Summary
In the field of epigenetic inheritance, delineating molecular mechanisms implicated in the transfer of paternal environmental conditions to descendants has been elusive. This protocol details how to track sperm chromatin intergenerationally. We describe mouse model design to probe chromatin states in single mouse sperm and techniques to assess pre-implantation embryo chromatin and gene expression. We place emphasis on how to obtain high-quality and quantifiable data sets in sperm and embryos, as well as highlight the limitations of working with low input.
For complete details on the use and execution of this protocol, please refer to Lismer et al. (2021).
Subject areas: Biophysics, Genomics, Sequencing, RNAseq, ChIPseq, Model Organisms, Gene Expression, Chromatin immunoprecipitation (ChIP)
Graphical Abstract
Highlights
-
•
Assessment of paternal environmental exposure effects on sperm chromatin by ChIP-seq
-
•
Tracking chromatin changes in sperm to embryos using ultra-low-input (ULI)-ChIP-seq
-
•
Identification of gene expression changes in embryos using low-input mRNA-seq
In the field of epigenetic inheritance, delineating molecular mechanisms implicated in the transfer of paternal environmental conditions to descendants has been elusive. This protocol details how to track sperm chromatin intergenerationally. We describe mouse model design to probe chromatin states in single mouse sperm and techniques to assess pre-implantation embryo chromatin and gene expression. We place emphasis on how to obtain high-quality and quantifiable data sets in sperm and embryos, as well as highlight the limitations of working with low input.
Before you begin
The following protocol can be used to assess the role of chromatin in the paternal transmission of any environmental exposure. This protocol can be applied to various environmental exposure models (e.g., diet, or toxicants), with variations in the developmental timing of exposure, mouse strains, and on any pre-implantation embryo stage. The methodologies shared here offer promising avenues to elucidate the complexity of altered chromatin state inheritance from sperm to the pre-implantation embryo.
Preparation of stock solutions
Timing: 1 day
| 1M CaCl2 | Dissolve 11.09 g of CaCl2 in 90 mL of ddH2O. Adjust volume to 100 mL. Autoclave and store at 20°C–25°C for up to 1 year. |
| 0.5M EDTA | Dissolve 18.6 g of EDTA in 90 mL of ddH2O. Stir solution with a magnetic stirrer and slowly add NaOH pellets until pH of the solution reaches 8.0. Adjust final volume to 100 mL with ddH2O. Autoclave and store at 20°C–25°C for up to 1 year. |
| 0.5M EGTA | Dissolve 19 g of EGTA in 90 mL of ddH2O. Stir solution with a magnetic stirrer and slowly add NaOH pellets until pH of the solution reaches 8.0. Adjust final volume to 100 mL with ddH2O. Autoclave and store at 20°C–25°C for up to 1 year. |
| 0.5M HEPES | Dissolve 23.83 g of HEPES in 190 mL ddH2O. Add HCl until the pH of the solution reaches 7.4. Adjust final volume to 200 mL with ddH2O. Autoclave and store at 20°C–25°C for up to 1 year. |
| 1M MgCl2 | Dissolve 9.52 g of MgCl2 in 90 mL ddH2O. Adjust final volume to 100 mL with ddH2O. Autoclave and store at 20°C–25°C for up to 1 year. |
| 1M MgSO4 | Dissolve 24.6 g of MgSO4 in 90 mL ddH2O. Adjust final volume to 100 mL with ddH2O. Autoclave and store at 20°C–25°C for up to 1 year. |
| 1M KCl | Dissolve 7.45 g of KCl in 90 mL ddH2O. Adjust final volume to 100 mL with ddH2O. Autoclave and store at 20°C–25°C for up to 1 year. |
| 10 mg/mL Proteinase K | Dissolve 10 mg of proteinase K in a solution composed of 940 μL ddH2O, 50 μL of 1M Tris-HCl (pH 8.0) and 10 μL of 1M CaCl2. Store the reconstituted Proteinase K in a −20°C freezer for 2–3 months. |
| 10 mg/mL RNase A | Dissolve 10 mg of RNase A in a solution composed of 975 μL ddH2O, 10 μL of 1M Tris-HCl (pH 8.0) and 3 μL of 5M NaCl. Store the reconstituted RNase A in a −20°C freezer for 2–3 months. |
| 3M Sodium acetate | Dissolve 40.8 g of sodium acetate in 90 mL ddH2O. Add glacial acetic acid until the pH of the solution reaches 5.2. Adjust final volume to 100 mL with ddH2O. Filter solution through a 0.22 μm filter and store at 20°C–25°C for up to 1 year. |
| 1M NaHCO3 | Dissolve 8.4 g of NaHCO3 in 90 mL ddH2O. Adjust final volume to 100 mL with ddH2O. Autoclave and store at 20°C–25°C for up to 1 year. |
| 5M NaCl | Dissolve 146.1 g of NaCl in 450 mL ddH2O. Adjust final volume to 500 mL with ddH2O. Autoclave and store at 20°C–25°C for up to 1 year. |
| 0.5M Sodium pyruvate | Dissolve 55 mg of sodium pyruvate in 1 mL ddH2O. Store 60 μL aliquots in a −20°C freezer. |
| 0.1M PMSF | Dissolve in 17.4 mg PMSF in 1 mL isopropanol. Store 50 μL aliquots in a −20°C for 2–3 months. |
Donner’s stock for sperm collection: Filter solution through a 0.22 μm filter and store in a 4°C fridge for up to 1 year
| Reagent | Final concentration | Amount |
|---|---|---|
| 5M NaCl | 135 mM | 5.4 mL |
| 1M KCl | 5 mM | 1 mL |
| 1M MgSO4 | 1 mM | 200 μL |
| 1M CaCl2 | 2 mM | 400 μL |
| 0.5M HEPES (pH 7.4) | 30 mM | 12 mL |
| ddH2O | N/A | 181 mL |
MNase Buffer Stock solution for sperm ChIP-seq: Autoclave solution and store at 20°C–25°C for up to 1 year
| Reagent | Final concentration | Amount |
|---|---|---|
| 1M Tris-HCl (pH 7.5) | 85 mM | 8.5 mL |
| 1M MgCl2 | 3 mM | 300 μL |
| 1M CaCl2 | 2 mM | 200 μL |
| ddH2O | N/A | 86 mL |
Buffer 1 Stock for sperm ChIP-seq: Filter solution through a 0.22 μm filter and at 20°C–25°C for up to 1 year
| Reagent | Final concentration | Amount |
|---|---|---|
| 1M Tris-HCl (pH 7.5) | 15.88 mM | 1.5 mL |
| 1M KCl | 61.14 mM | 6 mL |
| 1M MgCl2 | 5.26 mM | 500 μL |
| 0.5M EGTA | 0.11 mM | 20 μL |
| ddH2O | N/A | 87 mL |
Immunoprecipitation buffer for pre-implantation embryo ULI-ChIP-seq: Filter through a 0.22 μm filter and store at 20°C–25°C for up to 1 year
| Reagent | Final concentration | Amount |
|---|---|---|
| 1M Tris-HCl (pH 7.5) | 21.05 mM | 1 mL |
| 0.5M EDTA | 2.11 mM | 200 μL |
| 5M NaCl | 157.89 mM | 1.5 mL |
| 100× Triton | 1.05× | 500 μL |
| ddH2O | N/A | 44.3 mL |
Add protease inhibitor and PMSF fresh on day of experiment as indicated in step 1.b. of the ULI-ChIP-seq guidelines.
Exposing male mice to an environmental stressor
Timing: 2.5–4 months
-
1.
Expose male mice to an environmental stressor of interest (e.g., toxicant, diet, trauma…) for two full spermatogenic cycles beginning at weaning (i.e., 3 weeks of age).
Alternatives: Timing and duration of environmental exposure can vary as follows:
Exposure may begin in utero by exposing a pregnant female to the environmental exposure (Lambrot et al., 2013), at weaning during the first wave of spermatogenesis (Lismer et al., 2021), or at adulthood.
Exposure should last at least one full spermatogenic cycle (35 days in mice) (Oakberg, 1956) but can extend throughout multiple cycles.
CRITICAL: In Lismer et al., 2021, folate sufficient (FS) male mice were fed a diet with 2.0 mg of folate/kg, corresponding to the recommended folate amount for rodents (Reeves et al., 1993). Folate deficient (FD) mice were given a diet composed of 0.3 mg of folate/kg, consistent with the folate intake measured in folate deficient populations (Rogers et al., 2018). A complete depletion of dietary folate in mice has been shown to also result in DNA breaks (Swayne et al., 2012). Exposure to stressors at environmentally-relevant doses is recommended to obtain physiologically pertinent datasets. Exposing male mice to pharmacological doses may induce DNA breaks in germ cells and confound epigenetic inheritance studies.
Breeding exposed males to non-exposed superstimulated and superovulated females
Timing: variable depending on mouse facility size and number of personnel.
-
2.Determine the estrus phase of female mice that are between 6–9 weeks of age by performing a vaginal cytology (Figures 1A–1D) and (Byers et al., 2012):
-
a.Draw thumb-sized circles on glass slides using a hydrophobic marker.
-
b.With a sterile Pasteur pipette dropper, introduce approximatively 200 μL 1× PBS in the vaginal opening of a restrained female mouse. Aspirate and eject 1× PBS 3 times.
-
c.Place vaginal smear suspension in drawn circles.
-
d.Identify estrus phase by looking at slides under a microscope (Figures 1A–1D).
-
a.
-
3.
When in diestrus (Figure 1D), inject female intraperitoneally with 5 IU of PMSG diluted in chilled 0.9% NaCl. Super-stimulating in the diestrus phase will ensure that the female is in the estrus phase at time of breeding.
Note: the diestrus phase is evident by the enrichment of leukocyte cells in the vaginal smears (Figure 1D) and (Byers et al., 2012).
-
4.
48 h after superstimulation, superovulate females by intraperitoneal injection of 5 IU of hCG diluted in chilled 0.9% NaCl.
CRITICAL: Administer hormones in the evening between 16:00 to 18:00. Always be consistent with time of injection.
CRITICAL: Do not freeze-thaw hormone aliquots. Place in a −80°C freezer for no longer than 3 months.
-
5.
After hCG injection, place females with a male that has been exposed to an environmental stressor for 6–8 weeks (i.e., 9–11 weeks of age if environmental stressor began at weaning).
-
6.
The next morning, check females for vaginal plug. The presence of a plug is considered embryonic day E0.5.
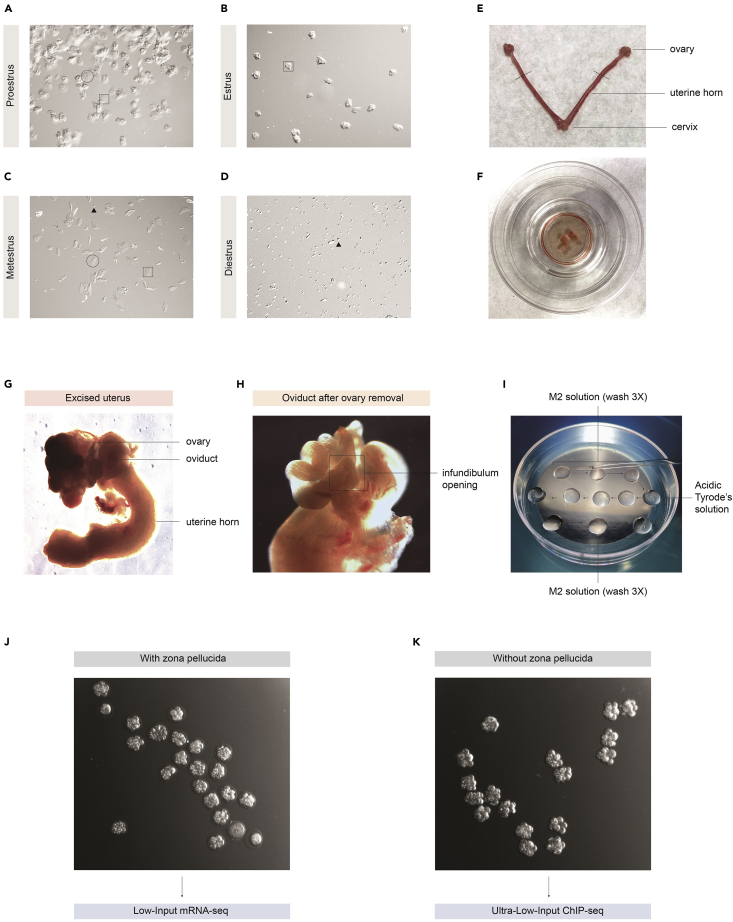
Figure 1.
8-cell embryo collection for ultra-low-input ChIP-seq and low-input mRNA-seq
(A–D) Vaginal smears of female mice in different estrus phases. Difference in cell type composition indicates the estrus phase. The three identifiable cells types are nucleated epithelial cells (circle), cornified epithelial cells (square), and leukocyte cells (arrow).
(E) Dissected uterus of female mouse after superstimulation and superovulation. Dashed lines indicate where the uterine horns should be excised.
(F) Cut left and right ovaries, oviducts, and segments of the uterus horn, in M2 solution of an IVF dish.
(G) Cut ovary, oviduct, and segment of the uterus horn under a dissection microscope.
(H) Oviduct after ovary removal. Infundibulum opening is shown (square).
(I) M2 solution and Acidic Tyrode’s solution 60 mm Petri dish set-up.
(J) 8-cell embryos and unfertilized ovaries before zona pellucida removal. At this stage, 8-cell embryos can be retrieved for Low-Input mRNA-seq.
(K) 8-cell embryos after zona pellucida removal. At this stage, 8-cell embryos can be retrieved for Ultra-Low-Input ChIP-seq.
Pre-implantation embryo collection for ULI-ChIP-seq and low-input mRNA-seq
Timing: 20–30 min per embryo collection
-
7.
Pull glass Pasteur pipettes using a Bunsen burner. Autoclave to sterilize.
-
8.
Prepare M2 solution by supplementing media with 1% Penicillin-Streptomycin. Thaw an aliquot of Acidic Tyrode’s solution. Warm solutions to 37°C.
-
9.Sacrifice female on embryonic day E2.5 to collect embryos at the 8-cell stage.Alternatives: The downstream Ultra-Low-Input ChIP-seq and Low-Input mRNA-seq experiments can also be used for different pre-implantation embryo stages. Precise timing of collection for pre-implantation embryo stages post-hCG injection is as follows (Zhang et al., 2016):
PN5 zygote: 27–28 h
Early 2-cell embryo: 30 h
Late 2-cell embryo: 43 h
4-cell embryo: 54–56 h
8-cell embryo: 68–70 h
- Blastocyst: 92–94 h
 CRITICAL: Always collect embryos at the same time of day and restrict collection time to a two-hour window to minimize variability.
CRITICAL: Always collect embryos at the same time of day and restrict collection time to a two-hour window to minimize variability.-
a.Remove uterus from pregnant female using 8.5 cm dissecting scissors (Figure 1E) and cut the uterus horns 1/3 below the oviduct to remove the uterine fundus and cervical regions (Figures 1E–1G). Wash ovaries, oviducts, and uterine horn segments in M2 solution to remove debris. Place excised uterus in 1 mL of M2 solution in an IVF 60 mm Petri dish (Figures 1F and 1G).Note: Hormone efficiency may vary between females (Luo et al., 2011). Indicators that hormone injections were effective are the redness and swelling of the uterus, as well as a high number of corpora lutea in the ovary.
- b.
-
a.
Note: A blunt-ended needle can be made by scraping the needle tip on sandpaper. Wash and sterilize needle after each embryo collection.
-
10.
Prepare a 60 mm Petri dish with 3 × 50 μL drops of M2 solution, 5 × 50 μL drops of Acidic Tyrode’s (see key resources table) and 3 × 50 μL drops of M2 solution (Figure 1I).
-
11.
Collect embryos with a pulled glass Pasteur pipette via mouth pipetting. Use filter tips to create a barrier between the experimenter and the M2 solution. Minimize amount of aspirated liquid when retrieving embryos to reduce carry-over of debris.
-
12.
Wash embryos by passing them through three drops of M2 solution (Figure 1I).
Note: If using embryos for the Low-Input mRNA-seq, the removal of the zona pellucida (steps 7–10) is not required (Figure 1J). After embryos have been washed 3 times in M2 solution and placed in an Eppendorf DNA Lo-Bind tube, add 20 μL of DNA/RNA Protection Reagent and flash freeze.
-
13.
Wash interior of pulled glass Pasteur pipette by aspirating Acidic Tyrode’s solution.
-
14.
Pass embryos through drops of Acidic Tyrode’s until removal of their zona pellucida (Figures 1I–1K). Minimize transfer of M2 solution into the Acidic Tyrode’s drops as this will decrease the Acidic Tyrode’s efficiency.
CRITICAL: When embryos are in Acidic Tyrode’s solution, do not allow them to touch the bottom of the Petri dish or they will stick to it and rupture upon removal. Furthermore, prolonged exposure to the Acidic Tyrode’s solution will result in embryo cell dissociation and may lead to cytolysis.
-
15.
After removal of the zona pellucida, wash embryos three times in M2 solution (Figure 1I).
-
16.
Using a new glass Pasteur pipette, aspirate all the embryos with as little M2 solution as possible. Cautiously transfer to an Eppendorf DNA Lo-Bind tube. Ensure that no bubbles are formed during the transfer process and that the glass Pasteur pipette tip does not break.
-
17.
Verify that embryos are not left in the glass Pasteur pipette by washing the interior of the pipette in a M2 solution drop.
-
18.
If using embryos for the ULI-ChIP-seq, add 20 μL of Nuclear Isolation Buffer to Eppendorf DNA Lo-Bind tube and flash freeze (Figure 1K).
CRITICAL: Perform embryo collection rapidly to maintain DNA and/or RNA integrity. Target times per female from moment of sacrifice to flash freezing of embryos are 15 min for ULI-ChIP-seq embryos and 10 min for Low-Input mRNA-Seq embryos.
Sperm collection
Timing: 3 h
-
19.
Prepare Donner’s solution from Donner’s stock (5 mL per mouse) and sterile filter with a 0.22 μm filter. For 50 mL (10 mice):
| Reagent | Final concentration | Amount |
|---|---|---|
| Donner’s stock | 1× | 47.5 mL |
| 1M NaHCO3 | 25 mM | 1,250 μL |
| 0.5M Sodium pyruvate | 1 mM | 100 μL |
| Sodium DL-Lactate Solution | 0.53% (vol/vol) | 265 μL |
| Bovine Serum Albumin (BSA) | 2% | 1 g |
-
20.
For each mouse, place 5 mL of Donner’s solution in a 60 mm Petri dish. Incubate at 37°C.
-
21.
For each mouse, place 5 mL of chilled 1× PBS in a 60 mm Petri dish. Place on ice.
-
22.
Sacrifice environmentally-exposed males after they have been used for breeding for a 2-week period. Prolonging duration of breeding may introduce variability regarding time exposed to the environmental stressor and age.
-
23.
Remove epididymis from males using 8.5 cm dissecting scissors and trim away epididymal adipose tissue (Figures 2A and 2B). Place epididymis in chilled 1× PBS of 60 mm Petri dishes.
CRITICAL: Do not collect sperm from epididymis with epididymitis which will appear inflamed and infected at the base of the cauda. Some mouse strains are more prone to developing epididymitis.
-
24.
Cut epididymis below the cauda (Figure 2B, dashed lines) and transfer caudas to a clean Parafilm square.
-
25.
Cut caudas 3–5 times using 8.5 cm dissecting scissors to release the sperm (Figure 2C).
-
26.
Place cut caudas in warmed Donner’s solution of 60 mm Petri dishes and incubate at 37°C with 20 rpm shaking for 1 h to allow for sperm to swim-out (Figure 2D).
-
27.
After swim-out incubation, filter Donner’s and sperm solution through 40 μm pore-size Nylon mesh filters and into a 50 mL Falcon tube (Figure 2E).
-
28.
Spin 50 mL Falcon tubes for 6 min at 2,600 × g and 4°C.
-
29.
Quickly remove supernatant without disrupting the pellet (Figure 2F).
-
30.
Gently resuspend sperm pellet in chilled 1× PBS and transfer to a 2 mL tube.
-
31.
Spin 2 mL tubes for 6 min at 2,600 × g and 4°C.
-
32.
Quickly remove supernatant without disrupting the pellet (Figure 2G).
-
33.
Gently resuspend sperm pellet in 200 μL Irvine Scientific Freezing Media (Yolk Buffer with Gentamicin Sulfate; see key resources table).
-
34.
Let tubes sit at 20°C–25°C for 5 min before placing in a −80°C freezer.
Notes: After swim-out, filtering, and 1× PBS wash, 7–10 million sperm cells from the cauda are expected for one C57BL6/J male mouse between 9 –11 weeks of age.
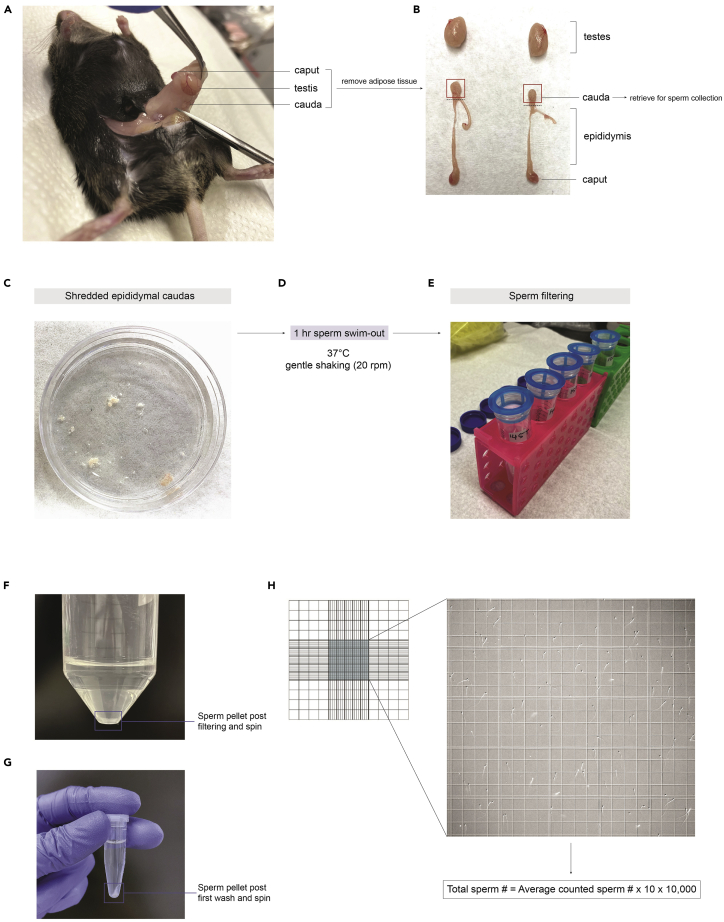
Figure 2.
Sperm collection and counting for ChIP-seq
(A) Identification of testis, caput, and cauda in a sacrificed mouse by pulling on the epididymal adipose tissue.
(B) Detatched testes, caputs, epididymis, and caudas, after removal from sacrificed male and cleaning of the adipose tissue. Dashed lines indicate the dissection cuts to make in order to retrieve the caudas.
(C) Shredded epididymal caudas (3–5 cuts with dissection scissors) in 37°C warmed Donner’s solution.
(D) Incubation parameters for sperm swim-out.
(E) Sperm filtering set-up using 40 μm filters and 50 mL Falcon tubes.
(F) Sperm pellet of a single mouse after first spin-down.
(G) Sperm pellet after first 1X PBS wash and spin-down.
(H) Hemocytometer diagram and example of sperm in the area to count.
Low-input and library preparation workspace sterilization
Timing: 2 h
CRITICAL: A sterile environment is required when working in low-input conditions and preparing libraries. Because of the low amount of starting material and high number of PCR amplification cycles, contamination is detrimental to final sequencing products (see troubleshooting problems 1 and 2). To minimize risk of contamination, these standard operating procedures should be adopted:
Label off a “sterile zone” of bench space where nothing that has not been thoroughly sterilized enters. Do not use this space to perform dissections.
Wipe all touched surfaces and all material that will be used in the experiments with 70% EtOH, 10% bleach, RNA-Zap and DNA-Away. This includes pens, chairs, pipettes.
Soak all pipette holders and trays in 10% bleach for 30 min before each new experiment and library preparation.
Use a new disposable lab coat for every new experiment and library preparation.
Use gloves that tighten around the mid-forearm to not have exposed skin. Frequently change your gloves and wipe with 70% EtOH, 10% bleach, RNA-Zap, DNA-Away. Change your gloves every time you touch something outside of your “sterile zone” (e.g., centrifuge, fridge door handle…), or pipette different adapters.
Use filtered pipette tips for Ultra-Low-Input ChIP-seq, Low-Input mRNA-seq, and all library preparations.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-histone 3 lysine 4 trimethylation | Cell Signaling Technology | Cat#9751; RRID:AB_2616028 |
| Rabbit polyclonal anti-histone 3 lysine 4 trimethylation | Diagenode | Cat#C15410003; RRID:AB_2616052 |
| Chemicals, peptides, and recombinant proteins | ||
| Bovine Serum Albumin (BSA), heat shock treated | Thermo Fisher | Cat#B16100 |
| Sodium DL-lactate solution | Sigma-Aldrich | Cat#L4263 |
| Sodium bicarbonate | Sigma-Aldrich | Cat#55761 |
| Sodium pyruvate | Sigma-Aldrich | Cat#P5280 |
| Micrococcal nuclease (MNase Nuclease S7) used for sperm ChIP-Seq | Roche | Cat#10107921001 |
| Complete Protease Inhibitor Cocktail | Roche | Cat#4693116001 |
| Dithiothreitol (DTT) | Bio-Shop | Cat#3483-12-3 |
| TERGITOL solution type NP-40 (NP-40) | Sigma-Aldrich | Cat#NP-40S |
| N-Ethylmaleimide (NEM) | Sigma-Aldrich | Cat#E3876 |
| RNAse A | Sigma-Aldrich | Cat#10109169001 |
| Proteinase K | Sigma-Aldrich | Cat#P2308 |
| Folligon Pregnant Mare Serum Gonadotropin (PMSG) | CDMV | Cat#103476 |
| Human chorionic gonadotropin (hCG) | EMD Millipore | Cat#230734 |
| Penicillin-Streptomycin | Thermo Fisher | Cat#15140122 |
| Acidic Tyrode’s Solution | MilliporeSigma | Cat#T1788 |
| Micrococcal nuclease (MNase) used for 8-cell embryo ULI-ChIP | New England Biolabs | Cat#M0247S |
| Tri-Reagent | MilliporeSigma | Cat#93289 |
| Phenol:chloroform:isoamyl alcohol mixture | Sigma-Aldrich | Cat#77617 |
| Chloroform | Sigma-Aldrich | Cat#288306 |
| Elution buffer | QIAGEN | Cat#1014609 |
| GenElute Linear Polyacrylamide (LPA) | MilliporeSigma | Cat#56575 |
| Phenylmethylsulfonyl fluoride (PMSF) | Sigma-Aldrich | Cat#329-98-6 |
| 20% Sodium dodecyl sulfate solution (SDS) | Thermo Fisher | Cat#BP1311-200 |
| DNA AWAY Surface Decontaminant | Thermo Fisher | Cat#21-236-28 |
| RNaseZAP | Sigma-Aldrich | Cat#R2020 |
| Critical commercial assays | ||
| 40 μM Micro-pore filter | Fisher Scientific | Cat#22363547 |
| 0.22 μM Membrane filter | Millipore | Cat#SLGPR33RS |
| BD PrecisionGlide 30G × ½ needles | BD | Cat#305106 |
| Freezing medium | Irvine Scientific | Cat#90128 |
| Protein A Dynabeads | Thermo Fisher | Cat#10002D |
| Protein G Dynabeads | Thermo Fisher | Cat#10003D |
| Eppendorf DNA LoBind Microcentrifuge Tubes | Fisher Scientific | Cat#13-698-791 |
| MaXtract High Density 1.5 mL Tubes | QIAGEN | Cat#129046 |
| Zymo Kit ChIP DNA Clean and Concentrator | Zymo Research | Cat#D5201 |
| Qiagen Ultralow Input Library Kit | QIAGEN | Cat#180495 |
| Agencourt AMPure XP Beads | Beckman Coulter | Cat#A63880 |
| M2 medium | MilliporeSigma | Cat#MR-015-D |
| Nuclear isolation buffer | Sigma-Aldrich | Cat#NUC-101 |
| DNA/RNA Protection Reagent | New England Biolabs | Cat#T2011L |
| NEBNext Single Cell / Low Input RNA Library Preparation Kit | New England Biolabs | Cat#E6420S |
| Deposited data | ||
| Mouse sperm H3K4me3 ChIP-seq | Lismer et al., 2021 | GSE135678 |
| Mouse 8-cell embryo H3K4me3 ULI-ChIP-seq | Lismer et al., 2021 | GSE135678 |
| Mouse 8-cell embryo mRNA-seq | Lismer et al., 2021 | GSE135678 |
| Experimental models: Organisms/strains | ||
| C57BL6/J wild-type male and female mice | Charles River Laboratories | N/A |
| Germ-cell-specific KDM1A overexpressing TG male mice on a C57BL6/J background | Siklenka et al. 2015 | N/A |
| Software and algorithms | ||
| Trimmomatic v0.38 | Bolger et al. 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| Bowtie2 v2.3.5 | Langmead et al., 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| SAMtools v1.7 | Li et al. 2009 | http://www.htslib.org |
| HISAT2 v2.1.0 | Kim et al., 2015 | https://daehwankimlab.github.io/hisat2/ |
| Stringtie v2.0.4 | Pertea et al. 2016 | https://ccb.jhu.edu/software/stringtie/ |
| DeepTools v3.5.0 | Ramírez et al., 2016 | https://deeptools.readthedocs.io/en/develop/ |
| Other | ||
| Folate-sufficient mouse chow | Harlan Laboratories | TD.01369 |
| Folate-deficient mouse chow | Harlan Laboratories | TD.01546 |
| DynaMag-2 magnetic rack | Thermo Fisher | Cat#12321D |
| BRAND® counting chamber BLAUBRAND® Neubauer improved | Sigma-Aldrich | Cat# BR717810 |
| Agilent 2100 Bioanalyzer | Agilent | Cat#G2939BA |
| Cell incubator-shaker | N/A | N/A |
| Dissecting microscope | N/A | N/A |
| Standard microscope | N/A | N/A |
| Benchtop centrifuge with refrigeration | N/A | N/A |
| Continuous flow centrifuge with refrigeration | N/A | N/A |
| Heat block | N/A | N/A |
| Walk-in 4°C fridge | N/A | N/A |
| Computer with pre-processing power | N/A | N/A |
Materials and equipment
Equipment
• Cell incubator-shaker
• Dissecting microscope
• Standard microscope
• Benchtop centrifuge with refrigeration
• Continuous flow centrifuge with refrigeration
• Heat block
• DynaMag-2 Magnetic rack
• BRAND® counting chamber BLAUBRAND® Neubauer improved
• Walk-in 4°C fridge
• Agilent 2100 Bioanalyzer (or alternatively a Tape Station): Agilent High Sensitivity DNA Kit (https://www.agilent.com/cs/library/usermanuals/public/G2938-90322_HighSensitivityDNAKit_QSG.pdf) and Agilent RNA 6000 Pico Kit (https://www.agilent.com/cs/library/usermanuals/public/RNA-6000-Pico_QSG.pdf)
• Computer with pre-processing power
Step-by-step method details
Chromatin immunoprecipitation and library preparation on single mouse sperm for H3K4me3
Timing: 5 days
This step describes how to perform a sperm-specific native ChIP-sequencing protocol on the sperm of individual mice for H3K4me3, using 8 million sperm cells, based on Hisano et al., 2013. We highlight our methodologies for mononucleosome isolation post-ChIP, as well as our library preparation procedures.
-
1.Antibody preparation (Day 1)
-
a.Prepare 0.5% Bovine Serum Albumin (BSA) solution fresh in 1× PBS: 0.25 g of BSA in 50 mL of chilled 1× PBS. Sterile filter and keep solution on ice.
-
b.Resuspend Dynabeads Protein A by inverting stock until slurry becomes uniform. Per mouse, pipet 50 μL of Dynabeads for chromatin immunoprecipitation and 50 μL of Dynabeads for pre-clearing into a 1.5 mL tube.
-
c.Place Dynabeads on a magnetic rack and slowly remove liquid without disrupting Dynabeads.
-
d.Remove Dynabeads from magnetic rack and wash with 100 μL of chilled 0.5% BSA solution. Minimize retention of Dynabeads in pipette tip by pipetting slowly.
-
e.Place Dynabeads on a magnetic rack and slowly remove liquid without disrupting the Dynabeads.
-
f.Repeat washing steps d and e twice more for a total of three washes.
-
g.After the final wash, resuspend Dynabeads in 80 μL of 0.5% BSA solution for each ChIP tube and 100 μL of 0.5% BSA solution for each pre-clear tube. Distribute 80 μL of the resuspended Dynabeads to 0.2 mL ChIP tubes or 100 μL of resuspended Dynabeads to 0.2 mL input tubes.Note: Before the chromatin immunoprecipitation, the digested chromatin will be pre-cleared in a Dynabeads Protein A 0.5% BSA solution without any antibody. This step removes molecules from the chromatin solution that bind to the Dynabeads non-specifically.
-
h.Add 20 μL H3K4me3 Cell Signaling antibody (Cat#9751) to each ChIP tube (equivalent to 5 μg of antibody per tube).
-
i.Place tubes on a rotator at 4°C. Ensure that the tubes undergo the largest possible revolution. Incubate for > 8 h.
-
a.
CRITICAL: Incubate in 0.2 mL tubes to prevent Dynabeads from drying out on the rim of a 1.5 mL tube.
Alternatives: This protocol can be used to probe other histone marks in sperm. However, it is important to optimize antibody concentration used in order to obtain high signal-to-noise ratios and library complexities (see troubleshooting problems 3 and 4). Optimization can be done by performing the described sperm ChIP-seq experiment using multiple antibody titrations, and subsequently performing low-depth sequencing (approximatively 10 million reads). The amount of antibody to use relies on two factors:
The breadth of the peak in sperm: a broad mark (e.g., H3K9me3) will require more antibody.
The efficiency of the antibody in sperm: this needs to be tested experimentally and may vary by cell type.
-
2.Sperm preparation and counting (Day 1)
 CRITICAL: Design all ChIP experiments to include each experimental group in every batch. For example, if you have 20 samples and 4 experimental groups (Lismer et al., 2021), perform 5 different ChIP experiments with each batch composed of the 4 experimental groups. Batch effects can occur in sequencing experiments and data should be analyzed to identify the influences experimental day by a principal component clustering analysis. If identified, batch effects can be bioinformatically corrected (Leek et al., 2012) if a balanced experimental design is used (Lismer et al., 2021).
CRITICAL: Design all ChIP experiments to include each experimental group in every batch. For example, if you have 20 samples and 4 experimental groups (Lismer et al., 2021), perform 5 different ChIP experiments with each batch composed of the 4 experimental groups. Batch effects can occur in sequencing experiments and data should be analyzed to identify the influences experimental day by a principal component clustering analysis. If identified, batch effects can be bioinformatically corrected (Leek et al., 2012) if a balanced experimental design is used (Lismer et al., 2021).-
a.Thaw sperm from different experimental groups on ice.
-
b.Add 500 mL of chilled 1× PBS per tube and gently pipette.
-
c.Spin tubes at 2,600 × g for 6 min at 4°C.
-
d.Confirm the presence of a pellet.
-
e.Without disrupting the pellet, remove the supernatant and gently resuspend in 1 mL of chilled 1× PBS.
-
f.Repeat washing steps c–e once more.
-
g.Ensure that sperm is well resuspended in chilled 1× PBS.
-
h.In new tubes, prepare two 1:10 sperm dilutions in ddH2O for each sample: 10 μL sperm and 1× PBS solution and 90 μL ddH2O.
-
i.Add 10 μL of diluted sperm to both counting chambers of a Neubauer Improved counting chamber.
-
j.Allow sperm to settle in the hemocytometer for 1 min.
-
k.Count sperm in the 25 middle squares of the hemocytometer grid (Figure 2H).
-
l.Extrapolate the total number of sperm per group in 1 mL of 1× PBS tube: Total sperm # = Average of two sperm counts × dilution factor (10) × 10,000.
 CRITICAL: Ensure that sperm tubes are kept on ice during the counting process.
CRITICAL: Ensure that sperm tubes are kept on ice during the counting process. CRITICAL: The sperm counting step will determine the downstream MNase digestion parameters therefore obtaining accurate and reproducible results is fundamental for this experiment. We recommend practicing the counting step beforehand to achieve counting consistency before using actual experimental sperm. If both sperm count replicates vary by more than 10 sperm counts, make two new 1:10 sperm dilutions in ddH2O and count two diluted sperm replicates again (see troubleshooting problem 5).
CRITICAL: The sperm counting step will determine the downstream MNase digestion parameters therefore obtaining accurate and reproducible results is fundamental for this experiment. We recommend practicing the counting step beforehand to achieve counting consistency before using actual experimental sperm. If both sperm count replicates vary by more than 10 sperm counts, make two new 1:10 sperm dilutions in ddH2O and count two diluted sperm replicates again (see troubleshooting problem 5). -
m.If tubes have over 8 million sperm cells in them, remove enough volume to specifically obtain 8 million sperm cells per tube. Add chilled 1× PBS to obtain a final total volume of 1 mL per tube.
-
a.
Alternatives: If experimental groups have less than 8 million sperm cells, adjust to obtain uniform sperm numbers across groups, and modify downstream MNase digestion parameters (step 5). We do not recommend using less than 6 million sperm cells as we have never tested it and therefore cannot reliably comment on data quality or read statistics.
-
3.Dithiothreitol (DTT) pre-treatment (Day 1)
 CRITICAL: Ensure that benchtop centrifuge is at 20°C–25°C.
CRITICAL: Ensure that benchtop centrifuge is at 20°C–25°C.-
a.Prepare 1M DTT solution fresh: 77.13 mg DTT in 500 μL ddH2O.
-
i.Add 50 μL of 1M DTT solution to each sample and incubate for 2 h at 20°C–25°C to release the protamines and relax the sperm chromatin before MNase digestion.
-
i.
-
b.5 min before the end of the DTT treatment, prepare a fresh 1M NEM solution: 125.13 mg NEM in 1 mL ddH2O.Note: NEM is difficult to dissolve. The solution should be incubated at 50°C for 5 min and vortexed.
-
c.For each tube, add 120 μL from the upper phase of 1M NEM solution while it is still on the heat block. Pipette up and down 20 times to mix well.
 CRITICAL: Do not take the oily bottom phase of the 1M NEM solution. In our hands, doing so will prevent the DTT quenching.
CRITICAL: Do not take the oily bottom phase of the 1M NEM solution. In our hands, doing so will prevent the DTT quenching. -
d.Incubate solution at 20°C–25°C for 30 min to quench the DTT.
-
a.
-
4.Chromatin preparation (Day 1)
-
a.Prepare 100 mM DTT solution fresh (for Complete Buffer 1.1): 15.4 g DTT in 1 mL ddH2O.
-
b.Prepare Complete Buffer 1.1 Solution using Buffer 1 Stock. For 1 mL:
Reagent Final concentration Amount Buffer 1 Stock solution 1× 950 μL 100 mM DTT solution 0.5 mM 5 μL Sucrose 0.3M 0.103 g Note: 100 μL of Complete Buffer 1.1 Solution per 2 million sperm cells will be required (50 μL of Complete Buffer 1.1 + 50 μL to prepare Complete Buffer Detergent 1.2). -
c.Spin sperm at 2,600 × g for 6 min at 20°C–25°C.
 CRITICAL: After the DTT pre-treatment, washes with 1× PBS must be carried at 20°C–25°C and not at 4°C or the sperm pellet will become brittle and not resuspend.
CRITICAL: After the DTT pre-treatment, washes with 1× PBS must be carried at 20°C–25°C and not at 4°C or the sperm pellet will become brittle and not resuspend. -
d.Carefully discard supernatant.
-
e.Gently resuspend pellet in 1 mL of 1× PBS.
-
f.Spin sperm at 2,000 × g for 5 min at 20°C–25°C.
-
g.Carefully discard supernatant.
-
h.Gently resuspend sperm pellet in Complete Buffer 1.1 solution using 50 μL of Complete Buffer 1.1 Solution per 2 million sperm cells. For 8 million sperm cells, resuspend in 200 μL Complete Buffer 1.1 solution.
-
i.Vigorously pipette up and down 20 times. Ensure that sperm is well resuspended.
-
j.Aliquot sperm in 50 μL volumes per tube. For 8 million sperm cells, 4 aliquot tubes will be made per experimental sample.
-
k.Make 10% NP-40 stock solution fresh: 100 μL NP-40 + 900 μL ddH2O (for Complete Buffer 1.2).Note: NP-40 is very viscous and should be pipetted slowly to obtain accurate measurements.
-
l.Make 10% sodium deoxycholate (DOC) stock solution fresh: 10 mg DOC in 100 μL ddH2O (for Complete Buffer 1.2).
-
m.Prepare Complete Buffer 1.2 Solution: 50 μL of Complete Buffer 1.2 Solution is required per sperm aliquot tube. For 500 μL:
Reagent Final concentration Amount Complete Buffer 1.1 solution N/A 425 μL 10% NP-40 solution 1.25% 25 μL 10% DOC solution 1% 50 μL  CRITICAL: Vortex for 1 min to ensure that the NP-40 and DOC are entirely dissolved.
CRITICAL: Vortex for 1 min to ensure that the NP-40 and DOC are entirely dissolved. -
n.Add 50 μL of Complete Buffer 1.2 solution to each sperm aliquot tube.
-
o.Vigorously pipette up and down 20 times.
-
p.Incubate on ice for 25 min.
-
a.
-
5.MNase digestion (Day 1)
 CRITICAL: Always titrate MNase with every new batch of MNase stock on practice sperm prior to the ChIP experiment to confirm enzyme efficiency. Once MNase is resuspended, make 7 μL aliquots (30 IU/μL) and store in a −20°C freezer. Do not freeze-thaw aliquots.
CRITICAL: Always titrate MNase with every new batch of MNase stock on practice sperm prior to the ChIP experiment to confirm enzyme efficiency. Once MNase is resuspended, make 7 μL aliquots (30 IU/μL) and store in a −20°C freezer. Do not freeze-thaw aliquots.-
a.Prepare MNase Buffer Complete solution. For 1 mL (to process 10 aliquots of 2 million sperm):
Reagent Final concentration Amount MNase Buffer Stock solution 1× 995 μL Sucrose 0.3M 0.1027 g MNase Stock aliquot 150 IU of MNase (15 IU/2 million sperm cells are required) 5 μL Note: 100 μL of MNase Buffer Complete will be required per 2 milllion sperm cells. CRITICAL: Always transport MNase on ice and keep MNase Buffer Complete solution on ice. Do not reuse a thawed aliquot.
CRITICAL: Always transport MNase on ice and keep MNase Buffer Complete solution on ice. Do not reuse a thawed aliquot. -
b.Add 100 μL of MNase Buffer Complete solution to each tube and mix vigorously by pipetting up and down 20 times.
-
c.Incubate at 37°C for exactly 5 min. Use 30 s intervals between experimental tubes or a multichannel pipette to keep MNase digestion time consistent.
 CRITICAL: MNase digestion starts when the tube enters the heat block. Absolute consistency in the timing of MNase digestion to ensure consistent mononucleosome digestion across experimental groups and batches. A slight variation in digestion time can lead to batch effects. Uniformity and efficiency of MNase digestion across samples in one batch is verified using an Agilent Bioanalyzer low input DNA chip (step 10.k.)
CRITICAL: MNase digestion starts when the tube enters the heat block. Absolute consistency in the timing of MNase digestion to ensure consistent mononucleosome digestion across experimental groups and batches. A slight variation in digestion time can lead to batch effects. Uniformity and efficiency of MNase digestion across samples in one batch is verified using an Agilent Bioanalyzer low input DNA chip (step 10.k.) -
d.Stop MNase digestion by adding 2 μL of 0.5 M EDTA. Pipette up and down vigorously. Vortex tubes on maximum setting for 5 s.
-
e.Place tubes on ice for 15 min.
-
f.Spin tubes at 17,000 × g for 10 min at 20°C–25°C.
-
g.Combine supernatants from aliquot tubes from the same mouse sperm. Pipette supernatant carefully to not aspirate debris and protamines present in the pellet. Keep samples on ice. Each experimental group should yield approximatively 800 μL.
-
h.Dissolve one tablet of protease inhibitor in 2 mL of ddH2O. Freeze 50 μL aliquots in a −20°C freezer.
-
i.Add 50 μL of protease inhibitor cocktail to each 800 μL of pooled supernatants.
-
a.
-
6.Chromatin Dynabeads™ pre-clearing (Day 1)Note: Pre-clearing digested chromatin removes molecules that non-specifically bind to Dynabeads Protein A, which is aimed to reduce background and increase specificity.
-
a.Prepare Combined Buffer solution fresh and place on ice. For 1 mL:
Reagent Final concentration Amount Buffer 1 Stock solution 0.5× 475 μL MNase Buffer Stock solution 0.5× 475 μL Sucrose 0.3M 0.103 g Note: 500 μL of Combined Buffer solution will be required per experimental sample. -
b.Place Dynabeads from pre-cleared tubes on a magnetic rack and slowly remove liquid without disrupting Dynabeads.
-
c.Remove Dynabeads from magnetic rack and resuspend in 100 μL of chilled 0.5% BSA solution.
-
d.Place Dynabeads on a magnetic rack and slowly remove liquid without disrupting the Dynabeads.
-
e.Repeat washing steps d and e twice more for a total of three washes.
-
f.Resuspend Dynabeads in 50 μL of chilled Combined Buffer solution per tube.
-
g.Add pre-clearing Dynabeads resuspended in Combined Buffer solution to sperm chromatin.
-
h.Rotate at 4°C for 1.5 h.
-
a.
Note: If using a new antibody or studying a different histone modification, sequencing the input should not be omitted.
-
7.Chromatin immunoprecipitation (Day 1)
-
a.After incubation, place Dynabeads from antibody tubes from step 1.h. on a magnetic rack and slowly remove liquid without disrupting Dynabeads.
-
b.Remove Dynabeads from magnetic rack and resuspend in 100 μL of chilled Combined Buffer.
-
c.Place Dynabeads on a magnetic rack and slowly remove liquid without disrupting the Dynabeads.
-
d.Repeat washing steps d and e twice more for a total of three washes.
-
e.Resuspend Dynabeads-Antibody complex in 100 μL of chilled Combined Buffer solution.
-
f.Place Dynabeads from pre-clear tubes from step 6.h. on a magnetic rack. To retrieve an input aliquot, remove 10 μL of solution and place in an input tube. Store input tubes a −80°C freezer.
-
g.Carefully transfer the remaining pre-cleared chromatin to Dynabeads-Antibody complex tubes.
-
h.Place the tubes on a rotator at 4°C. Ensure that the tubes undergo the largest revolution possible. Incubate 12–14 h.
-
a.
-
8.Washing Dynabeads-Antibody complex (Day 2)
 CRITICAL: Keep samples on ice as much as possible throughout the washes.
CRITICAL: Keep samples on ice as much as possible throughout the washes.-
a.Prepare Wash Buffer A fresh. Autoclave the buffer and chill in a 4°C fridge before beginning the washes. For 10 mL:
Reagent Final concentration Amount 1M Tris-HCl (pH 7.5) 50 mM 500 μL 0.5M EDTA 10 mM 200 μL 5M NaCl 75 mM 150 μL ddH2O N/A 9,150 μL -
b.Prepare Wash Buffer B fresh. Autoclave the buffer and chill in a 4°C fridge before beginning the washes. For 10 mL:
Reagent Final concentration Amount 1M Tris-HCl (pH 7.5) 50 mM 500 μL 0.5M EDTA 10 mM 200 μL 5M NaCl 125 mM 250 μL ddH2O N/A 9,050 μL -
c.Spin ChIP tubes at 400 × g for 1 min at 4°C.
-
d.Place tubes on a magnetic rack and carefully discard supernatant.
-
e.Remove tubes from magnetic rack and resuspend beads in 1 mL of chilled Wash Buffer A.
-
f.Incubate tubes at 4°C for 5 min on a rotator.
-
g.Spin tubes at 400 × g for 1 min at 4°C.
-
h.Place tubes on a magnetic rack and carefully discard supernatant.
-
i.Remove tubes from magnetic rack and resuspend beads in 1 mL of Wash Buffer B.
-
j.Incubate tubes at 4°C for 5 min on a rotator.
-
k.Repeat washing steps e and f with chilled Wash Buffer B once more.
-
l.Remove tubes from magnetic rack and resuspend beads in 1 mL of Wash Buffer B. Transfer mixtures to Eppendorf DNA LoBind tubes.
-
m.Incubate on a rotator for 5 min at 4°C.
-
n.Spin tubes at 400 × g for 1 min at 4°C.
-
o.Place tubes on magnetic rack and carefully discard supernatant.
-
a.
-
9.Elution (Day 2)Note: Prepare ChIP Elution buffer during incubations of step 8.
-
a.Prepare 1M DTT solution fresh: 77.13 mg DTT in 500 μL ddH2O.
-
b.Prepare 1M NaHCO3 solution fresh: 84.01 mg NaHCO3 in 1 mL ddH2O
-
c.Prepare ChIP Elution Buffer solution fresh. For 2 mL:
Reagent Final concentration Amount TE Buffer N/A 1,750 μL 1M DTT solution 5 mM 10 μL 1M NaHCO3 solution 100 mM 200 μL 10% SDS solution 0.2% 40 μL -
d.Heat ChIP Elution Buffer solution to 65°C. Maintaining the elution buffer at this temperature will increase elution efficiency.
-
e.For the ChIP tubes, add 125 μL of warmed ChIP Elution Buffer solution to washed Dynabeads-Antibody complex. Mix well by pipetting up and down 20 times. Vortex on maximum setting for 10 s and perform a quick spin.
-
f.For the input tubes, add 250 μL ChIP Elution Buffer solution to the tubes. Mix well by pipetting up and down 20 times. Vortex on maximum setting for 10 s and perform a quick spin.
-
g.Heat tubes at 65°C for 10 min with 400 × g shaking. For the ChIP tubes, vortex on maximum setting for 5 s every 4 min to resuspend nucleosomes. Perform a quick spin after every vortex.Note: There are no Dynabeads in the input tubes therefore the tubes can be left on the heat block for the full duration of the two ChIP elutions.
-
h.After elution, spin tubes at 400 × g for 1 min at 20°C–25°C and place on a magnetic rack.
-
i.Transfer ChIP elution products to new Eppendorf DNA LoBind tubes.
-
j.Repeat elution step for ChIP tubes e,g.–i and combine second elution products with appropriate Eppendorf DNA LoBind tubes.
-
k.Add 5 μL of 10 mg/mL RNase to all tubes (input and ChIP tubes) and incubate for 30 min at 37°C with 400 × g shaking.
-
l.Add 5 μL of 10 mg/mL Proteinase K to all tubes (input and ChIP tubes) and incubate 12–14 h at 55°C with 400 × g shaking.
-
a.
-
10.
DNA cleanup and concentration (Day 3)
DNA cleanup and concentration can be performed using the Zymo Research ChIP DNA Clean and Concentrator Kit (https://files.zymoresearch.com/protocols/_d5201_d5205_chip_dna_clean_concentrator_kit.pdf) with slight modifications:Note: Perform this step for the input and ChIP tubes if sequencing an input.-
a.In a 1.5 mL tube, add 1,250 μL of ChIP DNA Binding Buffer to each tube (5 volumes per each volume of sample).
-
b.Transfer mixture to Zymo-Spin Column in a Collection Tube.
-
c.Centrifuge at 10,000 × g for 30 s and discard flow-through. Steps c and d will need to be repeated because the volume in each tube exceeds the Zymo-Spin Column capacity.
-
d.Add 200 μL Wash Buffer to the column. Rotate pipette around interior of tube to wash walls of the Collection Tubes.
-
e.Centrifuge at 10,000 × g for 30 s and discard flow-through.
-
f.Transfer columns to new 1.5 mL tubes and add 15 μL of warmed Elution Buffer directly to the column matrix. Incubate at 20°C–25°C for 1 min.
-
g.Centrifuge at 10,000 × g for 1 min to elute DNA.
-
h.For the same tube, add 15 μL of warmed Elution Buffer directly to the column matrix for a second elution. Incubate at 20°C–25°C for 1 min.
-
i.Centrifuge at 10,000 × g for 1 min to elute DNA a second time.
-
j.Assess and quantify immunoprecipitation product on an Agilent Bioanalyzer low input DNA chip.
-
a.
Pause point: Eluted samples can be stored in a −80°C freezer for up to 1 year before mononucleosome isolation.
-
11.Immunoprecipitated mononucleosome isolation (Day 4)Note: Perform this step for the input and ChIP tubes if sequencing an input.
 CRITICAL: Removing non-mononucleosomal DNA fragments ensures specific amplification of mononucleosome fragments during library preparation. Determine cleaning parameters for each sample based on Bioanalyzer electropherogram reports. See https://www.broadinstitute.org/genome-sequencing/broadillumina-genome-analyzer-boot-camp for a thorough explanation on how varying Ampure XP Beads volume to solution volume ratio selects for different sizes of DNA. Isolate mononucleosomes as follows:
CRITICAL: Removing non-mononucleosomal DNA fragments ensures specific amplification of mononucleosome fragments during library preparation. Determine cleaning parameters for each sample based on Bioanalyzer electropherogram reports. See https://www.broadinstitute.org/genome-sequencing/broadillumina-genome-analyzer-boot-camp for a thorough explanation on how varying Ampure XP Beads volume to solution volume ratio selects for different sizes of DNA. Isolate mononucleosomes as follows:-
a.If samples do not have any high or low molecular weight DNA (Figure 3A): perform a non-stringent negative selection followed by a positive selection before library preparation.
-
i.Resuspend Ampure XP beads by inverting stock solution until slurry becomes uniform.
-
ii.Warm Qiagen Elution Buffer to 65°C.
-
iii.Adjust sample volume to 50 μL with Qiagen Elution Buffer.
-
iv.Add 27.5 μL Ampure XP beads to each sample (0.55×) and mix thoroughly by pipetting up and down 20 times.
-
v.Incubate at 20°C–25°C for 5 min.
-
vi.Place tubes on a magnetic rack and wait for Ampure XP beads to pellet.
-
vii.Gently retrieve the 77.5 μL of supernatant (negative selection) and place supernatant in a new tube.
-
viii.Add 50 μL of Ampure XP beads per sample (1×) and mix thoroughly by pipetting up and down 20 times.
-
ix.Incubate at 20°C–25°C for 10 min.
-
x.Place tubes on a magnetic rack and wait for Ampure XP beads to pellet.
-
xi.Remove supernatant without disrupting beads.
-
xii.Wash beads twice with 200 μL of freshly prepared 80% ethanol.
-
xiii.Air dry beads. Beads are dry when they are no longer glossy and when no liquid is visible in the tube.
-
xiv.Elute DNA in 25 μL of warmed Elution Buffer and vortex on high setting for 10 s.
-
xv.Incubate at 20°C–25°C for 10 min.
-
xvi.Place tubes on a magnetic rack and wait for Ampure XP beads to pellet.
-
xvii.Gently retrieve 25 μL of eluted DNA and place in a new tube.
-
xviii.Assess and quantify cleaned product on an Agilent Bioanalyzer low-input DNA chip.
-
i.
-
b.If samples have high molecular weight (di- or trinucleosomes due to inefficient MNase digestion) (Figures 3C and 3D) (see troubleshooting problem 5): perform a stringent negative selection followed by a positive selection before library preparation.
-
i.Resuspend Ampure XP beads by inverting stock solution until slurry becomes uniform.
-
ii.Warm Qiagen Elution Buffer to 65°C.
-
iii.Adjust sample volume to 50 μL with Qiagen Elution Buffer.
-
iv.Add 42.5 μL Ampure XP beads to each sample (0.85×) and mix thoroughly by pipetting up and down 20 times.
-
v.Incubate at 20°C–25°C for 5 min.
-
vi.Place tubes on a magnetic rack and wait for Ampure XP beads to pellet.
-
vii.Gently retrieve the 92.5 μL of supernatant (negative selection) and place supernatant in a new tube.
-
viii.Add 50 μL of Ampure XP beads per sample (1×) and mix thoroughly by pipetting up and down 20 times.
-
ix.Incubate at 20°C–25°C for 10 min.
-
x.Place tubes on a magnetic rack and wait for Ampure XP beads to pellet.
-
xi.Remove supernatant without disrupting beads.
-
xii.Wash beads twice with 200 μL of freshly prepared 80% ethanol.
-
xiii.Air dry beads. Beads are dry when they are no longer glossy and no liquid is visible in the tube.
-
xiv.Elute DNA in 25 μL of warmed Elution Buffer and vortex on high setting for 10 s.
-
xv.Incubate at 20°C–25°C for 10 min.
-
xvi.Place tubes on a magnetic rack and wait for Ampure XP beads to pellet.
-
xvii.Gently retrieve 25 μL of eluted DNA and place in a new tube.
-
xviii.Assess and quantify cleaned product on an Agilent Bioanalyzer low-input DNA chip.
-
i.
-
c.If samples have low molecular weight (overdigestion, short DNA fragments) (Figures 3D and 3E) (see troubleshooting problem 5): perform a stringent positive selection before library preparation.
-
i.Resuspend Ampure XP beads by inverting stock solution until slurry becomes uniform.
-
ii.Warm Qiagen Elution Buffer to 65°C.
-
iii.Adjust sample volume to 50 μL with Qiagen Elution Buffer.
-
iv.Add 80 μL Ampure XP beads to each sample (1.6×) and mix thoroughly by pipetting up and down 20 times.
-
v.Incubate at 20°C–25°C for 10 min.
-
vi.Place tubes on a magnetic rack and wait for Ampure XP beads to pellet.
-
vii.Remove supernatant without disrupting beads.
-
viii.Wash beads twice with 200 μL of freshly prepared 80% ethanol.
-
ix.Air dry beads. Beads are dry when they are no longer glossy and no liquid is visible in the tube.
-
x.Elute DNA in 25 μL of warmed Elution Buffer and vortex on high setting for 10 s.
-
xi.Incubate at 20°C–25°C for 10 min.
-
xii.Place tubes on a magnetic rack and wait until all Ampure XP beads have pelleted.
-
xiii.Gently retrieve 25 μL of eluted DNA and place in a new tube.
-
xiv.Assess and quantify cleaned product on an Agilent Bioanalyzer low input DNA chip.
-
i.
-
a.
CRITICAL: When performing the Ampure XP bead 80% ethanol washes, leave tubes on magnetic stand. Do not resuspend beads in 80% ethanol.
CRITICAL: When drying out the Ampure XP beads, ensure that the beads do not over-dry and crack. This will lead to loss of DNA.
Note: If samples have both high and low molecular weights, we recommend performing the 0.85× negative selection, 1× positive selection first, followed by the 1.6× positive selection.
Pause point: Cleaned samples can be stored in the −80°C freezer for up to 1 year before library preparation.
-
12.
Library preparation (Day 5)
Note: Perform this step for the input and ChIP tubes if sequencing an input.
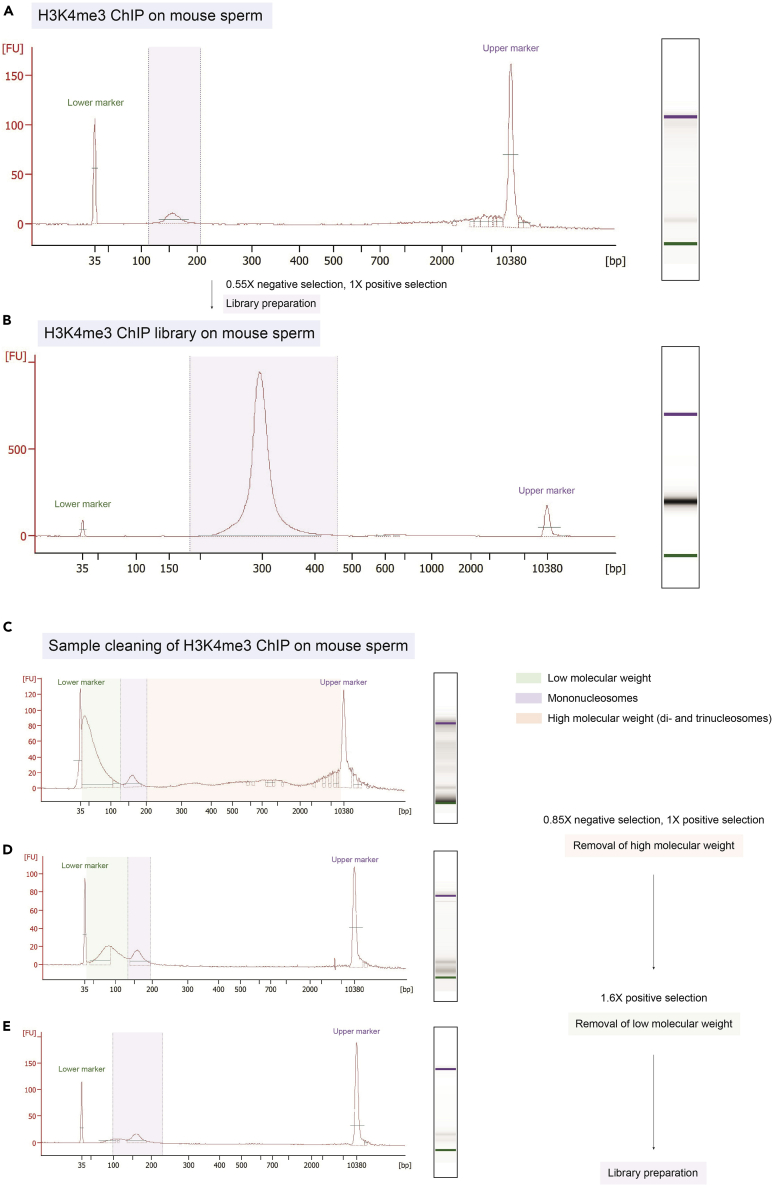
Figure 3.
Sperm ChIP-seq Bioanalyzer expected outcomes and mononucleosome isolation
(A) Gold-standard H3K4me3 ChIP Bioanalyzer electropherogram on 8 million mouse sperm from a single male. Perform a low-stringent mononucleosome isolation (0.55× negative selection, 1× positive selection) before library preparation.
(B) Gold-standard H3K4me3 ChIP library Bioanalyzer electropherogram on 8 million mouse sperm from a single male following 10 amplification cycles during library preparation.
(C) Example of an H3K4me3 ChIP Bioanalyzer electropherogram on million mouse sperm from a single male with low (green box) and high (orange box) molecular weight DNA that should be removed before library preparation.
(D) Bioanalyzer electropherogram from (C) after removal of high molecular weight (following a 0.85× negative size selection and 1× positive size selection).
(E) Bioanalyzer electropherogram from (D) after removal of low molecular weight (following a 1.6× positive size selection).
Prepare libraries using the Qiagen UltraLow Input Library Kit using the manufacturer’s instructions (https://www.qiagen.com/us/resources/resourcedetail?id=e1e88a40-bf6b-431a-9697-799e14103467&lang=en). For 8 million sperm cells, we use a 1:10 dilution for adapters (step 4) and perform 10 PCR library amplification cycles (step 21) (Figure 3B). Adapter dilution and PCR cycle number can vary depending on input quantity (see user manual).
CRITICAL: Before beginning library preparation, adjust all samples to obtain the same DNA concentration in each tube.
CRITICAL: Minimizing the number of PCR amplification cycles is important to reduce PCR duplicates. Verify with your sequencing platform the minimum and maximum concentration of DNA required for sequencing. Perform enough PCR amplification cycles to have a final DNA concentration just above the minimum DNA concentration required for sequencing.
Ultra-low-input ChIP-sequencing on 8-cell pre-implantation embryos and library preparation
Timing: 3 days
Alternatives: The steps of the ULI-ChIP-seq described below are specifically optimized for 250 pre-implantation embryo cells (equivalent to 32 × 8-cell embryos) targeting H3K4me3 (Diagenode, Cat#C15410003). The original ULI-ChIP-seq protocol from Brind’Amour et al., 2015, describes in detail the parameters to use if working with 1,000 to 1 million cells. These parameter differences include MNase digestion time and incubation temperature, amount of antibody needed per reaction, and number of PCR amplification cycles used during library preparation. Depending on cell type, cell number, and antibody used, optimization may be required.
-
13.Antibody preparation (Day 1)
-
a.Dissolve one tablet of protease inhibitor in 2 mL of ddH2O. Freeze 50 μL aliquots in a −20°C freezer.
-
b.Prepare Complete Immunoprecipitation buffer fresh and place on ice. For 3 mL:
Reagent Final concentration Amount Immunoprecipitation Buffer 1× 2,850 μL Protease inhibitor 1× 120 μL 0.1M PMSF 1 mM 30 μL -
c.Resuspend Dynabeads Protein A and Protein G by inverting stocks until slurries become uniform. For each sample, add 5 μL of Dynabeads Protein A and 5 μL of Dynabeads Protein G in 0.2 mL tubes. Half of the Dynabeads mixture will be used for the pre-clearing step and half will be used for the immunoprecipitation step.Note: The combination of Dynabeads Protein A and Protein G increases removal of non-specific binding than the use of Dynabeads Protein G alone. Because the Dynabeads are in excess, using both Protein A and Protein G Dynabeads does not hinder the immunoprecipitation.
-
d.Place Dynabeads on a magnetic rack and slowly remove liquid without disrupting Dynabeads.
-
e.Remove Dynabeads from magnetic rack and wash with 200 μL of chilled Complete Immunoprecipitation Buffer. Minimize retention of Dynabeads in pipette tip by pipetting slowly.
-
f.Place Dynabeads on a magnetic rack and slowly remove liquid without disrupting the Dynabeads.
-
g.Repeat washing steps d and e once more.
-
h.Remove Dynabeads from magnetic rack and perform a third wash with 200 μL of chilled Complete Immunoprecipitation Buffer.
-
i.Ensure that the Dynabeads are well resuspended in Complete Immunoprecipitation buffer. Transfer 100 μL of Dynabeads and Complete Immunoprecipitation buffer to a new 0.2 mL tube. The first tube will be used for the immunoprecipitation step and the second tube will be used for the pre-clearing step.
-
j.Place Dynabeads on a magnetic rack and slowly remove liquid without disrupting the Dynabeads.
-
k.Add 100 μL Complete Immunoprecipitation buffer to the pre-clear tube and 99 μL Complete Immunoprecipitation buffer to the immunoprecipitation tube.
-
l.Dilute Diagenode H3K4me3 antibody (Cat#C15410003; concentration 1.4 μg/μL): 1 μL antibody in 13 μL Complete Immunoprecipitation buffer. Mix well by pipetting up and down 20 times.Note: Each ULI-ChIP tube will use 0.125 μg of H3K4me3 antibody. This amount of H3K4me3 antibody to use has been optimized in Hanna et al. (2018) for 250 cells.Alternatives: As for the sperm ChIP-seq, this protocol can be used to probe other histone marks in pre-implantation embryos. Again, it is important to optimize antibody concentration used in order to obtain high signal-to-noise ratios and library complexities (see troubleshooting problems 3 and 4).
-
m.Add 1.25 μL of diluted antibody to each immunoprecipitation tube and mix well.
-
n.Place tubes on a rotator at 4°C. Ensure that the tubes undergo the largest possible revolution. Incubate for > 8 h.
-
a.
CRITICAL: Incubate in 0.2 mL tubes to prevent Dynabeads from drying out on the rim of a 1.5 mL tube.
-
14.Sample pooling (Day 1)Note: To obtain enough starting material (approximatively 250 cells), samples will most likely have to be pooled from different tubes of frozen embryos belonging to the same experimental group.
-
a.Thaw embryo tubes on ice.
-
b.Add 20 μL of nuclear isolation buffer to the first tube and mix by swerving the pipette tip in the tube.
-
c.Gently transfer the resuspended solution from the first tube to the second tube that belongs to the same experimental group.
-
d.Add 20 μL of nuclear isolation buffer to the second tube and mix by swerving the pipette tip in the tube.
-
e.Repeat steps c and d for as many tubes needing to be pooled.
-
f.Once pooling is complete, pulse spin tubes at 13,000 × g for 10 s.
-
g.Circle on the tube with a thin black marker where the embryos are expected to be pelleted based on the position of the tube in the centrifuge.
-
h.Using a tube with 10 μL of ddH2O as a reference, carefully remove enough solution in pooled tube to obtain a final volume of approximatively 10 μL. Pipette slowly and away from the expected pellet to not aspirate the embryos.
-
a.
-
15.MNase digestion (Day 1)
-
a.Prepare the following solutions fresh:
-
i.10% sodium deoxycholate (DOC): 10 mg DOC in 100 μL ddH2O
-
ii.200 mM DTT: 30.8 mg DTT in 1 mL ddH2O
-
iii.50% PEG 6000: 50 mg PEG 6000 in 100 μL ddH2O
-
iv1% Triton / 1% DOC: 100 μL 10× Triton + 100 μL 10% DOC in 800 μL ddH2O
-
i.
-
b.After samples have been pooled, add 1 μL of 1% Triton / 1% DOC detergent solution to each tube.
-
c.Mix vigorously by pipetting up and down 20 times.
-
d.Place tubes back on ice.
-
e.Prepare MNase master mix on ice. One sample requires 40 μL of MNase master mix:
Reagent Final concentration Amount 50% PEG 6000 6.24% 75 μL 10× MNase Digestion buffer 1.25× 75 μL 200 mM DTT 1.86 mM 5.6 μL MNase enzyme (NEB; 2,000 IU / μL) 3.33 IU / μL 1 μL ddH2O N/A 444.4 μL Note: The MNase used in the ULI-ChIP-seq procedure is from NEB whereas the MNase used in the sperm ChIP-seq procedure is from Roche (see key resources table). The 10× MNase Digestion buffer is supplied when purchasing NEB MNase enzyme. CRITICAL: always transport MNase on ice block and only add to master mix when ready to proceed. Keep master mix on ice.
CRITICAL: always transport MNase on ice block and only add to master mix when ready to proceed. Keep master mix on ice. -
f.Mix master mix thoroughly by pipetting up and down 20 times, and lightly vortexing. Perform a quick spin down.
-
g.Add 40 μL of MNase master mix to each tube and mix vigorously by pipetting up and down 20 times.
-
h.Incubate at 21°C for exactly 7.5 min. Use 30 s intervals between experimental tubes or a multichannel pipette to keep MNase digestion time consistent.
-
i.Stop MNase digestion by adding 5.5 μL of 100 mM EDTA to each tube of embryos at designated intervals. Pipette up and down vigorously. Vortex tubes on maximum setting for 5 s.
-
j.Add 4 μL of 1% Triton / 1% DOC detergent to each tube.
-
k.Place tubes on ice for 30 min.
-
a.
-
16.Digested chromatin manipulation (Day 1)
-
a.After incubation on ice, vortex tubes on the medium setting of a vortexer for 30 s.
-
b.Add 181.5 μL of chilled Complete Immunoprecipitation buffer per tube (total volume should be 242 μL and digested chromatin should take < 25% of the immunoprecipitated volume).
-
c.Place tubes on a rotator at 4°C. Ensure that the tubes undergo the largest possible revolution. Incubate for 1 h.
-
d.After incubation, vortex tubes on the medium setting of a vortexer for 30 s.
-
a.
-
17.Chromatin Dynabeads pre-clearing (Day 1)
 CRITICAL: For the following steps (steps 5–9), do not mix by pipetting up and down unless otherwise indicated. Doing so may result in losing sample getting in the pipette tip. Mix by flicking tubes.
CRITICAL: For the following steps (steps 5–9), do not mix by pipetting up and down unless otherwise indicated. Doing so may result in losing sample getting in the pipette tip. Mix by flicking tubes.-
a.Place Dynabeads from pre-cleared tubes on a magnetic rack and slowly remove liquid without disrupting Dynabeads.
-
b.Remove tubes from magnetic rack and add digested chromatin to pre-clear tubes.
-
c.Mix by flicking tubes 20 times and perform a quick spin down.
-
d.Rotate at 4°C for 1 h.
-
a.
-
18.Chromatin immunoprecipitation (Day 1)
-
a.After incubation, place Dynabeads from antibody tubes on a magnetic rack and slowly remove liquid without disrupting Dynabeads.
-
b.Pellet Dynabeads from pre-clear tubes. Remove 24 μL of solution from each tube (approximatively 10% of the total volume) and place in an input tube. Store input tubes in a −80°C freezer.
-
c.Carefully transfer the remaining pre-cleared chromatin to the Dynabeads:antibody complex tubes.
-
d.Place the tubes on a rotator at 4°C. Ensure that the tubes undergo the largest revolution possible. Incubate for 14–16 h.
-
a.
-
19.Washing Dynabeads-Antibody complex (Day 2)
 CRITICAL: Ensure Low Salt and High Salt Wash buffers are made fresh and chilled before use. Keep samples on ice as much as possible.
CRITICAL: Ensure Low Salt and High Salt Wash buffers are made fresh and chilled before use. Keep samples on ice as much as possible.-
a.Prepare Low Salt Wash Buffer fresh, sterile filter using a 0.22 μm filter, and place on ice. For 50 mL:
Reagent Final concentration Amount 1M Tris-HCl (pH 7.5) 20 mM 1,000 μL 0.5M EDTA 2 mM 200 μL 5M NaCl 150 mM 1,500 μL 10× Triton 1× 5,000 μL 20% SDS 0.1% 250 μL ddH2O N/A 42.05 mL -
b.Prepare High Salt Wash Buffer filter, sterile filter using a 0.22 μm filter, and place on ice. For 50 mL:
Reagent Final concentration Amount 1M Tris-HCl (pH 7.5) 20 mM 1,000 μL 0.5M EDTA 2 mM 200 μL 5M NaCl 500 mM 5,000 μL 10× Triton 1× 5,000 μL 20% SDS 0.1% 250 μL ddH2O N/A 48.55 mL -
c.Prepare ChIP Elution Buffer fresh (used in step 8). For 1 mL:
Reagent Final concentration Amount 1M NaHCO3 100 mM 100 μL 20% SDS 1% 50 μL ddH2O N/A 850 μL -
d.After incubation, place tubes on a magnetic rack and carefully discard supernatant.
-
e.Remove tubes from magnetic rack and add 200 μL of chilled Complete Immunoprecipitation buffer to each tube.
-
f.Mix by flicking tubes 20 times and perform a quick second spin down.
-
g.Place tubes on magnetic rack and carefully discard supernatant.
-
h.Remove tubes from magnetic rack and add 200 μL of chilled Low Salt Wash buffer to each tube.
-
i.Mix by flicking tubes 20 times and perform a quick spin down.
-
j.Place tubes on magnetic rack and carefully discard supernatant.
-
k.Remove tubes from magnetic rack and add 200 μL of chilled High Salt Wash buffer to each tube.
-
l.Mix by flicking tubes 20 times and perform a quick spin down.
-
m.Place tubes on magnetic rack and carefully discard supernatant.
-
n.Repeat washing steps i and j with chilled High Salt Wash buffer once more and transfer mixtures to Eppendorf DNA LoBind tubes.
-
o.Place tubes on magnetic rack and carefully discard supernatant.
-
a.
-
20.Elution (Day 2)
-
a.Add 30 μL ChIP Elution buffer to washed Dynabeads-Antibody complex. Mix well by flicking tubes 20 times. Vortex on maximum setting for 10 s and perform a quick spin.
-
b.Heat tubes at 65°C for 1.5 h with 400 × g shaking. Vortex on maximum setting for 5 s every 15 min to resuspend nucleosomes. Perform a quick spin after every vortex.
-
a.
-
21.DNA purification (Day 2)
-
a.Thaw input tubes from step 18.b. on ice and add 176 μL of Qiagen Elution Buffer to each tube.
-
b.Place ChIP tubes from step 20 on a magnetic rack and carefully transfer the supernatant (i.e: the eluted chromatin) to a new tube.
-
c.To maximize eluted chromatin retrieval, remove ChIP tubes from magnetic rack and add 170 μL Qiagen Elution Buffer to each tube. Vortex tubes on maximum setting for 30 s.
-
d.Place tubes on a magnetic rack and carefully transfer the supernatant to the corresponding eluted chromatin tube.
-
e.Add 1 μL of of 10 mg/mL RNase A and incubate for 30 min at 37°C.
-
f.Add 1 μL of of 10 mg/mL Proteinase K and incubate for 1 h at 55°C.
-
g.During incubation, prepare 3M sodium acetate: 246 mg in 1 mL ddH2O.
-
h.Pre-spin phase lock tubes (Qiagen MaXtract High Density, see key resources table) at 13,000 × g for 30 s at 20°C–25°C.
-
i.Add input and immunoprecipitated solutions to separate phase lock tubes.
-
j.Add 200 μL phenol:chloroform:isoamyl alcohol 25:24:1 to each sample. Mix by inverting tubes 20 tubes and vortex on medium setting for 5 s.
-
k.Spin tubes at 13,000 × g for 5 min at 20°C–25°C.
-
l.Take out upper phase and place in a new tube while ensuring to not touch gel with the tip of the pipette.
-
m.Wash upper phase with 200 μL chloroform to remove any traces of phenol and mix by inverting tubes 20 times.
-
n.Spin tubes at 13,000 × g for 5 min at 20°C–25°C.
-
o.During the spin, prepare tubes with 20 μL of 3M sodium acetate + 1 μL GenElute Linear Polyacrylamide (LPA).Note: GenElute LPA is an efficient neutral carrier that is used with ethanol to enhance the precipitation of picrogram quantities of DNA.
-
p.Remove supernatant without taking the bottom layer and transfer to the tubes prepared in step n.
 CRITICAL: Excess salt at the interphase from the ChIP sample might be visible. Remove the upper phase slowly to not disrupt the salts. After the chloroform wash, it is critical to not contaminate the pipette tip with salts or with the bottom phase of the mixture as this will compromise the elution step. Leaving out a small volume of the upper phase to avoid this is recommended.
CRITICAL: Excess salt at the interphase from the ChIP sample might be visible. Remove the upper phase slowly to not disrupt the salts. After the chloroform wash, it is critical to not contaminate the pipette tip with salts or with the bottom phase of the mixture as this will compromise the elution step. Leaving out a small volume of the upper phase to avoid this is recommended. -
q.Mix well by flicking tubes 20 times.
-
r.Add 550 μL of chilled pure ethanol and invert tubes 20 times.
-
s.Allow DNA to precipitate for 3 h at −20°C.
-
t.After precipitation, spin down DNA at 13,000 × g for 30 min at 20°C–25°C.
-
u.Take out supernatant and promptly squirt 200 μL of freshly prepared 70% ethanol onto the pellet so that it detaches from the surfaces of the tube wall.
-
v.Remove the 70% ethanol without disrupting or dissociating the pellet. Use a 10 μL pipette when most of the 70% ethanol is removed to eliminate any remnant drops in the tube.
-
w.Allow pellet to dry at 20°C–25°C. Pellet is dry when it is fully transparent.
 CRITICAL: Ensure that the pellet does not over-dry or it will not dissolve properly in Qiagen Elution buffer. Mark where the pellet is located with a marker outside the tube wall to be able to locate it better when it becomes transparent.
CRITICAL: Ensure that the pellet does not over-dry or it will not dissolve properly in Qiagen Elution buffer. Mark where the pellet is located with a marker outside the tube wall to be able to locate it better when it becomes transparent. -
x.Add 30 μL Qiagen Elution buffer to pellet and let sit for 3 min at 20°C–25°C to rehydrate the pellet. Resuspend the pellet by light vortexing and by pipetting up and down 20 times.
-
a.
Note: Due to the low amount of starting material, the entire ULI-ChIP experiment is performed blind (i.e., no pellets will be visible). Pipetting with care is critical.
CRITICAL: Move directly to library preparation and do not freeze samples after ULI-ChIP elution. Freezing samples will cause significant loss of material.
-
22.
Library preparation (Day 3)
Prepare libraries using the Qiagen UltraLow Input Library Kit using the manufacturer’s instructions (https://www.qiagen.com/us/resources/resourcedetail?id=e1e88a40-bf6b-431a-9697-799e14103467&lang=en). For approximatively 250 cells (equivalent to 30–32 × 8-cell embryos), we use a 1:1000 dilution for adapters (step 4) and perform 18 PCR library amplification cycles (step 21) (Figures 4A–4C).
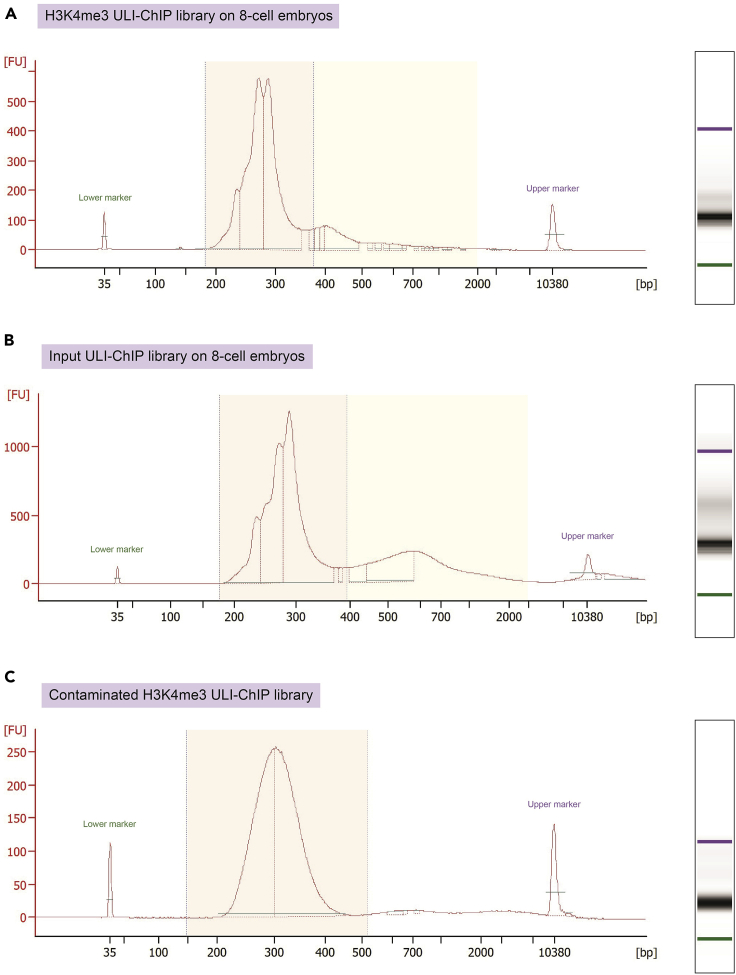
Note: The Agilent Bioanalyzer is not sensitive enough to detect mononucleosomes after the ULI-ChIP. Only run samples on the Bioanalyzer after library preparation. We recommend running the input on the Bioanalyzer alongside the ChIP. Input DNA quantity should be at least half of the total ChIP DNA quantity (Figures 4A and 4B).
Figure 4.
Ultra-low-input ChIP-seq Bioanalyzer expected outcomes
(A) H3K4me3 ULI-ChIP Bioanalyzer electropherogram on approximatively 250 8-cell embryo cells. Mononucleosome peak shown in orange box and undigested chromatin denoted by yellow box.
(B) Input ULI-ChIP Bioanalyzer electropherogram on approximatively 250 8-cell embryo cells. Mononucleosome peak shown in orange box and undigested chromatin denotated by yellow box.
(C) Example of a contaminated H3K4me3 ULI-ChIP library. Note that the center of the electropherogram peak is located at around 300 bp when it should be found between 270–290 bp.
Low-input mRNA-seq on pre-implantation embryos
Timing: 2–3 days
This step describes how to isolate total RNA on approximatively 100 pre-implantation 8-cell embryo cells and build messenger RNA libraries for mRNA-Seq.
CRITICAL: RNA degrades rapidly and obtaining high quality RNA with a RIN > 8 is necessary for mRNA-Seq because the first step for library preparation relies on a poly(A) tail selection (see troubleshooting problems 2 and 3).
-
23.Total RNA extraction (Day 1)Notes: The use of column-based methods for low-input extractions is not recommended because they lead to increased RNA loss.
 CRITICAL: Volume in tubes with embryo cells should comprise less than 10% of total Tri-Reagent volume.
CRITICAL: Volume in tubes with embryo cells should comprise less than 10% of total Tri-Reagent volume.-
a.Thaw tubes with embryo cells in DNA/RNA Protection Reagent on ice and pool embryos if necessary. In Lismer et al., we only pooled embryos that were sired by the same males.
-
b.Add 750 μL of Tri-Reagent. Mix up and down 10 times with a pipette to completely lyse cells.
-
c.Leave at 20°C–25°C for 5 min.
-
d.Add 150 μL of chloroform.
-
e.Vortex thoroughly on medium setting for 15 s.
-
f.Spin tubes at 13,000 × g and 4°C for 15 min.
-
g.Remove the top aqueous phase while leaving some behind to avoid contamination by interphase (DNA) and organic phase (protein, lipids). Ensure that pipette tip does not come into contact with these bottom phases.
-
h.Add 1 μL of GenElute Linear Polyacrylamide (LPA) to retrieved aqueous phases and mix vigorously by inverting the tubes 10 times and pulse vortexing.
-
i.Precipitate the RNA using 500 μL of ice-cold isopropanol. Precipitate in a −20°C freezer for 2.5 h.
-
j.Spin tubes at 13,000 × g and 4°C for 30 min.
-
k.Rapidly remove and discard liquid.
-
l.Wash pellet with freshly prepared 70% ethanol. Circle with a thin marker the location of the pellet on the tube because the pellet becomes transparent after it dries.
-
m.Once the pellet is translucent and liquid is no longer visible in the tube, resuspend the pellet in 12 μL of ddH2O.
-
n.Verify total RNA integrity and quantify using the Agilent RNA 6000 Pico Kit (Figure 5A).
-
a.
CRITICAL: Move directly to library preparation and do not freeze samples after total RNA extraction. Freezing samples will cause significant loss of material due to RNA degradation.
-
24.
Library preparation (Day 2–3)
Figure 5.
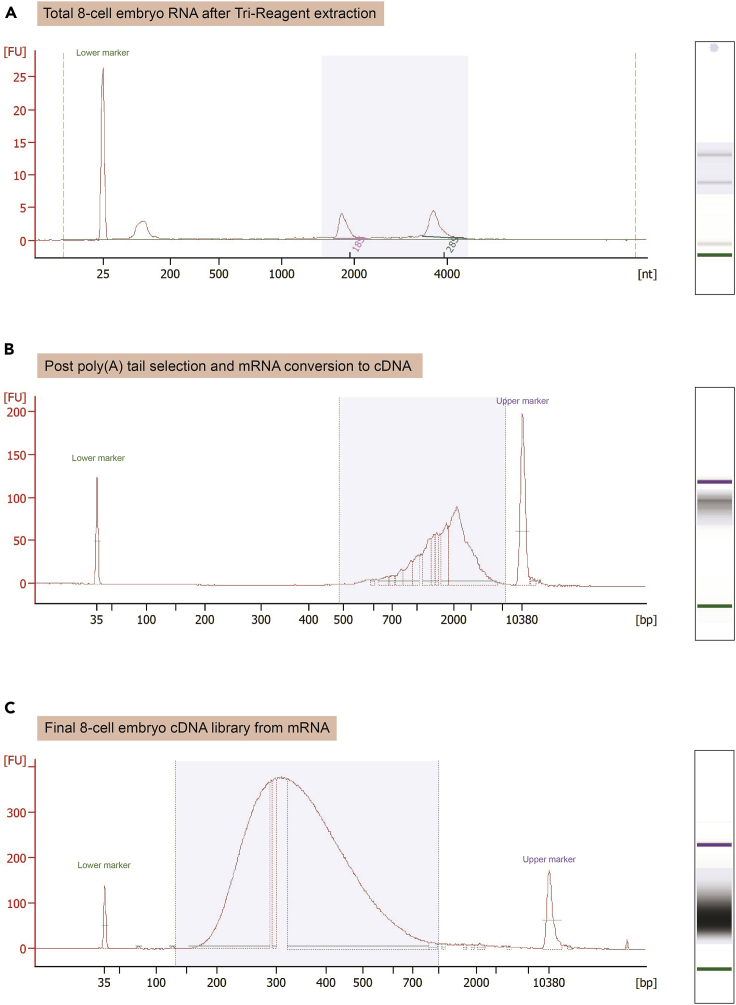
Low-input mRNA-seq Bioanalyzer expected outcomes
(A) Total RNA Bioanalyzer electropherogram on approximatively 100 × 8-cell embryos after Tri-Reagent RNA extraction.
(B) Bioanalyzer electropherogram of non-fragmented cDNA after poly(A) tail selection and mRNA conversion from embryo mRNA.
(C) Final Bioanalyzer electropherogram of fragmented, amplified, and cleaned cDNA from embryo mRNA.
For library preparation, use NEBNext Single Cell/Low Input RNA Library Prep Kit for Illumina following manufacturer’s instructions in “Protocol for Low Input RNA” for input < 5 ng total RNA per reaction (https://international.neb.com/protocols/2018/04/25/protocol-for-low-input-rna-cdna-synthesis-amplification-and-library-generation). For approximatively 100 cells (equivalent to 12–14 × 8-cell embryos), we perform 15 amplification cycles at the cDNA Amplification Step (step 2.4) (Figure 5B) and 8 amplification cycles for the PCR Enrichment Step (step 2.10) (Figure 5C).
CRITICAL: In Section 2.2, use 11 μl of total RNA per sample and remove ddH2O from Section 2.3 (Reverse Transcription and Template Switching).
Note: In Lismer et al., DNase treatment was not used for 100 cells because it led to too much material loss. The poly(A) tail selection was sufficient to generate clean mRNA libraries that did not contain genomic DNA contamination.
Pause point: We do not recommend pausing and freezing mRNA or cDNA until completion of the cDNA amplification by PCR (Section 2.4).
Expected outcomes
For ChIP-seq on sperm
For 8 million sperm cells from one C57BL6/J male mouse a 2–3 ng of mononucleosomal DNA yield is anticipated before library preparation (Figures 3A and 3C). After mononucleosome isolation, expected yield ranges from 1.5–2.5 ng of mononucleosomal DNA (Figures 3D and 3E). Center of mononucleosome fragments should be located at around 150–160 bp on Bioanalyzer electropherogram (Figures 3A and 3C–3E). After library preparation on isolated mononucleosomes, Electropherogram peaks should be narrow with a center shifted to around 270–290 bp (150–160 bp mononucleosome fragments + 120 bp Qiagen adapters) (Figure 3B).
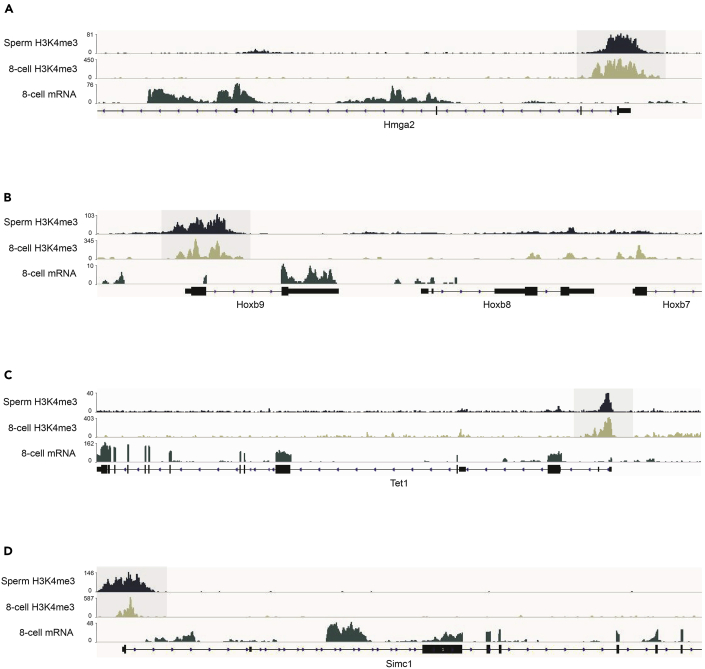
For ultra-low-input ChIP-seq on 8-cell embryos
For approximatively 250 8-cell embryo cells (32 pooled 8-cell embryos), center of electropherogram peaks after library preparation should be centered at around 270–290 bp (150–160 bp mononucleosome fragments + 120 bp Qiagen adapters) (Figure 4A). Note that mononucleosome signal after library preparation for the ULI-ChIP is not at smooth as expected for the sperm ChIP (Figures 3B and 4A). This is a caveat of working with an ultra-low input number of cells and having to use more amplification cycles during the library preparation.
For low-input mRNA-seq on 8-cell embryos
For approximatively 110 8-cell embryo cells (14 pooled 8-cell embryos), approximatively 2.5 ng of total RNA is expected per sample with a RIN > 8 (Figure 5A). After cDNA amplification (step 2.4), 7–12 ng of cDNA is expected (Figure 5B). After PCR Enrichment (step 2.10), 150–200 ng of cDNA is anticipated (Figure 5C). These numbers can vary depending on the number of input cells and how transcriptionally active the chosen cell type is. The electropherogram peak center for final cDNA product should be between 300–350 bp (Figure 5C).
Note: Final library concentration should be confirmed to be sufficient for your sequencing platform (1 ng/μL for Illumina HiSeq).
Note: Use paired-end sequencing for pre-implantation embryo data to improve read mappability in low-input samples.
Note: See Figures 6A–6D for Integrative Genome Viewer snapshots of sequencing datasets.
Figure 6.
Representative examples of obtained sequencing datasets
(A–D) Integrative Genome Viewer screenshots of folate sufficient wildtype sperm ChIP-seq (approximatively 8 million sperm cells), 8-cell embryo ULI-ChIP-seq (approximatively 30 pooled 8-cell embryos i.e., 250 cells), and 8-cell embryo mRNA-seq (approximatively 12 pooled 8-cell embryos i.e. 100 cells) datasets at the (A) Hmga2, (B) Hoxb9, (C) Tet1, and (D) Simc1 genes. Sperm and 8-cell embryo H3K4me3 peaks are indicated by gray boxes. 8-cell embryo mRNA-seq reads are shown in green.
Quantification and statistical analysis
Pre-processing sperm ChIP-sequencing data
Trim reads using Trimmomatic in single-end mode (Bolger et al., 2014) (TruSeq3-SE.fa:1:30:15 TRAILING:25 MINLEN:30). Map reads to the latest assembly of the mouse genome downloaded from UCSC Genome Browser (ex: mm10, NCBI Build 38) (Haeussler et al., 2018) with Bowtie2 (Langmead and Salzberg, 2012) in single-end mode using the standard settings. Use SAMtools (Li et al., 2009) to sort and convert SAM files into BAM files.
Pre-processing 8-cell embryo ULI-ChIP-sequencing data
Trim reads using Trimmomatic (Bolger et al., 2014) in paired-end mode (TruSeq3-PE.fa:1:30:15 TRAILING:25 MINLEN:30). Map reads to the latest assembly of the mouse genome downloaded from UCSC Genome Browser (ex: mm10, NCBI Build 38) (Haeussler et al., 2018) with Bowtie2 (Langmead and Salzberg, 2012) in paired-end mode using the standard settings. Use SAMtools (Li et al., 2009) to sort and convert SAM files into BAM files.
Pre-processing 8-cell embryo mRNA-sequencing data
Trim reads using Trimmomatic (Bolger et al., 2014) in paired-end mode (TruSeq3-PE.fa:1:30:15 TRAILING:25 MINLEN:30). Map reads to the latest assembly of the mouse genome downloaded from UCSC Genome Browser (ex: mm10, NCBI Build 38) (Haeussler et al., 2018) with Hisat2 (Kim et al., 2015) in paired-end mode using the standard settings. Use SAMtools (Li et al., 2009) to sort and convert SAM files into BAM files. Call transcripts using Stringtie (Pertea et al., 2016).
Generating bigwig files
Generate bigwig coverage tracks from aligned reads using DeepTools2 (Ramírez et al., 2016). For visualization, normalize reads using the Reads Per Kilobase Million function (RPKM).
Sequencing read statistic ranges from sperm ChIP-seq datasets (single-end 50 bp illumina HiSeq 4000)
| Experiment | Percentage of overall alignment rate | Number of aligned reads | Number of uniquely mapped reads | Number of reads aligned > one time |
|---|---|---|---|---|
| H3K4me3 Sperm ChIP-Seq | 96%–98% | 25,000,000–45,000,000 | 17,000,000–30,000,000 | 7,000,000–16,000,000 |
Sequencing read statistic ranges from 8-cell embryo ULI-ChIP-seq and 8-cell embryo mRNA-seq datasets (paired-end 100 bp illumina HiSeq 4000)
| Experiment | Percentage of overall alignment rate | Total number of aligned reads | Number of concordantly aligned reads | Number of concordantly uniquely mapped reads | Number of concordantly mapped reads aligned > one time | Number of discordantly aligned reads |
|---|---|---|---|---|---|---|
| H3K4me3 ULI-ChIP-Seq | 55%–80% | 38,000,000–48,000,000 | 37,000,000–47,000,000 | 30,000,000–39,000,000 | 7,000,000–9,000,000 | 900,000–1,200,000 |
| Low-Input mRNA-Seq | 75%–80% | 20,000,000–40,000,000 | 15,000,000–25,000,000 | 14,000,000–24,000,000 | 1,000,000–2,500,000 | 5,000,000–10,000,000 |
Limitations
Recognizing the caveats of antibody-based techniques
Antibody non-specific binding: using an antibody with high non-specific binding may lead to high background and low signal-to-noise ratio.
Antibody cross-reactivity: antibody cross-reactivity between related histone marks has been shown to occur in ChIP experiments (Shah et al., 2018).
Understanding the challenges of working with low input
Reprogramming event or limitations of low input conditions? Performing an immunoprecipitation experiment on rare cell populations does not capture the entire chromatin landscape and peaks of low histone densities are not efficiently captured (Brind'Amour et al., 2015). Conclusions regarding whether the absence of a peak corresponds to a reprogramming event or the limitations of the ULI-ChIP-seq cannot be drawn.
How low can you go? Lowering the amount of input material will decrease library complexity and genome-wide coverage of the pre-implantation embryo chromatin landscape. The antibody efficacy will play an important role in determining the minimum number of cells to use.
Troubleshooting
Problem 1: ULI-ChIP-seq Bioanalyzer electropherogram peak is not centered around the proper molecular weight (step 22)
This likely indicates the amplification of a contamination and absence of mononucleosomal chromatin (Figure 4C). Working in low-input conditions facilitates the incidence of sample contamination which may include bacterial, human, or adapter contaminations.
Potential solution
Sterilize your work environment as described in “Low-input and library preparation considerations.”
Remake all your buffers according to Brind'Amour et al., 2015.
If the problem persists, sequence sample at low-depth to determine whether the contaminating reads come from adapters, bacteria, or human.
Problem 2: RNA extraction on pre-implantation embryos leads to low RIN values
Low RIN values on pre-implantation embryo total mRNA will result in an inefficient library preparation and thus poor library complexity (step 23. n.)
Potential solution
Sterilize your work environment as described in “Low-input and library preparation considerations.” Always work in RNase-free conditions.
When collecting embryos, limit collection time to under 10 min to prevent RNA degradation.
Flash freeze 8-cell embryos in a stabilizing agent (e.g., DNA/RNA Protection Reagent, or directly in Tri-Reagent).
Problem 3: Poor library complexity is obtained after sequencing
This problem occurs when the number of uniquely mapped reads and genome-wide sequencing coverage are low (after sequencing of steps 12, 22, and 24)
Potential solution
For ChIP-seq or ULI-ChIP-Seq, determine whether more antibody needs to be used.
Aim to average 25 million reads per sperm sample and 45 million reads per embryo sample. Embryo samples are more likely to contain reads that do not map to the mouse genome that need to be discarded.
Use the minimum number of PCR amplification cycles during library preparation to obtain a final DNA yield appropriate for your sequencing platform.
Problem 4: ChIP-seq and ULI-ChIP-seq data sets have a lot of background noise
A high amount of background noise will decrease the signal to background ratio and prevent robust peak calling in ChIP-seq experiments (after sequencing of steps 12 and 22)
Potential solution
Always make wash buffers fresh.
Clean samples before library preparation with Ampure XP beads to remove high and low molecular weight and isolate mononucleosomes.
Perform the immunoprecipitation with the minimum required amount of antibody. Using too much antibody results in non-specific binding and subsequent background noise.
Problem 5: Overdigestion and/or underdigestion of the sperm chromatin (step 5)
Poor sperm chromatin digestion will be visible on a Bioanalyzer electropherogram before library preparation. Chromatin overdigestion involves an accumulation of DNA at low molecular weight (Figures 3C and 3D). An underdigestion of the chromatin is observable by the presence of di- and tri-nucleosomal DNA at high molecular weight (Figure 3C).
Potential solution
If a distinct mononucleosome peak is visible, clean low and/or high molecular weight with Ampure XP beads as indicated in step 11.
Verify that the MNase was aliquoted correctly and perform a gradient MNase digestion on practice sperm. Each MNase batch may slightly vary.
Always keep MNase on ice and do not freeze-thaw aliquots.
Mix MNase very well when introducing solution in tubes to allow for a homogenous distribution of the MNase in each sample.
Ensure that sperm counts were performed properly. Over-counting will lead to an underdigestion of the sperm chromatin whereas under-counting will lead to an overdigestion.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sarah Kimmins (sarah.kimmins@mcgill.ca).
Materials availability
This study did not generate any new reagents.
Data and code availability
The datasets generated during this study are available at GEO:GSE135678.
Acknowledgments
We thank McGill University mouse facility personnel, the Génome Québec Innovation Center. This research was funded by the Canadian Institute of Health Research (S.K. #358654).
Author contributions
A.L. performed the experiments; S.K., A.L., R.L., C.L., and V.D. developed the methodology and created the models. S.K., R.L., C.L., and V.D. supervised. S.K. and A.L. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Ariane Lismer, Email: ariane.lismer@mail.mcgill.ca.
Sarah Kimmins, Email: sarah.kimmins@mcgill.ca.
References
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brind'Amour J., Liu S., Hudson M., Chen C., Karimi M.M., Lorincz M.C. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat Commun. 2015;6:6033. doi: 10.1038/ncomms7033. [DOI] [PubMed] [Google Scholar]
- Byers S.L., Wiles M.V., Dunn S.L., Taft R.A. Mouse estrous cycle identification tool and images. PLoS One. 2012;7:e35538. doi: 10.1371/journal.pone.0035538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M., Zweig A.S., Tyner C., Speir M.L., Rosenbloom K.R., Raney B.J., Lee C.M., Lee B.T., Hinrichs A.S., Gonzalez J.N. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2018;47:D853–D858. doi: 10.1093/nar/gky1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna C.W., Taudt A., Huang J., Gahurova L., Kranz A., Andrews S., Dean W., Stewart A.F., Colomé-Tatché M., Kelsey G. MLL2 conveys transcription-independent H3K4 trimethylation in oocytes. Nat. Struct. Mol. Biol. 2018;25:73–82. doi: 10.1038/s41594-017-0013-5. [DOI] [PubMed] [Google Scholar]
- Hisano M., Erkek S., Dessus-Babus S., Ramos L., Stadler M.B., Peters A.H. Genome-wide chromatin analysis in mature mouse and human spermatozoa. Nat. Protoc. 2013;8:2449–2470. doi: 10.1038/nprot.2013.145. [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrot R., Xu C., Saint-Phar S., Chountalos G., Cohen T., Paquet M., Suderman M., Hallett M., Kimmins S. Low paternal dietary folate alters the mouse sperm epigenome and is associated with negative pregnancy outcomes. Nat. Commun. 2013;4:2889. doi: 10.1038/ncomms3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lismer A., Dumeaux V., Lafleur C., Lambrot R., Brind’Amour J., Lorincz M.C., Kimmins S. Histone H3 lysine 4 trimethylation in sperm is transmitted to the embryo and associated with diet-induced phenotypes in the offspring. Dev. Cell. 2021;56:671–686.e6. doi: 10.1016/j.devcel.2021.01.014. [DOI] [PubMed] [Google Scholar]
- Luo C., Zuñiga J., Edison E., Palla S., Dong W., Parker-Thornburg J. Superovulation strategies for 6 commonly used mouse strains. J. Am. Assoc. Lab. Anim. Sci. 2011;50:471–478. [PMC free article] [PubMed] [Google Scholar]
- Oakberg E.F. Duration of spermatogenesis in the mouse and timing of stages of the cycle of the seminiferous epithelium. Am. J. Anat. 1956;99:507–516. doi: 10.1002/aja.1000990307. [DOI] [PubMed] [Google Scholar]
- Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves P.G., Nielsen F.H., Fahey G.C., Jr. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr. 1993;123:1939–1951. doi: 10.1093/jn/123.11.1939. [DOI] [PubMed] [Google Scholar]
- Rogers L.M., Cordero A.M., Pfeiffer C.M., Hausman D.B., Tsang B.L., De-Regil L.M., Rosenthal J., Razzaghi H., Wong E.C., Weakland A.P. Global folate status in women of reproductive age: a systematic review with emphasis on methodological issues. Ann. NY Acad. Sci. 2018;1431:35–57. doi: 10.1111/nyas.13963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R.N., Grzybowski A.T., Cornett E.M., Johnstone A.L., Dickson B.M., Boone B.A., Cheek M.A., Cowles M.W., Maryanski D., Meiners M.J. Examining the roles of H3K4 methylation states with systematically characterized antibodies. Mol. Cell. 2018;72:162–177.e7. doi: 10.1016/j.molcel.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne B.G., Kawata A., Behan N.A., Williams A., Wade M.G., Macfarlane A.J., Yauk C.L. Investigating the effects of dietary folic acid on sperm count, DNA damage and mutation in BALB/C mice. Mutat. Res. 2012;737:1–7. doi: 10.1016/j.mrfmmm.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Zhang B., Zheng H., Huang B., Li W., Xiang Y., Peng X., Ming J., Wu X., Zhang Y., Xu Q. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature. 2016;537:553–557. doi: 10.1038/nature19361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during this study are available at GEO:GSE135678.