SUMMARY
In addition to driving specific gene expression profiles, transcriptional regulators are becoming increasingly recognized for their capacity to modulate chromatin structure. GATA6 is essential for the formation of definitive endoderm; however, the molecular basis defining the importance of GATA6 to endoderm commitment is poorly understood. The members of the GATA family of transcription factors have the capacity to bind and alter the accessibility of chromatin. Using pluripotent stem cells as a model of human development, we reveal that GATA6 is integral to the establishment of the endoderm enhancer network via the induction of chromatin accessibility and histone modifications. We additionally identify the chromatin-modifying complexes that interact with GATA6, defining the putative mechanisms by which GATA6 modulates chromatin architecture. The identified GATA6-dependent processes further our knowledge of the molecular mechanisms that underpin cell-fate decisions during formative development.
Graphical abstract
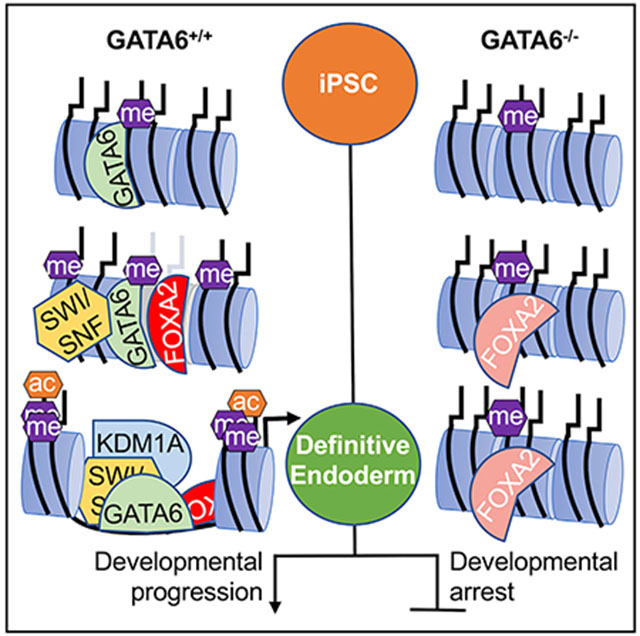
In brief
Using a hiPSC model of early development, Heslop et al. report that GATA6 is essential for chromatin patterning during definitive endoderm formation. The authors additionally identify the chromatin remodeling complexes that interact with GATA6 and that endoderm-enriched transcription factor binding profiles are significantly perturbed in the absence of GATA6.
INTRODUCTION
The importance of cell fate decisions in human development and disease is well accepted; however, the subcellular processes that dictate cell fate are less well defined. Human-induced pluripotent stem cells (hiPSCs) can be expanded indefinitely and differentiated to human somatic cell types by recapitulating known developmental signaling cascades. In combination with genome editing techniques (Liu et al., 2020; Ran et al., 2013), hiPSC culture models allow investigation into the mechanisms that orchestrate early human development at high resolution across the genome (Heslop and Duncan, 2019).
The members of the GATA family of transcription factors recognize the consensus sequence (A/T)GATA(A/G) (Molkentin, 2000). Of the GATA factors, GATA4 and GATA6 have known importance for endodermal-derived cell types, including hepatocytes (Zhao et al., 2005), intestinal epithelium (Beuling et al., 2012), pancreas (Carrasco et al., 2012; Watt et al., 2007; Xuan et al., 2012), and lung (Keijzer et al., 2001). The misregulation of these factors can have large-scale impact, including organ malformation, metabolic diseases, and cancer progression (De Franco et al., 2013; Zheng and Blobel, 2010). Previous work by us and others has shown that GATA6 is required for human PSC-derived definitive endoderm formation. GATA6 depletion results in developmental arrest, with a significant reduction of endodermal markers and cell death (Chia et al., 2019; Fisher et al., 2017; Shi et al., 2017; Tiyaboonchai et al., 2017). Studies of GATA6 haploinsufficiency using hiPSCs further revealed the dose-dependent requirement for GATA6 during pancreatic development (Chia et al., 2019; Shi et al., 2017; Tiyaboonchai et al., 2017). Interestingly, exogenous GATA6 expression in hiPSCs has also been used to induce the exit from pluripotency and differentiation toward endodermal lineages (Guye et al., 2016). Although GATA6 is known to be critical for development of the endoderm and its derivatives, the molecular mechanisms through which GATA6 mediates endodermal fate are poorly characterized.
A subset of transcription factors, commonly described as pioneer factors, are recognized as critical directors of cell fate. Pioneer factors bind to non-permissive regions of chromatin and alter accessibility through stepwise histone modifications, methylation changes, and nucleosome remodeling (Mayran and Drouin, 2018). The induced modifications make specific genomic loci permissive or refractive to the binding of transcription factors that control the onset of gene expression (Mayran and Drouin, 2018). The first report of developmental pioneer activity during endoderm development focused on the interaction of FOXA and GATA factors at an Alb gene enhancer (Cirillo et al., 2002). Continued efforts have identified a growing number of transcriptional regulators with differing capacities for binding nucleosomal DNA and modulating chromatin accessibility (Fernandez Garcia et al., 2019). In the context of development, the regulation of gene expression by controlling chromatin structure is important in defining the competency of a cell to adopt a specific fate.
GATA factors are emerging as important regulators of chromatin accessibility. GATA3 has been found to bind nucleosomal DNA and induce chromatin accessibility through the recruitment of SWI/SNF chromatin remodeling complexes in breast cancer cells (Takaku et al., 2016; Tanaka et al., 2020). GATA6 was recently shown to function as a pioneer factor in cardiac development, with perturbations in chromatin accessibility identified when hiPSCs with clinically relevant GATA6 mutations were differentiated to cardiac myocytes (Sharma et al., 2020). Several reports have identified GATA motifs to be enriched in chromatin that becomes accessible during definitive endoderm formation (Cernilogar et al., 2019; Genga et al., 2019; Lee et al., 2019; Meers et al., 2019); however, the active contributions of GATA factors to the altered accessibility at these loci have not been investigated.
Based on this information, we hypothesized that GATA6 controls chromatin accessibility during definitive endoderm formation. Mechanistic investigation revealed that GATA6 is required for chromatin patterning during the differentiation of hiPSCs to definitive endoderm. The impact on chromatin architecture is multifaceted and dynamic, with GATA6 having roles in both the opening of endodermal chromatin, and in the selective nature of enhancer decommissioning that occurs as the endoderm commits to a hepatic fate. Moreover, we identify multiple chromatin remodeling complexes that interact with GATA6, establishing the likely mechanisms through which GATA6 modulates chromatin accessibility. We conclude that GATA6 is a central regulator of the chromatin remodeling events that define definitive endoderm fate and establish its competency for further developmental progression.
RESULTS
GATA sites are the most significantly enriched motifs in chromatin with increased accessibility during endoderm formation
To understand the dynamics of chromatin accessibility during human endoderm formation and hepatic specification, we used the differentiation of PSCs as a model. We have previously described a differentiation protocol that recapitulates the major stages of hepatocyte formation (Si-Tayeb et al., 2010a). Using the early stages of the protocol (Figure 1A), we differentiated human PSCs (day 0) to a hepatic endoderm fate (day 8). The approach generates high-purity mesendoderm, definitive endoderm, and hepatic endoderm cells, as defined by the robust expression (>85% of cells) of stage-specific markers (Figures 1A and S1A). Differentially accessible regions across the genome were identified at 48-h intervals throughout the differentiation process by an assay for transposase accessible chromatin with high-throughput sequencing (ATAC-seq) (Table S1). As expected, investigation of the chromatin accessibility profile revealed substantial remodeling of chromatin, particularly during definitive endoderm formation, with 6,119 sites increasing and 3,286 sites decreasing in accessibility (Figures 1B and S1B). The later stages of differentiation showed a reduction in novel chromatin remodeling events, with 1,772 sites increasing and 1,475 decreasing in accessibility between day 4 and day 8 (Figures 1B and S1B). Interestingly, loss of accessibility appears to be a rapid response, with reduced accessibility more commonly observed 1–2 days after changes in growth factor supplementation (days 0–2 and days 4–6). Conversely, we observed greater variability in gain-of-accessibility events, with similar numbers of sites showing both rapid (1–2 days) or slower onset (3–4 days) chromatin opening after initial growth factor induction (Figure 1B).
Figure 1. Differentially accessible chromatin during endoderm formation and hepatic specification.
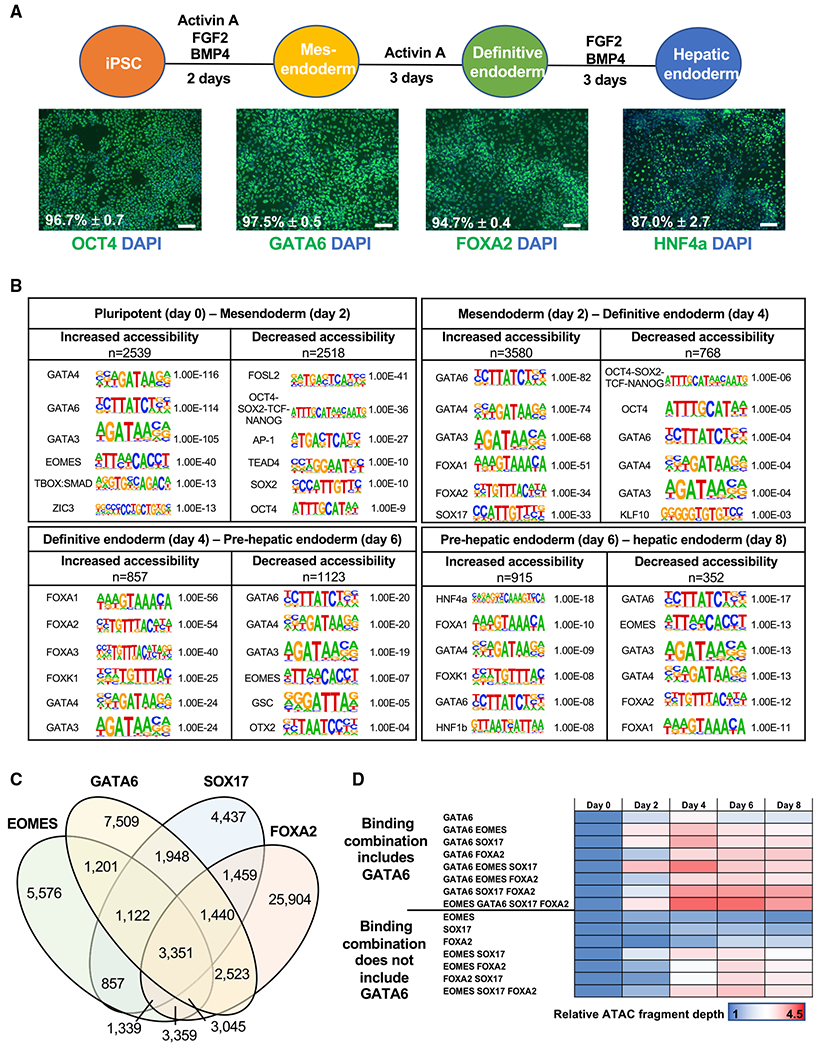
(A) Differentiation protocol used to study endoderm formation and hepatic specification. immunofluorescence analysis of stage specific markers. Scale bars, 100 μm. Positive cell population: flow cytometry analysis, n = 3, mean ± SD. See Figure S1A for histograms.
(B) Transcription factor motif enrichment at differentially accessible chromatin at 48-h intervals. n = sequences analyzed. p values: hypergeometric enrichment test using HOMER default background. See Table S1.
(C) Venn diagram showing the sites co-bound by GATA6, EOMES, SOX17, and FOXA2 in definitive endoderm using previously published ChIP-seq datasets (Tsankov et al., 2015).
(D) ATAC-seq fragment depth during endoderm formation and hepatic specification relative to day 0 values at the identified combinations of transcription factor binding.
Related to Figure S1D and Table S2.
To identify the putative drivers of the changes to chromatin accessibility at each time point, we identified the transcription factor motifs enriched at regions of differentially accessible chromatin (Figure 1B). As motif analysis commonly identifies numerous members of the same transcription factor family, which share similar consensus binding sequences, we used RNA sequencing (RNA-seq) analysis to identify the presence of mRNA encoding the cognate transcription regulators in the differentiation model (Figure S1C). We found that as the cells shifted from a pluripotent to a mesendodermal fate between day 0 and day 2, the binding motifs that were the most significantly enriched at regions of increased chromatin accessibility included GATA4/6 (Figure 1B). Motifs associated with the pluripotent state, such as OCT4, SOX2, NANOG, and AP-1, became significantly less accessible (Figure 1B). Enrichment of GATA motifs continues as the mesendoderm commits to an endodermal fate, between days 2 and 4 of differentiation, along with motifs bound by other known regulators of endodermal fate, such as FOXA and SOX17 (Figure 1B). Importantly, these results are consistent with previously published reports of motif enrichment during endoderm formation (Cernilogar et al., 2019; Genga et al., 2019; Lee et al., 2019; Li et al., 2019). As the endoderm transitions toward a hepatic fate, between days 4 and 6, GATA and FOXA continue to be enriched at regions of chromatin with increased accessibility (Figure 1B). Finally, during the formation of hepatic progenitor cells, between days 6 and 8, regions of increased accessibility are enriched for key activators of hepatic gene expression, including HNF4A and HNF1B (Figure 1B). Comparisons of heatmaps representing the RNA-seq and ATAC-seq datasets revealed how chromatin accessibility changes reflect the induction or loss of gene expression of the respective transcriptional regulator (Figure S1C).
Motif analysis alone cannot determine whether a binding site is occupied. Indeed, recent work has identified significant cell type-specific binding profiles of transcription factors, including GATA and FOXA (Donaghey et al., 2018). Therefore, we used chromatin immunoprecipitation sequencing (ChIP-seq) analysis in pluripotent and definitive endoderm samples to directly identify those transcription factors that occupied their target sequences. ATAC-seq datasets were aligned with published ChIP-seq datasets (Tsankov et al., 2015). For pluripotent cells, we examined OCT4, SALL4, SOX2, and NANOG, and for endoderm, we examined Brachyury (T), EOMES, SMAD2/3, GATA6, SOX17, FOXA2, HNF1B, and HNF4A. Using this combined approach, we identified each transcription factor’s binding loci that aligned with regions of differentially accessible chromatin during definitive endoderm formation and hepatic specification (Table S2). The results agreed with our motif analysis, demonstrating that EOMES, GATA6, SOX17, and FOXA2 commonly bind at sites of increased chromatin accessibility during definitive endoderm formation (Figure 1B; Table S2). Investigation of ATAC-seq density across the different combinations of EOMES, GATA6, SOX17, and FOXA2 revealed that loci bound by any combination of transcription factors had greater increases in chromatin accessibility than did those bound by a single factor (Figures 1C, S1D, and S1E). The results also indicate that all four factors contribute to chromatin patterning during endoderm formation. Importantly, analyses of the sites occupied by different transcription factor combinations revealed that loci that included GATA6 occupancy were consistently associated with greater increases in chromatin accessibility, compared to loci that were not bound by GATA6 (Figure 1D).
Cumulatively, these data show that regions of increased chromatin accessibility are most significantly associated with loci bound by GATA4/6 during definitive endoderm formation. Our previously published work revealed that GATA6, but not GATA4, is important for the generation of definitive endoderm (Fisher et al., 2017). Gene and protein expression analysis of the GATA and FOXA factors confirmed that GATA6 achieves maximal expression prior to GATA4 and FOXA1/2 (Figures S2A–S2C). Given the close correlation of GATA binding sites with accessible DNA and the relatively high level of GATA6 expression at early stages of endoderm formation, we focused specifically on the role of GATA6 in the patterning of DNA during endoderm formation and hepatic specification.
Increased chromatin accessibility and histone modifications at GATA6-bound sites are GATA6 dependent
To assess whether increased chromatin accessibility is dependent on GATA6 function, GATA6−/− iPSC lines were derived using CRISPR-Cas9. We generated two independent compound heterozygous iPSC lines harboring frameshift mutations in either exon 2 (GATA6Ex2Δ31/Δ38; p.[Ser208AlafsTer75];[Asn214SerfsTer72]) or exon 4 (GATA6Ex4Δ1/Δ2; p.[Glu460ArgfsTer12];[Ala459ArgfsTer2]) (Figure 2A). Western blot analysis confirmed that full-length GATA6 protein was undetectable in both of the GATA6−/− iPSC lines at day 2 of differentiation (Figure 2B); however, we detected low levels of a truncated GATA6 protein in the GATA6Ex2Δ31/Δ38 cells (Figure S2D). As previously reported, GATA6−/− pluripotent cells were significantly diminished in their capacity to form definitive endoderm (Chia et al., 2019; Fisher et al., 2017; Shi et al., 2017; Tiyaboonchai et al., 2017), with lineage-enriched markers significantly reduced (Figure 2C). In keeping with the truncated GATA6 protein retaining some functional capacity in GATA6Ex2Δ31/Δ38 cells, a more significant phenotype was observed in GATA6Ex4Δ1/Δ2 cells (Figure 2C); therefore, we focused our analysis on GATA6 exon 4 targeted cells. A doxycycline-inducible wild-type GATA6 cDNA was introduced to generate GATA6Ex4Δ1/Δ2;ind GATA6 pluripotent cells (Figures 2D and S2E). In this system, the presence of GATA6 protein is strictly dependent on doxycycline, which allows rescue of GATA6 expression within the same genomic background (Figures 2E and 2F). GATA6 was induced from day 1 to 4, based on the onset of GATA6 protein expression in GATA6+/+ cells (Figures 2G and S2C). Importantly, RNA-seq and flow cytometry analysis confirmed that the reduced capacity for definitive endoderm commitment in the absence of GATA6 could be significantly, but incompletely, rescued by inducing expression of GATA6 cDNA during the differentiation of GATA6Ex4Δ1/Δ2;ind GATA6 iPSCs (Figures 2H and S2F; Table S3).
Figure 2. Generation of GATA6−/− pluripotent stem cell lines.
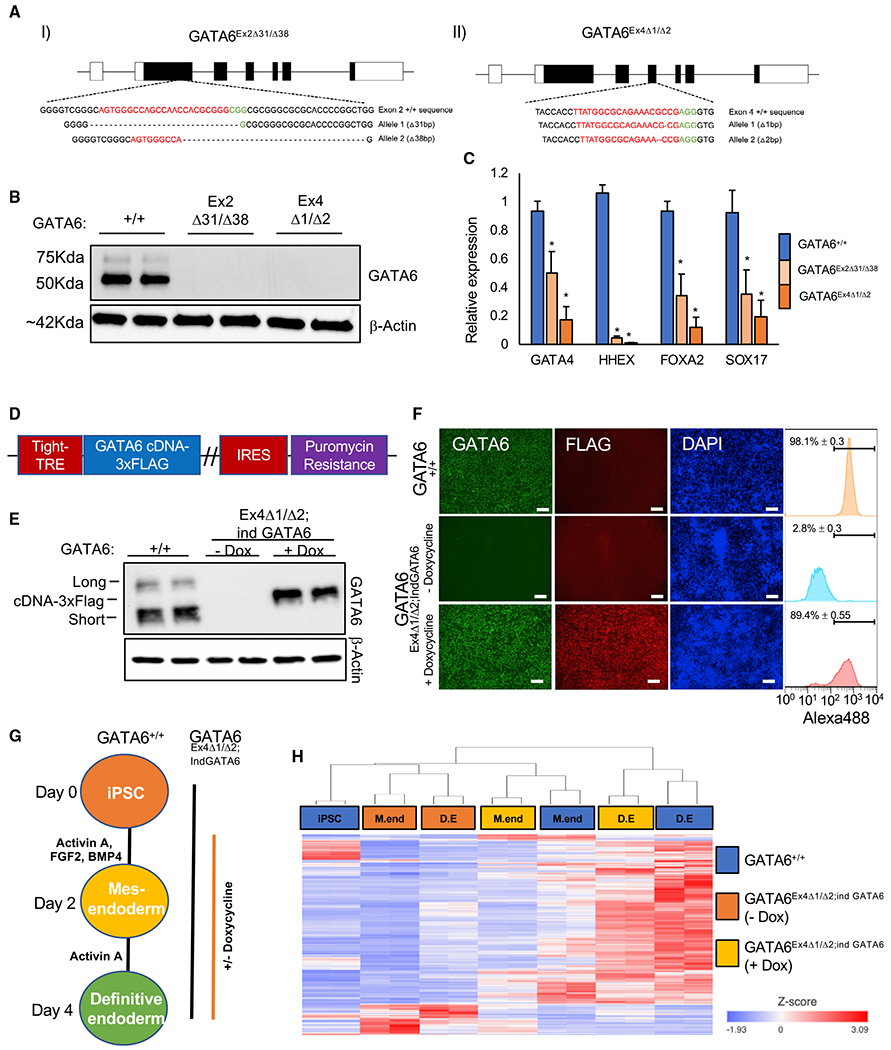
(A) CRISPR-Cas9-derived indels in K3 pluripotent stem cells using guide RNAs (gRNAs) targeting (I) exon 2 and (II) exon 4 of the GATA6 gene. Red denotes gRNA sequence; green denotes PAM sequence.
(B) Western blot analysis of GATA6 and β-actin in GATA6+/+, GATA6Ex2Δ31/Δ38 and GATA6Ex4Δ1/Δ2 cell lines at day 2 of differentiation.
(C) Graph showing qRT-PCR analysis of definitive endoderm-enriched mRNAs in definitive endoderm normalized to the housekeeping mRNA RPL13a. Mean values, n = 3, mean ± SD. Student’s t test: *p < 0.05.
(D) Doxycycline-inducible GATA6-3xFLAG cDNA plasmid.
(E) Western blot analysis of GATA6 and β-actin protein expression in GATA6+/+ and GATA6Ex4Δ1/Δ2;ind GATA6 pluripotent cells with and without doxycycline at day 2 of differentiation. Long, short, and exogenous short 3× FLAG epitope-tagged GATA6 isoforms are indicated.
(F) Immunofluorescence analysis of GATA6, FLAG, and DAPI in GATA6+/+ and GATA6Ex4Δ1/Δ2;ind GATA6 cells with and without doxycycline at day 2 of differentiation. Scale bars, 100 μm. GATA6-positive population: flow cytometry analysis, n = 3, mean ± SD.
(G) Differentiation protocol used to analyze GATA6 function.
(H) Heatmap with hierarchical clustering of GATA6-dependent genes during definitive endoderm formation in GATA6+/+ and GATA6Ex4Δ1/Δ2;ind GATA6 pluripotent cells with and without doxycycline. GATA6-dependent genes: false discovery rate (FDR) < 0.05: GATA6+/+ versus GATA6Ex4Δ1/Δ2;ind GATA6 (—dox), and GATA6Ex4Δ1/Δ2;ind GATA6 (—dox) versus GATA6Ex4Δ1/Δ2;ind GATA6 (+dox) at days 2 and 4 using gene-specific analysis (GSA; RNA-seq, n = 2).
To investigate whether the sites of increased chromatin accessibility during endoderm formation were dependent on GATA6, we performed ATAC-seq analysis at days 2 (mesendoderm) and 4 (definitive endoderm) on GATA6+/+ and GATA6Ex4Δ1/Δ2;ind GATA6 cells with and without exogenous GATA6 expression (Figure 2G; Table S1). Alignment of ATAC-seq and definitive endoderm GATA6 ChIP-seq datasets (Fisher et al., 2017) established whether GATA6 was bound to the identified regions of remodeled chromatin. Across all GATA6-bound sites, the ATAC-seq fragment depth in GATA6+/+ cells showed an increase in average chromatin accessibility during definitive endoderm formation (Figure 3A). In GATA6−/− cells, chromatin opening was repressed, demonstrating that increases in chromatin accessibility during endoderm formation were dependent on GATA6 expression (Figure 3A). Analyses of the accessibility profiles at all GATA6-bound regions revealed that most GATA6 binding events (43,256 sites) occurred within inaccessible chromatin, determined as regions of chromatin with accessibility levels below the threshold for peak detection (Figure S3A). Conversely, 23,325 sites overlapped with accessible genomic regions. Within the subset of accessible sites bound by GATA6, we identified 3,420 GATA6-dependent regions of chromatin accessibility. GATA6-dependent accessibility was defined as loci occupied by GATA6 that increase in accessibility during definitive endoderm formation in GATA6+/+ cells, but are only accessible in GATA6Ex4Δ1/Δ2;ind GATA6 cells when exogenous GATA6 cDNA expression is induced (Figure S3A). Interestingly, the rate of GATA6-dependent chromatin remodeling was variable, with regions of rapid and slow onset chromatin accessibility identified (Figures S3A and S3B).
Figure 3. Identification of GATA6-dependent chromatin accessibility and histone modifications.

(A) ATAC-seq fragment depth during definitive endoderm formation at sites bound by GATA6 in definitive endoderm, n = 66, 581 sites; (i) GATA6+/+ pluripotent cells days 0–4; (ii and iii) GATA6+/+ and GATA6Ex4Δ1/Δ2;ind GATA6 cells with and without doxycycline at (ii) mesendoderm and (iii) definitive endoderm.
(B) ATAC-seq profiles aligned with GATA6-bound loci in definitive endoderm at genomic loci neighboring genes with known roles during endoderm formation and specification. GATA6−/− refers to GATA6Ex4Δ1/Δ2;ind GATA6 cells.
(C) Venn diagram of genes that neighbor GATA6-dependent changes in chromatin accessibility, definitive endoderm GATA6-dependent genes (mRNA), and genes with increased mRNA expression during hepatic specification. See also Table S3.
(D) Heatmap of GATA6 (blue) and H3K4me1 (red) at GATA6 binding sites during endoderm formation in GATA6+/+ and GATA6Ex4Δ1/Δ2;ind GATA6 pluripotent cells with and without doxycycline. Signal intensity represents ChIP-seq density. Heatmaps are split based on chromatin accessibility profile during definitive endoderm formation. Related to Figures S3A and S3B. GATA6−/− refers to GATA6Ex4Δ1/Δ2;ind GATA6 cells.
(E) ATAC-seq and H3K4me1 profiles during endoderm formation at the AFP gene loci in GATA6+/+ and GATA6Ex4Δ1/Δ2;ind GATA6 cells with and without doxycycline; GATA6−/− refers to GATA6Ex4Δ1/Δ2;ind GATA6 cells.
The GATA6-dependent regions of chromatin accessibility included putative regulatory regions of genes whose expression was enriched during endoderm and hepatic formation, such as HNF1B, COL4A1, and SOX17 (Figures 3B and S3C). A subset of the identified chromatin domains with GATA6-dependent accessibility in definitive endoderm neighbored genes with significantly increased mRNA expression during hepatic specification. Therefore, the accessibility of GATA6-bound enhancers is established prior to the induction of neighboring gene expression (Figures 3C and S3D–S3F; Table S3). Interestingly, regions of increased accessibility in GATA6Ex4Δ1/Δ2;ind GATA6 cells without doxycycline were associated with mesendodermal and pluripotent gene expression profiles, indicating a failure of GATA6-deficient cells to commit to the endodermal lineage from preceding developmental stages (Figure S3G).
H3K4me1 is a histone modification associated with poised or active enhancer regions. We therefore addressed whether GATA6 influenced H3K4me1 modification of occupied sites during generation of the endoderm. Using H3K4me1 ChIP-seq, we first observed that GATA motifs were the most significantly enriched sites within regions of increased H3K4me1 accumulation during formation of the definitive endoderm ((Figure S4A). Regions of reduced H3K4me1 levels were enriched for pluripotent transcription factor motifs, such as OCT4, SOX2, and NANOG (Figure S4A). We next examined the impact of GATA6 expression on H3K4me1 modifications during endoderm formation. H3K4me1 showed a significant, GATA6-dependent increase at GATA6-bound loci (Figure 3D). Conversely, loss of H3K4me1 was not dependent on GATA6 (Figure S4B). At sites of rapid (accessible in mesendoderm) and slow (accessible in definitive endoderm) GATA6-dependent chromatin remodeling, most H3K4me1 deposition is achieved within the mesendoderm cell populations (Figure 3D). Importantly, H3K4me1 deposition was a more widespread effect of GATA6 binding than changes in accessibility, with sites lacking detectable chromatin accessibility also showing GATA6-dependent increases in H3K4me1 deposition between pluripotent and definitive endoderm stages (Figures 3D, S4C, and S4D). For example, a GATA6-occupied region upstream of the AFP gene accumulated H3K4me1 between day 0 (iPSC) and day 2 (mesendoderm) in a GATA6-dependent manner that preceded detectable chromatin accessibility (Figure 3E). Based on these cumulative data, we conclude that GATA6 is required for establishing domains of poised chromatin that can presage chromatin opening.
GATA6-dependent chromatin accessibility dynamics reflect co-bound transcription factor availability
GATA6 does not appear to function equivalently at all bound sites. Our ATAC-seq analysis revealed that most GATA6-binding events occupy inaccessible DNA throughout our hepatic differentiation protocol (Figure S3A). Therefore, it is important to understand what differentiates the GATA6 sites that increase in chromatin accessibility from those that remain refractive. Recent work has demonstrated that the co-localization of additional transcription factors is a key determinant of a pioneer factor’s ability to increase chromatin accessibility at specific sites (Donaghey et al., 2018; Mayran et al., 2019). In Figure 1, we compared the binding sites of EOMES, GATA6, SOX17, and FOXA2 in definitive endoderm and found that GATA6 often co-localizes with these transcription factors (Figure 1C). Moreover, the sites bound by GATA6 along with additional transcription factors commonly aligned with regions of DNA that increase in accessibility (Figure 1D). Therefore, we hypothesized that co-localization of the endoderm-enriched transcription factors define the GATA6-bound sites that can increase in accessibility.
During hepatic differentiation, the expression and availability of the endoderm-enriched transcription factors is dynamic. If transcription factor co-binding does enhance GATA6-dependent chromatin remodeling, we would expect the gain of accessibility to occur at different rates, dependent on the availability of the stabilizing protein during differentiation. Consistent with this model, we identified GATA6-dependent chromatin remodeling that occurred rapidly in mesendoderm (day 2; rapid) or more slowly in the definitive endoderm cell populations (day 4; slow onset) (Figure S3A). We next used motif enrichment analysis to identify the binding sites that are co-enriched with GATA6 at regions of GATA6-dependent chromatin remodeling at different stages of differentiation (Figure 4A; Table S4). The motif analyses revealed that loci associated with rapid GATA6-dependent accessibility (day 2) were significantly enriched for T-BOX:SMAD and SOX17 binding sites, when compared to GATA6-bound sites that remain inaccessible (Figure 4A; Table S4). Regions of GATA6 binding with slow-onset chromatin accessibility (definitive endoderm; day 4) were more significantly enriched for FOXA2 binding motifs (Figure 4A; Table S4). HNF1B and HNF4A motifs were enriched at GATA6-occupied loci that do not become accessible until hepatic specification (Table S4). Interestingly, GATA6 binding sites resistant to detectable increases in accessibility throughout our hepatic specification protocol had significant enrichment for the pancreatic enriched transcription factor PDX1 binding motif (Figures 4A and S4E; Table S4). Importantly, the co-binding sites enriched within regions of increased chromatin accessibility at each stage of hepatic differentiation correlated with the onset of mRNA expression encoding the identified co-regulators (Figure 4B). These datasets indicate that GATA6 chromatin remodeling kinetics are influenced by the dynamic availability of additional transcription factors during endoderm formation that stabilize GATA6 occupancy (Figure S4F).
Figure 4. Endoderm transcription factors are co-enriched at sites of GATA6-dependent chromatin accessibility.
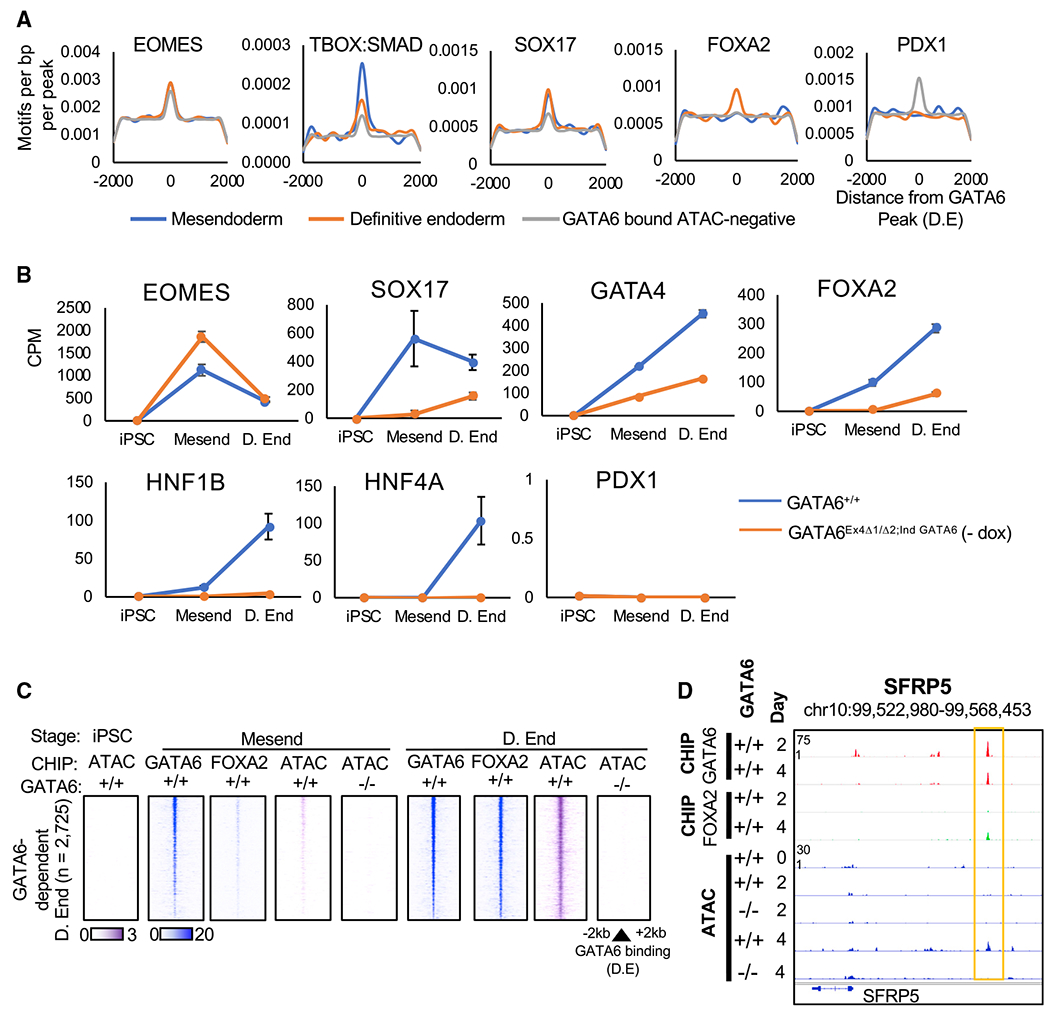
(A) Histograms showing density of specific motifs and their proximity to the GATA6 binding loci in definitive endoderm at sites with or without GATA6-dependent increases in chromatin accessibility. See also Table S4.
(B) mRNA expression data of co-regulators in GATA6+/+ and GATA6Ex4Δ1/Δ2;ind GATA6 cells during definitive endoderm formation; RNA-seq normalized by counts per million (CPM, n = 2, mean ± SD).
(C) Heatmaps depicting temporal GATA6 and FOXA2 ChIP-seq intensity (blue) and ATAC-seq (purple) at sites of slow-onset GATA6-dependent chromatin remodeling.
(D) GATA6 and FOXA2 ChIP-seq data aligned with ATAC-seq profiles during definitive endoderm formation at the SFRP5 gene loci in GATA6+/+ cells.
GATA6 binding precedes and is required for FOXA2 binding and chromatin remodeling during endoderm formation
Pioneer factors are defined by their capacity to bind inaccessible regions of DNA and induce chromatin remodeling. Our data show that GATA6 is required for a significant number of loci to gain accessibility during endoderm formation, and that GATA6 binds to these sites in definitive endoderm. However, similar results could be obtained if GATA6 binds to the identified sites only after the region has been made accessible by an independent pioneer factor whose expression is dependent on GATA6. Indeed, many of the regulators with binding sites co-enriched with GATA6 at regions of increased accessibility have significantly diminished mRNA expression in GATA6−/− cells (Figure 4B). Therefore, we aimed to establish whether GATA6 binds to regions of GATA6-dependent chromatin remodeling before the identified co-localized transcription factors, and whether GATA6 occupancy precedes the induction of accessibility.
We decided to focus on slow-onset chromatin remodeling events, as we predicted that the extended time course would allow us to identify sequential binding and remodeling events at higher resolution. Moreover, slow-onset chromatin remodeling sites were co-enriched for the FOXA2 binding motif. FOXA2 is a paradigm pioneer factor (Heslop and Duncan, 2020; Iwafuchi et al., 2020), and its expression is significantly reduced in GATA6Ex4Δ1/Δ2 cells (Figure 4B). Therefore, FOXA2 could bind and induce accessibility at these sites, permitting GATA6 to bind thereafter. We therefore established the dynamic binding profile of GATA6 and FOXA2 to determine whether GATA6 binds prior to FOXA2 and whether the region is accessible at the point of first engagement by GATA6. Using ChIP-seq analyses in GATA6+/+ cells, we identified the binding sites of GATA6 and FOXA2 in mesendoderm and definitive endoderm. We found that, at co-bound loci, GATA6 occupancy most commonly precedes FOXA2 binding (Figure S4G). At sites occupied by both transcription factors in definitive endoderm, GATA6 binding could be readily detected within mesendoderm and definitive endoderm cell populations, whereas FOXA2 binding was not robustly observable until definitive endoderm formation (Figures 4C, 4D, and S4G). Importantly, comparing the ChIP-seq to ATAC-seq data revealed that GATA6 binding predominantly preceded significant FOXA2 occupancy and chromatin accessibility at regions of GATA6-dependent chromatin remodeling (Figures 4C, 4D, S4G, and S4H). Moreover, in the absence of GATA6, the increases in chromatin accessibility do not occur (Figures 4C and 4D).
Our results demonstrate that GATA6 binding most commonly precedes that of FOXA2; however, as FOXA2 expression is diminished in GATA6−/− cells, the reduced chromatin accessibility identified at GATA6-FOXA2 co-bound loci in GATA6−/− definitive endoderm may still be solely attributable to the loss of FOXA2. Therefore, we aimed to establish whether regions of GATA6-dependent chromatin accessibility could be rescued by FOXA2 expression, in the absence of GATA6. We introduced a doxycycline-inducible FOXA2 cDNA into GATA6Ex4Δ1/Δ2 pluripotent cells (Figures 5A and S5A). The doxycycline-inducible pluripotent cell line allowed for the dose-dependent exogenous expression of FOXA2, thereby re-establishing robust FOXA2 expression in GATA6Ex4Δ1/Δ2;ind FOXA2 cells. The functional activity of the exogenous FOXA2 was confirmed by a luciferase assay using the TTR promoter region, which contains several FOXA2 binding sites (Figures S5B and S5C) (Costa and Grayson, 1991). We next performed ATAC-seq analysis to determine whether exogenous FOXA2 could restore wild-type definitive endoderm chromatin accessibility in GATA6Ex4Δ1/Δ2;ind FOXA2 cells. Exogenous FOXA2 expression was induced during endoderm formation and the accessibility of chromatin was measured in definitive endoderm cell populations (Figures 5B and 5C). The analysis revealed that exogenous FOXA2 expression was unable to restore the GATA6+/+ chromatin accessibility profile in GATA6Ex4Δ1/Δ2;ind FOXA2 cells (Figure 5D); however, limited increases in accessibility were detected as sites enriched for ZIC, MAFA, and JUNB binding sites (Figure S5D). Importantly, exogenous FOXA2 was also unable to rescue mRNA and protein expression of definitive endoderm and foregut-enriched markers in the GATA6Ex4Δ1/Δ2;ind FOXA2 cells (Figures S5E and S5F).
Figure 5. Exogenous FOXA2 expression does not restore definitive endoderm chromatin accessibility in GATA6−/− cells.
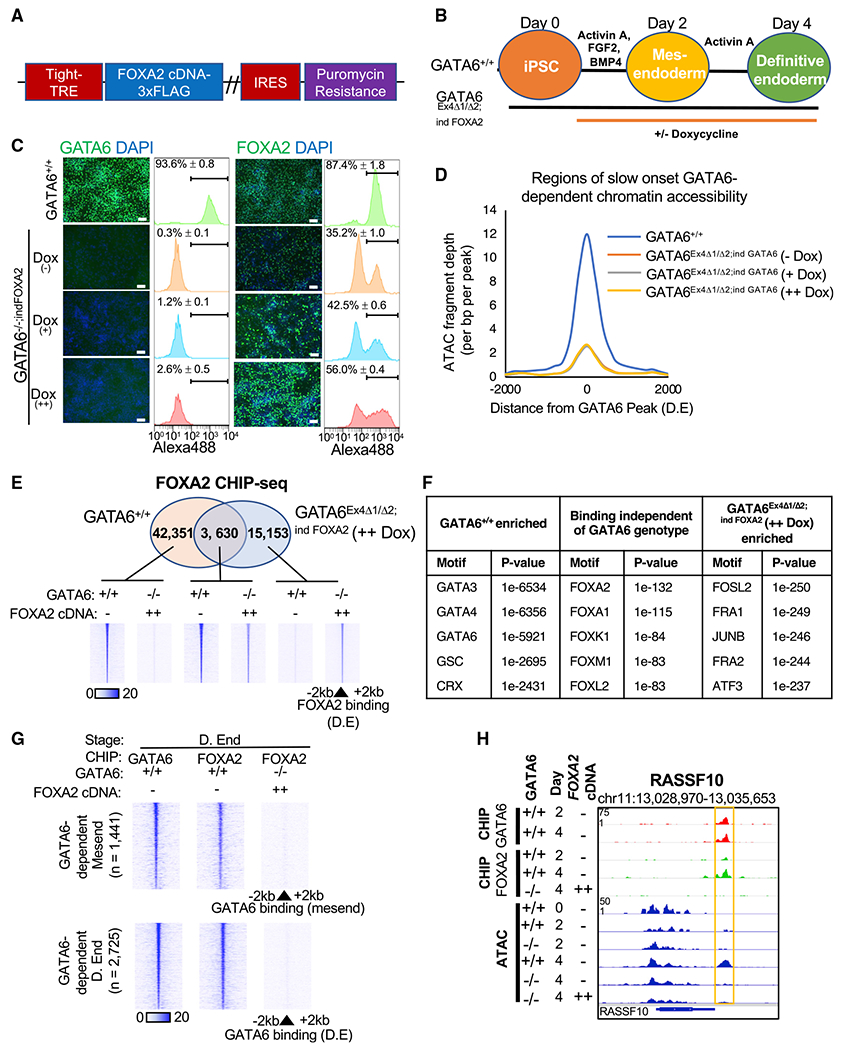
(A) Doxycycline-inducible FOXA2-3× FLAG cDNA vector.
(B) FOXA2 overexpression differentiation model.
(C) Immunofluorescence analysis of GATA6, FOXA2, and DAPI in GATA6+/+ and GATA6Ex4Δ1/Δ2;ind FOXA2 cells with and without doxycycline at day 4 of differentiation. Scale bars, 100 μm. Positive cell population: flow cytometry, n = 3, mean ± SD.
(D) Graph showing ATAC-seq fragment depth GATA6+/+ and GATA6Ex4Δ1/Δ2;ind FOXA2 cells with increasing doxycycline supplementation at sites of slow-onset GATA6-dependent chromatin remodeling in definitive endoderm; n = 2,404 regions.
(E) Venn diagram of loci bound by FOXA2 in GATA6+/+ and GATA6Ex4Δ1/Δ2;ind FOXA2 (++dox) cells at day 4 of differentiation with heatmaps of FOXA2 occupancy for each identified subset.
(F) Transcription factor motifs enriched within each subset of FOXA2 bound loci. p values: hypergeometric motif enrichment over the alternate subsets of FOXA2-bound loci.
(G) Heatmaps depicting GATA6 and FOXA2 ChIP-seq intensity at sites of GATA6-dependent chromatin remodeling in GATA6+/+ and GATA6Ex4Δ1/Δ2;ind FOXA2 (++dox) in mesendoderm and definitive endoderm populations.
(H) GATA6 and FOXA2 ChIP-seq data aligned with ATAC-seq profiles during definitive endoderm formation at the RASSF10 gene loci in GATA6+/+ and GATA6Ex4Δ1/Δ2;ind FOXA2 cells (++dox); GATA6−/− refers to GATA6Ex4Δ1/Δ2;ind FOXA2 cells.
We reasoned that the inability of exogenous FOXA2 to induce chromatin remodeling and mRNA expression of target genes in GATA6Ex4Δ1/Δ2;ind FOXA2 cells was due to an impaired ability for FOXA2 to engage with definitive endoderm-enriched binding sites. Therefore, we investigated whether the absence of GATA6 influenced the FOXA2 binding profile during endoderm formation. We performed FOXA2 ChIP-seq analysis on GATA6+/+ and GATA6Ex4Δ1/Δ2;ind FOXA2 (++dox) cells in definitive endoderm. Our results demonstrated that most FOXA2 binding sites identified in wild-type cells were significantly diminished in FOXA2 occupancy in the absence of GATA6 (Figures 5E and S5G). Consistent with the ATAC-seq results, FOXA2 preferentially occupied binding sites that were co-enriched for JUNB motifs in GATA6Ex4Δ1/Δ2;ind FOXA2 (++ dox) cells (Figure 5F). Therefore, in the absence of GATA6, FOXA2 cannot significantly engage with, or induce accessibility at, the majority of binding sites it usually occupies within wild-type definitive endoderm (Figures 5G and 5H). The results are consistent with FOXA2 overexpression studies in mouse embryonic stem cells (ESCs) and human fibroblasts (Cernilogar et al., 2019; Donaghey et al., 2018). Moreover, these datasets demonstrate that GATA6 occupancy is a prerequisite for FOXA2 binding at regions of chromatin remodeling and that GATA6 is required to establish accessibility at loci that define definitive endoderm formation and competency.
RIME analysis reveals activating and repressive chromatin remodeling complexes as part of the GATA6 interactome
To understand the mechanisms underlying GATA6-dependent chromatin remodeling, we used rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) to identify proteins that interact with chromatin-bound GATA6 (Figures 6A, 6B, and S6A; Table S5). RIME analysis of GATA6+/+ definitive endoderm revealed that the transcription factor SALL4 was the most enriched protein over IgG controls (Figure 6A). GATA6-SALL4 interactions have not previously been reported during definitive endoderm formation and may add further complexity to the transcription factor network that dictates endoderm commitment.
Figure 6. RIME analysis of GATA6 interacting proteins.
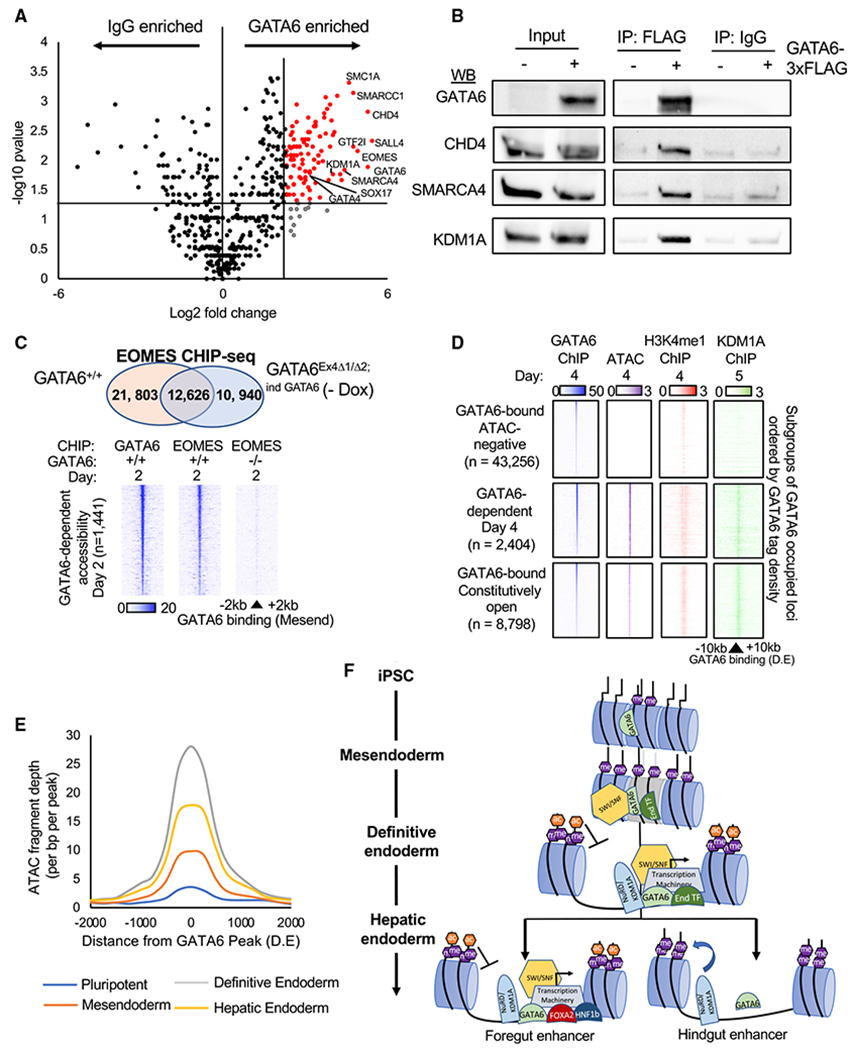
(A) Volcano plot of RIME-detected proteins with more than one spectral count and more than one unique peptide fragment. GATA6 interactome: 5-fold enriched over IgG controls, p < 0.05, n = 2. See Table S5.
(B) Validation of RIME proteins by western blot analysis of GATA6, CHD4, SMARCA4, and KDM1A following immunoprecipitation of GATA6-3× FLAG protein in GATA6Ex4Δ1/Δ2;ind GATA6 mesendoderm cultures differentiated with and without doxycycline.
(C) Venn diagram depicting overlap of peaks from EOMES ChIP-seq completed in GATA6+/+ and GATA6Ex4Δ1/Δ2;ind GATA6 cells without doxycycline differentiated to mesendoderm (upper panel). Heatmaps of EOMES and GATA6 ChIP-seq intensity in GATA6+/+ and GATA6Ex4Δ1/Δ2;ind GATA6 cells without doxycycline at regions of rapid GATA6-dependent chromatin remodeling (day 2) (lower panel).
(D) Published KDM1A ChIP-seq (Vinckier et al., 2020) from similar differentiation stage (day 5; foregut) overlapped with definitive endoderm GATA6 binding sites. GATA6 (blue), KDM1A (green), H3K4me1 (red), and ChIP-seq and ATAC-seq (purple) intensity displayed at subgroups of GATA6-bound sites with different endoderm accessibility profiles.
(E) Temporal profile of ATAC-seq data fragment depth during the hepatic specification at sites of GATA6-dependent chromatin accessibility. See Table S6.
(F) Proposed model of GATA6-dependent chromatin modulation during endoderm formation and specification. End transcription factors (TFs): EOMES, SOX17, and FOXA2.
Established endoderm-enriched transcription factors were also identified as GATA6-interacting proteins, such as SOX17 and GATA4 (Figure 6A; Table S5). FOXA2 and HNF1B were moderately enriched over IgG controls, but fell below the greater than one unique protein fragments required for inclusion in the downstream analysis (Table S5). Interestingly, EOMES was established as a high-confidence GATA6-interacting protein (Figure 6A). EOMES and GATA6 were recently found to co-operatively drive key parts of the early endoderm transcriptional profile (Chia et al., 2019). Therefore, we performed ChIP-seq for EOMES in mesendoderm cell populations, reflecting the earliest point that both GATA6 and EOMES are robustly expressed in our differentiation model (Figure S1C). By comparing binding sites occupied by EOMES in GATA6+/+ and GATA6Ex4Δ1/Δ2 cells, we identified the EOMES bound loci in wild-type cells that were GATA6-dependent and GATA6-independent (Figures 6C, S6B, and S6C). Using these datasets, we concluded that GATA6 is required for EOMES to occupy a significant proportion of the sites bound in GATA6+/+ cells; importantly, these included regions of GATA6-dependent chromatin remodeling (Figure 6C).
Significantly, the GATA6 interactome featured numerous proteins with well-established roles in chromatin remodeling and enhancer activation (Figure S6D). The identified proteins included members of the SWI/SNF chromatin modulating complex, such as SMARCA4 and SMARCC1 (Figures 6A, 6B, and S6D), and the histone acetyltransferases CBP/P300 (Table S5). Consistent with our findings, previous studies have shown that SMARCA4 interacts with other GATA factors (Takaku et al., 2016). Despite GATA6-dependent increase in H3K4me1 accumulation, we did not detect any interactions between GATA6 and a known H3K4 methyltransferase. WDR5 was enriched over IgG controls in our RIME analysis and the interaction was validated by co-immunoprecipitation (coIP) (Table S5; Figure S6E). In complex with ASH2L, WDR5 commonly links transcription factors to methyltransferase complexes (Cenik and Shilatifard, 2021). Further investigation revealed that ASH2L mRNA expression is GATA6-dependent during definitive endoderm formation (Figure S6E). Therefore, transcriptional activation of ASH2L and recruitment of methyltransferase proteins via interactions with WDR5 likely contribute to the GATA6-dependent accumulation of H3K4me1 at GATA6-bound loci.
NuRD protein complex members were also highly enriched within the GATA6 interactome (Figure S6D). The NuRD complex is most commonly associated with repressive chromatin remodeling; however, we observed relatively few GATA6-bound loci in definitive endoderm that reduced in accessibility (Table S6). KDM1A, a H3K4/H3K9 demethylase component of the NuRD complex, was significantly enriched within the GATA6 interactome and validated by coIP (Figures 6A and 6B). Using previously published ChIP-seq data for KDM1A in a similar foregut differentiation model (Vinckier et al., 2020), we identified the putative GATA6-KDM1A co-bound sites (Figure 6D). Interestingly, GATA6-KDM1A binding was enriched at euchromatic regions, such as accessible enhancers. KDM1A binding has previously been reported at active enhancers (Whyte et al., 2012). At these loci, the H3K4 demethylase activity of KDM1A is inhibited by histone acetylation or interactions with transcription factors (Alabdi et al., 2020; Shi et al., 2005). In keeping with these reports, we did not observe demethylation of H3K4me1 at GATA6-bound loci co-enriched for KDM1A (Figure 6D). KDM1Acan also demethylate the inhibitory modification H3K9me1/2; however, we did not identify a specific loss of H3K9me2 signal at loci of GATA6-dependent chromatin remodeling (Figure S6F).
GATA6-bound enhancers with reduced accessibility during hepatic specification are enriched for alternative lineage motifs
In Figure 1B, we established that a significant subset of loci with GATA6 motifs reduce accessibility during the transition of the endoderm to a hepatic fate. Therefore, we investigated whether accessible enhancers bound by GATA6/KDM1A in definitive endoderm undergo decommissioning during lineage commitment. In agreement with this model, the average accessibility of GATA6-dependent chromatin opening events that occur during definitive endoderm formation decline during hepatic specification (Figure 6E). Using ChIP-seq datasets (Loh et al., 2014), we confirmed that the transient accessibility at these sites is accompanied by the gain and subsequent loss of the active enhancer histone marks H3K27ac and H3K4me2 (Figure S6G).
We next investigated whether the GATA6-bound regions whose accessibility declines during hepatic specification could be related to the concurrent reduction in the expression of endoderm-enriched transcription factors. We compared the subsets of GATA6-bound euchromatic regions in definitive endoderm that maintained or declined in accessibility during hepatic specification for motif enrichment. Across all definitive endoderm euchromatic loci bound by GATA6, FOXA and HNF1B motifs were enriched in the regions that maintained accessibility during hepatic specification, whereas pluripotent and T-BOX motifs, including EOMES, were enriched in regions that declined in accessibility (Table S6). Interestingly, a subset of GATA6-dependent transiently accessible loci was moderately enriched for the motifs of non-foregut lineage master regulators: NKX2.1 (lung), CDX2 (hindgut) and PAX8 (thyroid) (Figure S6H).
In summary, this study has established GATA6 as essential for chromatin remodeling events that occur during early human development and defined specific molecular mechanisms that underpin the importance of GATA6 during the commitment of pluripotent cells to definitive endoderm.
DISCUSSION
Several groups have reported that the dynamic changes to chromatin accessibility that occur during definitive endoderm formation are enriched for GATA4/6 binding sites (Cernilogar et al., 2019; Gengaet al., 2019; Lee et al., 2019). In this study, we reveal that GATA6 binds to regions of DNA with low accessibility and is required for the remodeling of local chromatin structure at the occupied sites. Together, the induced modifications significantly contribute to the fate and competency of the endoderm.
Based on the cumulative data presented, we propose a model (Figure 6F) where GATA6 and other endoderm-enriched transcription factors bind thousands of loci throughout the genome. At sites of low chromatin accessibility, GATA6 binds and recruits H3K4me1 methyltransferases via WDR5/ASH2L and SWI/SNF complexes through direct interaction with SMARCA4. Together, the bound complexes induce chromatin remodeling, resulting in nucleosome repositioning or eviction. The complexes can be recruited more readily at sites of stabilizing transcription factor co-occupancy, such as EOMES and SOX17 in early endoderm. At later stages of endoderm formation, nucleosome eviction is further facilitated by the GATA6-dependent binding of FOXA2. In the absence of GATA6, co-bound transcription factors “sample” the sites at a significantly diminished, sub-threshold level. Following nucleosome eviction, enhancer/promoter activation is propagated by P300/CBP recruitment, inducing H3K27/H3K9 acetylation. Active transcription attracts the binding of the NuRD-KDM1A complex. The presence of hyperacetylated histones and active transcription factors, including GATA6, repress the inhibitory mechanisms of the NuRD-KDM1A complex. NuRD-KDM1A repression is maintained until the endodermal transcription factor network expression declines during hepatic specification. If the endoderm-specific transcription factors co-bound with GATA6, such as EOMES, are not replaced by hepatic-enriched regulators, including HNF1B and HNF4A, the dose-dependent repression of the NuRD-KDM1A complex also declines, permitting activation. Rapid enhancer decommissioning of non-hepatic enhancers follows via histone deacetylation, H3K4me1/2 demethylation, and repressive chromatin remodeling, thereby driving hepatic lineage commitment. Further studies are required to confirm whether the identified interactions between GATA6 and chromatin remodeling complexes are essential to the opening and closing of chromatin.
As we have reported previously, the loss of GATA6 expression results in a significant decrease in GATA4 expression (Fisher et al., 2017). While GATA4 depletion does not result in a comparable phenotype, exogenous GATA4 expression has been shown to rescue SOX17/FOXA2 expression in GATA6−/− definitive endoderm (Tiyaboonchai et al., 2017). Therefore, a dose-dependent redundancy exists between GATA4 and GATA6. Consequently, the full contribution of GATA factors to endoderm formation is likely masked by the diminished, but continued, presence of GATA4 in GATA6−/− cells. Interestingly, a small percentage of GATA6−/− cells retain the capacity to become SOX17/FOXA2 positive when differentiated toward definitive endoderm. We predict that this subset of GATA6−/− cells achieves the minimal threshold level of GATA4 expression that is sufficient to compensate for the absence of GATA6 during endoderm formation. Single-cell ATAC-seq analysis is required to ascertain whether the accessibility profile of the SOX17/FOXA2-positive GATA6−/− cells is also significantly perturbed. Interestingly, a recent study did find that CXCR4-positive GATA6−/− cells favored specification toward pulmonary, rather than hepatic, cell fates (Liao et al., 2018). Our results identified GATA6-dependent definitive endoderm chromatin accessibility that neighbored critical regulators of hepatic fate and preceded the robust mRNA expression of the corresponding genes during hepatic specification. The direct contribution of altered enhancer accessibility to neighboring gene expression will require further empirical assessment; however, the cumulative results suggest that GATA6-dependent chromatin remodeling is required to establish the competency of definitive endoderm for foregut commitment.
GATA6 haploinsufficiency is a condition that causes a range of developmental abnormalities in humans, including pancreatic agenesis and neonatal diabetes (De Franco et al., 2013). The identification of the PDX1 binding motif, a transcription factor required for pancreatic specification, at GATA6 binding sites in definitive endoderm is therefore provocative. The GATA6-PDX1 sites showed increased H3K4me1 deposition during definitive endoderm formation, but no detectable changes in chromatin accessibility. Similar low occupancy pioneer factor binding in nucleosomal DNA has been defined as “sampling” (Donaghey et al., 2018). Sampled sites commonly relate to loci bound by the same factor at greater density in alternate lineages. Interestingly, H3K4me1 deposition was recently found to prevent repressive DNA methylation modifications, establishing and preserving a poised enhancer state (Alabdi et al., 2020). Our results indicate the GATA6 sampling of loci co-enriched for PDX1 motifs, and the associated accumulation of H3K4me1, may have afunctional role in poising the sites for activation during later pancreatic specification. Importantly, transcription factor binding that temporally precedes chromatin opening has also been identified in vivo (Zaret, 2020). Thus, neonatal diabetes and other pathophysiologies associated with GATA6 haploinsufficiency may be underpinned by improper pre-patterning during endoderm formation, reducing developmental competency.
In summary, we have shown that GATA6 is required for the remodeling of chromatin structure and modifications that coalesce to define definitive endoderm fate. These roles are varied and include ensuring that chromatin is receptive to the onset of gene expression, interactions with critical histone modifying enzymes, and facilitating the selective decommissioning of enhancers as cells differentiate. We anticipate that the molecular mechanisms through which GATA6 acts are fundamental to controlling cell fate, significantly adding to our knowledge of developmental orchestration.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Stephen A. Duncan (duncanst@musc.edu).
Materials availability
All reagents generated in this study will be made available upon request to the lead contact.
Data and code availability
ATAC-seq, RNA-seq, and ChIP-seq datasets generated during this study are deposited in the GEO database under the GEO ID code: GEO: GSE156021. Previously published datasets used as part of this study are available in the GEO database. See key resources table for details.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | |||
|---|---|---|---|---|---|
| Antibodies | |||||
| GATA6 | Cell Signaling | 5851; RRID:AB_10705521 | |||
| GATA4 | Santa Cruz | SC-1237; RRID:AB_2108747 | |||
| FOXA2 | RnD | AF2400; RRID: AB_2294104 | |||
| SOX17 | RnD | AF1924; RRID:AB_355060 | |||
| OCT4 | Santa Cruz | SC-9081; RRID:AB_2167703 | |||
| HNF4A | Santa Cruz | SC-6556; RRID:AB_2117025 | |||
| EOMES | Cell Signaling | 66325; RRID: Unknown | |||
| CHD4 | Cell Signaling | 12011; RRID:AB_2734702 | |||
| SMARCA4 | Cell Signaling | 49360’ RRID:AB_2728743 | |||
| KDM1A | Cell Signaling | 2184; RRID:AB_2070132 | |||
| WDR5 | Cell Signaling | 13105; RRID:AB_2620133 | |||
| FLAG | Sigma-Aldrich | F1804; RRID:AB_262044 | |||
| Normal Mouse IgG | Sigma-Aldrich | 12-371; RRID:AB_145840 | |||
| H3K4me1 | Diagenode | C15410194; RRID:AB_2637078 | |||
| H3K9me2 | Abcam | ab1220; RRID:AB_449854 | |||
| H3K9me3 | Abcam | ab8898; RRID:AB_306848 | |||
| β-Actin | Sigma-Aldrich | A1978; RRID:AB_476692 | |||
| Bacterial and virus strains | |||||
| pzbFGF BL21 Star E. coli | Ludwig et al., 2006 | N/A | |||
| Chemicals, peptides, and recombinant proteins | |||||
| mTesR | Ludwig et al., 2006 | N/A | |||
| E-cadherin | Nagaoka et al., 2010 | N/A | |||
| RPMI 1640 | ThermoFisher Scientific | 22400089 | |||
| Geltrex | ThermoFisher Scientific | A1413302 | |||
| B-27 Supplement (50X), serum free | ThermoFisher Scientific | 17504044 | |||
| B-27 Supplement (50x), minus insulin | ThermoFisher Scientific | A1895601 | |||
| Activin A Recombinant Human Protein | ThermoFisher Scientific | PHC9563 | |||
| FGF-Basic (AA 10-155) Recombinant Human Protein |
ThermoFisher Scientific | PHG0023 | |||
| Purified recombinant zebrafish FGF-Basic | Ludwig et al., 2006 | N/A | |||
| BMP4 Recombinant Human Protein | ThermoFisher Scientific | PHC9533 | |||
| Formaldehyde 16%(w/v) | ThermoFisher Scientific | 28906 | |||
| Puromycin | Sigma-Aldrich | P9620 | |||
| Doxycycline hyclate | Sigma-Aldrich | D9891 | |||
| ROCK inhibitor Y27632 | StemRD | 146986-50-7 | |||
| DAPI | Sigma-Aldrich | D1388 | |||
| Critical commercial assays | |||||
| RNeasy mini Kit | QIAGEN | 74106 | |||
| TURBO DNA-free™ Kit | ThermoFisher/Ambion | AM1907 | |||
| M-MLV Reverse Transcriptase | ThermoFisher/Invitrogen | 28025-013 | |||
| TaqMan Gene Expression | ThermoFisher/Applied Biosystems | 4369016 | |||
| Pierce BCA protein assay kit | ThermoFisher Scientific | 23227 | |||
| Pierce Magnetic CHIP kit | ThermoFisher Scientific | 26157 | |||
| Transcription Factor Staining Buffer Set | ThermoFisher Scientific/eBioscience | 00-5523-00 | |||
| QuickExtract™ DNA extraction solution | Epicenter | QE09050 | |||
| Herculase II Fusion DNA Polymerase | Agilent Technologies | 600679 | |||
| Taq Polymerase | Sigma-Aldrich/Roche | 11146173001 | |||
| Deposited data | |||||
| ATAC-seq data | This paper | GEO: GSE156021 | |||
| RNA-seq data | This paper | GEO: GSE156021 | |||
| ChIP-seq: H3K4me1 | This paper | GEO: GSE156021 | |||
| ChIP-seq: H3K9me2 | This paper | GEO: GSE156021 | |||
| ChIP-seq H3K9me3 | This paper | GEO: GSE156021 | |||
| ChIP-seq GATA6 | This paper | GEO: GSE156021 | |||
| ChIP-seq FOXA2 | This paper | GEO: GSE156021 | |||
| ChIP-seq: EOMES | This paper | GEO: GSE156021 | |||
| Experimental models: Cell lines | |||||
| IPSC-K3 | Si-Tayeb et al., 2010b | N/A | |||
| IPSC-K3 GATA6Ex2Δ31/Δ38 | This paper | N/A | |||
| IPSC-K3 GATA6Ex4Δ1/Δ2 | This paper | N/A | |||
| IPSC-K3 GATA6Ex4Δ1/Δ2;indGATA6 | This paper | N/A | |||
| IPSC-K3 GATA6Ex4Δ1/Δ2;indFOXA2 | This paper | N/A | |||
| Oligonucleotides | |||||
| See Table S7 for oligonucleotides used | Integrated DNA Technologies | N/A | |||
| Recombinant DNA | |||||
| PX459 pSPCas9(BB)-2A-Puro vector | Ran et al., 2013 | Addgene Plasmid #62988 | |||
| pTET-Tight-GATA6-3xFLAG | Fisher et al., 2017 | N/A | |||
| pTET-Tight-FOXA2-3xFLAG | This paper | N/A | |||
| pGL4.20-TTR-3kb | This paper / Promega | E6751 | |||
| Software and algorithms | |||||
| TIDE | (Brinkman et al., 2014); https://tide.nki.nl/ | Version 2.0.1 | |||
| PANTHER classification system | (Mi et al., 2013); http://www.pantherdb.org/ | Version 15 | |||
| Graphpad with PRISM | https://www.graphpad.com/ | Version 8.4.2 | |||
| FlowJo | https://www.flowjo.com/ | Version 10.5.0 | |||
| EaSEQ | (Lerdrup et al., 2016); https://easeq.net/ | Version 1.111 | |||
| FastQC | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | Version 0.11.7 | |||
| Partek Flow | https://www.partek.com/partek-flow/ | Version 9.0.20.0526 | |||
| STAR | Partek Flow | Version 2.6.1d | |||
| BOWTIE2 | Partek Flow | Version 2.2.5 | |||
| MACS2 | Partek Flow | Version 2.1.1 | |||
| HOMER | (Heinz et al., 2010); http://homer.ucsd.edu/homer/ | Version 4.10 | |||
| Other | |||||
| Raw data (GATA6-birA ChIP-seq) | Fisher et al., 2017 | GEO: GSE77360 | |||
| Raw data (Endoderm TF ChIP-seq) | Tsankov et al., 2015 | GEO: GSE61475 | |||
| Raw data (KDM1A ChIP-seq) | Vinckier et al., 2020 | GEO: GSE104840 | |||
| Raw data (H3K4me2 and H3K27ac ChIP-seq) | Loh et al., 2014 | GEO: GSE52658 | |||
| Raw data (PDX1 ChIP-seq, pancreatic progenitor) | Lee et al., 2019 | GEO: GSE114102 | |||
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Human K3 pluripotent stem cells were generated from human foreskin fibroblasts and characterized previously (Si-Tayeb et al., 2010b). All K3 and K3-derived pluripotent cells were cultured in low oxygen conditions (37°C, 4% O2, 5% CO2) on E-cadherin-IgG Fc fusion protein matrix (Nagaoka et al., 2010), in mTeSR culture medium supplemented with zebrafish FGF (40ng/ml) (Ludwig et al., 2006). Cell culture medium was changed daily. Mycoplasma screening was completed at regular intervals.
METHOD DETAILS
CRISPR-Cas9 targeting of human pluripotent stem cells
CRISPR guide RNAs (gRNAs) targeting exon 2 or 4 of the GATA6 gene were chosen based on previously published examples of successful GATA6 CRISPR/Cas9-based gene editing (Shi et al., 2017; Tiyaboonchai et al., 2017). The guide RNA sequences (Table S7) were cloned into the PX459 pSPCas9(BB)-2A-Puro vector (Ran et al., 2013). GATA6+/+ K3 cells were seeded on to a Geltrex-coated (ThermoFisher/GIBCO, CA, #A1413302) 10cm2 tissue culture plate at 75% confluency. The following day, 30μg of the CRISPR/Cas9 plasmid with gRNA was introduced into the K3 pluripotent cells using Lipofectamine 3000 (ThermoFisher, CA, #L3000015), following the manufacturer’s protocol. After 24 hours, 1 μg/ml of puromycin (Sigma Aldrich, MO, #P9620) was supplemented into culture media for 48 hours. The surviving cell colonies were expanded and samples of each collected for analysis of genomic DNA using QuickExtract DNA extraction solution (Epicenter, WI, #QE09050). The CRISPR/Cas9 targeted regions were amplified using Herculase Fusion Polymerase (Agilent, CA, #600675) or Taq polymerase (Sigma Aldrich, MO, #11146173001) and run on Novex® 4%-20% TBE gels (ThermoFisher/Invitrogen, CA, #EC6225) to detect INDELs. Identified potential INDELs were confirmed by analyzing the amplicons using TIDE analysis and nucleotide sequencing (Brinkman et al., 2014).
GATA6 and FOXA2 cDNA rescue vector integration
The GATA6-3xflag cDNA doxycycline-inducible vector has been previously published (Fisher et al., 2017). The FOXA2-3xFLAG codon-optimized sequence was synthesized by GeneArt (ThermoFisher, CA) and inserted into the same vector backbone as used for the pTET-tight-GATA6-3xFLAG plasmid. Sequence was confirmed by sequencing and restriction digests. Each vector was linearized by PVU1 (New England Biolabs, MA, #R0150L) restriction digest for 2 hours at 37°C. The linearized vectors were purified and concentrated by ethanol precipitation and re-suspended in sterile PBS at a concentration of 1 μg/ul. Using a BTX electroporator (NY, ECM 630), GATA6−/−K3 pluripotent cells were electroporated with 30μg of the GATA6-3xFLAG or FOXA2-3xFLAG plasmids, respectively. Electroporated cells were seeded on Geltrex-coated plates with 10μM ROCK inhibitor Y27632 (StemRD, CA, #146986-50-7) for 24 hours. After 48 hours, puromycin was added to the media at 1μg/ml and maintained in the culture medium thereafter. Puromycin-resistant clones were expanded and functional integration of the respective plasmids confirmed by immunofluorescence analysis of FLAG and GATA6/FOXA2 expression with and without doxycycline (Sigma Aldrich, MO, #D9891) supplementation.
Differentiation of pluripotent stem cells
Pluripotent stem cells were seeded on Geltrex-coated tissue culture plates. After 24 hours, the cells were subjected to hepatic differentiation as previously described (Mallanna and Duncan, 2013). Briefly, iPSCs were cultured in RPMI medium supplemented with 1x B27 minus insulin (ThermoFisher Scientific, NY, #A1895601), Activin A(100ng/ml; ThermoFisher Scientific, NY, #PHC9563), fibroblast growth factor 2 (FGF2 20ng/ml; ThermoFisher Scientific, NY, #PHG0023), and bone morphogenetic protein 4 (BMP4,10ng/ml; ThermoFisher Scientific, NY, #PHC9533) for 2 days. RPMI supplemented with 1x B27 minus insulin and Activin A (100ng/ml) was used for a further 3 days to form definitive endoderm. Subsequently, the definitive endoderm was specified toward a hepatic fate for 3 days by supplementing RPMI culture medium with 1x B27 (ThermoFisher Scientific, NY, #17504044), BMP4 (20ng/ml) and FGF2 (10ng/ml).
Immunofluorescence
Cells were washed once with PBS and then fixed with 4% paraformaldehyde (w/v; Santa Cruz, TX, #sc-281692) for 30 minutes. Fixed cells were washed three times and then permeabilized by incubating with 0.5% Triton X-100 for 15 minutes. Samples were blocked with 3% BSA (w/v) in PBS for 1 hour. Primary antibodies were incubated overnight 4°C in 1% BSA (w/v). The following day, samples were washed three times with 1% BSA (w/v) incubated with DAPI (1 μg/ml; Sigma Aldrich, MO, #D1388) and Alexafluor conjugated secondary antibodies (ThermoFisher Scientific, NY) for 2 hours at room temperature. Samples were washed three times with PBS before image capture using the ZOE fluorescent cell imager (BioRad, CA). Control and experiment wells were processed identically. Antibodies and the corresponding dilutions used listed in Table S8.
Western blotting
Cells were washed once with PBS. Whole cell lysate samples were collected using 1x RIPA buffer (Millipore, MA, #20-188) with HALT protease inhibitor cocktail (ThermoFisher Scientific, NY, #78443). Total protein content was measured using BCA assay (ThermoFisher/Pierce, IL, #23227). 5–50 μg total protein was separated by SDS—PAGE using 4%–15% Mini-protean TGX stain-free precast gels (BioRad, CA, #4568184), and transferred to PVDF membranes using the Trans-Blot Turbo Transfer System (BioRad, CA, #1704155). Membranes were blocked with 5% milk or BSA (w/v) in TBS with 0.1% tween (TBS-T), followed by overnight incubation with 1% milk or BSA in TBS-T at 4°C. The following day, membranes were washed with 1% milk or BSA (w/v) in TBS-T. HRP-conjugated secondary antibodies were then added and incubated for 2 hours at 1:5000 dilution. After washing, Clarity chemiluminescent reagent (BioRad, CA, #1705061) was added for 5 minutes before visualization on the Chemidoc Touch gel imager (BioRad, CA). Antibodies used for western blotting are listed in Table S8.
Co-immunoprecipitation
GATA6Ex4Δ1/Δ2:ind GATA6 cells were differentiated to mesendoderm (day 2). Between days 1 and 2, the cells were cultured in the absence or presence of 10ng/ml doxycycline to induce GATA6–3xFLAG expression at endogenous levels. At Day 2, cells were washed once with PBS and lysed with IP lysis buffer (ThermoFisher/Pierce, IL, #87787) with HALT proteinase inhibitor cocktail (ThermoFisher/Pierce, IL, #78443) and Universal Nuclease (ThermoFisher/Pierce, IL, #88701). The sample was incubated at room temperature for 10 minutes before centrifugation to remove cell debris. The lysate was pre-cleared with pre-washed Protein G Dynabeads (ThermoFisher Scientific, NY, #10004D) for 2 hours at 4°C. An input sample was taken and the remaining lysate split equally and incubated with either 1 μg of FLAG M2 (Sigma-Aldrich, MO, #F1804) or 1 μg whole mouse IgG (Sigma-Aldrich, MO, #12–371) antibodies overnight at 4°C. The following day, pre-washed Protein G Dynabeads were added and the mixture was incubated for 2 hours at 4°C. The beads were then washed 5 times with IP lysis buffer and eluted in 1x Lamelli buffer (BioRad, CA, #1610747) at 95°C for 10 minutes. Samples were then analyzed by western blot. Antibodies used for co-IP listed in Table S8.
Flow cytometry
Samples were prepared using a Transcription Factor Staining Buffer Set (ThermoFisher/eBioscience, CA, #00–5523-00). Briefly, iPSCs were differentiated to the stage of interest. Cells were then detached using 0.25% trypsin (ThermoFisher Scientific, NY, #25200056). The cell pellet was washed twice with PBS. Cells were then fixed in fixation buffer (ThermoFisher/eBioscience) for 30 minutes at room temperature. Cells were washed once with permeabilization buffer (ThermoFisher/eBioscience) and spun at 500xg for 5 minutes. The cell pellet was resuspended in permeabilization buffer containing the primary antibody and incubated for 1 hour at room temperature. Cells were washed once with permeabilization buffer and spun at 500xg for 5 minutes. Cells pellets were resuspended in permeabilization buffer containing Alexafluor 488 secondary antibody (ThermoFisher Scientific, NY) and incubated for 30 minutes in the dark at room temperature. Cells were washed once before resuspension in permeabilization buffer and analysis using the Guava EasyCyte Mini (Millipore, MA). Data analysis was completed using FlowJo software (Version 10.5.0; FlowJo, OR) with appropriate controls. Antibodies used listed in Table S8.
Quantitative real-time PCR analysis
Cultured cells were washed once with PBS. RNA samples were then collected using RLT buffer from the RNeasy mini Kit and RNA was isolated following the manufacturer’s protocol (QIAGEN, #74106). The TURBO DNA-free Kit (ThermoFisher/Ambion, NY, #AM1907) was subsequently used to remove contaminating genomic DNA. cDNA was synthesized using M-MLV Reverse Transcriptase (ThermoFisher/Invitrogen, NY, #28025-013). Quantitative real-time PCR (qRT-PCR) analysis was performed using TaqMan® Gene Expression assay (ThermoFisher/Applied Biosystems, NY, #4369016) or Power SYBR Green PCR assay systems (Thermo Fisher/Applied Biosystems, NY, #4367659). qRT-PCR analysis was conducted using a CFX384 real-time PCR machine (BioRad, CA). The primers used for qRT-PCR analysis are listed in Table S7.
Luciferase activity experiments
A3kb TTR promoter region was inserted into pGL4.20 Firefly Luciferase plasmid (Promega, WI, #E6751). The pGL4.20-TTR-3kb and pGL4.20 with no insert were ethanol precipitated and resuspended in sterile PBS at 1 μg/ul. GATA6Ex4Δ1/Δ2;ind FOXA2 cells were cultured with or without doxycycline for 24 hours. Using a BTX electroporator (NY, ECM 630), the cells were electroporated with 30μg of the pGL4.20-TTR-3kb or pGL4.20 plasmid. Electroporated cells were seeded on Geltrex-coated plates with 10μM ROCK inhibitor Y27632 (StemRD, CA, #146986-50-7) for 24 hours, with or without doxycycline. The following day, cells were lysed and analyzed for luciferase luminescence using the Luciferase Assay System (Promega, WI, #E1500). Luminescence was detected using a Synergy HTX Microplate reader (BioTek, VT) and normalized by the total protein content of each sample.
RNA-seq analysis
RNA was isolated and DNase treated as described in the qRT-PCR method section. Following treatment with TURBO DNase, samples were re-purified using the RNeasy kit clean-up protocol. RNA purity was confirmed by Nanodrop (ThermoFisher, CA, #ND-ONE-W). Samples were submitted to BGI, who performed ribosomal depletion, library preparation and sequencing on the BGISEQ-500 platform. 100bp paired-end reads were generated for each sample and FASTQ files provided.
ATAC-seq analysis
Cells were differentiated to the time points of interest and then frozen with 10% DMSO. Samples were submitted to QuickBiology, CA who completed the subsequent processing and analysis steps using established experimental pipelines (Buenrostro et al., 2015). 25 million 150bp paired-end reads were generated using the Illumina HiSeq 4000 platform (Illumina Inc., CA) and FASTQ files provided.
ChIP-seq analysis
Cells were cross-linked with 1% formaldehyde (ThermoFisher/Pierce, IL, #28906) for 10 minutes at room temperature. The cross-linking reaction was subsequently quenched by adding 125mM glycine and incubating at room temperature for 5 minutes. Cells were then washed twice with ice cold PBS, collected by scraping into PBS contained HALT protease and phosphatase inhibitors (ThermoFisher/Pierce, IL, #78440) and pelleted. Chromatin was isolated using MNase enzymatic digestion following the manufacturers protocol (Magnetic CHIP kit, ThermoFisher/Pierce, IL, #26157). Briefly, cell nuclei were first isolated using a membrane lysis buffer. MNase was then added to the isolated nuclei at optimized concentrations and incubated at 37°C for 15 minutes. The enzymatic reaction was stopped with EDTA and digested chromatin was then extracted by brief sonication. Chromatin isolated from 4 individual differentiations were pooled for each condition. Pooled chromatin was incubated with primary antibodies overnight at 4°C. The following day, protein A/G magnetic beads were added to the sample-antibody mixture and incubated at 4°C for 2 hours. Beads were then washed three times in a low salt wash buffer, and once in a high salt wash buffer, before reverse cross-linking in elution buffer. Eluted samples were subjected to proteinase K digestion and the DNA isolated using DNA-binding columns. Samples were submitted to Beijing Genomics Institute (BGI), who performed library preparation and sequencing on the BGISEQ-500 platform. 50bp single end reads were generated and provided as FASTQ files. Table S8 details the antibodies and concentrations used for ChIP-seq analysis.
RIME analysis
Approximately 2x107 K3 GATA6+/+ cells were differentiated to day 4 of the hepatic differentiation protocol. Definitive endoderm cells were cross-linked with 1% formaldehyde for 10 minutes at room temperature. The cross-linking reaction was subsequently quenched by adding 125mM glycine and incubating at room temperature for 5 minutes. Cells were scraped and collected into tubes before centrifuging at 800 x g for 10 minutes. Cells were washed by removing supernatant and adding PBS with 0.5% IGEPAL-CA630 (Sigma Aldrich, MO, #I302) and repeating the centrifugation. Cells were resuspended in PBS-IGEPAL with 1mM PMSF (Sigma Aldrich, MO, #P7626) and the centrifugation repeated once more. Cell pellets were snap frozen. RIME analysis was performed by Active Motif, CA., according to published protocols (Mohammed et al., 2016). GATA6 and IgG antibodies were used to immunoprecipitate 150 μg cross-linked chromatin, followed by mass-spectrometry to identify the peptide fragments. The bioinformatic data provided identified proteins which showed 5-fold enrichment over IgG controls, > 1 spectral count and > 1 unique peptide fragments.
QUANTIFICATION AND STATISTICAL ANALYSIS
Alignment, annotation, and quantification
All raw sequencing FASTQ files were analyzed by FastQC software (Version: 0.11.7). RNA-seq datasets were aligned to the hg19 reference genome using STAR (Version 2.6.1d). Aligned reads were filtered to remove duplicate alignments and mismatches. RNA-seq aligned reads were annotated and normalized by counts per million using Partek Flow software (Partek, MO). ChIP-seq FASTQ files were aligned the hg19 reference genome using Bowtie2 (Version: 2.2.5). Uniquely mapped reads were then used for peak calling by MACS2 (Version: 2.1.1) using the broad peaks setting for histone modifications and default settings for transcription factors. Partek Flow software was used to annotate and quantify peaks. ATAC-seq FASTQ files were trimmed using Partek Flow to remove adaptor sequences. Contaminating mtDNA sequences were removed using Bowtie2 pre-alignment tools (Version: 2.2.5). Trimmed reads were aligned the hg19 reference genome using Bowtie2 (Version: 2.2.5). Uniquely mapped reads were then used for peak calling by MACS2 (Version: 2.1.1) using settings adapted for ATAC-seq samples:-nomodel-shift —100-extsize 200. Partek Flow software was used to annotate and quantify peaks. The HOMER (Version 4.10) mergePeaks script with literal overlap settings ‘-d given’ was used to establish overlapping binding sites/accessible chromatin between datasets. The ‘-venn’ function was used to identify the number of overlapping sites in each group.
Differential analysis of samples
RNA-seq: Differential analysis of normalized RNA-seq data was completed using gene-specific analysis (GSA) in the Partek Flow software suite. GSA analysis calculates the sample distribution of each gene and accordingly applies the appropriate statistical test with multiple correction. GATA6-dependent genes were identified as those which were differentially expressed between the GATA6+/+ and GATA6−/− cells, and could be rescued by exogenous expression of GATA6 cDNA (FDR < 0.05). Heatmap with Euclidean hierarchical clustering was generated using Partek Flow default settings with GATA6-dependent genes. Differential analysis of CHIP- and ATAC-seq quantified peaks was completed using GSA analysis. Peaks were identified as differential between groups using the following statistical cut offs: p < 0.05 and 1.5-fold change. Two component PCA analysis was generated using Partek Flow software using default settings with normalized ATAC-seq values.
Graphs and statistical analysis
Combinational analysis of RNA-, ATAC- and ChIP-seq analysis was completed using HOMER software. The HOMER script findMotifsGenome.pl was used with settings ‘-200bp -mask’ to identify motif sequences enriched in differentially accessible chromatin during differentiation. When comparing between subsets of binding sites, the comparative dataset was used as background with the ‘-bg’ function to remove motifs equally present in all experimental groups. P values calculated by HOMER using hypergeometric enrichment test over background.
The annotatePeaks.pl script was used to identify the nearest transcription start site at regions with GATA6-dependent changes in chromatin accessibility. The identified genes were compared to genes with GATA6-dependent mRNA expression profiles to establish the transcriptional effect of GATA6-dependent changes in chromatin accessibility. The annotatePeaks.pl script was also used to analyze the fragment depth of CHIP or ATAC-seq samples at subsets of transcription factor binding sites.
Heatmaps were created using EaSEQ software (Version 1.111). The HOMER script makeUCSCfile was used to generate bedGraph files with default parameters. Bedgraph files were uploaded to the EaSEQ software. Genomic sites of interest were also uploaded as .txt files. Heatmaps were created using the EaSEQ Heatmap function. Heatmaps for comparisons were processed identically. Statistical comparisons of tag densities were completed using GraphPad Prism software (Version 8.4.2). Tag densities were generated for each sample using the HOMER annotatePeaks.pl script and uploaded to GraphPad software. Comparisons of tag densities at the same sites between samples were completed using the paired Wilcoxon rank signed rank test. Multiple paired testing was completed using Friedman’s ANOVA with Dunn’s multiple comparisons. Direct comparisons of different genomic loci within the same sample were completed using the non-paired Mann-Whitney U Test. Testing across multiple non-paired groups was completed using Kruskall-Wallis ANOVA with Dunn’s multiple correction testing.
Supplementary Material
Highlights.
Newly accessible chromatin in definitive endoderm is enriched for GATA motifs
Definitive endoderm patterning is significantly perturbed in GATA6−/− cells
GATA6 interacts with multiple chromatin remodeling complexes
GATA6-dependent patterning commonly precedes the onset of gene expression
ACKNOWLEDGMENTS
This work was supported by funds from the National Institutes of Health (DK102716, DK119728, DK123704, GM130457, and CA138313) and the American Liver Foundation (2018 Charles Trey MD Memorial Postdoctoral Research to J.A.H.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109145.
DECLARATION OF INTERESTS
S.A.D. is the founder of Grùthan Biosciences, LLC. The remaining authors declare no competing interests.
REFERENCES
- AlAbdi L, Saha D, He M, Dar MS, Utturkar SM, Sudyanti PA, McCune S, Spears BH, Breedlove JA, Lanman NA, and Gowher H (2020). Oct4-mediated inhibition of Lsd1 activity promotes the active and primed state of pluripotency enhancers. Cell Rep. 30, 1478–1490.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuling E, Aronson BE, Tran LMD, Stapleton KA, ter Horst EN, Vissers LATM, Verzi MP, and Krasinski SD (2012). GATA6 is required for proliferation, migration, secretory cell maturation, and gene expression in the mature mouse colon. Mol. Cell. Biol. 32, 3392–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman EK, Chen T, Amendola M, and van Steensel B (2014). Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ (2015). ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco M, Delgado I, Soria B, Martin F, and Rojas A (2012). GATA4 and GATA6 control mouse pancreas organogenesis. J. Clin. Invest. 122, 3504–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenik BK, and Shilatifard A (2021). COMPASS and SWI/SNF complexes in development and disease. Nat. Rev. Genet. 22, 38–58. [DOI] [PubMed] [Google Scholar]
- Cernilogar FM, Hasenöder S, Wang Z, Scheibner K, Burtscher I, Sterr M, Smialowski P, Groh S, Evenroed IM, Gilfillan GD, et al. (2019). Pre-marked chromatin and transcription factor co-binding shape the pioneering activity of Foxa2. Nucleic Acids Res. 47, 9069–9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia CY, Madrigal P, Denil SLIJ, Martinez I, Garcia-Bernardo J, El-Khairi R, Chhatriwala M, Shepherd MH, Hattersley AT, Dunn NR, and Vallier L (2019). GATA6 cooperates with EOMES/SMAD2/3 to deploy the gene regulatory network governing human definitive endoderm and pancreas formation. Stem Cell Reports 12, 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, and Zaret KS (2002). Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9, 279–289. [DOI] [PubMed] [Google Scholar]
- Costa RH, and Grayson DR (1991). Site-directed mutagenesis of hepatocyte nuclear factor (HNF) binding sites in the mouse transthyretin (TTR) promoter reveal synergistic interactions with its enhancer region. Nucleic Acids Res. 19, 4139–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Franco E, Shaw-Smith C, Flanagan SE, Shepherd MH, Hattersley AT, and Ellard S; International NDM Consortium (2013). GATA6 mutations cause a broad phenotypic spectrum of diabetes from pancreatic agenesis to adult-onset diabetes without exocrine insufficiency. Diabetes 62, 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaghey J, Thakurela S, Charlton J, Chen JS, Smith ZD, Gu H, Pop R, Clement K, Stamenova EK, Karnik R, et al. (2018). Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat. Genet. 50, 250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez Garcia M, Moore CD, Schulz KN, Alberto O, Donague G, Harrison MM, Zhu H, and Zaret KS (2019). Structural features of transcription factors associating with nucleosome binding. Mol. Cell 75, 921–932.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JB, Pulakanti K, Rao S, and Duncan SA (2017). GATA6 is essential for endoderm formation from human pluripotent stem cells. Biol. Open 6, 1084–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genga RMJ, Kernfeld EM, Parsi KM, Parsons TJ, Ziller MJ, and Maehr R (2019). Single-cell RNA-sequencing-based CRISPRi screening resolves molecular drivers of early human endoderm development. Cell Rep. 27, 708–718.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guye P, Ebrahimkhani MR, Kipniss N, Velazquez JJ, Schoenfeld E, Kiani S, Griffith LG, and Weiss R (2016). Genetically engineering self-organization of human pluripotent stem cells intoa liver bud-like tissue using Gata6. Nat. Commun. 7, 10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineagedetermining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop JA, and Duncan SA (2019). The use of human pluripotent stem cells for modeling liver development and disease. Hepatology 69, 1306–1316. [DOI] [PubMed] [Google Scholar]
- Heslop JA, and Duncan SA (2020). FoxA factors: The chromatin key and doorstop essential for liver development and function. Genes Dev. 34, 1003–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi M, Cuesta I, Donahue G, Takenaka N, Osipovich AB, Magnuson MA, Roder H, Seeholzer SH, Santisteban P, and Zaret KS (2020). Gene network transitions in embryos depend upon interactions between a pioneer transcription factor and core histones. Nat. Genet. 52, 418–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijzer R, van Tuyl M, Meijers C, Post M, Tibboel D, Grosveld F, and Koutsourakis M (2001). The transcription factor GATA6 is essential for branching morphogenesis and epithelial cell differentiation during fetal pulmonary development. Development 128, 503–511. [DOI] [PubMed] [Google Scholar]
- Lee K, Cho H, Rickert RW, Li QV, Pulecio J, Leslie CS, and Huangfu D (2019). FOXA2 is required for enhancer priming during pancreatic differentiation. Cell Rep. 28, 382–393.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerdrup M, Johansen JV, Agrawal-Singh S, and Hansen K (2016). An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat. Struct. Mol. Biol. 23, 349–357. [DOI] [PubMed] [Google Scholar]
- Li QV, Dixon G, Verma N, Rosen BP, Gordillo M, Luo R, Xu C, Wang Q, Soh CL, Yang D, et al. (2019). Genome-scale screens identify JNK-JUN signaling as a barrier for pluripotency exit and endoderm differentiation. Nat. Genet. 51,999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CM, Mukherjee S, Tiyaboonchai A, Maguire JA, Cardenas-Diaz FL, French DL, and Gadue P (2018). GATA6 suppression enhances lung specification from human pluripotent stem cells. J. Clin. Invest. 128, 2944–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JT, Corbett JL, Heslop JA, and Duncan SA (2020). Enhanced genome editing in human iPSCs with CRISPR-CAS9 by co-targeting ATP1a1. PeerJ 8, e9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh KM, Ang LT, Zhang J, Kumar V, Ang J, Auyeong JQ, Lee KL, Choo SH, Lim CYY, Nichane M, et al. (2014). Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell 14, 237–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig TE, Bergendahl V, Levenstein ME, Yu J, Probasco MD, and Thomson JA (2006). Feeder-independent culture of human embryonic stem cells. Nat. Methods 3, 637–646. [DOI] [PubMed] [Google Scholar]
- Mallanna SK, and Duncan SA (2013). Differentiation of hepatocytes from pluripotent stem cells. Curr. Protoc. Stem Cell Biol. 26, 1G.4.1–1G.4.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayran A, and Drouin J (2018). Pioneer transcription factors shape the epigenetic landscape. J. Biol. Chem. 293, 13795–13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayran A, Sochodolsky K, Khetchoumian K, Harris J, Gauthier Y, Bemmo A, Balsalobre A, and Drouin J (2019). Pioneer and nonpioneer factor cooperation drives lineage specific chromatin opening. Nat. Commun. 10, 3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meers MP, Janssens DH, and Henikoff S (2019). Pioneer factor-nucleosome binding events during differentiation are motif encoded. Mol. Cell 75, 562–575.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, and Thomas PD (2013). PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41 (Database issue), D377–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed H, Taylor C, Brown GD, Papachristou EK, Carroll JS, and D’Santos CS (2016). Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat. Protoc. 11, 316–326. [DOI] [PubMed] [Google Scholar]
- Molkentin JD (2000). The zinc finger-containing transcription factors GATA-4, -5, and-6. Ubiquitously expressed regulators of tissue-specific gene expression. J. Biol. Chem. 275, 38949–38952. [DOI] [PubMed] [Google Scholar]
- Nagaoka M, Si-Tayeb K, Akaike T, and Duncan SA (2010). Culture of human pluripotent stem cells using completely defined conditions on a recombinant E-cadherin substratum. BMC Dev. Biol. 10, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J,Agarwala V, Scott DA, and Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Wasson LK, Willcox JAL, Morton SU, Gorham JM, De-Laughter DM, Neyazi M, Schmid M, Agarwal R, Jang MY, et al. ; Pediatric Cardiac Genomics Consortium (2020). GATA6 mutations in hiPSCs inform mechanisms for maldevelopment of the heart, pancreas, and diaphragm. eLife 9, 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, and Shi Y (2005). Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell 19, 857–864. [DOI] [PubMed] [Google Scholar]
- Shi Z-D, Lee K, Yang D, Amin S, Verma N, Li QV, Zhu Z, Soh C-L, Kumar R, Evans T, et al. (2017). Genome editing in hPSCs reveals GATA6 Haploinsufficiency and a genetic interaction with GATA4 in human pancreatic development. Cell Stem Cell 20, 675–688.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, North PE, Dalton S, and Duncan SA (2010a). Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology 51, 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K, Noto FK, Sepac A, Sedlic F, Bosnjak ZJ, Lough JW, and Duncan SA (2010b). Generation of human induced pluripotent stem cells by simple transient transfection of plasmid DNA encoding reprogramming factors. BMC Dev. Biol. 10, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaku M, Grimm SA, Shimbo T, Perera L, Menafra R, Stunnenberg HG, Archer TK, Machida S, Kurumizaka H, and Wade PA (2016). GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol. 17, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Takizawa Y, Takaku M, Kato D, Kumagawa Y, Grimm SA, Wade PA, and Kurumizaka H (2020). Interaction of the pioneer transcription factor GATA3 with nucleosomes. Nat. Commun. 11, 4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiyaboonchai A, Cardenas-Diaz FL, Ying L, Maguire JA, Sim X, Jobaliya C, Gagne AL, Kishore S, Stanescu DE, Hughes N, et al. (2017). GATA6 plays an important role in the induction of human definitive endoderm, development of the pancreas, and functionality of pancreatic β cells. Stem Cell Reports 8, 589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov AM, Gu H, Akopian V, Ziller MJ, Donaghey J, Amit I, Gnirke A, and Meissner A (2015). Transcription factor binding dynamics during human ES cell differentiation. Nature 518, 344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinckier NK, Patel NA, Geusz RJ, Wang A, Wang J, Matta I, Harrington AR, Wortham M, Wetton N, Wang J, et al. (2020). LSD1-mediated enhancer silencing attenuates retinoic acid signalling during pancreatic endocrine cell development. Nat. Commun. 11, 2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt AJ, Zhao R, Li J, and Duncan SA (2007). Development of the mammalian liver and ventral pancreas is dependent on GATA4. BMC Dev. Biol. 7, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, and Young RA (2012). Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482, 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan S, Borok MJ, Decker KJ, Battle MA, Duncan SA, Hale MA, Macdonald RJ, and Sussel L (2012). Pancreas-specific deletion of mouse Gata4 and Gata6 causes pancreatic agenesis. J. Clin. Invest. 122, 3516–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS (2020). Pioneer transcription factors initiating gene network changes. Annu. Rev. Genet. 54, 367–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Watt AJ, Li J, Luebke-Wheeler J, Morrisey EE, and Duncan SA (2005). GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol. Cell. Biol. 25, 2622–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R, and Blobel GA (2010). Gata transcription factors and cancer. Genes Cancer 1, 1178–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ATAC-seq, RNA-seq, and ChIP-seq datasets generated during this study are deposited in the GEO database under the GEO ID code: GEO: GSE156021. Previously published datasets used as part of this study are available in the GEO database. See key resources table for details.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | |||
|---|---|---|---|---|---|
| Antibodies | |||||
| GATA6 | Cell Signaling | 5851; RRID:AB_10705521 | |||
| GATA4 | Santa Cruz | SC-1237; RRID:AB_2108747 | |||
| FOXA2 | RnD | AF2400; RRID: AB_2294104 | |||
| SOX17 | RnD | AF1924; RRID:AB_355060 | |||
| OCT4 | Santa Cruz | SC-9081; RRID:AB_2167703 | |||
| HNF4A | Santa Cruz | SC-6556; RRID:AB_2117025 | |||
| EOMES | Cell Signaling | 66325; RRID: Unknown | |||
| CHD4 | Cell Signaling | 12011; RRID:AB_2734702 | |||
| SMARCA4 | Cell Signaling | 49360’ RRID:AB_2728743 | |||
| KDM1A | Cell Signaling | 2184; RRID:AB_2070132 | |||
| WDR5 | Cell Signaling | 13105; RRID:AB_2620133 | |||
| FLAG | Sigma-Aldrich | F1804; RRID:AB_262044 | |||
| Normal Mouse IgG | Sigma-Aldrich | 12-371; RRID:AB_145840 | |||
| H3K4me1 | Diagenode | C15410194; RRID:AB_2637078 | |||
| H3K9me2 | Abcam | ab1220; RRID:AB_449854 | |||
| H3K9me3 | Abcam | ab8898; RRID:AB_306848 | |||
| β-Actin | Sigma-Aldrich | A1978; RRID:AB_476692 | |||
| Bacterial and virus strains | |||||
| pzbFGF BL21 Star E. coli | Ludwig et al., 2006 | N/A | |||
| Chemicals, peptides, and recombinant proteins | |||||
| mTesR | Ludwig et al., 2006 | N/A | |||
| E-cadherin | Nagaoka et al., 2010 | N/A | |||
| RPMI 1640 | ThermoFisher Scientific | 22400089 | |||
| Geltrex | ThermoFisher Scientific | A1413302 | |||
| B-27 Supplement (50X), serum free | ThermoFisher Scientific | 17504044 | |||
| B-27 Supplement (50x), minus insulin | ThermoFisher Scientific | A1895601 | |||
| Activin A Recombinant Human Protein | ThermoFisher Scientific | PHC9563 | |||
| FGF-Basic (AA 10-155) Recombinant Human Protein |
ThermoFisher Scientific | PHG0023 | |||
| Purified recombinant zebrafish FGF-Basic | Ludwig et al., 2006 | N/A | |||
| BMP4 Recombinant Human Protein | ThermoFisher Scientific | PHC9533 | |||
| Formaldehyde 16%(w/v) | ThermoFisher Scientific | 28906 | |||
| Puromycin | Sigma-Aldrich | P9620 | |||
| Doxycycline hyclate | Sigma-Aldrich | D9891 | |||
| ROCK inhibitor Y27632 | StemRD | 146986-50-7 | |||
| DAPI | Sigma-Aldrich | D1388 | |||
| Critical commercial assays | |||||
| RNeasy mini Kit | QIAGEN | 74106 | |||
| TURBO DNA-free™ Kit | ThermoFisher/Ambion | AM1907 | |||
| M-MLV Reverse Transcriptase | ThermoFisher/Invitrogen | 28025-013 | |||
| TaqMan Gene Expression | ThermoFisher/Applied Biosystems | 4369016 | |||
| Pierce BCA protein assay kit | ThermoFisher Scientific | 23227 | |||
| Pierce Magnetic CHIP kit | ThermoFisher Scientific | 26157 | |||
| Transcription Factor Staining Buffer Set | ThermoFisher Scientific/eBioscience | 00-5523-00 | |||
| QuickExtract™ DNA extraction solution | Epicenter | QE09050 | |||
| Herculase II Fusion DNA Polymerase | Agilent Technologies | 600679 | |||
| Taq Polymerase | Sigma-Aldrich/Roche | 11146173001 | |||
| Deposited data | |||||
| ATAC-seq data | This paper | GEO: GSE156021 | |||
| RNA-seq data | This paper | GEO: GSE156021 | |||
| ChIP-seq: H3K4me1 | This paper | GEO: GSE156021 | |||
| ChIP-seq: H3K9me2 | This paper | GEO: GSE156021 | |||
| ChIP-seq H3K9me3 | This paper | GEO: GSE156021 | |||
| ChIP-seq GATA6 | This paper | GEO: GSE156021 | |||
| ChIP-seq FOXA2 | This paper | GEO: GSE156021 | |||
| ChIP-seq: EOMES | This paper | GEO: GSE156021 | |||
| Experimental models: Cell lines | |||||
| IPSC-K3 | Si-Tayeb et al., 2010b | N/A | |||
| IPSC-K3 GATA6Ex2Δ31/Δ38 | This paper | N/A | |||
| IPSC-K3 GATA6Ex4Δ1/Δ2 | This paper | N/A | |||
| IPSC-K3 GATA6Ex4Δ1/Δ2;indGATA6 | This paper | N/A | |||
| IPSC-K3 GATA6Ex4Δ1/Δ2;indFOXA2 | This paper | N/A | |||
| Oligonucleotides | |||||
| See Table S7 for oligonucleotides used | Integrated DNA Technologies | N/A | |||
| Recombinant DNA | |||||
| PX459 pSPCas9(BB)-2A-Puro vector | Ran et al., 2013 | Addgene Plasmid #62988 | |||
| pTET-Tight-GATA6-3xFLAG | Fisher et al., 2017 | N/A | |||
| pTET-Tight-FOXA2-3xFLAG | This paper | N/A | |||
| pGL4.20-TTR-3kb | This paper / Promega | E6751 | |||
| Software and algorithms | |||||
| TIDE | (Brinkman et al., 2014); https://tide.nki.nl/ | Version 2.0.1 | |||
| PANTHER classification system | (Mi et al., 2013); http://www.pantherdb.org/ | Version 15 | |||
| Graphpad with PRISM | https://www.graphpad.com/ | Version 8.4.2 | |||
| FlowJo | https://www.flowjo.com/ | Version 10.5.0 | |||
| EaSEQ | (Lerdrup et al., 2016); https://easeq.net/ | Version 1.111 | |||
| FastQC | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | Version 0.11.7 | |||
| Partek Flow | https://www.partek.com/partek-flow/ | Version 9.0.20.0526 | |||
| STAR | Partek Flow | Version 2.6.1d | |||
| BOWTIE2 | Partek Flow | Version 2.2.5 | |||
| MACS2 | Partek Flow | Version 2.1.1 | |||
| HOMER | (Heinz et al., 2010); http://homer.ucsd.edu/homer/ | Version 4.10 | |||
| Other | |||||
| Raw data (GATA6-birA ChIP-seq) | Fisher et al., 2017 | GEO: GSE77360 | |||
| Raw data (Endoderm TF ChIP-seq) | Tsankov et al., 2015 | GEO: GSE61475 | |||
| Raw data (KDM1A ChIP-seq) | Vinckier et al., 2020 | GEO: GSE104840 | |||
| Raw data (H3K4me2 and H3K27ac ChIP-seq) | Loh et al., 2014 | GEO: GSE52658 | |||
| Raw data (PDX1 ChIP-seq, pancreatic progenitor) | Lee et al., 2019 | GEO: GSE114102 | |||


