Abstract
Proper migration of neurons is one of the most important aspects of early brain development. After neuronal progenitors are born in their respective germinal niches, they must migrate to their final locations to form precise neural circuits. A majority of migrating neurons move by associating and disassociating with glial fibers, which serve as scaffolding for the developing brain. Cerebellar granule neurons provide a model system for examination of the mechanisms of neuronal migration in dissociated and slice culture systems; the ability to purify these cells allows migration assays to be paired with genetic, molecular, and biochemical findings. CGNs migrate in a highly polarized fashion along radial glial fibers, using a two-stroke nucleokinesis cycle. The PAR polarity complex of PARD3, PARD6, and an atypical protein kinase C (aPKC) regulate several aspects of neuronal migration. The PAR polarity complex regulates the coordinated movements of the centrosome and soma during nucleokinesis, and also the stability of the microtubule cytoskeleton during migration. PAR proteins coordinate actomyosin dynamics in the leading process of migrating neurons, which are required for migration. The PAR complex also controls the cell-cell adhesions made by migrating neurons along glial cells, and through this mechanism regulates germinal zone exit during prenatal brain development. These findings suggest that the PAR complex coordinates the movement of multiple cellular elements as neurons migrate and that further examination of PAR complex effectors will not only provide novel insights to address fundamental challenges to the field but also expand our understanding of how the PAR complex functions at the molecular level.
Keywords: Cerebellum, Neuronal migration, Centrosome, Nucleokinesis, PAR complex
1. Introduction
In the developing brain, immature neurons must migrate from the proliferative germinal zones to their final destinations (Hatten and Heintz 1995; Marín et al. 2010; Manzini and Walsh 2011; Métin et al. 2008; Millen and Gleeson 2008; Vallee et al. 2009). They are guided along their way by association and disassociation with glial fibers that act as neuronal migration tracts (Hatten 1990; O’Rourke et al. 1992). Neurons throughout the brain migrate by saltatory motion, in which the highly dynamic forward extension of the leading process is followed by somal translocation (Edmondson and Hatten 1987). This two-step motion is a highly orchestrated process involving coordination of the actin and microtubule cytoskeletons and associated motor proteins (Trivedi and Solecki 2011; Bellion et al. 2005; Kawauchi and Hoshino 2007; Valiente and Marín 2010). The correct orientation and migration of these cells is fundamental to the proper formation of neural circuits. Errors in neuronal migration and germinal zone exit are implicated in developmental and cognitive disorders such as lissencephaly, mental retardation, epilepsy, and pediatric cancers (Métin et al. 2008; Kato and Dobyns 2003; Ross and Walsh 2001). The molecular mechanisms of neuronal migration provide insight into the progression and treatment of these diseases.
The cerebellar granule neuron (CGN), the most common cerebellar neuron, has been used as a model for studies of polarity and migration. CGNs are born prenatally in the rhombic lip of the developing brain and form a secondary germinal zone in the external granule layer (EGL) of the developing cerebellum (Fig. 7.1) (Gregory et al. 1988; Rakic 1971; Ryder and Cepko 1994). In the EGL, immature proliferative granule neuron progenitors (GNPs) follow tangential migration paths parallel to the surface of the developing brain. CGNs begin to terminally differentiate between postnatal days 6–8 (P6–8). This process comprises germinal zone exit, axon extension, transition from tangential to radial migration (perpendicular to the cerebellar surface) along Bergmann glia, and arrival at their final positions in the internal granule layer (IGL) (Rakic 1972; Komuro and Rakic 1998). As radial migration continues through to P15, the EGL disappears as all CGNs have evacuated this transient germinal zone and have migrated into the IGL.
Fig. 7.1. Germinal zone migration in the developing cerebellum.
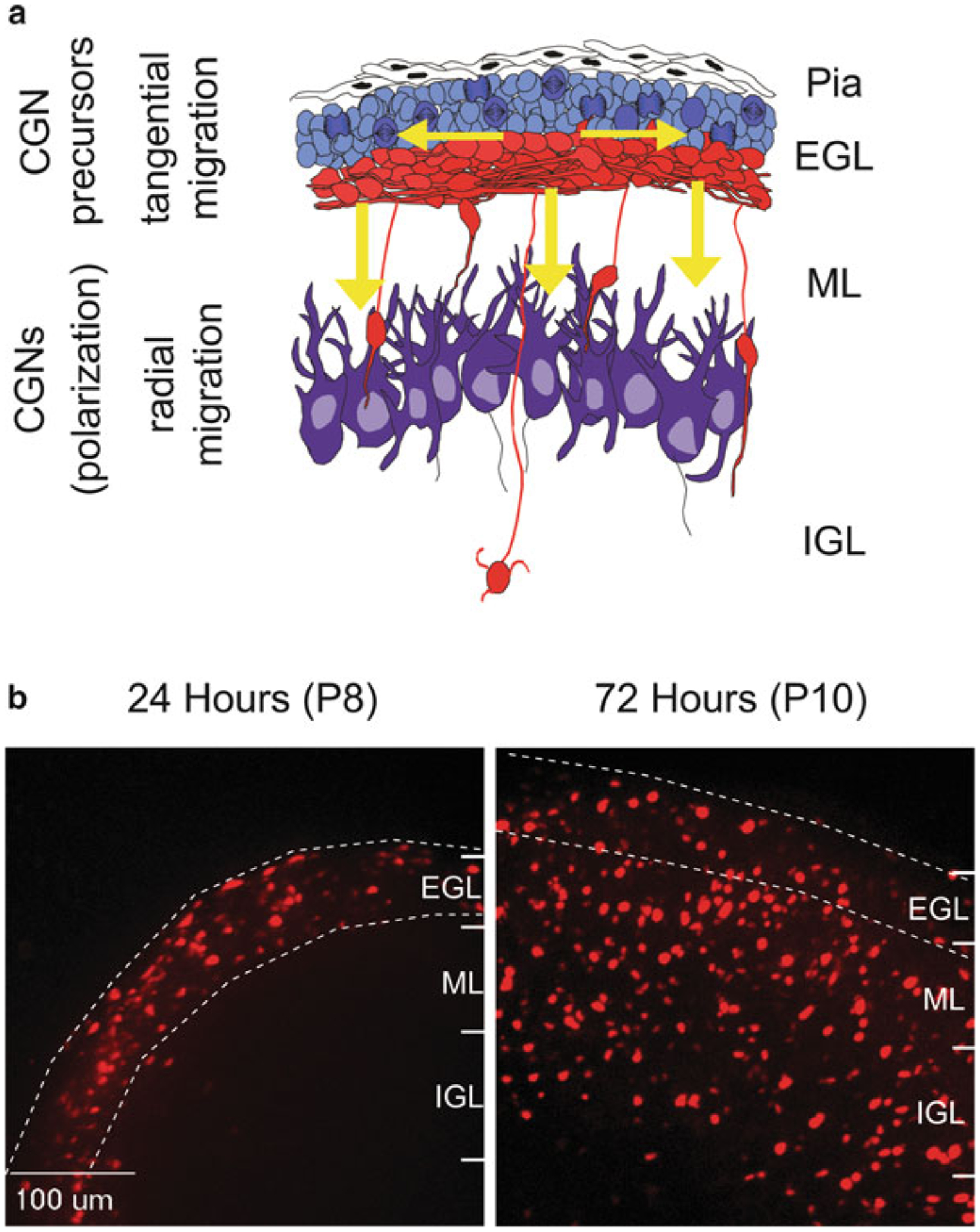
(a) Cerebellar granule neuron precursors (cGNPs) migrate tangentially (horizontal arrows) within the External Granule Layer (EGL). They then transition to a radial migration mode (vertical arrows) and migrate along glial fibers through the Molecular Layer (ML) and into the Internal Granule Layer (IGL). (b) Cereballarslice cultures electroporated with CGN-specific H2B-mCherry nuclei to track neuronal migration. At postnatal day 8 (P8, 24 h post electroporation), H2B-mCherry labeled CGNs migrate tangentially through the EGL. By P10 (72 h post electroporation) most CGNs have evacuated the EGL and migrated radially into the ML and IGL
Advances in microscopy have allowed ever more detailed views of the morphology of migrating neurons in both dissociated culture and slice imaging systems. CGNs have provided a prototypic model for examination of neuronal migration, progressing from studies of fixed cells to high temporal–resolution live imaging assays of migrating cells. First, electron microscopy of the developing cerebellum in Rhesus macaques showed the migration of individual CGNs perpendicular to the surface of the brain along radial fibers later identified as Bergmann glia, with leading and trailing processes extending from their elongated soma (Rakic 1971, 1972). At the junctions of migrating CGNs, electron microscopy identified interstitial densities, or regions of the cell in which submembranous cytoskeletal elements attach to microtubules, thereby anchoring the cytoskeleton to a point at which forward force can be generated from cell-cell contacts (Gregory et al. 1988). Subsequent time-lapse imaging revealed that CGNs are highly polarized, having dynamic leading and trailing processes, while the nucleus occupies most of the somal volume. This polarity facilitates nuclear movement as a crucial aspect of saltatory CGN migration (Edmondson and Hatten 1987; Rivas and Hatten 1995; Solecki et al. 2004).
As with all neurons, the dynamic leading processes of CGNs are guided by extracellular cues but their movements are not synchronized with those of the neuronal soma (Edmondson and Hatten 1987). Polarized somal and organelle movement during CGN migration provided a foundation for understanding the basis of the saltatory movement cycle, in which the soma moves at an average rate of 33 ± 20 μm/h (Edmondson and Hatten 1987). Interestingly, forward movement of vesicles precedes somal movement, implying that specializations in cellular structures occur prior to somal movement. This concept was expanded with the observation that the centrosome enters the leading process prior to somal translocation, in what is termed the two-stroke motility cycle (Fig. 7.2) (Solecki et al. 2009). Strikingly, the saltatory timing first observed in early differential interference contrast (DIC) microscopy studies matches the two-stroke motility cycle of centrosome and soma (Solecki et al. 2009). This original observation of the mechanisms of CGN migration was expanded to apply to several other neuronal subtypes (Bellion et al. 2005; Schaar and McConnell 2005; Tsai et al. 2007; Sakakibara et al. 2013; Yanagida et al. 2012; Yang et al. 2012; Shinohara et al. 2012). Recently it has been shown a cytoplasmic dilation develops within the leading processes of subventricular-zone neurons before nuclear translocation, similar to the morphologic change seen during actin and microtubule enrichment of the leading processes of CGNs (Schaar and McConnell 2005; Rivas and Hatten 1995). Thus in vitro studies of CGN morphology provide a cellular context for understanding the large-scale migration patterns within the developing brain.
Fig. 7.2. The two-stroke nucleokinesis cycle of migrating neurons.
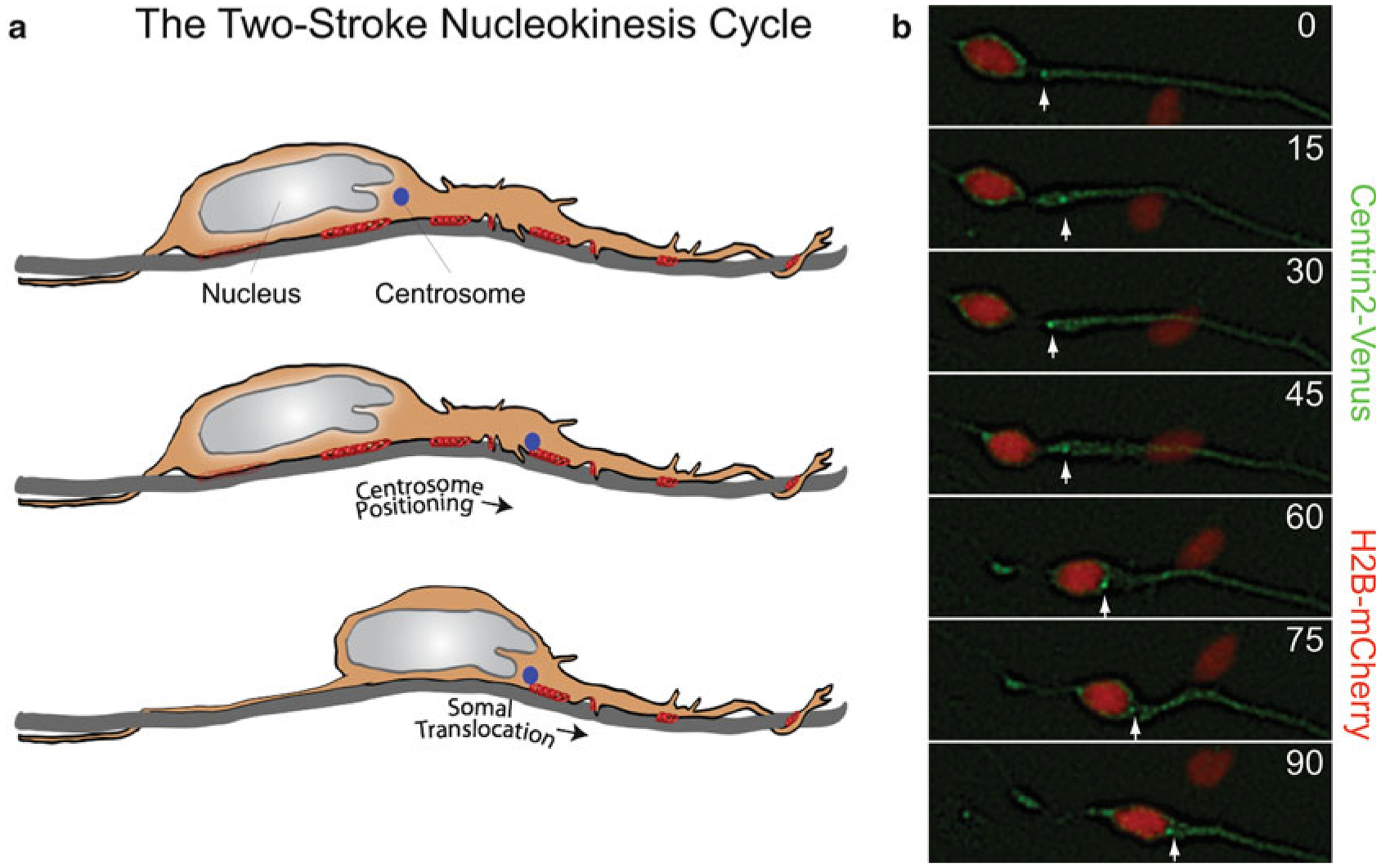
(a) In the two stroke nucleokinesis cycle, the centrosome is positioned into the neuronal leading process before somal translocation. (b) Time-lapse imaging of a migrating CGN whose centrosome is labeled with Centrin2-Venus (green) (white arrow) and whose nucleus is labeled with H2B-mCherry (red). Centrosome positioning occurs 0 and 30 min, and somal translocation occurs between 45 and 90 min
Advances in ex vivo imaging have shown complex alterations in the morphology of migrating CGNs as they transit different environments and interact with different cell types. The shapes of radially migrating CGNs change as they pass through different layers of the developing cerebellum. The growth cone of the leading tip of migrating neurons has dynamic filopodia and lamellipodia, which are dynamic extensions that form and retract as the neuron samples its environment and moves forward (Gregory et al. 1988). In the molecular layer of the cerebellum, CGNs assume an extended shape as they move rapidly along Bergmann glia, while they assume a more rounded shape as they transiently and slowly migrate through the Purkinje cell layer. As CGNs enter the IGL, the cell body again assumes an extended shape for rapid movement independent of Bergmann glia (Komuro and Rakic 1998). Observation of tangentially migrating CGNs shows that in the EGL their velocity is dependent on their position. Their most rapid rate of tangential migration occurs in the center of the EGL, where they maintain short leading and trailing processes. As they move to the bottom of the EGL, their tangential migration velocity slows and they extend longer leading and trailing processes. CGNs slowly migrate out of the EGL upon reaching its interface with the molecular layer and begin radial migration into the molecular layer (Komuro et al. 2001). Because the multiple modes of CGN migration involve region-specific rates and morphologies, the motor systems and cytoskeletal regulation mechanisms that regulate these different types of migration are of great interest.
The cytoskeletons of migrating neurons are dynamic, changing within the different migration environments. The leading processes of migrating neurons are enriched in microtubules and actin, which extend toward a tubulin cage surrounding the nucleus (Rivas and Hatten 1995). Regulation of the microtubule cytoskeleton is a driving factor in neuronal migration. The microtubule array of migrating neurons is highly polarized, as growing microtubule “plus” tips extend into the leading process and depolymerizing “minus” ends are oriented toward the nucleus (Rakic et al. 1996). The genetics of human neuronal migration disorders further highlight that microtubule cytoskeleton and its associated motor protein dynein are regulators of neuronal migration. The cytoplasmic dynein motor protein is a polypeptide of 12 subunits, comprising two identical heavy chains that contain the AAA ATPase domains required for activity, two intermediate chains involved in cargo anchoring, and additional intermediate and light chains whose functions remain unclear (Cho and Vale 2012; Dujardin and Vallee 2002; Feng et al. 2000). Genetic analysis of lissencephaly identified mutations in the dynein adaptor protein Lissencephaly 1 (LIS1) (Reiner et al. 1993; Dujardin et al. 2003; Faulkner et al. 2000; Hirotsune et al. 1998; Smith et al. 2000). Lissencephaly also results from mutations in the Doublecortin (DCX) gene, which encodes a microtubule bundling protein and is expressed in migrating neurons (Kato and Dobyns 2003; Francis et al. 1999; Gleeson et al. 1999; Allen et al. 1998). Both LIS1 and dynein play roles in radial neuronal migration (Tsai et al. 2007; Tanaka et al. 2004; Smith et al. 2000; Shu et al. 2004) (see also Chap. 1). As more genes that participate in the regulation of neuronal migration and brain development are identified, additional genetic causes of cognitive and developmental brain disorders will be recognized.
Active migration of CGNs requires coordination between the microtubule and actin cytoskeletons and their associated motor proteins (Ridley et al. 2003). The leading processes of migrating neurons are enriched in actin, and disruption of the actin cytoskeleton with cytochalasin B is sufficient to inhibit migration, implicating actin subunit assembly in migration (Rivas and Hatten 1995; Le Clainche and Carlier 2008). Actin-based motility is dependent on the myosin family of motor proteins. Myosin II contains two heavy chains that constitute the head and tail domains of the protein and four light chains that bind to the heavy chains (Vallee et al. 2009). Phosphorylation of myosin II by myosin light chain kinase or myosin heavy chain kinase is required for ATP hydrolysis, which drives motor function (Kamm and Stull 2001; Moussavi et al. 1993). Nonmuscle myosin IIb, the main myosin expressed in the developing brain, was identified as important to neuronal migration when mutation in the motor domain of nonmuscle myosin heavy chain IIb was observed to disrupt CGN migration and cerebellar foliation (Ma et al. 2004; Vicente-Manzanares et al. 2009; Rochlin et al. 1995). Actomyosin enrichment of the leading process suggests this compartment may be the main site for actin cytoskeletal dynamics in migrating neurons (Rivas and Hatten 1995; Le Clainche and Carlier 2008).
Migrating CGNs encounter multiple microenvironments and make several types of cell-cell contact as they migrate from the EGL to the IGL (Komuro and Rakic 1998). In CGNs migrating along glial fibers, the dynamic leading process is observed to wrap around Bergmann glia, and junctional adhesion molecule (JAM)-mediated adhesions are shown to form at cell-cell contacts (Famulski et al. 2010). As the neurons encounter different cell types, their modes of migration and their adhesions change accordingly (Hatten 1990; Fishman and Hatten 1993). Astrotactin provides a receptor system for CGN migration along astroglia, and the integrin β1 receptor promotes migration along laminin fibers (Edmondson et al. 1988; Fishell and Hatten 1991; Fishman and Hatten 1993). Increased astrotactin expression, identified as a general feature of migratory cells, is noted in migratory CGNs in the EGL of the cerebellum (Zheng et al. 1996). In vitro assays of CGN migration along glial membrane- and laminincoated fibers mirrored the saltatory nucleokinesis cycle observed in slice migration assays; however, brief, limited migration was observed on collagen and fibronectin fibers (Fishman and Hatten 1993). Individual cell surface receptors have been identified by in vitro migration assays as a requirement for neuronal migration, but it is unclear which combination of receptors is used and how they are anchored to the cytoskeleton in the different migration modes in the developing cerebellum.
Cell biology and genetic studies have created a basic framework to explore how neurons migrate from a GZ to their final laminar positions. However, several challenges remain : (1) Current migration models show inconsistencies, how will these differences be resolved? (2) How will the ever increasing array of cytoskeletal regulators be woven into an integrated model of neuronal migration? (3) What mechanisms control migration initiation and migration mode during GZ exit?
As all migrating cells are polarized (i.e., have spatially defined cytoskeletal organizations that are globally coordinated to execute complex motility programs), we will address these three major challenges by examining how polarity signaling globally organizes the neuronal cytoskeleton rather than by the reductionist approach of studying single cytoskeletal components in isolation. The best characterized cell polarity signaling molecules are the evolutionarily conserved partitioning defective (PAR) proteins (Kemphues et al. 1988). The PARD3 and PARD6 adaptor proteins form a complex containing atypical PKC and the CDC42 or Rac1 Rho GTPases (Joberty et al. 2000; Lin et al. 2000). This ternary complex is critical for tight junction formation, mitotic spindle orientation, cell migration and axon specification (Munro 2006; Barnes and Polleux 2009; Nance and Zallen 2011). This chapter will discuss the multiple roles of these proteins in neuronal migration through (1) organelle structure and movement, (2) coordination of cytoskeletal dynamics and associated motors, and (3) interaction with cell-cell focal adhesions. As studies of the PARD3/PARD6/aPKC complex (the PAR complex) progress, the individual roles of the PAR proteins in the centrosome, nucleus, actomyosin cytoskeleton, and focal adhesions are becoming clearer. PAR signaling has been shown to control the two-stroke nucleokinesis cycle of centrosome motion followed by somal translocation (Solecki et al. 2004). During the nucleokinesis cycle, the PAR complex has been shown to regulate myosin II activation and the actin cytoskeleton (Solecki et al. 2009). The role of PAR proteins and their regulation of focal adhesion turnover through the seven in absentia homolog (SIAH) E3 ubiquitin ligase (Famulski et al. 2010) has introduced PAR signaling as being regulated by protein degradation. The question of how PAR controls focal adhesions leads us to investigate how polarity complexes are related to the neuronal cytoskeleton and how these two dynamic structures control cell adhesion and migration.
Taken together, the available evidence indicates that the dynamic PAR complex plays key roles in nucleokinesis and adhesion control. We will now discuss in detail the role of PAR protein in each of these processes in the following sections of the chapter.
2. The PAR Polarity Complex and Microtubule Cytoskeletal Regulation
2.1. Cerebellar Granule Neurons Migrate with Coordinated Organelle Movements
Neurons migrate via a coordinated two-stroke motion of the centrosome and nucleus. Time-lapse imaging of actively migrating CGNs shows that in the majority of migrating neurons, forward movement of the centrosome is followed by somal translocation (Solecki et al. 2004). As centrosome movement precedes nuclear movement, models have been proposed in which the centrosome acts as a microtubule organizing center, projecting microtubules rearward to the perinuclear tubulin cage and “pulling” the nucleus forward through a dynein-mediated process. A competing model shows microtubules from the nuclear tubulin cage extending past the centrosome and anchoring in the membrane of the leading process (Higginbotham and Gleeson 2007; Tsai and Gleeson 2005). The relationship between centrosome positioning and nuclear translocation may differ among migration modes, as in vitro migration assays of CGNs identify a subset of neurons in which the nucleus overtakes the centrosome during active migration (Umeshima et al. 2007). Electron microscopy of CGNs has shown that microtubules extend from the nuclear cage forward to both the centrosome and the leading process membrane, although the anchor point for the microtubule cytoskeleton in the leading process remains unclear. As described in the next section, the PARD6 component of the PAR complex plays an important role in not only regulating the structure the tubulin cage but also the saltatory cadence of centrosome and somal motility.
2.2. PARD6 Signaling Controls Centrosome Positioning and Microtubule Dynamics
Using high temporal–resolution live imaging techniques, Solecki and colleagues (2004) demonstrated that the forward movements of the centrosome and the nucleus are tightly coordinated in migrating neurons. Photobleaching experiments showed the microtubule cytoskeleton to be highly dynamic. Overexpression of PARD6α in granule neurons inhibits neurite extension and disintegration of the perinuclear tubulin cage, showing that PARD6α controls the microtubule dynamics of migrating neurons (Solecki et al. 2004). Disruption of PARD6α signaling also uncoupled the movements of the centrosome and nucleus and prevented migration of granule neurons along Bergmann glial fibers. By using Venus-labeled PARD6α, Solecki et al. (2004) showed PARD6α to colocalize with γ-tubulin and therefore to be a component of the centrosome. PARD6α shows a relationship to centrosome structure, as overexpression of PARD6α reduced levels of centrosomal γ-tubulin (Solecki et al. 2004).
The mechanism by which PARD6 mechanistically controls centrosome positioning and migration has recently been clarified in non-neuronal systems such as epithelial cells. PARD6α siRNA disrupts the microtubule cytoskeleton in epithelial cells (Kodani et al. 2010). PARD6 is also a controlling element of the mitotic spindle, as RNAi of either PARD6α or PARD6γ causes multipolar spindle formation and mitotic failure in epithelial cells (Kodani et al. 2010; Dormoy et al. 2013). In epithelial cell wound healing assays, PARD6γ RNAi-depleted cells were unable to migrate (Dormoy et al. 2013). As overexpression of PARD6α uncouples centrosomal and nuclear movement and disrupts migration, it remains unclear whether the centrosome was acting as an organizer of polarity and migration or a reporter of cellular mechanisms that control migration in these studies.
The PAR polarity complex may play both structural and signaling roles at the centrosome in migrating neurons. In epithelial cells, PARD6α interacts with the centriolar components PCM-1 and dynactin subunit p150Glued, as shown through colocalization and immunoprecipitation studies (Kodani et al. 2010). The recruitment of PARD6α to the centrosome requires intact microtubules and dynein, as PARD6α was dispersed from the centrosome when microtubules were destabilized by nocodazole treatment and when dynein function was inhibited by overexpression of dynactin subunit p150Glued (Kodani et al. 2010; Young et al. 2000). Depletion of PARD6α by RNAi reduced microtubule-dependent recruitment of the centrosome proteins pericentrin, PCM-1, centrin, ninein, Cep170, and CPAP, showing that PARD6α promotes centrosome protein recruitment (Kodani et al. 2010). PARD6γ has been found to be a component of the mother centriole (Dormoy et al. 2013) and is required for recruitment of centrosome proteins such as PARD6α and p150Glued. Interestingly, recruitment of PARD6γ to the mother centriole is microtubule-independent and requires the C-terminus of the PARD6γ protein (Dormoy et al. 2013). These findings show that PARD6 plays both structural and recruitment roles in the centrosome. It is currently unclear where PARD6 lies in the hierarchy of centrosome protein assembly.
2.3. PARD3 Regulates Centrosome Protein Recruitment and Orientation
PARD3 plays roles other than those of PARD6 in regulating centrosomal dynamics. PARD3 associates with dynein, as shown by co-immunoprecipitation of PARD3 with dynein light intermediate chain 2 (Schmoranzer et al. 2009). Dynein is required for assembly of γ-tubulin on centrosomes (Young et al. 2000), supporting the role of the PAR complex in proper centrosome assembly (Fig. 7.3). Removal of PARD3 prevents correct centrosome positioning in relation to the nucleus in epithelial cells (Schmoranzer et al. 2009). Like PARD6, PARD3 plays roles in both directed migration and organelle positioning, and PARD3 RNAi depletion inhibits migration of epithelial cells in wound healing assays (Schmoranzer et al. 2009). Depletion of PARD3 results in increased microtubule dynamics at cell-cell contacts, showing that PARD3 plays a role in microtubule stability in epithelial cells (Schmoranzer et al. 2009). Additional studies have shown PARD3 to stabilize and bundle microtubules both in vitro and in hippocampal neurons (Chen et al. 2013). The role of the PAR complex at the centrosome, as observed in several migrating cell types, and its link to dynein, add to our understanding of regulation of the cytoskeleton and neuronal migration by the PAR polarity complex.
Fig. 7.3. PAR proteins and cytoplasmic Dynein directed minus-end transport.
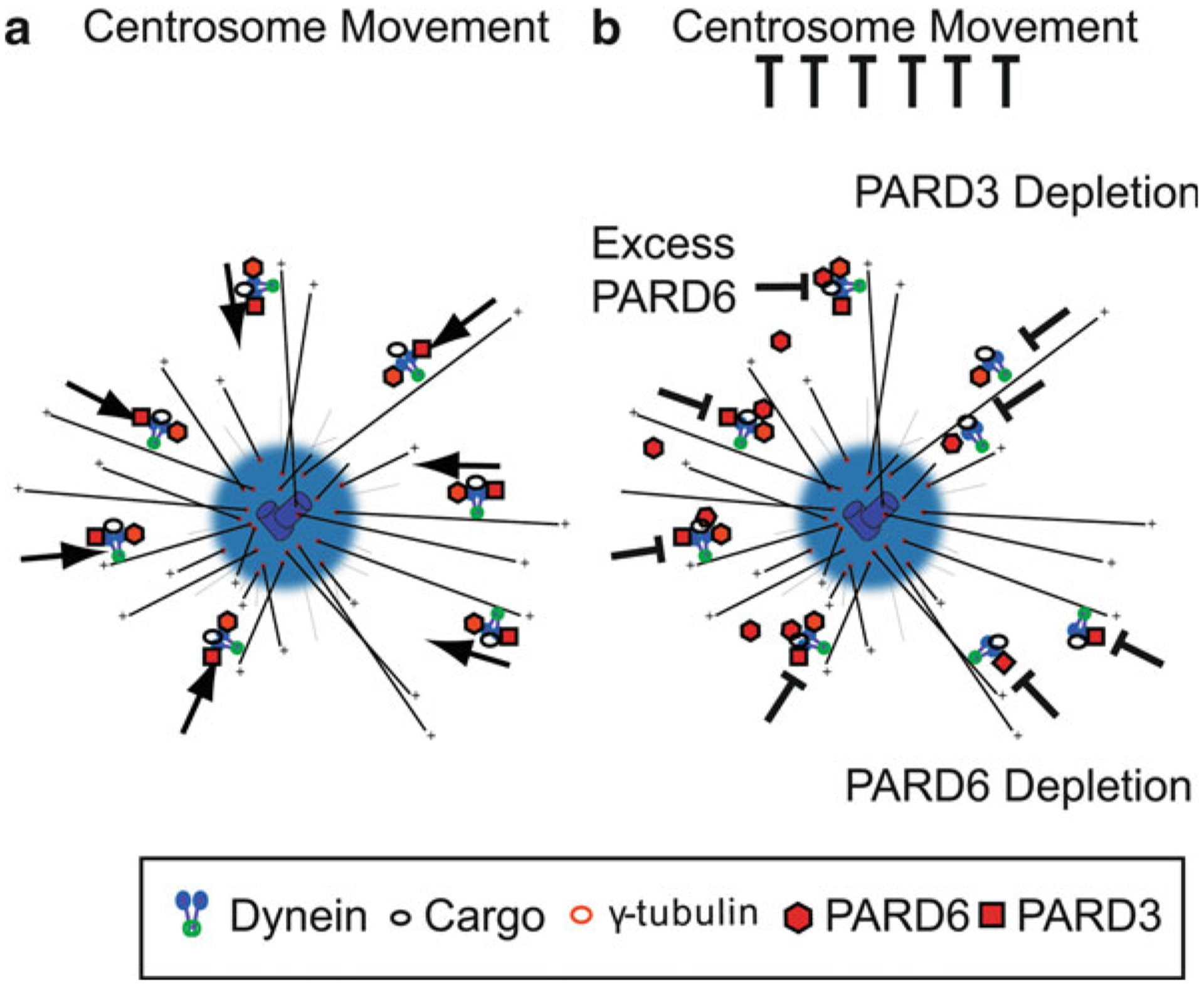
(a) Model of Dynein-directed minus-end transport of centrosome components mediated by PAR. This mechanism is responsible for proper centrosome motility. (b) Disruption of PAR protein components may result in inhibition of dynein mediated centrosome assembly and centrosome motility by PARD6 overexpression (Solecki et al. 2004), PARD6 RNAi (Kodani et al. 2010), and PARD3 depletion (Schmoranzer et al. 2009)
The PAR polarity complex and its individual components have been shown to play roles in centrosome structure, protein recruitment, and motility in addition to regulating the dynamics of the microtubule cytoskeleton. After examining the role of the PAR polarity complex in controlling migration through microtubule-based mechanisms, we will explore the role of this complex in regulating the actin cytoskeleton.
3. PAR Complex Regulation of Myosin II Motors
3.1. PARD6α Regulates Myosin II Dynamics in Migrating CGNs
Identification of PAR polarity proteins as regulators of the microtubule cytoskeleton during neuronal migration led to examination of other cytoskeletal elements in migrating neurons. While many migration studies focused on force generation by microtubule-dynein systems, Solecki and colleagues (2009) examined the role of leading-process actomyosin in migrating CGNs. Two opposing models have been proposed for the mechanism by which actomyosin contributes to force generation in neuronal migration: (1) a dynamic forward reach-and-pull model, in which leading-process actomyosin contraction pulls the neuron forward and (2) a rearward contraction model, in which actomyosin contraction at the rear of the cell pushes the migrating neuron forward (Fig. 7.4) (Trivedi and Solecki 2011; Martini and Valdeolmillos 2010; Tsai et al. 2007). Time-lapse microscopy and photobleaching/photoactivation experiments show that leading-process actin is highly dynamic in migrating neurons, and pharmacological stabilization of the actin cytoskeleton or inhibition of the myosin II motor reduces leading-process dynamics, disrupts the two-stroke nucleokinesis cycle, and halts migration of CGNs (Solecki et al. 2009). The centrosome is central to the nucleokinesis cycle, and both actin and myosin light-chain kinase were found to accumulate at the centrosome in the leading edge of migrating neurons. The importance of myosin II to neuronal migration was shown by pharmacological inhibition of the myosin II motor with blebbistatin, which halted centrosome motion and the two-stroke nucleokinesis cycle (Solecki et al. 2009). The PAR complex is a key regulator of actomyosin dynamics in the leading process. In previous studies, overexpression of PARD6α was shown to inhibit neuronal migration (Solecki et al. 2004). The same group (Solecki et al. 2009) later reported that reduced myosin II activation in cells overexpressing PARD6α was one mechanism of migration inhibition. Overexpression of PARD6α or the truncated IQ motif of PARD6α significantly reduced leading-process actin turnover in migrating CGNs (Solecki et al. 2009), showing for the first time that the PAR complex can control myosin II through direct interaction (Fig. 7.5). Co-immunoprecipitation studies revealed that full-length PARD6α binds to myosin light chain and myosin light chain kinase and that overexpression of the PARD6α IQ domain inhibits myosin light chain binding to PARD6α (Solecki et al. 2009). In other studies in C. elegans embryos, cortical flow of actin and non-muscle myosin II transported the PARD3/PARD6/aPKC complex to the anterior of the cell, maintaining polarity (Munro et al. 2004). Myosin IIb-deficient fibroblasts show polarity defects and increased levels of cytosolic PARD3 and PARD6 (Solinet et al. 2011). The mechanism of this relationship remains unclear, although it is possible that myosin II controls proper localization and stabilization of the PAR polarity complex. These results show that the PAR polarity complex regulates actomyosin contractility in the leading process of migrating neurons via PARD6α.
Fig. 7.4. Actomyosin pulling models for Glial-guided neuronal migration.
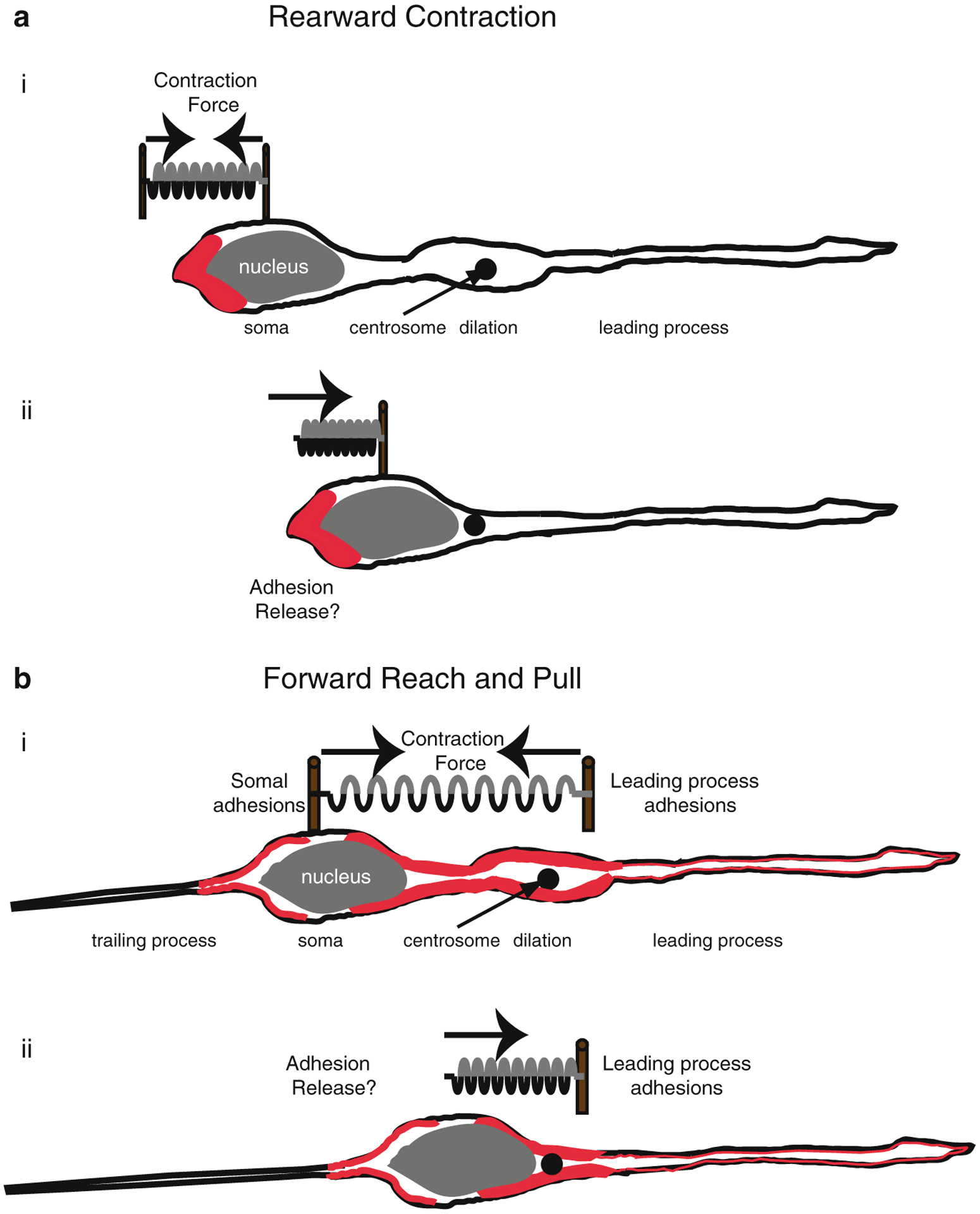
(a) Rearward Contraction model. (i) Prior to somal movement, actomyosin (red) is heavily enriched at the cell rear. (ii) During somal movement myosin II squeezing at the rear is thought to “push” the cell body forward. (b) Reach and Pull model. (i) Prior to somal movement, actomyosin (red) is heavily enriched in the leading process from the cytoplasmic dilation to the neuronal soma. Given a muscle-like contraction of the F-actin array by myosin II, a taut spring effectively describes the forces produced when leading process and somal actomyosin anchoring (i.e., adhesions) are balanced before somal movement: one force vector points from the leading process back towards the soma whereas another force vector points from the soma towards the dilation (the future direction of somal movement). (ii) Once somal adhesions release, as described in (Gregory et al. 1988), actomyosin tension generated in the leading process primes somal movement towards the cytoplasmic dilation (Reproduced with permission of (Trivedi and Solecki 2011))
Fig. 7.5. Model of Par6α interaction with the Myosin II motor complex and the Myosin cycle.
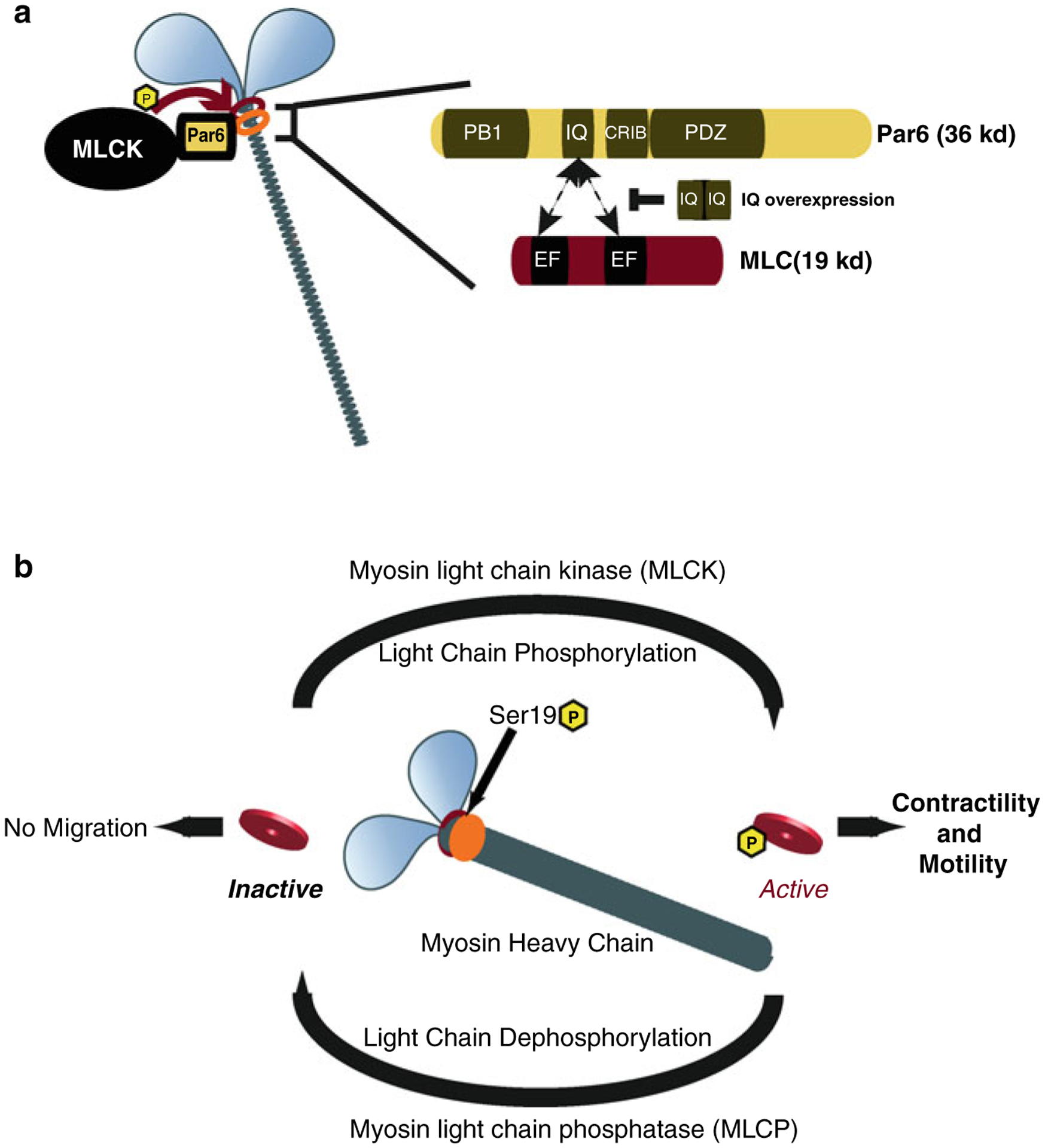
(a) Par6α binds to both MLC and MLCK, key signaling nodes regulating actomyosin contractility. Inset : The PARD6-MLC interaction may be mediated by the IQ domain of Par6α (IQ Motif (aa 104–120) = AFASNSLQRRKKGLLLRPV) and the EF hand domains of MLC. (b) Myosin contractility is dependent on Myosin Light Chain (MLC) phosphorylation by Myosin Light Chain Kinase (MLCK) at Ser19 and is required for neuronal migration. De-phosphorylation of MLC by Myosin Light Chain Phosphatase (MLCP) results in MLC inactivity and lack of myosin contractility. MCL and MLCP activity cycles Myosin contractility in migratory cells ((a) Reproduced with permission of (Solecki et al. 2009))
3.2. Actomyosin Dynamics in Migrating Neurons
Further studies examining the dynamics of actin cytoskeletal elements in CGNs buttress the importance of leading-process actin. The forward flow of actin in the leading process plays several roles important to migration. He and coworkers (2010) examined the role of cytoskeletal components and motors in vitro in distinct regions of migrating rat CGNs. In their microdissection experiments, severing the distal leading tip of migrating neurons was sufficient to inhibit somal translocation, while a dynamic leading tip contributed to somal translocation by a distance of several cell-body lengths (He et al. 2010). By micropipetting actin-destabilizing drugs (cytochalasin D, latrunculin A) or actin-stabilizing drugs (jasplakinolide) into the vicinity of the leading processes of migrating neurons, they also showed that leading-process actin dynamics are required for somal translocation. Pharmacological inhibition of leading-process actin dynamics halted somal translocation; however, when the inhibitor was concentrated in the cell body area, it did not similarly inhibit somal translocation (He et al. 2010). The same group (He et al. 2010) also compared the roles of the microtubule cytoskeleton and of actin in the leading tip and found that the microtubule-destabilizing drug nocodazole did not halt somal translocation, but rather enhanced the rate of nuclear migration. Directed inhibition of myosin II by applying blebbistatin to the leading tip of migrating CGNs halted somal translocation, while blebbistatin treatment at the rear of the cell increased the rate of nuclear migration, demonstrating the importance of the myosin II motor (He et al. 2010). These findings showed that polarized activity of myosin II plays an important role in neuronal migration.
Wang and coworkers (2012) expanded on the role of leading-process actomyosin and its importance in active migration of neurons. They used antibody-coated quantum dots to track the movement of the membrane proteins VAMP2 and endogenous neurotrophin receptor TrkB in actively migrating mouse CGNs and showed that both proteins are non-randomly translocated (in a form of biased drift) in a myosin II–dependent manner toward the leading process. This non-random translocation was not identified in non-migratory cells, leading the authors to hypothesize that the biased forward drift of receptors may be involved in the guidance of migrating neurons. Taken together, these data support the forward flow of the F-actin cytoskeleton in the leading process of migrating neurons observed in our laboratory (Gupton and Waterman-Storer 2006; Vicente-Manzanares et al. 2007; Solecki et al. 2009) and highlight that forward flow is not just important for centrosome positioning but also regulates positioning of receptors within the leading process.
3.3. Potential Role of Actomyosin in Generating Leading Process Traction Forces
The organization of motor proteins and their function in migrating cells is highly regulated during migration. The role of myosin II in both the two-stroke nucleokinesis cycle (Solecki et al. 2009) and biased drift of surface receptors (Wang et al. 2012) provides insight as to how the regulation of actomyosin in the leading process controls migration. It is possible that the actin mechanisms involved in receptor transport in migrating neurons also play a role in the formation and maturation of cell-surface adhesion dynamics. Studies of myosin II in migrating epithelial cells provide an example of the possible roles of these motor proteins in migrating neurons. Active migration of epithelial cells requires the coordination of actin, myosin II, and focal adhesions (Gupton and Waterman-Storer 2006). Gupton and Waterman-Storer examined the migration of Ptk1 cells on various concentrations of extracellular matrix (ECM) and observed the effects of these concentrations on the actin cytoskeleton and cellular adhesion. Migration conditions were optimal at intermediate ECM concentrations, indicating that too much or too little adhesion limits cell migration. Higher ECM concentrations were associated with more pronounced focal adhesion density, yet migration was halted due to lack of focal adhesion turnover. Myosin II activity was highest at intermediate ECM concentrations, showing that migration is optimal when there is high myosin II activity, which is associated with efficient turnover and maturation of focal adhesions; these findings highlight the importance of actomyosin in active cell migration at the level of adhesion. The relation of myosin II activity to focal adhesion stability and maturation of adhesions illustrates how myosin II may control leading process traction. If the leading process of migrating neurons is analogous to the myosin II enriched lamellum of migrating fibroblasts, then myosin II motor activity may fine tune leading process adhesion to neuronal migration substrates. Current studies are further investigating the role of the PAR complex in the balance of actomyosin dynamics in the leading process of migrating neurons.
As actomyosin has a demonstrated role in the leading process of migrating neurons and in nucleokinesis, its interaction with PARD6α provides a mechanism linking polarity complexes with cytoskeletal motor systems in migrating neurons. The role of PARD6α in regulating microtubule dynamics in neurons also provides insight into the interaction of the actin and microtubule cytoskeletal systems in neuronal migration. A molecular clutch model has been proposed to allow transmission of polymerization-driven flow of myosin into traction (Mitchison and Kirschner 1988; Gardel et al. 2010). It is possible that a function of the PAR complex in the leading process of migrating CGNs is as a clutch between the myosin and microtubule networks to generate forward force on the cell and/or individual organelles. As both myosin II and the PAR complex have been shown to play an integral role in neuronal migration, the relationship between polarity, motor proteins, and cellular adhesions must be considered to further complete our understanding. We will next discuss the relation between PAR proteins and adhesion molecules.
4. The PAR Complex and Adhesion Mechanisms
4.1. CGN Migration Varies in Subsections of the Cerebellum
Neural progenitors proliferate in germinal zones of the brain and must then migrate to their final locations to establish proper neural circuits. A key factor in controlling germinal zone exit is the regulation of cell-cell contacts, especially those that occur as migrating neurons encounter different cell types within distinct regions of the developing cerebellum. During germinal zone exit, CGNs must migrate tangentially among other CGNs in the upper and middle layers of the external granule layer before they transition to the inner layer of the EGL (Komuro et al. 2001). The migration rates differ in these distinct regions of the EGL, suggesting that the motor and adhesion systems of migration may differ as well. Exiting the EGL and moving into the molecular layer, CGNs migrate along glial fibers from the EGL of the cerebellum to their final location in the IGL.
4.2. Antagonistic Interaction of PARD3 and SIAH
The mechanisms controlling the germinal-zone exit of migratory neuronal precursors have revealed novel insights into when and how CGNs initiate the first step in their journey to the IGL. Previously, two competing models were used to explain germinal zone exit: in one model it was thought that a new form of cell-cell adhesion was initiated upon movement of CGNs from the EGL into the molecular layer. The other model proposed that removal of a form of cell-cell adhesion maintained in the EGL allowed maturing CGNs to exit the germinal zone (Métin and Luccardini 2010). Examination of PARD3 function in germinal zone exit suggests that the first model may be active and regulated by polarity signaling cascades.
Famulski et al. identified PARD3 as a novel regulator of cell adhesion through interaction with the E3 ubiquitin ligase SIAH (Famulski et al. 2010). SIAH was identified from a yeast two-hybrid screen of PAR complex interaction partners. Evidence of physical interaction between these proteins led researchers to examine the amino acid sequence of PARD3. Examination of PARD3 motifs identified two SIAH degron recognition sequences as potential regulation sites of germinal zone exit. SIAH and PARD3 interact directly through the SIAH substrate-binding domain targeting the two SIAH-degron recognition sequences of PARD3 in an interaction that requires the catalytic SINA substrate binding domain of SIAH. Ubiquitination of PARD3 by SIAH results in PARD3 degradation by the proteasome, revealing an antagonistic interaction between PARD3 and SIAH (Famulski et al. 2010).
Expression analysis revealed a reciprocal expression profile of SIAH and PARD3. SIAH showed high expression in CGN progenitors, which was extinguished in differentiated CGNs in the developing cerebellum. In contrast, PARD3 was found to be expressed at low levels in the EGL and elevated levels in differentiating CGNs. Systematic necessity/sufficiency testing was then used to test whether these reciprocal expression profiles were functionally relevant to germinal zone exit. Ectopic expression of PARD3 in CGN precursors in the EGL, which normally express low levels of this polarity protein, was sufficient to induce precocious germinal zone exit, while gene silencing of PARD3 blocked migration, showing that its activity was necessary for immature CGNs to exit the EGL and migrate to their final destination (Fig. 7.6) (Famulski et al. 2010). In contrast, ectopic expression of SIAH inhibited germinal zone exit of CGNs and maintained tangential migration paths. Interestingly, co-expression of PARD3 with SIAH was sufficient to restore directed migration and germinal zone exit. Finally, SIAH silencing induced precocious germinal zone exit to a degree similar to that of PARD3 gain of function (Famulski et al. 2010). These results identified PARD3 and SIAH as novel regulators of the CGN migratory path and germinal zone exit through posttranslational modification of PARD3.
Fig. 7.6. Model of SIAH E3 Ligase regulation of germinal zone exit.
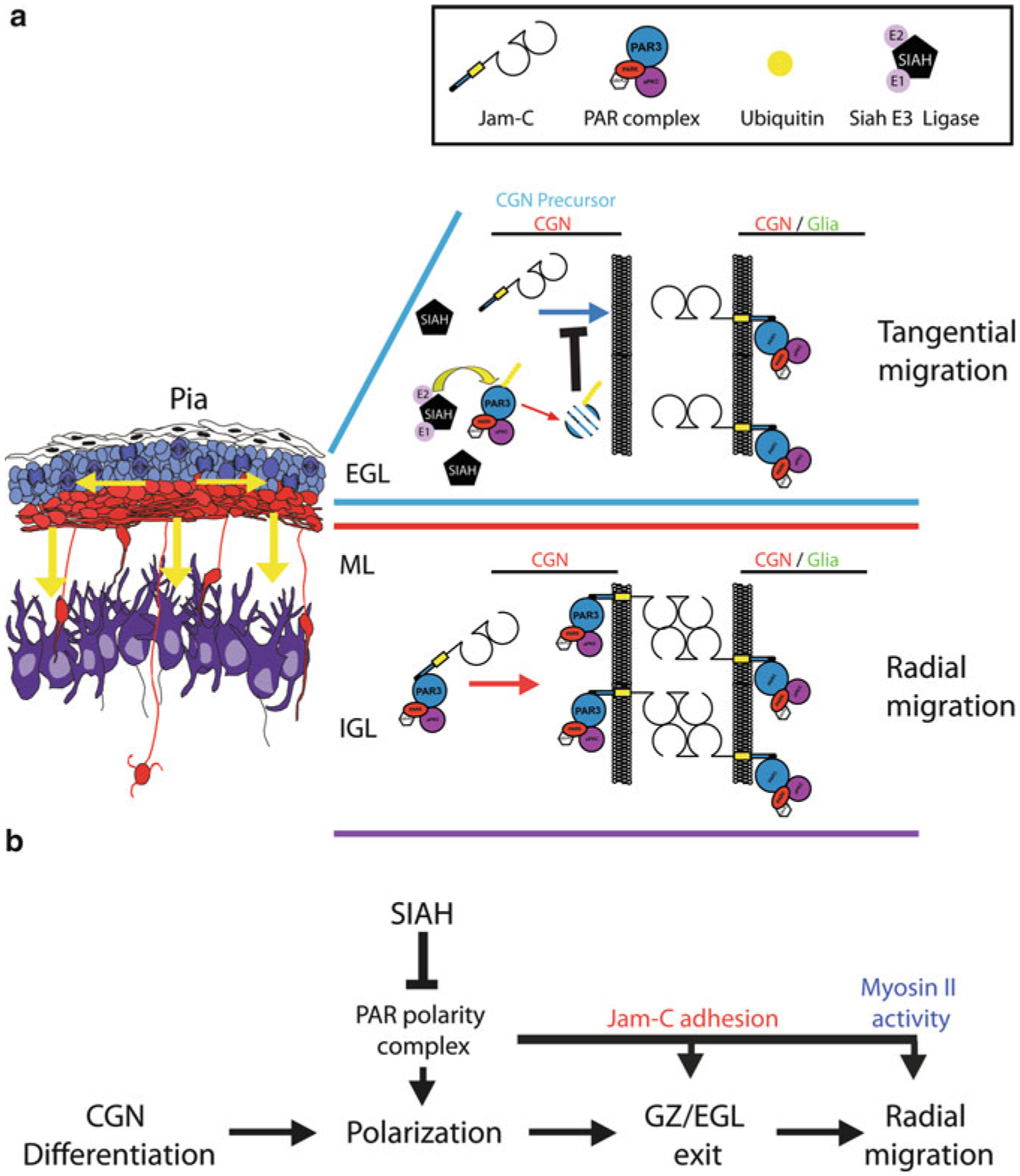
(a) During cerebellar development CGN precursors migrate tangentially within the EGL. Upon differentiation and polarization, CGNs exit the GZ/EGL and migrate radially to traverse the ML and assume their final position in the IGL. Within the developing postnatal cerebellum SIAH (E3 ubiquitin ligase) is highly expressed in the EGL, where it ubiquitinates PARD3A to target it for proteasome-mediated degradation. PARD3A degradation results in inactivation of the PAR polarity complex, thereby inhibiting recruitment of the JAM-C adhesion molecule to contacts between CGNs or CGN precursors and glial cells. The absence of JAM-C-mediated adhesion prevents GZ exit by restricting the radial migration of CGN precursors. (b) The PAR polarity complex is required for differentiated CGNs to polarize, exit the GZ via JAM-C-mediated adhesion, and migrate radially via activation of the myosin II motor. SIAH negatively regulates CGN polarization, GZ exit, and radial migration by inactivating the PAR polarity complex (Reproduced with permission of (Famulski et al. 2010))
PARD3 had previously been found to localize to cell-cell contacts and therefore to be essential for junction formation in epithelial cells (Hirose et al. 2002). Famulski and coworkers showed that PARD3 regulates germinal zone exit by interacting directly with junctional adhesion molecule C (JAM-C), a cell-cell adhesion molecule whose role in stabilizing cell-cell contacts is required for CGN migration to the IGL (Famulski et al. 2010). Using a JAM-C-pHluorin probe to observe JAM-C junctions in living cells, the authors characterized JAM-C junctions forming at cell-cell contacts in vivo in migrating neurons. SIAH gain of function dissolved JAM-C tight junctions in a manner that was rescued by PARD3, while SIAH silencing greatly enhanced JAM-C adhesion, suggesting that the antagonistic relationship between SIAH and PARD3 controls the surface levels of JAM-C. The critical importance of JAM-C adhesion for germinal zone exit was illustrated by JAM-C gain of function experiments: overexpression of a constitutively active version of JAM-C not only induced precocious germinal zone exit but also fully rescued a SIAH gain-of-function phenotype. Famulski and colleagues identified SIAH as an inhibitor of PARD3-dependent JAM-C adhesion, revealing how reduced polarity signaling within the EGL of the cerebellum prevents the onset of cell-cell contacts that are necessary for germinal zone exit. This was the first demonstration of the direct control of neuron–glial cell adhesion by a polarity complex in the developing nervous system.
4.3. Mechanisms of PAR-Mediated Cell Adhesion
The PAR polarity complex is indirectly related to mechanisms of cell-cell adhesion. The interactions of PAR in epithelial cell junctions and turnover may elucidate the role of PAR proteins in neuronal migration. Myosin II promotes junction formation in epithelial cells by strengthening remodeling adhesions (Vicente-Manzanares et al. 2009; Bertet et al. 2004). As PARD6α has been found to be a regulator of myosin II (Solecki et al. 2009), the PARD3-SIAH complex may be an additional mechanism by which the PAR polarity complex controls polarity and adhesion. Interactions between cell adhesions and cytoskeletal motor systems provide the context in which migrating cells generate force to propel themselves forward. Proteomic analysis identified dynein intermediate chain 2 (DIC2) as a phosphorylation target of aPKC of the PAR complex (Rosse et al. 2012). Regulation of DIC2 by aPKC controls focal adhesion turnover through interaction with focal adhesion complex member paxillin (Rosse et al. 2012). PARD3 was also implicated as a regulator of focal adhesion kinase (FAK) through mass-spectrometry identification of PARD3 binding partners in epithelial cells (Itoh et al. 2010; Xie et al. 2003). Reduction of PARD3 in epithelial cells inhibited adhesion-induced activation of FAK, implicating the PAR polarity complex in the regulation of focal adhesions. Interaction of the PAR complex with both the microtubule and actin cytoskeletons in migrating cells potentially links the two systems, allowing crosstalk between them. As disruption of PAR signaling uncouples the two-stroke nucleokinesis cycle and inhibits recruitment of centrosome proteins, it is possible that interaction between the PAR complex and myosin II (PARD6) and/or dynein (PARD3) is required for proper cytoskeletal rearrangement and is an integral component of neuronal motility.
5. Further Studies of the Compartmental Roles of PAR in Migrating Neurons
PAR proteins and the PARD3/PARD6/aPKC complex have been identified as key regulators of neuronal migration and germinal zone exit. The role of PAR proteins in discrete regions of the cell, interacting with the centrosome or specifically at cell adhesions, may differ from the roles of the PAR complex in migrating cells. Several challenges remain in understanding the mechanisms involved in neuronal migration, and they can be addressed by future studies of the PAR complex. By what mechanism does PARD6 regulate the microtubule and actomyosin cytoskeleton? As the PAR complex has been proposed to regulate actomyosin contraction in the leading edge of migrating neurons, there may be a connection between myosin and dynein motor–generated force in migrating neurons. Does PARD6 or the PAR complex directly link the actin and microtubule cytoskeletons in the leading process of migrating neurons as a molecular clutch? High-resolution co-localization studies of specific components of the PAR complex with cytoskeletal motor systems in the leading processes of actively migrating neurons may reveal such transient interactions. What additional signals control PARD3’s regulation of cell-cell adhesions in the transition from tangential to radial migration? Examining how PARD3 regulates adhesion systems other than JAM-C, and the cytoskeletal systems that use these adhesions to generate propulsive force, will provide a larger context for understanding how the neuronal cytoskeleton interacts with cell-cell adhesions.
Acknowledgements
We thank Sharon Naron, Niraj Trivedi, and Shalini Singh for editorial support in preparation of the manuscript. The Solecki Lab is funded by the American Lebanese Syrian Associated Charities (ALSAC), by grant #1-FY12-455 from the March of Dimes, and by grant 1R01NS066936 from the National Institute of Neurological Disorders and Stroke (NINDS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NINDS or the National Institutes of Health.
References
- Allen KM, Gleeson JG, Shoup SM, Walsh CA (1998) A YAC contig in Xq22. 3-q23, from DXS287 to DXS8088, spanning the brain-specific genes doublecortin (DCX) and PAK3. Genomics 52(2):214–218 [DOI] [PubMed] [Google Scholar]
- Barnes AP, Polleux F (2009) Establishment of axondendrite polarity in developing neurons. Annu Rev Neurosci 32:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellion A, Baudoin JP, Alvarez C, Bornens M, Metin C (2005) Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J Neurosci 25(24):5691–5699. doi:25/24/5691 [pii] 10.1523/JNEUROSCI.1030-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertet C, Sulak L, Lecuit T (2004) Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature 429(6992):667–671. doi: http://www.nature.com/nature/journal/v429/n6992/suppinfo/nature02590_S1.html [DOI] [PubMed] [Google Scholar]
- Chen S, Chen J, Shi H, Wei M, Castaneda-Castellanos David R, Bultje Ronald S, Pei X, Kriegstein Arnold R, Zhang M, Shi S-H (2013) Regulation of microtubule stability and organization by mammalian Par3 in specifying neuronal polarity. Dev Cell 24(1):26–40. doi: 10.1016/j.devcel.2012.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C, Vale RD (2012) The mechanism of dynein motility: insight from crystal structures of the motor domain. Biochim Biophys Acta 1823(1):182–191. doi: 10.1016/j.bbamcr.2011.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormoy V, Tormanen K, Sütterlin C (2013) Par6γ is at the mother centriole and controls centrosomal protein composition through a Par6α-dependent pathway. J Cell Sci 126(3):860–870. doi: 10.1242/jcs.121186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin DL, Vallee RB (2002) Dynein at the cortex. Curr Opin Cell Biol 14(1):44–49 [DOI] [PubMed] [Google Scholar]
- Dujardin DL, Barnhart LE, Stehman SA, Gomes ER, Gundersen GG, Vallee RB (2003) A role for cytoplasmic dynein and LIS1 in directed cell movement. J Cell Biol 163(6):1205–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson JC, Hatten ME (1987) Glial-guided granule neuron migration in vitro: a high-resolution time-lapse video microscopic study. J Neurosci 7(6):1928–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson J, Liem R, Kuster J, Hatten M (1988) Astrotactin: a novel neuronal cell surface antigen that mediates neuron-astroglial interactions in cerebellar microcultures. J Cell Biol 106(2):505–517. doi: 10.1083/jcb.106.2.505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famulski JK, Trivedi N, Howell D, Yang Y, Tong Y, Gilbertson R, Solecki DJ (2010) Siah regulation of Pard3A controls neuronal cell adhesion during germinal zone exit. Science 330(6012):1834–1838. doi: 10.1126/science.1198480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner NE, Dujardin DL, Tai CY, Vaughan KT, O’Connell CB, Wang Y, Vallee RB (2000) A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol 2(11):784–791 [DOI] [PubMed] [Google Scholar]
- Feng Y, Olson EC, Stukenberg PT, Flanagan LA, Kirschner MW, Walsh CA (2000) LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron 28(3):665. [DOI] [PubMed] [Google Scholar]
- Fishell G, Hatten ME (1991) Astrotactin provides a receptor system for CNS neuronal migration. Development 113(3):755–765 [DOI] [PubMed] [Google Scholar]
- Fishman R, Hatten M (1993) Multiple receptor systems promote CNS neural migration. J Neurosci 13(8):3485–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis F, Koulakoff A, Boucher D, Chafey P, Schaar B, Vinet MC, Friocourt G, McDonnell N, Reiner O, Kahn A, McConnell SK, Berwald-Netter Y, Denoulet PJC (1999) Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron 23(2):247–256 [DOI] [PubMed] [Google Scholar]
- Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM (2010) Mechanical integration of actin and adhesion dynamics in cell migration. Annu Rev Cell Dev Biol 26:315–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson JG, Lin PT, Flanagan LA, Walsh CA (1999) Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron 23(2):257–271. doi:S0896–6273(00)80778–3 [pii] [DOI] [PubMed] [Google Scholar]
- Gregory W, Edmondson J, Hatten M, Mason C (1988) Cytology and neuron-glial apposition of migrating cerebellar granule cells in vitro. J Neurosci 8(5):1728–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton SL, Waterman-Storer CM (2006) Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell 125(7):1361–1374 [DOI] [PubMed] [Google Scholar]
- Hatten ME (1990) Riding the glial monorail: a common mechanism for glial-guided neuronal migration in different regions of the developing mammalian brain. Trends Neurosci 13(5):179–184. doi: 10.1016/0166-2236(90)90044-B [DOI] [PubMed] [Google Scholar]
- Hatten ME, Heintz N (1995) Mechanisms of neural patterning and specification in the development cerebellum. Annu Rev Neurosci 18(1):385–408 [DOI] [PubMed] [Google Scholar]
- He M, Z Z-h, G C-b, Xia D, Y X-b (2010) Leading tip drives soma translocation via forward F-actin flow during neuronal migration. J Neurosci 30(32):10885–10898. doi: 10.1523/jneurosci.0240-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham HR, Gleeson JG (2007) The centrosome in neuronal development. Trends Neurosci 30(6):276–283 [DOI] [PubMed] [Google Scholar]
- Hirose T, Izumi Y, Nagashima Y, Tamai-Nagai Y, Kurihara H, Sakai T, Suzuki Y, Yamanaka T, Suzuki A, Mizuno K, Ohno S (2002) Involvement of ASIP/PAR-3 in the promotion of epithelial tight junction formation. J Cell Sci 115(12):2485–2495 [DOI] [PubMed] [Google Scholar]
- Hirotsune S, Fleck MW, Gambello MJ, Bix GJ, Chen A, Clark GD, Ledbetter DH, McBain CJ, Wynshaw-Boris A (1998) Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat Genet 19(4):333–339 [DOI] [PubMed] [Google Scholar]
- Itoh N, Nakayama M, Nishimura T, Fujisue S, Nishioka T, Watanabe T, Kaibuchi K (2010) Identification of focal adhesion kinase (FAK) and phosphatidylinositol 3-kinase (PI3-kinase) as Par3 partners by proteomic analysis. Cytoskeleton 67(5):297–308. doi: 10.1002/cm.20444 [DOI] [PubMed] [Google Scholar]
- Joberty G, Petersen C, Gao L, Macara IG (2000) The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol 2(8):531–539 [DOI] [PubMed] [Google Scholar]
- Kamm KE, Stull JT (2001) Dedicated myosin light chain kinases with diverse cellular functions. J Biol Chem 276(7):4527–4530. doi: 10.1074/jbc.R000028200 [DOI] [PubMed] [Google Scholar]
- Kato M, Dobyns WB (2003) Lissencephaly and the molecular basis of neuronal migration. Hum Mol Genet 12(Spec No 1):R89–R96 [DOI] [PubMed] [Google Scholar]
- Kawauchi T, Hoshino M (2007) Molecular pathways regulating cytoskeletal organization and morphological changes in migrating neurons. Dev Neurosci 30(1–3):36–46 [DOI] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS (1988) Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52(3):311–320 [DOI] [PubMed] [Google Scholar]
- Kodani A, Tonthat V, Wu B, Sütterlin C (2010) Par6α interacts with the dynactin subunit p150Glued and is a critical regulator of centrosomal protein recruitment. Mol Biol Cell 21(19):3376–3385. doi: 10.1091/mbc.E10-05-0430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro H, Rakic P (1998) Distinct modes of neuronal migration in different domains of developing cerebellar cortex. J Neurosci 18(4):1478–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro H, Yacubova E, Yacubova E, Rakic P (2001) Mode and tempo of tangential cell migration in the cerebellar external granular layer. J Neurosci 21(2):527–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clainche C, Carlier M-F (2008) Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev 88(2):489–513. doi: 10.1152/physrev.00021.2007 [DOI] [PubMed] [Google Scholar]
- Lin D, Edwards AS, Fawcett JP, Mbamalu G, Scott JD, Pawson T (2000) A mammalian PAR-3–PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat Cell Biol 2(8):540–547 [DOI] [PubMed] [Google Scholar]
- Ma X, Kawamoto S, Hara Y, Adelstein RS (2004) A point mutation in the motor domain of nonmuscle myosin II-B impairs migration of distinct groups of neurons. Mol Biol Cell 15(6):2568–2579. doi: 10.1091/mbc.E03-11-0836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzini MC, Walsh CA (2011) What disorders of cortical development tell us about the cortex: one plus one does not always make two. Curr Opin Genet Dev 21(3):333–339. doi: 10.1016/j.gde.2011.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín O, Valiente M, Ge X, Tsai L-H (2010) Guiding neuronal cell migrations. Cold Spring Harb Perspect Biol 2(2):a001834. doi: 10.1101/cshperspect.a001834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini FJ, Valdeolmillos M (2010) Actomyosin contraction at the cell rear drives nuclear translocation in migrating cortical interneurons. J Neurosci 30(25):8660–8670. doi: 10.1523/jneurosci.1962-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métin C, Luccardini C (2010) Ubiquitination inhibits neuronal exit. Science 330(6012):1754–1755. doi: 10.1126/science.1200475 [DOI] [PubMed] [Google Scholar]
- Métin C, Vallee RB, Rakic P, Bhide PG (2008) Modes and mishaps of neuronal migration in the mammalian brain. J Neurosci 28(46):11746–11752. doi: 10.1523/jneurosci.3860-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen KJ, Gleeson JG (2008) Cerebellar development and disease. Curr Opin Neurobiol 18(1):12–19. doi: 10.1016/j.conb.2008.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M (1988) Cytoskeletal dynamics and nerve growth. Neuron 1(9):761–772. doi: 10.1016/0896-6273(88)90124-9 [DOI] [PubMed] [Google Scholar]
- Moussavi RS, Kelley CA, Adelstein RS (1993) Phosphorylation of vertebrate nonmuscle and smooth muscle myosin heavy chains and light chains. Mol Cell Biochem 127–128:219–227 [DOI] [PubMed] [Google Scholar]
- Munro EM (2006) PAR proteins and the cytoskeleton: a marriage of equals. Curr Opin Cell Biol 18(1):86–94 [DOI] [PubMed] [Google Scholar]
- Munro E, Nance J, Priess JR (2004) Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans Embryo. Dev Cell 7(3):413–424. doi: 10.1016/j.devcel.2004.08.001 [DOI] [PubMed] [Google Scholar]
- Nance J, Zallen JA (2011) Elaborating polarity: PAR proteins and the cytoskeleton. Development 138(5): 799–809. doi: 10.1242/dev.053538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke NA, Dailey ME, Smith SJ, McConnell SK (1992) Diverse migratory pathways in the developing cerebral cortex. Science 258(5080):299–302 [DOI] [PubMed] [Google Scholar]
- Rakic P (1971) Neuron-glia relationship during granule cell migration in developing cerebellar cortex. A Golgi and electronmicroscopic study in Macacus rhesus. J Comp Neurol 141(3):283–312 [DOI] [PubMed] [Google Scholar]
- Rakic P (1972) Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol 145(1):61–83 [DOI] [PubMed] [Google Scholar]
- Rakic P, Knyihar-Csillik E, Csillik B (1996) Polarity of microtubule assemblies during neuronal cell migration. Proc Natl Acad Sci U S A 93(17):9218–9222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH (1993) Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature 364(6439):717–721 [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR (2003) Cell migration: integrating signals from front to back. Science 302(5651):1704–1709 [DOI] [PubMed] [Google Scholar]
- Rivas RJ, Hatten ME (1995) Motility and cytoskeletal organization of migrating cerebellar granule neurons. J Neurosci 15(2):981–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochlin MW, Itoh K, Adelstein RS, Bridgman PC (1995) Localization of myosin II A and B isoforms in cultured neurons. J Cell Sci 108(Pt 12):3661–3670 [DOI] [PubMed] [Google Scholar]
- Ross ME, Walsh CA (2001) Human brain malformations and their lessons for neuronal migration. Annu Rev Neurosci 24:1041–1070 [DOI] [PubMed] [Google Scholar]
- Rosse C, Boeckeler K, Linch M, Radtke S, Frith D, Barnouin K, Morsi AS, Hafezparast M, Howell M, Parker PJ (2012) Binding of dynein intermediate chain 2 to paxillin controls focal adhesion dynamics and migration. J Cell Sci 125(16):3733–3738. doi: 10.1242/jcs.089557 [DOI] [PubMed] [Google Scholar]
- Ryder EF, Cepko CL (1994) Migration patterns of clonally related granule cells and their progenitors in the developing chick cerebellum. Neuron 12(5):1011–1029. doi: 10.1016/0896-6273(94)90310-7 [DOI] [PubMed] [Google Scholar]
- Sakakibara A, Sato T, Ando R, Noguchi N, Masaoka M, Miyata T (2013) Dynamics of centrosome translocation and microtubule organization in neocortical neurons during distinct modes of polarization. Cereb Cortex [DOI] [PubMed] [Google Scholar]
- Schaar BT, McConnell SK (2005) Cytoskeletal coordination during neuronal migration. Proc Natl Acad Sci U S A 102(38):13652–13657. doi: 10.1073/pnas.0506008102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoranzer J, Fawcett JP, Segura M, Tan S, Vallee RB, Pawson T, Gundersen GG (2009) Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration. Curr Biol 19(13):1065–1074. doi: 10.1016/j.cub.2009.05.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara R, Thumkeo D, Kamijo H, Kaneko N, Sawamoto K, Watanabe K, Takebayashi H, Kiyonari H, Ishizaki T, Furuyashiki T (2012) A role for mDia, a Rho-regulated actin nucleator, in tangential migration of interneuron precursors. Nat Neurosci 15(3):373–380 [DOI] [PubMed] [Google Scholar]
- Shu T, Ayala R, Nguyen MD, Xie Z, Gleeson JG, Tsai LH (2004) Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron 44(2):263–277 [DOI] [PubMed] [Google Scholar]
- Smith DS, Niethammer M, Ayala R, Zhou Y, Gambello MJ, Wynshaw-Boris A, Tsai L-H (2000) Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat Cell Biol 2(11):767–775 [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME (2004) Par6alpha signaling controls glial-guided neuronal migration. Nat Neurosci 7(11):1195–1203 [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Trivedi N, Govek E-E, Kerekes RA, Gleason SS, Hatten ME (2009) Myosin II motors and F-Actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron 63(1):63–80. doi: 10.1016/j.neuron.2009.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinet S, Akpovi C, Garcia C, Barry A, Vitale M (2011) Myosin IIB deficiency in embryonic fibroblasts affects regulators and core members of the par polarity complex. Histochem Cell Biol 136(3):245–266. doi: 10.1007/s00418-011-0840-0 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, Gleeson JG (2004) Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol 165(5):709–721. doi: 10.1083/jcb.200309025 jcb.200309025 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi N, Solecki DJ (2011) Neuronal migration illuminated: a look under the hood of the living neuron. Cell Adh Migr 5(1):42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai LH, Gleeson JG (2005) Nucleokinesis in neuronal migration. Neuron 46(3):383–388 [DOI] [PubMed] [Google Scholar]
- Tsai JW, Bremner KH, Vallee RB (2007) Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci 10(8):970–979 [DOI] [PubMed] [Google Scholar]
- Umeshima H, Hirano T, Kengaku M (2007) Microtubule-based nuclear movement occurs independently of centrosome positioning in migrating neurons. Proc Natl Acad Sci U S A 104(41):16182–16187. doi: 10.1073/pnas.0708047104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiente M, Marín O (2010) Neuronal migration mechanisms in development and disease. Curr Opin Neurobiol 20(1):68–78 [DOI] [PubMed] [Google Scholar]
- Vallee RB, Seale GE, Tsai J-W (2009) Emerging roles for myosin II and cytoplasmic dynein in migrating neurons and growth cones. Trends Cell Biol 19(7):347–355. doi: 10.1016/j.tcb.2009.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Zareno J, Whitmore L, Choi CK, Horwitz AF (2007) Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J Cell Biol 176(5):573–580. doi: 10.1083/jcb.200612043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR (2009) Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol 10(11):778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, She L, Y-n S, X-b Y, Wen Y, M-m P (2012) Forward transport of proteins in the plasma membrane of migrating cerebellar granule cells. Proc Natl Acad Sci 109(51):E3558–E3567. doi: 10.1073/pnas.1219203110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Sanada K, Samuels BA, Shih H, Tsai LH (2003) Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell 114(4):469–482 [DOI] [PubMed] [Google Scholar]
- Yanagida M, Miyoshi R, Toyokuni R, Zhu Y, Murakami F (2012) Dynamics of the leading process, nucleus, and Golgi apparatus of migrating cortical interneurons in living mouse embryos. Proc Natl Acad Sci U S A 109(41):16737–16742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Sun Y, Zhang F, Zhu Y, Shi L, Li H, Xu Z (2012) POSH localizes activated Rac1 to control the formation of cytoplasmic dilation of the leading process and neuronal migration. Cell Rep 2(3):640–651 [DOI] [PubMed] [Google Scholar]
- Young A, Dictenberg JB, Purohit A, Tuft R, Doxsey SJ (2000) Cytoplasmic dynein-mediated assembly of pericentrin and γ tubulin onto centrosomes. Mol Biol Cell 11(6):2047–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Heintz N, Hatten ME (1996) CNS gene encoding astrotactin, which supports neuronal migration along glial fibers. Science 272(5260):417–419 [DOI] [PubMed] [Google Scholar]


