Abstract
Cancer cells reprogram lipid metabolism during their malignant progression, but limited information is currently available on the involvement of alterations in fatty acid synthesis in cancer development. We herein demonstrate that acetyl-CoA carboxylase 1 (ACC1), a rate-limiting enzyme for fatty acid synthesis, plays a critical role in regulating the growth and differentiation of leukemia-initiating cells. The Trib1-COP1 complex is an E3 ubiquitin ligase that targets C/EBPA, a transcription factor regulating myeloid differentiation, for degradation, and its overexpression specifically induces acute myeloid leukemia (AML). We identified ACC1 as a target of the Trib1-COP1 complex and found that an ACC1 mutant resistant to degradation because of the lack of a Trib1-binding site attenuated complex-driven leukemogenesis. Stable ACC1 protein expression suppressed the growth-promoting activity and increased ROS levels with the consumption of NADPH in a primary bone marrow culture, and delayed the onset of AML with increases in mature myeloid cells in mouse models. ACC1 promoted the terminal differentiation of Trib1-COP1–expressing cells and eradicated leukemia-initiating cells in the early phase of leukemic progression. These results indicate that ACC1 is a natural inhibitor of AML development. The upregulated expression of the ACC1 protein has potential as an effective strategy for cancer therapy.
Keywords: Oncology
Keywords: Cancer, Tumor suppressors, Ubiquitin-proteosome system
Introduction
Cancer cells reprogram metabolic pathways to support continuous proliferation under stress conditions that are characterized by high demands for energy production and the supply of nutrients including carbohydrates, proteins, and lipids (1). Although previous studies reported the importance of alterations in glucose and glutamine metabolism, limited information is currently available on the role of alterations in fatty acid metabolism in cancer development (2–4). The first rate-limiting step of fatty acid synthesis (FAS) is mediated by acetyl-CoA carboxylase 1 (ACC1), which catalyzes acetyl-CoA for conversion into malonyl-CoA in the cytosol. Malonyl-CoA is utilized by fatty acid synthase for de novo FAS accompanied by the consumption of large amounts of NADPH (5). An isoform of ACC1, ACC2 localizes at the mitochondrial outer membrane. ACC1 and ACC2 exhibit similar catalytic activities for the production of malonyl-CoA, but function in different metabolic pathways. ACC2-generated malonyl-CoA inhibits CPT-1 to prevent fatty acids from moving into the mitochondria to drive fatty acid oxidation (5). ACC1 provides the materials for FAS, which needs energy consumption, while ACC2 provides inhibitors of fatty acid oxidation, which suppress energy production, indicating that they regulate not only fatty acid homeostasis, but also the energy balance in cells.
ACC1 was initially considered to function as an oncoprotein because its RNAi-mediated knockdown and pharmacological inhibition suppressed cell growth in some prostate cancer cell lines and non–small cell lung cancer mouse models (6, 7). However, recent studies suggested that ACC1 acts as a tumor suppressor by regulating the energy balance and suppressing protein acetylation in solid tumor xenograft models (8, 9). ACC1 is functionally regulated at many levels during metabolic stress, including phosphorylation by AMP-activated protein kinase (AMPK), transcriptional regulation by sterol regulatory element–binding protein 1 (SREBP-1) transcription factors, and protein degradation by Trib3 (10–12). The enzymatic activity of ACC1 for FAS is inhibited by AMPK-mediated phosphorylation. AMPK targets numerous metabolic factors and restores energy homeostasis by sensing ATP consumption (13). The inhibition of ACC1 via AMPK facilitates anchorage-independent cell growth and xenografted tumor formation in lung cancer models (8), and promotes the metastasis of breast cancer cells through enhancements in acetyl-CoA and protein acetylation (9). Thus, ACC1 appears to be a key enzyme in tumorigenesis; however, the mechanisms by which alterations in ACC1 affect cell metabolism and contribute to cancer progression currently remain unclear.
Constitutive photomorphogenic 1 (COP1) is a highly conserved E3 ubiquitin ligase (14). Putative substrates of COP1 in mammals include transcription factors (c-Jun, ETV family members, p53, C/EBPA, TORC2, and FoxO1) and enzymes (ACC) (12, 15–20), implying roles for COP1 involved in cell growth, differentiation, and metabolism. COP1 exerts its effects through its substrates by direct interactions and with the aid of adaptor proteins, such as Tribbles, which confers diversity and specificity to the activity of COP1. The mammalian homologs of Tribbles, Trib1, Trib2, and Trib3, consist of a pseudokinase domain and consensus COP1-binding motif at the carboxyl terminus and function as an adaptor protein to recruit COP1 to its specific substrates for proteasome-mediated degradation (12, 21). The ligase complex of COP1 with Trib1 and Trib2, but not Trib3, targets C/EBPA for degradation, and its overexpression has been shown to specifically induce acute myeloid leukemia (AML) in a mouse model (18, 22–24). C/EBPA is a critical transcription factor regulating myeloid differentiation in normal hematopoiesis, and loss-of-function alterations in C/EBPA are one of the major causes of human AML (25). Regarding Trib3-mediated protein degradation, a Trib3-COP1 complex was previously shown to promote ACC ubiquitination and stimulate lipolysis during fasting as well as the loss of insulin signaling in adipose tissue (12). Although the amino acid sequences of the 3 Tribbles are highly conserved, it currently remains unclear whether a relationship exists between leukemogenesis and cell metabolism in Tribbles-COP1 complex activities.
In the present study, we investigated the role of ACC1 in myeloid leukemogenesis. We identified ACC1 as a substrate of the Trib1-COP1 complex and found that its degradation was a critical event in the initiation of metabolic reprogramming to support the energy demands for leukemic progression. We also found that the stabilization of ACC1 suppressed the self-renewal activity of leukemia-initiating cells and enforced their myeloid differentiation in AML mouse models, which protected mice from leukemic death. The present results provide an effective strategy for cancer therapy.
Results
The Trib1-COP1 complex targets ACC1 for proteasome-mediated degradation.
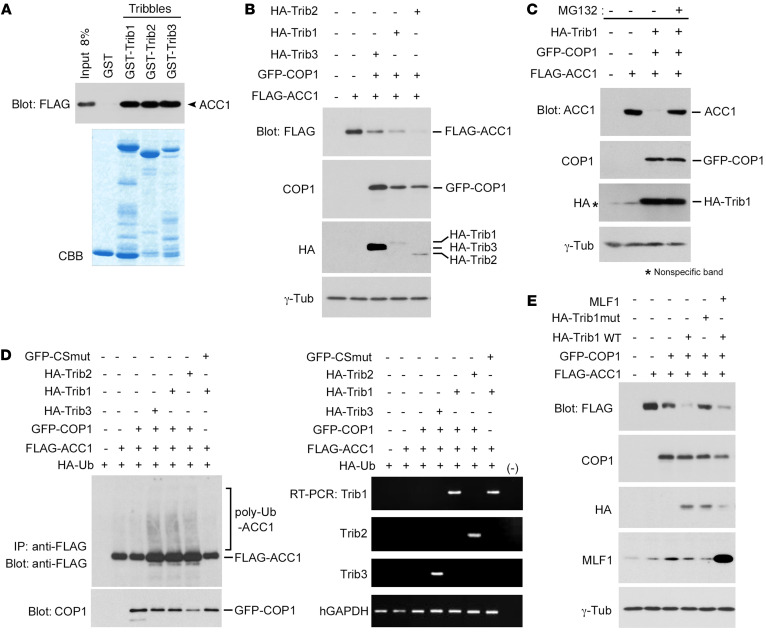
Tribbles are adaptor proteins that recruit COP1 to a specific substrate for degradation. By forming a complex with COP1, Trib1 and Trib2, but not Trib3, downregulate the expression of the C/EBPA protein and induce AML in mouse models. Since a Trib3-COP1 complex targets ACC1 for degradation and the amino acid sequences of the 3 Tribbles are highly conserved, we investigated the mechanisms by which ACC1 interacts with Trib1 and Trib2 in vitro. We performed a pull-down assay with glutathione S-transferase–tagged (GST-tagged) Tribbles produced in E. coli and FLAG-tagged ACC1 expressed in 293T cells, and found that ACC1 directly bound to not only Trib3, but also Trib1 and Trib2 (Figure 1A). We then investigated whether a Trib1-COP1 complex downregulated the expression level of the ACC1 protein in a proteasome-dependent manner. Consistent with previous findings, Trib3 coexpressed with COP1 induced a significant decrease in ACC1 protein levels in 293T cells. Trib1 and Trib2 coexpressed with COP1 more efficiently downregulated ACC1 protein expression (Figure 1B). A treatment with MG132, a specific chemical inhibitor of proteasomes, restored ACC1 protein levels in Trib1-COP1–cotransduced cells (Figure 1C), indicating that the Trib1-COP1 complex degrades ACC1 through the ubiquitin-proteasome system. We then examined whether Trib1 and COP1 induced the ubiquitination of ACC1. ACC1 coexpressed with Tribbles (Trib1, Trib2, and Trib3) and COP1 in cells was strongly polyubiquitinated, whereas the expression of COP1 alone and Trib1/CSmut, a ligase activity–deficient COP1 mutant, failed to induce ubiquitination (18) (Figure 1D). These results indicate that the ligase activity of the Trib1-COP1 complex is essential for ACC1 ubiquitination. We also investigated the role of Trib1-COP1 complex formation using a Trib1 mutant and the involvement of MLF1 (Figure 1E). All Tribbles have a COP1-binding motif ([D/E](x)xxVP[D/E]) at the carboxyl terminus (21). We constructed a Trib1 mutant that cannot bind to COP1 by replacing 3 residues of the COP1-binding motif (DQIVPE) with alanine (AQIAAE). The coexpression of the Trib1 mutant and COP1 failed to reduce ACC1 protein levels, indicating that the ability of Trib1 to bind to COP1 is crucial for the targeting of ACC1. We previously reported that MLF1 stabilized the C/EBPA protein by interfering with the formation of the Trib1-COP1 complex in the nucleus (26). However, the coexpression of MLF1 with Trib1 and COP1 did not affect ACC1 protein levels, suggesting that MLF1 stabilized the nuclear protein (C/EBPA), but not the cytoplasmic protein (ACC1).
Figure 1. Trib1 recruits COP1 to ACC1, and specifically induces proteasome-mediated degradation.
(A) Specific interactions between ACC1 and Tribble proteins in vitro. GST-control, GST-Trib1, GST-Trib2, and GST-Trib3 fusion proteins shown at the top were incubated with 293T cell lysates containing FLAG-tagged ACC1 proteins. Bound proteins were detected by immunoblotting with an antibody against a FLAG epitope. GST-fused proteins were visualized by Coomassie brilliant blue (CBB) staining to evaluate their amounts. (B) The ACC1 protein is downregulated by Trib1-COP1 and Trib2-COP1 as well as Trib3-COP1. 293T cells were transfected with vectors encoding FLAG-ACC1 and GFP-COP1 together with HA-tagged Tribbles (HA-Trib1, HA-Trib2, or HA-Trib3). Cell lysates were analyzed by immunoblotting with antibodies against a FLAG epitope, COP1, an HA epitope, and γ-tubulin. (C) A treatment with MG132 inhibits ACC1 degradation. 293T cells transfected with a combination of vectors encoding FLAG-ACC1, GFP-COP1, and HA-Trib1 were incubated in the presence and absence of MG132 and analyzed by immunoblotting with antibodies against ACC1, COP1, an HA epitope, and γ-tubulin. (D) All Tribbles-COP1 promote the ubiquitination of ACC1. 293T cells were transfected with a combination of vectors shown at the top. Cell lysates were analyzed by immunoprecipitation with an antibody against a FLAG epitope followed by immunoblotting with an antibody against a FLAG epitope, and by immunoblotting with an antibody against COP1. Total RNA was analyzed by semiquantitative RT-PCR (semi-qRT-PCR) using a pair of primers specific to Trib1, Trib2, Trib3, and human GAPDH. (E) ACC1 is degraded through the interaction with Trib1. 293T cells were transfected with a combination of vectors shown at the top. Cell lysates were analyzed by immunoblotting with antibodies against a FLAG epitope, COP1, an HA epitope, MLF1, and γ-tubulin.
Identification of the Tribbles-binding site in ACC1 required for its degradation.
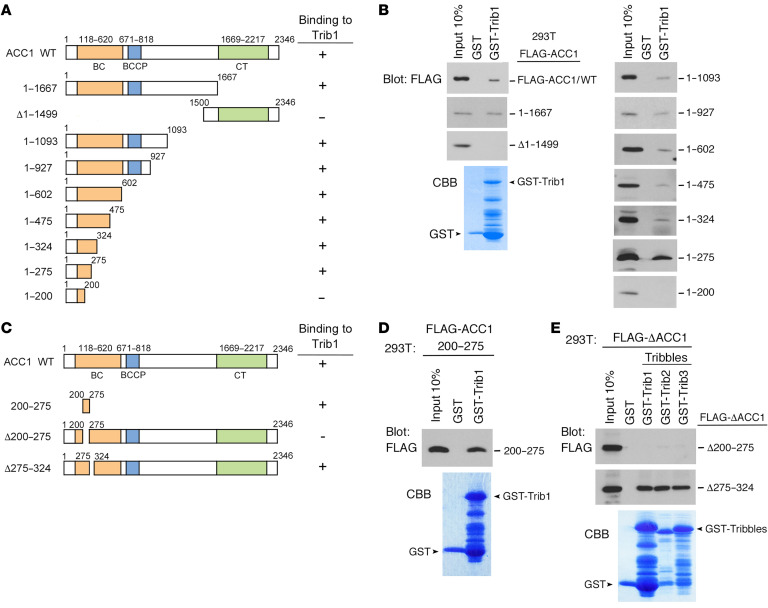
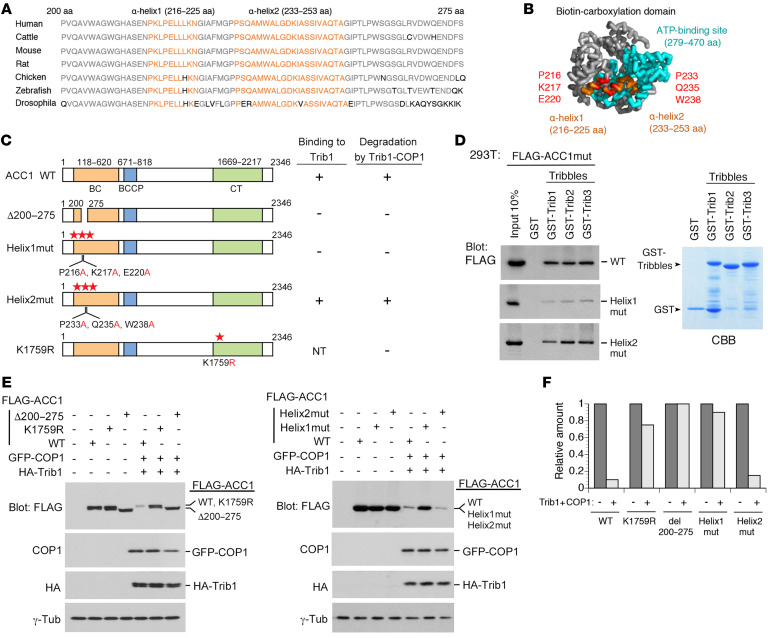
To elucidate the sequences of ACC1 required for the interaction with Trib1, we constructed a series of ACC1 deletion mutants and expressed them in 293T cells after FLAG tagging (Figure 2A). We performed a pull-down assay with these mutants and found that an ACC1 mutant containing 275 N-terminal amino acids (ACC1/1–275), but not that containing the 200 N-terminal amino acids (ACC1/1–200), bound to Trib1 in vitro (Figure 2, A and B). This result suggested that residues 200–275 of ACC1 are required for the interaction with Trib1. We constructed mutants containing only the putative binding site (ACC1/200–275) and lacking the region (ACC1/Δ200–275), and a mutant lacking the region next to residues 200–275 as a control (ACC1/Δ275–324) (Figure 2C). The pull-down assay showed that Trib1 bound to ACC1/200–275 and ACC1/Δ275–324, but not to ACC1/Δ200–275 (Figure 2, D and E). We also confirmed that Trib2 and Trib3 as well as Trib1 were unable to bind to ACC1/Δ200–275 (Figure 2E) under the condition that all Tribbles tightly bound to ACC1/Δ275–324, implying that ACC1 has a consensus-binding site to interact with all Tribbles. As shown in Figure 3, A and B, residues 200–275 of ACC1 is a highly conserved region from Drosophila to humans and contains two α-helices (Helix1: residues 216–225; Helix2: residues 233–253) located at the N-terminus upstream of the ATP-binding site (residues 279–470) in the biotin carboxylase (BC) domain. To examine whether these α-helices are important for ACC1 to bind to Tribbles, we constructed mutants of Helix1 and Helix2 (Helix1mut and Helix2mut) by replacing 3 residues located at the outer surface of the BC domain with alanine (Helix1mut: P216A, K217A, and E220A; Helix2mut: P233A, Q235A, and W238A) (Figure 3, B and C). We performed a pull-down assay with these mutants and found that the ability of Helix1mut to bind to Tribbles was significantly weaker than those of ACC1 wild type (WT) and Helix2mut (Figure 3D). We also constructed a K1759R mutant, which lacked one of the major ubiquitination sites of ACC1 (27) (Figure 3C).
Figure 2. Identification of the Tribbles-binding site in ACC1.
(A) Schematic representation of ACC1 deletion mutants. The results of Trib1 binding are summarized on the right. (B) GST-control and GST-Trib1 fusion proteins were incubated with 293T cell lysates containing FLAG-tagged ACC1WT and deletion mutant proteins. Bound proteins were detected by immunoblotting with an antibody against a FLAG epitope. The GST-Trib1–fused protein was visualized by CBB staining to evaluate its amount. (C) Schematic representation of ACC1 deletion mutants minimized for ACC1-Trib1 binding. The results of Trib1 binding are summarized on the right. (D and E) GST-control and GST-Trib1 fusion proteins were incubated with 293T cell lysates containing FLAG-ACC1/200–275 (D). GST-control and all GST-Tribbles (GST-Trib1, GST-Trib2, and GST-Trib3) fusion proteins were incubated with 293T cell lysates containing FLAG-ACC1/Δ200–275 and ACC1/Δ275–324 (E). Bound proteins were detected by immunoblotting with an antibody against a FLAG epitope. GST-Tribbles–fused proteins were visualized by CBB staining to evaluate their amounts.
Figure 3. Construction of ACC1 point mutants resistant to Trib1-COP1–mediated degradation.
(A) Sequence alignment of residues 200–275 of human ACC1 with compatible residues of various species. The region includes two α-helices (Helix1: residues 216–225; Helix2: residues 233–253) marked in orange and other conserved residues shown in gray. (B) Structure of the biotin carboxylase (BC) domain of ACC1. Helix1 and Helix2 (in orange) are positioned at the outside of the ATP-binding site (in blue). Mutants of Helix1 and Helix2 were constructed by replacement of 3 residues positioned at the interaction surfaces with alanine (Helix1mut: P216A, K217A, and E220A; Helix2mut: P233A, Q235A, and W238A; shown in red). This model is generated by the human ACC1 full crystal structure (PDB:6G2D) with PyMOL (http://www.pymol.org). (C) Schematic representation of ACC1 point mutants for the ACC1-Trib1 binding and ubiquitination sites. The results of Trib1 binding and degradation are summarized on the right. NT, not tested. (D) GST-control and all GST-Tribbles (GST-Trib1, GST-Trib2, and GST-Trib3) fusion proteins were incubated with 293T cell lysates containing FLAG-ACC1WT, Helix1mut, and Helix2mut. Bound proteins were detected by immunoblotting with an antibody against a FLAG epitope. GST-Tribbles–fused proteins were visualized by CBB staining to evaluate their amounts. (E and F) Helix1mut and K1759R are resistant to degradation. 293T cells were transfected with the combination of vectors shown at the top. Cell lysates were analyzed by immunoblotting with antibodies against a FLAG epitope, COP1, an HA epitope, and γ-tubulin (E). Relative amounts of proteins were quantified using ImageJ software (NIH) (F).
To investigate whether ACC1 protein levels inversely correlate with the ability of the ACC1 mutant to bind to Tribbles and whether the K1759R mutant is resistant to ubiquitination, we ectopically expressed ACC1WT, Δ200–275, Helix1mut, Helix2mut, and K1759R together with Trib1 and COP1 in 293T cells. We found that ACC1WT and Helix2mut were markedly downregulated, whereas Δ200–275, Helix1mut, and K1759R were stably detected, even in the presence of the Trib1-COP1 ligase complex (Figure 3, E and F). These results indicate that an α-helix (residues 216–225) and the lysine 1759 residue are critical for ACC1 degradation mediated by the Trib1-COP1 complex.
Stabilization of ACC1 suppresses the cellular growth advantage induced by the Trib1-COP1 complex and increases intracellular ROS levels and NADPH consumption.
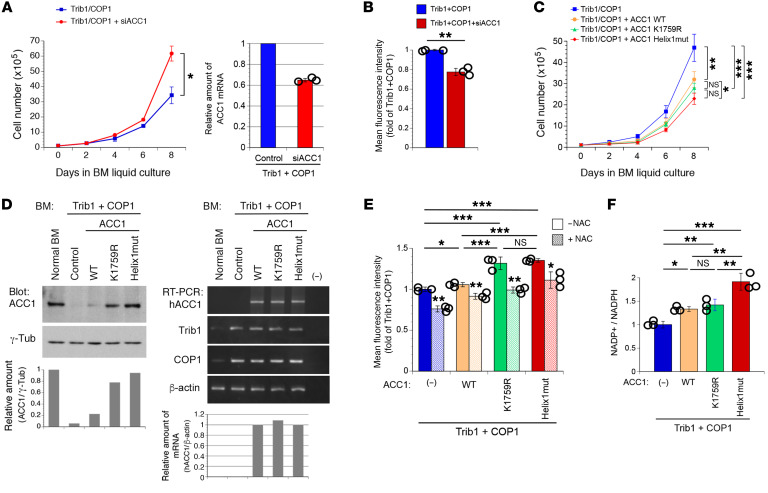
We investigated the effects of ACC1 on the Trib1-COP1–induced growth advantage in murine primary bone marrow (BM) cells in a liquid culture. Since the ectopically expressed Trib1-COP1 complex was expected to decrease endogenous ACC1 protein levels, we initially examined whether the additional downregulation of ACC1 promotes cellular proliferation and affects reactive oxygen species (ROS) levels, by monitoring metabolic stress in cells. We transduced primary BM cells with retroviruses expressing Trib1 (Trib1-IRES-GFP) and COP1 (COP1-IRES-GFP) in the presence or absence of ACC1-specific siRNA (siACC1) and sorted GFP-positive cells for further analyses. The knockdown of ACC1 accelerated Trib1-COP1–induced cell growth and significantly reduced intracellular ROS levels (Figure 4, A and B), suggesting that the downregulation of ACC1 enabled cells to tolerate oxidative stress and promote proliferation.
Figure 4. Stabilized ACC1 suppresses cell growth and increases ROS levels and NADPH consumption.
(A and B) Primary BM cells were infected with retroviruses expressing Trib1- and COP1-IRES-GFP in the presence and absence of ACC1-specific siRNA (siACC1). GFP-positive cells were sorted by flow cytometry and cultured in BM medium. Cell numbers were counted for growth curves (A). GFP-positive cells in A were transferred to IL-3–containing medium with low glucose (1 g/L), maintained for 3 days, and analyzed to measure ROS levels (B). P values were calculated with Student’s t test (*P < 0.05, **P < 0.01). Data are the average of 3 independent experiments (A and B) shown as mean ± SEM. (C–F) Primary BM cells were infected with retroviruses expressing Trib1- and COP1-IRES-GFP together with and without ACC1WT, K1759R, and Helix1mut. GFP-positive cells were sorted by flow cytometry and cultured in BM medium. Cell numbers were counted for growth curves (C). Cell lysates of GFP-positive cells in C were analyzed by immunoblotting with antibodies against ACC1 and γ-tubulin (D, left). Total RNA was analyzed by semi-qRT-PCR using a pair of primers specific to human ACC1 (hACC1) Trib1, COP1, and β-actin (D, right). Relative amounts of proteins (hACC1/γ-tubulin) and mRNAs (hACC1/β-actin) were quantified using ImageJ software (D, bottom panels). GFP-positive cells in C were transferred to IL-3–containing medium with low glucose (1 g/L), maintained in the absence and presence of 1 mM N-acetylcysteine (NAC) for 3 days, and analyzed to measure ROS levels (E) and the NADP+/NADPH ratio (F). P values were calculated with 1-way ANOVA with Tukey’s multiple-comparison post-test (*P < 0.05, **P < 0.01, ***P < 0.001). Data are the average of 3 independent experiments (C, E, and F) shown as mean ± SEM.
We hypothesized that the stabilization of the ACC1 protein suppresses Trib1-COP1–induced cell growth by balancing ROS generation associated with FAS. To test this, we used the ACC1 mutants K1759R and Helix1mut, which are resistant to Trib1-COP1–mediated degradation. We transduced primary BM cells with retroviruses expressing ACC1WT, K1759R, or Helix1mut together with Trib1 and COP1 and sorted GFP-positive cells for further experiments. The growth of cells expressing ACC1WT, K1759R, and Helix1mut was significantly slower than that of cells expressing Trib1-COP1 alone, and Helix1mut more strongly inhibited cell growth than ACC1WT (Figure 4C). This result implied that the stability of ACC1 proteins in these BM cells correlated with the suppression of Trib1-COP–induced cell growth. We examined ACC1 protein and mRNA levels in these cells (Figure 4D). We also confirmed coexpression of COP1 and ACC1 in all individual GFP-positive cells in culture by immunostaining with antibodies against COP1 and ACC1 (Supplemental Figure 1 and Supplemental Figure 2; supplemental material available online with this article; https://doi.org/10.1172/JCI141529DS1). Although normal BM cells expressed a significant amount of endogenous ACC1, primary BM cells transduced with Trib1-COP1 alone and those cotransduced with ACC1WT showed markedly reduced expression levels of both endogenous and ectopic proteins. Cells cotransduced with K1759R or Helix1mut exhibited higher expression levels (Figure 4D); however, the same amounts of ACC1 transcripts were detected among ACC1WT-, K1759R-, and Helix1mut-transduced cells (Figure 4D), which indicated that stabilization of the ACC1 protein due to its resistance to degradation contributed to the suppression of Trib1-COP1–induced cell growth.
To clarify whether the antiproliferative effects induced by ACC1 are associated with intracellular metabolic stress, we measured ROS levels and energy consumption (the NADP+/NADPH ratio) in transduced BM cells. The coexpression of ACC1WT, K1759R, or Helix1mut with Trib1 and COP1 induced increases in ROS levels in correlation with their expression levels. K1759R- and Helix1mut-transduced cells showed markedly higher ROS levels than those expressing ACC1WT (Figure 4E), and the treatment of these cells with the antioxidant N-acetylcysteine reduced their ROS levels. Since ACC1-mediated FAS consumes large amounts of energy in cells, the upregulation of ACC1 is expected to accelerate NADPH consumption, thereby increasing ROS levels. As anticipated, NADPH consumption by cells expressing ACC1WT, K1759R, or Helix1mut together with Trib1-COP1 was greater than that by cells expressing Trib1-COP1 alone, with Helix1mut-transduced cells showing the highest NADP+/NADPH ratio (Figure 4F). These results suggest that ACC1 degradation mediated by the Trib1-COP1 complex decreases NADPH energy consumption, which regenerates reduced glutathione (GSH) to eliminate ROS during leukemogenesis. The ectopic expression of degradation-resistant ACC1 mutants may rebalance oncogene-induced metabolic reprograming into a normal status.
Stabilization of ACC1 suppresses Trib1-COP1–induced leukemic progression and promotes cell differentiation.
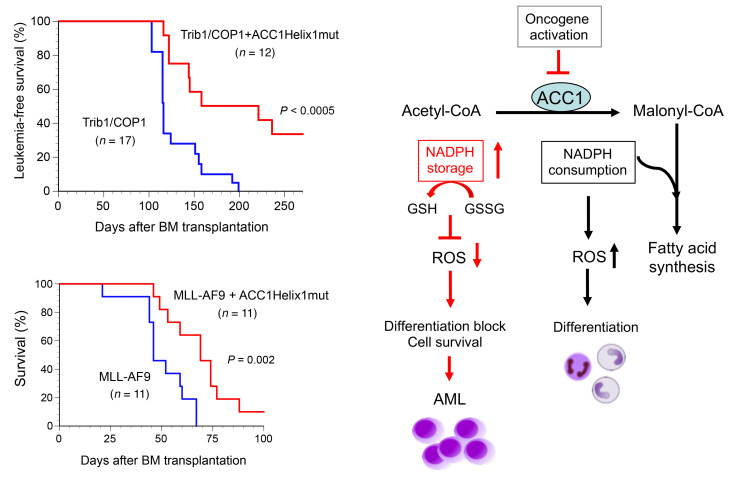
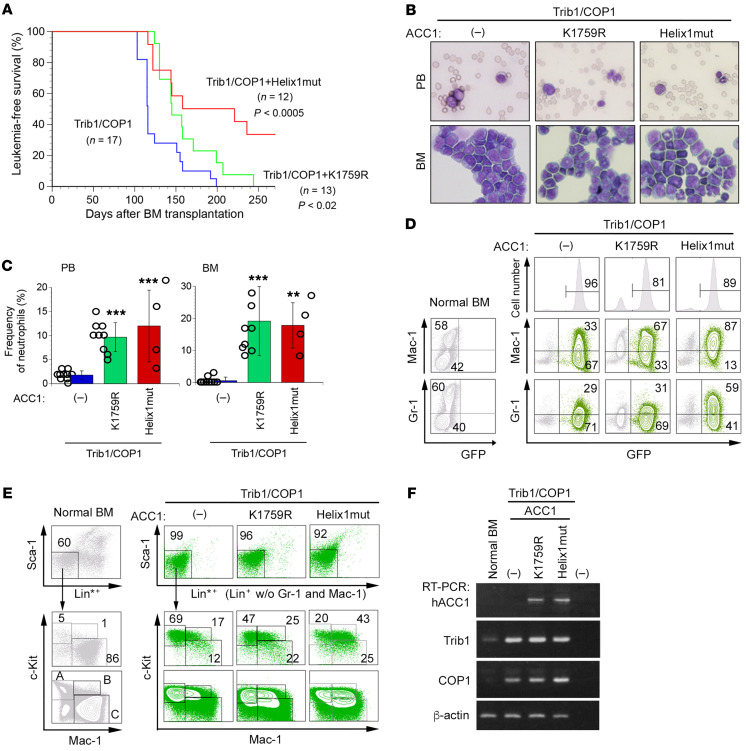
We investigated whether stable ACC1 expression affects Trib1-COP1–induced AML development in mouse models. We transduced primary BM cells with retroviruses expressing Trib1 (Trib1-IRES-GFP) and COP1 (COP1-IRES-GFP) together with the K1759R and Helix1 ACC1 mutants and intravenously injected them into sublethally irradiated mice. Mice with transplanted Trib1-COP1–transduced BM (Trib1-COP1 control mice) developed AML with complete penetrance 100–200 days after transplantation. In contrast, the onset of AML was significantly delayed in mice with transplanted K1759R- and Helix1mut-cotransduced BM, and more than one-third of mice with Helix1mut did not develop AML, by the time when all control mice and mice with K1759R-cotransduced BM had died of AML (Figure 5A). While all Trib1-COP1 control mice eventually exhibited leukocytosis with markedly increased immature myeloid blasts in the BM, we detected significant increases in mature neutrophils as well as immature cells in the BM of K1759R and Helix1mut mice (Figure 5, B and C). Consistent with this morphological observation, the majority of GFP-positive leukemic cells in Trib1-COP1 control mice were Mac-1lo/+Gr-1lo immature myeloid cells, whereas GFP-positive cells in Helix1mut mice showed an increase in differentiated Mac-1hiGr-1hi myeloid cells (Figure 5D).
Figure 5. Stabilized ACC1 suppresses leukemic progression and induces cell differentiation in AML.
Mice were transplanted with BM cells infected with retroviruses expressing Trib1- and COP1-IRES-GFP together with and without K1759R and Helix1mut and analyzed for several months after BM transplantation. (A) Myeloid leukemia–free survival curves of transplanted mice. Results are derived from 4 independent transfer experiments. P values versus Trib1-COP1 control mice were calculated with the log-rank test. (B) May-Grünwald-Giemsa–stained peripheral blood (PB) smears and cytospins of BM cells in AML mice. Original magnification, ×400. (C) Frequency of neutrophils in the PB and BM cells of Trib1/COP1 (n = 10), K1759R (n = 9), and Helix1mut (n = 4) mice. P values were calculated with 1-way ANOVA with Tukey’s multiple-comparison post-test (**P < 0.01, ***P < 0.001). Data are shown as mean ± SEM. (D) FACS analysis of GFP-positive BM cells for immature (Mac-1lo, Gr-1lo) and differentiated (Mac-1hi, Gr-1hi) granulocytes. The population of GFP-positive cells in BM is shown in the top panels. The distribution pattern in normal BM cells is shown in the left panels. (E) A detailed FACS analysis of GFP-positive BM cells for lineage-negative cells, excluding Mac-1 and Gr-1 (Lin*–: CD3–B220–TER-119–). Lin*–Sca-1– (Lin*: lineage marker without Mac-1 and Gr-1) BM cells were separated into 3 fractions: c-KithiMac-1– (fraction A: the CMP/GMP/MEP compartment), c-Kit+Mac-1+ (fraction B: committed myeloid progenitors), and c-Kitlo/–Mac-1+ (fraction C: differentiated myeloid cells). The distribution pattern in normal BM cells is shown in the left panels. (F) Total RNA extracted from GFP-positive BM cells was analyzed by semi-qRT-PCR using a pair of primers specific to hACC1, Trib1, COP1, and β-actin.
We examined whether the coexpression of K1759R and Helix1mut affects the stage of myeloid differentiation at which Trib1-COP1 blocks further differentiation and induces AML (26). We separated Lin*–Sca-1– (Lin*: lineage marker without Mac-1 and Gr-1) BM cells into 3 fractions (28): c-KithiMac-1– (fraction A, containing the CMP/GMP/MEP compartment), c-Kit+Mac-1+ (fraction B, containing committed myeloid progenitors), and c-Kitlo/–Mac-1+ (fraction C, containing differentiated myeloid cells), and analyzed their distribution patterns in normal BM cells (Figure 5E, fractions A–C) and transplanted BM cells (Figure 5E). While immature c-KithiMac-1– cells (fraction A) were dominant in Trib1-COP1 control mice, the number of mature c-Kit+Mac-1+ and c-Kit–Mac-1+ cells (fractions B and C) increased in Helix1mut mice (Figure 5E). We assessed the expression of transduced genes by reverse transcription PCR (RT-PCR) (Figure 5F), and the results obtained indicated that this was not due to the impaired expression of Trib1 and COP1. Therefore, the stabilization of ACC1 appears to partly induce terminal myeloid differentiation and suppresses AML development in mouse models.
ACC1 downregulation is required for self-renewal activity in Trib1-COP1–induced leukemia-initiating cells.
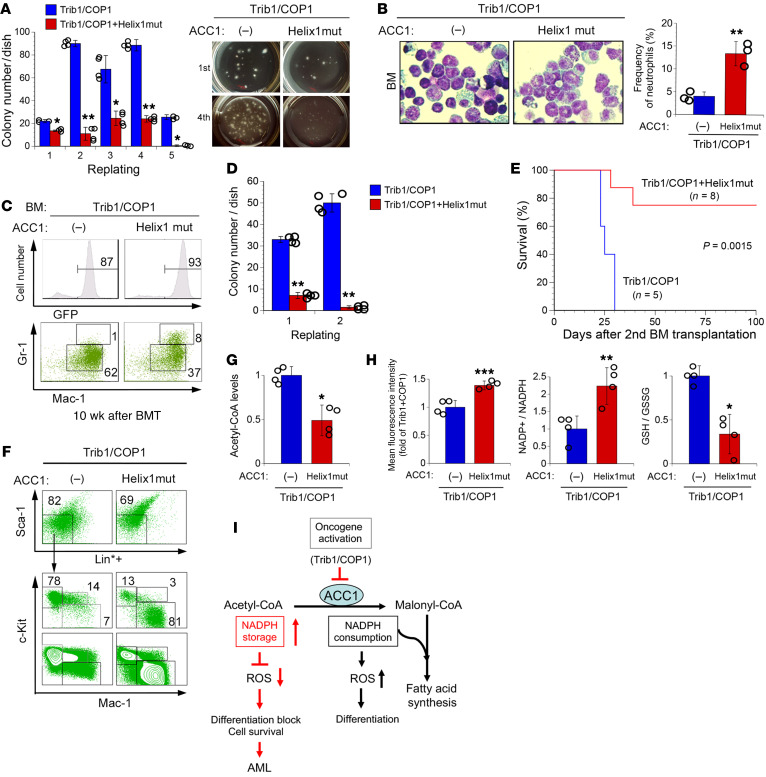
To assess whether ACC1 is involved in the self-renewal activity of Trib1-COP1–transduced leukemic cells, we performed in vitro colony formation assays with Trib1-COP1 control and Helix1mut-cotransduced BM cells. An equal number of GFP-positive cells from each culture was sorted and subjected to serial plating in methylcellulose. The number of colonies in Helix1mut-cotransduced cells was markedly lower than that in Trib1-COP1 control cells (Figure 6A), and the frequency of morphologically differentiated myeloid cells increased in Helix1mut-cotransduced cells (Figure 6B), suggesting that ACC1 degradation is critical for Trib1-COP1–expressing cells to gain self-renewal ability and block myeloid differentiation.
Figure 6. ACC1 stabilization induces the loss of self-renewal activity in leukemia-initiating cells.
(A and B) GFP-positive BM cells expressing Trib1/COP1 together with and without Helix1mut were sorted and plated in methylcellulose-based medium. Colony numbers were counted after 10 days and replated in fresh medium for serial colony assays (A, left panel). Plates from the first culture and the fourth serial culture are shown (A, right panel). May-Grünwald-Giemsa stain (original magnification, ×400) and frequency of neutrophils (n = 3) in colony-forming cells from the first culture (B). (C–H) BM cells from the early phase of Trib1-COP1 control mice and mice with Helix1mut-cotransduced BM (10 weeks after BM transplantation) were analyzed. (C) FACS analysis for immature (Mac-1+Gr-1lo) and differentiated (Mac-1hiGr-1hi) granulocytes. The population of GFP-positive cells in BM is shown in the top panels. (D) GFP-positive cells were sorted and plated for colony assays. (E) Kaplan-Meier survival curves of secondary transplanted mice. Sublethally irradiated mice received 1 × 105 GFP-positive BM cells each from two Trib1/COP1 and two Helix1mut mice. (F) A detailed FACS analysis of GFP-positive BM cells from secondary transplanted mice in E. Lin*–Sca-1– cells were separated into 3 fractions: c-KithiMac-1– (fraction A), c-Kit+Mac-1+ (fraction B), and c-Kitlo/–Mac-1+ (fraction C). (G and H) GFP-positive BM cells from the early phase of Trib1/COP1 (n = 4) and Helix1mut (n = 4) mice were analyzed to measure acetyl-CoA levels (G), ROS levels, the NADP+/NADPH ratio, and the GSH/GSSG ratio (H). (I) Proposed model of the ACC1-mediated pathway and effects on leukemia cells. P values were calculated by Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001) in A, B, D, G, and H and by log-rank test in E. Data are the average of 2 independent experiments with 4 dishes (A and D) and 4 independent experiments (G and H) shown as mean ± SEM.
To further elucidate the initial effects of ACC1 on Trib1-COP1–expressing leukemia-initiating cells (LICs), we analyzed the early phase of leukemic progression with mouse models 10–12 weeks after BM transplantation (BMT). In BM cells from an early phase, even though GFP-positive cells exhibited clonal expansion and constituted 87% and 93% of all mononuclear cells in Trib1-COP1 control mice and Helix1mut mice, respectively, the latter had more differentiated Mac-1hiGr-1hi myeloid cells than the former (Figure 6C), suggesting that Helix1mut has the ability to induce the myeloid differentiation of LICs. We then assessed the self-renewal activity of these cells by performing colony formation assays in a suspension culture. An equal number of GFP-positive cells from each BM of BMT mice was sorted and subjected to first plating and secondary replating in methylcellulose. The number of colonies in Helix1mut mice was markedly lower than that in Trib1-COP1 control mice, and cells from these colonies almost lost their self-renewal activity in the secondary replating (Figure 6D). To clarify whether Helix1mut prevents the establishment of Trib1-COP1–induced leukemic stem cells in vivo, we transplanted BM cells isolated from these mice into irradiated secondary recipients in order to investigate whether these cells maintain their leukemia-initiating ability in mice. All Trib1-COP1 control secondary recipients died of AML within 30 days, whereas the majority of Helix1mut secondary recipients failed to develop AML for at least 100 days (Figure 6E). In Trib1-COP1 secondary recipients, the immunophenotype of GFP-positive cells was similar to that in primary Trib1-COP1 mice, in which the immature c-KithiMac-1– population (fraction A) dominated with a marked decrease in the mature c-Kit–Mac-1+ population (fraction C). However, in Helix1mut mice, the immature c-KithiMac-1– population (fraction A) decreased with a marked increase in the mature c-Kit–Mac-1+ population (fraction C) (Figure 6F). These results indicated that stabilized ACC1 promotes the terminal differentiation of Trib1-COP1–expressing cells and eradicates LICs in the early phase of leukemic progression.
We further investigated whether stable ACC1 expression affects the intracellular metabolic status in Trib1-COP1–expressing LICs. A substantial number of GFP-positive cells from each BM of BMT mice was sorted and subjected to metabolic analyses. Since ACC1 consumes acetyl-CoA to generate malonyl-CoA as the first step of de novo FAS, we measured the levels of acetyl-CoA to evaluate the synthetic progression to malonyl-CoA in cells, and found that the levels of acetyl-CoA in Helix1mut mice were significantly lower than those in Trib1-COP1 control mice (Figure 6G). We also measured ROS levels, energy consumption (the NADP+/NADPH ratio), and ROS elimination activity (the GSH/GSSG ratio) in these cells, and found that Helix1mut coexpression induced an increase in ROS levels and the NADP+/NADPH ratio and a decrease in the GSH/GSSG ratio (Figure 6H), consistent with the observation in a primary BM culture (Figure 4, E and F). These results indicate that the stabilization of ACC1 upregulates ROS levels by promoting NADPH consumption and resultant reduction of GSH regeneration, and imply that ACC1-associated metabolic imbalance is one of the critical factors during the Trib1-COP1–induced development of LICs (Figure 6I).
ACC1 is downregulated in human AML and stable ACC1 expression suppresses MLL-AF9–induced AML in mouse models.
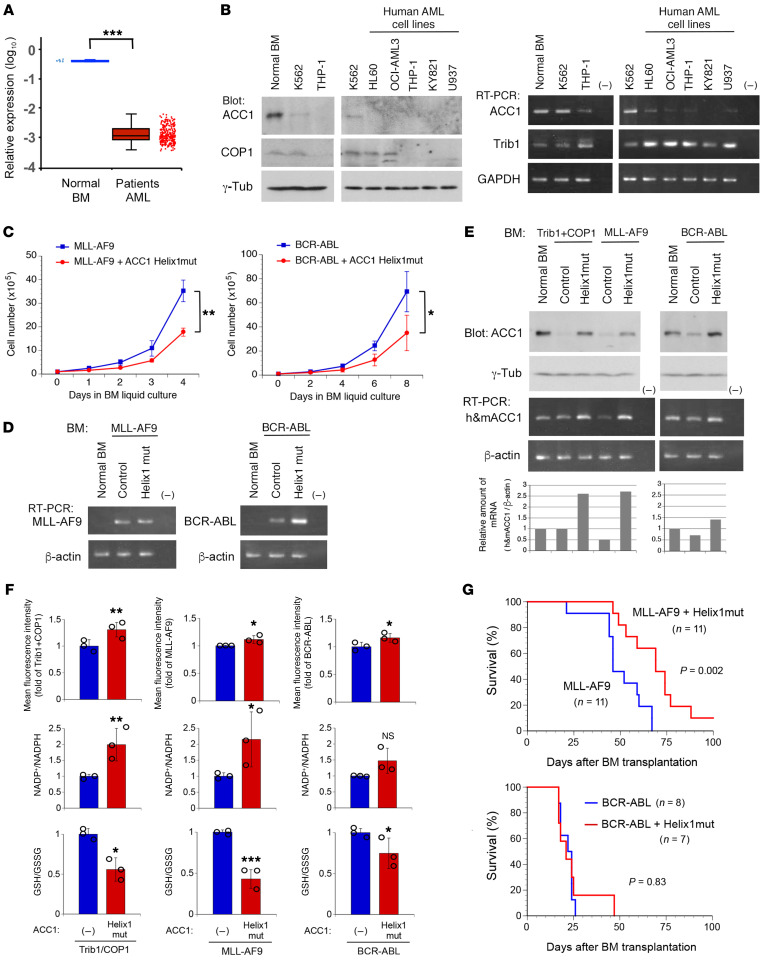
We next addressed 2 issues to validate our findings: (a) whether the downregulation of ACC1 is commonly observed in human AML, and (b) whether the stabilization of ACC1 suppresses AML development driven by other well-characterized oncogenes. To investigate whether the downregulation of ACC1 is common among human leukemia, we analyzed the mRNA levels of ACC1 in a GEO data set of 285 cases of AML compared with those in 9 healthy donors. We found that ACC1 transcripts were markedly suppressed in patients with AML compared with healthy donors (Figure 7A; P < 0.0001). We also examined protein and mRNA levels of ACC1 in various human hematopoietic cell lines derived from patients with leukemia. Normal BM cells and K562 cells, a chronic myeloid leukemia (CML) cell line carrying p210 BCR-ABL, showed a significant amount of ACC1 protein and transcripts with a lower expression of Trib1, whereas 5 AML cell lines, including THP-1, which carries an MLL-AF9 fusion oncogene, expressed undetectable levels of ACC1 protein and markedly lower levels of ACC1 transcripts, but contained markedly high levels of Trib1 (Figure 7B). These results indicate that the downregulation of ACC1 in mRNA or protein levels is a common feature among human AML.
Figure 7. ACC1 is downregulated in human AML and stabilized ACC1 partially suppresses MLL-AF9–induced AML.
(A) Microarray data analysis of the ACC1 expression in human AML. Gene expression data of normal BMs from healthy donors (n = 9) and AML patient samples (n = 285) were obtained from the GEO data sets. (B) ACC1 protein and mRNA levels in human leukemic cell lines. Cell lysates from normal BM, a CML cell line (K562), and AML cell lines (HL60, OCI-AML3, THP-1, KY821, and U937) were analyzed by immunoblotting with antibodies against ACC1, COP1, and γ-tubulin. Total RNA was analyzed by semi-qRT-PCR using a pair of primers specific to ACC1, Trib1, and GAPDH. (C–F) Primary BM cells were infected with retroviruses expressing MLL-AF9, BCR-ABL, and Trib1- and COP1-IRES-GFP together with and without ACC1 Helix1mut. Sorted GFP-positive cells were cultured in BM medium. Cell numbers were counted for growth curves (C). Total RNA extracted from GFP-positive cells in C was analyzed by semi-qRT-PCR using a pair of primers specific to MLL-AF9, BCR-ABL (D), human and mouse (h&m) ACC1 (E), and β-actin (D and E). Cell lysates of GFP-positive cells in C were analyzed by immunoblotting with antibodies against ACC1 and γ-tubulin (E). Relative amounts of mRNAs (h&mACC1/β-actin) were quantified using ImageJ software (E, bottom panels). GFP-positive cells in C were transferred to IL-3–containing medium with low glucose (1 g/L) and analyzed to measure ROS levels, the NADP+/NADPH ratio, and the GSH/GSSG ratio (F). Data are the average of 3 independent experiments (C and F) shown as mean ± SEM. (G) Survival curves of transplanted mice with BM expressing MLL-AF9-IRES-GFP and BCR-ABL-IRES-GFP together with and without ACC1 Helix1mut. Results are derived from 3 independent transfer experiments. P values were calculated with Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001) in A, C, and F and with log-rank test in G.
To examine whether the stabilization of ACC1 suppresses leukemia development induced by oncogenes other than Trib1-COP1, we chose leukemogenic mouse models driven by the MLL-AF9 and BCR-ABL chimeric oncogenes (29–31). We transduced primary BM cells with retroviruses expressing ACC1 Helix1mut together with MLL-AF9 (MLL-AF9-IRES-GFP) and BCR-ABL (BCR-ABL-IRES-GFP) and sorted GFP-positive cells for further experiments. The growth of cells coexpressing Helix1mut was significantly slower than that of cells expressing MLL-AF9 and BCR-ABL alone (Figure 7, C and D). Interestingly, primary BM cells transduced with MLL-AF9 alone showed markedly reduced expression levels of endogenous ACC1 protein and mRNA compared with those in normal BM cells, whereas cells transduced with BCR-ABL exhibited their moderate reductions (Figure 7E). Comparing cells expressing MLL-AF9 and Trib1-COP1, the expression levels of ACC1 protein were reduced in both cases, but the levels of ACC1 transcripts were markedly lower only in cells expressing MLL-AF9 (Figure 7E). These findings indicated that the expression of ACC1 is generally downregulated in human leukemia but in a specific manner depending on the driver of leukemogenesis.
To evaluate the intracellular metabolic status, we measured ROS levels, energy consumption (the NADP+/NADPH ratio), and ROS elimination activity (the GSH/GSSG ratio) in cells transduced with 3 different leukemic drivers. The coexpression of Helix1mut with MLL-AF9 increased ROS levels and the NADP+/NADPH ratio and decreased the GSH/GSSG ratio. These results are parallel to those with Trib1 and COP1 (Figure 7F). However, the coexpression of Helix1mut with BCR-ABL showed similar but much-limited effects on these ROS-associated activities (Figure 7F).
We investigated whether stable ACC1 expression affects leukemia development induced by MLL-AF9 and BCR-ABL in mouse models. All mice with transplanted MLL-AF9–transduced BM developed AML and died within 67 days after transplantation, while mice with transplanted BM transduced with MLL-AF9 together with Helix1mut showed a significantly prolonged survival (Figure 7G). In contrast, most of the mice with transplanted cells expressing BCR-ABL with or without Helix1mut died within 26 days (Figure 7G), which is consistent with previous reports with BCR-ABL mouse models (32). The coexpression of Helix1mut appeared to affect the survival in MLL-AF9–induced AML, but not that in BCR-ABL–induced CML.
Discussion
Three important results were obtained in the present study: (a) ACC1 and C/EBPA targeted by the Trib1-COP1 ligase complex are critical factors for degradation during myeloid leukemogenesis. (b) A shortage of the ACC1 enzyme eliminates ROS by maintaining high levels of NADPH through a decrease in FAS, a process that consumes NADPH, indicating that ACC1 dysregulation induces an energy imbalance in LICs. (c) The stabilization of ACC1 suppresses cell proliferation and promotes granulocyte differentiation in leukemia-initiating mice (Figure 6I). These results lead to further questions regarding the relationship between the reprogramming of fatty acid metabolism and tumorigenesis.
Cancer cells have a high demand for fatty acids as a source of energy, the biosynthesis of membranes, and signaling molecules for proliferation. Since ACC1 is a rate-limiting enzyme of de novo FAS in cells, the present result showing that ACC1 suppressed cell proliferation appears to be contradictory to cancer cells needing fatty acids as materials for membranes to support rapid cell division. Some pharmacological inhibitors of ACC (ACC1 and ACC2) suppress the growth of non–small cell lung cancers and fatty liver–related hepatocellular carcinomas (7, 33). However, de novo FAS is also a process that consumes a large amount of NADPH in cells. NADPH is a coenzyme that is indispensable for many biological reactions, such as replenishing GSH, which is a major ROS-scavenging system in cells (34). The inhibition of ACC-catalyzed reactions to FAS may result in an excessive supply of NADPH, which is utilized to reduce ROS levels for cell survival in cancer-initiating cells.
ACCs are substrates of AMPK, a master regulator of metabolism. The direct phosphorylation of ACC1 and ACC2 by AMPK controls cellular lipid metabolism. The former suppresses FAS in the cytosol, and the latter promotes fatty acid oxidation at the mitochondrial outer membrane (13). The inhibition of these ACCs maintains NADPH levels by blocking FAS, which consumes NADPH, and by inducing fatty acid oxidation to regenerate NADPH, thereby changing the major energy flux for use. Jeon et al. reported that the AMPK-ACC pathways regulated NADPH homeostasis to promote cell survival in solid tumors by reducing ROS activity and also that ACC mutants resistant to AMPK-mediated phosphorylation inhibited tumor growth (8). This is functionally consistent with the present results showing that the degradation of the ACC1 protein was one of the triggers for the onset of leukemia and that an ACC1 mutant resistant to Trib1-COP1–mediated degradation suppressed the progression of leukemia accompanied by increased ROS levels and a high NADP+/NADPH ratio.
The relationship observed between fatty acid metabolism and myeloid differentiation in leukemogenesis is an interesting result. We initially suspected that stabilized ACC1 suppresses Trib1-COP1–driven AML development, but did not expect it to induce terminal myeloid differentiation. However, recent findings support these results. AMPK protects MLL-AF9–driven myeloid LICs from metabolic stress in the BM. By interrupting glucose metabolism, the deletion of AMPK delays leukemogenesis exhibiting differentiated morphologies and depletes LICs by increasing ROS levels and DNA damage (35). Since AMPK is a negative regulator upstream of ACC1 and ACC2, stabilized ACC1 may behave in line with AMPK-deficient effects in myeloid LICs. Oxidative stress and DNA damage are considered to limit the self-renewal capacity of both normal hematopoietic stem cells and leukemic stem cells (36–38). Increased levels of ROS in hematopoietic progenitors and LICs are associated with myeloid differentiation. The histone methyltransferase MLL4 was shown to be required for the maintenance of MLL-AF9–driven leukemic stem cells by regulating transcriptional programs, such as FoxOs, and the GSH detoxification pathway to attenuate oxidative stress. The deletion of MLL4 enforces the myeloid differentiation of leukemic blasts with increased levels of ROS (38). Although the mechanisms by which the moderate upregulation of ROS induces cellular differentiation in both normal and cancer stem cells have not yet been elucidated in detail, ROS levels may be one of the factors influencing their destinies.
We herein demonstrated that the stabilization of ACC1 is a potential therapeutic strategy for cancer. We identified a binding site for Tribbles within the ACC1 protein. ACC1 binds to all Tribbles through an α-helical region (residues 216–225) in its N-terminal BC domain. The results obtained from mouse models revealed that stabilized ACC1 eliminated leukemic stem cells by promoting terminal differentiation. Therefore, a small-molecule inhibitor targeting the α-helical region of ACC1 is a promising tool for cancer therapy. It stabilizes ACC1 by preventing it from interacting with Tribbles and inhibiting its degradation. Although Trib3 does not induce AML, it is overexpressed in breast and colorectal cancers and correlates with metastatic activity (39, 40). Small-molecule inhibitors may be useful in the treatment of patients who have solid tumors and AML that strongly express Tribbles but who have low ACC1 protein levels, and may be more effective in treatments combined with conventional chemotherapies.
Despite the suppression of ACC1-catalyzed de novo FAS in Trib1-COP–driven leukemic cells, these cells acquired self-renewal and growth-promoting activities, implying that other mechanisms of fatty acid uptake, such as enhanced import via cell membrane transporters, contribute to the maintenance of cellular fatty acid levels for rapid proliferation during cancer progression. Increased mobilization from extracellular adipose tissues is another major source of fatty acids, and high levels of the fatty acid transporter CD36 in cancer stem cells have been associated with metastasis and chemotherapy resistance in various solid tumors and myeloid leukemias (41, 42). Moreover, normal BM is an adipocyte-rich microenvironment that tightly regulates the balance of normal hematopoietic maturation (43). In the early phase of leukemia initiation, fatty acid uptake from adipocytes enriched in BM may be sufficient for cell growth. The accumulation of NADPH through the inhibition of ACC1 activity to reduce ROS levels may be more important for LICs to survive rather than de novo FAS.
In conclusion, we herein demonstrated that the upregulated expression of the ACC1 protein suppressed the initiation of Trib1-COP1–driven myeloid leukemia by changing the energy flux from NADPH accumulation to consumption, which increased ROS levels, thereby inducing terminal myeloid differentiation. The present results clearly show that the fatty acid metabolic pathway is an effective target for cancer therapy.
Methods
Plasmid construction, cell culture, transfection, and retroviral production.
Complementary DNA (cDNA) fragments containing the entire coding sequence of human ACC1 were amplified by PCR using the K562 cDNA library as a template. ACC1 cDNAs (WT and mutants) were subcloned into FLAG-tagged expression vectors (Sigma-Aldrich) and the pMSCV retrovirus vector. Trib1 cDNAs were subcloned into the hemagglutinin-tagged expression vector (a gift from Junichi Fujisawa, Kansai Medical University, Osaka, Japan) and the pMSCV-IRES-GFP retrovirus vector (a gift from Owen Witte, Howard Hughes Medical Institute, UCLA, Los Angeles, California, USA). COP1 cDNAs were subcloned into the GFP-tagged expression vector by modification of the pIRES puro3 vector (BD Biosciences) and pMSCV-IRES-GFP retrovirus vector. pMSCV-neo-MLL-AF9 and pMSCV-p210BCR-ABL-IRES-GFP plasmids were provided by Akihiko Yokoyama (Tsuruoka Metabolomics Laboratory, National Cancer Center, Yamagata, Japan) and Owen Witte, respectively (44, 45). MLL-AF9 cDNA was subcloned into the pMSCV-IRES-GFP retrovirus vector. A vector for RNAi specific to mouse ACC1 was constructed based on the pSUPER RNAi system (pSUPER.retro.neo; Oligoengine). The siRNA sequence targeting ACC1 was 5′-GCAGATTGCCAACATCCTAGA-3′.
293T human embryonic kidney cells were cultured in DMEM supplemented with 10% FBS, 2 mM glutamine, 100 U/mL of penicillin, and 100 μg/mL of streptomycin and then transfected with expression vectors via the calcium phosphate–DNA precipitation method (46). Regarding viral production, the plasmid was cotransfected into 293T cells together with a plasmid encoding an ecotropic helper virus containing a defective virion-packaging (φ2) sequence. Culture supernatants containing retroviruses were harvested 48–72 hours after transfection and used for infection.
Human hematopoietic cell lines derived from CML (K562) and AMLs (HL60, OCI-AML3, THP-1, KY821, and U937) were maintained in RPMI 1640 medium supplemented with 10% FBS, 2 mM glutamine, 100 U/mL of penicillin, and 100 μg/mL of streptomycin. The K562, HL60, THP-1, KY821, and U937 cell lines were provided by Yoshinobu Matsuo (Fujisaki Cell Center, Okayama, Japan), and the OCI-AML3 cell line was purchased from the DSMZ cell lines bank (Germany).
BM transplantation and analyses.
BM cells were aseptically isolated from the femurs and tibiae of 8-week-old C57BL/6 mice (CLEA Japan Inc.) that had been intravenously injected with 5-fluorouracil (150 mg/kg of body weight; Kyowa Hakko Kogyo) 5 days before; incubated in BM medium (DMEM, 15% heat-inactivated FBS, 2 mM glutamine, 100 U/mL of penicillin, 100 μg/mL of streptomycin, 5% WEHI-3B conditioned medium, 6 ng/mL mouse IL-3, 10 ng/mL human IL-6, and 50 ng/mL mouse stem cell factor) (recombinant cytokines were from R&D Systems Inc.); and infected with a retroviral supernatant according to the spin infection procedure described previously in the presence of Polybrene (6 μg/mL) (47).
Infected, unsorted BM cells (0.5 × 106 to 1 × 106 cells) were injected into the veins of 8- to 10-week-old C57BL/6 mice that had been sublethally irradiated (10 Gy; M-150WE, SOFTEX) several hours before the injection. All mice were maintained in the Nara Institute of Science and Technology Animal Facility in accordance with the Institute’s guidelines. Blood samples were analyzed with an automated cell counter (F-820 analyzer, Sysmex) and also by inspection of blood smears after staining with May-Grünwald-Giemsa solution (Merck).
In the liquid culture, BM cells were cultured in BM medium for 7 days after retroviral infection, and GFP-positive populations in BM cells were isolated by cell sorting with FACSAria (BD Biosciences) and maintained in BM medium. Where indicated, cells were maintained in medium supplemented with 10% WEHI-3B conditioned medium as a source of IL-3. In the colony formation assay, sorted GFP-positive BM cells were plated at 2 × 103 cells per 35-mm dish onto methylcellulose-based medium containing mouse IL-3 (10 ng/mL), human IL-6 (10 ng/mL), mouse stem cell factor (SCF) (50 ng/mL), and human erythropoietin (3 U/mL) (MethoCult GF M3434, STEMCELL Technologies Inc.), and colonies were enumerated after 10 days. Cells for second to fourth plating were collected from methylcellulose-based medium, counted, and replated at 1 × 104 cells per 35-mm dish into fresh methylcellulose-based medium containing mouse IL-3 (10 ng/mL), human IL-6 (10 ng/mL), and mouse SCF (50 ng/mL) (MethoCult GF M3534).
Flow cytometric analyses.
Pharm Lyse (BD Biosciences) was used to remove red blood cells from the sample. BM and spleen cells were analyzed and sorted with FACSAria (BD Biosciences) after staining with antibodies against CD3 (145-2C11), B220 (RA3-6B2), TER-119 (TER-119), Mac-1 (CD11b, M1/70), Gr-1 (RB6-8C5), c-Kit (ACK2), and Sca-1 (D7) (all from eBioscience), which were conjugated with phycoerythrin (PE), PE-Cy5, allophycocyanin, Pacific blue, or biotin, and together with streptavidin-PE-Cy7.
Protein analyses.
Cell lysis, immunoprecipitation, gel electrophoresis, and immunoblotting were performed as described previously (48). A rabbit polyclonal antibody against COP1 and a mouse monoclonal antibody against MLF1 (3E9) were generated using bacterially produced polypeptides in our laboratory (48). Mouse monoclonal antibodies against a FLAG epitope (M5) and a hemagglutinin (HA) peptide epitope (12CA5) were purchased from WAKO and Roche, respectively. A mouse monoclonal antibody against γ-tubulin (GTU-88) and a rabbit polyclonal antibody against an HA epitope (HA.11) were obtained from Sigma-Aldrich and Berkeley Antibody Co., respectively. A rabbit polyclonal antibody against ACC1 (#4190) was purchased from Cell Signaling Technology.
In the in vitro binding assay, cDNAs encoding Trib1, Trib2, and Trib3 were amplified by PCR, confirmed by sequencing, and inserted into the vector pGEX (GE Healthcare), and Tribble proteins were expressed in bacteria and purified as described (48). Crude cell extracts containing a series of the FLAG-tagged ACC1 deletion mutant and point mutant proteins were prepared from 293T cells transfected with the FLAG-ACC1 mutant expression vectors. A binding assay was performed as described previously (48). Glutathione S-transferase (GST) proteins were quantitated by staining with Coomassie brilliant blue after separation by SDS-PAGE.
In the in vivo ubiquitination assay, 293T cells were transfected with a combination of vectors encoding FLAG-ACC1, GFP-COP1, GFP-COP1/CSmut, HA-Trib1, HA-Trib2, HA-Trib3, and HA-Ub, and harvested 2 days after transfection. Cells were lysed in SDS sample buffer (40 mM Tris-HCl, pH 6.8, 0.1 M DTT, 1% SDS, 10% glycerol, and 0.05% bromophenol blue). After boiling, cell lysates were diluted with EBC buffer (50 mM Tris-HCl, pH 7.5, 120 mM NaCl, 0.5% NP-40, and 1 mM EDTA containing 5 mg/mL of aprotinin, 1 mM PMSF, 0.1 mM NaF, 0.1 mM NaVO4, and 1 mM DTT) and immunoprecipitated with an antibody against a FLAG epitope followed by immunoblotting with an antibody against a FLAG epitope.
Measurement of intracellular ROS.
GFP-positive BM cells were stained using the Cellular Reactive Oxygen Species Detection Assay Kit (Deep Red Fluorescence, Abcam) according to the manufacturer’s instructions, and analyzed with a FACSCalibur flow cytometer (BD Biosciences).
NADPH assay and GSH assay.
The ratios of intracellular NADP+/NADPH and GSH/GSSG in GFP-positive BM cells were measured with the NADP/NADPH-Glo Assay Kit (Promega) and the GSH/GSSG-Glo Assay Kit (Promega) according to the manufacturer’s instructions. NADP+, NADPH, GSH, and GSSG levels were individually measured, and the ratio was then calculated.
Semiquantitative RT-PCR.
Total RNA was isolated using the ISOGEN reagent (Nippon Gene) and reverse-transcribed using RNase-free Superscript reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. Semiquantitative RT-PCR was performed within a linear range as described previously (49), and data were normalized to the expression levels of β-actin and GAPDH for each sample. We confirmed that reactions were quantitatively performed within a linear range by conducting several control experiments. The following oligonucleotide primers specific to mouse Trib1, Trib2, Trib3, COP1, and β-actin and human ACC1, GAPDH, MLL-AF9, and BCR-ABL were used: mouse Trib1, 5′-GGACTTTGGAGACATGCACTCCT-3′ (sense) and 5′-GACCAAAAGCGTATAGAGCATCACC-3′ (antisense); mouse Trib2, 5′-GCAACATCAACCAAATCACG-3′ (sense) and 5′-GCGTCTTCCAAACTCTCCAG-3′ (antisense); mouse Trib3, 5′-CAGGACAAGATGCGACGAGCTACAC-3′ (sense) and 5′-AGGTGGCCCTAGCCGTACAG-3′ (antisense); mouse COP1, 5′-AGGTTTCAGTGGGACCTCTC-3′ (sense) and 5′-GACCTTTGACCTCTGTCCTG-3′ (antisense); mouse β-actin, 5′-CTTCTACAATGAGCTGCGTGT-3′ (sense) and 5′-CAACGTCACACTTCATGATGG-3′ (antisense); human ACC1, 5′-AGCTGTAAGAGCTCATTTTGGAGG-3′ (sense) and 5′-TTCAGAAGTGTATGAGCAGGAAGG-3′ (antisense); human GAPDH, 5′-CCATCACCATCTTCCAGGAG-3′ (sense) and 5′-CCTGCTTCACCACCTTCTTG-3′ (antisense); MLL-AF9, 5′-GATGCCTTCCAAAGCCTACCTG-3′ (sense) and 5′-CTGTGAAGCTCTACCAGTTCATCT-3′ (antisense); BCR-ABL, 5′-GAAGTGTTTCAGAAGCTTCTCC-3′ (sense) and 5′-GTTTGGGCTTCACACCATTCC-3′ (antisense).
Bioinformatics analysis of human gene expression.
Human gene expression profiling data were obtained from the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/). For the ACC1 expression analysis in human leukemic progression, the GEO data sets, accession numbers GSE33075 (normal BMs) and GSE1159 (AML), were retrieved and analyzed statistically using R version 3.2.2 (R Foundation for Statistical Computing, Vienna, Austria).
Statistics.
Statistical analysis was performed by 2-tailed Student’s t tests with unpaired analysis or 1-way ANOVA with Tukey’s multiple-comparison post-test (*P < 0.05, **P < 0.01, ***P < 0.001), and data represent mean ± SEM, except for the survival curves in mice. P values in the survival curves were calculated with the log-rank test.
Study approval.
All animal experiments were conducted according to the protocols approved by the Nara Institute of Science and Technology Animal Care and Use Committee.
Author contributions
HI and NYK designed the experiments. HI, IN, and NYK performed the experiments. HI, IN, JK, and NYK analyzed the data. HI and NYK wrote the manuscript. JK edited the manuscript.
Supplementary Material
Acknowledgments
We thank Akihiko Yokoyama and Owen N. Witte for pMSCV-neo-MLL-AF9 and pMSCV-BCR-ABL-IRES-GFP retrovirus vectors. We thank Takashi Yokoyama for the technical assistance with the mouse models. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, and Culture of Japan (26440100, 17J07454, 17K07387, 20K06626).
Version 1. 06/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(12):e141529.https://doi.org/10.1172/JCI141529.
Contributor Information
Hidenori Ito, Email: ito.hidenori.ia0@bs.naist.jp.
Ikuko Nakamae, Email: ikuko-n@bs.naist.jp.
Jun-ya Kato, Email: jkata@bs.naist.jp.
Noriko Yoneda-Kato, Email: noriko-k@bs.naist.jp.
References
- 1.Zhu J, Thompson CB. Metabolic regulation of cell growth and proliferation. Nat Rev Mol Cell Biol. 2019;20(7):436–450. doi: 10.1038/s41580-019-0123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Currie E, et al. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18(2):153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vander Heiden MG, et al. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altman BJ, et al. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16(10):619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wakil SJ, Abu-Elheiga LA. Fatty acid metabolism: target for metabolic syndrome. J Lipid Res. 2009;50(suppl):S138–S143. doi: 10.1194/jlr.R800079-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brusselmans K, et al. RNA interference-mediated silencing of the acetyl-CoA-carboxylase-alpha gene induces growth inhibition and apoptosis of prostate cancer cells. Cancer Res. 2005;65(15):6719–6725. doi: 10.1158/0008-5472.CAN-05-0571. [DOI] [PubMed] [Google Scholar]
- 7.Svensson RU, et al. Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat Med. 2016;22(10):1108–1119. doi: 10.1038/nm.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeon SM, et al. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485(7400):661–665. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rios Garcia M, et al. Acetyl-CoA carboxylase 1-dependent protein acetylation controls breast cancer metastasis and recurrence. Cell Metab. 2017;26(6):842–855. doi: 10.1016/j.cmet.2017.09.018. [DOI] [PubMed] [Google Scholar]
- 10.Fullerton MD, et al. Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat Med. 2013;19(12):1649–1654. doi: 10.1038/nm.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011;13(4):376–388. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi L, et al. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science. 2006;312(5781):1763–1766. doi: 10.1126/science.1123374. [DOI] [PubMed] [Google Scholar]
- 13.Garcia D, Shaw RJ. AMPK: mechanisms of cellular energy sensing and restoration of metabolic balance. Mol Cell. 2017;66(6):789–800. doi: 10.1016/j.molcel.2017.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marine JC. Spotlight on the role of COP1 in tumorigenesis. Nat Rev Cancer. 2012;12(7):455–464. doi: 10.1038/nrc3271. [DOI] [PubMed] [Google Scholar]
- 15.Wertz IE, et al. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science. 2004;303(5662):1371–1374. doi: 10.1126/science.1093549. [DOI] [PubMed] [Google Scholar]
- 16.Vitari AC, et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature. 2011;474(7351):403–406. doi: 10.1038/nature10005. [DOI] [PubMed] [Google Scholar]
- 17.Dornan D, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429(6987):86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida A, et al. COP1 targets C/EBPα for degradation and induces acute myeloid leukemia via Trib1. Blood. 2013;122(10):1750–1760. doi: 10.1182/blood-2012-12-476101. [DOI] [PubMed] [Google Scholar]
- 19.Dentin R, et al. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature. 2007;449(7160):366–369. doi: 10.1038/nature06128. [DOI] [PubMed] [Google Scholar]
- 20.Kato S, et al. COP1 functions as a FoxO1 ubiquitin E3 ligase to regulate FoxO1-mediated gene expression. J Biol Chem. 2008;283(51):35464–35473. doi: 10.1074/jbc.M801011200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eyers PA. TRIBBLES: a twist in the pseudokinase tail. Structure. 2015;23(11):1974–1976. doi: 10.1016/j.str.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Keeshan K, et al. Tribbles homolog 2 inactivates C/EBPalpha and causes acute myelogenous leukemia. Cancer Cell. 2006;10(5):401–411. doi: 10.1016/j.ccr.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin G, et al. Trib1 and Evi1 cooperate with Hoxa and Meis1 in myeloid leukemogenesis. Blood. 2007;109(9):3998–4005. doi: 10.1182/blood-2006-08-041202. [DOI] [PubMed] [Google Scholar]
- 24.Dedhia PH, et al. Differential ability of Tribbles family members to promote degradation of C/EBPalpha and induce acute myelogenous leukemia. Blood. 2010;116(8):1321–1328. doi: 10.1182/blood-2009-07-229450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koschmieder S, et al. Dysregulation of the C/EBPalpha differentiation pathway in human cancer. J Clin Oncol. 2009;27(4):619–628. doi: 10.1200/JCO.2008.17.9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamae I, et al. Myeloid leukemia factor 1 stabilizes tumor suppressor C/EBPα to prevent Trib1-driven acute myeloid leukemia. Blood Adv. 2017;1(20):1682–1693. doi: 10.1182/bloodadvances.2017007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner SA, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10(10):M111.013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirstetter P, et al. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008;13(4):299–310. doi: 10.1016/j.ccr.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Krivtsov AV, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 30.Muntean AG, Hess JL. The pathogenesis of mixed-lineage leukemia. Annu Rev Pathol. 2012;7:283–301. doi: 10.1146/annurev-pathol-011811-132434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong S, Witte ON. Modeling Philadelphia chromosome positive leukemias. Oncogene. 2001;20(40):5644–5659. doi: 10.1038/sj.onc.1204638. [DOI] [PubMed] [Google Scholar]
- 32.Pear WS, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92(10):3780–3792. [PubMed] [Google Scholar]
- 33.Lally JSV, et al. Inhibition of Acetyl-CoA carboxylase by phosphorylation or the inhibitor ND-654 suppresses lipogenesis and hepatocellular carcinoma. Cell Metab. 2019;29(1):174–182. doi: 10.1016/j.cmet.2018.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trachootham D, et al. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8(7):579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 35.Saito Y, et al. AMPK protects leukemia-initiating cells in myeloid leukemias from metabolic stress in the bone marrow. Cell Stem Cell. 2015;17(5):585–596. doi: 10.1016/j.stem.2015.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nijnik A, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447(7145):686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- 37.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128(2):325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 38.Santos MA, et al. DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature. 2014;514(7520):107–111. doi: 10.1038/nature13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu JM, et al. TRIB3 supports breast cancer stemness by suppressing FOXO1 degradation and enhancing SOX2 transcription. Nat Commun. 2019;10(1):5720. doi: 10.1038/s41467-019-13700-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, et al. TRIB3 promotes lung cancer progression by activating β-catenin signaling. Eur J Pharmacol. 2019;863:172697. doi: 10.1016/j.ejphar.2019.172697. [DOI] [PubMed] [Google Scholar]
- 41.Pascual G, et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017;541(7635):41–45. doi: 10.1038/nature20791. [DOI] [PubMed] [Google Scholar]
- 42.Ye H, et al. Leukemic stem cells evade chemotherapy by metabolic adaptation to an adipose tissue niche. Cell Stem Cell. 2016;19(1):23–37. doi: 10.1016/j.stem.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyd AL, et al. Acute myeloid leukaemia disrupts endogenous myelo-erythropoiesis by compromising the adipocyte bone marrow niche. Nat Cell Biol. 2017;19(11):1336–1347. doi: 10.1038/ncb3625. [DOI] [PubMed] [Google Scholar]
- 44.Okuda H, et al. Cooperative gene activation by AF4 and DOT1L drives MLL-rearranged leukemia. J Clin Invest. 2017;127(5):1918–1931. doi: 10.1172/JCI91406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong S, et al. Cell context-specific effects of the BCR-ABL oncogene monitored in hematopoietic progenitors. Blood. 2003;101(10):4088–4097. doi: 10.1182/blood-2002-11-3376. [DOI] [PubMed] [Google Scholar]
- 46.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7(8):2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoneda-Kato N, Kato JY. Shuttling imbalance of MLF1 results in p53 instability and increases susceptibility to oncogenic transformation. Mol Cell Biol. 2008;28(1):422–434. doi: 10.1128/MCB.02335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoneda-Kato N, et al. Myeloid leukemia factor 1 regulates p53 by suppressing COP1 via COP9 signalosome subunit 3. EMBO J. 2005;24(9):1739–1749. doi: 10.1038/sj.emboj.7600656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsumoto N, et al. Elevated MLF1 expression correlates with malignant progression from myelodysplastic syndrome. Leukemia. 2000;14(10):1757–1765. doi: 10.1038/sj.leu.2401897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.