Abstract
Down syndrome (DS), caused by trisomy of chromosome 21, occurs in 1 of every 800 live births. Early defects in cortical development likely account for the cognitive impairments in DS, although the underlying molecular mechanism remains elusive. Here, we performed histological assays and unbiased single-cell RNA-Seq (scRNA-Seq) analysis on cerebral organoids derived from 4 euploid cell lines and from induced pluripotent stem cells (iPSCs) from 3 individuals with trisomy 21 to explore cell-type–specific abnormalities associated with DS during early brain development. We found that neurogenesis was significantly affected, given the diminished proliferation and decreased expression of layer II and IV markers in cortical neurons in the subcortical regions; this may have been responsible for the reduced size of the organoids. Furthermore, suppression of the DSCAM/PAK1 pathway, which showed enhanced activity in DS, using CRISPR/Cas9, CRISPR interference (CRISPRi), or small-molecule inhibitor treatment reversed abnormal neurogenesis, thereby increasing the size of organoids derived from DS iPSCs. Our study demonstrates that 3D cortical organoids developed in vitro are a valuable model of DS and provide a direct link between dysregulation of the DSCAM/PAK1 pathway and developmental brain defects in DS.
Keywords: Neuroscience, Stem cells
Keywords: Embryonic stem cells, Neurodevelopment, iPS cells
Introduction
Down syndrome (DS), caused by trisomy of chromosome 21, is the most frequent genetic cause of birth defects and cognitive abnormalities, with an occurrence of 1 of 800 live births (1). Decades of research using transgenic mouse models and postmortem human tissues have revealed that the characteristics of DS include reduced brain weight, cerebral atrophy, a thinner cortex, impaired neurogenesis, and altered cortical lamination (2–6). Moreover, in vitro culturing of neural progenitor cells (NPCs) isolated from DS fetuses revealed impaired proliferation (7) and reduced neurogenesis (8–10), which was informative in dissecting the mechanisms underlying early brain defects in DS. Nevertheless, the above results were based on inaccessible pathological specimens, so there is a need to better clarify the pathogenesis of DS by using a readily available model of human origin. The advent of human induced pluripotent stem cells (iPSCs) presents unprecedented opportunities to establish human cellular models for investigating neurodevelopmental diseases. Using human PSC technology, our previous reports demonstrated that DS iPSC–derived neurons had significant synaptic deficits (11) as well as impaired migration and neurite projections (12). Despite the accumulated knowledge in the past 2 decades regarding neural development in DS, the mechanisms underlying the impaired cortical development in DS are still unclear.
Recent advances in cerebral organoid culturing systems have opened new avenues to study human developmental disorders (13–24). Cerebral organoids are 3D structures reminiscent of human brain regions, including the cerebral cortex and subventricular zone. Recently, several independent studies established cerebral organoids to clarify the pathogenesis of microcephaly, autism, Miller-Dieker syndrome, and other neurodevelopmental disorders (14, 15, 20, 21, 25–29), but there have been no reports on the pathological mechanism underlying delayed cortical development in DS.
Among the protein-coding genes in HSA21 (human chromosome 21), DS cell adhesion molecule (DSCAM) encodes a cell adhesion molecule involved in neuronal generation, maturation, dendrite morphology, and neuronal wiring (30, 31), which are important for brain development. Triplication of the DSCAM gene deregulated the activity of p21-activated kinase 1 (PAK1) and phosphorylated PAK1 (p-PAK1), resulting in neuronal connectivity dysfunction in immortalized cells from trisomy 16 (Ts16) fetal mice (32). PAK1, a gene downstream of DSCAM, plays a role in cortical development by regulating the proliferation of neural progenitors (33). Nevertheless, it is unclear whether the DSCAM/PAK1 pathway regulates the development of the cerebral cortex in DS.
Here, we established 3D cerebral organoid culture to investigate the mechanisms associated with abnormal cerebral development in DS. To resolve confounding factors owing to the asynchronous and heterogeneous appearance of disease phenotypes in in vitro culture, we performed high-resolution single-cell transcriptomic analyses to uncover the cell-type–specific molecular pathology of DS. Our data showed that DS iPSC-derived cerebral organoids partially recapitulated the abnormalities observed in DS mouse models and postmortem DS brain samples, including a reduced proliferation rate and abnormal neurogenesis. Furthermore, we showed that a thinner cortex and proliferation deficits in DS organoids could be reversed by knocking out 1 locus of the 3 DSCAM alleles. Likewise, the small-molecule inhibitor FRAX486 could rescue the neuropathological phenotypes by regulating the expression of PAK1. Our findings may provide a potential target for prenatal intervention for DS.
Results
Single-cell RNA-Seq reveals altered neural development in trisomy 21 organoids.
To generate organoids from human PSCs, we modified the protocol of cerebral organoid generation described in previous reports (refs. 34–37, Figure 1A, and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI135763DS1). After continuous culturing for 30–60 days, the cerebral organoids showed stratified neuroepithelium-like architecture expressing neural progenitor markers on day 30 and cortical layer markers from days 50 to 70 (Supplemental Figure 1). Moreover, the cortical upper-layer markers could be examined after 100–115 days of differentiation (Supplemental Figure 1).
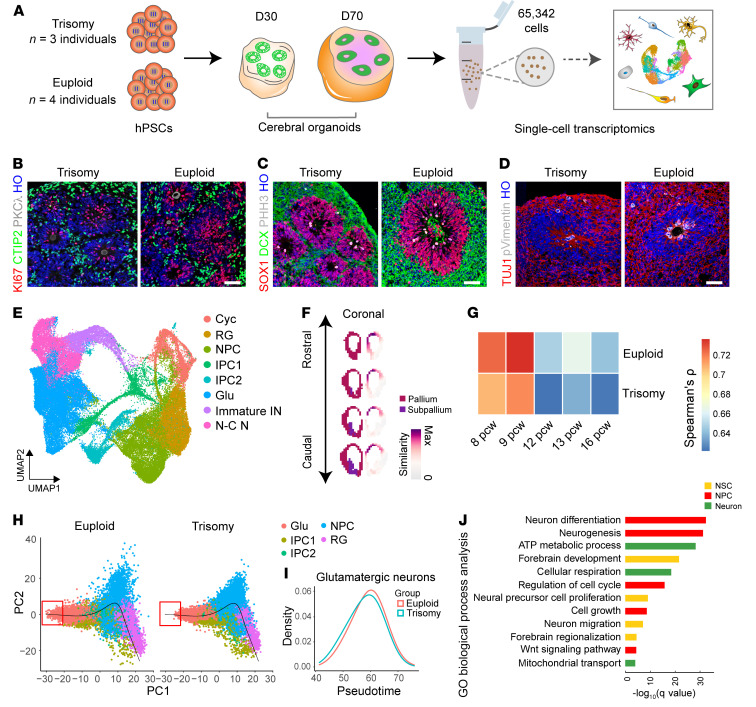
Figure 1. Single-cell transcriptional profiling of cerebral organoids derived from trisomy 21 and euploid hPSC lines.
(A) Schematic illustrating the generation and analysis of cerebral organoids from trisomy 21 and euploid (n = 3 human iPSC lines from individuals with trisomy 21; n = 4 euploid hPSC lines). D30, day 30; D70, day 70. (B) Immunofluorescence analysis of cerebral organoids 30 days after the initiation of differentiation shows the proliferation marker Ki67 (red), the early-born neuron marker CTIP2, and the adherens junction marker PKCλ (gray) in trisomy 21 and euploid organoids. HO, Hoechst staining. Scale bar: 50 μm. (C) Immunostaining for the neural progenitor marker SOX1 (red), the newborn neuron marker DCX (green), and the mitotic marker PHH3 (gray) on day 30 of differentiation in trisomy 21 and euploid organoids. Scale bar: 50 μm. (D) Representative images of organoids showing the neuronal marker TUJ1 (red) and the M-phase marker p-vimentin (gray). Scale bar: 50 μm. (E) UMAP plot of cell types detected in euploid (n = 5) and trisomy (n = 4) organoids. Cyc, cycling; RG, radial glial cells; IPC, intermediate progenitor cells; Glu, glutamatergic neurons; immature IN, immature inhibitory neurons; N-C N, noncommitted neurons. (F) VoxHunt spatial brain mapping of all the clusters in the cerebral organoids onto data from E13.5 mouse brains from the Allen Brain Institute. Coronal sections are shown with scaled similarity scores. Max, maximum. (G) Transcriptome correlation between day-30 organoids and developing neocortex samples from the BrainSpan project (pcw 8–16). The mean Spearman’s correlation coefficients (r) are indicated. (H) Developmental trajectories of the major cell types detected in day-30 trisomy 21 and euploid organoids. (I) Distributions of glutamatergic neurons in trisomy and euploid organoids over pseudotime. (J) GO analysis of DEGs across all cell types between trisomy 21 and euploid organoids. Selected GO terms with a FDR of less than 0.05 are shown. NSC, neural stem cell.
Next, we generated organoids from 3 iPSC lines from 3 patients with DS (DS1, 2DS3, and DSP), 3 euploid iPSC lines (IMR90-4, ihtc-03, and DS2U), and 1 human embryonic stem cell (hESC) line (H9) (Figure 1A). Similar to euploid organoids, trisomy 21 organoids displayed regionalization and cortical layers (Figure 1, B–D). To gain insight into the cell-type–specific transcriptomic changes in trisomy and euploid organoids, we profiled cerebral organoids (n = 65,342 cells from 7 human PSC [hPSC] lines) on day 30 and day 70 using 10× genomics chromium single-cell RNA-Seq (scRNA-Seq) (Figure 1A and Supplemental Table 3). Unbiased clustering identified 8 major cell types (Figure 1E), which were then annotated according to the expression of known cell-type markers (Supplemental Figure 2A). To verify the regional identity of cells in cerebral organoids, we mapped the scRNA-Seq data onto 3D ISH data from the Allen Brain Atlas by using the VoxHunt algorithm. We found that clusters in cerebral organoids highly mapped onto the dorsal forebrain of the E13.5 mouse brain (Figure 1F). In addition, our samples showed a maximum correlation with transcriptomes from post-conception week (pcw) 8 to pcw 16 samples of the developing human brain (Supplemental Figure 2B). We then compared our scRNA-Seq transcriptomic data with the BrainSpan database. Remarkably, organoids showed a significant positive correlation with postmortem fetal tissues at pcw 8 and 9 (Figure 1G). By comparing the proportion of cells in each cluster, we found that cell diversity was similar across lines and groups (Supplemental Figure 2C). The box plot between pseudotime and subclusters supported our speculation that developmental stage classification was positively correlated with pseudotime (Supplemental Figure 2D), while a delayed developmental state from the progenitor-to-neuron trajectory was found in trisomy 21 organoids in comparison with the developmental state of euploid organoids on day 30 (Figure 1, H and I). Gene ontology (GO) analysis showed that the significantly differentially expressed genes (DEGs) among all the clusters were enriched in biological processes such as neurogenesis, forebrain development, and neural precursor cell proliferation (Figure 1J). Consistent with this, pathway enrichment analyses indicated that neurogenesis-related pathways were markedly altered (Supplemental Figure 2E).
Taken together, these data show that neural development in DS cerebral organoids was significantly altered compared with that in euploid organoids.
Disrupted chromatin accessibility underlies impaired transcription in cerebral organoids derived from trisomy iPSCs.
Large genomic changes can result in the disruption of several aspects of chromatin conformation (38), such as the distribution of chromatin accessibility. To test whether global chromatin accessibility was affected in DS, we profiled the genome-wide chromatin accessibility of day-30 cerebral organoids using the assay for transposase-accessible chromatin by sequencing (ATAC-Seq). In general, the distribution of chromatin accessibility remained unchanged in DS cerebral organoids (Supplemental Figure 3A). However, we observed dramatic and widespread increases in chromatin accessibility across the promoter regions of chromosome 21 in DS, which conformed with the characteristics of trisomy 21 (Figure 2, A and B). Many regions on other chromosomes also showed significant changes in accessibility, with 1785 increased and 1695 decreased differentially open chromatin regions (dOCRs) identified in organoids derived from trisomy iPSCs compared with those derived from euploid iPSCs (Figure 2C). Among them, OCRs associated with the markers PAX6, GLPER, PTCH1, LMO1, VCAM1, and WNT7A showed a decrease in chromatin openness at their promoters in the trisomy samples (Supplemental Figure 3B). By identifying the transcription factor motifs enriched in the trisomy- and euploid-specific accessible regions, we found that many genes regulated by SOX2, lsl1, and Rfx5 had decreased accessibility in the trisomy samples (Supplemental Figure 3C). This result was consistent with our finding that genes near dOCRs were enriched in neurogenesis and nervous system development (Supplemental Figure 3D).
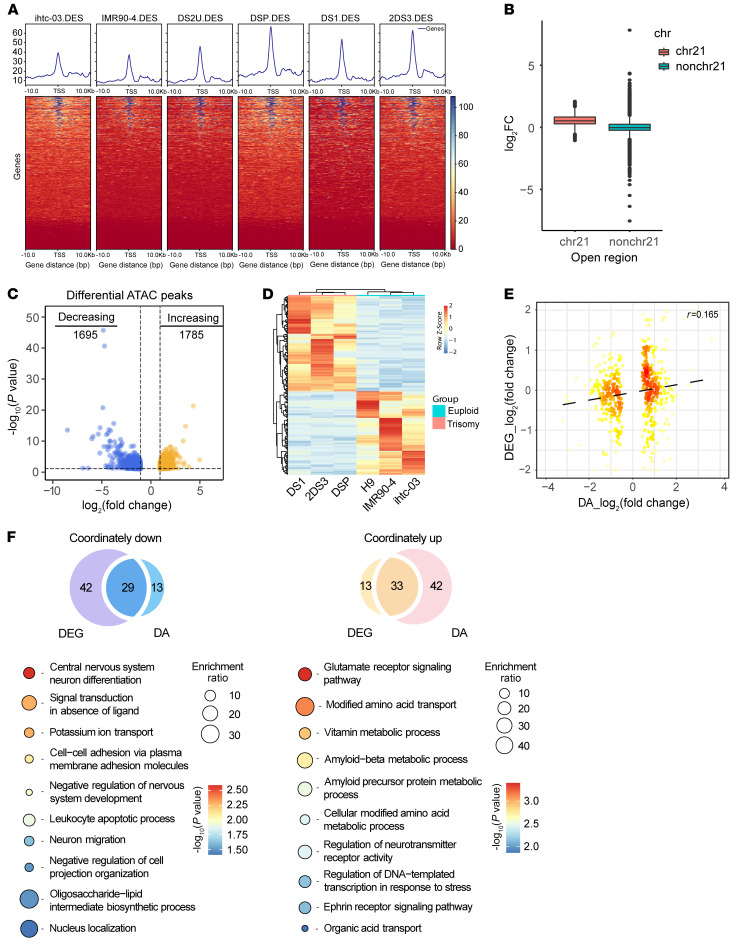
Figure 2. Transcriptional and ATAC-Seq analyses in cerebral organoids derived from trisomy 21 and euploid PSCs.
(A) Heatmaps of regions that are differentially accessible between trisomy 21 and euploid cells at promoter regions on chromosome 21. (B) Box plots for a log2 fold change of OCRs among chromosome 21 (chr21) and other chromosomes (nonchr21). (C) Volcano plot of differential ATAC peaks in trisomy 21 cerebral organoids. Differential ATAC peaks were identified by DESeq2. The color intensity represents the density of the points in the volcano plot. Increased ATAC sites are shown in orange (log2 fold change >1 and P < 0.05, n = 1785), and decreased ATAC sites are shown in blue (log2 fold change <–1 and P < 0.05, n = 1695). (D) Heatmap of transcriptome analysis shows 193 significantly DEGs in trisomy 21 organoids compared with euploid control organoids with a fold change of greater than 2 and q value of less than 0.05. DA, differentially accessible. (E) Correlations between gene expression (log2 fold change) and chromatin accessibility (log2 fold change). Pearson’s correlation coefficient r = 0.165. (F) GO analysis of the genes showing coordinately altered expression and accessibility between trisomy 21 and euploid organoids.
To examine whether disruption of chromatin accessibility would be responsible for transcriptomic changes, we performed bulk RNA-Seq analyses of day-30 cerebral organoids in the 2 groups. Differential expression profiling of 104 upregulated genes and 91 downregulated genes is displayed as a heatmap in Figure 2D (see also Supplemental Table 5). Consistent with the findings of the scRNA-Seq data, the DEGs between the trisomy- and euploid-derived organoids showed significant enrichment of genes related to nervous system development, cell proliferation, and neurogenesis (Supplemental Figure 3, E and F, and Supplemental Tables 6 and 7). Notably, the expression of Ki67 and PAX6, which are involved in regulating proliferation, was decreased in the trisomy organoids (Supplemental Figure 3, G and H). We then mapped the 20 differentially expressed proteins related to neurogenesis and 29 proteins related to cell proliferation using the STRING database (Supplemental Figure 3I). Not surprisingly, we observed a positive correlation between the change in a differentially accessible peak and the fold change in expression of its nearest gene (Spearman’s coefficient r = 0.165; Figure 2E). GO analyses of the coordinately upregulated and downregulated genes were enriched among biological processes such as central nervous system neuron differentiation, neuron migration, and glutamate receptor signaling (Figure 2F).
Taken together, these results suggested that at least part of the transcriptomic changes in DS are linked to the altered accessibility of their adjacent chromatin.
Reduced proliferation of NPCs results in smaller DS-derived cortical organoids.
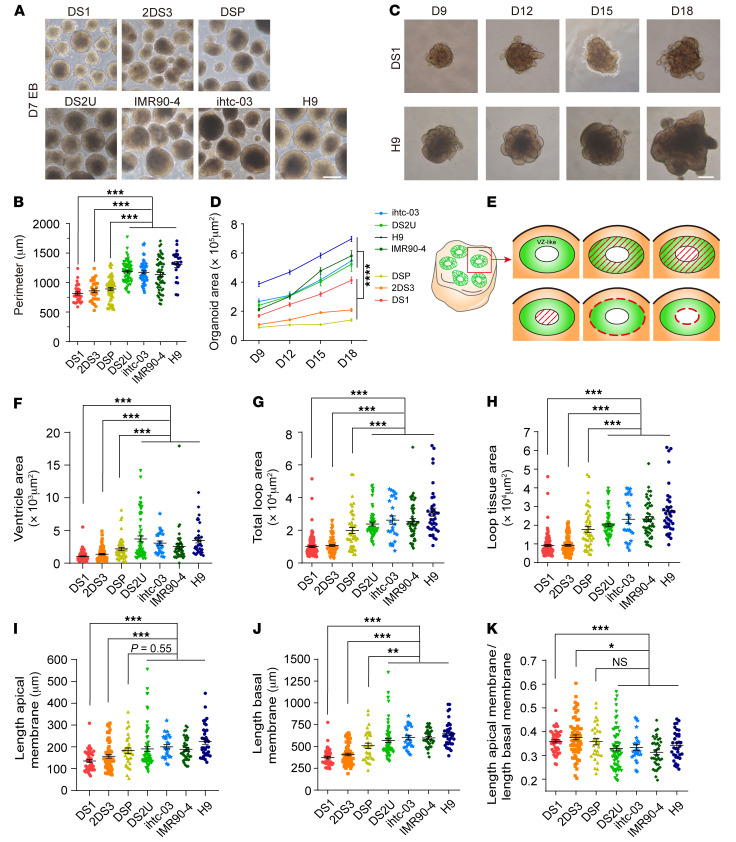
To verify the neuropathological phenotypes observed in the RNA-Seq data, we compared the proliferation of DS and control organoids. Strikingly, in contrast to euploid embryonic bodies (EBs) on day 7, trisomy EBs showed a reduced perimeter length (Figure 3, A and B). Moreover, the size of the trisomy 21 organoids was obviously smaller than that of the euploid organoids, and the former had significantly decreased expansion rates (Figures 3, C and D, and Supplemental Figure 4C). To further examine the abnormal structures in day-30 trisomy 21 organoids, we assessed the architecture of neuroepithelial loops with respect to the length of the apical and basal membranes, the diameter of the loops, and the size of the ventricle-like area, total loop area, and loop tissue area in the trisomy and euploid organoids (Figure 3E). We found that, compared with the euploid organoids, the trisomy 21 organoids had reductions in all parameters (Figure 3, F–K). This phenotype is consistent with previous observations of a reduction in brain size in patients with DS (2, 4–6).
Figure 3. Reduced size and expansion rates of organoids from patients with DS.
(A) Bright-field microscopic images of trisomy 21 and euploid EBs on day 7. Scale bar: 250 μm. (B) Quantification of EB perimeters on day 7. At least 25 EBs were analyzed for each cell line; n ≥3 independent experiments. Data represent the mean ± SEM. ***P < 0.001, by 1-way ANOVA followed by Dunnett’s multiple-comparison test. (C) Bright-field microscopic images of trisomy 21 and euploid organoids at different developmental time points. Scale bar: 250 μm. (D) Quantification of the organoid area of trisomy 21 and euploid organoids on days 9, 12, 15, and 17 after differentiation reflected a reduction in the expansion rate of trisomy 21 organoids compared with euploid organoids. Organoids (n ≥16) from 3 independent biological replicate experiments were analyzed for each cell line. Data represent the mean ± SEM. ****P < 0.0001, by 2-way ANOVA followed by Sidak’s multiple-comparison test. (E) Schematic overview of the different parameters of neuroepithelial loops in organoids 30 days after the induction of differentiation. Shown are loop tissue area (top middle), total loop area (top right), ventricle area (bottom left), basal membrane length (bottom middle), and apical membrane length (bottom right). (F–K) Quantification of a series of parameters in neuroepithelial loops of trisomy 21 and euploid organoids on day 30. Organoids (n ≥16) from 3 independent biological replicate experiments were analyzed for each cell line. Data represent the mean ± SEM. ***P < 0.001, by 1-way ANOVA followed by Dunnett’s multiple-comparison test.
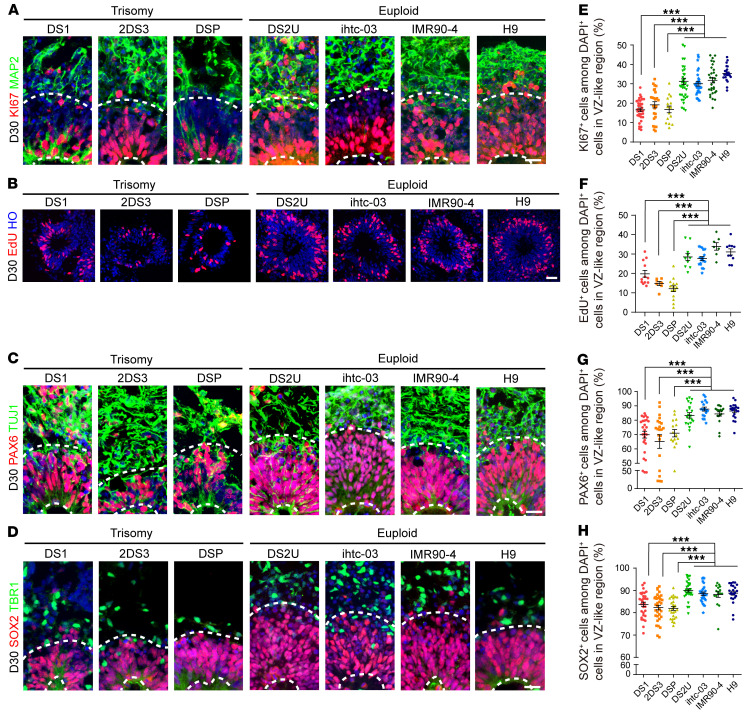
Defects in NPC proliferation during early neural development could influence the size of the forebrain (39, 40). To test whether altered proliferation of ventricular zone (VZ) NPCs may delay the organoid expansion of trisomy, we quantified the percentages of Ki67+, EdU+, and PAX6+ cells in the VZ-like region 30 days after the initiation of differentiation (Figure 4, A–C and E–G). In accordance with the reduction in organoid size, we found not only markedly decreased proliferation in the trisomy cortical VZ-like regions but also fewer SOX2+ progenitor cells located in the region (day 30: DS1, 83.76% ± 1%; 2DS3, 82.31% ± 1.02%; DSP, 81.96% ± 0.88%; DS2U, 90.01% ± 0.77%; ihtc-03, 88.53% ± 0.77%; IMR90-4, 88.16% ± 0.77%; and H9, 88.79% ± 0.90%; Figure 4, D and H), whereas the expression of apoptosis markers was not significantly altered at the same time point (Supplemental Figure 4, D and E). These results suggested that decreased proliferation of trisomy NPCs was responsible for the smaller size of the trisomy 21 organoids.
Figure 4. Proliferation studies of day-30 cerebral organoids.
(A–D) Immunofluorescence of Ki67+, EdU+, SOX2+, and PAX6+ proliferating radial glial progenitors, mature MAP2+, TUJ1+ neurons, and deep-layer VI TBR1+ excitatory neurons after 30 days of differentiation. Scale bar: 20 μm. (E–H) Quantification of the proportion of Ki67+, SOX2+, PAX6+, and EdU+ cells in trisomy 21 and euploid organoids after 30 days of differentiation. n = 15–42 VZ-like regions in at least 5 organoids per cell line. Data represent the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, by 1-way ANOVA followed by Dunnett’s multiple-comparison test.
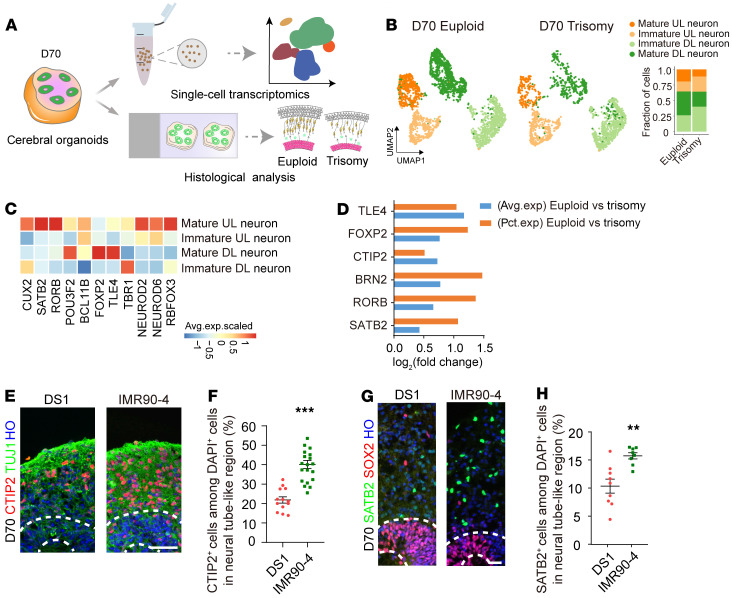
We next assessed the generation of different subtypes of cortical neurons using the validated markers 50 days after differentiation initiation. We found that the percentage of CTIP2+ cells was decreased in the DS group compared with the euploid control group (DS1, 17.01% ± 1.87%; IMR90-4, 31.1% ± 2.17%), whereas the percentage of TBR1+ cells was unchanged (Supplemental Figure 4, F–H). To further study the later developmental stages in DS, we performed scRNA-Seq and histological analysis of day-70 cerebral organoids (Figure 5A). Uniform manifold approximation and projection (UMAP) visualization of subclusters of glutamatergic neurons showed that the composition of cortical mature deep-layer and mature upper-layer neurons was dramatically decreased in the trisomy 21 organoids (Figure 5, B and C). Moreover, the proportion of cells expressing cortical upper-layer markers, such as BRN2 and SATB2, was decreased in trisomy glutamatergic neurons (Figure 5D). Indeed, our histology results showed that the percentages of CTIP2+ and SATB2+ cells were significantly decreased by day 70 in the DS-derived organoids, which was consistent with the gene expression profile (CTIP2: DS1, 21.87% ± 1.63%; IMR90-4, 40.13% ± 2.1%; SATB2: DS1, 9.23% ± 1.03%; IMR90-4, 15.75% ± 0.5%; Figure 5, E–H).
Figure 5. Neurogenesis studies of cerebral organoids.
(A) Schematic illustrating the single-cell transcriptomic and histological analysis of trisomy 21 and euploid organoids. (B) UMAP visualization of scRNA expression in glutamatergic neuron subclusters of trisomy 21 and euploid organoids after 70 days of in vitro differentiation. Chart on the right shows the comparisons of cell composition between the trisomy 21 and euploid organoids after 70 days of differentiation. DL, deep layer; UL, upper layer. (C) Average expression (avg.exp.) levels of representative markers in each glutamatergic neuron subcluster are shown according to scaled expression scores. (D) Histogram of the log2 fold change of average expression levels and percentage (pct.) of expression for significantly different genes in the glutamatergic neuron subcluster. (E) Images show decreased maturation of CTIP2+ neurons on day 70 in trisomy 21 organoids compared with euploid organoids. Scale bar: 50 μm. (F) Quantification of the proportion of CTIP2+ cells in both trisomy and euploid organoids on day 70. n = 13–20 neural tube–like regions in at least 5 organoids per cell line. Data represent the mean ± SEM. ***P < 0.001, by Student’s t test. (G) Images show decreased maturation of SATB2+ neurons on day 70 in trisomy 21 organoids compared with euploid organoids. Scale bar: 25 μm. (H) Quantification of the proportion of SATB2+ cells in trisomy and euploid organoids on day 70. n = 9–11 neural tube–like regions in at least 5 organoids per cell line. Data represent the mean ± SEM. **P < 0.01, by Student’s t test.
Taken together, these results suggested that decreased NPC proliferation might contribute to reduced neurogenesis in trisomy 21 organoids.
KD of DSCAM reverses proliferation defects in DS organoids.
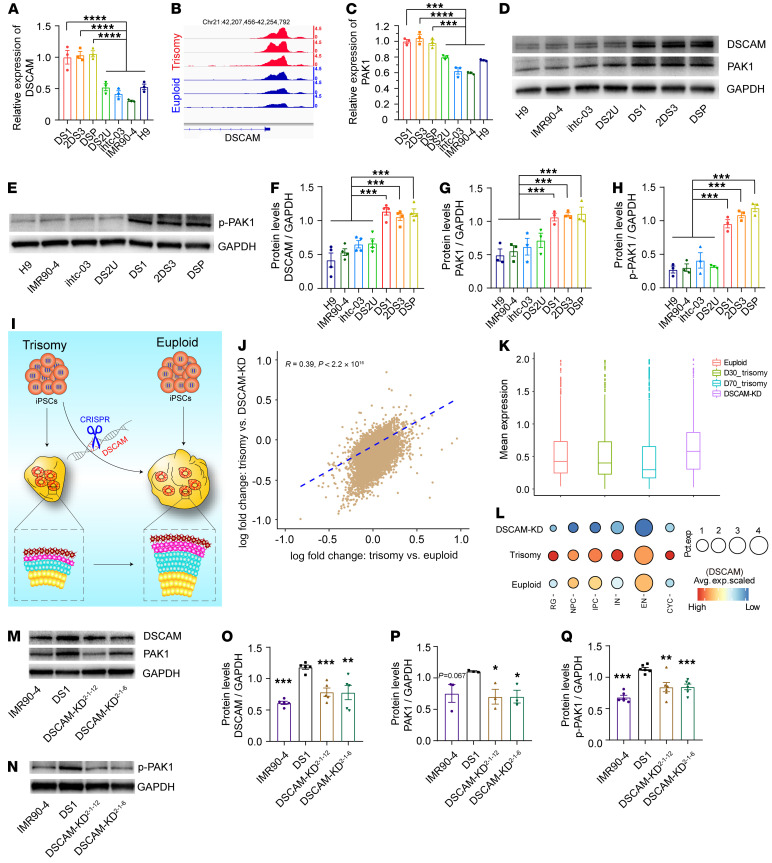
Previous studies have demonstrated that DSCAM is significantly overexpressed in postmortem tissues from individuals with DS (41) and animal models of DS (42–44). In line with previous studies, we found that DSCAM expression was increased in cells with trisomy 21, as assessed by quantitative real-time PCR (qPCR) (Figure 6A). Meanwhile, OCRs of the marker DSCAM also showed a marked increase in chromatin openness at their promoters in trisomy 21 organoids (Figure 6B). Strikingly, we observed that the protein levels of DSCAM in DS organoids were also significantly higher than those in the controls (Figure 6, D and F). PAK1 is the downstream effector of DSCAM signaling and plays a key role in proliferation and synaptic plasticity during the neurodevelopmental stage (32), which prompted us to probe the expression levels of PAK1. Indeed, qPCR analysis showed a higher level of PAK1 mRNA in trisomy cells than in euploid control cells (Figure 6C), which was consistent with the observed protein level (Figure 6, D and G). In addition, the protein level of p-PAK1 was substantially increased in DS organoids compared with euploid organoids (Figures 6, E and H). These results show that the DSCAM/PAK1 pathway was altered in trisomy 21 organoids.
Figure 6. DSCAM-KD rescues impaired DSCAM/PAK1 signaling in DS-derived cortical cultures.
(A) Relative expression levels of DSCAM in day-30 trisomy 21 and euploid organoids as assessed by qPCR. (B) Coverage maps of normalized ATAC-Seq signals from trisomy 21 and euploid organoids showing a differentially accessible (DA) peak near the DSCAM gene on chromosome 21. (C) Relative mRNA expression levels of PAK1 in day-30 trisomy 21 and euploid organoids as assessed by qPCR. (D and E) Detection of DSCAM, PAK1, and p-PAK1 expression in trisomy 21 and euploid organoids on day 30, as assessed by Western blotting. (F–H) Representative quantitation of relative DSCAM, PAK1, and p-PAK1 protein expression in trisomy 21 and euploid organoids. n ≥3 independent experiments. Data represent the mean ± SEM. ***P < 0.001, by 1-way ANOVA followed by Dunnett’s multiple-comparison test. (I) Schematic diagram illustrating the effects of DSCAM KD on proliferation and neurogenesis in trisomy 21 and euploid cerebral organoids. (J) Correlations of the changes in expression between trisomy 21 organoids and DSCAM-KD organoids and between trisomy 21 organoids and euploid organoids. Spearman’s correlation coefficient: r = 0.39. (K) Box plot showing the average expression level of the trisomy 21–associated DEGs that were downregulated among trisomy 21, euploid, and DSCAM-KD cerebral organoids. (L) Dot plot showing DSCAM expression levels in multiple clusters among trisomy 21, euploid, and DSCAM-KD cerebral organoids. The size of each circle reflects the percentage of cells in a cluster where DSCAM was detected, and the color intensity reflects the average expression level within each cluster. (M and N) Representative Western blots of DSCAM, PAK1, and p-PAK1 levels in trisomy 21 and DSCAM-KD organoids. (O–Q) Representative relative quantitation of DSCAM, PAK1, and p-PAK1 expression levels in trisomy 21 and DSCAM-KD organoids. n ≥3 independent experiments. Data represent the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, by 1-way ANOVA followed by Dunnett’s multiple-comparison test.
We then hypothesized that the neurogenesis defects in DS observed in vitro are attributable to altered expression of DSCAM and its downstream protein PAK1. To test this hypothesis, we used CRISPR/Cas9 genome editing to establish DSCAM-knockdown (DSCAM-KD) iPSCs from the DS1 iPSC line (Figure 6I and Supplemental Figure 5A), followed by differentiation of the DS and edited iPSC lines (referred to herein as DSCAM-KD2-1-6 and DSCAM-KD2-1-12) into cerebral cortical organoids. Karyotype analysis of DS and DSCAM-KD iPSC–derived organoids showed trisomy of HSA21 (Supplemental Figure 5B). scRNA expression profiling of DSCAM-KD cerebral organoids 30 days after the induction of differentiation showed that there was a positive correlation in the changes in expression between DSCAM-KD organoids and euploid organoids with respect to trisomy, which indicated successful restoration of the transcriptome in DSCAM-KD organoids (Figure 6J). In addition, we noticed that the mean expression levels of genes downregulated in DS were restored in the DSCAM-KD groups to levels similar to those in the euploid groups (Figure 6K). Further analysis showed that the mean expression level of DSCAM was markedly downregulated in subclusters of DSCAM-KD cerebral organoids compared with expression levels in DS1 cerebral organoids (Figure 6L).
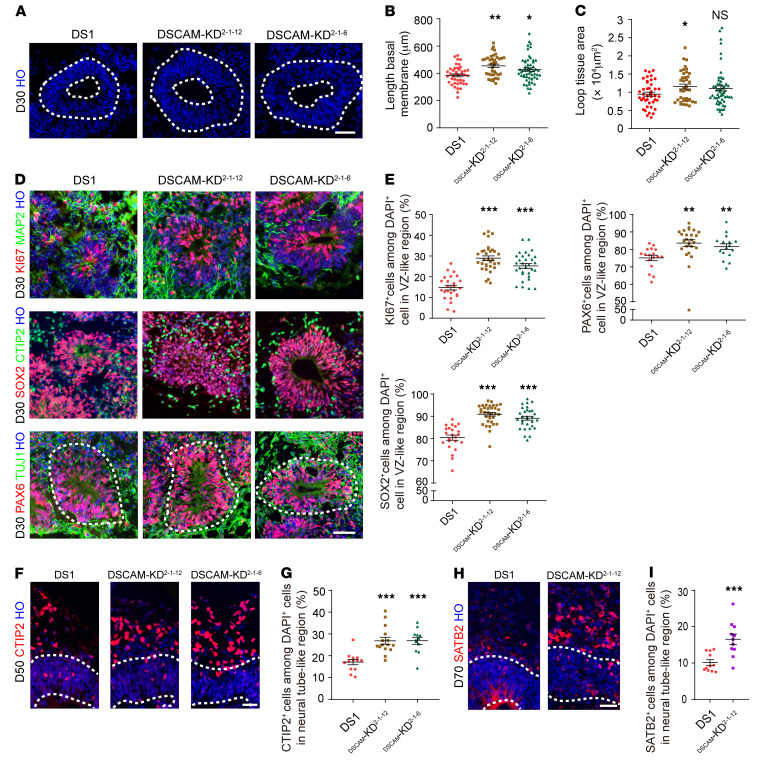
Specifically, the protein expression levels of DSCAM, PAK1, and p-PAK1 were decreased in the DSCAM-KD groups compared with expression levels in the DS1 group (Figure 6, M–Q). These results confirmed the efficient knockdown of DSCAM in the DSCAM-KD2-1-6 and DSCAM-KD2-1-12 groups, with a prominent and consistent reduction at both the mRNA and protein levels. Furthermore, we observed a partial rescue in all parameters in DSCAM-KD organoids compared with DS1 organoids (Figure 7, A–C, and Supplemental Figure 5, C–E).
Figure 7. Knocking down DSCAM rescues abnormal neurogenesis in DS-derived cortical cultures.
(A) Representative images of day-30 organoids stained with Hoechst, which show the quantitation of the different parameters. Scale bar: 50 μm. (B and C) Quantitation of the basal membrane length and loop tissue area in the neuroepithelial loops of trisomy 21 and euploid organoids after 30 days of differentiation. Organoids (n ≥15) from 3 independent biologic replicate experiments were analyzed for each cell line. Data represent the mean ± SEM. *P < 0.05 and **P < 0.01, by 1-way ANOVA followed by Dunnett’s multiple-comparison test. (D) Representative images of day-30 trisomy 21 and DSCAM-KD organoids stained for Ki67, SOX2, PAX6, MAP2, CTIP2, and TUJ1 expression. Scale bar, 50 μm. (E) Quantification of the proportion of Ki67+, SOX2+, and PAX6+ cells in day-30 trisomy 21 and DSCAM-KD organoids. n = 17–33 VZ-like regions in at least 10 organoids per cell line. Data represent the mean ± SEM. **P < 0.01 and ***P < 0.001, by 1-way ANOVA followed by Dunnett’s multiple-comparison test. (F and G) Immunocytochemical staining and quantification of CTIP2+ cells in both trisomy 21 and DSCAM-KD organoids after 50 days of differentiation. n = 13–15 neural tube–like regions in at least 7 organoids per cell line. Data represent the mean ± SEM. ***P < 0.001, by 1-way ANOVA followed by Dunnett’s multiple-comparison test. Scale bar: 35 μm. (H and I) Immunocytochemical staining and quantification of the proportion of SATB2+ cells in both trisomy and DSCAM-KD organoids on day 70. n = 11 neural tube–like regions in at least 6 organoids per cell line. Data represent the mean ± SEM. ***P < 0.001, by Student’s t test. Scale bar: 35 μm.
Next, we examined whether a reduction in DSCAM gene dosage in DS1 organoids could rescue the proliferation deficits observed in dorsal neural progenitors. Remarkably, we found that there was increased expression of Ki67, PAK6, and SOX2 in the VZ-like region in the DSCAM-KD groups (Figure 7, D and E), suggesting enhanced proliferation upon KD of DSCAM. Importantly, the reduced DSCAM gene dosage in the trisomy 21 organoids restored the numbers of CTIP2+ neurons (DS1: 17.11% ± 1.20%; DSCAM-KD2-1-12: 26.86% ± 1.62%; DSCAM-KD2-1-6: 26.88% ± 1.62%) and SATB2+ neurons (DS1: 10.14% ± 0.76%; DSCAM-KD2-1-12: 16.54% ± 1.44%) to numbers similar to those in the euploid organoids (Figure 7, F–I).
To confirm the effects of DSCAM in DS, we further downregulated DSCAM in DS1 by performing a CRISPR interference (CRISPRi) experiment (Supplemental Figure 6, A–D). The expression levels of DSCAM, PAK1, and p-PAK1 were robustly increased in DSCAM-KD cerebral organoids (Supplemental Figure 6, E–G). Furthermore, the perimeter of day-7 EBs, the size of the VZ in the organoids, cell proliferation, and cortical neurogenesis were effectively elevated in the CRISPRi-based DSCAM-KD groups (Supplemental Figure 7, A–H).
Taken together, these results demonstrated that the proliferation and neurogenesis deficits in trisomy 21 organoids can be rescued by regulating the DSCAM gene.
FRAX486 rescued the abnormal proliferation and neurogenesis of DS organoids.
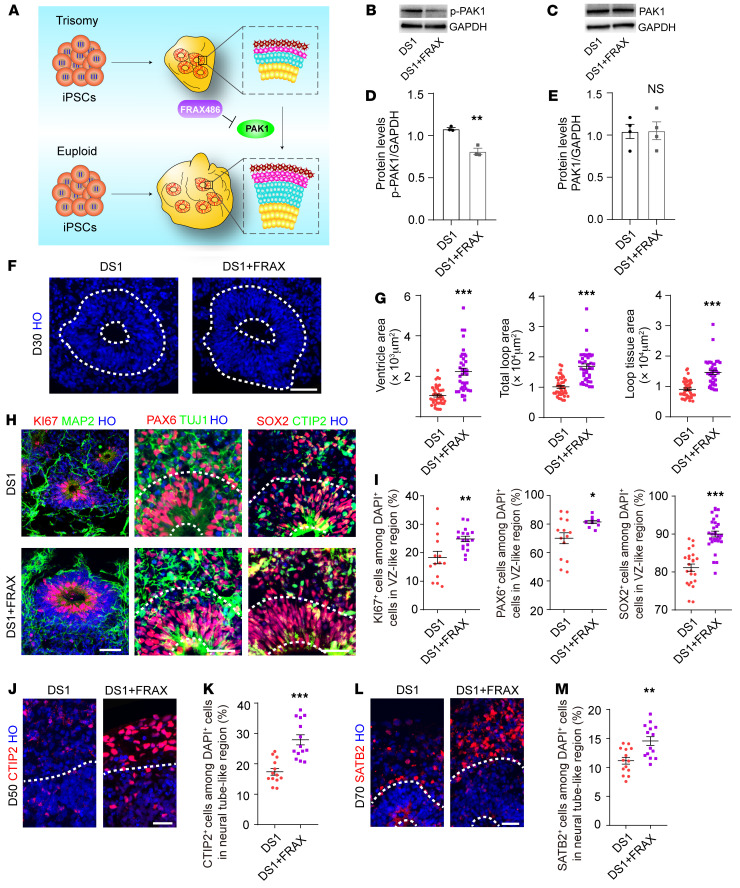
Considering that the DSCAM/PAK1 pathway is involved in neurodevelopment and is altered in DS (32, 33), we investigated whether the defects could be rescued by applying small molecules targeting this pathway (Figure 8A). Indeed, pretreatment of DS1 organoids with FRAX486, an inhibitor that regulates the phosphorylation of PAK1, reduced the protein levels of p-PAK1 (Figures 8, B and D), without changing the expression levels of total PAK1 (Figure 8, C and E). Of note, we did not find significant changes in the protein levels of PAK1 or p-PAK1 in euploid organoids that were pretreated with FRAX486 (Supplemental Figure 8, A–H). Furthermore, we tested whether inhibition of PAK1 with FRAX486 could ameliorate the defective neurogenesis of DS organoids. Indeed, we observed a partial rescue of the abnormal architecture of neuroepithelial loops (Figure 8, F and G, and Supplemental Figure 8, I–K). In addition, FRAX486 effectively increased the proliferation of NPCs after 30 days of differentiation (Ki67: DS1, 18.32% ± 2.19%; DS1 plus FRAX486, 24.85% ± 0.95%; PAX6: DS1, 70.05% ± 3.84%; DS1 plus FRAX486, 81.50% ± 1.13%; and SOX2: DS1, 81.14% ± 0.97%; DS1 plus FRAX486, 90.01% ± 0.77%; Figure 8, H and I). Furthermore, PAK1 inhibition via FRAX486 treatment subsequently increased the expression of CTIP2 (50 days after the initiation of differentiation, DS1: 17.37% ± 1.11%; DS1 plus FRAX486: 27.92% ± 1.67%) and SATB2 (70 days after the initiation of differentiation, DS1, 11.17% ± 0.6%; DS1 plus FRAX486, 14.57% ± 0.75%), suggesting the rescue of neurogenesis (Figure 8, J–M).
Figure 8. Downregulation of PAK1 rescues proliferation and neurogenesis deficits in DS.
(A) Schematic of PAK1 correction. (B and C) Western blot detection of PAK1 and p-PAK1 expression in trisomy 21 organoids 30 days after treatment with FRAX486 (FRAX). (D and E) Western blot analysis of PAK1 and p-PAK1 expression in trisomy 21 organoids 30 days after treatment with FRAX486. n ≥3 independent experiments. Data represent the mean ± SEM. **P < 0.01, by Student’s t test. (F) Images showing outlines of the different parameters in trisomy 21 organoids and rescued organoids on day 30. Scale bar: 50 μm. (G) Quantitation of the different parameters of neuroepithelial loops in trisomy 21 and rescued organoids. Organoids (n ≥10) from 3 independent biological replicate experiments were analyzed for each cell line. Data represent the mean ± SEM. ***P < 0.001, by Student’s t test. (H) Representative images of trisomy 21 and rescued organoids stained for Ki67, SOX2, PAX6, MAP2, CTIP2, and TUJ1. Scale bars: 20 μm. (I) Proportion of Ki67+, SOX2+, and PAX6+ cells in trisomy 21 and rescued organoids on day 30. n = 10–30 VZ-like regions in at least 6 organoids per cell line. Data represent the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, by Student’s t test. (J and K) Immunocytochemical staining and quantification of the proportion of CTIP2+ cells in day-50 trisomy 21 and rescued organoids. n = 13–14 neural tube–like regions in at least 7 organoids per cell line. Data represent the mean ± SEM. ***P < 0.001, by Student’s t test. Scale bar: 35 μm. (L and M) Immunocytochemical staining and quantification of the proportion of SATB2+ cells in day-70 trisomy 21 and rescued organoids. n = 9–14 neural tube–like regions in at least 6 organoids per cell line. Data represent the mean ± SEM. **P < 0.01, by Student’s t test. Scale bar: 35 μm.
Discussion
In this study, we evaluated cortical developmental defects in DS using a patient iPSC–derived cerebral organoid model. The DS organoids showed reduced proliferation in the VZ, decreased neuron distribution in the cortical plate, and smaller cerebral organoids. Transcriptomic and Western blot results revealed that DSCAM and PAK1 may contribute to these defects. Moreover, genetic correction and treatment with small-molecule inhibitor targeting of the DSCAM/PAK1 pathway rescued the neurogenesis deficits.
A recent study using DS iPSCs reported that overexpression of OLIG2 in DS cells may cause overproduction of subclass-specific GABAergic interneurons (primarily CR+ and SST+ neurons), which might contribute to intellectual disabilities related to trisomy 21 (45); however, this work did not mention cortical development in DS. Early studies using postmortem tissue from individuals with DS have demonstrated that these individuals had a 20%–50% reduction in neurons in the granular layers (2, 46, 47). In addition, subsequent studies on DS fetal brains described fewer neurons (20%–50% less than the controls) and a significant reduction in brain weight at pcw 15 (6, 48). These observations suggested impaired neurogenesis in the cortex during prenatal neurodevelopment. In line with this hypothesis, we demonstrated reduced proliferation of progenitors and a subsequent reduction in the number of cortical neurons expressing markers corresponding to layers IV and II (Figures 4 and 5). These observations provide evidence of impaired neurogenesis of cortical neurons that may underlie the reduced cortical size in the brains of individuals with DS.
Many attempts have been made to investigate HSA21 genes associated with neurological defects in DS (49, 50). However, the molecular mechanisms underlying smaller brain sizes remain unknown. Our transcript profiling indicated that multiple neurogenesis pathways were altered in the cerebral organoids derived from DS iPSCs. We focused on the DSCAM pathway and showed that DSCAM expression was upregulated in DS iPSC–derived cerebral organoids. DSCAM is critical for neurodevelopment in flies and mice (32, 51), but its role in human brain development is poorly understood. Strikingly, our Western blot analysis revealed increased DSCAM expression in cerebral organoids derived from iPSCs from patients with DS. Overexpression of DSCAM in neurons from a mouse model of DS could deregulate the activity of its downstream gene PAK (32). Indeed, we found abnormal activation of PAK1 and p-PAK1 in the DS groups, which may be partially responsible for the neurogenesis defects in DS.
Recently, DSCAM has received increasing attention because of its implication in neurodevelopment, but the function of DSCAM in NPCs is not entirely known. Prior work has revealed that knocking down DSCAM increases the complexity of proximal dendritic branching and inhibits axon growth in mouse cortical neurons (52). However, the role of DSCAM in cortical neurogenesis in primates has not been studied. Its downstream gene PAK1 has been reported to play an essential role in regulating actin cytoskeleton dynamics, dendritic spine morphogenesis, and cortical neurogenesis in a mouse model (33, 53). Here, we used both genetic correction and small-molecule inhibitors to rescue NPC proliferation and neurogenesis defects in DS organoids. Our work indicated that the DSCAM/PAK1 pathway may play a role in regulating proliferation and neurogenesis, which might be associated with the abnormal development of DS neurons.
Our cerebral organoid models provide a new avenue to study the early developmental deficits in DS. By using transcriptomic analysis, CRISPR-based gene correction, and small-molecule intervention, we demonstrated that the malformation of cortical development in DS was attributed to reduced neuronal proliferation of progenitors and was accompanied by neurogenesis deficits. Among them, dysfunction of the DSCAM/PAK1 pathway has been shown to play a critical role in the pathogenesis of DS. On the one hand, our current analyses provide candidates for identifying therapeutic targets and screening drugs for the treatment of DS. Moreover, DSCAM and PAK1 are potential therapeutic targets to reverse abnormal neurodevelopment and improve postnatal cognitive function in DS.
Methods
iPSC culturing and generation of organoids.
Trisomy 21 iPSC lines (DS1, 2DS3, and DSP) and euploid iPSC lines (IMR90-4, H9, DS2U and ihtc-03) were used in this study (a detailed list is provided in Supplemental Table 1). DS iPSC lines were generated from 2 patients with DS as reported by Weick et al. (11) or from an individual with mosaic DS. The euploid iPSC lines DS2U (the euploid control was from the same patient with DS1), wild-type IMR90-4 (WiCell agreement no. 17-W0063), ihtc-03 (established in our laboratory), and the hESC line H9 (WiCell agreement no. 16-W0060) served as euploid controls. iPSC lines were maintained under feeder-free conditions by coating the culture plates with vitronectin (Thermo Fisher Scientific) as described in our previous studies (35). After 5–7 days of culturing in E8 medium (Thermo Fisher Scientific), hPSCs were dissociated with EDTA (Lonza) for 1–2 minutes at 37°C and seeded in a 6-well plate at a density of 1 × 105 cells per well. Detachment of iPSCs to obtain embryoid bodies (EBs) required dispase (Thermo Fisher Scientific) to begin the process of neural differentiation; the bodies were then cultured in neural induction medium containing N2 supplement (Thermo Fisher Scientific), nonessential amino acids (MEM-NEAA, Thermo Fisher Scientific), and DMEM/F12 (Thermo Fisher Scientific) for 7 days. On day 7, EBs were resuspended in Matrigel (Corning), which was pipetted cold in 3 mm dimples on a sheet of Parafilm that was sterilized by UV light for 30 minutes. These droplets solidified at 37°C and were subsequently removed from the Parafilm and grown in differentiation medium that was changed every 5 days.
Genome editing.
DS iPSC lines with DSCAM KD were generated by CRISPR/Cas9. Exon 1 of DSCAM was selected for the guide RNA (gRNA) design according to the CRISPR online design tool at http://crispr.mit.edu/ The DSCAM gRNA sequences of a pair of oligonucleotides for the targeting site were as follows: forward, 5′-CAGGCGATGAAAGACGTGAAATGT-3′; reverse, 5′-AACCATGAGAGGCAATGTTG-3′. After transformation and extraction of the plasmid, 15 μg gRNA-containing plasmid was transferred to 50,000 DS1 cells using an electroporation apparatus (Lonza). After 24 hours of electroporation, puromycin was added to the cell cultures for 2 days. Three to 4 days after electroporation, single clones were picked and reseeded in a 24-well plate. KD of the DSCAM gene was verified by sequencing, and 2 iPSC lines — DSCAM-KD2-1-12 and DSCAM-KD2-1-6 — were obtained. As KD groups, DSCAM-KD2-1-12 and DSCAM-KD2-1-6 were used to verify the molecular mechanism related to the DSCAM/PAK1 pathway during cortical development.
Immunostaining.
Organoids were fixed with 4% paraformaldehyde for 2 hours in an Eppendorf tube. After they were washed with PBS for 10 minutes 3 times, the organoids were submerged in 20% sucrose in PBS overnight at 4°C. After the organoids sank to the bottom of the tube, the soaking solution was replaced by 30% sucrose in PBS at 4°C. Organoids were embedded in OCT compound and cryosectioned at 10 μm. Tissue sections were then used for immunostaining. For immunohistochemical analysis, sections were washed with PBS 3 times and then blocked and permeabilized in 1% Triton (Bio-Link) and 5% donkey serum (MilliporeSigma) in PBS. Organoids were incubated at 4°C overnight in primary antibodies (see detailed list in Supplemental Table 2) diluted in 0.2% Triton and 5% donkey serum. On the second day, organoids were incubated for 1 hour at 20°C in secondary antibodies (see detailed list in Supplemental Table 2) diluted in 5% donkey serum. After treatment with primary and secondary antibodies, three 10-minute washes in PBS were performed. Coverslips were mounted for fluorescence imaging. Images were acquired using an Eclipse 80i fluorescence microscope (Nikon).
EdU Click-iT assay.
Organoids were kept in neural induction medium with EdU (10 μM, Life Technologies, Thermo Fisher Scientific) for 2 hours. Then, the neural induction medium was discarded before the organoids were fixed with 4% paraformaldehyde for 2 hours at room temperature. After they were washed twice with 3% BSA, the organoids were maintained in 0.5% Triton for 20 minutes at 20°C, after which an EdU Click-iT assay was performed according to the manufacturer’s instructions (Invitrogen, Thermo Fisher Scientific, C10338), followed by immunostaining.
Western blot analysis.
Organoids were lysed in RIPA buffer containing protease and a protease inhibitor cocktail (Roche). Proteins were loaded onto gels (SurePage) and separated by SDS-PAGE with 100 V electrophoresis. Then, proteins were transferred onto polyvinylidene fluoride membranes at 300 mA for 2 hours and blocked in 5% skim milk for 2 hours at room temperature. Primary antibodies (listed in Supplemental Table 2) were incubated overnight at 4°C before the membranes were washed with 8× PBST solution 5 times for 8 minutes the next day. The secondary antibodies were incubated with the membranes on a shaker for 2 hours at room temperature. After the incubation was completed, the secondary antibody was decanted, and the membranes were again washed 5 times with 1× PBST for 8 minutes. Anti-GAPDH was used as an internal reference and loading control. HRP-conjugated IgG was used as the secondary antibody, and the ECL system was used for detection of the protein bands. The luminol substrate solutions A and B were mixed at a volume of 1:1 and then added to the surface of the membrane in the dark. After 1 minute, the protein bands were exposed over a time gradient.
Bulk RNA-Seq, ATAC-Seq, and bioinformatics analysis.
Following a standard protocol, total RNA from day-30 trisomy 21 and euploid organoids was extracted using a TRIzol reagent kit (Invitrogen, Thermo Fisher Scientific). RNA integrity was checked on an Agilent 2100 BioAnalyzer to conduct quality control (Agilent Technologies). Library construction and high-throughput RNA-Seq were performed with the HiSeq 4000 sequencing platform (Illumina). RNA-Seq reads were aligned to the human reference genome (GRCh37/hg19) using HISAT2 software (version 2.1.0.) (54).
The gene abundances were calculated and normalized as transcripts per million (TPM). We determined DEGs between the trisomy 21 and euploid groups using the DESeq2 (55) package (version 1.30.0). Enriched GO terms were identified with MGI Gene Ontology Term Finder (http://www.informatics.jax.org/gotools/MGI_Term_Finder.html). A log2 fold change of 1 or higher and a P value of less than 0.05 were used as thresholds. Protein-protein interaction (PPI) network analysis of the differentially expressed proteins was performed using STRING version 10.0a software.
For ATAC-Seq, organoids were dissociated into a single-cell suspension. Approximately 50,000 single cells from each group were used for nuclei preparation. First, cells were spun for 5 minutes at 500g and 4°C and then washed once with 50 μL cold 1× PBS buffer before another centrifugation step for 5 minutes at 500g and 4°C. Next, cells were lysed in 50 μL cold lysis buffer (10 mM Tris-HCl, pH 7.4; 10 mM NaCl; 3 mM MgCl2 and 0.1% IGEPAL CA-630) on ice, and nuclei were pelleted by centrifugation at 500g for 10 minutes. Nuclei were collected and resuspended in 50 μL transposase reaction mix from a Nextera DNA Sample Preparation Kit (Illumina), followed by incubation at 37°C for 30 minutes to carry out the Tn-5 transposition reaction. Next, DNA fragments were purified using a MinElute PCR Purification Kit (QIAGEN). Transposed DNA fragments were then amplified using the following PCR conditions: 1 cycle at 72°C for 5 minutes and 98°C for 30 seconds, followed by 10 cycles at 98°C for 10 seconds, 63°C for 30 seconds, and 2°C for 2 minutes. The resulting ATAC-Seq libraries were purified (MinElute Kit, QIAGEN) and 150 bp paired-end sequenced on the Illumina Nova 6000 platform to a depth of 4.0 × 107 reads.
ATAC-Seq data were then processed after minor modifications (including quality control, trimming, filtering, aligning, and peak calling). In brief, FastQC (version 0.11.7, Babraham Bioinformatics, http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc) was used to evaluate the quality of the sequencing data, and reads with a Phred quality score greater than 30 were used for downstream analysis. For the purpose of obtaining clean data with minimal background noise, we removed and trimmed the adaptor sequences using Trimmomatic. Subsequently, the remaining clean reads were aligned to the hg38 reference genome using Burrows-Wheeler Aligner (BWA) software. SAMtools (version 1.3.1) was applied to filter out multiple mapped reads, and BED tools were used to filter out mitochondrial reads.
We applied Homer software (version 4.6; findPeaks-style dnase) and macs2 software with a Q value cutoff of less than 0.05 to identify peaks, determine peak position and distribution on the genome, identify peak-associated genes, and discover de novo binding motifs.
Differential peaks between trisomy 21 organoids and euploid organoids were identified by DESeq2 (55), with the thresholds of a log2 fold change of greater than 1 and a P value of less than 0.05. Genome-wide normalized signal coverage tracks were created by bamCoverage in deepTools (version 3.3.0) and were visualized in the Integrative Genomics Viewer (IGV version 2.5.0). GO enrichment analysis of the genes associated with ATAC-Seq peaks was performed using clusterProfiler (version 3.10.1; ref. 56). Fisher’s exact test was applied to identify the significant GO categories, and the FDR was adopted to correct the P values.
Dissociation of brain organoids and scRNA-Seq.
Day-30 and day-70 organoids derived from trisomy 21, euploid, and DSCAM-KD iPSC lines were prepared for scRNA-Seq (sample information is provided in Supplemental Table 3). Briefly, 5–7 cerebral organoids were dissociated into a single-cell suspension via incubation with 1 mL tryp-LE (Life Technologies, Thermo Fisher Scientific) for 35 minutes at 37°C with gentle agitation every 5–8 minutes, followed by 3 washes with 2% FBS in DPBS and gentle titration using a P200 pipette. A single-cell suspension was subsequently collected into 1.5 mL microtubes at a cell density of 1000 cells/μL, and approximately 12,000 cells in each channel were loaded onto a Chromium Single Cell 3′ Chip (10× Genomics, PN-120236) and processed through a Chromium controller to generate single-cell gel beads in emulsions (GEMs) according to the manufacturer’s instructions. Captured cells were lysed, and the released RNA was barcoded through reverse transcription in individual GEMs. Reverse transcription was performed on a S1000 Touch Thermal Cycler (Bio-Rad) at 53°C for 45 minutes, followed by 85°C for 5 minutes and a final hold at 4°C. cDNA was generated and then amplified, and quality was assessed using an Agilent 4200 (Agilent Technologies). scRNA-Seq libraries were prepared with the Chromium Single Cell 3′ Library & Gel Bead Kit V3 (10× Genomics, 1000075) and then sequenced on an Illumina NovaSeq 6000, with a sequencing depth of at least 750,000 reads per cell and a paired-end 150 bp (PE150) reading strategy.
Data analysis for scRNA-Seq.
Reads were aligned to the hg38 human reference genome, and gene-level unique molecular identifier (UMI) counts were obtained using Cell Ranger (version 3.1.0). The expression matrix was processed with Seurat (version 3.1.5; ref. 57). The criteria to select cells for subsequent analysis were as follows: 547 < UMIs per cell < 18040, 398 < detected genes < 4849, and a mitochondrial transcript proportion < 0.1731563. SCTransform normalization was applied to each Seurat object to control confounding sources of variations such as sequencing depth and mitochondrial fraction (58). In addition, the integration of single-cell data was done to correct the batch effect. Expression matrices were summarized by the top 14 principal components. Visualization of the transcriptomic profiles was conducted by UMAP. The Louvian modularity optimization algorithm was applied to iteratively group cells into clusters. Cell clusters were annotated to known biological cell types using canonical cell marker genes. Identification of DEGs in trisomy 21 organoids compared with euploid organoids in each cluster was performed using the MAST (version 1.14.0) package (59). The following model was fit with MAST: zlm (~ group + time, sca, ebayes = TRUE).
Multiple hypothesis testing corrections were performed using Bonferroni and Holm corrections (60). GO enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment of DEGs were performed using ToppGene (https://toppgene.cchmc.org/) (61), and the results were visualized with R. Developmental trajectory analysis was performed using slingshot (version 1.4.0, Bioconductor) with default parameters (62). Unbiased spatial mapping of all clusters was performed using VoxHunt. The transcriptome profiles were compared using BrainSpan, the largest data set containing the postmortem developmental human brain atlas (63).
Cell lines.
The H9 and IMR90-4 cell lines were obtained via a WiCell agreement (H9: NO.16-W0060; IMR90-4: NO.17-W0063); DS1, 2DS3, DSP, and DS2U cell lines were gifts of the Bhattacharyya laboratory (Department of Cell and Regenerative Biology, School of Medicine and Public Health, University of Wisconsin, Madison, Wisconsin, USA). The ihtc-03 cell line was established in our laboratory (35).
Generation of DSCAM-KD iPSC lines using CRISPRi.
The CRISPRi dual vector was packaged in lentivirus to establish the CRISPRi-based DSCAM-KD human iPSC line. First, the lentiviral vector containing dCas9-KRAB used for this study was modified from the lentiCRISPR vector (no. 61425) obtained from Addgene, with the dCas9-VP64 cassette replaced by dCas9-KRAB. We then designed sgRNAs targeting the DNA region from –50 to 300 bp near the transcription start site (TSS) of the DSCAM gene and used a CRISPR web tool (http://crispr.mit.edu/) to minimize off-target effects of the sgRNAs. The location of the TSS was determined using NCBI’s GenBank database (https://www.ncbi.nlm.nih.gov/nuccore/NM_001271534.3). Three gRNAs targeting DSCAM as well as a nontargeting negative control gRNA were selected (Supplemental Table 4) using the online CRISPR design tool at BioinfoGP (https://bioinfogp.cnb.csic.es/tools/breakingcas/); these sequences were cloned into the hU6-sgRNA-SV40-EGFP vector (GeneChem Technologies, GV371).
For the dual-vector multiplex experiments, human iPSCs from DS1 were first infected with lentivirus expressing dCas9-KRAB three days after mechanical passage for 24 hours. Selection was applied 7 days after infection with blasticidin (1 μg/mL) in E8 medium in the presence of the ROCK inhibitor Y27632 (2 μM) (STEMCELL Technologies) and then persistently cultured in the presence of 0.2 μg/mL blasticidin 13 days after infection to maintain the selection, which was continued for 5 weeks until stable colonies appeared. A second transduction in clones was performed using lentivirus encoding either targeting or scrambled sgRNA. Twenty-four hours after lentiviral infection, the cell culture medium was replaced with E8 medium supplemented with ROCK inhibitor (STEMCELL Technologies). Fourteen days after infection, cells were dissociated with Accutase treatment for 10 minutes at 37°C to create a single-cell suspension; this suspension was then transferred to a 5 mL flow cytometry tube with a strainer cap before FACS purification could be completed on a BD FACS Aria Fusion instrument. Clones were then expanded into larger vessel formats and used for further experiments, including functional those for CRISPRi activity and organoid differentiation.
qPCR.
All RNA samples were extracted using a TRIzol kit (Thermo Fisher Scientific). One microgram of total RNA from each sample was reverse transcribed into cDNA and then subjected to qPCR using the SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific). The primers used for qPCR were as follows: DSCAM forward primer, CCAGGCTCAGGTAATCTCA and reverse primer, AGCATAGTCTGTGTTCCGA; PAK1 forward primer, CAGCCCCTCCGATGAGAAATA, and reverse primer, CAAAACCGACATGAATTGTGTGT.
Cytogenetic analysis.
Cerebral organoids were collected for cell culturing, followed by karyotyping. GTG banding was performed according to a standard protocol. Karyotypes were determined from G-banding analysis using a standard protocol according to the International System for Human Cytogenomic Nomenclature (ISCN) 2016 nomenclature.
Quantification of neural tubes in organoids and statistical analysis.
Organized portions of the organoids around the VZ-like structure were the target area for quantification of neural tubes in organoids. The image was rotated such that the VZ-like region was horizontal. Then, a box was defined with a specific width but flexible height to cover the entire stratified region from the apical surface of the VZ to the top surface of the organoid. All Hoechst-stained cells and cells expressing the target markers within that box were counted for statistical analysis. Organoids from the same experiment and from the same clone, but from independent experiments, were used as technical replicates. The data were averaged to obtain a single value. Average data from different biological replicates from different individuals were used to determine the average and standard error.
Data availability.
The raw data of scRNA-Seq data, bulk RNA-Seq data, and ATAC-Seq data used in this study have been deposited in the NCBI’s Sequence Read Achieve (SRA) (accession number SRR14243996-SRR14244067).
Statistics.
All data represent the mean ± SEM. Statistical analyses and graphing were done using GraphPad Prism, version 8 (GraphPad Software). The statistical methods relevant to each figure are described in the figure legends. Statistical comparisons between 2 groups were performed using the Student’s t test. Other statistical analyses were performed using 1-way ANOVA with Dunnett’s multiple-comparison test. P values of less than 0.05 were considered to indicate a significant difference between groups.
Study approval.
Verbal and written consent for the generation and differentiation of the ihtc-03 cell line was obtained from the donor. This study was approved by the ethics committee of Nanjing Medical University ([2016]NO.326).
Author contributions
XYT and YL designed the experiments. XYT, LX, ML, JW, AB, and YL wrote the manuscript. XYT and LX performed the experiments, with technical assistance from XG, QZ, CYL, DW, KHF, MX, XH, SW, XW, XYT, YH, and XYZ. ML, JW, and YH performed RNA-Seq and scRNA-Seq analyses. ML, XYT, YW, and YH performed ATAC-Seq analyses. XYT and YL performed additional data analyses. YL directed the project.
Supplementary Material
Acknowledgments
This study was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDA16010306); the National Natural Science Foundation of Chinese Grants (81922022, 91849117, 81471301, 82022021, 81971189, and 81701320); the National Key Research and Development Program of China (2019YFA0802703); the Jiangsu Outstanding Young Investigator Program (BK20160044); and the Jiangsu Province’s Innovation Program.
Version 1. 05/04/2021
In-Press Preview
Version 2. 06/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(12):e135763.https://doi.org/10.1172/JCI135763.
Contributor Information
Xiao-Yan Tang, Email: xytang5227@163.com.
Lei Xu, Email: leoshju@njmu.edu.cn.
Jingshen Wang, Email: wangjs@njmu.edu.cn.
Yuan Hong, Email: justhongyuan@njmu.edu.cn.
Yuanyuan Wang, Email: wangyuanyuan@njmu.edu.cn.
Qian Zhu, Email: zhuqian@njmu.edu.cn.
Da Wang, Email: dawang@njmu.edu.cn.
Xin-Yue Zhang, Email: zhangxinyue_yuzu@126.com.
Chun-Yue Liu, Email: liuchunyue@njmu.edu.cn.
Kai-Heng Fang, Email: fkh543026800@gmail.com.
Xiao Han, Email: xhan1988@njmu.edu.cn.
Shihua Wang, Email: 792697323@qq.com.
Xin Wang, Email: wangx@xmu.edu.cn.
Min Xu, Email: xumin@njmu.edu.cn.
Anita Bhattacharyya, Email: Bhattacharyy@Waisman.Wisc.Edu.
Xing Guo, Email: guox@njmu.edu.cn.
Mingyan Lin, Email: linmingyan@njmu.edu.cn.
Yan Liu, Email: yanliu@njmu.edu.cn.
References
- 1.Hibaoui Y, et al. Modelling and rescuing neurodevelopmental defect of Down syndrome using induced pluripotent stem cells from monozygotic twins discordant for trisomy 21. EMBO Mol Med. 2014;6(2):259–277. doi: 10.1002/emmm.201302848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golden JA, Hyman BT. Development of the superior temporal neocortex is anomalous in trisomy 21. J Neuropathol Exp Neurol. 1994;53(5):513–520. doi: 10.1097/00005072-199409000-00011. [DOI] [PubMed] [Google Scholar]
- 3.Chakrabarti L, et al. Defects in embryonic neurogenesis and initial synapse formation in the forebrain of the Ts65Dn mouse model of Down syndrome. J Neurosci. 2007;27(43):11483–11495. doi: 10.1523/JNEUROSCI.3406-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schapiro MB, et al. Serial quantitative CT analysis of brain morphometrics in adult Down’s syndrome at different ages. Neurology. 1989;39(10):1349–1353. doi: 10.1212/WNL.39.10.1349. [DOI] [PubMed] [Google Scholar]
- 5.Coyle JT, et al. The neurobiologic consequences of Down syndrome. Brain Res Bull. 1986;16(6):773–787. doi: 10.1016/0361-9230(86)90074-2. [DOI] [PubMed] [Google Scholar]
- 6.Wisniewski KE. Down syndrome children often have brain with maturation delay, retardation of growth, and cortical dysgenesis. Am J Med Genet Suppl. 1990;7:274–281. doi: 10.1002/ajmg.1320370755. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, et al. OLIG2 over-expression impairs proliferation of human Down syndrome neural progenitors. Hum Mol Genet. 2012;21(10):2330–2340. doi: 10.1093/hmg/dds052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bahn S, et al. Neuronal target genes of the neuron-restrictive silencer factor in neurospheres derived from fetuses with Down’s syndrome: a gene expression study. Lancet. 2002;359(9303):310–315. doi: 10.1016/S0140-6736(02)07497-4. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharyya A, et al. A critical period in cortical interneuron neurogenesis in down syndrome revealed by human neural progenitor cells. Dev Neurosci. 2009;31(6):497–510. doi: 10.1159/000236899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattacharyya A, Svendsen CN. Human neural stem cells: a new tool for studying cortical development in Down’s syndrome. Genes Brain Behav. 2003;2(3):179–186. doi: 10.1034/j.1601-183X.2003.00025.x. [DOI] [PubMed] [Google Scholar]
- 11.Weick JP, et al. Deficits in human trisomy 21 iPSCs and neurons. Proc Natl Acad Sci U S A. 2013;110(24):9962–9967. doi: 10.1073/pnas.1216575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huo HQ, et al. Modeling Down syndrome with patient iPSCs reveals cellular and migration deficits of GABAergic neurons. Stem Cell Reports. 2018;10(4):1251–1266. doi: 10.1016/j.stemcr.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe M, et al. Self-organized cerebral organoids with human-specific features predict effective drugs to combat Zika virus infection. Cell Rep. 2017;21(2):517–532. doi: 10.1016/j.celrep.2017.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mariani J, et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell. 2015;162(2):375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadoshima T, et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci U S A. 2013;110(50):20284–20289. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pasca AM, et al. Human 3D cellular model of hypoxic brain injury of prematurity. Nat Med. 2019;25(5):784–791. doi: 10.1038/s41591-019-0436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amin ND, Pasca SP. Building models of brain disorders with three-dimensional organoids. Neuron. 2018;100(2):389–405. doi: 10.1016/j.neuron.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Pasca SP. Assembling human brain organoids. Science. 2019;363(6423):126–127. doi: 10.1126/science.aau5729. [DOI] [PubMed] [Google Scholar]
- 20.Bershteyn M, et al. Human iPSC-derived cerebral organoids model cellular features of lissencephaly and reveal prolonged mitosis of outer radial glia. Cell Stem Cell. 2017;20(4):435–449. doi: 10.1016/j.stem.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iefremova V, et al. An organoid-based model of cortical development identifies non-cell-autonomous defects in Wnt signaling contributing to Miller-Dieker syndrome. Cell Rep. 2017;19(1):50–59. doi: 10.1016/j.celrep.2017.03.047. [DOI] [PubMed] [Google Scholar]
- 22.Qian X, et al. Sliced human cortical organoids for modeling distinct cortical layer formation. Cell Stem Cell. 2020;26(5):766–781. doi: 10.1016/j.stem.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowers M, et al. FASN-dependent lipid metabolism links neurogenic stem/progenitor cell activity to learning and memory deficits. Cell Stem Cell. 2020;27(1):98–109. doi: 10.1016/j.stem.2020.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Yoon KJ, et al. Zika-virus-encoded NS2A disrupts mammalian cortical neurogenesis by degrading adherens junction proteins. Cell stem cell. 2017;21(3):349–358. doi: 10.1016/j.stem.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan TA, et al. Neuronal defects in a human cellular model of 22q11.2 deletion syndrome. Nat Med. 2020;26(12):1888–1898. doi: 10.1038/s41591-020-1043-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnstone M, et al. Reversal of proliferation deficits caused by chromosome 16p13.11 microduplication through targeting NFκB signaling: an integrated study of patient-derived neuronal precursor cells, cerebral organoids and in vivo brain imaging. Mol Psychiatry. 2019;24(2):294–311. doi: 10.1038/s41380-018-0292-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, et al. Loss of NARS1 impairs progenitor proliferation in cortical brain organoids and leads to microcephaly. Nat Commun. 2020;11(1):4038. doi: 10.1038/s41467-020-17454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang Y, et al. Dysregulation of BRD4 function underlies the functional abnormalities of MeCP2 mutant neurons. Mol Cell. 2020;79(1):84–98.e9. doi: 10.1016/j.molcel.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miura Y, et al. Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat Biotechnol. 2020;38(12):1421–1430. doi: 10.1038/s41587-020-00763-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia YL, et al. Expression and significance of DSCAM in the cerebral cortex of APP transgenic mice. Neurosci Lett. 2011;491(2):153–157. doi: 10.1016/j.neulet.2011.01.028. [DOI] [PubMed] [Google Scholar]
- 31.Chen BE, et al. The molecular diversity of Dscam is functionally required for neuronal wiring specificity in Drosophila. Cell. 2006;125(3):607–620. doi: 10.1016/j.cell.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 32.Perez-Nunez R, et al. Overexpressed Down Syndrome cell adhesion molecule (DSCAM) deregulates P21-activated Kinase (PAK) activity in an in vitro neuronal model of Down syndrome: consequences on cell process formation and extension. Neurotox Res. 2016;30(1):76–87. doi: 10.1007/s12640-016-9613-9. [DOI] [PubMed] [Google Scholar]
- 33.Pan X, et al. PAK1 regulates cortical development via promoting neuronal migration and progenitor cell proliferation. Mol Brain. 2015;8:36. doi: 10.1186/s13041-015-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9(10):2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan F, et al. Induction of human somatostatin and parvalbumin neurons by expressing a single transcription factor LIM homeobox 6. Elife. 2018;7:e37382. doi: 10.7554/eLife.37382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, et al. Directed differentiation of forebrain GABA interneurons from human pluripotent stem cells. Nat Protoc. 2013;8(9):1670–1679. doi: 10.1038/nprot.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, et al. Medial ganglionic eminence-like cells derived from human embryonic stem cells correct learning and memory deficits. Nat Biotechnol. 2013;31(5):440–447. doi: 10.1038/nbt.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, et al. Local and global chromatin interactions are altered by large genomic deletions associated with human brain development. Nat Commun. 2018;9(1):5356. doi: 10.1038/s41467-018-07766-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caviness VS, Jr, et al. Numbers, time and neocortical neuronogenesis: a general developmental and evolutionary model. Trends Neurosci. 1995;18(9):379–383. doi: 10.1016/0166-2236(95)93933-O. [DOI] [PubMed] [Google Scholar]
- 40.Inglis-Broadgate SL, et al. FGFR3 regulates brain size by controlling progenitor cell proliferation and apoptosis during embryonic development. Dev Biol. 2005;279(1):73–85. doi: 10.1016/j.ydbio.2004.11.035. [DOI] [PubMed] [Google Scholar]
- 41.Saito Y, et al. The developmental and aging changes of Down’s syndrome cell adhesion molecule expression in normal and Down’s syndrome brains. Acta Neuropathol. 2000;100(6):654–664. doi: 10.1007/s004010000230. [DOI] [PubMed] [Google Scholar]
- 42.Alves-Sampaio A, et al. NMDA-mediated regulation of DSCAM dendritic local translation is lost in a mouse model of Down’s syndrome. J Neurosci. 2010;30(40):13537–13548. doi: 10.1523/JNEUROSCI.3457-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amano K, et al. Dosage-dependent over-expression of genes in the trisomic region of Ts1Cje mouse model for Down syndrome. Hum Mol Genet. 2004;13(13):1333–1340. doi: 10.1093/hmg/ddh154. [DOI] [PubMed] [Google Scholar]
- 44.Guedj F, et al. Analysis of adult cerebral cortex and hippocampus transcriptomes reveals unique molecular changes in the Ts1Cje mouse model of down syndrome. Brain Pathol. 2015;25(1):11–23. doi: 10.1111/bpa.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu R, et al. OLIG2 drives abnormal neurodevelopmental phenotypes in human iPSC-based organoid and chimeric mouse models of Down syndrome. Cell Stem Cell. 2019;24(6):908–926.e8. doi: 10.1016/j.stem.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wisniewski KE, et al. Evidence of arrest of neurogenesis and synaptogenesis in brains of patients with Down’s syndrome. N Engl J Med. 1984;311(18):1187–1188. doi: 10.1056/NEJM198411013111818. [DOI] [PubMed] [Google Scholar]
- 47.Ross MH, et al. Down’s syndrome: is there a decreased population of neurons? Neurology. 1984;34(7):909–916. doi: 10.1212/WNL.34.7.909. [DOI] [PubMed] [Google Scholar]
- 48.Guihard-Costa AM, et al. Biometry of face and brain in fetuses with trisomy 21. Pediatr Res. 2006;59(1):33–38. doi: 10.1203/01.pdr.0000190580.88391.9a. [DOI] [PubMed] [Google Scholar]
- 49.Korbel JO, et al. The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc Natl Acad Sci U S A. 2009;106(29):12031–12036. doi: 10.1073/pnas.0813248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lyle R, et al. Genotype-phenotype correlations in Down syndrome identified by array CGH in 30 cases of partial trisomy and partial monosomy chromosome 21. Eur J Hum Genet. 2009;17(4):454–466. doi: 10.1038/ejhg.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamakawa K, et al. DSCAM: a novel member of the immunoglobulin superfamily maps in a Down syndrome region and is involved in the development of the nervous system. Hum Mol Genet. 1998;7(2):227–237. doi: 10.1093/hmg/7.2.227. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L, et al. DSCAM and DSCAML1 regulate the radial migration and callosal projection in developing cerebral cortex. Brain Res. 2015;1594:61–70. doi: 10.1016/j.brainres.2014.10.060. [DOI] [PubMed] [Google Scholar]
- 53.Dolan BM, et al. Rescue of fragile X syndrome phenotypes in Fmr1 KO mice by the small-molecule PAK inhibitor FRAX486. Proc Natl Acad Sci U S A. 2013;110(14):5671–5676. doi: 10.1073/pnas.1219383110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim D, et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Love MI, et al. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu G, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20(1):296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finak G, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Puoliväli T, et al. Influence of multiple hypothesis testing on reproducibility in neuroimaging research: A simulation study and Python-based software. J Neurosci Methods. 2020;337:108654. doi: 10.1016/j.jneumeth.2020.108654. [DOI] [PubMed] [Google Scholar]
- 61.Chen J, et al. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(web server issue):W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Street K, et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19(1):477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478(7370):483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of scRNA-Seq data, bulk RNA-Seq data, and ATAC-Seq data used in this study have been deposited in the NCBI’s Sequence Read Achieve (SRA) (accession number SRR14243996-SRR14244067).