Graphical abstract
Keywords: Mitochondria, mtDNA, Gene editing
Abstract
Mitochondria, as the energy factory of cells, participate in metabolism processes and play a critical role in the maintenance of human life activities. Mitochondria belong to semi-automatic organelles, which have their own genome different from nuclear genome. Mitochondrial DNA (mtDNA) mutations can cause a series of diseases and threaten human health. However, an effective approach to edit mitochondrial DNA, though long-desired, is lacking. In recent years, gene editing technologies, represented by restriction endonucleases (RE) technology, zinc finger nuclease (ZFN) technology, transcription activator-like effector nuclease (TALEN) technology, CRISPR system and pAgo-based system have been comprehensively explored, but the application of these technologies in mitochondrial gene editing is still to be explored and optimized. The present study highlights the progress and limitations of current mitochondrial gene editing technologies and approaches, and provides insights for development of novel strategies for future attempts.
1. Mitochondria and mitochondrial diseases
1.1. Mitochondria
Mitochondria, as a kind of semi-autonomous organelle, widely exist in eukaryotic cells, which are the main places of aerobic respiration. They provide energy for cell activities through electron transport chain and oxidative phosphorylation system. Meanwhile, mitochondria participate in Krebs cycle, β-oxidation, lipid and cholesterol synthesis and other important metabolic pathways [1]. They are also involved in apoptosis and production of oxygen free radicals [2], [3], [4].
Mitochondria have their independent genome distinct from nuclear genome, which are substantially similar to bacterial genome. Mitochondrial DNA (mtDNA) is an important part of the genome [5]. Mitochondrial DNA sequencing demonstrated that the two strands of human mtDNA are divided into heavy chain and light chain, which contain 16,569 base pairs, encoding 2 rRNAs, 22 tRNAs and 13 protein subunits [5], [6], [7], [8]. Mitochondrial proteins (encoded by mitochondrial DNA) differ among species and tissues. The transcription and translation of mtDNA are completely controlled by the nuclear DNA coding factors through binding to the unique noncoding region (1 kb D-loop) of mitochondrial genome [7]. The peptides synthesized by 13 mtDNA genes interact with more than 60 nuclear coding peptides to form mitochondrial respiratory chain, which is required for aerobic cell metabolism [9]. Therefore, mitochondrial function depends on the interaction of nuclear and mitochondrial genes. Any abnormal nuclear or mitochondrial genome may lead to mitochondrial diseases.
1.2. Mitochondrial diseases
Inherited mitochondrial diseases are characterized by oxidative phosphorylation defects, which are caused by mutations in nuclear DNA (nDNA) and mtDNA genes encoding structural mitochondrial proteins or proteins involved in mitochondrial function [4], [10], [11]. These are the most common genetic metabolic disorders and one of the most common forms of genetic neural disorders [12]. Although the mitochondrial genome is small, mtDNA is a significant cause for genetic diseases. When the heteroplasmy, meaning the coexistence of mutant mtDNA and the wild-type mtDNA, reaches a threshold level, certain mitochondrial diseases will appear [13]. In recent years, with the further study of mtDNA, it has been found that mitochondrial genes are closely associated with some human diseases, such as Leigh syndrome [14], [15], [16], NARP syndrome (Neuron, Ataxia, and Retinitis Pigmentosa) [17], [18] and LHON(Leber Hereditary Optic Neuropathy) [19], [20]. At present, considerable progress has been made in the understanding of the basic mitochondrial genetics and the relationship between gene mutations and disease phenotypes, as well as in the identification of acquired mtDNA mutations in aging and cancer (Table 1). By now, several mitochondrial gene mutations and the related mitochondrial diseases still have no feasible or efficient treatments. With the development of gene editing technologies, it is promising to eliminate mitochondrial mutations and develop gene therapy of mitochondrial diseases.
Table 1.
Mitochondrial DNA Base Substitution Diseases: Coding and Control Region Point Mutations with confirmed Status (https://www.mitomap.org/foswiki/bin/view/MITOMAP/MutationsCodingControlCfrm).
| Locus | Position | Disease | Nucleotide Change |
|---|---|---|---|
| MT-ND1 | 3376 | LHON MELAS overlap | G-A |
| 3460 | LHON | G-A | |
| 3635 | LHON | G-A | |
| 3697 | MELAS/LS/LDYT/BSN | G-A | |
| 3700 | LHON | G-A | |
| 3733 | LHON | G-A | |
| 3890 | Progressive Encephalomyopathy/LS/Optic Atrophy | G-A | |
| 3902 | EXIT + myalgia/severe LA + cardiac/3-MGA aciduria | ACCTTGC-GCAAGGT | |
| 4171 | LHON/Leigh-like phenotype | C-A | |
| MT-CO1 | 7445 | SNHL | A-G |
| MT-ATP8/6 | 8528 | Infantile cardiomyopathy | T-C |
| MT-ATP6 | 8851 | BSN/Leigh syndrome | T-C |
| 8969 | Mitochondrial myopathy, lactic acidosis and sideroblastic anemia (MLASA)/IgG nephropathy | G-A | |
| 8993 | NARP/Leigh Disease/MILS/other | T-C | |
| 8993 | NARP/Leigh Disease/MILS/other | T-G | |
| 9035 | Ataxia syndromes | T-C | |
| 9155 | MIDD, renal insufficiency | A-G | |
| 9176 | Leigh Disease/Spastic Paraplegia | T-G | |
| 9176 | FBSN/Leigh Disease | T-C | |
| 9185 | Leigh Disease/Ataxia syndromes/NARP-like disease | T-C | |
| 9205 | Encephalopathy/Seizures/Lacticacidemia | TA-del | |
| MT-ND3 | 10,158 | Leigh Disease/MELAS | T-C |
| 10,191 | Leigh Disease/Leigh-like Disease/ESOC | T-C | |
| 10,197 | Leigh Disease/Dystonia/Stroke/LDYT | G-A | |
| MT-ND4L | 10,663 | LHON | T-C |
| MT-ND4 | 11,777 | Leigh Disease | C-A |
| 11,778 | LHON/Progressive Dystonia | G-A | |
| MT-ND5 | 12,706 | Leigh Disease | T-C |
| 13,042 | Optic neuropathy/ retinopathy/ LD | G-A | |
| 13,051 | LHON | G-A | |
| 13,094 | Ataxia + PEO/MELAS, LD, LHON, myoclonus, fatigue | T-C | |
| 13,513 | Leigh Disease/MELAS/LHON-MELAS Overlap Syndrome/negative association w Carotid Atherosclerosis | G-A | |
| 13,514 | Leigh Disease/MELAS/Ca2+ downregulation | A-G | |
| MT-ND6 | 14,459 | LDYT/Leigh Disease/dystonia/carotid atherosclerosis risk | G-A |
| 14,482 | LHON | C-A | |
| 14,482 | LHON | C-G | |
| 14,484 | LHON | T-C | |
| 14,487 | Dystonia/Leigh Disease/ataxia/ptosis/epilepsy | T-C | |
| 14,495 | LHON | A-G | |
| 14,568 | LHON | C-T | |
| MT-CYB | 14,849 | EXIT/Septo-Optic Dysplasia | T-C |
| 14,864 | MELAS | T-C | |
| 15,579 | Multisystem Disorder, EXIT | A-G |
| Notes: | |||
|---|---|---|---|
| LHON | Leber Hereditary Optic Neuropathy | MM | Mitochondrial Myopathy |
| AD | Alzheimer’s Disease | LIMM | Lethal Infantile Mitochondrial Myopathy |
| ADPD | Alzheimer’s Disease and Parkinson’s Disease | MMC | Maternal Myopathy and Cardiomyopathy |
| NARP | Neurogenic muscle weakness, Ataxia, and Retinitis Pigmentosa; alternate phenotype at this locus is reported as Leigh Disease | FICP | Fatal Infantile Cardiomyopathy Plus, a MELAS-associated cardiomyopathy |
| MELAS | Mitochondrial Encephalomyopathy, Lactic Acidosis, and Stroke-like episodes | LDYT | Leber's hereditary optic neuropathy and Dystonia |
| MERRF | Myoclonic Epilepsy and Ragged Red Muscle Fibers | MHCM | Maternally inherited Hypertrophic Cardiomyopathy |
| CPEO | Chronic Progressive External Ophthalmoplegia | KSS | Kearns Sayre Syndrome |
| DM | Diabetes Mellitus | DMDF | Diabetes Mellitus + Deafness |
| CIPO | Chronic Intestinal Pseudo obstruction with myopathy and Ophthalmoplegia | DEAF | Maternally inherited Deafness or aminoglycoside-induced Deafness |
| PEM | Progressive encephalopathy | SNHL | Sensorineural Hearing Loss |
1.3. Transport of proteins into mitochondria
Mitochondria, with bilayer membrane structure, encompass two compartments—intermembrane space (IMS) and matrix [21]. The function relies on the assembly of about 1000 resident proteins, 99% of which are synthesized in the cytoplasm as precursor proteins and transferred into four different sites of mitochondria through translocation complexes [22]. So far, many studies have been carried out on the mechanism of mitochondrial membrane penetration of proteins. Precursor proteins are first synthesized in the cytoplasm with targeting information, which can be recognized by specific receptor protein on the outer membrane of mitochondria [23]. About 60% of these proteins carry cleavable targeting signals, which are removed by mitochondrial processing peptidase after introduction [23]. Other mitochondrial proteins contain different types of non-removable targeting signals, such as transmembrane segments [23]. The process that precursors enter matrix through bilayer membrane involves three internal protein complexes to mediate membrane translocation and sorting of precursor proteins [24]. The general entrance pore of the outer membrane is formed by the TOM complex, which is responsible for the initial translocation of more than 90% of mitochondrial precursor proteins from the cytoplasm to the membrane space [25]. TOM40 is the pivotal protein, which forms the center of the channel. Other TOM proteins, such as TOM20 and TOM70, assist in the recognition of targeting signals [25], [26], [27]. Various TOM proteins coordinate with each other to maintain the stability and function of TOM complex [25], [26]. Nevertheless, the underlying mechanism of its mediation is still unclear, and the channel structure is not fully determined [25]. In addition, the abundant cytoplasmic chaperones, such as heat shock protein 70 (HSP70), have been proved to combine mitochondrial precursor proteins in a non-specific manner to prevent misfolding and aggregation before translocation [21], [23], [27]. It has been reported that a large number of cytoplasmic factors also promote the transport of precursor proteins to mitochondria, which is the key to targeting of mitochondrial precursor proteins [21], [23], [27]. TIM complex, mainly mediated by TIM23, takes part in protein transport to matrix [21], [23], [27]. A recent study identified a new targeting pathway called ER-SURF, discovering that endoplasmic reticulum (ER) surface also plays an important and positive role in ribosome to mitochondrial targeting. However, the details of this pathway remain to be clarified [21], [23].
2. Mitochondrial gene editing technology
Mitochondrial transplantation, a novel therapeutic intervention to treat mitochondrial diseases, is simple and rapid [28]. However, in order to maintain the long-term therapeutic effect, mitochondrial transplantation should be carried out many times, and the methods of mitochondrial isolation, the mitochondrial sources, the route of administration, and doses all influence theperformance of mitochondrial transplantation [29]. Several studies have concluded that immune response occurs after mitochondrial transplantation, and the establishment of a method permitting mitochondria to be stored in the long term is a vital issue [29]. In addition, given that the occurrence of mitochondrial diseases is mainly due to the mutation of mtDNA up to a certain level, resulting in functional disorders, gene editing technology specifically targeting mitochondrial genes will become a potential approach to treat mitochondrial diseases. This method can reduce the proportion of mutant mtDNA by modifying the mutated mitochondrial gene to help solve mitochondrial diseases (Fig. 1). In the long run, comparing with mitochondrial transplantation, mitochondrial gene editing technology has wide application prospects. At present, the main methods for mitochondrial gene editing are as follows (restriction endonucleases (RE) technology, zinc finger nuclease (ZFN) technology, transcription activator like effector nuclease (ALEN) technology and CRISPR/Cas9 system) (Fig. 2A,B,C, Table 2, , Table 3).
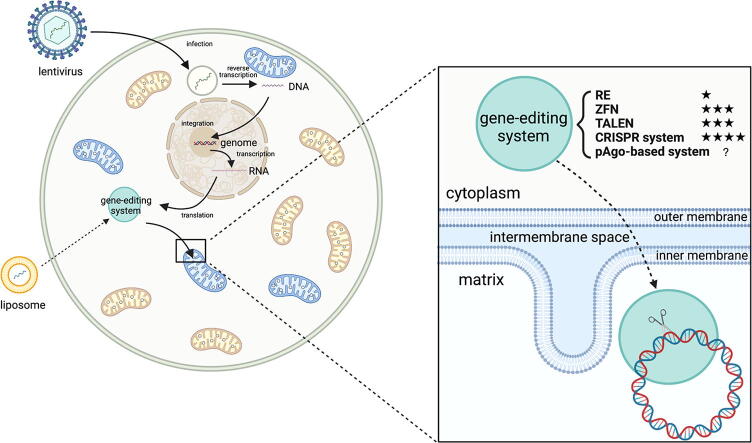
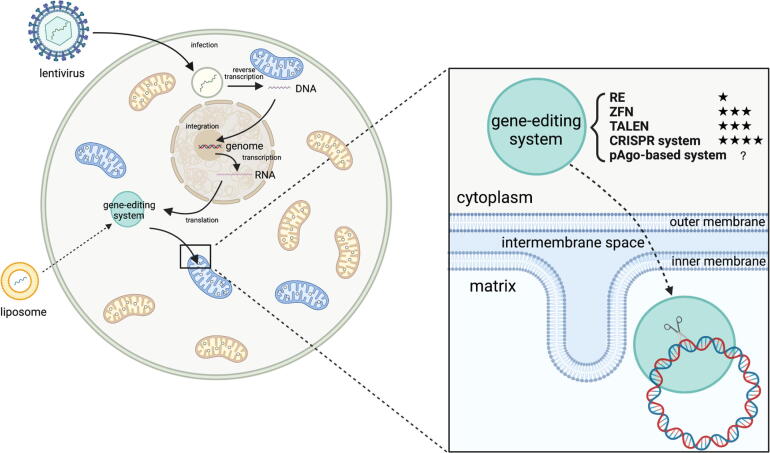
Fig. 1.
Graphical summary of gene editing system delivery into the mitochondria. Protein or RNA component can be imported through lentivirus transfection and nuclear genome expression while modified by MTS or RP-Loop. DNA component can be imported through liposome transfection or other methods. The blue-color mitochondria stand for mitochondria containing mutated mtDNA (target site of gene editing system) while the yellow-color mitochondria stand for wild-type mitochondria. (RE:Restriction endonuclease; ZFN:Zinc Finger Nuclease; TALEN:Transcription Activator-Like Effectors Nuclease; CRISPR:Clustered regularly interspaced short palindromic repeats; pAgo:prokaryotic Argonaute proteins). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
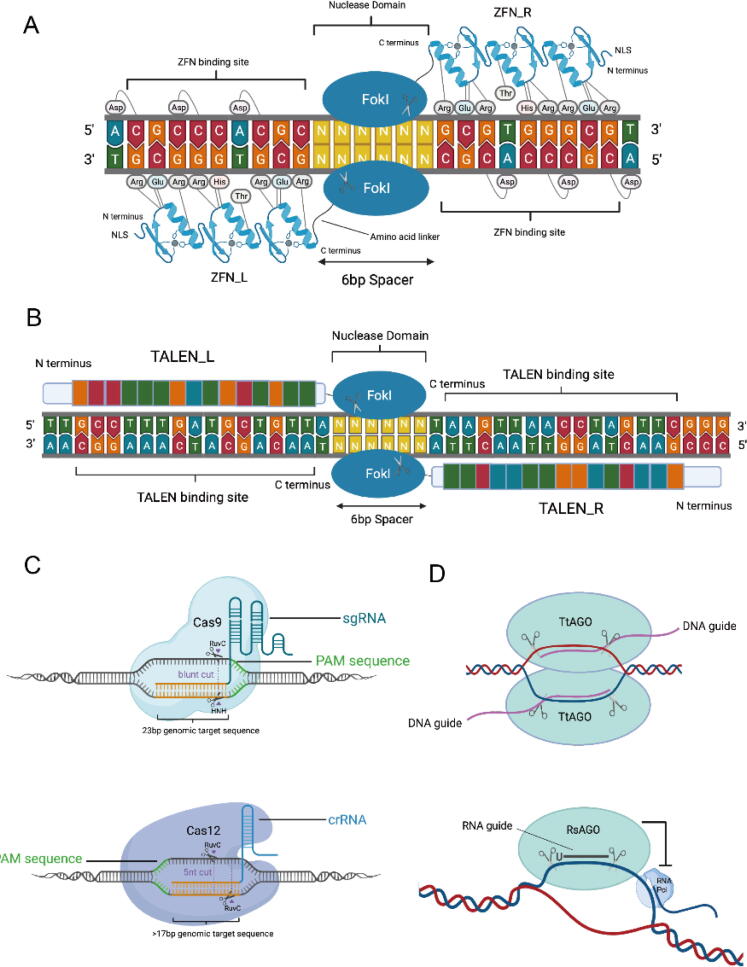
Fig. 2.
Several methods of gene editing. (A) Graphical abstract of the mechanism of ZFN. (B) Graphical abstract of the mechanism of TALEN. (C) Graphical abstract of the mechanism of CRISPR/Cas9 and CRISPR/Cas12 system. (D) Graphic abstract of the mechanism of pAgo-based system, taking TtAgo(DNA-guide) and RsAgo(RNA-guide) as examples.
Table 2.
Comparison of several gene editing methods.
| Characteristics/types | RE [30] | ZFN [33] | TALEN [44] | CRISPR-Cas [53], [54], [55] | CRISPR-Cpf [59], [60], [61], [62] |
|---|---|---|---|---|---|
| Recognition method | RE self-recognition | ZF array protein recognition | TALE array protein recognition | Guide RNA recognition | Mature crRNA recognition |
| Cleavage method | RE | FokⅠ dimer | FokⅠ dimer | Cas protein monomer | Cpf1 protein |
| Recognition sequence features | Depends on restriction enzyme types | Recognizing 9–18 bp from two sides of double chain respectively with 3 bp as a unit | Recognizing 14–20 bp from two sides of double chain respectively, recognizing one-to-one with T on the 0th base | Recognizing ~20 bp single chain, 3 'terminal sequence is NGC | Recognizing multiple sites, the 3 'end is rich in t sequence, forming sticky ends |
| Application scope | DNA, RNA | Only DNA | Only DNA | DNA, RNA | DNA, RNA |
| Advantage | High recognition accuracy and cleavage efficiency | Low immunogenicity and easy transportation | High specificity, easy transportation and operation | High specificity, easy operation and short cycle | High specificity, easy operation and wide target |
| Disadvantage | Limited target positions, no artificial design | Poor specificity and miss target | Cumbersome operation and miss target | Miss target, PAM dependent | Miss target, PAM dependent and low activity |
Table 3.
Cases of different mitochondrial gene editing technologies.
| Gene editing approaches | Cell type | Principle | Target | Outcome | Reference |
|---|---|---|---|---|---|
| RE | Enucleated Skin fibroblasts | Restriction nuclease SmaI modified by MTS | m.8993 T>G mutation | Decreasing the ratio of mutated mtDNA | [31] |
| Simian virus 40 (SV40) immortalized NZB/BALB fibroblasts | Restriction enzyme ApaLI modified by 5′and 3′ UTR sequences of ATP5b | BALB/NZB mtDNA | Preventing the transmission of mitochondrial genomes | [32] | |
| ZFN | COS-7, 143B (TK-) wild-type cells and 143B (TK-) NARP cybrid cells; | ZFP modified by MTS and NES | m.8993 T>G mutation | Cleavage of target site | [39] |
| cell lines 143B (TK-) and 143B (TK-) NARP cybrids; Flp-In TREx HEK293 cell-line | Single-chain ZFNs conjugating two FokI nuclease domains, together with an N-terminal mitochondrial targeting sequence | m.8993 T>G mutation | ZFNs are efficiently transported into mitochondria and cleave dsDNA at predicted sites adjacent to the mutation, increasing the proportion of wt mtDNA molecules in the cell | [37] | |
| H39 HOS; HOS 143B | Replace homodimer FokI with heterodimer in mtZFNs | m.8993 T>G mutation; common deletion (CD) | reduction in mutant mtDNA haplotype load, and subsequent repopulation of wild-type mtDNA restored mitochondrial respiratory function in a CD cybrid cell model | [38] | |
| Human osteosarcoma 143B | Precisely controll the expression of mtZFNs by using iterative editing strategy and HammerHead Ribozyme (HHR) | m.8993 T>G mutation | Realizing the nearly complete transformation of mtDNA heterogeneity to wild type | [42] | |
| m.5024C>T mouse embryonic fibroblast (MEF) | Using systemically administered mtZFNs delivered by adeno-associated virus | m.5024C>T tRNAAla mutation | Specific elimination of mutant mtDNA across the heart, coupled to a reversion of molecular and biochemical phenotypes | [36] | |
| TALEN | Human osteosarcoma cells | SOD2 mitochondria localization signal(MLS) modified TALENs | m.14459G>A mutation; common deletion(CD) | Permanent reductions in deletion or point mutant mtDNA in patient–derived cells | [44] |
| MELAS-iPSCs | Platinum Gate TALEN construction system | m.13513G>A mutation | The m.13513G>A heteroplasmy level in MELAS-iPSCs was decreased in the short term by transduction of G13513A-mpTALEN | [64] | |
| Fusions of an osteosarcoma cell line and dermal fibroblasts | hybrid molecules mitoTev-TALEs from T4 phage with MTS | m.8344A>G mutation | Shifting the mtDNA ratio toward the wild type; Improvement of oxidative phosphorylation function | [65] | |
| Fusions of an osteosarcoma cell line and dermal fibroblasts | MLS modified TALENs | m.8344A>G tRNA Lys; m.13513G>A ND5 mutation | Reduction of the levels of the targeted pathogenic mtDNAs; Recovery of respiratory capacity and oxidative phosphorylation enzymes activity | [66] | |
| MELAS-iPSCs; porcine oocytes | MTS modified TALENs | m.3243A>G mutation | elimination of the m.3243A>G mutation in MELAS-iPSCs; reduction in the human m.3243A>G mtDNA mutation in porcine oocytes | [67] | |
| NZB/BALB oocytes; artificial mammalian oocytes | TALENs modified by MTS | NZB mtDNA; m.14459G>A mutation | Preventing the transmission of mitochondrial genomes; Specific reduction of human mutated mitochondrial genomes responsible for mitochondrial diseases in mammalian oocytes | [32] | |
| BTA calli; SW18 which was originally created by asymmetric cell fusion between a rapeseed ‘Westar’ as the recipient and a CMS radish as the donor | MLS modified TALENs | CMS-associated genes (orf79 and orf125) | Knock out of CMS-associated genes (orf79 and orf125), strongly suggesting that these genes are causes of CMS | [47] | |
| E6-E7 gene from the human papilloma virus immortalized MEFs derived from the heteroplasmic tRNAAla m.5024C>T mouse | MLS modified TALENs; Intramuscular, intravenous, and intraperitoneal injections of AAV9-mitoTALE | m.5024C>T mutation | Muscle and heart were efficiently transduced and showed a robust reduction in mutant mtDNA, which was stable over time; The molecular defect, namely a decrease in transfer RNAAla levels, was restored by the treatment | [46] | |
| CRISPR | HEK-293 T | Using MLS to import Cas9 together with gRNA into mitochondria | Cox1; Cox3 | Building a mtDNA mutation cell model | [68] |
| Zebrafish embryo of the AB strain; HEK-293 T | Modifing Cas9 protein with MTS in zebrafish to insert DNA fragments by HR after generating DSB on target site | Human mitochondrial genes ND1 and ND4 and two sites of zebrafish mitochondrial gene Dloop | Reduction mtDNA copy number in both human cells and zebrafish; An exogenous single-stranded DNA arm was knocked into the targeting loci accurately, and could be steadily transmitted to F1 generation of zebrafish | [69] | |
| Clostridium reinhardtii wild-type line (CC-125); Yeast strains: MCC109ρ0, MCC125, CUY563, NB80 | directly introduced the “editing plasmid” into the yeast and chloroplast of Arabidopsis thaliana by microinjection, then inserted DNA fragments by HR after generating DSB on target site | psaA | Confirming donor DNA insertion at the target sites facilitated by homologous recombination only in the presence of Cas9/gRNA activity in yeast mitochondria and Chlamydomonas chloroplasts | [70] |
2.1. Restriction endonuclease (RE)
Restriction endonuclease (RE) is to identify and adhere to specific DNA sequences of nucleotide, then cut the phosphodiester bond at specific site of each DNA strand. RE has the advantages of high recognition accuracy, high cutting efficiency and the ability to cut at certain sites. However, it also has some limitations. For example, it can only identify a limited number of nucleic acid sequences and cannot be designed artificially [30].
RE has the advantage of simple operation process, and is first applied in mitochondrial gene editing [31]. The restriction endonuclease SmaI, modified by additional mitochondrial targeting signals (MTS), can be transferred into the mitochondria after expression in the nuclear, significantly reducing the proportion of mutated mtDNA. In addition, introduced restriction endonuclease ApaLI modified by MTS and 5′ and 3′ UTR sequences of ATP5b into oocytes and single-celled embryos also substantially reduced the proportion of BALB mtDNA [32].These findings demonstrate that the delivery of the engineered endonuclease gene to the corresponding patient is promising in reducing the proportion of mtDNA mutation regarding the RE recognition. However, RE only recognizes limited sequences and cannot meet the repair needs for huge mitochondrial gene mutations, dampening the application of gene editing technology in mitochondria.
2.2. Zinc finger nuclease (ZFN)
ZFN consists of two parts, zinc finger DNA-binding domain which is responsible for recognition of specific DNA sequence, and the restriction nuclease FokI domain which mediates DNA cutting and makes fragment insertion or frameshift mutations of target gene through DNA damage repair mechanism. The zinc-finger DNA binding domain is composed of three groups of zinc-finger proteins, each of which includes about 30 amino acids connected to a single zinc atom and bound to 3 bp of DNA. It should be noted that the cutting domain is non-specific and performs its function only when it exists as a dimer, which requires the design of both 5 'to 3′ and 3 'to 5′ domains [33]. ZFN technology for DNA has been routinely used to design nuclear genomes to add, modify, or destroy genes [34], [35].
Up until now, ZFN technology has been applied to edit mitochondrial genes. MTS and nuclear export signal (NES) are used to help ZFN protein target mitochondria, reducing cytotoxicity caused by damage in nuclear genes to some extent [36], [37], [38], [39], and a pair of monomers targeting mtDNA are designed — one of which binds to the wild-type sequence, while the other binding sequence contains mutation sites. The homologous dimers of nucleic acid enzyme FokI can cut mtDNA and produce linear DNA. Due to the lack of non-homologous end joining (NHEJ) repair pathway towards DNA double strand break in mitochondria, the damaged DNA will degrade [40]. But this method is less efficient and easily off-target. Subsequently, a single-strand ZFN protein with double FokI is designed, which has higher accuracy and efficiency, simplifying the process. However, further studies pointed out that single-strand ZFN protein with dual FokI also has various limitations, such as the inability to identify mutations with deletion of long fragments like common deletion (CD). Meanwhile, the absence of dimer restriction enables this single-stranded ZFN protein to maintain activated nuclease activity, which may have potential cytotoxicity [38]. Gammage et al. attempted to improve the traditional dimer ZFN [38]. They used heterologous rather than homologous dimers to reduce the incidence of off-target, and adjusted the order of NES, label protein and DNA binding proteins in order to reduce the mutual interference between different components, thus improving the stability. This method indeed successfully increased five times of wild-type mtDNA proportion, higher than the efficiency of single ZFN protein with double FokI editing (wild type mtDNA proportion increased two times). It also enables the tool to identify CD — two monomers designed to identify wild-type sequences on each side of the deletion sequence. When deletion mutations occur, the heterodimer FokI is close enough to perform nuclease activity.
Due to the extremely high demand for mitochondria in muscle, brain cells and the specific symptoms appearing when the mitochondrial gene mutation reaches a certain proportion, such as movement disorders, retinitis pigmentosa, cell models which contains mitochondrial mutations associated with these diseases are selected for further experiments [36]. After knocked out by mtZFN, the proportion of mutated genes in mitochondria was significantly reduced, which brought hope for the gene therapy of mitochondrial diseases. However, at the same time, the number of normal genes rather than mutated mitochondrial genes also showed partially decrease. In addition, the use of mtZFN in cells with high levels of mitochondrial gene mutation may cause significantly decrease of mitochondria number which may trigger cell death [41]. Hence, they tried to reduce the knockout efficiency and use iterative knockout strategies. They also attempted to use hammer head shape ribozymes (HHR) tools to control the expression level of mtZFN. Apart from proof in the genetic level, the mutant gene knockout was also verified in organelle and cell level, ATP production capacity and energy charge were improved to a certain extent after modifying the mtZFN. The amount of citric acid and aconitic acid, which are two important products in mitochondrial metabolism, had also promoted, further illustrating the effectiveness of mtZFN [42].
In order to further verify the feasibility of mtZFN, in vivo studies were conducted [36]. Mice model with Cardiac myocyte m.5024C>T tRNA (Ala) mutant was selected for the experiment [36]. After associated virus tail intravenous injection, mtZFN expression was confirmed in heart cell, and the level of pyruvic acid and aspartic acid rose while lactic acid level dropped [36]. This showed that the ratio of mitochondrial respiratory energy generation increased in cell energy supply while that of the glycolytic pathway was reduced, thus proving the validity of the mtZFN in vivo [36].
However, mtZFN has a rather low editing efficiency while iterative editing strategy takes a long time, because it may take about a week to carry out mtZFN for once. Besides, each of zinc-finger protein group which includes about 30 amino acids connected to a single zinc atom will bind to 3 bp of DNA, which may cause the potential inaccuracy.
2.3. Transcription activator-like effectors nuclease (TALEN)
TALEN is a gene targeted manipulation technique derived from plant pathogenic bacteria. The amino acid sequence of the DNA binding domain in TAL protein which derived from plant bacteria Xanthomonas sp has a strong correspondence with the nucleic acid sequence of the target site [43]. One module unit recognizes one base [44], which is simple and highly specific. By combining various modules, the target specific knockout or endogenous gene expression regulation can be performed on any nucleotide sequence [44]. So far, it has been successfully applied in many species such as human, rat, mouse, pig, sheep, zebrafish, Arabidopsis and yeast [43]. Its basic principle is roughly the same as that of ZFN, which is also composed of recognition domain and cleavage domain [44]. Cleavage domain also consists of non-specific nuclease FokI, which only plays an effect in the formation of dimers [43]. However, its DNA recognition domain is different, which is composed of TAL proteins in series (generally about 20) [44]. Each TAL protein recognizes and binds to a corresponding base, which ensures its higher specificity [45].
TALEN technology has also been used in mitochondrial gene editing [46]. MTS of superoxide dismutase (SOD) is modified on the TALEN monomer protein amino end, so that TALEN related proteins could be targeted into mitochondria to play effect [46]. Since the nucleic acid enzyme FokI plays its effect only in dimers, this group focused on fragment missing mutation in mtDNA. They designed TALEN DNA identification areas on both sides of the missing fragment, so that when the loss of fragment occurred, FokI can be close enough to form dimers and play the cutting effect of nuclease. This resulted in the degradation of mutated mtDNA and the transformation of mtDNA heteroplasmy to wild type. In addition, monomer protein chain from TALEN is designed to recognize m.5024C>T mutation in order to achieve the purpose of knockout [47]. In addition, the latest study reported that two kind of proteins in TALEN could be replaced by one protein — compact TALEN (cTALEN) [48]. In addition, mito-TALEN is also applied to block the intergenerational transmission of mitochondrial diseases [32]. mito-TALEN is also used to reduced human mutated mtDNA levels responsible for LHON, and NARP in mammalian oocytes. Due to the characteristics of matrilineal inheritance in mitochondria and the inability to reproduce in the early stage of embryo development, the number of mitochondria in oocytes was greatly reduced, which affected implantation in the uterus. Therefore, mito-TALEN provides a novel insight for blocking the intergenerational transmission of mitochondrial diseases, avoiding the operational complexity and ethical issues associated with the “three-parent baby” [49].
Based on TALE technology, DddA (Double-stranded DNA Deaminase toxin A), which can catalyze the conversion of cytosine (C) into uracil (U) on DNA double strand but is toxic to mammalian cells, was divided into two inactive halves called split-DddA [50]. They fused split-DddA with the TALE protein and uracil glycosylase inhibitors to assemble an RNA free DddA derived cytosine base editor (DdCBE), which catalyzed the conversion of C•G to T•A in human mtDNA, and they proved its high targeting specificity and editing accuracy [50]. Only when the two TALE proteins are combined with mtDNA, will the two split-DddA semi molecules reassemble together, restoring the activity of the catalytic base editing [50]. Notably, the use of UGI can protect U from the effect of glycosylase, so that it will avoid being recognized by DNA damage repair mechanism which will make it cut off from DNA and replaced by C [51]. It is found that the addition of UGI increased the editing efficiency by approximately 8-fold [50]. Most interestingly, the system with MTS can also accomplish the accurate editing of mtDNA, providing an unprecedented tool for the study of mitochondrial genetic diseases [50], [52]. In addition, mitochondrial DNA mutation model related to diseases in human cells can be successfully constructed by using this system [50]. However, The DdCBE system relying on DddA can only effectively edit the C base next to T in the genome, which has great limitations [50].
The DNA binding domain in TAL protein has a one-to-one correspondence with the nucleic acid sequence of the target site, which ensures the high accuracy. However, this also brings mito-TALEN excessive molecular weight, which builds a strong barrier for virus packaging, cell and mitochondria import.
2.4. CRISPR system
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated(Cas)protein system was originally discovered in bacteria and archaea. It is an immune defense system formed in the course of long-term evolution to resist invading viruses and other foreign DNA invasions [53], [54]. CRISPR is composed of many short and conservative repeat sequence regions (Repeats) and spacer regions (Spacers) [45]. Repeats contain palindrome sequence, which can form hairpin structure, while spacers, foreign DNA sequences captured by bacteria, are special [54], [55]. The upstream leader is considered to be the promoter of CRISPR sequence [54], [55]. A polymorphic family gene Cas exists at upstream of the sequence [54], [55]. When foreign DNA invades, the Cas protein will play an effect to cut it into short sequences, and then be inserted as a spacer between the conserved repeat sequences, which contains palindrome sequences, which can form hairpin structures [54], [55]. When the same foreign DNA invades again, the spacer will be used as a template to transcribe CRISPR RNA (crRNA), which combines with transactivating crRNA (tracrRNA) to guide Cas to cut the invading foreign DNA [45]. It should be noted that identifying a specific sequence and mediating the cleavage also needs to satisfy the presence of protospacer adjacent motif (PAM) after the homologous sequence, which limits the sites available for editing. In fact, as a gene editing tool, the CRISPR/CAS system itself mediates only DNA double-strand breaks (DSB), and subsequent cell DNA repair completes gene editing at specific sites, which includes nonhomologous end-joining (NHEJ) and homology-directed repair (HDR). NHEJ will add or delete a few bases to the DSB site during repair, thereby causing the gene to produce frameshift mutations, achieving the purpose of knocking out the gene, while HDR mediates error-free DNA when there is a homologous donor DNA as a template for repair and insertion of foreign fragments.
According to the sequence and structure of the Cas protein, CRISPR/Cas system is divided into three types: I, II, and III. Among them, the most widely used is the type II CRISPR/Cas system that only requires one Cas protein, Cas9, because of the ability of Cas9 to co-locate RNA, DNA and the tremendous transformation potential [56]. In actual use, guide RNA (gRNA), which includes the necessary structural components of crRNA and tracrRNA, is more convenient and efficient. The CRISPR/Cas system has shown great potential in many application fields, such as genome editing, transcription regulation, virus detection, etc. The CRISPR/Cas9 system has been proven to edit the genomes of mammals, zebrafish, mice, fruit flies and other animals. Additionally, catalytically defective Cas9 (dCas9) can bind to a protein domain that activates or inhibits transcription, and can perform precise and stable transcriptional control of target genes under the guidance of gRNA [57]. Furthermore, the CRISPR/Cas13-based SHERLOCK technology can effectively detect the virus and is also suitable for the detection of the 2020 new coronavirus in a short period of time [58].
Rapid progress has been made using CRISPR system in nuclear gene editing. However, for relatively smaller operation objects such as mtDNA, it is difficult to operate the CRISPR Cas9 system. Moreover, mitochondria lack the ability to import RNA, which prevents the gRNA-Cas9 complex from recognizing the targeted sequence. How to introduce this RNA into mitochondria, and how to ensure the successful orientation to mutated gene after designing gRNA? All these problems limit the feasibility and effectiveness of the application of CRISPR cas9 system in mitochondrial gene editing.
Generally, TALEN is superior to ZFN as it no longer needs to recognize DNA sequences through base triplets, and the biunique recognition gives higher specificity and freedom in choosing target sequences. Besides, TALEN also has the advantages of simpler operation and shorter cycle. The most important part in gene editing is DNA binding. ZFN and TALEN both utilize protein as a guide to achieve targeted editing which needs complex and time-consuming protein engineering, while CRISPR-Cas system is led by gRNA which is relatively cheap and easy to design and produce. In the field of mitochondrial gene editing, what should be considered is the ability of RNA and protein to get through mitochondrial membrane, the size of the occupied space and the operability of transportation into mitochondrial membrane.
Recently, a new type of CRISPR system, CRISPR-Cpf1 system [59], was discovered and defined as the second generation of gene editing technology [59]. It replaces Cas9 protein with Cpf1 protein, which has not only the similar genome editing efficiency as Cas9 [60], but also more advantages than Cas9. Cpf1 system merely needs one RNA for gene editing [59], and it can process precursor CRISPR-derived RNA (pre-crRNA) by itself, then target and cleave DNA specifically by using processed crRNA [61]. Cpf1 enzyme is also smaller than standard Cas9 and is easier to transmit to cells and tissues [59]. When cutting DNA, Cpf1 system formed sticky end instead of flat end, which has more accuracy and lower off-target effect, though there is still a certain off-target effects in this system [60]. Besides, Cpf1 incision is far away from the recognition site, which means that it can still play an effect even the target gene changes at the cleavage site, providing multiple opportunities to complete the editing [61]. Cpf1 can guide RNAs to edit multiple genes or multiple sites of a single gene at the same time, improving the efficiency of gene knockout, which is in favor of editing many proteins at the same time in combination screening [62]. It expands the selection range of gene editing target sites further by using a simple transport system to command CRISPR effector protein and guide RNAs [61], and it has been widely considered with great potential in gene therapy applications.
Although Cpf1 is superior to Cas9, it has a lower activity, and due to the relative fixation of PAM sequence, its target selection is limited [63]. It remains to be explored on how to improve the activity and expand the target range to have a broader gene editing effect. As a new gene editing technology, the operation of the CRISPR-Cpf1 system is simpler and more accurate, which can also achieve more effective editing for the small mitochondrial genes, but the application of this technology to mitochondrial gene editing has not been reported yet. In the future, CRISPR-Cpf1 system may become an effective way for mitochondrial gene editing.
3. Mitochondrial gene editing by CRISPR Cas9 system
It was first reported in 2015 that CRISPR/cas9 system was successfully applied to mitochondrial gene editing [70]. However, the findings were controversy and was also questioned by another group [71].
Firstly, the mechanism of the introduction of gRNA to mitochondria remains elusive. The 20 ribonucleotides stem ring series come from the H1 RNA (RNase P enzyme RNA component) [72], which is attached to the injected cytoplasmic RNA, can successfully target RNA into mitochondria. This method is effective for both the non-coding RNA, such as tRNA, and mRNA that encodes proteins, which shed light on importing gRNA to mitochondria in CRISPR /Cas9 system. Gammage et al. summarized the existing RNA import theory and its refute [71]. They denied the existence of RNA components in mtRNase P and believed that RNase P and RNase MRP mainly plays a role in the nucleus. They also questioned the role of mitochondrial ribosomes in assisting rRNA and PNPase in cytoplasmic RNA transport [56]. It has also been reported that two domains of yeast cytoplasmic tRNALys(CUU), F-arm and D-hairpin [73], as well as some domains of 5S RNA, may also help RNA to enter mitochondria [74].
Due to the lack of NHEJ repair pathway while the existence of homologous recombination (HR) repair pathway in mitochondria [75], DNA sequence insertion in mtDNA was attempted and succeeded in human and zebrafish mitochondria [69], yeast mitochondria and Arabidopsis chloroplasts [70], utilizing the CRISPR- Cas9 system to produce double strand break (DSB) and exogenous DNA to support homologous fragments. In the past, the mitochondria penetration of DNA faced great difficulties, because of the membrane potential, pH and the phagocytosis of bacteriophages [76]. However foreign DNA has been successfully introduced into mitochondria of yeast by biolistic bombardment of cells with DNA-coated tungsten particles, electroporation, DQAsome complexes and TAT [76]. Another study utilizes MTS to modify adeno associated virus capsid VP2 and succeeds in transporting ND4 gene into mitochondria, which provides a new method for mitochondrial delivery of DNA sequence [75]. Other mitochondria delivery systems for drugs or proteins are developed through metal–organic framework (MOF) or triphenylphosphonium (TPP) and cell-penetrating poly(disulfide)s (CPD) modified biodegradable silica nanoparticles (BS–NPs), which are potential for mitochondria delivery of exogenous DNA or RNA component [77], [78].
In the study of Bian et al, they modified Cas9 with MTS and transcript in vitro, then microinjected gRNA and product of in-vitro transcription into cytoplasm. They also introduced exogenous DNA vector containing homologous arm by ViaFect transfection [69]. Surprisingly, exogenous ssDNA with short homologous arm was knocked into the targeting loci accurately, and this mutagenesis could be steadily transmitted to F1 generation of zebrafish, which indicates ssDNA and gRNA can be transported into mitochondria without modification and is consistent with the recent report published by others [79]. Then mito-Cas9 protein can specifically target mtDNA and reduce mtDNA copy number in both human cells and zebrafish [69].
Another group of researchers introduce “Edit Plasmids”, which contains Cas9 gene, gRNA express sequence and homologous sequence, into yeast mitochondria and Chlamydomonas chloroplasts by microprojectile transformation method [70]. The result shows donor DNA insertion at the target sites facilitated by homologous recombination, which is similar with the work in zebrafish [69].
However, the existence of an endogenous mechanism for nucleic acid import into mammalian mitochondria, a prerequisite for mitochondrial CRISPR/Cas9 gene editing, remains controversial [71].
4. pAgo-based system
Argonaute proteins were first identified in eukaryotes, but homologous prokaryotic Argonaute proteins (pAgos) have also been found in archaea and bacteria [80]. Argonaute proteins constitute a highly diverse family of nucleic acid-guided proteins and exhibit the potential in genome editing [81]. Eukaryotic Argonaute proteins mediate RNA-guided RNA interference while pAgos generally utilize DNA guides to target complementary DNA sequences to protect their hosts against invading DNA [82], [83]. For example, the cyanobacterium Synechococcus elongates (SeAgo) is a DNA-guided nuclease preferentially acting on single-stranded DNA targets, with non-specific guide-independent activity observed for double-stranded substrates [84]. Clostridium butyricum (CbAgo) targets multicopy genetic elements and induces DNA interference between homologous sequences and triggers DNA degradation. The loading of CbAgo with small DNA guides depends on both its intrinsic endonuclease activity and the double-strand break repair machinery [85]. Interestingly, both Methanocaldococcus jannaschii (MjAgo) and Thermus thermophilus (TtAgo) possess two modes of action: the canonical guide-dependent endonuclease activity and a non-guided DNA endonuclease activity [86], [87], [88], [89], [90], [91]. In addition, CbAgo and Limnothrix rosea (LrAgo) can act as DNA-guided DNA nucleases at physiological temperatures, expanding their potential use as a tool in genomic applications [92], [93]. In particular, some pAgos like Rhodobacter sphaeroides (RsAgo) acquire guides from plasmid-derived mRNA to interfere the plasmid DNA, reducing transcription of reporter genes and plasmid content [81], [83], [94].
Recently, several pAgos have been identified to enhance the positive selection process of RecA mediated DNA strand exchange through the interaction of its PIWI domain and recombinase A (RecA), which guides the homologous recombination of target sequences [82,95]. Due to the fact that mitochondrial DNA maintain renewal and replication, it is reasonable to introduce pAgos homologous arms to target the mutation sequence in mitochondria. The homologous arm should be designed as a normal mtDNA sequence with a certain length to guide the transformation of mutated DNA sequence to normal DNA in a new round of replication process. As long as the mutated DNA is transformed into normal DNA, the damaged mitochondria may be rescued and recovered. However, exploiting pAgo-based system to edit mitochondrial DNA requires further explored (Fig. 2D).
5. Conclusions
The collective findings from the studies reviewed herein the main methods for mitochondrial gene editing. So far, though the traditional gene editing methods such as RE, ZFN and TALEN have made some progress in mitochondrial gene editing, a complete, specific and systematic operation technology of mitochondrial gene editing has not been established yet. Using modified CRISPR/Cas9 system has partially solved the problem of gene orientation, but there also exists certain defects. Until now, there is no sufficient and reasonable experimental evidence to support the conclusion. However, based on these studies, researchers can try to improve CRISPR/Cas9 and other gene editing techniques, like pAgo-based system, to edit specific mitochondrial genes. Whether the abnormal mitochondria are cleared after the adapted CRISPR/Cas9 system breaks mtDNA, or the mutant mtDNA fragments are repaired directly by pAgo-based gene editing techniques to restore the normal function of the mitochondria, the mitochondrial dysfunction will be well rescued. If the problems of inefficient delivery of guide RNA and Cas9 enzyme complexes into the mitochondria and low editing specificity can be ameliorated. The adapted CRISPR/Cas9 system and the pAgo-based gene editing system may become the most promising ways for mitochondrial gene editing. These will surely provide insights and approaches for the diagnosis and treatment of inherited mitochondrial disorder in the future.
6. Author statement
Y. Xiang designed and supervised the study; X. Y., J.J. and Z.L. performed the comprehensively references investigation and analysis. Y. Xiang, J. L, X. Y., J.J. and Z.L. wrote the manuscripts. Y., J.J. and Z.L. contributed equally to this work.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Y. Xiang received support from National Outstanding Youth Science Fund Project of National Natural Science Foundation of China (81822048), Fundamental Research Funds for the Central Universities of China (22120200064), and the Frontier Science Research Center for Stem Cells, Ministry of Education.
References
- 1.Schapira A.HV. Mitochondrial diseases. Lancet. 2012;379(9828):1825–1834. doi: 10.1016/S0140-6736(11)61305-6. [DOI] [PubMed] [Google Scholar]
- 2.DiMauro S., Schon E.A. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348(26):2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 3.Ghelli A., Zanna C., Porcelli A.M., Schapira A.H.V., Martinuzzi A., Carelli V. Leber's hereditary optic neuropathy (LHON) pathogenic mutations induce mitochondrial-dependent apoptotic death in transmitochondrial cells incubated with galactose medium. J Biol Chem. 2003;278(6):4145–4150. doi: 10.1074/jbc.M210285200. [DOI] [PubMed] [Google Scholar]
- 4.Wallace DC. Mitochondrial diseases in man and mouse. Science (1999). [DOI] [PubMed]
- 5.Allen J.F., Raven J.A., Andersson G.E., Karlberg O., Canbäck B., Kurland C.G. On the origin of mitochondria: a genomics perspective. Philos Trans R Soc Lond. 2003;358(1429):165–179. doi: 10.1098/rstb.2002.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H.L., Coulson A.R., Drouin J. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 7.Barshad G., Marom S., Cohen T., Mishmar D. Mitochondrial DNA transcription and its regulation: an evolutionary perspective. Trends Genet. 2018;34(9):682–692. doi: 10.1016/j.tig.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Barchiesi A., Vascotto C. Transcription, processing, and decay of mitochondrial RNA in health and disease. Int J Mol Sci. 2019;20(9):2221. doi: 10.3390/ijms20092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chinnery P.F., Turnbull D.M. Mitochondrial DNA and disease. Lancet. 1999;354(SI):S17–S21. doi: 10.1016/s0140-6736(99)90244-1. [DOI] [PubMed] [Google Scholar]
- 10.DiMauro S., Davidzon G. Mitochondrial DNA and disease. Ann Med. 2005;37(3):222–232. doi: 10.1080/07853890510007368. [DOI] [PubMed] [Google Scholar]
- 11.Greaves L.C., Reeve A.K., Taylor R.W., Turnbull D.M. Mitochondrial DNA and disease. J Pathol. 2012;226:274–286. doi: 10.1002/path.3028. [DOI] [PubMed] [Google Scholar]
- 12.Suomalainen A., Battersby B.J. Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nat Rev Mol Cell Biol. 2018;19(2):77–92. doi: 10.1038/nrm.2017.66. [DOI] [PubMed] [Google Scholar]
- 13.Taylor R.W., Turnbull D.M. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6(5):389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelnena et al. Complete mtDNA sequencing reveals mutations m.9185T > C and m.13513G > A in three patients with Leigh syndrome. [DOI] [PubMed]
- 15.Yanping Mitochondrial DNA mutations in late-onset Leigh syndrome. J Neurol. 2018 doi: 10.1007/s00415-018-9014-5. [DOI] [PubMed] [Google Scholar]
- 16.Sofou K., de Coo I.F.M., Ostergaard E., Isohanni P., Naess K., De Meirleir L. Phenotype-genotype correlations in leigh syndrome: New insights from a multicentre study of 96 patients. J Med Genet. 2018;55(1):21–27. doi: 10.1136/jmedgenet-2017-104891. [DOI] [PubMed] [Google Scholar]
- 17.Mordel P. A 2 bp deletion in the mitochondrial ATP 6 gene responsible for the NARP (neuropathy, ataxia, and retinitis pigmentosa) syndrome. Biochem Biophys Res Commun. 2017 doi: 10.1016/j.bbrc.2017.10.066. S0006291X17320442. [DOI] [PubMed] [Google Scholar]
- 18.Kytvuori L. A novel mutation m.8561C>G in MT-ATP6/8 causing a mitochondrial syndrome with ataxia, peripheral neuropathy, diabetes mellitus, and hypergonadotropic hypogonadism. J Neurol. 2016;263:2188–2195. doi: 10.1007/s00415-016-8249-2. [DOI] [PubMed] [Google Scholar]
- 19.Carelli V., Carbonelli M., Coo I.F.D., Kawasaki A., Barboni P. International consensus statement on the clinical and therapeutic management of leber hereditary optic neuropathy. J Neuro-Ophthalmol. 2017;37:371. doi: 10.1097/WNO.0000000000000570. [DOI] [PubMed] [Google Scholar]
- 20.Wallace D., Singh G., Lott M., Hodge J., Schurr T., Lezza A. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science. 1988;242(4884):1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 21.Hansen K.G., Herrmann J.M. Transport of proteins into mitochondria. Protein J. 2019;38(3):330–342. doi: 10.1007/s10930-019-09819-6. [DOI] [PubMed] [Google Scholar]
- 22.Araiso Y., Tsutsumi A., Qiu J., Imai K., Shiota T., Song J. Structure of the mitochondrial import gate reveals distinct preprotein paths. Nature. 2019;575(7782):395–401. doi: 10.1038/s41586-019-1680-7. [DOI] [PubMed] [Google Scholar]
- 23.Becker T., Song J., Pfanner N. Versatility of preprotein transfer from the cytosol to mitochondria. Trends Cell Biol. 2019;29(7):534–548. doi: 10.1016/j.tcb.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt S., Strub A., Voos W. Protein translocation into mitochondria. Biol Signals Receptors. 2001;10(1-2):14–25. doi: 10.1159/000046873. [DOI] [PubMed] [Google Scholar]
- 25.Tucker K., Park E. Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution. Nat Struct Mol Biol. 2019;1–9 doi: 10.1038/s41594-019-0339-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Callegari, S., Cruz-Zaragoza, L. D. & Rehling, P. From TOM to the TIM23 complex – handing over of a precursor. Biol Chem 401, 709-721, doi:doi:10.1515/hsz-2020-0101 (2020). [DOI] [PubMed]
- 27.Neupert W., Herrmann J.M. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76(1):723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 28.McCully J.D., Cowan D.B., Emani S.M., del Nido P.J. Mitochondrial transplantation: From animal models to clinical use in humans. Mitochondrion. 2017;34:127–134. doi: 10.1016/j.mito.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Yamada Y., Ito M., Arai M., Hibino M., Tsujioka T., Harashima H. Challenges in promoting mitochondrial transplantation therapy. Int J Mol Sci. 2020;21(17):6365. doi: 10.3390/ijms21176365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pingoud, A., Wilson, G. G. & Wende, W. Type II restriction endonucleases — a historical perspective and more. Nucleic Acids Res 44, 8011-8011 (2016). [DOI] [PMC free article] [PubMed]
- 31.Tanaka M. Gene therapy for mitochondrial disease by delivering restriction endonuclease SmaI into mitochondria. J Biomed Sci. 2002;9:534–541. doi: 10.1159/000064726. [DOI] [PubMed] [Google Scholar]
- 32.Reddy, P. et al. Selective Elimination of Mitochondrial Mutations in the Germline by Genome Editing. (2015). [DOI] [PMC free article] [PubMed]
- 33.Mani M., Kandavelou K., Dy F.J., Durai S., Chandrasegaran S. Design, engineering, and characterization of zinc finger nucleases. Biochem Biophys Res Commun. 2005;335(2):447–457. doi: 10.1016/j.bbrc.2005.07.089. [DOI] [PubMed] [Google Scholar]
- 34.Papworth M., Kolasinska P., Minczuk M. Designer zinc-finger proteins and their applications. Gene. 2006;366(1):27–38. doi: 10.1016/j.gene.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 35.Sundar Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005 doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gammage P.A., Viscomi C., Simard M.-L., Costa A.S.H., Gaude E., Powell C.A. Genome editing in mitochondria corrects a pathogenic mtDNA mutation in vivo. Nat Med. 2018;24(11):1691–1695. doi: 10.1038/s41591-018-0165-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michal M., Papworth M.A., Miller J.C., Murphy M.P., Aaron K. Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA. Nucleic Acids Res. 2008;36:3926–3938. doi: 10.1093/nar/gkn313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gammage P.A., Rorbach J., Vincent A.I., Rebar E.J., Minczuk M. Mitochondrially targeted ZFNs for selective degradation of pathogenic mitochondrial genomes bearing large-scale deletions or point mutations. EMBO Mol Med. 2014;6:458–466. doi: 10.1002/emmm.201303672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minczuk M, Papworth MA, Kolasinska P, Murphy MP, Klug, A. Sequence-specific modification of mitochondrial DNA using a chimeric zinc finger methylase. [DOI] [PMC free article] [PubMed]
- 40.Alexeyev M, Shokolenko I, Wilson G, Ledoux S. The maintenance of mitochondrial DNA integrity--critical analysis and update. Cold Spring Harbor Persp Biol 5, a012641 (2013). [DOI] [PMC free article] [PubMed]
- 41.Gammage PA. et al. Near-complete elimination of mutant mtDNA by iterative or dynamic dose-controlled treatment with mtZFNs. (2016). [DOI] [PMC free article] [PubMed]
- 42.Bacman S.R., Williams S.L., Pinto M., Peralta S., Moraes C.T. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat Med. 2013;19(9):1111–1113. doi: 10.1038/nm.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bacman S.R., Williams S.L., Pinto M., Peralta S., Moraes C.T. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat Med. 2013;13 doi: 10.1038/nm.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaj T., Gersbach C.A., Barbas C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bacman S.R., Kauppila J.H.K., Pereira C.V., Nissanka N., Miranda M., Pinto M. MitoTALEN reduces mutant mtDNA load and restores tRNAAla levels in a mouse model of heteroplasmic mtDNA mutation. Nat Med. 2018;24(11):1696–1700. doi: 10.1038/s41591-018-0166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kazama T., Okuno M., Watari Y., Yanase S., Koizuka C., Tsuruta Y.u. Curing cytoplasmic male sterility via TALEN-mediated mitochondrial genome editing. Nat Plants. 2019;5(7):722–730. doi: 10.1038/s41477-019-0459-z. [DOI] [PubMed] [Google Scholar]
- 47.Hsu P., Lander E., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leslie, M. 'Old' genome editors might treat mitochondrial diseases. ence 361, 1302 (2018). [DOI] [PubMed]
- 49.Mok B.Y., de Moraes M.H., Zeng J., Bosch D.E., Kotrys A.V., Raguram A. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature. 2020;583(7817):631–637. doi: 10.1038/s41586-020-2477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aushev M., Herbert M. Mitochondrial genome editing gets precise. Nature. 2020;583(7817):521–522. doi: 10.1038/d41586-020-01974-6. [DOI] [PubMed] [Google Scholar]
- 52.Mali P., Esvelt K.M., Church G.M. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10(10):957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 54.Horvath P., Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Ence. 2010;327(5962):167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 55.Wang G., Chen H.-W., Oktay Y., Zhang J., Allen E.L., Smith G.M. PNPASE regulates RNA import into mitochondria. Cell. 2010;142(3):456–467. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anzalone A.V., Koblan L.W., Liu D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol. 2020;38(7):824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 57.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14(10):2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zetsche B., Gootenberg J., Abudayyeh O., Slaymaker I., Makarova K., Essletzbichler P. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163(3):759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lei, C. et al. The CCTL (Cpf1-assisted Cutting and Taq DNA ligase-assisted Ligation) method for efficient editing of large DNA constructs in vitro. Nucleic Acids Res, e74-e74 (2017). [DOI] [PMC free article] [PubMed]
- 60.Zetsche B., Heidenreich M., Mohanraju P., Fedorova I., Kneppers J., DeGennaro E.M. Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35(1):31–34. doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhong G., Wang H., Li Y., Tran M.H., Farzan M. Cpf1 proteins excise CRISPR RNAs from mRNA transcripts in mammalian cells. Nat Chem Biol. 2017;13(8):839–841. doi: 10.1038/nchembio.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haihua SaCas9 requires 5'-NNGRRT-3' PAM for sufficient cleavage and possesses higher cleavage activity than SpCas9 or FnCpf1 in human cells. Biotechnol J. 2018 doi: 10.1002/biot.201800080. [DOI] [PubMed] [Google Scholar]
- 63.Yahata N, Matsumoto Y, Omi M, Yamamoto N, Hata R. TALEN-mediated shift of mitochondrial DNA heteroplasmy in MELAS-iPSCs with m.13513G>A mutation. Sci Rep 7, 15557, doi: 10.1038/s41598-017-15871-y (2017). [DOI] [PMC free article] [PubMed]
- 64.Pereira C.V. mitoTev-TALE: a monomeric DNA editing enzyme to reduce mutant mitochondrial DNA levels. EMBO Mol Med. 2018;10 doi: 10.15252/emmm.201708084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hashimoto M., Bacman S.R., Peralta S., Falk M.J., Chomyn A., Chan D.C. MitoTALEN: a general approach to reduce mutant mtDNA loads and restore oxidative phosphorylation function in mitochondrial diseases. Mol Therapy J Am Soc Gene Therapy. 2015;23(10):1592–1599. doi: 10.1038/mt.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Y.i., Wu H., Kang X., Liang Y., Lan T., Li T. Targeted elimination of mutant mitochondrial DNA in MELAS-iPSCs by mitoTALENs. Protein Cell. 2018;9(3):283–297. doi: 10.1007/s13238-017-0499-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jo A., Ham S., Lee G.H., Lee Y.-I., Kim S., Lee Y.-S. Efficient mitochondrial genome editing by CRISPR/Cas9. Biomed Res Int. 2015;2015:1–10. doi: 10.1155/2015/305716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bian W.-P., Chen Y.-L., Luo J.-J., Wang C., Xie S.-L., Pei D.-S. Knock-in strategy for editing human and zebrafish mitochondrial DNA using mito-CRISPR/Cas9 system. ACS Synth Biol. 2019;8(4):621–632. doi: 10.1021/acssynbio.8b00411. [DOI] [PubMed] [Google Scholar]
- 69.Yoo B.C., Yadav N.S., Orozco E.M., Sakai H. Cas9/gRNA-mediated genome editing of yeast mitochondria and Chlamydomonas chloroplasts. PeerJ. 2020;8 doi: 10.7717/peerj.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gammage PA, Moraes CT, Minczuk M. Mitochondrial genome engineering: the revolution may not Be CRISPR-Ized. (2018). [DOI] [PMC free article] [PubMed]
- 71.G. et al. Correcting human mitochondrial mutations with targeted RNA import. Proc Natl Acad Ences 109 (2012). [DOI] [PMC free article] [PubMed]
- 72.Tonin Y., Heckel A.-M., Vysokikh M., Dovydenko I., Meschaninova M., Rötig A. Modeling of antigenomic therapy of mitochondrial diseases by mitochondrially addressed RNA targeting a pathogenic point mutation in mitochondrial DNA. J Biol Chem. 2014;289(19):13323–13334. doi: 10.1074/jbc.M113.528968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smirnov A., Tarassov I., Mager-Heckel A.-M., Letzelter M., Martin R.P., Krasheninnikov I.A. Two distinct structural elements of 5S rRNA are needed for its import into human mitochondria. RNA (New York) 2008;14(4):749–759. doi: 10.1261/rna.952208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu H., Koilkonda R.D., Chou T.-H., Porciatti V., Ozdemir S.S., Chiodo V. Gene delivery to mitochondria by targeting modified adenoassociated virus suppresses Leber's hereditary optic neuropathy in a mouse model. Proc Natl Acad Sci U S A. 2012;109(20):E1238–E1247. doi: 10.1073/pnas.1119577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Torchilin V.P. Recent approaches to intracellular delivery of drugs and DNA and organelle targeting. Annu Rev Biomed Eng. 2006;8(1):343–375. doi: 10.1146/annurev.bioeng.8.061505.095735. [DOI] [PubMed] [Google Scholar]
- 76.Yuan P., Mao X., Wu X., Liew S.S., Li L., Yao S.Q. Mitochondria-targeting, intracellular delivery of native proteins using biodegradable silica nanoparticles. Angew Chem Int Ed. 2019;131(23):7739–7743. doi: 10.1002/anie.201901699. [DOI] [PubMed] [Google Scholar]
- 77.Haddad S., Abánades Lázaro I., Fantham M., Mishra A., Silvestre-Albero J., Osterrieth J.W.M. Design of a functionalized metal-organic framework system for enhanced targeted delivery to mitochondria. J Am Chem Soc. 2020;142(14):6661–6674. doi: 10.1021/jacs.0c00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Loutre R., Heckel A.M., Smirnova A., Entelis N., Tarassov I. Can mitochondrial DNA be CRISPRized: pro and contra: can mitochondrial DNA be crisprized. Int Union Biochem Mol Biol Life. 2018;70 doi: 10.1002/iub.1919. [DOI] [PubMed] [Google Scholar]
- 79.Swarts DC. et al. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res 43, 5120-5129, doi:10.1093/nar/gkv415 (2015). [DOI] [PMC free article] [PubMed]
- 80.Hegge J.W., Swarts D.C., van der Oost J. Prokaryotic Argonaute proteins: novel genome-editing tools? Nat Rev Microbiol. 2018;16(1):5–11. doi: 10.1038/nrmicro.2017.73. [DOI] [PubMed] [Google Scholar]
- 81.Swarts D.C. Prokaryotic argonautes function beyond immunity by unlinking replicating chromosomes. Cell. 2020;182(6):1381–1383. doi: 10.1016/j.cell.2020.08.037. [DOI] [PubMed] [Google Scholar]
- 82.Lisitskaya L., Aravin A.A., Kulbachinskiy A. DNA interference and beyond: structure and functions of prokaryotic Argonaute proteins. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-07449-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olina A. et al. Genome-wide DNA sampling by Ago nuclease from the cyanobacterium Synechococcus elongatus. [DOI] [PMC free article] [PubMed]
- 84.Kuzmenko A., Oguienko A., Esyunina D., Yudin D., Petrova M., Kudinova A. DNA targeting and interference by a bacterial Argonaute nuclease. Nature. 2020;587(7835):632–637. doi: 10.1038/s41586-020-2605-1. [DOI] [PubMed] [Google Scholar]
- 85.Zander A., Willkomm S., Ofer S., van Wolferen M., Egert L., Buchmeier S. Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii. Nat Microbiol. 2017;2(6) doi: 10.1038/nmicrobiol.2017.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Swarts D.C., Jore M.M., Westra E.R., Zhu Y., Janssen J.H., Snijders A.P. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014;507(7491):258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Swarts D.C., Szczepaniak M., Sheng G., Chandradoss S.D., Zhu Y., Timmers E.M. Autonomous generation and loading of DNA guides by bacterial argonaute. Mol Cell. 2017;65(6):985–998.e6. doi: 10.1016/j.molcel.2017.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Willkomm S., Oellig C.A., Zander A., Restle T., Keegan R., Grohmann D. Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein. Nat Microbiol. 2017;2(6) doi: 10.1038/nmicrobiol.2017.35. [DOI] [PubMed] [Google Scholar]
- 89.Sashital D.G. Prokaryotic argonaute uses an all-in-one mechanism to provide host defense. Mol Cell. 2017;65(6):957–958. doi: 10.1016/j.molcel.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 90.Jolly S.M., Gainetdinov I., Jouravleva K., Zhang H., Strittmatter L., Bailey S.M. Thermus thermophilus argonaute functions in the completion of DNA replication. Cell. 2020;182(6):1545–1559.e18. doi: 10.1016/j.cell.2020.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kuzmenko A., Yudin D., Ryazansky S., Kulbachinskiy A., Aravin A.A. Programmable DNA cleavage by Ago nucleases from mesophilic bacteria Clostridium butyricum and Limnothrix rosea. Nucleic Acids Res. 2019;47:5822–5836. doi: 10.1093/nar/gkz379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hegge JW. et al. DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute. Nucleic Acids Res 47, 5809-5821, doi:10.1093/nar/gkz306 (2019). [DOI] [PMC free article] [PubMed]
- 93.van der Oost J., Swarts D., Jore M. Prokaryotic Argonautes – variations on the RNA interference theme. Microbial Cell. 2014;1(5):158–159. doi: 10.15698/mic2014.05.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fu, L. et al. The prokaryotic Argonaute proteins enhance homology sequence-directed recombination in bacteria. Nucleic Acids Res, 7 (2019). [DOI] [PMC free article] [PubMed]