32 peptidase family. Acta Crystallogr D Biol Crystallogr, 71(Pt 12), 2505–2512. doi: 10.1107/S1399004715019549 [DOI] [PubMed] [Google Scholar]
- Shibata T, Hishida T, Kubota Y, Han YW, Iwasaki H, & Shinagawa H (2005). Functional overlap between RecA and MgsA (RarA) in the rescue of stalled replication forks in Escherichia coli. Genes Cells, 10(3), 181–191. doi: 10.1111/j.1365-2443.2005.00831.x [DOI] [PubMed] [Google Scholar]
- Simionato MR, Tucker CM, Kuboniwa M, Lamont G, Demuth DR, Tribble GD, & Lamont RJ (2006). Porphyromonas gingivalis genes involved in community development with Streptococcus gordonii. Infect Immun, 74(11), 6419–6428. doi:IAI.00639–06 [pii] 10.1128/IAI.00639-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar SL, & Asma S (2000). Smoking-attributable periodontitis in the United States: findings from NHANES III. National Health and Nutrition Examination Survey. J Periodontol, 71(5), 743–751. [DOI] [PubMed] [Google Scholar]
- Wang R, & Li H (2012). The mysterious RAMP proteins and their roles in small RNA-based immunity. Protein Sci, 21(4), 463–470. doi: 10.1002/pro.2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SG, & Dessen A (2014). Structure of a bacterial alpha2-macroglobulin reveals mimicry of eukaryotic innate immunity. Nat Commun, 5, 4917. doi: 10.1038/ncomms5917 [DOI] [PubMed] [Google Scholar]
- Wright CJ, Xue P, Hirano T, Liu C, Whitmore SE, Hackett M, & Lamont RJ (2014). Characterization of a bacterial tyrosine kinase in Porphyromonas gingivalis involved in polymicrobial synergy. Microbiologyopen, 3(3), 383–394. doi: 10.1002/mbo3.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Mengin-Lecreulx D, Liu XW, Patin D, Farr CL, Grant JC, … Wilson IA (2015). Insights into Substrate Specificity of NlpC/P60 Cell Wall Hydrolases Containing Bacterial SH3 Domains. MBio, 6(5), e02327–02314. doi: 10.1128/mBio.02327-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Noiri Y, Yamaguchi M, Asahi Y, Maezono H, Kuboniwa M, … Ebisu S (2013). The sinR ortholog PGN_0088 encodes a transcriptional regulator that inhibits polysaccharide synthesis in Porphyromonas gingivalis ATCC 33277 biofilms. PLoS One, 8(2), e56017. doi: 10.1371/journal.pone.0056017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon JJ, Grossi SG, Machtei EE, Ho AW, Dunford R, & Genco RJ (1996). Cigarette smoking increases the risk for subgingival infection with periodontal pathogens. J Periodontol, 67(10 Suppl), 1050–1054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


