Supplemental Digital Content is available in the text.
Keywords: clinical notes, critical care, machine learning, mortality, natural language processing
Abstract
OBJECTIVES:
To evaluate whether different approaches in note text preparation (known as preprocessing) can impact machine learning model performance in the case of mortality prediction ICU.
DESIGN:
Clinical note text was used to build machine learning models for adults admitted to the ICU. Preprocessing strategies studied were none (raw text), cleaning text, stemming, term frequency-inverse document frequency vectorization, and creation of n-grams. Model performance was assessed by the area under the receiver operating characteristic curve. Models were trained and internally validated on University of California San Francisco data using 10-fold cross validation. These models were then externally validated on Beth Israel Deaconess Medical Center data.
SETTING:
ICUs at University of California San Francisco and Beth Israel Deaconess Medical Center.
SUBJECTS:
Ten thousand patients in the University of California San Francisco training and internal testing dataset and 27,058 patients in the external validation dataset, Beth Israel Deaconess Medical Center.
INTERVENTIONS:
None.
MEASUREMENTS AND MAIN RESULTS:
Mortality rate at Beth Israel Deaconess Medical Center and University of California San Francisco was 10.9% and 7.4%, respectively. Data are presented as area under the receiver operating characteristic curve (95% CI) for models validated at University of California San Francisco and area under the receiver operating characteristic curve for models validated at Beth Israel Deaconess Medical Center. Models built and trained on University of California San Francisco data for the prediction of inhospital mortality improved from the raw note text model (AUROC, 0.84; CI, 0.80–0.89) to the term frequency-inverse document frequency model (AUROC, 0.89; CI, 0.85–0.94). When applying the models developed at University of California San Francisco to Beth Israel Deaconess Medical Center data, there was a similar increase in model performance from raw note text (area under the receiver operating characteristic curve at Beth Israel Deaconess Medical Center: 0.72) to the term frequency-inverse document frequency model (area under the receiver operating characteristic curve at Beth Israel Deaconess Medical Center: 0.83).
CONCLUSIONS:
Differences in preprocessing strategies for note text impacted model discrimination. Completing a preprocessing pathway including cleaning, stemming, and term frequency-inverse document frequency vectorization resulted in the preprocessing strategy with the greatest improvement in model performance. Further study is needed, with particular emphasis on how to manage author implicit bias present in note text, before natural language processing algorithms are implemented in the clinical setting.
Clinical note text contains valuable information that may not be fully captured anywhere else in the electronic health record (EHR) (1). Because reading note text and extracting information has been resource intensive, incorporating text into large clinical studies has historically been challenging (2). Over the last 10 years, natural language processing (NLP) has increasingly been used to automate information extraction from note text (3). Incorporating note text from the EHR into statistical models has improved the prediction of some important clinical outcomes, including in critical care (4–9). NLP is increasingly used in medicine to study clinical outcomes, augment clinical decision support, and assist with clinical research (3, 10–12). Because NLP is more frequently being used in the clinical domain and because application of clinical knowledge is integral to creating robust NLP models, it is important for clinicians to understand how NLP models are developed (1).
An important component of NLP model development is preparation of text for analysis (referred to as “preprocessing”) (13). Preprocessing can include several steps to transform raw note text data into data that is ready for inclusion in statistical models. However, how much or how little preprocessing is done is at the discretion of the investigator (14). Little is known about the impact of the choice among alternative preprocessing strategies on model performance in the prediction of important critical care outcomes. The objective of this study is to evaluate whether different preprocessing strategies can impact machine learning model performance in the ICU. To demonstrate this, we show results for different preprocessing strategies when text is used to predict inhospital mortality. We do this first for penalized logistic regression models, but to give a sense of the robustness of these findings, also give summary results for two other artificial intelligence analytic approaches, feed forward neural networks, and random forest classification.
METHODS
Study Cohort
We analyzed note text of patients greater than or equal to 18 years with an ICU length of stay greater than 4 hours who were admitted to the ICU at the University of California San Francisco (UCSF) between December 22, 2011, and May 29, 2017, or the ICU at Beth Israel Deaconess Medical Center (BIDMC) between June 9, 2100, and October 25, 2205. BIDMC dates appear to be in the future because they are masked for de-identification purposes. Note text data were extracted from BIDMC via the Medical Information Mart for Intensive Care III database and from the EHR at UCSF (15). Only the first admission to the ICU during the study period for each patient was included. Note text for 12 hours prior to ICU admission to 24 hours after ICU admission were included. Clinical notes written in the ICU by physicians, nurse practitioners, physician assistants, or registered nurses were included. For each patient, we combined all notes in the time frame to form a single document, or corpus, for the patient. To improve processing times, a random selection of 10,000 patients (from all patients admitted to the adult ICU at UCSF during the study period) was included. All patients from the study population at BIDMC were included during model validation. This study was approved by the UCSF Institutional Review Board (No. 12-08609), which waived the requirement for informed consent.
Preprocessing
We describe commonly used methods to preprocess note text data (Fig. 1). Text cleaning, stemming, term frequency-inverse document frequency (TF-IDF) vectorization, and creation of n-grams are all forms of preprocessing. All preprocessing was done using the Python Version 3 Natural Language Toolkit (NLTK) package, version 3.4.5, a commonly used program for NLP (16, 17).
Figure 1.
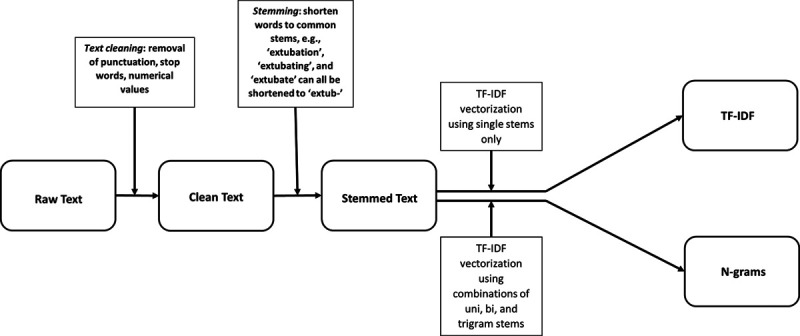
Preprocessing note text pathway. TF-IDF = term frequency-inverse document frequency.
Cleaning Text
We first transformed raw text to clean text. In NLP, “text cleaning” usually refers to removal of punctuation and numerical values from notes, as well as removal of words that are expected to have little predictive value because they are so common (these words are called “stop words”) (14). Examples of stop words are “a,” “an,” “and,” “for,” “it,” and “the” (18). Because the NLTK package is commonly used, we initially used the default list of stop words in the package. Supplemental Table 1 (http://links.lww.com/CCX/A656) contains the full list of the NLTK stop words. Numerical values in notes reflect a wide variety of concepts from vital signs to phone numbers. Numerical values may be removed because numbers on their own do not provide meaning. For example, the number 60 has very different meaning when it is preceded by “heart rate” versus “systolic blood pressure” versus followed by “days in the hospital.” Additionally, other numbers such as phone numbers can be associated with inappropriate significance. For example, a phone number could be highly associated with inhospital mortality if the number was for the coroner’s office, but one would not want to include that number in a model to predict mortality.
Stemming
We then transformed the cleaned text by stemming it. The goal of stemming is to consolidate words that have the same meaning to their stem or root base, so that all the predictive power of the underlying concept is captured in a single predictor variable, rather than being spread across multiple predictors (14). Without stemming, words such as “ventilator,” “ventilating,” and “ventilated” would be considered as three discrete terms that are not related to each other. With stemming, all three words could be truncated to a stem such as “ventil,” and the predictive power of the concept “the patient needs mechanical ventilation” can all be assigned to the single stem, rather than spread out across different terms that mean the same thing clinically.
Count Vectorization
Count vectorization is a method used to transform text into numeric outputs (19). Count vectorization counts the number of times a particular term is written in a given document. We restricted the terms counted in each document to the 1,000 terms that were most commonly used across the text of all UCSF patients.
Term Frequency-Inverse Document Frequency Vectorization
TF-IDF vectorization is a method to adjust for how commonly a term is found in note text. TF-IDF is a calculated numerical weight. Term frequency is calculated as how often a term appears in the entire corpus for an individual patient divided by how much was written about the patient (measured by how many terms there are in the corpus for that patient) (20). In NLP, term frequency is usually multiplied by inverse document frequency (IDF). IDF represents how commonly a particular term is found across all patients and is calculated in this case as the log of the number of patients (since each patient has a single document, their corpus), divided by the number of patients with the term. Therefore, if every patient has the term (an example in the ICU might be “IV,” since nearly all ICU patients have an IV catheter), then the ratio of all patients to all patients with the term is near 1. Because the log of 1 is 0, the term IV would be zeroed out and would not be included as a predictor in the statistical model. Rare terms that are present in few documents will have an IDF closer to 1 (21). When the TF and IDF are multiplied together, the numerical weight for a particular term is generated. The mathematical equations used to calculate TF-IDF are shown in Supplemental Figure 1 (http://links.lww.com/CCX/A655). The numerical weight for each term is then incorporated into statistical models. Once text is transformed into numeric outputs, the text is referred to as “featurized data.”
N-Grams
N-grams represent a string of n words. For example, single words are unigrams and two words are bigrams (5). Investigators may consider use of n-grams for a variety of reasons. For instance, it may be expected that two- or three-word phrases may capture important risk factors or differences among risk factors. For example, “liver failure” may be more predictive than “liver” and “failure” when treated as separate entities. Alternatively, “acute lymphocytic leukemia” may carry different risk of inhospital mortality than “chronic lymphocytic leukemia,” and the investigators may be concerned that such distinctions would get lost treating each word separately (in which case there would be a single coefficient generated by the model for the word “leukemia” that would merge the risk of acute and chronic leukemias). In addition, using n-grams allows for incorporation of negation terms such as (“not septic”) into a model (5, 22). The default list of stop words in the NLTK package included words such as “don’t” and “weren’t.” For our n-grams analysis, we did not remove stop words that implied negation in medical text even though they were on the NLTK list of usual stop words. Thus, we included terms such as “no” and “wasn’t” in text. The list of negation words that we included for the n-grams analysis, even though they are on the NLTK stop word list, is shown in Supplemental Table 2 (http://links.lww.com/CCX/A656).
Statistical Analysis
We developed mortality prediction models using note text that had undergone different types of preprocessing. The “Raw Text” model used note text directly extracted from the EHR as predictors of inhospital mortality. The “Clean Text” model removed punctuation, stop words, and numerical values from notes. The “Stemming” model used clean text that underwent stemming. The “TF-IDF” model used Clean Text that underwent stemming and TF-IDF vectorization. Last, the n-gram model used note text that used clean text (with a modified stop word list), underwent stemming, and TF-IDF vectorization that used unigram, bi-gram, and tri-gram term combinations (Fig. 1).
Model Development and Validation
We used penalized logistic regression (L1 penalized/“Least Absolute Shrinkage and Selection Operator” and L2 penalized/“Ridge”) to model the association between inhospital mortality and different types of preprocessed note text. Penalized logistic regression was used because current widely accepted mortality prediction models use logistic regression and because logistic regression allows easy interpretation of the association of individual terms or stems with the outcome (4, 23). Count vectorization was used to associate a numerical value with terms in the Raw Text, Clean Text, and Stemming models. TF-IDF vectorization was used in the TF-IDF and n-gram models. We included the top 1,000 features with the highest count vectorization or TF-IDF vectorization values calculated in each model. Beta-coefficients for each term were calculated. Because current literature has demonstrated that machine learning models may perform better than standard statistical methods in predicting mortality, we also used random forests and feed forward neural networks to model inhospital mortality using different types of preprocessed note text (8).
Models were trained on UCSF data and then validated on both UCSF and BIDMC data. This was done so that models developed at UCSF could be externally validated on unseen data from another institution (BIDMC). Using data from another hospital not involved in training the models is a robust method to demonstrate that changes in model performance are not due to noise or chance alone and also shows that the model is generalizable to patients at other institutions (Supplemental Fig. 2, http://links.lww.com/CCX/A655). We assessed model performance by calculating the area under the receiver operating characteristic curve (AUROC) to determine discrimination for each model. We expected model performance to decrease when models developed at UCSF were validated on BIDMC data because model overfitting is common for models trained and validated on the same data and because BIDMC was hypothesized to have different patterns of clinical documentation than UCSF.
Ten-fold cross validation on the UCSF data was used to determine CIs for the AUROC. Ten-fold cross validation is used to train and validate machine learning models. The dataset is randomly divided into 10 equal parts. Nine of the subsets are used to train the data and the 10th subset is used to validate the data. The subsets are then reshuffled and the subset that was used for validation is incorporated into the training data and one of the subsets used for training is used as the new validation dataset. This reshuffling occurs 10 times until all subsets have been used for both training and validation. Once the cross validation is complete, the AUROC is averaged and the CI is computed (24). Models were also trained using 10-fold cross validation for hyper-parameter estimation in both UCSF and BIDMC datasets.
We assessed model calibration with calibration curves for all statistical models. Samples were divided in ten deciles for calibration according to their predicted mortality probabilities. For each decile, means of predicted and observed death were obtained. For each observed mean, the 95% CI was also computed. Models developed on UCSF data were calibrated on BIDMC data.
Programming
The Scikit-Learn package in Python Version 3 was used to create the logistic regression, random forests, the feedforward neural network (multilayer perceptron) models, and the calibration curves.
RESULTS
There were 27,058 patients admitted to BIDMC and 10,000 patients sampled from UCSF. Inhospital mortality rate at BIDMC and UCSF was 10.9% and 7.4%, respectively. The average number of notes written per patient was 3.5 and 7.7 at BIDMC and UCSF, respectively. Table 1 describes note characteristics and Table 2 describes demographic characteristics.
TABLE 1.
Note Demographics
| Variable | University of California San Francisco | Beth Israel Deaconess Medical Center |
|---|---|---|
| Number of notes per patient | 7 (5–9), 7.7 | 3 (2–4), 3.5 |
| Number of words per note in raw note text | 235 (60–748), 496 | 194 (96–345), 285 |
| Number of words per note after text cleaning | 141 (36–443), 294 | 117 (57–198), 165 |
| Number of unique words per note after text cleaning | 107 (32–285), 178 | 96 (50–155), 125 |
| Total number of unique words across all patient notes after text cleaning | 100,509 | 145,675 |
Ranges are median (interquartile range), mean.
TABLE 2.
Patient Cohort Characteristics
| Variable | University of California San Francisco (n = 10,000) | Beth Israel Deaconess Medical Center (n = 27,058) |
|---|---|---|
| Male (%) | 52.5 | 58 |
| Age (yr) | 60 (47–69) | 64 (51–76) |
| Mortality (%) | 7.4 | 10.9 |
| Length of ICU stay in survivors (d) | 2 (1–3) | 2 (1–4) |
| Length of ICU stay in patients that died | 2.6 (1.1–6.1) | 3.3 (1.4–7.7) |
| Type of ICU (%) | ||
| Combined medical and surgical | 7.9 | NA |
| Medical | 17.8 | 33.9 |
| Surgical | 18.1 | 29.1 |
| Neurologic | 39.6 | NA |
| Cardiac | 16.6 | 37 |
NA = not available.
Ranges are median (interquartile range).
Each step with preprocessing note text resulted in small improvements in model performance when testing models built at UCSF on BIDMC data (Table 3). Models built and tested on UCSF data for the prediction of inhospital mortality improved from the raw note text model (AUROC, 0.84; CI, 0.80–0.89) to the TF-IDF model (AUROC, 0.89; CI, 0.85–0.94). Models validated on BIDMC data had a similar increase in model performance from raw note text (AUROC at BIDMC: 0.72) to the TF-IDF model (AUROC at BIDMC: 0.83) but an expected decrease in model performance overall (Table 3). Creation of n-grams did not improve model performance (AUROC at BIDMC: 0.80).
TABLE 3.
Impact of Preprocessing Strategies on Logistic Regression Model Performance
| Variable | AUROC (95th% CI) for UCSF Model Validated on UCSF Data | AUROC for UCSF Model Validated on Beth Israel Deaconess Medical Center |
|---|---|---|
| Raw text | 0.84 (0.80–0.89) | 0.72 |
| Cleaned text | 0.85 (0.79–0.91) | 0.75 |
| Cleaned and stemmed text | 0.84 (0.79–0.90) | 0.77 |
| Term frequency-inverse document frequency | 0.89 (0.85–0.94) | 0.83 |
| 1–3 n-grams | 0.90 (0.86–0.93) | 0.80 |
AUROC = area under the receiver operating characteristic curve, UCSF = University of California San Francisco.
Preprocessing resulted in increases in AUROC not just for penalized logistic regression but also for feed forward neural network and random forest analyses (Supplemental Tables 3 and 4, http://links.lww.com/CCX/A656). Feed forward neural network models had the best model discrimination using raw text (AUROC at BIDMC: 0.76). Penalized logistic regression models experienced the greatest gain in model performance on text that underwent TF-IDF vectorization when compared with raw text. For all models built at UCSF, AUROC decreased when applied to BIDMC data (Table 3 and Supplemental Table 3, http://links.lww.com/CCX/A656), but the decline in AUROC was smaller for TF-IDF models than raw text models.
Calibration curves for the penalized logistic regression models showed that models using TF-IDF vectorization had the best model calibration. The TF-IDF model had the best calibration for patients with low predicted mortalities, while the n-grams model had the best model calibration for patients with higher predicted mortality (Supplemental Fig. 3, http://links.lww.com/CCX/A655). Models using the feed forward neural network had the poorest model calibration when compared with models developed using penalized logistic regression and random forest (Supplemental Figs. 4 and 5, http://links.lww.com/CCX/A655).
Terms most strongly associated with inhospital mortality and survival are shown in Figure 2 and Supplemental Table 5 (http://links.lww.com/CCX/A656). The following terms are stems of words. The terms “Pupils” and “Famili” were strongly associated with inhospital mortality in all models TF-IDF beta-coefficient 2.89 and 2.77, respectively). “Extub” and “Anesthesia” were strongly associated with survival in all models (beta-coefficient –1.72 and –1.67, respectively) (Fig. 2). “Death” was present in all models but was most strongly associated with inhospital mortality in the raw text model (beta-coefficient 0.67). “IV prn” was the bi-gram most strongly associated with inhospital mortality, while “IV continuous” was the bi-gram most strongly associated with survival.
Figure 2.
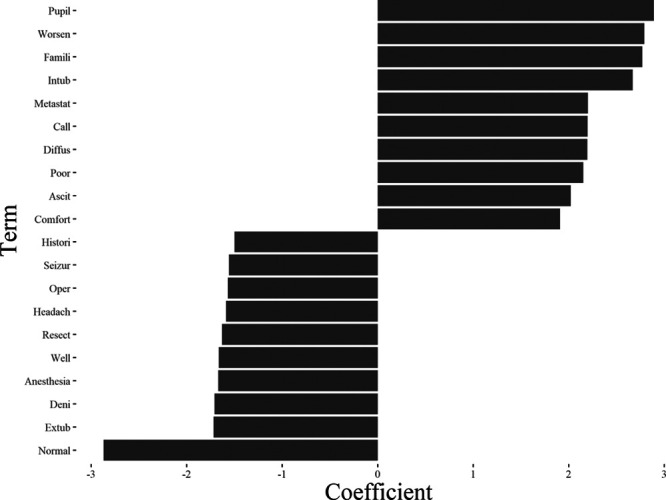
Top beta-coefficients associated with mortality and survival in the term frequency-inverse document frequency model. Beta-coefficients less than 0 are associated with survival and coefficients greater than 0 are associated with mortality.
DISCUSSION
Preprocessing note text improved model discrimination for the prediction of inhospital mortality for adults admitted to the ICU. Each step in preprocessing note text resulted in small improvements in model performance when testing models built at UCSF on BIDMC data. After preprocessing of text, models developed using single institution note text data using TF-IDF vectorization had AUROCs of 0.79–0.83 when applied at another academic institution.
As measured by AUROC, model discrimination in this study is only slightly inferior to reported performance of models that used structured data (e.g., laboratory values and vital signs) in their predictions as well (4, 25). We hypothesize that this is because clinicians’ words in their notes capture much of the predictive information in structured data. For example, we found the presence of the stem “pupil” was predictive of inhospital mortality, and this likely correlates highly with having a low Glasgow Coma Score (because clinicians do not check or report pupillary reflexes on patients unless their Glasgow Coma Score is very low).
Given terms like “pupil” and “hemorrhag” are strongly associated with mortality in the clinical literature, we expected the finding that these stems were associated with inhospital mortality in all of the models (26, 27). Extubation was strongly associated with survival, which reflects that patients who are able to separate from mechanical ventilation have a higher odds of survival (28). “Anesthesia” and “resect” were strongly associated with survival, and these terms may select for a cohort of patients stable enough to receive anesthesia for an operation. However, association of these terms with mortality cannot be generalized to other datasets. This is because documentation styles may vary in ways that are associated with outcome. For example, if clinicians at another institute wrote out the phrase "pupils equal round and reactive" instead of "PERRL," (the abbreviation of the above phrase), pupils would be associated with survival rather than with mortality.
The purpose of the study was to understand the methodologic impact of preprocessing strategies on model development. Because these models were intended only for research, factors associated with bias in note text were not explored. Development of fair, unbiased NLP models is critical when the intention is to apply models in the clinical setting. Models that use NLP on note text in the clinical setting without consideration of potential implicit bias among clinicians pose significant ethical concerns. Using clinical insights and opinions in note text as the only basis for a prediction algorithm may inadvertently create a self-affirming algorithm where verbiage from note writers may unintentionally perpetuate or worsen disparities. Consideration should also be given to determine whether particular terms should be excluded from note text prior to model development. For example, further investigation is needed to assess the impact of social determinants of health on mortality prediction and whether such factors should be included in mortality prediction models (29).
Several terms that represent clinician recognition of likelihood of inhospital mortality in notes (such as “death” and “famili”) were found in every model (30, 31). This raises the question of whether terms that suggest clinician concern for inhospital mortality risk should be treated as stop words (i.e., should be removed) in mortality prediction models. Removal of these words may decrease model performance. However, since including those words leads to some of the risk patients face being attributed to those words, keeping them in may lower the risk attributed to other relevant clinical factors.
Identifying optimal parameters needed to preprocess note text is needed to develop the best predictive models. The largest improvement in model performance occurred between use of raw text and text that underwent TF-IDF vectorization. TF-IDF vectorization may be particularly important in preprocessing clinical note in contexts where institutions or individual clinicians routinely use templates, smart phrases, or cut and paste technology when writing notes (32). Clusters of note text with near duplication are common. These clusters of notes can influence term frequency calculations, but this effect may be mitigated by multiplying by the inverse of document frequency (33).
Addition of word combinations (n-grams) that capture risk factors expressed as a phrase (“acute lymphocytic leukemia”) or that reflect negation (“not septic”) did not meaningfully improve model performance. This may be because single words or stems capture most of the risk. For example, the risk difference between “acute lymphocytic leukemia” and “chronic lymphocytic leukemia” may already be captured in a model that only uses “leukemia” because the treatments are different. That is, the model may interpret “vincristine” used to treat acute lymphocytic leukemia to convey higher risk than “fludarabine” used to treat chronic lymphocytic leukemia, so the difference in risk by type of leukemia may be captured by treatment terms in the text. Alternatively, the risk may be captured in part by complications, such as “aspergillus,” that are more common in the course of acute lymphocytic leukemia.
There may also be reasons identifying negation through n-grams is not a very important determinant of AUROC using TF-IDF. In general, terms that are negated will only be negated once or a small number of times, while conditions that are present will be mentioned many times. Thus, negated terms usually will have low term frequency even if they are negated once. Therefore, failing to capture the negation will not change that the term is not a prominent feature of the patient’s presentation.
Our study has important limitations. Because we validated our data with eight ICU’s in only two institutions, we cannot know how those models would perform on note text from other ICUs. We also had to restrict our sample size to 10,000 patients because of the limited computing time available to analyze note text. In addition, methodologies that improved model prediction of inhospital mortality in adults admitted to the ICU cannot be assumed to improve the prediction of other outcomes, such as length of stay or illness severity. Structured data (such as laboratory values and vital signs) were not included in model development because we did not aim to build a superior mortality prediction model to those that have been published. Instead, the focus of the study was to demonstrate the methodological impact of text preprocessing strategies on inhospital mortality model performance alone. Because patients who died within the first 24 hours of admission were not excluded from the study, reported model performance may be better than if these patients were excluded from the study. However, because improvement in model performance was shown with addition of preprocessing techniques in all statistical models, exclusion of this subpopulation would likely not have changed the observed findings that preprocessing algorithms matter in the prediction of inhospital mortality.
CONCLUSIONS
Differences in preprocessing algorithms applied to note text impacted model discrimination in the prediction of inhospital mortality for adults admitted to the ICU. Completing a preprocessing pathway including cleaning, stemming, and TF-IDF vectorization resulted in the preprocessing method with the greatest improvement in model performance. N-grams did not meaningfully improve model performance. Consideration of how to manage implicit bias present in note text prior to clinical implementation of NLP algorithms is important. Further study is needed to develop fair NLP algorithms before they can be applied in the clinical setting. Additionally, consideration should be given to how personal clinician sentiment may drive model prediction. For example, if clinician charting is optimistic with respect to outcome, the sentiment clinicians express may be captured in the model and thus create a “self-fulfilling prophecy” of note text generating a probability of survival that could be misinterpreted as solely objective. Helping clinicians understand preprocessing strategies may facilitate their participation in the development and eventual clinical integration of NLP-based predictive models.
Supplementary Material
Footnotes
This study was approved by the University of California San Francisco Institutional Review Board (No. 12-08609), which waived the need for informed consent.
Data from Beth Israel Deaconess Medical Center is freely available at https://mimic.physionet.org/. Data from University of California San Francisco is not publicly available as it contains protected health information.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccejournal).
Dr. Butte is a co-founder and consultant to Personalis and NuMedii; he is a consultant to Samsung, Mango Tree Corporation, and in the recent past, 10x Genomics, Helix, Pathway Genomics, and Verinata (Illumina); he has served on paid advisory panels or boards for Geisinger Health, Regenstrief Institute, Gerson Lehrman Group, AlphaSights, Covance, Novartis, Genentech, Merck, and Roche; he is a shareholder in Personalis and NuMedii; he is a minor shareholder in Apple, Facebook, Alphabet (Google), Microsoft, Amazon, Snap, Snowflake, 10x Genomics, Illumina, Nuna Health, Assay Depot (Scientist.com), Vet24seven, Regeneron, Sanofi, Royalty Pharma, Pfizer, BioNTech, AstraZeneca, Moderna, Biogen, Twist Bioscience, Pacific Biosciences, Editas Medicine, Invitae, and Sutro, and several other nonhealth-related companies and mutual funds; and he has received honoraria and travel reimbursement for invited talks from Johnson and Johnson, Roche, Genentech, Pfizer, Merck, Lilly, Takeda, Varian, Mars, Siemens, Optum, Abbott, Celgene, AstraZeneca, AbbVie, Westat, several investment and venture capital firms, and many academic institutions, medical or disease specific foundations and associations, and health systems. Dr. Butte receives royalty payments through Stanford University, for several patents and other disclosures licensed to NuMedii and Personalis. Dr. Butte’s research has been funded by National Institutes of Health (NIH), Northrop Grumman (as the prime on an NIH contract), Genentech, Johnson and Johnson, Food and Drug Administration, Robert Wood Johnson Foundation, Leon Lowenstein Foundation, Intervalien Foundation, Priscilla Chan and Mark Zuckerberg, the Barbara and Gerson Bakar Foundation, and in the recent past, the March of Dimes, Juvenile Diabetes Research Foundation, California Governor’s Office of Planning and Research, California Institute for Regenerative Medicine, L’Oreal, and Progenity. The remaining authors have disclosed that they do not have any potential conflicts of interest.
REFERENCES
- 1.Sanchez-Pinto LN, Luo Y, Churpek MM. Big data and data science in critical care. Chest. 2018; 154:1239–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assale M, Dui LG, Cina A, et al. The revival of the notes field: Leveraging the unstructured content in electronic health records. Front Med (Lausanne). 2019; 6:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaggal VC, Elayavilli RK, Mehrabi S, et al. Toward a learning health-care system - knowledge delivery at the point of care empowered by big data and NLP. Biomed Inform Insights. 2016; 8(Suppl 1):13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marafino BJ, Park M, Davies JM, et al. Validation of prediction models for critical care outcomes using natural language processing of electronic health record data. JAMA Netw Open. 2018; 1:e185097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marafino BJ, Davies JM, Bardach NS, et al. N-gram support vector machines for scalable procedure and diagnosis classification, with applications to clinical free text data from the intensive care unit. J Am Med Inform Assoc. 2014; 21:871–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marafino BJ, Boscardin WJ, Dudley RA. Efficient and sparse feature selection for biomedical text classification via the elastic net: Application to ICU risk stratification from nursing notes. J Biomed Inform. 2015; 54:114–120 [DOI] [PubMed] [Google Scholar]
- 7.Marafino BJ, Dudley RA, Shah NH, et al. Accurate and interpretable intensive care risk adjustment for fused clinical data with generalized additive models. AMIA Jt Summits Transl Sci Proc. 2018; 2017:166–175 [PMC free article] [PubMed] [Google Scholar]
- 8.Weissman GE, Hubbard RA, Ungar LH, et al. Inclusion of unstructured clinical text improves early prediction of death or prolonged ICU stay. Crit Care Med. 2018; 46:1125–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parreco J, Hidalgo A, Kozol R, et al. Predicting mortality in the surgical intensive care unit using artificial intelligence and natural language processing of physician documentation. Am Surg. 2018; 84:1190–1194 [PubMed] [Google Scholar]
- 10.Pruitt P, Naidech A, Van Ornam J, et al. A natural language processing algorithm to extract characteristics of subdural hematoma from head CT reports. Emerg Radiol. 2019; 26:301–306 [DOI] [PubMed] [Google Scholar]
- 11.Castro VM, Dligach D, Finan S, et al. Large-scale identification of patients with cerebral aneurysms using natural language processing. Neurology. 2017; 88:164–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alsawas M, Alahdab F, Asi N, et al. Natural language processing: Use in EBM and a guide for appraisal. Evid Based Med. 2016; 21:136–138 [DOI] [PubMed] [Google Scholar]
- 13.Yim WW, Yetisgen M, Harris WP, et al. Natural language processing in oncology: A review. JAMA Oncol. 2016; 2:797–804 [DOI] [PubMed] [Google Scholar]
- 14.Uysal AK, Gunal S. The impact of preprocessing on text classification. Inf Process Manag. 2014; 50:104–112 [Google Scholar]
- 15.Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a Freely Accessible Critical Care Database. Scientific Data 2016. Available at: https://www.nature.com/articles/sdata201635. Accessed May 25, 2021 [DOI] [PMC free article] [PubMed]
- 16.Van Rossum G, Drake FL: Python 3 Reference Manual. Scotts Valley, CA, CreateSpace, 2009 [Google Scholar]
- 17.Bird S, Klein E, Loper E: Natural Language Processing With Python. Sebastapol, CA, O’Reilly Media Inc, 2009 [Google Scholar]
- 18.Alajmi A, Saad EM, Darwish RR. Toward an ARABIC stop-words list generation. Int J Comp Appl. 2012; 46:8–13 [Google Scholar]
- 19.Singh P. Natural language processing. : Machine Learning With PySpark. New York, NY, Apress, 2019, pp 191–218 [Google Scholar]
- 20.Stephen TW, Hongfang L, Dingcheng L. Unified medical language system term occurrences in clinical notes: A large-scale corpus analysis. J Am Med Informatics Assoc. 2012; 19:e149–e156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen E: Text Mining and Network Analysis of Digital Libraries in R. Elsevier, 2013 [Google Scholar]
- 22.Brown P, Della Pietra V, de Souza P. Class-based n-gram models of natural language. Comput Linguist. 1992; 18:467–479 [Google Scholar]
- 23.Zimmerman JE, Kramer AA, McNair DS, Malila FM. Acute Physiology and Chronic Health Evaluation (APACHE) IV: Hospital mortality assessment for today’s critically ill patients. Crit Care Med. 2006; 34:1297–1310 [DOI] [PubMed] [Google Scholar]
- 24.Stone M. Cross-validation: A review. Stat A J Theor Appl Stat. 1978; 9:127–139 [Google Scholar]
- 25.Kuzniewicz MW, Vasilevskis EE, Lane R, et al. Variation in ICU risk-adjusted mortality: Impact of methods of assessment and potential confounders. Chest. 2008; 133:1319–1327 [DOI] [PubMed] [Google Scholar]
- 26.Lieberman JD, Pasquale MD, Garcia R, et al. Use of admission Glasgow Coma Score, pupil size, and pupil reactivity to determine outcome for trauma patients. J Trauma. 2003; 55:437–442; discussion 442–443 [DOI] [PubMed] [Google Scholar]
- 27.Shaheen AA, Kaplan GG, Myers RP. Weekend versus weekday admission and mortality from gastrointestinal hemorrhage caused by peptic ulcer disease. Clin Gastroenterol Hepatol. 2009; 7:303–310 [DOI] [PubMed] [Google Scholar]
- 28.Camp SL, Stamou SC, Stiegel RM, et al. Can timing of tracheal extubation predict improved outcomes after cardiac surgery? HSR Proc Intensive Care Cardiovasc Anesth. 2009; 1:39–47 [PMC free article] [PubMed] [Google Scholar]
- 29.McCradden MD, Joshi S, Anderson JA, et al. Patient safety and quality improvement: Ethical principles for a regulatory approach to bias in healthcare machine learning. J Am Med Inform Assoc. 2020; 27:2024–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blinderman CD, Billings JA. Comfort care for patients dying in the hospital. N Engl J Med. 2015; 373:2549–2561 [DOI] [PubMed] [Google Scholar]
- 31.Lilley EJ, Lindvall C, Lillemoe KD, et al. Measuring processes of care in palliative surgery: A novel approach using natural language processing. Ann Surg. 2018; 267:823–825 [DOI] [PubMed] [Google Scholar]
- 32.Weis JM, Levy PC. Copy, paste, and cloned notes in electronic health records: Prevalence, benefits, risks, and best practice recommendations. Chest. 2014; 145:632–638 [DOI] [PubMed] [Google Scholar]
- 33.Gabriel RA, Kuo TT, McAuley J, et al. Identifying and characterizing highly similar notes in big clinical note datasets. J Biomed Inform. 2018; 82:63–69 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


