Abstract
mRNA display is a robust in vitro selection technique allows the selection of protein libraries of trillions of variants. mRNA display relies upon a covalent linkage between a protein and its encoding mRNA molecule; the power of the technique stems from the stability of this link, and the large degree of control over experimental conditions afforded to the researcher. This chapter describes the major advantages that make the mRNA display technique superior to comparable in vivo and in vitro methods.
Keywords: Protein engineering, In vitro selection, In vitro evolution, Directed evolution, Enzyme, Unnatural amino acids
1. Introduction
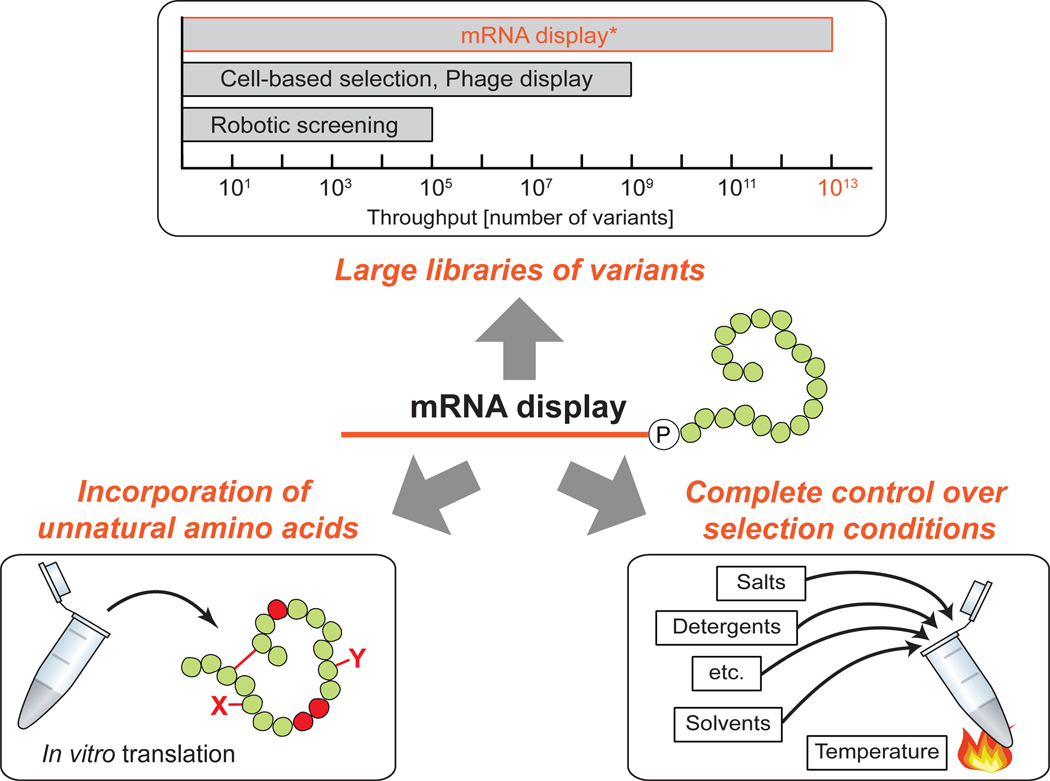
mRNA display is an in vitro selection and evolution technique that enables screening of trillions of protein variants for desired functions in a single experiment. The key feature of this technique is the covalent linkage of an mRNA with its encoding protein which is established during in vitro translation [1, 2]. The stable genotype-phenotype linkage renders the protein directly amplifiable and enables the enrichment of mRNA-displayed protein variants with desired properties. This article will describe the advantages of the mRNA display technology in comparison to other in vivo and in vitro screening techniques (Fig. 1.). Specifically, we will discuss the value of interrogating more protein variants than is possible with most other techniques (1), the benefits of an in vitro method that frees protein expression from cellular constraints (2), allows the incorporation of unnatural amino acids in the protein variants (3), and permits protein screening under an unlimited range of desirable conditions (4). We will conclude by discussing the current limitations of the technique and how they are being overcome, as well as future prospects of mRNA display.
Fig. 1.
Advantages of mRNA display comprise the use of very large libraries of protein variants, the ability to readily incorporate unnatural amino acids and perform selections under a wide range of conditions. * Ribosome display uses libraries of similar size.
2. The numbers game – Advantages of large libraries
Directed evolution is a proven protein engineering method to generate proteins with modified properties by mimicking the natural process of mutagenesis and selection [3, 4]. The chance of identifying a protein variant with a desired function from a library of mutants is proportional to the number of protein variants that can efficiently be tested. Numerous methods have been developed to readily create large numbers of protein variants including techniques such as random mutagenesis by error-prone PCR, DNA shuffling, or the use of random synthetic DNA [5–10]. The generation of genetic diversity is therefore not the limiting aspect of a directed evolution approach, but instead the throughput of the subsequent screening step represents the bottleneck.
The following thought experiment will demonstrate the importance of library size in directed evolution approaches. Let us assume that in a typical binding pocket of a protein or active site of an enzyme there are about a dozen amino acids which line the cavity and form the first shell of contact to the ligand. Many of these residues (and more throughout the protein) will strongly influence the binding affinity or enzymatic activity. A common engineering approach to modify these properties is therefore to use site-saturation mutagenesis of these first shell residues to generate diversity for a directed evolution experiment [9–11]. A simple calculation shown in Table 1 exemplifies the dilemma the researcher quickly faces. The numbers demonstrate that classic cell-based screening methods with a throughput of 102-104 allow exhaustive sampling for only about up to 3 positions, and higher-throughput FACS-based screening or in vivo selection methods for up to 6 positions (Table 2). In reality, this problem can be further exacerbated several fold due to the redundancy of the genetic code. Several engineering teams have attempted to somewhat lessen this numbers problem by synthesizing so called “smart libraries”, which sample a limited number of codons to reduce library size while mostly maintaining the chemical diversity [10–12]. Unfortunately, these approaches only slightly delay the combinatorial “explosion” of possible variants. In any case, for smart libraries or not so smart ones, the chances of finding a protein variant with a potentially rare new or improved property increases proportionally with the number of variants interrogated.
Table 1.
Library size required to sample all possible variants in site-saturation mutagenesis
| Positions randomized | Number of possible variants |
|---|---|
| 1 | 20 |
| 3 | 8.0 × 103 |
| 6 | 6.4 × 107 |
| 10 | 1.0 × 1013 |
Table 2.
Differences in throughput for protein screening and selection techniques
| Technique | Typical library size | References |
|---|---|---|
| Classic cell-based screen | 102–104 | [3] |
| Robotic screen | 105 | [98, 99] |
| FACS-based screen | 107 | [100] |
| In vivo selection | 106–108 | [17–19, 101] |
| Cell-surface display | [16] | |
| Eukaryote | 106–107 | |
| Prokaryote | 108–1010 | |
| Phage display | ~109 | [20] |
| Ribosome display | 1012–1014 | [15] |
| mRNA display | 1012–1014 | [23] |
mRNA display allows the use of protein libraries exceeding 1013 variants, which is several orders of magnitude larger than all in vivo methods and most in vitro techniques (Table 2) [13]. In contrast, cell-surface display and other selection methods that require in vivo expression of the protein library [9, 14–16] are limited by the transformation efficiency, which can be as high as 1010 for Escherichia coli [17] and 107 for Saccharomyces cerevisiae [18] (Table 2). However, more typical in vivo libraries contain only about 106-108 unique sequences [19]. The widely used phage display technology has been reported to screen up to 109-1012 sequences [20–22], but again 109 is more common because the technique is also limited by in vivo transformation into E. coli for propagation [20]. mRNA display, on the other hand, is performed entirely in vitro and therefore the number of sequences is not limited by a transformation step [13, 23, 24].
mRNA display selections with trillions of variant sequences have been successfully performed to isolate proteins with de novo functions, thereby demonstrating that large libraries can yield potentially rare protein variants [8, 25–27]. The generation of de novo functions, e.g. to instill binding or catalytic capability into a protein originally lacking those properties entirely, is still considered a great challenge in protein engineering [28]. The first example of mRNA display producing de novo proteins was the isolation of artificial ATP-binding proteins from a completely random polypeptide library [8]. In addition to yielding specific and tight binders, this work also determined for the first time the distribution of functional proteins in protein sequence space by identifying one ATP binder for each 1011 random protein sequences. Subsequently, mRNA display also yielded artificial ATP-binding proteins from a library based on an unrelated protein scaffold [29]. Furthermore, de novo enzymes that catalyze a reaction unobserved in nature were produced by mRNA display from a library based on a non-catalytic protein scaffold [13, 26]. During the selection process for these de novo enzymes, surprisingly, the original scaffold was abandoned and instead a new fold was formed that lacks classic secondary structure motifs and, like the catalyzed reaction, has not been found in nature [30]. The large library size in mRNA display also enables the screening of an entire natural proteome, for example to identify protein-protein [31–33] or protein-drug interactions [34, 35], and to isolate kinase [36] or protease substrates [37, 38]. The length of polypeptides that have been successfully screened ranges from small peptides of <10 residues [1, 39] to proteins of ~650 residues [33, 40]. The display efficiency and thereby the size of library produced per volume of translation reaction slightly increases with decreasing size of the displayed peptide [31]. To date, mRNA display has produced a wide range of engineered polypeptides including peptide inhibitors, antibody mimics, and antibody fragments (Fab) [40–44].
Methods for screening large libraries for function are an integral part of the modern protein engineer’s toolbox and in vitro techniques such as mRNA display allow the highest variety of sequences to be tested. Besides large library size, further benefits of mRNA display include a high level of control over protein translation and the conditions under which the selections can be performed. These advantages will be described in detail in the following sections.
3. What’s in a cell? - Benefits of in vitro protein expression
All steps of the mRNA display procedure are performed entirely in vitro including protein expression. This fact alleviates many of the potential issues associated with in vivo protein production. The heterologous expression of proteins in E. coli or other cell-based systems can lead to the accumulation of protein aggregates that are detrimental to cell viability [45, 46]. Foreign, high copy number proteins can disrupt normal metabolism or lead to toxic products [47, 48]. Alternatively, the heterologous proteins themselves can in some cases be toxic to the cell [46, 49]. Cell-free protein expression removes both the need to maintain the viability of the cell and the danger of other cell components confounding a selection by acting as competing enzymes, potential inhibitors, or alternative substrates.
Alternative display methods, cell-surface and phage display, include an in vivo translation step and are therefore additionally limited by the requirement for transport of a folded protein across a cell membrane [16] or the release of a phage from a host cell [14, 21]. This effectively constitutes an additional undesired selection bias for proteins that can be successfully transported for display. The mRNA display technique has no such requirement.
Cell-based methods will likely degrade poorly folded protein library variants, but the experimenter has no real means to influence this process. In contrast, in vitro selections provide the option to fine-tune the removal of poorly folded proteins by selective protease treatment [50].
Cell-free protein translation in mRNA display is typically performed using eukaryotic cell lysates from rabbit or wheat germ [13] allowing a wider range of post-translational modification than would be possible with bacterial in vivo expression. Protein expression conditions can be controlled even more tightly with the use of the fully chemically-defined PURE translation system (Protein synthesis Using Recombinant Elements) [51, 52]. The PURE translation system is reconstituted from the purified components necessary for E. coli translation such as tRNAs, aminoacyl-tRNA synthetases, amino acids, ribosomes and release factors [53].
4. Expanding the protein alphabet with non-standard amino acids
The use of the PURE cell-free translation system also enables the facile incorporation of diverse non-standard amino acids (NSAA) into mRNA-displayed proteins, thereby vastly expanding the chemical properties of the selected proteins. NSAAs are either rare, naturally occurring amino acids different from the standard 20 residues, or completely synthetic amino acids that introduce novel chemistries to engineered proteins [54, 55]. NSAAs have been used to enhance protein stability, catalysis or detection [54–59]; enable subsequent protein modification via bio-orthogonal click chemistry [54, 59]; and generate biologically active peptides for pharmaceutical use. To date, more than 150 different NSAAs have been genetically encoded in proteins [60]. While NSAAs can be incorporated by different approaches both in vitro and in some cases in vivo [56], the PURE cell-free system is the most versatile method as it can use multiple orthogonal incorporation techniques in parallel by altering the composition of a user-defined translation system [54, 61]. mRNA display in combination with PURE can therefore readily realize the full potential of the ever-expanding repertoire of amino acid building blocks [54–56, 62].
Protein libraries containing NSAAs have been selected by mRNA display for a wide range of activities. The first such proof-of-principle mRNA display selection incorporated biocytin, a biotin derivative of lysine. The biotin-labeled peptides were then enriched by selection against a streptavidin agarose matrix [39]. In separate work, four-base codons were used to incorporate biocytin and other NSAAs to select for novel streptavidin-binding peptides [63]. Other groups have screened for inhibitors against Breast Cancer Associated protein 1 (BRCA1) [64], or improved the protease stability of peptides through unnatural methylation [65].
The PURE system has also been used in conjunction with mRNA display to introduce synthetic amino acids that allow further modification by click chemistry. Methionine was replaced with an alkyne-modified glycine, which enabled the post-translational modification with azide-modified high mannose glycans. These mRNA-displayed glycopeptides were then selected to tightly bind to a HIV broadly neutralizing antibody [66]. Although this is the only published combination of mRNA display and click chemistry to date, this principle could be used to immobilize an mRNA-displayed peptide to a surface for purification, trap a reaction substrate or product, or attach to a binding partner in solution.
NSAA incorporation has been harnessed to select macrocyclic peptide binders from mRNA-displayed libraries of up to 1013 variants [63, 67–78]. Circularized peptides are favored in drug screening due to their increased proteolytic stability, improved membrane permeability, and reduced conformational flexibility that can lead to tighter target binding [35, 79–81]. An overview of peptide cyclization strategies can be found in several reviews [35, 79–82]. One of these mRNA-displayed macrocyclic peptide libraries contained twelve NSAAs and only eight standard amino acids, yielding binders to thrombin with a low nanomolar dissociation constant [74]. NSAAs can also be used to perform the peptide cyclization itself. This has been achieved by a number of different means, including the formation of a thioether bond between a chloroacetyl-analog of tyrosine and a cysteine side chain [75, 83]; or by using 4-selenalysine that was subsequently converted to dehydroalanine and cyclized with a cysteine residue to form a linkage similar to natural lantipeptides [73]. In a further study, an altered mRNA display protocol, RaPID (Random nonstandard Peptide Integrated Discovery) [71, 83], produced macrocyclic peptides that functioned as isoform-selective inhibitors of Protein Kinase B [75]. These examples demonstrate that the in vitro translation feature of mRNA display enables the experimenter to harness the continuing progress in non-standard amino acid incorporation and promises to greatly expand the potential for protein engineering in both fundamental research and for practical applications.
5. Complete control over selection conditions due to robust in vitro format
Cell-based selections are by necessity limited to conditions that are compatible with cell growth and replication, while in vitro selections by mRNA display enable the researcher to control selection conditions freely. Numerous parameters such as pH, temperature, and ionic strength can be precisely controlled. The format also allows easy inclusion of additional components such as reaction substrates, binding targets, solvents or other chemicals. Unlike within the crowded cytosol of a cell where millimolar concentrations of potentially competing proteins, solutes and other factors could confound selection outcomes, all components of an in vitro selection system are entirely controlled by the experimenter.
In order to identify “winning” sequences by any protein screening or selection technique, each protein variant must be linked to its genetic information. For example, each gene variant is contained inside the cell or viral capsid for cell-based screening and phage display, respectively. Compared to the size of the protein variant molecule, a cell or a phage is a very large entity and will therefore alter the properties of the displayed protein. In mRNA display, this essential gene-protein link is dramatically miniaturized to a simple covalent bond between the protein and its encoding mRNA via the small molecule puromycin. Therefore, the bias that an attached cell or phage might exert on a protein selection is reduced to the bare necessity – the mRNA itself. In addition, this stable covalent linkage between the protein and mRNA further facilitates the use of stringent selection conditions. The ribosome display technology, which otherwise has many favorable features in common with mRNA display such as large library size and the in vitro format, uses the whole ribosome to keep mRNA and protein together in a non-covalent and therefore less stable manner [15]. The small size of the linkage used in mRNA display – relative to cell, phage, or ribosome display constructs – likely increases the chances of a displayed peptide maintaining the same physicochemical properties as the respective unfused peptide. For example, a library of mRNA-displayed fusions was selected for the ability to infiltrate cancer cells. It was subsequently demonstrated that the identified peptides alone had the same cell-penetrating properties as the mRNA-displayed fusions [84, 85]. The functionality of isolated peptides is therefore not significantly hindered by the presence of the fused mRNA, and peptide variants selected by mRNA display are accurately represented.
The stability of the covalent linkage between mRNA and protein also allows a wide variety of conditions to be used during a selection. For example, in order to evolve artificial ATP binders towards greater structural stability, a selection was carried out in the presence of increasing concentrations of the chemical denaturant guanidine hydrochloride [86, 87]. Furthermore, an artificial RNA ligase enzyme, originally selected at room temperature, was evolved for increased thermostability by performing the selection at elevated temperature. The resulting thermostable ligase variant had a melting temperature of 72°C, which was 35 degrees more stable than the most closely related variant selected at ambient temperature [88].
Cell-based screening and selection systems can yield false positive results due to the unintentional up-regulation of multicopy suppressors [89] with promiscuous activity that is similar to the desired activity. Nearly 40% of all E. coli proteins are estimated to be promiscuous [90]. Therefore, selections for enzymatic function through complementation of auxotrophic strains are complicated by the risk of chromosomal mutations leading to the overexpression of existing weakly promiscuous enzymes [91, 92] or by the library variant itself acting as a transcription factor to the same effect. The tight control over selection conditions afforded by the mRNA display method completely avoids such issues.
A typical round of an mRNA display selection takes approximately 2–3 days depending on selection conditions and purification steps [13, 93]. Recently however, this time has been drastically reduced to about two hours per round. An optimized version of the mRNA display protocol successfully accomplished six rounds of selection in just fourteen hours thanks to a one-pot transcription, translation, and puromycin coupling reaction [94]. This mRNA display version was termed TRAP display: transcription−translation coupled with association of puromycin linker. Furthermore, coupling mRNA display with continuous magnetic flow separation and subsequent high throughput sequencing has enabled the selection of IgG binders with nanomolar affinities in a single round [95]. These examples demonstrate that the in vitro format and the small, covalent gene-protein linkage in mRNA display provide great freedom to choose selection conditions and selection stringency.
6. Addressing potential concerns with mRNA display
mRNA display is a robust method for the selection of functional proteins from large libraries. While this technique compares favorably with other selection methods in many aspects, we will here also address potential concerns associated with the technique and ways to overcome them.
RNA stability:
A common question about mRNA display involves the perceived sensitivity of RNA to degradation. This question likely originates from people’s experiences isolating long RNA from lysed cells. However, due to the in vitro nature of the entire mRNA display procedure, RNA nuclease degradation is simply not an issue. Simple precautions such as using nuclease-free chemicals, wearing gloves during experiments and protecting against dust contamination are usually sufficient to avoid degradation [13]. Only when mRNA-displayed proteins are to be incubated with biological specimens does mRNA degradation become a concern due to the inherent presence of nucleases. Nevertheless, recent work on selecting cell-penetrating peptides showed that up to 60% of the mRNA-displayed library could be recovered intact after the incubation with cells [85]. Moreover, through a modification of the mRNA display procedure, cDNA-displayed protein libraries can be produced which avoid mRNA stability concerns altogether [96, 97].
Monomeric vs. dimeric proteins:
The design of the mRNA display approach results in a single polypeptide chain covalently linked to its encoding mRNA. Therefore, mRNA display has been limited in principle to the selection of monomeric proteins. But, recently, a new strategy was developed to address this limitation. The polypeptides of the heavy and light chains of an antibody Fab fragment were separately mRNA-displayed and then mixed to form heterodimers. After affinity selection, the heterodimers were individually encapsulated in water-in-oil emulsions and the winning genes were recovered by overlap-extension PCR [44].
7. Conclusions
In vitro selection of polypeptides by mRNA display has proven to be an excellent tool for peptide design, protein engineering, and the investigation of protein interactions. The ability to search very large libraries of candidate sequences dramatically outperforms all in vivo methods and most other in vitro selection techniques. The convenient option of incorporating unnatural amino acids in the protein libraries expands the possibilities of obtaining new structures and activities that have no natural precedent and could not be selected by other means. This avenue is being heavily pursued to generate macrocyclic peptide therapeutics with unnatural modifications, opening the way to more potent protein inhibitors. The covalent genotype-phenotype linkage in mRNA display not only allows easy recovery and identification of the selected candidates but also, due to its robustness, enables the use of a wide variety of selection conditions, many of them incompatible with in vivo methods. This can be especially valuable in enzyme engineering in order to evolve soluble, folded, thermostable proteins that are also compatible with organic solvents, extreme pH, or high salt concentrations.
In summary, if you want to search the largest possible libraries of protein variants under the widest range of selection conditions – the mRNA display technology should be your method of choice.
Acknowledgements
The authors thank Fredarla Miller for comments on the manuscript. We gratefully acknowledge financial support from the US National Institutes of Health GM108703, AI113406, the US National Aeronautics and Space Administration NNX14AK29G, the Simons Foundation, and the Biocatalysis Initiative of the BioTechnology Institute at the University of Minnesota.
References
- 1.Roberts RW and Szostak JW, RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc Natl Acad Sci U S A, 1997. 94(23): p. 12297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nemoto N, et al. , In vitro virus: bonding of mRNA bearing puromycin at the 3’-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. FEBS Lett, 1997. 414(2): p. 405–8. [DOI] [PubMed] [Google Scholar]
- 3.Packer MS and Liu DR, Methods for the directed evolution of proteins. Nat Rev Genet, 2015. 16(7): p. 379–94. [DOI] [PubMed] [Google Scholar]
- 4.Lane MD and Seelig B, Advances in the directed evolution of proteins. Curr Opin Chem Biol, 2014. 22: p. 129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cadwell RC and Joyce GF, Randomization of genes by PCR mutagenesis. Genome Research, 1992. 2(1): p. 28–33. [DOI] [PubMed] [Google Scholar]
- 6.Stemmer WP, Rapid evolution of a protein in vitro by DNA shuffling. Nature, 1994. 370(6488): p. 389–91. [DOI] [PubMed] [Google Scholar]
- 7.Baker M, Protein engineering: navigating between chance and reason. Nat Methods, 2011. 8(8): p. 623–6. [DOI] [PubMed] [Google Scholar]
- 8.Keefe AD and Szostak JW, Functional proteins from a random-sequence library. Nature, 2001. 410(6829): p. 715–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bornscheuer U. and Kazlauskas RJ, Survey of protein engineering strategies. Curr Protoc Protein Sci, 2011. Chapter 26: p. Unit26 7. [DOI] [PubMed] [Google Scholar]
- 10.Tee KL and Wong TS, Polishing the craft of genetic diversity creation in directed evolution. Biotechnol Adv, 2013. 31(8): p. 1707–21. [DOI] [PubMed] [Google Scholar]
- 11.Currin A, et al. , Synthetic biology for the directed evolution of protein biocatalysts: navigating sequence space intelligently. Chem Soc Rev, 2015. 44(5): p. 1172–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reetz MT, Kahakeaw D, and Lohmer R, Addressing the numbers problem in directed evolution. Chembiochem, 2008. 9(11): p. 1797–804. [DOI] [PubMed] [Google Scholar]
- 13.Seelig B, mRNA display for the selection and evolution of enzymes from in vitro-translated protein libraries. Nat Protoc, 2011. 6(4): p. 540–52. [DOI] [PubMed] [Google Scholar]
- 14.Sidhu SS, et al. , Phage display for selection of novel binding peptides. Methods Enzymol, 2000. 328: p. 333–63. [DOI] [PubMed] [Google Scholar]
- 15.Pluckthun A, Ribosome display: a perspective. Methods Mol Biol, 2012. 805: p. 3–28. [DOI] [PubMed] [Google Scholar]
- 16.Smith MR, Khera E, and Wen F, Engineering novel and improved biocatalysts by cell surface display. Ind. Eng. Chem. Res, 2015. 54(16): p. 4021–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dower WJ, Miller JF, and Ragsdale CW, High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res, 1988. 16(13): p. 6127–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawai S, Hashimoto W, and Murata K, Transformation of Saccharomyces cerevisiae and other fungi: methods and possible underlying mechanism. Bioeng Bugs, 2010. 1(6): p. 395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golynskiy MV, et al. , In vitro evolution of enzymes. Methods Mol Biol, 2013. 978: p. 73–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derda R, et al. , Diversity of phage-displayed libraries of peptides during panning and amplification. Molecules, 2011. 16(2): p. 1776–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebrahimizadeh W. and Rajabibazl M, Bacteriophage vehicles for phage display: biology, mechanism, and application. Curr Microbiol, 2014. 69(2): p. 109–20. [DOI] [PubMed] [Google Scholar]
- 22.Staniszewska M, et al. , A phage display-based approach to investigate abnormal neovessels of the retina. Invest Ophthalmol Vis Sci, 2012. 53(8): p. 4371–9. [DOI] [PubMed] [Google Scholar]
- 23.Cotten SW, et al. , mRNA display-based selections using synthetic peptide and natural protein libraries. Methods Mol Biol, 2012. 805: p. 287–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts RW, Totally in vitro protein selection using mRNA-protein fusions and ribosome display. Curr Opin Chem Biol, 1999. 3(3): p. 268–73. [DOI] [PubMed] [Google Scholar]
- 25.Cho G, et al. , Constructing high complexity synthetic libraries of long ORFs using in vitro selection. J Mol Biol, 2000. 297(2): p. 309–319. [DOI] [PubMed] [Google Scholar]
- 26.Seelig B. and Szostak JW, Selection and evolution of enzymes from a partially randomized non-catalytic scaffold. Nature, 2007. 448(7155): p. 828–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cotten SW, et al. , Selection of proteins with desired properties from natural proteome libraries using mRNA display. Nat Protoc, 2011. 6(8): p. 1163–82. [DOI] [PubMed] [Google Scholar]
- 28.Golynskiy MV and Seelig B, De novo enzymes: from computational design to mRNA display. Trends Biotechnol, 2010. 28(7): p. 340–5. [DOI] [PubMed] [Google Scholar]
- 29.Cho GS and Szostak JW, Directed evolution of ATP binding proteins from a zinc finger domain by using mRNA display. Chem Biol, 2006. 13(2): p. 139–47. [DOI] [PubMed] [Google Scholar]
- 30.Chao FA, et al. , Structure and dynamics of a primordial catalytic fold generated by in vitro evolution. Nat Chem Biol, 2013. 9(2): p. 81–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H. and Liu R, Advantages of mRNA display selections over other selection techniques for investigation of protein-protein interactions. Expert Rev Proteomics, 2011. 8(3): p. 335–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen X, et al. , Identification of novel SHPS-1-associated proteins and their roles in regulation of insulin-like growth factor-dependent responses in vascular smooth muscle cells. Mol Cell Proteomics, 2009. 8(7): p. 1539–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen X, et al. , Scanning the human proteome for calmodulin-binding proteins. Proc Natl Acad Sci U S A, 2005. 102(17): p. 5969–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S. and Roberts RW, A novel strategy for in vitro selection of peptide-drug conjugates. Chem Biol, 2003. 10(3): p. 233–9. [DOI] [PubMed] [Google Scholar]
- 35.Josephson K, Ricardo A, and Szostak JW, mRNA display: from basic principles to macrocycle drug discovery. Drug Discovery Today, 2014. 19(4): p. 388–399. [DOI] [PubMed] [Google Scholar]
- 36.Olson CA, et al. , mRNA display selection of a high-affinity, modification-specific phospho-IkappaBalpha-binding fibronectin. ACS Chem Biol, 2008. 3(8): p. 480–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valencia CA, et al. , mRNA-display-based selections for proteins with desired functions: a protease-substrate case study. Biotechnol Prog, 2008. 24(3): p. 561–9. [DOI] [PubMed] [Google Scholar]
- 38.Baggio R, et al. , Identification of epitope-like consensus motifs using mRNA display. J Mol Recognit, 2002. 15(3): p. 126–34. [DOI] [PubMed] [Google Scholar]
- 39.Li S, Millward S, and Roberts R, In vitro selection of mRNA display libraries containing an unnatural amino acid. J Am Chem Soc, 2002. 124(34): p. 9972–3. [DOI] [PubMed] [Google Scholar]
- 40.Sumida T, Doi N, and Yanagawa H, Bicistronic DNA display for in vitro selection of Fab fragments. Nucleic Acids Res, 2009. 37(22): p. e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson DS, Keefe AD, and Szostak JW, The use of mRNA display to select high-affinity protein-binding peptides. Proc Natl Acad Sci U S A, 2001. 98(7): p. 3750–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ja WW, et al. , Extension of Drosophila melanogaster life span with a GPCR peptide inhibitor. Nat Chem Biol, 2007. 3(7): p. 415–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu L, et al. , Directed evolution of high-affinity antibody mimics using mRNA display. Chem Biol, 2002. 9(8): p. 933–42. [DOI] [PubMed] [Google Scholar]
- 44.Sumida T, Yanagawa H, and Doi N, In vitro selection of fab fragments by mRNA display and gene-linking emulsion PCR. J Nucleic Acids, 2012. 2012: p. 371379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hannig G. and Makrides SC, Strategies for optimizing heterologous protein expression in Escherichia coli. Trends Biotechnol, 1998. 16(2): p. 54–60. [DOI] [PubMed] [Google Scholar]
- 46.de Marco A, et al. , Native folding of aggregation-prone recombinant proteins in Escherichia coli by osmolytes, plasmid- or benzyl alcohol-overexpressed molecular chaperones. Cell Stress Chaperones, 2005. 10(4): p. 329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng CY, et al. , Production of 2,3-butanediol in Saccharomyces cerevisiae by in silico aided metabolic engineering. Microb Cell Fact, 2012. 11: p. 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sikkema J, de Bont JA, and Poolman B, Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev, 1995. 59(2): p. 201–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schlegel S, et al. , Optimizing heterologous protein production in the periplasm of E. coli by regulating gene expression levels. Microb Cell Fact, 2013. 12: p. 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Golynskiy MV, Haugner JC 3rd, and Seelig B, Highly diverse protein library based on the ubiquitous (β/α)8 enzyme fold yields well-structured proteins through in vitro folding selection. Chembiochem, 2013. 14(13): p. 1553–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimizu Y, Kanamori T, and Ueda T, Protein synthesis by pure translation systems. Methods, 2005. 36(3): p. 299–304. [DOI] [PubMed] [Google Scholar]
- 52.Shimizu Y, et al. , Cell-free translation reconstituted with purified components. Nat Biotechnol, 2001. 19(8): p. 751–5. [DOI] [PubMed] [Google Scholar]
- 53.Li J, et al. , Improved cell-free RNA and protein synthesis system. PLoS One, 2014. 9(9): p. e106232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rogers JM and Suga H, Discovering functional, non-proteinogenic amino acid containing, peptides using genetic code reprogramming. Org Biomol Chem, 2015. [DOI] [PubMed] [Google Scholar]
- 55.Zhang WH, Otting G, and Jackson CJ, Protein engineering with unnatural amino acids. Curr Opin Struct Biol, 2013. 23(4): p. 581–7. [DOI] [PubMed] [Google Scholar]
- 56.Young TS and Schultz PG, Beyond the canonical 20 amino acids: expanding the genetic lexicon. J Biol Chem, 2010. 285(15): p. 11039–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hammerling MJ, et al. , Bacteriophages use an expanded genetic code on evolutionary paths to higher fitness. Nat Chem Biol, 2014. 10(3): p. 178–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kolev JN, et al. , Enhancing the efficiency and regioselectivity of P450 oxidation catalysts by unnatural amino acid mutagenesis. Chembiochem, 2014. 15(7): p. 1001–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hinz FI, Dieterich DC, and Schuman EM, Teaching old NCATs new tricks: using noncanonical amino acid tagging to study neuronal plasticity. Curr Opin Chem Biol, 2013. 17(5): p. 738–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dumas A, et al. , Designing logical codon reassignment – Expanding the chemistry in biology. Chem. Sci, 2015. 6(1): p. 50–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Forster AC, Weissbach H, and Blacklow SC, A simplified reconstitution of mRNA-directed peptide synthesis: activity of the epsilon enhancer and an unnatural amino acid. Anal Biochem, 2001. 297(1): p. 60–70. [DOI] [PubMed] [Google Scholar]
- 62.Liu CC and Schultz PG, Adding new chemistries to the genetic code. Annu Rev Biochem, 2010. 79: p. 413–44. [DOI] [PubMed] [Google Scholar]
- 63.Muranaka N, Hohsaka T, and Sisido M, Four-base codon mediated mRNA display to construct peptide libraries that contain multiple nonnatural amino acids. Nucleic Acids Res, 2006. 34(1): p. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.White ER, et al. , Peptide library approach to uncover phosphomimetic inhibitors of the BRCA1 C-terminal domain. ACS Chem Biol, 2015. 10(5): p. 1198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frankel A, Millward SW, and Roberts RW, Encodamers: unnatural peptide oligomers encoded in RNA. Chem Biol, 2003. 10(11): p. 1043–50. [DOI] [PubMed] [Google Scholar]
- 66.Horiya S, et al. , Directed evolution of multivalent glycopeptides tightly recognized by HIV antibody 2G12. J Am Chem Soc, 2014. 136(14): p. 5407–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Millward SW, Takahashi TT, and Roberts RW, A general route for post-translational cyclization of mRNA display libraries. J Am Chem Soc, 2005. 127(41): p. 14142–3. [DOI] [PubMed] [Google Scholar]
- 68.Seebeck FP and Szostak JW, Ribosomal synthesis of dehydroalanine-containing peptides. J Am Chem Soc, 2006. 128(22): p. 7150–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Millward SW, et al. , Design of cyclic peptides that bind protein surfaces with antibody-like affinity. ACS Chem Biol, 2007. 2(9): p. 625–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kawakami T, Murakami H, and Suga H, Messenger RNA-programmed incorporation of multiple N-methyl-amino acids into linear and cyclic peptides. Chem Biol, 2008. 15(1): p. 32–42. [DOI] [PubMed] [Google Scholar]
- 71.Yamagishi Y, et al. , Natural product-like macrocyclic N-methyl-peptide inhibitors against a ubiquitin ligase uncovered from a ribosome-expressed de novo library. Chem Biol, 2011. 18(12): p. 1562–70. [DOI] [PubMed] [Google Scholar]
- 72.Ma Z. and Hartman MC, In vitro selection of unnatural cyclic peptide libraries via mRNA display. Methods Mol Biol, 2012. 805: p. 367–90. [DOI] [PubMed] [Google Scholar]
- 73.Hofmann FT, Szostak JW, and Seebeck FP, In vitro selection of functional lantipeptides. J Am Chem Soc, 2012. 134(19): p. 8038–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schlippe YV, et al. , In vitro selection of highly modified cyclic peptides that act as tight binding inhibitors. J Am Chem Soc, 2012. 134(25): p. 10469–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hayashi Y, Morimoto J, and Suga H, In vitro selection of anti-Akt2 thioether-macrocyclic peptides leading to isoform-selective inhibitors. ACS Chem Biol, 2012. 7(3): p. 607–13. [DOI] [PubMed] [Google Scholar]
- 76.Menegatti S, et al. , mRNA display selection and solid-phase synthesis of Fc-binding cyclic peptide affinity ligands. Biotechnol Bioeng, 2013. 110(3): p. 857–70. [DOI] [PubMed] [Google Scholar]
- 77.Tanaka Y, et al. , Structural basis for the drug extrusion mechanism by a MATE multidrug transporter. Nature, 2013. 496(7444): p. 247–51. [DOI] [PubMed] [Google Scholar]
- 78.Kawakami T, Ishizawa T, and Murakami H, Extensive reprogramming of the genetic code for genetically encoded synthesis of highly N-alkylated polycyclic peptidomimetics. J Am Chem Soc, 2013. 135(33): p. 12297–304. [DOI] [PubMed] [Google Scholar]
- 79.Bashiruddin NK and Suga H, Construction and screening of vast libraries of natural product-like macrocyclic peptides using in vitro display technologies. Curr Opin Chem Biol, 2015. 24: p. 131–8. [DOI] [PubMed] [Google Scholar]
- 80.Bhat A, Roberts LR, and Dwyer JJ, Lead discovery and optimization strategies for peptide macrocycles. Eur J Med Chem, 2015. 94: p. 471–9. [DOI] [PubMed] [Google Scholar]
- 81.Passioura T, et al. , Selection-based discovery of druglike macrocyclic peptides. Annu Rev Biochem, 2014. 83: p. 727–52. [DOI] [PubMed] [Google Scholar]
- 82.Ito K, Passioura T, and Suga H, Technologies for the synthesis of mRNA-encoding libraries and discovery of bioactive natural product-inspired non-traditional macrocyclic peptides. Molecules, 2013. 18(3): p. 3502–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morimoto J, Hayashi Y, and Suga H, Discovery of macrocyclic peptides armed with a mechanism-based warhead: isoform-selective inhibition of human deacetylase SIRT2. Angew Chem Int Ed Engl, 2012. 51(14): p. 3423–7. [DOI] [PubMed] [Google Scholar]
- 84.Higa M, et al. , Identification of a novel cell-penetrating peptide targeting human glioblastoma cell lines as a cancer-homing transporter. Biochem Biophys Res Commun, 2015. 457(2): p. 206–12. [DOI] [PubMed] [Google Scholar]
- 85.Lee JH, et al. , Screening of cell-penetrating peptides using mRNA display. Biotechnol J, 2012. 7(3): p. 387–96. [DOI] [PubMed] [Google Scholar]
- 86.Chaput JC and Szostak JW, Evolutionary optimization of a nonbiological ATP binding protein for improved folding stability. Chemistry & Biology, 2004. 11(6): p. 865–874. [DOI] [PubMed] [Google Scholar]
- 87.Smith MD, et al. , Structural insights into the evolution of a non-biological protein: importance of surface residues in protein fold optimization. PLoS One, 2007. 2(5): p. e467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morelli A, Haugner J, and Seelig B, Thermostable artificial enzyme isolated by in vitro selection. PLoS One, 2014. 9(11): p. e112028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Patrick WM, et al. , Multicopy suppression underpins metabolic evolvability. Mol Biol Evol, 2007. 24(12): p. 2716–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nam H, et al. , Network context and selection in the evolution to enzyme specificity. Science, 2012. 337(6098): p. 1101–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yip SH and Matsumura I, Substrate ambiguous enzymes within the Escherichia coli proteome offer different evolutionary solutions to the same problem. Mol Biol Evol, 2013. 30(9): p. 2001–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Miller BG and Raines RT, Reconstitution of a defunct glycolytic pathway via recruitment of ambiguous sugar kinases. Biochemistry, 2005. 44(32): p. 10776–83. [DOI] [PubMed] [Google Scholar]
- 93.Barendt PA, et al. , Streamlined protocol for mRNA display. ACS Comb Sci, 2013. 15(2): p. 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ishizawa T, et al. , TRAP display: a high-speed selection method for the generation of functional polypeptides. J Am Chem Soc, 2013. 135(14): p. 5433–40. [DOI] [PubMed] [Google Scholar]
- 95.Olson CA, et al. , Single-round, multiplexed antibody mimetic design through mRNA display. Angew Chem Int Ed Engl, 2012. 51(50): p. 12449–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kurz M, et al. , cDNA-protein fusions: covalent protein-gene conjugates for the in vitro selection of peptides and proteins. Chembiochem, 2001. 2(9): p. 666–72. [DOI] [PubMed] [Google Scholar]
- 97.Yamaguchi J, et al. , cDNA display: a novel screening method for functional disulfide-rich peptides by solid-phase synthesis and stabilization of mRNA-protein fusions. Nucleic Acids Res, 2009. 37(16): p. e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Agresti JJ, et al. , Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc Natl Acad Sci U S A, 2010. 107(9): p. 4004–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wojcik M, et al. , High-throughput screening in protein engineering: Recent advances and future perspectives. Int J Mol Sci, 2015. 16(10): p. 24918–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aharoni A, et al. , High-throughput screening of enzyme libraries: thiolactonases evolved by fluorescence-activated sorting of single cells in emulsion compartments. Chem Biol, 2005. 12(12): p. 1281–9. [DOI] [PubMed] [Google Scholar]
- 101.Cobb RE, Sun N, and Zhao H, Directed evolution as a powerful synthetic biology tool. Methods, 2013. 60(1): p. 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]