Abstract
Osteosarcoma (OS) is the most common type of primary bone tumor in children and adults. Dangshen (Codonopsis pilosula) is a traditional Chinese medicine commonly used in the treatment of OS worldwide. However, the molecular mechanisms of Dangshen in OS remain unclear. Hence, in this study, we aimed to systematically explore the underlying mechanisms of Dangshen in the treatment of OS. Our study adopted a network pharmacology approach, focusing on the identification of active ingredients, drug target prediction, gene collection, gene ontology (GO) enrichment, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment, and other network tools. The network analysis identified 15 active compounds in Dangshen that were linked to 48 possible therapeutic targets related to OS. The results of the gene enrichment analysis show that Dangshen produces a therapeutic effect in OS likely by regulating multiple pathways associated with DNA damage, cell proliferation, apoptosis, invasion, and migration. Based on the network pharmacology approach, we successfully predicted the active compounds and their respective targets. In addition, we illustrated the molecular mechanisms that mediate the therapeutic effect of Dangshen in OS. These findings may aid in the development of novel targeted therapies for OS in the future.
1. Introduction
Osteosarcoma (OS) is the most common type of cancer of the bones. It is a malignant tumor that primarily affects the long bones (e.g., legs), but it can also start in other bones. OS is rarely diagnosed in patients under five years of age, and the bimodal age-incidence curve peaks during the second decade of life (10–20 years old) and late adulthood (>40 years old) [1, 2]. As of 2019, approximately 560 children and adolescents are affected each year in the United States [1, 3], with a global incidence of 3.4 cases per one million people. Most OS patients present with metastatic disease, which contributes to its high morbidity and mortality rates worldwide. As of 2020, the standard treatment for OS is systemic chemotherapy [4], as the tumor is often resistant to radiation therapy. Surgical resection may be an option for patients diagnosed with locally noninvasive disease [5]. Most patients undergo multifaceted treatments that include preoperative chemotherapy, postoperative chemotherapy, surgical resection, and radiation therapy in rare cases [6]. Detectable metastases are present in only 20% of patients, and most of the remaining 80% of patients have undetectable micrometastases [7]. This makes it challenging to monitor disease progression and treatment response, which is why many physicians rely on long-term systemic chemotherapy [8]. However, the systemic chemotherapeutics needed to control the disease have serious adverse effects that further hinder the effective treatment of patients in the clinic [9].
In recent years, some hospitals have assessed the efficacy of traditional Chinese medicines as long-term treatments for OS. In some cases, orally administered Shenqi could decrease the growth, metastasis, and the number of chemotherapy-related side effects significantly in patients, especially those who received systemic chemotherapy [10, 11]. The major component of the Shenqi oral preparation is Dangshen, also known as (Codonopsis pilosula). Dangshen belongs to the family of Campanulaceae, a precious plant that grows at altitudes of 2000 meters in southern China [12]. The dry roots of this plant have been used for thousands of years in traditional Chinese medicine to treat qi and blood deficiencies, the loss of appetite, respiratory symptoms (e.g., cough, asthma, and shortness of breath), and cardiovascular problems (e.g., palpitations) [13]. Dangshen displays a variety of pharmacological effects on the circulatory system, immune system, digestive system, endocrine system, and reproductive system [14]. Dangshen has been shown to inhibit cancer growth in S180 tumor-bearing mice, while enhancing the immune response, increasing spleen weight, promoting lymphocyte proliferation, and increasing natural killer (NK) cell activity [15, 16].
DNA damaging therapies are widely used as trigger molecules to study the signaling pathways of OS [17]. There has been a remarkable undertaking of investigations into the different signaling pathways involved in the pathogenesis of OS. Many signaling pathways, such as Wnt, PI3K/AKT, and JAK/STAT, reflect their specific roles in OS [18]. However, conventional research methods have been unable to fully elucidate the mechanisms of action. Nevertheless, the integration of bioinformatics and network pharmacology provides a practical approach to explore and verify the mechanisms of action [19]. Network pharmacology can systematically reveal the active components in drug molecules. In addition, network pharmacology can be used to predict the relationship between drug components and gene targets [20]. Therefore, in this study, we aimed to use network pharmacology to uncover the mechanisms by which Dangshen produces therapeutic effects in patients with OS, along with the associated signaling pathways.
2. Materials and Methods
2.1. Chemical Compounds in Dangshen
A flowchart of the study design is shown in Figure 1. The components of Dangshen were searched in the Traditional Chinese Medicine Systems Pharmacology (TCMSP) (http://tcmspw.com/tcmsp.php) database [21] and Traditional Chinese Medicines Integrated Database (TCMID) (http://www.megabionet.org/tcmid/) [22]. TCMSP provides comprehensive information about components in Chinese herbs, while TCMID provides information on all aspects of traditional Chinese medicines, including herbs and herbal ingredients. Oral bioavailability (OB), which is the percentage of an orally administered drug that reaches the systemic circulation, reflects the degree of absorption and utilization of drugs in the body [23]. The drug-likeness (DL) value reflects the structural similarity between the compound and drug molecule, so the DL compounds are more likely to display suitable pharmacodynamic and pharmacokinetic properties [24]. Therefore, we selected the candidate compounds based on OB and DL properties. As suggested by the TCMSP database, OB ≥ 30% and DL ≥ 0.18 were used as the screening criteria, and the compounds whose OB ≥ 30% and DL ≥ 0.18 were selected for subsequent experiments [25]. We searched the oral bioavailability of all the compounds of Dangshen on PubMed. If the OB data of some compounds were previously reported in related experiments, the real-world data of OB were used instead of silicon data, and the highest reported OB value was adopted. Otherwise, silicon data were used for OB. The TCMSP database calculated OB values by using OBioavail1.1. This model shows good potential in facilitating the prediction of oral bioavailability and can be applied in drug design.
Figure 1.
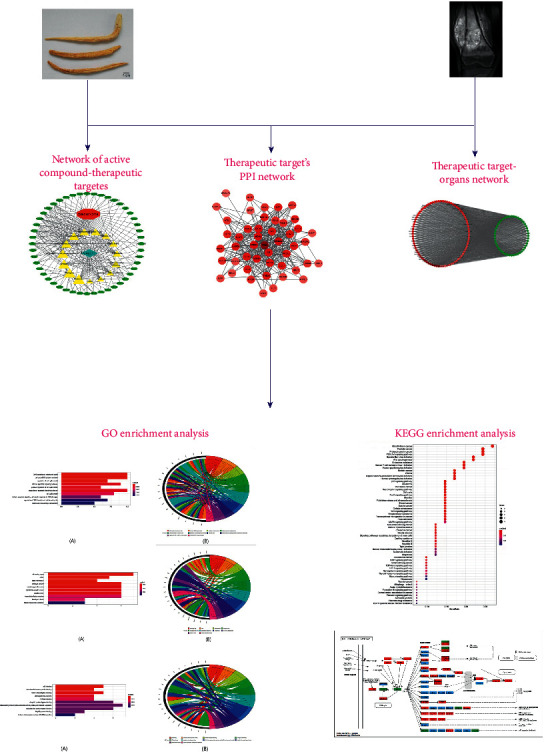
Schematic illustration showing the network pharmacology study of Dangshen (Codonopsis pilosula) for the treatment of osteosarcoma (OS).
2.2. Compounds of Dangshen and Their Targets
PubChem (https://pubchem.ncbi.nlm.nih.gov/) is an open chemistry database of the National Institutes of Health (NIH) [26]. This database serves as an important source of chemical information, including chemical structures, biological activities, chemical and physical properties, and safety [27]. We imported the compounds filtered from Dangshen into PubChem and obtained their 3D molecular structure files in SDF format. Structural information is necessary for predicting the targets of compounds, so the compounds without precise structural details were removed from the analysis.
PharmMapper (http://www.lilab-ecust.cn/pharmmapper/check.html) is a freely accessed web server that uses the pharmacophore mapping approach to identify potential small-molecule targets [28, 29]. We imported the 3D structural files in SDF format into PharmMapper and selected the pharmacophore model with a pKd value ≥ 6.0. In our study, the top matched 50 targets were selected as the potential targets of each compound.
2.3. Collection of Gene Targets in OS
The human genes associated with OS were gathered from OMIM (Online Mendelian Inheritance in Man, https://omim.org/) and GeneCards (https://www.genecards.org/). OMIM is an authoritative and comprehensive database of human genes and genetic phenotypes [30], while GeneCards is an integrative database that provides information on all predicted and annotated human genes [31]. The search term “osteosarcoma” was used to retrieve the OS targets from both databases.
2.4. Therapeutic Targets of Dangshen in OS
We screened the active compounds of Dangshen and obtained their target genes. We also gathered the OS-related genes. The potential therapeutic targets were identified from the shared genes mentioned above.
2.5. Protein-Protein Interaction (PPI) Data
The therapeutic targets were imported into the STRING database to obtain their interaction relationship. STRING (https://string-db.org/, version 11.0) is a database that contains known and predicted protein-protein interactions, and it collects the information using bioinformatics strategies [32]. The species were limited to “Homo sapiens,” and the PPIs with confidence scores >0.4 were selected for this study.
2.6. Target Organs
Data about the organ targets were collected from the BioGPS (http://biogps.org) database. BioGPS is an extensible and customizable genetic annotation portal that enables researchers to acquire distributed genetic annotation resources [33]. To reveal the underlying mechanisms of Dangshen in OS, median gene expression levels were used as the standard to screen for organs with high expression of the therapeutic targets.
2.7. Network Construction
The network of active compounds and therapeutic targets was constructed by linking the compounds and therapeutic targets to understand the complex interactions between the compounds of Dangshen and the therapeutic targets of OS. The network of therapeutic targets and organs was established by linking therapeutic targets and their distribution in organs to clarify the relationship between the therapeutic targets and organs with increased expression of the target. The therapeutic targets' PPI network was built by linking the therapeutic targets to their interacting targets. Next, Cytoscape version 3.7.2 (http://www.cytoscape.org/) was used to present the networks mentioned above, which is a software program for network visualization [34]. Lastly, NetworkAnalyzer [35] was used to calculate three topological parameters of each node in the network, including the degree, betweenness centrality, and closeness centrality [36].
2.8. GO and KEGG Pathway Enrichment Analysis
To learn more about the role of therapeutic targets involved in the biological process (BP), cell component (CC), and molecular function (MF), we used the Gene Ontology (GO) database (http://geneontology.org/) to clarify the possible biological mechanisms [37]. The Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.kegg.jp/) is a database for extracting biological information about functional classification, annotation, and enriched pathways of various genes [38]. In this study, we used an R-package-Bioconductor clusterProfiler to perform the GO and KEGG enrichment analysis. The R-package-Bioconductor clusterProfiler is widely used to automate the biological term classification and enrichment analysis of gene clusters [39].
3. Results
3.1. Chemical Compounds of Dangshen
Using the keyword search in TCMSP and TCMID, a total of 134 components of Dangshen were identified, including flavonoids, steroids, alkaloids, glycosides, and triterpenes. According to the OB and DL characteristics of the ingredients, 25 screened compounds were chosen for the next experiments. As structural information is essential for predicting the targets of a compound, ten compounds without 3D structural information were discarded. Finally, 15 compounds were determined as possible active compounds whose characteristics are listed in Table 1.
Table 1.
Characteristics of the active ingredients.
| Compound | MF | Structure | MW | OB (%) | DL | HL |
|---|---|---|---|---|---|---|
| Stigmasterol | C29H48O | fx1 | 412.77 | 43.83 | 0.76 | 5.57 |
| Stigmast-7-enol | C29H50O | fx2 | 414.79 | 37.42 | 0.75 | 6.28 |
| Luteolin | C15H10O6 | fx3 | 286.25 | 32.00 | 0.25 | 15.94 |
| 11-Hydroxyrankinidine | C20H24N2O4 | fx4 | 356.46 | 40.00 | 0.66 | 10.80 |
| Perlolyrine | C16H12N2O2 | fx5 | 264.30 | 65.95 | 0.27 | 12.62 |
| Glycitein | C16H12O5 | fx6 | 284.28 | 50.48 | 0.24 | 16.32 |
| Spinasterol | C29H48O | fx7 | 412.77 | 42.98 | 0.76 | 5.32 |
| Frutinone A | C16H8O4 | fx8 | 264.24 | 65.90 | 0.34 | 19.10 |
| Poriferasta-7,22E-dien-3beta-ol | C29H48O | fx9 | 412.77 | 42.98 | 0.76 | 5.48 |
| 7-Methoxy-2-methyl isoflavone | C17H14O3 | fx10 | 266.31 | 42.56 | 0.20 | 16.89 |
| 5-α-Stigmastan-3,6-dione | C29H48O2 | fx11 | 428.77 | 33.12 | 0.79 | 5.19 |
| 3-β-Hydroxymethyllenetanshiquinone | C18H14O4 | fx12 | 294.32 | 32.16 | 0.41 | 22.51 |
| Zinc03978781 | C29H48O | fx13 | 412.77 | 43.83 | 0.76 | 5.79 |
| Taraxerol | C30H50O | fx14 | 426.80 | 38.40 | 0.77 | 2.07 |
| Stigmasterone | C29H46O | fx15 | 410.75 | 45.40 | 0.76 | 5.65 |
3.2. Dangshen Compound Targets
We obtained the top 50 matched targets for each potential active compound from PharmMapper. These targets were regarded as the potential targets of Dangshen (Supplementary Table S1).
3.3. Collection of Gene Targets for OS
“Osteosarcoma” was used as the keyword to retrieve the OS targets from OMIM and GeneCards databases. A total of 2,079 genes were retrieved from the two databases (Supplementary Table S2).
3.4. Therapeutic Targets of Dangshen for OS
The targeted genes of Dangshen and OS were obtained. Using the shared genes described above, 48 possible therapeutic targets were obtained, and the features are listed in Table 2.
Table 2.
Characteristics of the 48 therapeutic targets.
| Target | Name | Degree | Betweenness centrality | Closeness centrality |
|---|---|---|---|---|
| TP53 | Cellular tumor antigen p53 | 40 | 0.19161047 | 0.87037037 |
| HSP90AA1 | Heat shock protein HSP 90-alpha | 33 | 0.08748708 | 0.77049180 |
| CCND1 | G1/S-specific cyclin-D1 | 30 | 0.05438164 | 0.73437500 |
| AR | Androgen receptor | 29 | 0.08088557 | 0.72307692 |
| MDM2 | E3 ubiquitin-protein ligase Mdm2 | 28 | 0.05726816 | 0.70149254 |
| ERBB2 | Receptor tyrosine-protein kinase erbB-2 | 28 | 0.05040772 | 0.71212121 |
| IGF1R | Insulin-like growth factor-1 receptor | 27 | 0.04394018 | 0.70149254 |
| DICER1 | Endoribonuclease Dicer | 24 | 0.03867798 | 0.66197183 |
| CCNE1 | G1/S-specific cyclin-E1 | 19 | 0.01018846 | 0.61842105 |
| THBS1 | Thrombospondin-1 | 19 | 0.01547887 | 0.61038961 |
| SOD2 | Superoxide dismutase [Mn], mitochondrial | 18 | 0.01677310 | 0.61038961 |
| XRCC6 | ATP-dependent DNA helicase 2 subunit 1 | 16 | 0.02818547 | 0.59493671 |
| MAPK9 | Mitogen-activated protein kinase 9 | 16 | 0.01100001 | 0.59493671 |
| E2F1 | Transcription factor E2F1 | 15 | 0.00460006 | 0.58750000 |
| MUC1 | Mucin-1 | 15 | 0.01251038 | 0.58024691 |
| CUL1 | Cullin-1 | 14 | 0.02049293 | 0.56626506 |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 | 14 | 0.04760817 | 0.58750000 |
| B2M | Beta-2-microglobulin | 14 | 0.01143842 | 0.58024691 |
| MPO | Myeloperoxidase | 14 | 0.01314265 | 0.57317073 |
| HDAC6 | Histone deacetylase 6 | 12 | 0.00411776 | 0.56626506 |
| MAP2K2 | Dual specificity mitogen-activated protein kinase kinase 2 | 12 | 0.00552768 | 0.55294118 |
| EZR | Ezrin | 12 | 0.00234926 | 0.57317073 |
| PAX6 | Paired box protein Pax-6 | 12 | 0.00636814 | 0.55952381 |
| TP73 | Tumor protein p73 | 10 | 0.00045360 | 0.54022989 |
| BMPR2 | Bone morphogenetic protein receptor type 2 | 10 | 0.00217280 | 0.55294118 |
| RPS3 | 40S ribosomal protein S3 | 9 | 0.00443058 | 0.53409091 |
| HDAC8 | Histone deacetylase 8 | 9 | 0.00168382 | 0.54022989 |
| POLB | DNA polymerase β | 9 | 0.00620249 | 0.54651163 |
| FOLH1 | Glutamate carboxypeptidase 2 | 9 | 0.00471413 | 0.54651163 |
| SMAD1 | Mothers against decapentaplegic homolog 1 | 8 | 0.00048566 | 0.52808989 |
| ATIC | Bifunctional purine biosynthesis protein PURH | 8 | 0.00650462 | 0.52222222 |
| S100A6 | Protein S100-A6 | 8 | 0.00018501 | 0.53409091 |
| NR3C2 | Mineralocorticoid receptor | 7 | 0.00397370 | 0.52808989 |
| EIF2AK2 | Interferon-induced, double-stranded RNA-activated protein kinase | 7 | 0.00917844 | 0.52808989 |
| RPA1 | Replication protein A 70 kDa DNA-binding subunit | 6 | 0.00034097 | 0.51648352 |
| SATB2 | DNA-binding protein SATB2 | 5 | 0.00026981 | 0.49473684 |
| CDCA8 | Borealin | 5 | 0 | 0.49473684 |
| FLI1 | Friend leukemia integration 1 transcription factor | 5 | 0.00061402 | 0.50000000 |
| NHP2L1 | Human recombinant protein P01 | 4 | 0.00020331 | 0.47000000 |
| CAMK2A | Calcium/calmodulin-dependent protein kinase type II α chain | 4 | 0 | 0.46078431 |
| HSD11B1 | Corticosteroid 11-β-dehydrogenase isozyme 1 | 4 | 0.00067245 | 0.44761905 |
| ASS1 | Argininosuccinate synthase | 4 | 0 | 0.49473684 |
| FAP | Seprase | 4 | 0.00011563 | 0.44339623 |
| CANT1 | Soluble calcium-activated nucleotidase 1 | 3 | 0.00036617 | 0.44761905 |
| BCL9 | B-cell CLL/lymphoma 9 protein | 3 | 0 | 0.48453608 |
| CSDE1 | Cold shock domain-containing protein E1 | 3 | 0.00053191 | 0.41592920 |
| BRD7 | Bromodomain-containing protein 7 | 2 | 0 | 0.47959184 |
| SRGAP2 | SLIT-ROBO rho GTPase-activating protein 2 | 1 | 0 | 0.37301587 |
3.5. Active Compound-Therapeutic Target Network
The active compound-therapeutic target network is depicted in Figure 2. This network demonstrates the complicated relationship between the compounds and therapeutic targets, including 65 total nodes (15 compound nodes, 48 therapeutic target nodes, one Dangshen node, and one OS node) and 204 edges. In Figure 2, the therapeutic targets are represented by green ovals, Dangshen is represented by a blue quadrangle, OS is represented by a red hexagon, active compounds are represented by yellow triangles, and the sizes of compound nodes were proportional to their degree. The three with the highest degree of the compound nodes were Frutinone A (degree = 16), Perlolyrine (degree = 14), and Glycitein (degree = 13). The three compounds were more likely to show significant therapeutic activity against OS.
Figure 2.
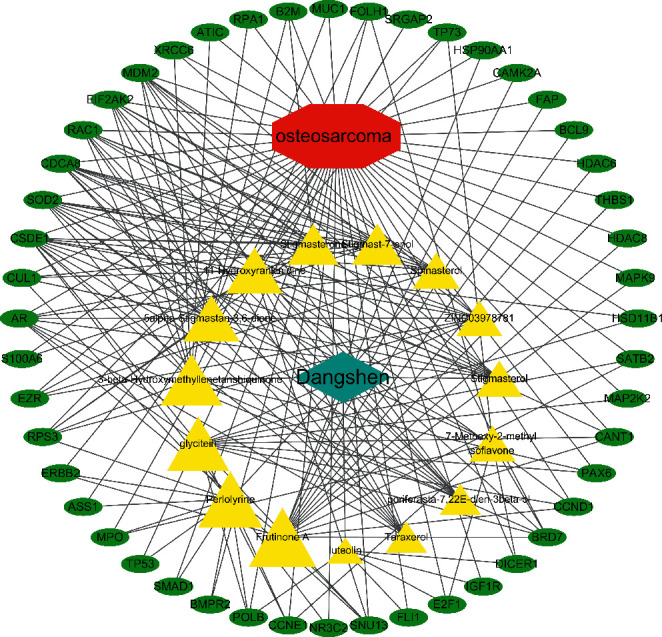
Active compounds-therapeutic targets network. Yellow triangles represent the active compounds from Dangshen, while green ovals represent the therapeutic targets. The size of the triangles is directly proportional to their degree.
3.6. Therapeutic Target-PPI Network
The PPI network of the therapeutic targets is shown in Figure 3, including 48 nodes and 304 edges. NetworkAnalyzer was employed to calculate three topological features of the 48 targets to identify the key nodes in the network (Table 2). The median values of the degree, node betweenness, and closeness were 10, 0.047, and 0.549, respectively. The nodes with “degree >10,” “node betweenness >0.047,” and “node closeness >0.63” were considered to be the key targets. Hence, 20 genes were identified as central targets of Dangshen against OS, including TP53, HSP90AA1, CCND1, AR, ERBB2, MDM2, IGF1R, DICER1, CCNE1, SOD2, among others.
Figure 3.
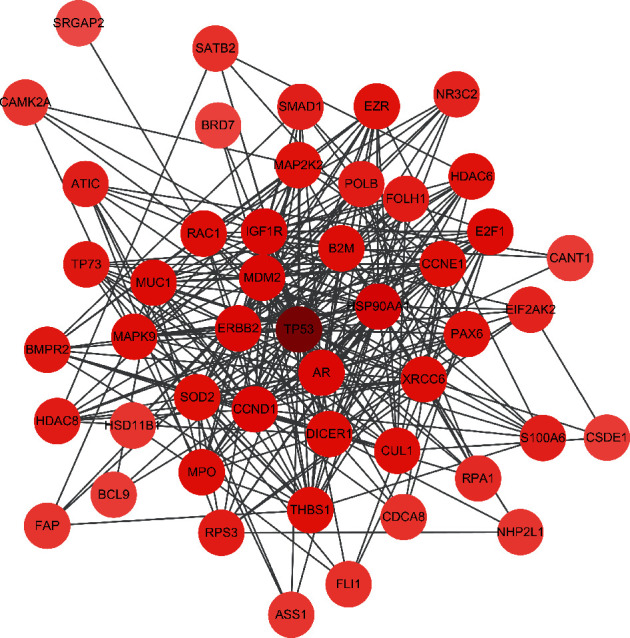
PPI network of therapeutic targets. Hexagons represent the therapeutic targets, and the color shade of hexagons is directly proportional to their degree.
3.7. Therapeutic Target-Organ Network
The organs with high expression of each therapeutic target were collected via BioGPS (Supplementary Table S3). The therapeutic target-organ network, shown in Figure 4, is used to delineate the relationship between therapeutic targets and the organs that highly express these targets, including 132 nodes (58 therapeutic target nodes and 84 organ nodes) and 2,031 edges. The color shade of the organ node is proportional to its degree, as shown in Figure 4. These findings demonstrate that many therapeutic targets are highly expressed in tissues, such as the thyroid, retina, pituitary, and pineal gland, and on the surface of antigens, including CD33, CD34, and CD56.
Figure 4.

Therapeutic target-organs network. Red ovals represent the tissues with high expression levels of the targets, and the color shade of the red ovals is directly proportional to the degree. Green ovals represent therapeutic targets.
3.8. GO and KEGG Pathway Enrichment
To illuminate the complex mechanisms of Dangshen against OS, we conducted analyses of the GO biological process (BP), cell component (CC), and molecular function (MF) for the 48 therapeutic targets. The top ten biological processes, cell components, and molecular functions are shown in Figures 5(a), 6(a), and 7(a), respectively. The relationship between the genes and biological processes, cell component, and molecular function targets is depicted in Figures 5(b), 6(b), and 7(b), respectively. The details of the GO enrichment analysis of BP, CC, and MF are listed in Supplementary Tables S4–S6, respectively.
Figure 5.
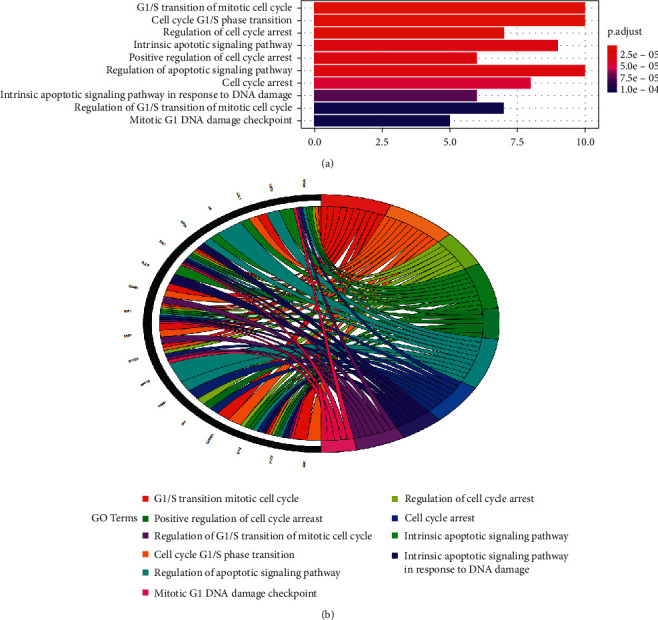
Top ten significant biological process (BP) entries. (a): GO enrichment analysis of therapeutic targets for biological process. (b): Relationship between the therapeutic targets and biological process.
Figure 6.
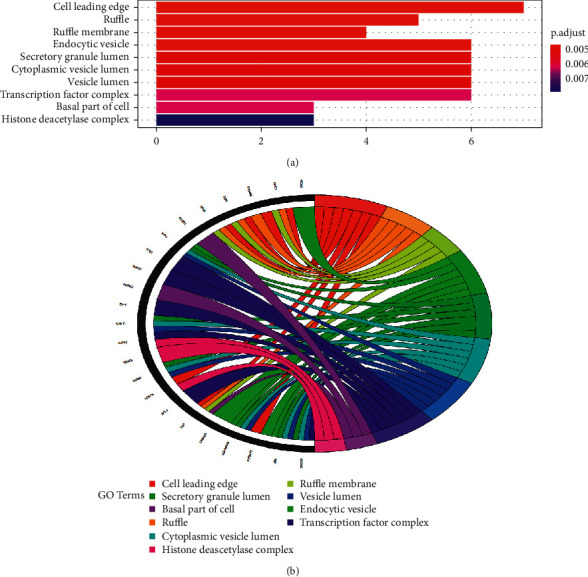
Top ten significant cell component (CC entries). (a): GO enrichment analysis of therapeutic targets for cell components. (b) Relationship between the therapeutic targets and cell components.
Figure 7.
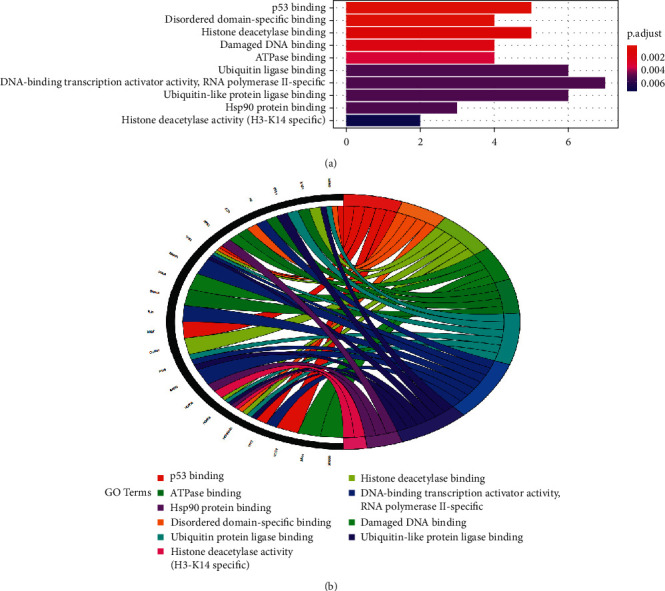
Top ten significant molecular function (MF) entries. (a): GO enrichment analysis of therapeutic targets for molecular function. (b) Relationship between the therapeutic targets and molecular function.
KEGG pathway enrichment analysis was performed to explore the underlying mechanisms of Dangshen against OS further. As shown in Supplementary Table S7 and Figure 8, there are 69 primary pathways that participate in Dangshen against OS with p < 0.05. These 69 pathways involve human diseases, pathophysiological mechanisms, and signaling pathways. The top ten significantly enriched signaling pathways include the p53 signaling pathway, PI3K-Akt signaling pathway, neurotrophin signaling pathway, FoxO signaling pathway, Wnt signaling pathway, ErbB signaling pathway, TGF-β signaling pathway, HIF-1 signaling pathway, sphingolipid signaling pathway, and MAPK signaling pathway. Many therapeutic targets are involved in these signaling pathways. Figure 9 depicts a concept map containing Dangshen and OS targets in the P53 signaling pathway, further demonstrating that Dangshen regulates key targets in this signaling pathway.
Figure 8.
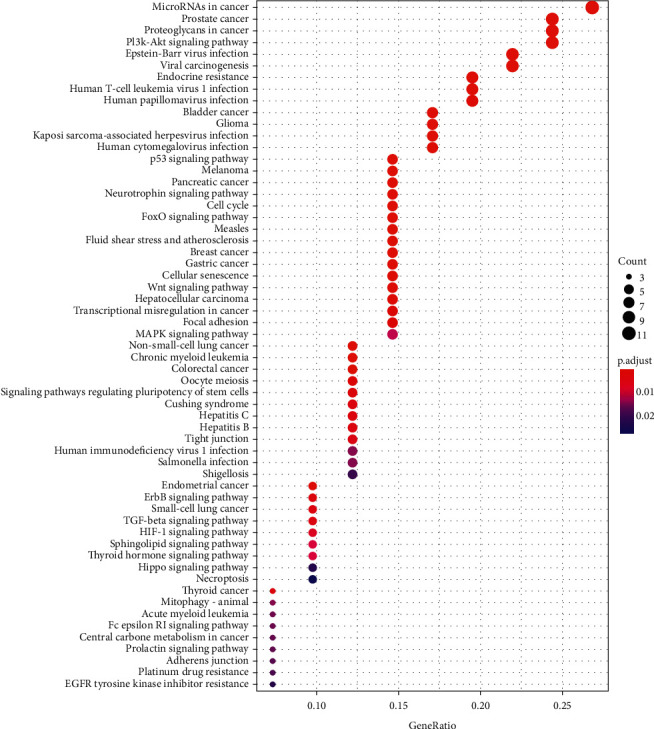
KEGG enrichment analysis for therapeutic targets.
Figure 9.
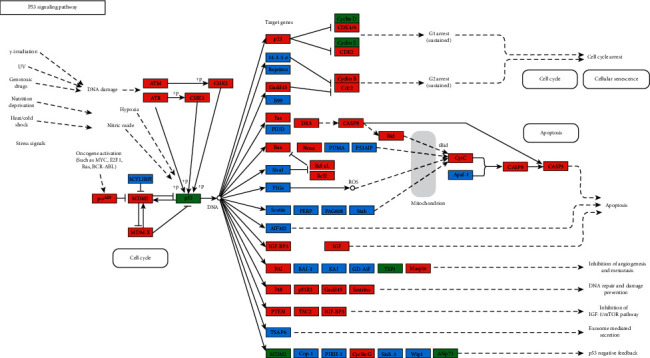
Modulation of the p53 signaling pathway by Dangshen. Targets associated with OS are in red and green, targets of Dangshen are in green, and other proteins in the pathway are in blue.
4. Discussion
Osteosarcoma (OS) is the most common primary bone tumor found in the clinic [40]. It is characterized by high metastatic rates, poor prognoses, and high mortality rates [41]. Dangshen (Codonopsis pilosula) is a well-known herbal medicine, and traditional Chinese medicine (TCM) preparation, based on Dangshen, which has shown high efficacy in the treatment of OS [10, 11]. However, its pharmacological mechanisms remain unclear. In the present study, we used network pharmacology to explore the potential active compounds and underlying mechanisms of Dangshen against OS.
After applying the screening methods, we identified 15 active compounds and 48 potential therapeutic targets. The active compounds of Dangshen likely treat OS by regulating these targets. We identified two active compounds, stigmasterol and luteolin, that have been studied previously for their efficacy against OS. Stigmasterol is a phytosterol, which has been shown to exert anticancer, antipyretic, and immune-modulating properties [42–44]. Previously, Trouillas et al. showed that stigmasterol could decrease the proliferation of OS cells [45]. Luteolin is a flavonoid found in vegetables and fruits. It can inhibit the proliferation and induce the apoptosis of OS cells by effectively downregulating the expression of BCL-2, caspase-3, and survivin proteins levels, while upregulating BAX protein levels [46]. In addition, it can induce autophagy in U2OS cells and enhance the sensitivity of these cells to doxorubicin-mediated autophagy signaling [47].
From the therapeutic target-PPI network, the following targets showed larger degree values: TP53, HSP90AA1, CCND1, AR, ERBB2, MDM2, IGF1R, DICER1, CCNE1, and SOD2. These targets may play a major role in the therapeutic effect of Dangshen against OS. Over 70% of OS cases show structural variants or mutations in the TP53 gene [48]. TP53 is a transcription factor that stabilizes following genotoxic stress and induces the transcription of genes associated with cell apoptosis, cycle arrest, and metabolism; thereby, suppressing the development and progression of tumors [49, 50]. HSP90AA1, a 90-kDa heat shock protein [51], is an important target for cancer treatment because it can stabilize several cancer-related client proteins essential for tumor progression, such as AKT, PIM1, and HIF1A [52]. Some studies found that, in tumor biopsies, the absence of HSP90AA1 may serve as a biomarker of favorable outcomes [53, 54]. CCND1 is a member of the cyclin family that encodes cyclin-D1. In addition, it plays a key role in cell cycle regulation [55]. There is substantial evidence showing that CCND1 plays an important role in the development of human cancers [56], including the migration and metastasis of OS [57]. The ERBB family of tyrosine kinases plays an important role in cell cycle regulation, cell proliferation, and cell movement [58]. Tumors that overexpress ERBB2 are less likely to respond to anticancer therapies [59]. Previously, Abdou et al. reported on the overexpression of ERBB2 in OS and its adverse prognostic features, including higher tumor grades [60]. In addition, Wang et al. reported that chimeric anti-caspase-6 and anti-ERBB2 antibodies reduced the metastatic potential of human OS cells [61]. These findings suggest that the therapeutic effect of Dangshen against OS is primarily mediated by cell apoptosis, cell cycle arrest, and the inhibition of tumor cell migration and metastasis.
Next, we performed the GO enrichment analysis and KEGG pathway enrichment analysis of the therapeutic targets. Based on the GO terms, the therapeutic targets showed a strong correlation with the biological processes, such as the G1/S transition of mitotic cell cycle, cell cycle G1/S phase transition, regulation of cell cycle arrest, and the intrinsic apoptotic signaling pathway; cell components, such as cell leading edge, ruffle, ruffle membrane, and endocytic vesicle; and molecular functions, such as p53 binding, disordered domain specific binding, histone deacetylase binding, damaged DNA binding, and ATPase binding. Hence, the mechanism of action for Dangshen may include biological processes, molecular functions, and various cellular components. For example, imbalanced cell cycle regulation is characteristic of tumor cells, and functional defects in cell cycle checkpoints lead to genetic changes that lead to tumor development and progression [62, 63]. In addition, the G1/S phase transition is the target of many anticancer drugs [64]. Apoptosis is a form of cell death that occurs upon receipt of internal or external death signals [65, 66]. In addition, Chaiyawat et al. reported that reduced expression of histone deacetylase-2 of HDAC2 is associated with dismal patient outcomes in OS [67]. Furthermore, Sun et al. reported that histone deacetylase-2 may stimulate the ATM/p53 pathway, leading to DNA damage-mediated cell death in human OS cells [68]. In addition, Cao et al. found that overexpression of histone deacetylase-4 promotes the proliferation and invasion of OS cells [69].
Based on the KEGG terms, the therapeutic targets for Dangshen against OS were primarily associated with the p53 signaling pathway, PI3K-Akt signaling pathway, FoxO signaling pathway, Wnt signaling pathway, and ErbB signaling pathway. P53 plays a critical role in cell cycle checkpoints regulation, DNA damage, and prevention of nonmalignant cells from developing malignant phenotypes [70, 71]. In addition, p53 is an essential regulator of epithelial-mesenchymal transition (EMT) [72], as it promotes the reversal of mesenchymal cells to the epithelial cell phenotype, which reduces the migration and invasion of cells [73]. Many anticancer drugs regulate the p53 signaling pathway. For example, theabrownin triggers DNA damage and induces apoptosis in U2OS cells via p53 signaling activation [74]. Activation of the PI3K-Akt signaling pathway is also associated with cell proliferation and apoptosis of OS cells [75, 76], and the downregulation of AKT reduces cyclin-D1 levels, preventing cells from cycling from G1 to S [77]. The reduced expression of cyclin-D1 also leads to the inhibition of cell proliferation [78]. Simultaneously, AKT downregulates the expression of two essential proteins responsible for apoptosis, caspase-3 and caspase-8 [79]. Abnormal Wnt/β-catenin signaling is closely related to the formation, metastasis, and apoptosis of many cancers [80]. The upregulation of Wnt/β-catenin signaling was recently observed in OS [81]. As such, the WIF-1 protein, encoded by Wnt inhibitory factor-1 gene, is an important regulatory factor in the Wnt signaling pathway [82]. The WIF-1 gene combines with the Wnt protein to prevent Wnt signaling [83]. Previously, Li et al. reported on the downregulation of WIF-1 in OS cells [84]. Hence, the KEGG analysis revealed that Dangshen produces anticancer effects in OS through the regulation of several proteins, including MDM2, TP53, RAC1, ERBB2, and CCND1, which are all important mediators of various cellular signaling pathways. In addition, most therapeutic targets play their roles in multiple signaling pathways. In addition, most of the therapeutic targets play essential roles in multiple signaling pathways.
Network pharmacology is an analytical method still in development worldwide. However, the method has some inherent flaws. For example, it heavily relies on existing resources of the databases, so it cannot analyze the compounds, targets, or mechanisms that have not been previously explored. Moreover, its predicted active ingredients, targets, and mechanisms of action are all purely theoretical, and there is a lack of experimental verification. Therefore, further clinical investigations are needed.
5. Conclusions
In this study, we explored the therapeutic mechanisms of Dangshen against OS through a network pharmacology approach. The therapeutic properties of Dangshen against OS arise from the regulation of biological pathways involved in the proliferation, apoptosis, invasion, and migration of cells, along with DNA damage. We believe these findings demonstrate the importance of understanding traditional Chinese medicines. The current study relied on data mining and analysis, and further clinical investigations are needed to verify the therapeutic mechanisms of Dangshen against OS.
Acknowledgments
This work was supported by the Science and Technology Development Project of Jilin Province (20190201081JC) and Health Special Project of Jilin Province (JLSWSRCZX2020-0053).
Data Availability
The data sets generated and analyzed during the present study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Supplementary Materials
Description of Supplementary Table S1: the top 50 matched targets for each potential active compound of Dangshen were gathered from PharmMapper, and these targets were regarded as the potential targets of Dangshen. The relationship between the compounds and their matched targets is listed in Supplementary Table S1. Description of Supplementary Table S2: the genes related to osteosarcoma were collected from GeneCards and OMIM databases. The genes related to osteosarcoma are listed in Supplementary Table S2. Description of Supplementary Table S3: the organs with high expression of each therapeutic target were collected via BioGPS. The relationship between the therapeutic targets and their high expressed organs is listed in Supplementary Table S3. Description of Supplementary Table S4: we used the Gene Ontology (GO) database to clarify the possible biological mechanisms. The details of the GO enrichment analysis of biological process are listed in Supplementary Table S4. Description of Supplementary Table S5: we used the Gene Ontology (GO) database to clarify the possible biological mechanisms. The details of the GO enrichment analysis of cell component are listed in Supplementary Table S5. Description of Supplementary Table S6: we used the Gene Ontology (GO) database to clarify the possible biological mechanisms. The details of the GO enrichment analysis of molecular function are listed in Supplementary Table S6. Description of Supplementary Table S7: we used the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to extract biological information about functional classification, annotation, and enriched pathways of therapeutic targets. The details of the KEGG enrichment analysis are listed in Supplementary Table S7. .
References
- 1.Moore D. D., Luu H. H. Osteosarcoma. Cancer Treatment and Research. 2014;162:65–92. doi: 10.1007/978-3-319-07323-1_4. [DOI] [PubMed] [Google Scholar]
- 2.Meyers P. A., Gorlick R. Osteosarcoma. Pediatric Clinics of North America. 1997;44(4):973–989. doi: 10.1016/s0031-3955(05)70540-x. [DOI] [PubMed] [Google Scholar]
- 3.Ottaviani G., Jaffe N. The epidemiology of osteosarcoma. In: Jaffe N., Bruland O. S., Bielack S., editors. Pediatric and Adolescent Osteosarcoma. Boston, MA, USA: Springer; 2010. [Google Scholar]
- 4.Ferrari S., Serra M. An update on chemotherapy for osteosarcoma. Expert Opinion on Pharmacotherapy. 2015;16(18):2727–2736. doi: 10.1517/14656566.2015.1102226. [DOI] [PubMed] [Google Scholar]
- 5.Toki S., Kobayashi E., Yoshida A., et al. A clinical comparison between dedifferentiated low-grade osteosarcoma and conventional osteosarcoma. The Bone & Joint Journal. 2019;101(6):745–752. doi: 10.1302/0301-620x.101b6.bjj-2018-1207.r1. [DOI] [PubMed] [Google Scholar]
- 6.Ferguson J. L., Turner S. P. Bone cancer: diagnosis and treatment principles. American Family Physician. 2018;98(4):205–213. [PubMed] [Google Scholar]
- 7.Sasaki R., Osaki M., Okada F. MicroRNA-based diagnosis and treatment of metastatic human osteosarcoma. Cancers. 2019;11(4):p. 553. doi: 10.3390/cancers11040553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Messerschmitt P. J., Garcia R. M., Abdul-Karim F. W., Greenfield E. M., Getty P. J. Osteosarcoma. Journal of the American Academy of Orthopaedic Surgeons. 2009;17(8):515–527. doi: 10.5435/00124635-200908000-00005. [DOI] [PubMed] [Google Scholar]
- 9.Glasser D. B., Lane J. M., Huvos A. G., Marcove R. C., Rosen G. Survival, prognosis, and therapeutic response in osteogenic sarcoma. The memorial hospital experience. Cancer. 1992;69(3):698–708. doi: 10.1002/1097-0142(19920201)69:3<698::aid-cncr2820690317>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 10.Kuang T., Liu Y. Application of Shenqi fuzheng injection in 30 cases of osteosarcoma with high dose neoadjuvant chemotherapy. Journal of Chinese Oncology. 2009;15(05):473–474. [Google Scholar]
- 11.Hu Y., Li H., Zuo Y. Effect of Shenqi Fuzheng Injection on immune function in patients with osteosarcoma high-dose chemotherapy. China Continuing Medical Education. 2018;10(05):149–151. [Google Scholar]
- 12.Su J.-S., Qin F.-Y., Liu Y., Zhang Y. Four new polyynes from Codonopsis pilosula collected in Yunnan province, China. Natural Product Research. 2020:1–8. doi: 10.1080/14786419.2020.1712390. [DOI] [PubMed] [Google Scholar]
- 13.Fu Y.-P., Li L.-X., Zhang B.-Z., et al. Characterization and prebiotic activity in vitro of inulin-type fructan from Codonopsis pilosula roots. Carbohydrate Polymers. 2018;193:212–220. doi: 10.1016/j.carbpol.2018.03.065. [DOI] [PubMed] [Google Scholar]
- 14.Zou Y.-F., Zhang Y.-Y., Fu Y.-P., et al. A polysaccharide isolated from codonopsis pilosula with immunomodulation effects both in vitro and in vivo. Molecules. 2019;24(20):p. 3632. doi: 10.3390/molecules24203632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J., Hu L., Wu H., et al. Codonopsis pilosula polysaccharides on tumor-bearing mice immune response and anti-tumor effect. Chinese Journal of Cancer Prevention and Treatment. 2015;22(17):1357–1362. [Google Scholar]
- 16.Wang H., Lin H., Tan J., et al. Codonopsis pharmacological effects and clinical application research progress. World Latest Medicine Information. 2019;19(07):21–22+4. [Google Scholar]
- 17.Hattinger C. M., Patrizio M. P., Luppi S., et al. Current understanding of pharmacogenetic implications of DNA damaging drugs used in osteosarcoma treatment. Expert Opinion on Drug Metabolism & Toxicology. 2019;15(4):299–311. doi: 10.1080/17425255.2019.1588885. [DOI] [PubMed] [Google Scholar]
- 18.Angulo P., Kaushik G., Subramaniam D., et al. Natural compounds targeting major cell signaling pathways: a novel paradigm for osteosarcoma therapy. Journal of Hematology & Oncology. 2017;10(1):p. 10. doi: 10.1186/s13045-016-0373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain B., Raj U., Varadwaj P. K. Drug target interplay: a network-based analysis of human diseases and the drug targets. Current Topics in Medicinal Chemistry. 2018;18(13):1053–1061. doi: 10.2174/1568026618666180719160922. [DOI] [PubMed] [Google Scholar]
- 20.Shao L., Zhang B. J. C. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chinese Journal of Natural Medicines. 2013;11(2):110–120. doi: 10.1016/S1875-5364(13)60037-0. [DOI] [PubMed] [Google Scholar]
- 21.Ru J., Li P., Wang J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics. 2014;6(1):p. 13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue R., Fang Z., Zhang M., Yi Z., Wen C., Shi T. TCMID: traditional Chinese Medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Research. 2013;41:D1089–D1095. doi: 10.1093/nar/gks1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karami Z., Saghatchi Zanjani M. R., Nasihatsheno N., Hamidi M. Improved oral bioavailability of repaglinide, a typical BCS Class II drug, with a chitosan-coated nanoemulsion. Journal of Biomedical Materials Research. 2019;108 doi: 10.1002/jbm.b.34426. [DOI] [PubMed] [Google Scholar]
- 24.Lipinski C. A. Drug-like properties and the causes of poor solubility and poor permeability. Journal of Pharmacological and Toxicological Methods. 2000;44(1):235–249. doi: 10.1016/s1056-8719(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 25.Gong B., Kao Y., Zhang C., Zhao H., Sun F., Gong Z. Exploring the pharmacological mechanism of the herb pair “HuangLian-GanJiang” against colorectal cancer based on network pharmacology. Evidence-Based Complementary and Alternative Medicine. 2019;2019:12. doi: 10.1155/2019/2735050.2735050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S., Thiessen P. A., Bolton E. E., et al. PubChem substance and compound databases. Nucleic Acids Research. 2016;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S., Chen J., Cheng T., et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Research. 2019;47:D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X., Shen Y., Wang S., et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Research. 2017;45:W356–W360. doi: 10.1093/nar/gkx374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X., Pan C., Gong J., Liu X., Li H. Enhancing the enrichment of pharmacophore-based target prediction for the polypharmacological profiles of drugs. Journal of Chemical Information and Modeling. 2016;56(6):1175–1183. doi: 10.1021/acs.jcim.5b00690. [DOI] [PubMed] [Google Scholar]
- 30.Amberger J. S., Hamosh A. Searching online mendelian inheritance in man (OMIM): a knowledgebase of human genes and genetic phenotypes. Current Protocols in Bioinformatics. 2017;58:1–12. doi: 10.1002/cpbi.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stelzer G., Rosen N., Plaschkes I., et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Current Protocols in Bioinformatics. 2016;54:1–33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 32.Szklarczyk D., Gable A. L., Lyon D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu C., Jin X., Tsueng G., Afrasiabi C., Su A. I. BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Research. 2016;44:D313–D316. doi: 10.1093/nar/gkv1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smoot M. E., Ono K., Ruscheinski J., Wang P.-L., Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Jong H., Geiselmann J., Hernandez C., Page M. Genetic Network Analyzer: qualitative simulation of genetic regulatory networks. Bioinformatics. 2003;19(3):336–344. doi: 10.1093/bioinformatics/btf851. [DOI] [PubMed] [Google Scholar]
- 36.Raman K., Damaraju N., Joshi G. K. The organisational structure of protein networks: revisiting the centrality-lethality hypothesis. Systems and Synthetic Biology. 2014;8(1):73–81. doi: 10.1007/s11693-013-9123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ning Z., Jiang Z. GOVis, a gene ontology visualization tool based on multi-dimensional values. Protein & Peptide Letters. 2010;17(5):675–680. doi: 10.2174/092986610791112675. [DOI] [PubMed] [Google Scholar]
- 38.Xing Z., Chu C., Chen L., Kong X. The use of gene ontology terms and KEGG pathways for analysis and prediction of oncogenes. Biochimica et biophysica acta. 2016;1860:2725–2734. doi: 10.1016/j.bbagen.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 39.Yu G., Wang L.-G., Han Y., He Q.-Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simpson E., Brown H. L. Understanding osteosarcomas. Journal of the American Academy of Physician Assistants. 2018;31(8):15–19. doi: 10.1097/01.jaa.0000541477.24116.8d. [DOI] [PubMed] [Google Scholar]
- 41.Grohar P. J., Janeway K. A., Mase L. D., Schiffman J. D. Advances in the treatment of pediatric bone sarcomas. American Society of Clinical Oncology Educational Book. 2017;37:725–735. doi: 10.14694/edbk_175378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Etsebeth S., Albrecht C., Pegel K. Beta-sitosterol and beta-sitosterol glucoside stimulate human peripheral blood lymphocyte proliferation: implications for their use as an immunomodulatory vitamin combination. International Immunopharmacology. 1996;18(12) doi: 10.1016/s0192-0561(97)85551-8. [DOI] [PubMed] [Google Scholar]
- 43.Bouic P. J., Lamprecht J. H. Plant sterols and sterolins: a review of their immune-modulating properties. Alternative Medicine Review: A Journal of Clinical Therapeutic. 1999;4(3):170–177. [PubMed] [Google Scholar]
- 44.Chen W.-P., Yu C., Hu P.-F., Bao J.-P., Tang J.-L., Wu L.-D. Stigmasterol blocks cartilage degradation in rabbit model of osteoarthritis. Acta biochimica Polonica. 2012;59(4) doi: 10.18388/abp.2012_2088. [DOI] [PubMed] [Google Scholar]
- 45.Trouillas P., Corbière C., Liagre B., Duroux J.-L., Beneytout J.-L. Structure-function relationship for saponin effects on cell cycle arrest and apoptosis in the human 1547 osteosarcoma cells: a molecular modelling approach of natural molecules structurally close to diosgenin. Bioorganic & Medicinal Chemistry. 2005;13(4):1141–1149. doi: 10.1016/j.bmc.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y., Kong D., Wang X., Dong X., Tao Y., Gong H. Molecular mechanisms of luteolin induced growth inhibition and apoptosis of human osteosarcoma cells. Iranian Journal of Pharmaceutical Research: IJPR. 2015;14(2):531–8. [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang B., Yu X., Xia H. The flavonoid luteolin enhances doxorubicin-induced autophagy in human osteosarcoma U2OS cells. International Journal of Clinical and Experimental Medicine. 2015;8(9):7.15190 [PMC free article] [PubMed] [Google Scholar]
- 48.Thoenen E., Curl A., Iwakuma T. J. P. TP53 in Bone and Soft Tissue Sarcomas. Pharmacology & Therapeutics. 2019;202 doi: 10.1016/j.pharmthera.2019.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ranjan A., Iwakuma T. Non-canonical cell death induced by p53. International Journal of Molecular Sciences. 2016;17(12):p. 2068. doi: 10.3390/ijms17122068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lane D., Levine A. p53 research: the past thirty years and the next thirty years. Cold Spring Harbor Perspectives in Biology. 2010;2(12) doi: 10.1101/cshperspect.a000893.a000893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang C., Huang D., Ma C., et al. Identification of pathogenic genes and transcription factors in osteosarcoma. Pathology & Oncology Research. 2019;26:1–8. doi: 10.1007/s12253-019-00645-w. [DOI] [PubMed] [Google Scholar]
- 52.Taipale M., Jarosz D. F., Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nature Reviews Molecular Cell Biology. 2010;11(7):515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 53.Ruiz M. I. G., Floor K., Roepman P., et al. Integration of gene dosage and gene expression in non-small cell lung cancer, identification of HSP90 as potential target. PLoS One. 2008;3(3) doi: 10.1371/journal.pone.0001722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng Q., Chang J. T., Geradts J., et al. Amplification and high-level expression of heat shock protein 90 marks aggressive phenotypes of human epidermal growth factor receptor 2 negative breast cancer. Breast Cancer Research. 2012;14(2):p. R62. doi: 10.1186/bcr3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao M., Xu P., Liu Z., et al. Dual roles of miR-374a by modulated c-Jun respectively targets CCND1-inducing PI3K/AKT signal and PTEN-suppressing Wnt/β-catenin signaling in non-small-cell lung cancer. Cell Death & Disease. 2018;9(2):78–17. doi: 10.1038/s41419-017-0103-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56.Chen D.-G., Zhu B., Lv S.-Q., et al. Inhibition of EGR1 inhibits glioma proliferation by targeting CCND1 promoter. Journal of Experimental & Clinical Cancer Research. 2017;36(1):p. 186. doi: 10.1186/s13046-017-0656-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Z., Wang C., Prendergast G., Pestell R. G. Cyclin D1 functions in cell migration. Cell Cycle. 2006;5(21):2440–2442. doi: 10.4161/cc.5.21.3428. [DOI] [PubMed] [Google Scholar]
- 58.Pavlenko I. A., Zavalishina L. E., Povilaitite P. E. HER2/neu gene amplification as a mechanism of clonal heterogeneity in breast cancer. Arkhiv Patologii. 2019;81(6):p. 49. doi: 10.17116/patol20198106149. [DOI] [PubMed] [Google Scholar]
- 59.Gutierrez C., Schiff R., medicine L. HER2: biology, detection, and clinical implications. Archives of Pathology & Laboratory Medicine. 2011;135(1):55–62. doi: 10.5858/2010-0454-rar.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abdou A. G., Kandil M., Asaad N. Y., et al. The prognostic role of Ezrin and HER2/neu expression in osteosarcoma. Applied Immunohistochemistry & Molecular Morphology. 2016;24(5):355–363. doi: 10.1097/pai.0000000000000197. [DOI] [PubMed] [Google Scholar]
- 61.Wang L.-F., Zhou Y., Xu Y.-M., et al. A caspase-6 and anti-HER2 antibody chimeric tumor-targeted proapoptotic molecule decreased metastasis of human osteosarcoma. Cancer Investigation. 2009;27(7):774–780. doi: 10.1080/07357900802427935. [DOI] [PubMed] [Google Scholar]
- 62.Li B., Zhou P., Xu K., et al. Metformin induces cell cycle arrest, apoptosis and autophagy through ROS/JNK signaling pathway in human osteosarcoma. International Journal of Biological Sciences. 2020;16(1):74–84. doi: 10.7150/ijbs.33787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stewart Z. A., Westfall M. D., Pietenpol J. A. Cell-cycle dysregulation and anticancer therapy. Trends in Pharmacological Sciences. 2003;24(3):139–145. doi: 10.1016/s0165-6147(03)00026-9. [DOI] [PubMed] [Google Scholar]
- 64.Catanzaro D., Ragazzi E., Vianello C., Caparrotta L., MJNpc M. Effect of quercetin on cell cycle and cyclin expression in ovarian carcinoma and osteosarcoma cell lines. SAGE Journal. 2015;10(8) doi: 10.1177/1934578x1501000813. [DOI] [PubMed] [Google Scholar]
- 65.Zimmermann K. C., Bonzon C., Green D. R. The machinery of programmed cell death. Pharmacology & Therapeutics. 2001;92(1):57–70. doi: 10.1016/s0163-7258(01)00159-0. [DOI] [PubMed] [Google Scholar]
- 66.Tanuma S.-I., Shibui Y., Oyama T., Uchiumi F., Abe H. Targeting poly (ADP-ribose) glycohydrolase to draw apoptosis codes in cancer. Biochemical Pharmocology. 2019;167 doi: 10.1016/j.bcp.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 67.Chaiyawat P., Pruksakorn D., Phanphaisarn A., Teeyakasem P., Klangjorhor J., Settakorn J. Expression patterns of class I histone deacetylases in osteosarcoma: a novel prognostic marker with potential therapeutic implications. Modern Pathology. 2018;31(2):264–274. doi: 10.1038/modpathol.2017.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun D., Yu M., Li Y., et al. Histone deacetylase 2 is involved in DNA damage‐mediated cell death of human osteosarcoma cells through stimulation of the ATM/p53 pathway. FEBS Open Bio. 2019;9(3):478–489. doi: 10.1002/2211-5463.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cao K., Wang H., Fang Y., et al. Histone deacetylase 4 promotes osteosarcoma cell proliferation and invasion by regulating expression of proliferating cell nuclear antigen. Frontiers in Oncology. 2019;9 doi: 10.3389/fonc.2019.00870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cao J., Liu X., Yang Y., et al. Decylubiquinone suppresses breast cancer growth and metastasis by inhibiting angiogenesis via the ROS/p53/Bai1 signaling pathway. Angiogenesis. 2020;23:1–14. doi: 10.1007/s10456-020-09707-z. [DOI] [PubMed] [Google Scholar]
- 71.Kashyap V. K., Dan N., Chauhan N., et al. VERU-111 suppresses tumor growth and metastatic phenotypes of cervical cancer cells through the activation of p53 signaling pathway. Cancer Letters. 2020;470:64–74. doi: 10.1016/j.canlet.2019.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee S.-H., Lee S.-J., Jung Y. S., et al. Blocking of p53-Snail binding, promoted by oncogenic K-Ras, recovers p53 expression and function. Neoplasia. 2009;11(1):22–IN6. doi: 10.1593/neo.81006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Powell E., Piwnica-Worms D., Piwnica-Worms H. Contribution of p53 to metastasis. Cancer Discovery. 2014;4:405–414. doi: 10.1158/2159-8290.CD-13-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jin W., Zhou L., Yan B., et al. Theabrownin triggersDNAdamage to suppress human osteosarcoma U2OScells by activating p53 signalling pathway. Journal of Cellular and Molecular Medicine. 2018;22(9):4423–4436. doi: 10.1111/jcmm.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang T., Gong X., Jiang R., Li H., Du W., Kuang G. Ferulic acid inhibits proliferation and promotes apoptosis via blockage of PI3K/Akt pathway in osteosarcoma cell. American Journal of Translational Research. 2016;8(2):968–80. [PMC free article] [PubMed] [Google Scholar]
- 76.Cjijoph Z.-Z. Berberine induced apoptosis of human osteosarcoma cells by inhibiting phosphoinositide 3 kinase/protein kinase B (PI3K/Akt) signal pathway activation. Iranian Journal of Public Health. 2016;45(5):p. 578. [PMC free article] [PubMed] [Google Scholar]
- 77.Resnitzky D., Reed S. I., biology c. Different roles for cyclins D1 and E in regulation of the G1-to-S transition. Molecular and Cellular Biology. 1995;15(7):3463–3469. doi: 10.1128/mcb.15.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kline C. L. B., Van den Heuvel A. P. J., Allen J. E., Prabhu V. V., Dicker D. T., El-Deiry W. S. ONC201 kills solid tumor cells by triggering an integrated stress response dependent on ATF4 activation by specific eIF2α kinases. Science Signaling. 2016;9(415) doi: 10.1126/scisignal.aac4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu Y. W., Yang T., Zhao L., et al. Activation of Adenosine 2A receptor inhibits neutrophil apoptosis in an autophagy-dependent manner in mice with systemic inflammatory response syndrome. Scientific Reports. 2016;6(1):33614–33626. doi: 10.1038/srep33614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim S.-M., Kim E.-M., Ji K.-Y., et al. TREM2 acts as a tumor suppressor in colorectal carcinoma through wnt1/β-catenin and erk signaling. Cancers. 2019;11(9):p. 1315. doi: 10.3390/cancers11091315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin C. H., Ji T., Chen C.-F., Hoang B. H. Wnt signaling in osteosarcoma. Advances in Experimental Medicine and Biology. 2014;804:33–45. doi: 10.1007/978-3-319-04843-7_2. [DOI] [PubMed] [Google Scholar]
- 82.Malinauskas T., Aricescu A. R., Lu W., Siebold C., Jones E. Y. Modular mechanism of Wnt signaling inhibition by Wnt inhibitory factor 1. Nature Structural & Molecular Biology. 2011;18(8):886–893. doi: 10.1038/nsmb.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tang Q., Zhao H., Yang B., et al. WIF-1 gene inhibition and Wnt signal transduction pathway activation in NSCLC tumorigenesis. Oncology Letters. 2017;13(3):1183–1188. doi: 10.3892/ol.2017.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li W., Meng Z., Zou T., et al. MiR-374a activates Wnt/β-catenin signaling to promote osteosarcoma cell migration by targeting WIF-1. Pathology & Oncology. 2018;26:1–7. doi: 10.1007/s12253-018-0556-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Supplementary Table S1: the top 50 matched targets for each potential active compound of Dangshen were gathered from PharmMapper, and these targets were regarded as the potential targets of Dangshen. The relationship between the compounds and their matched targets is listed in Supplementary Table S1. Description of Supplementary Table S2: the genes related to osteosarcoma were collected from GeneCards and OMIM databases. The genes related to osteosarcoma are listed in Supplementary Table S2. Description of Supplementary Table S3: the organs with high expression of each therapeutic target were collected via BioGPS. The relationship between the therapeutic targets and their high expressed organs is listed in Supplementary Table S3. Description of Supplementary Table S4: we used the Gene Ontology (GO) database to clarify the possible biological mechanisms. The details of the GO enrichment analysis of biological process are listed in Supplementary Table S4. Description of Supplementary Table S5: we used the Gene Ontology (GO) database to clarify the possible biological mechanisms. The details of the GO enrichment analysis of cell component are listed in Supplementary Table S5. Description of Supplementary Table S6: we used the Gene Ontology (GO) database to clarify the possible biological mechanisms. The details of the GO enrichment analysis of molecular function are listed in Supplementary Table S6. Description of Supplementary Table S7: we used the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to extract biological information about functional classification, annotation, and enriched pathways of therapeutic targets. The details of the KEGG enrichment analysis are listed in Supplementary Table S7. .
Data Availability Statement
The data sets generated and analyzed during the present study are available from the corresponding author upon reasonable request.


