Abstract
Antibodies have proven to be central in the development of diagnostic methods over decades, moving from polyclonal antibodies to the milestone development of monoclonal antibodies. Although monoclonal antibodies play a valuable role in diagnosis, their production is technically demanding and can be expensive. The large size of monoclonal antibodies (150 kDa) makes their re-engineering using recombinant methods a challenge. Single-domain antibodies, such as “nanobodies,” are a relatively new class of diagnostic probes that originated serendipitously during the assay of camel serum. The immune system of the camelid family (camels, llamas, and alpacas) has evolved uniquely to produce heavy-chain antibodies that contain a single monomeric variable antibody domain in a smaller functional unit of 12–15 kDa. Interestingly, the same biological phenomenon is observed in sharks. Since a single-domain antibody molecule is smaller than a conventional mammalian antibody, recombinant engineering and protein expression in vitro using bacterial production systems are much simpler. The entire gene encoding such an antibody can be cloned and expressed in vitro. Single-domain antibodies are very stable and heat-resistant, and hence do not require cold storage, especially when incorporated into a diagnostic kit. Their simple genetic structure allows easy re-engineering of the protein to introduce new antigen-binding characteristics or attach labels. Here, we review the applications of single-domain antibodies in laboratory diagnosis and discuss the future potential in this area.
Keywords: Single-domain antibodies, Nanobodies, Monoclonal antibodies, Laboratory diagnosis
IMMUNOASSAYS IN GENERAL
Ligand binding assays are fundamental in laboratory medicine for measuring analytes and biomarkers. This class of assays exploits the binding reaction between an analyte or biomarker and a specific affinity reagent. In immunoassays, the affinity reagent is an antibody.
Immunoassays form the mainstay for protein biomarker measurements, and numerous proteins can be measured in healthy and diseased states. Some target proteins are abundantly present (>10 mg/mL), whereas others are found at very low concentrations (<1 pg/mL) in clinical samples. The development of a suitable immunoassay depends on the availability of the protein antigen and the generation of an immune response in the host animal and the subsequent production of antibodies. Owing to the inherent diversity of an immune response and the structure and binding affinity of different antibodies for the same antigen, antibodies used in one assay or platform behave differently from those used in another, unless they are the same clone of monoclonal antibody. There are numerous examples of variable results between different platforms for the same analyte, such as thyroid stimulating hormone and cancer antigen 19-9 [1, 2]. Post-translational modifications of a protein analyte might be another factor that could affect reactivity with an antibody and lead to variable results. The challenges of using conventional antibodies in the laboratory have been outlined by Goodman [3].
However, there are many problems with immunoassays. In general, antibodies can originate from polyclonal or monoclonal sources. Although they are easy to produce, polyclonal antibodies are variable by nature, and there can be batch-to-batch variations in sera. Polyclonal antibodies have the advantage of being able to recognize multiple epitopes of a complex antigen, but inconsistency in production has hindered their use. The development of monoclonal antibodies was a milestone in the evolution of ligand-based assays. Monoclonal antibodies recognize a single epitope and can be produced in a pure and homogeneous form indefinitely from a hybridoma. Although monoclonal antibodies have a valuable role in diagnosis, their production is technically demanding and can be expensive. Moreover, the size of monoclonal antibodies (150 kDa) makes their re-engineering using recombinant methods a challenge. There is thus a need to develop new robust and reliable antibody probes for laboratory diagnosis. Conventional antibodies or complementary nucleic acid sequences represent the most common form of probes for the detection of various target molecules. Through the years, there have been attempts to reduce antibodies into fragments, either via enzymatic digestion methods (e.g., using pepsin or papain) or via recombinant engineering methods, such as those employing fragment antigen binding (Fab), single chain variable fragment (ScFv), and fragment variable (Fv) [4]. The discovery of naturally occurring heavy chain-only antibodies (HCAbs) in camelids heralded a new era in antibody engineering [5, 6].
SINGLE-DOMAIN ANTIBODIES: GENERATION AND PROPERTIES
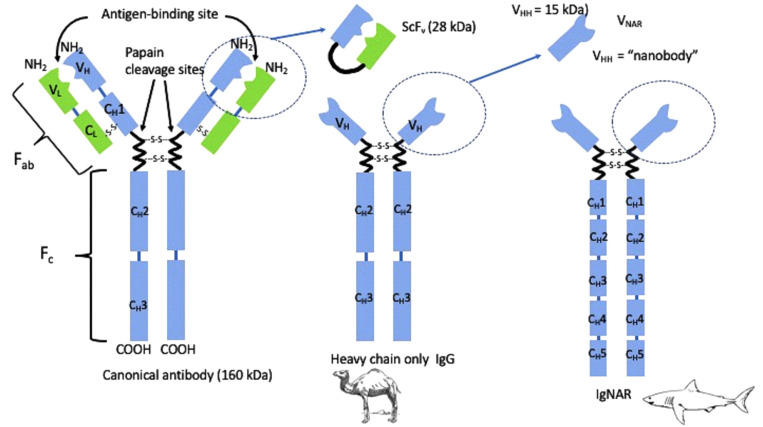
The classical/canonical antibody in vertebrates contains two identical heavy and two identical light chains (Fig. 1). The immune system of camelids (camels, dromedaries, llamas, guanacos, vicuñas, and alpacas) has evolved uniquely to produce dimeric HCAb of approximately 90 kDa that lack light chains and the CH1 domain (the first constant heavy chain domain) (Fig. 1). Among mammals, only members of the Camelidae family produce endogenous functional heavy-chain-only IgG. Interestingly, a similar biological phenomenon is observed in sharks [6, 7]. Cartilaginous fish, including nurse (Ginglymostoma cirratum), wobbegong (Orectolobus maculates), and dogfish (Squalus acanthias and Mustelus canis) sharks, also remarkably produce functional heavy-chain-only immunoglobulins (HCIgs), named Ig new antigen receptor (IgNARs). The IgNAR of cartilaginous fish is also a homodimer of two heavy chains with one variable domain and five constant domains. In cartilaginous fish, the IgNARs account only for approximately 5% of total Igs. Various camelids express different proportions of HCAb, ranging from 50–75% in camels to approximately 20–40% in llama.
Fig. 1.
Comparison of the canonical Ig structure with that of heavy-chain-only antibodies.
Abbreviations: VHH, variable heavy chain-only antibodies; CH, constant region of heavy chain; VL, variable region of light chain; VNAR, variable domain of immunoglobulin new antigen receptor; Ig-NAR, immunoglobulin new antigen receptor; ScFv, single chain variable fragment.
The serendipitous discovery of the natural occurrence of the unique, functional, homodimeric, HCIg in camelid serum resulted from a laboratory course for graduate students at the Vrije Universiteit Brussels, Belgium [5]. Unlike the canonical mammalian antibody, HCIg contains a single intact antigen-binding domain (variable domain of heavy chain of HCIg, VHH). The counterpart to the VHH in cartilaginous fish is variable domain of IgNAR (V-NAR). This single-domain fragment contains only two hypervariable loop structures participating in antigen binding, whereas the VHH contains three hypervariable loops for antigen recognition. It has been postulated that the absence of the CH1 in these HCIgs resulted from the loss of a splice consensus signal at the 5´ end of the “CH1-hinge” intron during evolution [8, 9]. This altered splicing results in the joining of the CH2 domain of HCAb to the variable domain through a “hinge” region, which is unique to this class of antibodies [5].
There are two distinct types of hinges in the heavy chain-only IgG of camelids: the short hinge and the long hinge [5, 10]. The VHH resembles conventional VH domains but is distinct in sequence and structure in that its sequence contains a few critical amino acid substitutions in the region that normally interacts with the variable light chain domain (VL) [5, 11-14]. These substitutions of large hydrophobic amino acids (in VH) with smaller, hydrophilic amino acids (in VHH) are responsible for the soluble behavior of the VHH and its function in the absence of a VL partner.
Thus, these variable antigen-binding domains (VHH and V-NAR) are fully functional within a small 12–15 kDa unit [15]. The VHH contains approximately 120 amino acid residues, encoded by a gene of 360 bp. This gene can be cloned and subcloned easily. A protein size of approximately 13 kDa is well within the limits of bacterial expression and is comparable with ScFv (25 kDa), Fab fragments (57 kDa), and the intact IgG (150 kDa).
VHH molecules form the basis to generate small, recombinant, autonomous single-domain antigen-binding fragments. The term “nanobody” was coined by the company Ablynx (Ghent, Belgium) in 2003 as a trademark, but it is now being generally used to describe small, recombinant, autonomous single-domain antigen-binding proteins, because of their small size (13 kDa; 2.5 nm in diameter and 4 nm in length). These nanobodies comprise a relatively new class of diagnostic probes. The patent on the use of fragments derived from the HCAb originally filed by Vrije Universiteit Brussel expired in 2014, resulting in increased interest in the commercialization of nanobodies [15].
The process of generating nanobodies begins with the immunization of a camelid. Following the development of an immune response, B-lymphocytes are isolated either from the peripheral blood or from lymph nodes and used to isolate mRNA. The mRNA is then used in reverse transcription to produce cDNA and perform PCR with VHH-specific primers to amplify the VHH gene region, which is then cloned into a phage display expression vector to generate a library. Only primers for VHH are required, as opposed to the entire HCIg, because VHH alone contains the fully functional antigen-binding domain of the HCIg.
The process of animal immunization can also be bypassed through the construction of a naive or synthetic cDNA library. Thus, it is also possible to obtain VHHs from non-immune libraries [16], but immune libraries have greater diversity and usually yield VHHs with higher affinities [17]. Reactive VHHs that might bind weakly to the target can be modified by random or site-directed mutagenesis to isolate higher affinity binders.
Using the solid phase immobilized protein antigen (e.g., on magnetic beads), phage-expressing reactive nanobodies can be isolated through repeated cycles of screening and amplification in bacterial culture. Phage inserts can then be subcloned into bacterial or yeast expression vectors. The VHH gene can also be altered to a desired affinity and specificity by random mutagenesis or can be fused to a variety of short-peptide immunoassay tags (i.e., His6 [six histidine], c-myc [derived from the c-myc proto-oncogene], HA [derived from influenza hemagglutinin], Avi [biotin ligase target], ALFA [small α-helix tag], and C-tag [4-amino acid peptide]) or various enzymes (horseradish peroxidase, alkaline phosphatase, or hemolysin). The nanobody construct can be produced ad infinitum in microbial cultures in the range of mg to g/L, a highly efficient yield for protein production. Target-recognizing nanobody clones can then be assayed for reactivity and utility in a variety of immunoassays, including sandwich or competitive ELISA, chemiluminescent enzyme immunoassay (CLEIA), bioluminescent enzyme immunoassay (BLEIA), lateral flow assay (LFA), and microfluidic and electrochemical devices for use in point-of-care testing, among others.
A typical nanobody has affinities (equilibrium dissociation constant) in the nanomolar to picomolar range, making them highly suited for application in ligand-binding assays [10]. They can bind to antigens with comparable affinity to that of conventional IgG, even though they lack the light chains that make up the antigen-binding region in IgG [18]. Since the functional part of the entire nanobody molecule is smaller than that of a conventional mammalian antibody, recombinant engineering and protein expression in vitro using bacterial production systems is much simpler using nanobodies. The entire nanobody gene can be cloned and expressed in vitro. It was later discovered in 1997 that nanobodies retain binding properties after long incubations (two weeks) at 37°C [19]. Moreover, these antibodies are very stable and heat-resistant at even higher temperatures, meaning that cold storage is not required, especially if these are incorporated into a diagnostic kit. Their resistance to pH extremes gives nanobodies the potential to be developed into oral drugs [20]. Furthermore, the simple structure of the genes encoding them allows re-engineering of the antigenic characteristics of nanobodies to “humanize” them for therapeutic applications. The following characteristics make nanobodies potentially useful for developing reagents for laboratory diagnosis:
Low cost of production: the small size of nanobodies enables easier production and high yields in moderate volumes of bacterial culture. Expression can be periplasmic or cytoplasmic. Periplasmic expression allows disulfide bridges to form and the purification of proteins from periplasmic extracts at yields of 1–20 mg/L on average [21]. Cytoplasmic expression produces much higher yields at 60–200 mg/L.
Easy tailoring to meet the application requirements (i.e., to improve specificity and affinity for broadening detection possibilities): the genes encoding nanobodies can be easily re-engineered to select for altered binding properties or epitope tagging for an immunoassay configuration. Single-domain antibodies bind to targets with comparable affinity to that of many conventional antibodies, sometimes with dissociation constants in the low picomolar range [22]. A myriad of methods is available to increase the affinity or avidity of any given nanobody. The availability of cDNA for a particular nanobody allows easy insertion of protein tags or labels using standard recombination methods.
Robustness and long shelf live: nanobodies are exceptionally heat stable in comparison with Igs and ScFv fragments and can thus be easily shipped at most ambient temperatures [18, 23]. Their melting temperatures (Tm) can be as high as 80°C, and some can be engineered to have a Tm of even up to 90°C. Some VHHs, but not all, can refold and renature to 100% after denaturation.
Targeting cryptic or hidden epitopes: the small size of a nanobody allows it to enter antigen-binding sites in protein pockets and cavities that might not be accessible to conventional antibodies [24]. VHHs can bind to a variety of epitopes, from enzyme active sites [25, 26] to small molecules or haptens [27, 28].
Low immunogenicity [29, 30] and rapid blood clearance: the small size of a nanobody allows it to be freely filtered in the glomerulus, facilitating excretion. Therefore, for non-invasive in vivo imaging or therapeutic applications, if a nanobody is tagged with an anti-cancer molecule, residual toxicity will be minimized as the nanobody gets excreted. Nanobodies have low immunogenicity due to their sequence similarity with human IgG [31].
THREE-DIMENSIONAL STRUCTURE OF NANOBODIES IN COMPLEX WITH ANTIGENS
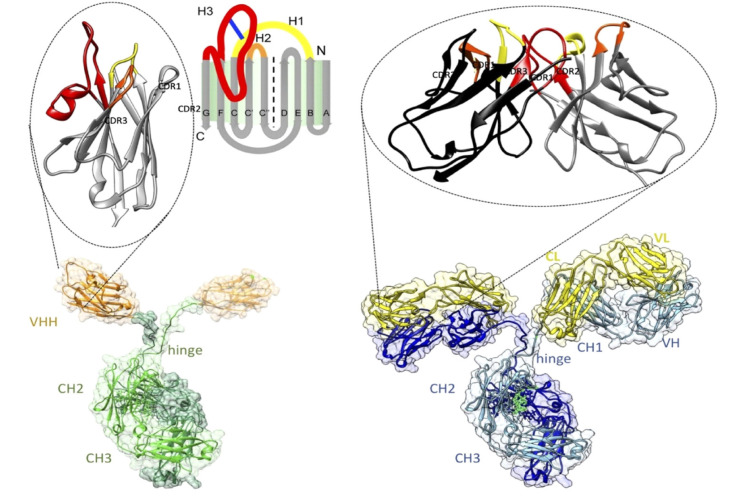
The average affinity of a nanobody for its antigen is approximately 6 nM, which is comparable with the affinity of the monomeric antigen-binding sites (Fab or ScFv) of conventional antibodies for their antigens [32]. The structures of numerous nanobodies have been revealed by X-ray crystallography [32]. VHH contains an IgV fold with nine β-strands and a conserved disulfide bond between Cys23 and Cys104 (International ImMunoGeneTics [IMGT] information system numbering; Fig. 2). The V domain contains three hypervariable loops linked by four conserved framework regions. The paratopes on nanobodies are enriched with aromatic residues similar to conventional antibodies. The interactions between nanobodies and antigens are mediated by the three complementarity-determining region (CDR) loops and dominated by the CDR3 loop. The antigen epitopes tend to be more rigid and concave and are also enriched with aromatic residues. In contrast, conventional antibodies use six CDR loops (three in the variable domain of the heavy chain and three in the variable domain of the light chain) for antigen binding [32]. These structural insights are instrumental for the rational-design engineering of nanobodies into more potent affinity reagents [33].
Fig. 2.
Architectures of homodimeric heavy‐chain antibodies (bottom left) with the Ag‐binding single‐domain VHH enlarged on top and classical heterotetrameric antibodies (bottom right) with the Ag‐binding variable fragments (comprising VH and VL domains) enlarged on top. Adapted from Muyldermans (2021, ref 33). (Permission under Creative Commons Attribution license) (https://doi.org/10.1111/febs.15515).
Abbreviations: VHH, variable heavy chain-only antibodies; CH, constant region of heavy chain; VL, variable region of light chain; CDR, complementarity-determining region.
APPLICATION OF NANOBODIES AND ASSAY PERFORMANCE
The question regarding which immunoassay applications benefit most from the substitution of classical antibodies with nanobodies remains. Nanobodies have been introduced in a wide variety of laboratory diagnostic techniques, mainly those for infectious diseases (Table 1). Considering their low cost of production and robust behavior (thermoresistance, long shelf life even in the absence of a cold chain), nanobodies are expected to become a preferred affinity reagent in future affordable LFAs. Nanobodies are perhaps less applicable for the “pregnancy test” type of applications but more applicable to assays that are used to monitor infectious diseases in animals (both farm and wild-life animals) living in remote areas. Efforts were made to monitor trypanosome and dengue fever using nanobodies [34, 35].The lack of applicability to techniques, such as a pregnancy/HCG detection test, relates to the long-established footprint of conventional monoclonal antibody usage, and investment in HCG detection will make it difficult for nanobodies to replace monoclonal antibodies for these techniques.
Table 1.
Selected examples of the use of nanobodies (and V-NAR) in disease diagnosis
| Antigen (target protein or small molecule) | Application | Method | Reference |
|---|---|---|---|
| Detection of parasite or fungal infection | |||
| Malarial apical membrane antigen-1 | Plasmodium falciparum | Immunofluorescence (V-NAR) | [55] |
| Trypanosomal pyruvate kinase | Trypanosoma congolense | ELISA and LFA | [35] |
| Paraflagellar rod protein | All trypanosome species (Trypanosoma evansi, Trypanosoma congolense, Trypanosoma brucei, Trypanosoma vivax) | ELISA and immunofluorescence | [56] |
| Glycosomal aldolase | Trypanosoma congolense | ELISA | [57, 58] |
| Iron superoxide dismutase 1, tryparedoxin 1, nuclear transport factor 2 | Leishmania infantum | ELISA | [59] |
| Fasciola excretory secretory protein | Fasciola hepatica | ELISA | [60] |
| Taenia solium 14 kDa diagnostic glycoprotein | Taenia solium | Sandwich ELISA | [61] |
| Excretory secretory protein | Toxocara canis | Sandwich ELISA and electrochemical magnetosensor | [49, 50, 62] |
| Alternaria mycotoxin tenuazonic acid | Alternaria | CLEIA and BLEIA | [63] |
| Detection of bacterial infection | |||
| Chaperonin GroEL | Brucella | ELISA | [20] |
| Flagella | Campylobacter jejuni or Campylobacter coli | Fluorescence microscopy/immunoblotting | [64, 65] |
| Listeria monocytogenes | Sandwich ELISA | [66] | |
| Shiga toxin type 2 (B domain) | Shigella dysenteriae | Sandwich ELISA | [67] |
| Cholera toxin | Vibrio cholerae | V-NAR Sandwich ELISA | [68] |
| Detection of viral infection | |||
| Dengue virus type 2 NS1 protein | Dengue virus | LFA | [34] |
| H5N1 | Influenza H5N1 | Double nanobody Sandwich ELISA | [69] |
| E2/E3E2 envelope protein | Western equine encephalitis virus | Sandwich ELISA | [70] |
| FMDV 3ABC protein and synthetic peptides | Foot-and-mouth disease virus | Competitive ELISA | [71] |
| PEDV N protein | Porcine epidemic diarrhea virus | Blocking ELISA | [72] |
| Ebola virus nuclear protein | Zaire Ebola virus | V-NAR ELISA | [73] |
| Detection of small toxic molecules | |||
| Caffeine | Caffeine contamination | Competitive ELISA | [27] |
| Biphenyl 2,3-dioxygenase | Oil refinery waste treatment | Western blot | [36] |
| Parathion | Organophosphorous pesticide detection | One-step direct competitive fluorescent immunoassay | [37] |
| 3-Phenoxybenzoic acid | Detection of pyrethroid insecticides in urine | One-step direct competitive fluorescent immunoassay | [38] |
| Tetrabromobisphenol | Flame retardant | Competitive ELISA | [40] |
| Dicamba | Contamination with selective herbicide | CLEIA | [39] |
| Detection of human disease or malignancy | |||
| Human glycophorin A (CD235a) | Anti-HIV-1 p24 antibodies | Anti-CD235a VHH fused to HIV-1 p24–agglutination for HIV diagnosis | [74] |
| Alpha-fetoprotein | Cancer biomarker | ELISA and immuno-PCR | [75] |
| Pancreatic secretory zymogen-granule glycoprotein 2 | Crohn’s disease | ELISA and immunohistochemistry | [76] |
| CEA | Cancer biomarker | Biosensor for cancer biomarker | [44] |
| Procalcitonin | Serum marker for bacterial infections | Electrochemiluminescence | [43] |
| Growth hormone | Anti-doping assay | Sandwich ELISA | [48] |
| CD22 | B-cell malignancies/leukemia | ELISA and FACS | [77] |
| hPSA | Prostate cancer | Sandwich ELISA/Surface plasma resonance-based assay | [45, 46] |
| CD38 | Soluble CD38 in multiple myeloma | Sandwich assay | [78] |
| Human β-2-microglobulin | Amyloid disease | Fluorescence immunostaining | [79] |
| In vivo non-invasive imaging | |||
| EGFR | Tumor solid burden | SPECT/microCT | [80] |
| CD33 | Acute myeloid leukemia | Non-invasive imaging (PET/SPECT) | [81] |
| HER2 | Breast cancer | [54] | |
| MMR | Tumor-associated macrophages | SPECT/microCT and PET | [82, 83] |
| PSMA | Prostate cancer | SPECT/microCT | [84] |
| CD20 | Non-Hodgkin lymphoma | Theranostic | [85, 86] |
| DPP6 | Pancreatic endocrine cells | SPECT/CT | [87] |
| CA IX | Hypoxic ductal carcinoma | Molecular fluorescence imaging | [88] |
| Clec4F and Vsig4 | Kupffer cells, acute hepatitis, staging of liver pathogenesis | Immunohistochemistry and SPECT | [89] |
| CRIg | Rheumatoid arthritis/joint inflammation | SPECT/CT | [90, 91] |
| VCAM1 | Atherosclerotic lesions | SPECT imaging | [92] |
| Fibronectin | Breast cancer, melanoma | PET/CT | [93] |
Abbreviations: LFA, lateral flow assay; SPECT, single-photon emission computed tomography; CT, computed tomography; CLEIA, chemiluminescence enzyme immunoassay; BLEIA, bioluminescent enzyme immunoassay; MMR, macrophage mannose receptor; V-NAR, variable domain of immunoglobulin new antigen receptor. PEDV, porcine epidemic diarrhea virus; FMDV, foot-and-mouth disease virus; VHH, variable heavy chain-only antibodies (also known as single-domain antibodies); HIV, human immunodeficiency virus; PCR, polymerase chain reaction; FACS, fluorescent activated cell sorting; hPSA, human prostate specific antigen; EGFR, epidermal growth factor receptor; PET, positron emission tomography; CA IX, carbonic anhydrase 9; PSMA, prostate-specific membrane antigen; NS1, non-structural protein 1.
In addition to LFAs, nanobodies have been assessed for the detection of various targets in ELISA-based methods. Although the generation of nanobodies against haptens is challenging, nanobodies against multiple small organic compounds have been identified [27, 36-39]. These nanobodies, combined with sensitive detection technologies or in competitive ELISA formats, appear to be a valuable tool for monitoring contaminants in soil or on food. For this, first, contaminating herbicides, fungicides, or insecticides must be extracted from soil or food. Many of these contaminants are hydrophobic and poorly soluble in aqueous solutions, and their extraction in dimethyl sulfoxide, methanol, acetone, and other organic solvents is not compatible with the proteinaceous probes. However, stable nanobodies appear to be resistant to exposure to such non-physiological solutes [40-42].
The diagnostic sensitivities of LFAs and standard sandwich ELISA for particularly difficult-to-measure targets (i.e., glycosylated, biomarkers involved in variable assemblies or when present at extremely low concentrations), seem to be inadequate. In such cases, it might be necessary to switch to more sensitive detection methods, such as CLEIA or BLEIA, or more sophisticated new diagnostic instruments, such as (magneto-) electrochemical sensors [39, 43, 44]. The flexible format and small size of nanobodies allows for tailoring and adaptation for directional coupling at a high density on magnetic beads or on the sensor layer of novel biosensors [45, 46]. For example, the prostate specific antigen (PSA) sandwich immunosensor has a detection limit of 0.08 ng/mL and range of 0.1–100 ng/mL while the surface plasmon resonance assay for PSA has a detection limit of 0.3 ng/mL, well below the clinical-detection lower limit of 4 ng/mL [45, 46]. This contrasts with the higher detection limits of various automated platforms using conventional antibodies [47]. Similar advantages in sensitivity were seen with a growth hormone assay utilizing a nanobody, wherein a detection range of 0.5–110 ng/mL was achieved [48]. Our efforts to design better performing assays culminated in a recent highly reproducible sensitive and specific assay for Toxocara excretory secretory components [49, 50].
CONCLUSIONS
Although many efforts have been undertaken in academic institutions to introduce nanobodies in standard ELISAs to monitor biomarkers for human health (Table 1), it will be extremely difficult to substitute the well-established classical monoclonal antibodies in these applications (refer to the HCG test example). Nevertheless, there might be one notable exception, specifically in vivo non-invasive imaging [51]. The small size of monomeric nanobodies allows them to rapidly extravasate from veins and diffuse evenly into tissues to reach their targets. At the same time, excess nanobodies are rapidly cleared from the blood via the kidneys. Hence, radionuclide-labeled nanobodies injected intravenously distribute throughout the body, whereas kidney clearance removes excess free nanobody, with a minor fraction accumulating at the diseased sites in the body, based on the specificity of the nanobody. If extended half-life is desired to improve imaging, a nanobody can be coupled to other proteins [52, 53]. The lesions or spots of diseased, infected, or inflamed tissue will be loaded with labeled nanobodies that can be monitored via PET or single-photon emission computed tomography whole body scans. This highly promising non-invasive imaging technique has been used in mouse studies with nanobodies against cancer biomarkers and for sites of inflammation (Table 1). A phase I study has been reported for anti-HER2 nanobodies in breast cancer patients [54]. As illustrated in this review, increasing future applications of nanobodies for diagnostic purposes are likely in the clinical laboratory, particularly in the context of infectious disease, as well as for imaging.
Footnotes
AUTHOR CONTRIBUTIONS
Pillay TS conceived the idea for the review article and produced the first draft. Muyldermans S reviewed and modified the first draft extensively for the final submission.
CONFLICTS OF INTEREST
None declared
RESEARCH FUNDING
Funding was provided by the National Research Foundation, South Africa.
REFERENCES
- 1.Rawlins ML, Roberts WL. Performance characteristics of six third-generation assays for thyroid-stimulating hormone. Clin Chem. 2004;50:2338–44. doi: 10.1373/clinchem.2004.039156. [DOI] [PubMed] [Google Scholar]
- 2.La'ulu SL, Roberts WL. Performance characteristics of five automated CA 19-9 assays. Am J Clin Pathol. 2007;127:436–40. doi: 10.1309/H52VET3M6P7GYWG1. [DOI] [PubMed] [Google Scholar]
- 3.Goodman SL. The antibody horror show: an introductory guide for the perplexed. N Biotechnol. 2018;45:9–13. doi: 10.1016/j.nbt.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 4.O'Kennedy R, Roben P. Antibody engineering: an overview. Essays Biochem. 1991;26:59–75. doi: 10.1243/03093247V261075. [DOI] [PubMed] [Google Scholar]
- 5.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–8. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 6.Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature. 1995;374:168–73. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 7.Roux KH, Greenberg AS, Greene L, Strelets L, Avila D, McKinney EC, et al. Structural analysis of the nurse shark (new) antigen receptor (NAR): molecular convergence of NAR and unusual mammalian immunoglobulins. Proc Natl Acad Sci U S A. 1998;95:11804–9. doi: 10.1073/pnas.95.20.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen VK, Hamers R, Wyns L, Muyldermans S. Loss of splice consensus signal is responsible for the removal of the entire C(H)1 domain of the functional camel IGG2A heavy-chain antibodies. Mol Immunol. 1999;36:515–24. doi: 10.1016/S0161-5890(99)00067-X. [DOI] [PubMed] [Google Scholar]
- 9.Woolven BP, Frenken LG, van der Logt P, Nicholls PJ. The structure of the llama heavy chain constant genes reveals a mechanism for heavy-chain antibody formation. Immunogenetics. 1999;50:98–101. doi: 10.1007/s002510050694. [DOI] [PubMed] [Google Scholar]
- 10.van der Linden R, de Geus B, Stok W, Bos W, van Wassenaar D, Verrips T, et al. Induction of immune responses and molecular cloning of the heavy chain antibody repertoire of Lama glama. J Immunol Methods. 2000;240:185–95. doi: 10.1016/S0022-1759(00)00188-5. [DOI] [PubMed] [Google Scholar]
- 11.Vu KB, Ghahroudi MA, Wyns L, Muyldermans S. Comparison of llama VH sequences from conventional and heavy chain antibodies. Mol Immunol. 1997;34:1121–31. doi: 10.1016/S0161-5890(97)00146-6. [DOI] [PubMed] [Google Scholar]
- 12.Harmsen MM, Ruuls RC, Nijman IJ, Niewold TA, Frenken LG, de Geus B. Llama heavy-chain V regions consist of at least four distinct subfamilies revealing novel sequence features. Mol Immunol. 2000;37:579–90. doi: 10.1016/S0161-5890(00)00081-X. [DOI] [PubMed] [Google Scholar]
- 13.Decanniere K, Muyldermans S, Wyns L. Canonical antigen-binding loop structures in immunoglobulins: more structures, more canonical classes? J Mol Biol. 2000;300:83–91. doi: 10.1006/jmbi.2000.3839. [DOI] [PubMed] [Google Scholar]
- 14.Muyldermans S, Atarhouch T, Saldanha J, Barbosa JA, Hamers R. Sequence and structure of VH domain from naturally occurring camel heavy chain immunoglobulins lacking light chains. Protein Eng. 1994;7:1129–35. doi: 10.1093/protein/7.9.1129. [DOI] [PubMed] [Google Scholar]
- 15.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–97. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 16.Verheesen P, Roussis A, de Haard HJ, Groot AJ, Stam JC, den Dunnen JT, et al. Reliable and controllable antibody fragment selections from Camelid non-immune libraries for target validation. Biochim Biophys Acta. 2006;1764:1307–19. doi: 10.1016/j.bbapap.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen VK, Desmyter A, Muyldermans S. Functional heavy-chain antibodies in Camelidae. Adv Immunol. 2001;79:261–96. doi: 10.1016/S0065-2776(01)79006-2. [DOI] [PubMed] [Google Scholar]
- 18.van der Linden RH, Frenken LG, de Geus B, Harmsen MM, Ruuls RC, Stok W, et al. Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim Biophys Acta. 1999;1431:37–46. doi: 10.1016/S0167-4838(99)00030-8. [DOI] [PubMed] [Google Scholar]
- 19.Arbabi Ghahroudi M, Desmyter A, Wyns L, Hamers R, Muyldermans S. Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS Lett. 1997;414:521–6. doi: 10.1016/S0014-5793(97)01062-4. [DOI] [PubMed] [Google Scholar]
- 20.Abbady AQ, Al-Daoude A, Al-Mariri A, Zarkawi M, Muyldermans S. Chaperonin GroEL a Brucella immunodominant antigen identified using Nanobody and MALDI-TOF-MS technologies. Vet Immunol Immunopathol. 2012;146:254–63. doi: 10.1016/j.vetimm.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Saerens D, Stijlemans B, Baral TN, Nguyen Thi GT, Wernery U, Magez S, et al. Parallel selection of multiple anti-infectome Nanobodies without access to purified antigens. J Immunol Methods. 2008;329:138–50. doi: 10.1016/j.jim.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Sockolosky JT, Dougan M, Ingram JR, Ho CC, Kauke MJ, Almo SC, et al. Durable antitumor responses to CD47 blockade require adaptive immune stimulation. Proc Natl Acad Sci U S A. 2016;113:E2646–54. doi: 10.1073/pnas.1604268113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dumoulin M, Conrath K, Van Meirhaeghe A, Meersman F, Heremans K, Frenken LG, et al. Single-domain antibody fragments with high conformational stability. Protein Sci. 2002;11:500–15. doi: 10.1110/ps.34602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lauwereys M, Arbabi Ghahroudi M, Desmyter A, Kinne J, Hölzer W, De Genst E, et al. Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. EMBO J. 1998;17:3512–20. doi: 10.1093/emboj/17.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desmyter A, Decanniere K, Muyldermans S, Wyns L. Antigen specificity and high affinity binding provided by one single loop of a camel single-domain antibody. J Biol Chem. 2001;276:26285–90. doi: 10.1074/jbc.M102107200. [DOI] [PubMed] [Google Scholar]
- 26.Desmyter A, Transue TR, Ghahroudi MA, Thi MH, Poortmans F, Hamers R, et al. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat Struct Biol. 1996;3:803–11. doi: 10.1038/nsb0996-803. [DOI] [PubMed] [Google Scholar]
- 27.Ladenson RC, Crimmins DL, Landt Y, Ladenson JH. Isolation and characterization of a thermally stable recombinant anti-caffeine heavy-chain antibody fragment. Anal Chem. 2006;78:4501–8. doi: 10.1021/ac058044j. [DOI] [PubMed] [Google Scholar]
- 28.Spinelli S, Frenken LG, Hermans P, Verrips T, Brown K, Tegoni M, et al. Camelid heavy-chain variable domains provide efficient combining sites to haptens. Biochemistry. 2000;39:1217–22. doi: 10.1021/bi991830w. [DOI] [PubMed] [Google Scholar]
- 29.Kijanka M, Dorresteijn B, Oliveira S, van Bergen en Henegouwen PM. Nanobody-based cancer therapy of solid tumors. Nanomedicine (Lond) 2015;10:161–74. doi: 10.2217/nnm.14.178. [DOI] [PubMed] [Google Scholar]
- 30.Vincke C, Loris R, Saerens D, Martinez-Rodriguez S, Muyldermans S, Conrath K. General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J Biol Chem. 2009;284:3273–84. doi: 10.1074/jbc.M806889200. [DOI] [PubMed] [Google Scholar]
- 31.Klarenbeek A, El Mazouari K, Desmyter A, Blanchetot C, Hultberg A, de Jonge N, et al. Camelid Ig V genes reveal significant human homology not seen in therapeutic target genes, providing for a powerful therapeutic antibody platform. MAbs. 2015;7:693–706. doi: 10.1080/19420862.2015.1046648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zavrtanik U, Lukan J, Loris R, Lah J, Hadži S. Structural basis of epitope recognition by heavy-chain camelid antibodies. J Mol Biol. 2018;430:4369–86. doi: 10.1016/j.jmb.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Muyldermans S. A guide to: generation and design of nanobodies. FEBS J. 2021;288:2084–102. doi: 10.1111/febs.15515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fatima A, Wang H, Kang K, Xia L, Wang Y, Ye W, et al. Development of VHH antibodies against dengue virus type 2 NS1 and comparison with monoclonal antibodies for use in immunological diagnosis. PLoS One. 2014;9:e95263. doi: 10.1371/journal.pone.0095263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinto Torres JE, Goossens J, Ding J, Li Z, Lu S, Vertommen D, et al. Development of a nanobody-based lateral flow assay to detect active Trypanosoma congolense infections. Sci Rep. 2018;8:9019. doi: 10.1038/s41598-018-26732-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fraile S, Jiménez JI, Gutiérrez C, de Lorenzo V. NanoPad: an integrated platform for bacterial production of camel nanobodies aimed at detecting environmental biomarkers. Proteomics. 2013;13:2766–75. doi: 10.1002/pmic.201300009. [DOI] [PubMed] [Google Scholar]
- 37.Zhang YQ, Xu ZL, Wang F, Cai J, Dong JX, Zhang JR, et al. Isolation of Bactrian camel single domain antibody for parathion and development of one-step dc-FEIA method using VHH-alkaline phosphatase fusion protein. Anal Chem. 2018;90:12886–92. doi: 10.1021/acs.analchem.8b03509. [DOI] [PubMed] [Google Scholar]
- 38.Huo J, Li Z, Wan D, Li D, Qi M, Barnych B, et al. Development of a highly sensitive direct competitive fluorescence enzyme immunoassay based on a nanobody-alkaline phosphatase fusion protein for detection of 3-phenoxybenzoic acid in urine. J Agric Food Chem. 2018;66:11284–90. doi: 10.1021/acs.jafc.8b04521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huo J, Barnych B, Li Z, Wan D, Li D, Vasylieva N, et al. Hapten synthesis, antibody development, and a highly sensitive indirect competitive chemiluminescent enzyme immunoassay for detection of dicamba. J Agric Food Chem. 2019;67:5711–9. doi: 10.1021/acs.jafc.8b07134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim HJ, McCoy MR, Majkova Z, Dechant JE, Gee SJ, Tabares-da Rosa S, et al. Isolation of alpaca anti-hapten heavy chain single domain antibodies for development of sensitive immunoassay. Anal Chem. 2012;84:1165–71. doi: 10.1021/ac2030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, Bever CR, Majkova Z, Dechant JE, Yang J, Gee SJ, et al. Heterologous antigen selection of camelid heavy chain single domain antibodies against tetrabromobisphenol A. Anal Chem. 2014;86:8296–302. doi: 10.1021/ac5017437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang JR, Wang Y, Dong JX, Yang JY, Zhang YQ, Wang F, et al. Development of a simple pretreatment immunoassay based on an organic solvent-tolerant nanobody for the detection of carbofuran in vegetable and fruit samples. Biomolecules. 2019;9:576. doi: 10.3390/biom9100576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Sun Y, Elseviers J, Muyldermans S, Liu S, Wan Y. A nanobody-based electrochemiluminescent immunosensor for sensitive detection of human procalcitonin. Analyst. 2014;139:3718–21. doi: 10.1039/c4an00626g. [DOI] [PubMed] [Google Scholar]
- 44.Liu JL, Raghu D, Anderson GP, Goldman ER, Christodoulides JA, Raphael MP. Improving biosensing activity to carcinoembryonic antigen with orientated single domain antibodies. Heliyon. 2017;3:e00478. doi: 10.1016/j.heliyon.2017.e00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saerens D, Frederix F, Reekmans G, Conrath K, Jans K, Brys L, et al. Engineering camel single-domain antibodies and immobilization chemistry for human prostate-specific antigen sensing. Anal Chem. 2005;77:7547–55. doi: 10.1021/ac051092j. [DOI] [PubMed] [Google Scholar]
- 46.Liu X, Wang D, Chu J, Xu Y, Wang W. Sandwich pair nanobodies, a potential tool for electrochemical immunosensing serum prostate-specific antigen with preferable specificity. J Pharm Biomed Anal. 2018;158:361–9. doi: 10.1016/j.jpba.2018.06.021. [DOI] [PubMed] [Google Scholar]
- 47.Bock JL, Klee GG. How sensitive is a prostate-specific antigen measurement? How sensitive does it need to be? Arch Pathol Lab Med. 2004;128:341–3. doi: 10.5858/2004-128-341-HSIAPA. [DOI] [PubMed] [Google Scholar]
- 48.Murad H, Assaad JM, Al-Shemali R, Abbady AQ. Exploiting nanobodies in the detection and quantification of human growth hormone via phage-sandwich enzyme-linked immunosorbent assay. Front Endocrinol (Lausanne) 2017;8:115. doi: 10.3389/fendo.2017.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morales-Yánez F, Trashin S, Hermy M, Sariego I, Polman K, Muyldermans S, et al. Fast one-step ultrasensitive detection of Toxocara canis antigens by a nanobody-based electrochemical magnetosensor. Anal Chem. 2019;91:11582–8. doi: 10.1021/acs.analchem.9b01687. [DOI] [PubMed] [Google Scholar]
- 50.Morales-Yánez F, Trashin S, Sariego I, Roucher C, Paredis L, Chico M, et al. Electrochemical detection of Toxocara canis excretory-secretory antigens in children from rural communities in Esmeraldas Province, Ecuador: association between active infection and high eosinophilia. Parasit Vectors. 2020;13:245. doi: 10.1186/s13071-020-04113-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaneycken I, D'Huyvetter M, Hernot S, De Vos J, Xavier C, Devoogdt N, et al. Immuno-imaging using nanobodies. Curr Opin Biotechnol. 2011;22:877–81. doi: 10.1016/j.copbio.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 52.Hutt M, Färber-Schwarz A, Unverdorben F, Richter F, Kontermann RE. Plasma half-life extension of small recombinant antibodies by fusion to immunoglobulin-binding domains. J Biol Chem. 2012;287:4462–9. doi: 10.1074/jbc.M111.311522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Unverdorben F, Färber-Schwarz A, Richter F, Hutt M, Kontermann RE. Half-life extension of a single-chain diabody by fusion to domain B of staphylococcal protein A. Protein Eng Des Sel. 2012;25:81–8. doi: 10.1093/protein/gzr061. [DOI] [PubMed] [Google Scholar]
- 54.Keyaerts M, Xavier C, Heemskerk J, Devoogdt N, Everaert H, Ackaert C, et al. Phase I study of 68Ga-HER2-nanobody for PET/CT assessment of HER2 expression in breast carcinoma. J Nucl Med. 2016;57:27–33. doi: 10.2967/jnumed.115.162024. [DOI] [PubMed] [Google Scholar]
- 55.Nuttall SD, Humberstone KS, Krishnan UV, Carmichael JA, Doughty L, Hattarki M, et al. Selection and affinity maturation of IgNAR variable domains targeting Plasmodium falciparum AMA1. Proteins. 2004;55:187–97. doi: 10.1002/prot.20005. [DOI] [PubMed] [Google Scholar]
- 56.Obishakin E, Stijlemans B, Santi-Rocca J, Vandenberghe I, Devreese B, Muldermans S, et al. Generation of a nanobody targeting the paraflagellar rod protein of trypanosomes. PLoS One. 2014;9:e115893. doi: 10.1371/journal.pone.0115893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Odongo S, Sterckx YG, Stijlemans B, Pillay D, Baltz T, Muyldermans S, et al. An anti-proteome nanobody library approach yields a specific immunoassay for Trypanosoma congolense diagnosis targeting glycosomal aldolase. PLoS Negl Trop Dis. 2016;10:e0004420. doi: 10.1371/journal.pntd.0004420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pinto J, Odongo S, Lee F, Gaspariunaite V, Muyldermans S, Magez S, et al. Structural basis for the high specificity of a Trypanosoma congolense immunoassay targeting glycosomal aldolase. PLoS Negl Trop Dis. 2017;11:e0005932. doi: 10.1371/journal.pntd.0005932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abeijon C, Dilo J, Tremblay JM, Viana AG, Bueno LL, Carvalho SFG, et al. Use of VHH antibodies for the development of antigen detection test for visceral leishmaniasis. Parasite Immunol. 2018;40:e12584. doi: 10.1111/pim.12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barreto T, Alfonso Y, Lafaye P, García Lazaro MDP, Agueda Perez L, Herrera-Velit P, et al. Single-chain antibodies from alpaca for the detection of Fasciola hepatica antigens. Rev Peru Med Exp Salud Publica. 2018;35:573–80. doi: 10.17843/rpmesp.2018.354.3101. [DOI] [PubMed] [Google Scholar]
- 61.Deckers N, Saerens D, Kanobana K, Conrath K, Victor B, Wernery U, et al. Nanobodies, a promising tool for species-specific diagnosis of Taenia solium cysticercosis. Int J Parasitol. 2009;39:625–33. doi: 10.1016/j.ijpara.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 62.Morales-Yanez FJ, Sariego I, Vincke C, Hassanzadeh-Ghassabeh G, Polman K, Muyldermans S. An innovative approach in the detection of Toxocara canis excretory/secretory antigens using specific nanobodies. Int J Parasitol. 2019;49:635–45. doi: 10.1016/j.ijpara.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 63.Wang F, Li ZF, Yang YY, Wan DB, Vasylieva N, Zhang YQ, et al. Chemiluminescent enzyme immunoassay and bioluminescent enzyme immunoassay for tenuazonic acid mycotoxin by exploitation of nanobody and nanobody-nanoluciferase fusion. Anal Chem. 2020;92:11935–42. doi: 10.1021/acs.analchem.0c02338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Riazi A, Strong PC, Coleman R, Chen W, Hirama T, van Faassen H, et al. Pentavalent single-domain antibodies reduce Campylobacter jejuni motility and colonization in chickens. PLoS One. 2013;8:e83928. doi: 10.1371/journal.pone.0083928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hussack G, Riazi A, Ryan S, van Faassen H, MacKenzie R, Tanha J, et al. Protease-resistant single-domain antibodies inhibit Campylobacter jejuni motility. Protein Eng Des Sel. 2014;27:191–8. doi: 10.1093/protein/gzu011. [DOI] [PubMed] [Google Scholar]
- 66.Tu Z, Chen Q, Li Y, Xiong Y, Xu Y, Hu N, et al. Identification and characterization of species-specific nanobodies for the detection of Listeria monocytogenes in milk. Anal Biochem. 2016;493:1–7. doi: 10.1016/j.ab.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 67.Melli LJ, Zylberman V, Hiriart Y, Lauche CE, Baschkier A, Pardo R, et al. Development and evaluation of a novel VHH-based immunocapture assay for high-sensitivity detection of Shiga toxin Type 2 (Stx2) in stool samples. J Clin Microbiol. 2020;58:e01566–19. doi: 10.1128/JCM.01566-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu JL, Anderson GP, Delehanty JB, Baumann R, Hayhurst A, Goldman ER. Selection of cholera toxin specific IgNAR single-domain antibodies from a naïve shark library. Mol Immunol. 2007;44:1775–83. doi: 10.1016/j.molimm.2006.07.299. [DOI] [PubMed] [Google Scholar]
- 69.Zhu M, Gong X, Hu Y, Ou W, Wan Y. Streptavidin-biotin-based directional double Nanobody sandwich ELISA for clinical rapid and sensitive detection of influenza H5N1. J Transl Med. 2014;12:352. doi: 10.1186/s12967-014-0352-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu JL, Shriver-Lake LC, Zabetakis D, Goldman ER, Anderson GP. Selection of single-domain antibodies towards western equine encephalitis virus. Antibodies (Basel) 2018;7:44. doi: 10.3390/antib7040044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gelkop S, Sobarzo A, Brangel P, Vincke C, Romão E, Fedida-Metula S, et al. The development and validation of a novel nanobody-based competitive ELISA for the detection of foot and mouth disease 3ABC antibodies in cattle. Front Vet Sci. 2018;5:250. doi: 10.3389/fvets.2018.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma Z, Wang T, Li Z, Guo X, Tian Y, Li Y, et al. A novel biotinylated nanobody-based blocking ELISA for the rapid and sensitive clinical detection of porcine epidemic diarrhea virus. J Nanobiotechnology. 2019;17:96. doi: 10.1186/s12951-019-0531-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goodchild SA, Dooley H, Schoepp RJ, Flajnik M, Lonsdale SG. Isolation and characterisation of Ebolavirus-specific recombinant antibody fragments from murine and shark immune libraries. Mol Immunol. 2011;48:2027–37. doi: 10.1016/j.molimm.2011.06.437. [DOI] [PubMed] [Google Scholar]
- 74.Habib I, Smolarek D, Hattab C, Grodecka M, Hassanzadeh-Ghassabeh G, Muyldermans S, et al. V(H)H (nanobody) directed against human glycophorin A: a tool for autologous red cell agglutination assays. Anal Biochem. 2013;438:82–9. doi: 10.1016/j.ab.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 75.Chen J, He QH, Xu Y, Fu JH, Li YP, Tu Z, et al. Nanobody medicated immunoassay for ultrasensitive detection of cancer biomarker alpha-fetoprotein. Talanta. 2016;147:523–30. doi: 10.1016/j.talanta.2015.10.027. [DOI] [PubMed] [Google Scholar]
- 76.Schlör A, Holzlöhner P, Listek M, Griess C, Butze M, Micheel B, et al. Generation and validation of murine monoclonal and camelid recombinant single domain antibodies specific for human pancreatic glycoprotein 2. N Biotechnol. 2018;45:60–8. doi: 10.1016/j.nbt.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 77.Faraji F, Tajik N, Behdani M, Shokrgozar MA, Zarnani AH, Shahhosseini F, et al. Development and characterization of a camelid single-domain antibody directed to human CD22 biomarker. Biotechnol Appl Biochem. 2018;65:718–25. doi: 10.1002/bab.1654. [DOI] [PubMed] [Google Scholar]
- 78.Li T, Li SL, Fang C, Hou YN, Zhang Q, Du X, et al. Nanobody-based dual epitopes protein identification (DepID) assay for measuring soluble CD38 in plasma of multiple myeloma patients. Anal Chim Acta. 2018;1029:65–71. doi: 10.1016/j.aca.2018.04.061. [DOI] [PubMed] [Google Scholar]
- 79.Huang C, Li D, Ren J, Ji F, Jia L. Generation and application of fluorescent anti-human beta2-microglobulin VHH's via amino modification. Molecules. 2019;24:2600. doi: 10.3390/molecules24142600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gainkam LO, Keyaerts M, Caveliers V, Devoogdt N, Vanhove C, Van Grunsven L, et al. Correlation between epidermal growth factor receptor-specific nanobody uptake and tumor burden: a tool for noninvasive monitoring of tumor response to therapy. Mol Imaging Biol. 2011;13:940–8. doi: 10.1007/s11307-010-0428-4. [DOI] [PubMed] [Google Scholar]
- 81.Romão E, Krasniqi A, Maes L, Vandenbrande C, Sterckx YG, Stijlemans B, et al. Identification of nanobodies against the acute myeloid leukemia marker CD33. Int J Mol Sci. 2020;21:310. doi: 10.3390/ijms21010310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Movahedi K, Schoonooghe S, Laoui D, Houbracken I, Waelput W, Breckpot K, et al. Nanobody-based targeting of the macrophage mannose receptor for effective in vivo imaging of tumor-associated macrophages. Cancer Res. 2012;72:4165–77. doi: 10.1158/0008-5472.CAN-11-2994. [DOI] [PubMed] [Google Scholar]
- 83.Blykers A, Schoonooghe S, Xavier C, D'Hoe K, Laoui D, D'Huyvetter M, et al. PET imaging of macrophage mannose receptor-expressing macrophages in tumor stroma using 18F-Radiolabeled camelid single-domain antibody fragments. J Nucl Med. 2015;56:1265–71. doi: 10.2967/jnumed.115.156828. [DOI] [PubMed] [Google Scholar]
- 84.Evazalipour M, D'Huyvetter M, Tehrani BS, Abolhassani M, Omidfar K, Abdoli S, et al. Generation and characterization of nanobodies targeting PSMA for molecular imaging of prostate cancer. Contrast Media Mol Imaging. 2014;9:211–20. doi: 10.1002/cmmi.1558. [DOI] [PubMed] [Google Scholar]
- 85.Krasniqi A, D'Huyvetter M, Xavier C, Van der Jeught K, Muyldermans S, Van Der Heyden J, et al. Theranostic radiolabeled anti-CD20 sdAb for targeted radionuclide therapy of non-Hodgkin lymphoma. Mol Cancer Ther. 2017;16:2828–39. doi: 10.1158/1535-7163.MCT-17-0554. [DOI] [PubMed] [Google Scholar]
- 86.Krasniqi A, Bialkowska M, Xavier C, Van der Jeught K, Muyldermans S, Devoogdt N, et al. Pharmacokinetics of radiolabeled dimeric sdAbs constructs targeting human CD20. N Biotechnol. 2018;45:69–79. doi: 10.1016/j.nbt.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 87.Balhuizen A, Massa S, Mathijs I, Turatsinze JV, De Vos J, Demine S, et al. A nanobody-based tracer targeting DPP6 for non-invasive imaging of human pancreatic endocrine cells. Sci Rep. 2017;7:15130. doi: 10.1038/s41598-017-15417-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van Brussel AS, Adams A, Vermeulen JF, Oliveira S, van der Wall E, Mali WP, et al. Molecular imaging with a fluorescent antibody targeting carbonic anhydrase IX can successfully detect hypoxic ductal carcinoma in situ of the breast. Breast Cancer Res Treat. 2013;140:263–72. doi: 10.1007/s10549-013-2635-6. [DOI] [PubMed] [Google Scholar]
- 89.Zheng F, Sparkes A, De Baetselier P, Schoonooghe S, Stijlemans B, Muyldermans S, et al. Molecular imaging with Kupffer cell-targeting nanobodies for diagnosis and prognosis in mouse models of liver pathogenesis. Mol Imaging Biol. 2017;19:49–58. doi: 10.1007/s11307-016-0976-3. [DOI] [PubMed] [Google Scholar]
- 90.Zheng F, Perlman H, Matthys P, Wen Y, Lahoutte T, Muyldermans S, et al. Specificity evaluation and disease monitoring in arthritis imaging with complement receptor of the Ig superfamily targeting Nanobodies. Sci Rep. 2016;6:35966. doi: 10.1038/srep35966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zheng F, Put S, Bouwens L, Lahoutte T, Matthys P, Muyldermans S, et al. Molecular imaging with macrophage CRIg-targeting nanobodies for early and preclinical diagnosis in a mouse model of rheumatoid arthritis. J Nucl Med. 2014;55:824–9. doi: 10.2967/jnumed.113.130617. [DOI] [PubMed] [Google Scholar]
- 92.Broisat A, Hernot S, Toczek J, De Vos J, Riou LM, Martin S, et al. Nanobodies targeting mouse/human VCAM1 for the nuclear imaging of atherosclerotic lesions. Circ Res. 2012;110:927–37. doi: 10.1161/CIRCRESAHA.112.265140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jailkhani N, Ingram JR, Rashidian M, Rickelt S, Tian C, Mak H, et al. Noninvasive imaging of tumor progression, metastasis, and fibrosis using a nanobody targeting the extracellular matrix. Proc Natl Acad Sci U S A. 2019;116:14181–90. doi: 10.1073/pnas.1817442116. [DOI] [PMC free article] [PubMed] [Google Scholar]