Abstract
For research on tendon injury, many different animal models are utilized; however, the extent to which these species simulate the clinical condition and disease pathophysiology has not yet been critically evaluated. Considering the importance of inflammation in tendon disease, this study compared the cellular and molecular features of inflammation in tenocytes of humans and four common model species (mouse, rat, sheep, and horse). While mouse and rat tenocytes most closely equalled human tenocytes’ low proliferation capacity and the negligible effect of inflammation on proliferation, the wound closure speed of humans was best approximated by rats and horses. The overall gene expression of human tenocytes was most similar to mice under healthy, to horses under transient and to sheep under constant inflammatory conditions. Humans were best matched by mice and horses in their tendon marker and collagen expression, by horses in extracellular matrix remodelling genes, and by rats in inflammatory mediators. As no single animal model perfectly replicates the clinical condition and sufficiently emulates human tenocytes, fit-for-purpose selection of the model species for each specific research question and combination of data from multiple species will be essential to optimize translational predictive validity.
Subject terms: Experimental models of disease, Preclinical research, Translational research, Cell biology
Introduction
Animal models are cornerstones of biomedical and translational medicine research. They are used when it is unethical or impractical to study the target species to explore basic pathophysiological mechanisms, to evaluate safety and efficacy of new treatment approaches, and to decide whether novel therapeutic candidates warrant the economic and moral costs of clinical development1–7. For 90% of new treatment strategies, however, translation from basic science to the clinic fails, mainly because clinical trials show them to be inefficient (52%) or unsafe (24%) during phases II and III4,5,8. Such translational failures cost animal lives, strain clinical trial volunteers, and burden biomedical research, the pharmaceutical industry and health care systems. So far, attempts to optimize translational success have mainly focused on internal validity flaws such as methodological shortcomings in animal and clinical trials, publication bias, or overoptimistic conclusions about efficacy. Yet another key factor, the external validity, or generalizability, of animal models has received little attention4,5,8–13. Common problems of external validity include species differences in disease pathophysiology, common confounding comorbidities and the selection of outcome measures5. An animal model should sufficiently emulate aetiology, pathophysiology, symptomatology and response to therapeutic interventions of the target species to allow extrapolation5,11. As no single animal model perfectly recapitulates the clinical realm, fit-for-purpose validation and selection of the most appropriate model species is essential10–13. Unfortunately, for musculoskeletal disorders, such as tendinopathy, in depth validation studies of animal models beyond structural and biomechanical similarities are largely lacking.
Tendinopathy, a disabling overuse injury, is the most common musculoskeletal complaint for which patients seek medical attention14. It is prevalent in both occupational and athletic settings, afflicting 25% of the adult population, and accounting for 30–50% of all sport injuries15–18. Major tendons experiencing high loads are most commonly affected, especially the weight-bearing and energy-storing Achilles tendon, which routinely experiences loads of up to 12.5 times the weight of the individual6,19. Many intrinsic and extrinsic factors, including age, body weight and physical loading, influence the aetiopathogenesis of tendinopathy. Overload and repetitive strain lead to accumulation of microdamage and concurrent inflammatory, dysregulated reparative and degenerative processes, causing clinical symptoms, e.g., activity-related pain, focal tendon tenderness and swelling, and functional limitations. Overt clinical symptoms such as pain are preceded by tendinous matrix remodelling, an inflammatory cellular process mediated in part by metalloproteinase enzymes20,21. Due to its low cellularity, vascularity and metabolic rate, a tendon’s response to injury is inefficient, requiring lengthy periods of recuperation and often resulting in a fibrovascular scar. Scar tissue has significantly inferior biomechanical properties than the original tendon tissue and is prone to re-injury16,22–24. Current treatment options are mostly palliative and fail to restore the functional properties of injured tendons16,22–24. Tendinopathy thus has a significant adverse impact on quality of life and costs individuals and the society an estimated annual expense of $30 billion25,26. This is driving research efforts into unravelling the molecular mechanisms of tendinopathy and developing targeted regenerative therapies. Of particular interest in this context are the cellular and molecular processes orchestrating inflammation in tendinopathy and the mechanisms governing the development of chronic inflammation that fails to resolve in persistently symptomatic patients27.
Tendon injury induces a local inflammatory response, characterized by immune cell infiltration and the expression of pro‐inflammatory mediators, which in turn reduce collagen production and induce vasodilation, angiogenesis, and matrix metalloproteinase (MMPs) expression28–31. Furthermore, the inflammatory milieu can modify tenocyte physiology by increasing metabolic activity and inducing an activated, proinflammatory phenotype with inflammation memory and the capacity for endogenous production of cytokines such as TNF-α, IL-1β, IL-6, IL-10, VEGF and TGF-β29,30,32. While the initial inflammatory response is essential to start the healing process, sustained inflammatory conditions contribute to dysregulated matrix remodelling and fibrovascular scarring during healing18,31. Chronic inflammation thus drives tendon degeneration before tearing or any other clinical signs of tendinopathy, impairs healing after injury and promotes the development of tendinopathy14.
While human tendon tissue can typically be procured only from individuals with advanced pathology, animal models provide the opportunity to obtain tissue during all stages of tendinopathy to study organ, cellular and molecular changes over the entire course of the disease. In animal models, consistent and repeatable injuries can be induced, evaluated and treated, while controlling for potential confounding influences23,33,34. Since no species has been established as the gold standard for tendinopathy research, many induced and spontaneous animal models ranging from small rodents (mice, rats) to large animals (sheep, horses) are utilized19,35–43. While the biomechanical properties of the various species are well established, their ability to simulate the pathophysiology of human tendon disease, including the molecular behaviour of key genes and pathways, has not been critically evaluated yet and detailed analyses of species- specific differences in cytokine expression and regulation as well as of tenocytes susceptibility to cytokines are still lacking44.
Considering the importance of inflammation in tendon disease29,33,45,46, this study compares the cellular and molecular features of inflammation in tenocytes of humans and four common model species (mouse, rat, sheep, and horse) to aid in the evidence-based selection of fit-for-purpose translational animal models for tendon research. Mice and rats are included due to their prevalent use as laboratory animals and availability of species-specific molecular tools6,47. Larger animals are used increasingly as translational models due to their more comparable tendon dimensions and biomechanics23,48–50. Horses present an attractive model of human tendinopathy since their superficial digital flexor tendon is a weight-bearing and energy-storing tendon analogous to the human Achilles tendon, which is similarly prone to naturally occuring tendon disease with high recurrence rates9,51,52. Furthermore equine ageing proceeds similarly to humans39,51,52. Sheep are included because features of clinical tendinopathy of horses could be emulated also in ovine induced models51,53–57. In particular, the ovine intra‐synovial tendon lesion model mimics the clinical intra‐synovial tendon disease of humans and horses more accurately than small animal extra‐synovial models, e.g., with respect to histology and gene expression, to similarities in the biomechanical environment and to failure of lesions to heal51,53–57.
Results
Morphology
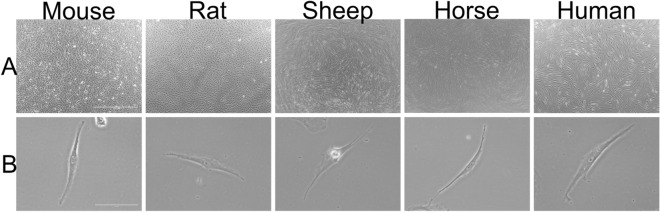
Tenocytes from all five species shared common characteristics with a fusiform appearance, adherence to the flask and similar dimensions (Fig. 1): human tenocytes measured 177.8 ± 40.1 μm (mean ± s.d.) in length and 20.2 ± 4.4 μm in width, mouse 163.7 μm (± 23.6) × 19.4 μm (± 3.3), rat 182.5 (± 26.6) × 24.2 (± 8.0), sheep 206.3 (± 45.5) × 28.9 (± 8.5) and horse 193.2 (± 8.4) × 15.4 (± 1.4). In high confluency, tenocytes from human, sheep and horse showed similar morphology, creating cell bundles arranged in a storiform pattern (Fig. 1A), while tenocytes from mouse and rat had a more scattered appearance with a random orientation.
Figure 1.
Micrographs of tenocytes derived from the Achilles tendon in the mouse, rat, sheep and human or the superficial digital flexor tendon in the horse. (A) shows the tenocytes at a magnification of 40x (scale bar: 1000 μm), while (B) was taken at a 400 × magnification (scale bar: 100 μm).
Proliferation assay
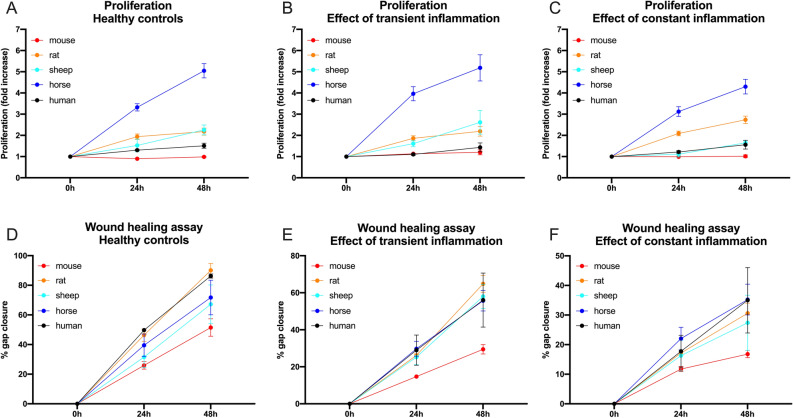
Proliferation, migration and gene expression of tenocytes of all five species were compared under standard culture conditions (healthy control) as well as under transient (24 h) and constant exposure to inflammatory stimuli (Fig. 2). Under healthy conditions (Fig. 3, Table 1), equine tenocytes had the highest proliferation rates while murine cells had the lowest. Human tenocytes exhibited the second lowest proliferation capacity with an inability to double the cell amount over the 48 h observation period. Sheep and rat tenocytes were in the middle. From Fig. 3, it can be seen that slopes are quite variable among species, but within species variation is low. Therefore, the slopes of tendon cells of all four model species are significantly different from those of humans (p < 0.001; to correct for multiple testing using Bonferroni with four comparisons, the nominal significance levels, 0.05, 0.01, and 0.001, are set to the corrected levels: 0.0125, 0.002, 0.0002, respectively), even for only three biological replicates per species. Under constant exposure to inflammatory stimulation (10 ng/ml IL1β and 10 ng/ml TNFα), the proliferation of sheep tenocytes decreased significantly and fell to about human levels, i.e., the difference in the proliferation slopes between healthy and constantly inflamed sheep decreased significantly (p < 0.01). All other differences in slopes between healthy, constantly and transiently (only 24 h inflammatory stimulation) inflamed conditions were not significant.
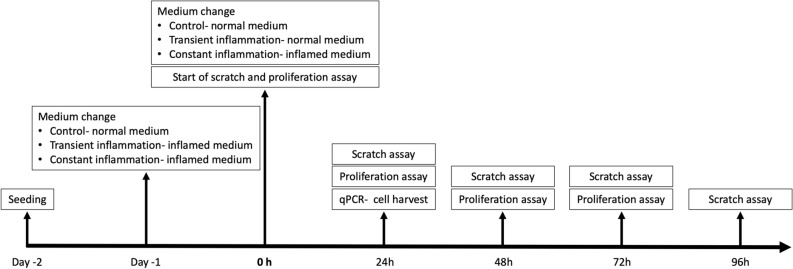
Figure 2.
Study timeline detailing the experimental protocol. Gene expression, proliferation and migration of tenocytes of all five species (human, mouse, rat, sheep, horse) were compared under standard culture conditions (healthy control) as well as under transient (24 h) and constant exposure to inflammatory stimuli (10 ng/ml IL1β and 10 ng/ml TNFα). After 24-h exposure to inflammation, the transient inflammation group received fresh culture medium, while for the constant inflammation group fresh medium was again supplemented with inflammatory factors.
Figure 3.
The proliferation capacity (A–C) of tenocytes from 5 different mammalian species (mouse, rat, sheep, horse, and human) under healthy (Ctr/A), transient (TI/B) and constant (CI/C) inflammatory condition is illustrated as fold increase over the course of 2 days (indicated as mean ± SEM calculated from three biological replicates). For pairwise comparisons and significance values see Table 1. A wound healing assay (D-F) was used to determine the migratory capacity of tenocytes from five different mammalian species under healthy (D), transient (E) and constant (F) inflammatory conditions (indicated as mean ± SEM calculated from three biological replicates). For pairwise comparisons and significance values see Table 1.
Table 1.
The first column corresponds to the healthy situation, the second and third to transiently and constantly inflamed, respectively.
| Species | Healthy | Transient inflammation | Constant inflammation | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Prolif. slope | Std. error | p-value | slope: diff. to healthy | Std. error | p-value | slope: diff. to healthy | Std. error | p-value | ||
| Proliferation | Human | 0.1027 | 0.0168 | 1.75e-09 *** | − 0.0167 | 0.0237 | 0.4801 | 0.0089 | 0.0237 | 0.7074 |
| Slope: diff. to human | St. error | p-value | slope: diff. to human | St. error | p-value | slope: diff. to human | St. error | p-value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | − 0.1250 | 0.0237 | 1.95e-07 *** | 0.0508 | 0.0335 | 0.1301 | − 0.0301 | 0.0335 | 0.3691 | |
| Rat | 0.0933 | 0.0237 | 9.35e-05 *** | − 0.0038 | 0.0335 | 0.9103 | 0.0420 | 0.0335 | 0.2103 | |
| Sheep | 0.1169 | 0.0237 | 1.10e-06 *** | 0.0585 | 0.0335 | 0.0815 | − 0.0878 | 0.0335 | 0.00900 * | |
| Horse | 0.3000 | 0.0237 | < 2e-16 *** | 0.0233 | 0.0335 | 0.4868 | − 0.0640 | 0.0335 | 0.0568 |
| Migration slope | Std. Error | p-value | slope: diff. to healthy | St. error | p-value | slope: diff. to healthy | St. error | p-value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Migration | Human | − 0.0792257 | 0.0032 | < 2e-16 *** | 0.0277 | 0.0045 | 2.76e-09 *** | 0.0475 | 0.0045 | < 2e-16 *** |
| Slope: diff. to human | Std. Error | p-value | slope: diff. to human | St. error | p-value | slope: diff. to human | St. error | p-value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | 0.0311503 | 0.0045 | 3.07e-11 *** | − 0.0075 | 0.0064 | 0.2452 | − 0.0156 | 0.0064 | 0.0154 | |
| Rat | 0.0001854 | 0.0046 | 0.9155 | − 0.0063 | 0.0064 | 0.3287 | 0.0035 | 0.0065 | 0.5938 | |
| Sheep | 0.0185550 | 0.0045 | 5.46e− 05 *** | − 0.0188 | 0.0064 | 0.00374 * | − 0.0112 | 0.0064 | 0.0811 | |
| Horse | 0.0132563 | 0.0045 | 0.00375 ** | − 0.0123 | 0.0064 | 0.0567 | − 0.0140 | 0.0064 | 0.0302 |
First rows (human) correspond to tests for non-zero slopes of the proliferation and migration curves in humans, i.e., either non-zero proliferation or non-zero gap closure in humans. Second to fifth rows (different animal species) correspond to tests of differences of the animal models to humans in the slopes of the proliferation and migration curves of tenocytes. Calculations used an ANCOVA. Means, standard errors and p-values are reported. To correct for multiple testing with four comparisons using Bonferroni, the nominal significance levels (0.05, 0.01, and 0.001) are set to the corrected levels (0.0125, 0.002, 0.0002, respectively). P-values are marked with stars from * (significant) to *** (highly significant) using this correction.
Wound healing (scratch) assay
Gap closure was significantly different (in all cases p < 0.001) between all conditions, fastest for healthy and slowest for constantly inflamed cells of all species (Fig. 3, Table 1). Murine tenocytes showed the slowest wound healing for all conditions. There was no statistically significant difference in migration speed between healthy and transient inflammatory conditions in tenocytes of any species and between healthy and continuously inflamed conditions only for mouse and rat cells (p < 0.05).
Under healthy conditions, gap closure of tenocytes from all species except rats was significantly different (in all cases p < 0.003) from humans with cells from rats showing the fastest wound healing (gap closure at 48 h mean 90.08% ± 8.01% s.d.), closely followed by humans (gap closure at 48 h mean 86.25% ± 2.47% s.d.) and cells from mice the slowest (gap closure at 48 h mean 51.49% ± 10.23% s.d.). Under transient inflammation rat tenocytes again were fastest (gap closure at 48 h mean 64.81% ± 7.84% s.d.), with ovine (gap closure at 48 h mean 58% ± 11.17% s.d.), human (gap closure at 48 h mean 56.06% ± 25.27% s.d.) and equine tenocytes (gap closure at 48 h mean 55.71% ± 9.6% s.d.) following with similar wound healing rates, while murine tendon cells again were slowest (gap closure at 48 h mean 29.46% ± 4.36% s.d.). Under constant inflammation, equine tenocytes were fastest (gap closure at 48 h mean 35.23% ± 8.98% s.d.) followed closely by human tendon cells (gap closure at 48 h mean 34.99% ± 19.12% s.d.). The change in migration speed compared to healthy was significantly different from human tenocytes for ovine tendon cells under transient and equine and murine tenocytes under constant inflammation (Table 1).
Quantitative PCR
The species show variable approximations of human expression levels among functional gene groups and conditions (Tables 2 and 3, Figs. 4 and 5, suppl. Figure 1 and 2, suppl. table 1)58. A univariate Analysis of Variance (ANOVA) demonstrated significant differences between each species and humans in many genes relevant for tendon function and inflammatory response (Table 2). Remarkably, healthy tenocytes of all four species show significant differences to humans in their expression of Col1, the main tendon matrix component, of collagenase MMP13 and of the key inflammatory mediator COX2. Similarly, significant differences from humans in the COX2 expression of transiently inflamed tenocytes are evident for all four species and in IL6 expression for all species except rats. In contrast, no significant differences to humans were seen in MMP1 expression for any species and the osteogenic marker ALP was only significantly different in healthy murine tenocytes. NFkB expression exhibited a significant difference only in healthy horse tendon cells and p53 in healthy horse and rat tenocytes.
Table 2.
Mean difference to human and p-values of the gene expression calculated with ANOVA of healthy, transiently inflamed and constantly inflamed tenocytes of the four animal model species.
| SCX | TNC | TNM | COL1 | COL3 | COL5 | MMP1 | MMP3 | MMP13 | IL6 | COX2 | NFKB | P53 | ALP | FAK | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Healthy | Mouse | Diff | 2.65 | − 0.74 | − 1.05 | − 3.03 | 2.54 | − 2.17 | − 9.34 | 9.45 | 8.50 | 4.65 | 9.40 | − 0.20 | − 0.30 | 3.14 | − 0.25 |
| Adj p | 0.230 | 0.811 | 0.951 | 0.038 | 0.122 | 0.609 | 0.064 | 0.002 | 0.000 | 0.005 | 0.000 | 0.999 | 0.961 | 0.032 | 0.995 | ||
| Rat | Diff | 5.28 | 2.84 | − 1.37 | 3.95 | 3.80 | 4.62 | − 2.97 | 10.24 | 5.24 | 0.54 | 11.30 | 3.00 | 4.47 | 1.85 | 4.72 | |
| Adj p | 0.008 | 0.014 | 0.882 | 0.008 | 0.015 | 0.067 | 0.851 | 0.001 | 0.013 | 0.978 | 0.000 | 0.053 | 0.000 | 0.287 | 0.000 | ||
| Sheep | Diff | 2.39 | 3.79 | − 3.52 | − 7.91 | − 0.50 | 0.41 | − 4.71 | 9.12 | 9.47 | − 0.16 | 13.61 | − 0.72 | − 0.36 | − 0.14 | 3.95 | |
| Adj p | 0.313 | 0.002 | 0.201 | 0.000 | 0.981 | 0.998 | 0.539 | 0.003 | 0.000 | 1.000 | 0.000 | 0.931 | 0.927 | 1.000 | 0.001 | ||
| Horse | Diff | 1.49 | − 0.24 | − 3.11 | − 4.72 | 0.41 | 1.71 | − 2.93 | 1.94 | 5.37 | − 3.67 | 15.65 | 3.79 | 3.52 | − 1.11 | 2.11 | |
| Adj p | 0.711 | 0.996 | 0.296 | 0.002 | 0.991 | 0.780 | 0.857 | 0.793 | 0.012 | 0.022 | 0.000 | 0.014 | 0.000 | 0.720 | 0.055 | ||
| Transient inflammation | Mouse | Diff | − 1.22 | − 0.69 | − 2.59 | − 2.62 | − 3.06 | − 3.07 | 1.23 | − 5.08 | − 4.96 | − 4.02 | − 4.15 | − 1.24 | − 0.59 | 0.46 | − 0.46 |
| Adj p | 0.515 | 0.870 | 0.048 | 0.177 | 0.017 | 0.010 | 0.984 | 0.011 | 0.001 | 0.001 | 0.000 | 0.180 | 0.803 | 0.990 | 0.963 | ||
| Rat | Diff | − 2.94 | − 1.42 | − 2.85 | − 2.93 | − 4.43 | − 3.57 | − 0.66 | − 3.22 | − 1.56 | − 0.98 | − 4.84 | − 0.01 | − 0.34 | − 1.54 | − 1.48 | |
| Adj p | 0.019 | 0.342 | 0.029 | 0.114 | 0.001 | 0.004 | 0.999 | 0.124 | 0.412 | 0.537 | 0.000 | 1.000 | 0.967 | 0.563 | 0.299 | ||
| Sheep | Diff | − 0.59 | − 0.20 | 0.59 | − 0.12 | − 1.84 | − 1.49 | 0.39 | − 6.54 | − 4.19 | − 2.83 | − 6.20 | − 0.52 | 0.16 | 1.38 | 1.31 | |
| Adj p | 0.930 | 0.998 | 0.937 | 1.000 | 0.189 | 0.291 | 1.000 | 0.002 | 0.004 | 0.007 | 0.000 | 0.841 | 0.998 | 0.651 | 0.404 | ||
| Horse | Diff | − 0.77 | 1.44 | − 0.34 | 1.40 | − 0.36 | − 0.95 | 3.97 | − 0.32 | 0.15 | 3.25 | − 6.63 | − 0.16 | 1.38 | 2.22 | 0.98 | |
| Adj p | 0.840 | 0.330 | 0.992 | 0.691 | 0.989 | 0.671 | 0.498 | 0.999 | 1.000 | 0.003 | 0.000 | 0.997 | 0.152 | 0.249 | 0.658 | ||
| Constant inflammation | Mouse | Diff | − 1.23 | − 0.51 | − 3.33 | − 1.89 | − 3.30 | − 2.52 | 1.64 | − 4.82 | − 3.41 | − 4.54 | − 3.96 | − 1.33 | − 0.09 | 2.83 | 0.22 |
| Adj p | 0.911 | 0.949 | 0.166 | 0.361 | 0.003 | 0.501 | 0.977 | 0.014 | 0.209 | 0.021 | 0.094 | 0.586 | 1.000 | 0.093 | 0.998 | ||
| Rat | Diff | − 3.27 | − 1.76 | − 4.86 | − 3.80 | − 7.16 | − 5.29 | 4.75 | − 1.96 | 3.26 | 1.13 | − 1.71 | 0.13 | 0.18 | − 1.09 | − 1.35 | |
| Adj p | 0.238 | 0.177 | 0.028 | 0.020 | 0.000 | 0.039 | 0.509 | 0.490 | 0.241 | 0.866 | 0.727 | 1.000 | 0.999 | 0.797 | 0.412 | ||
| Sheep | Diff | − 2.01 | − 0.63 | − 1.41 | 0.56 | − 1.81 | − 2.94 | 1.51 | − 6.73 | − 2.93 | − 0.42 | − 7.68 | − 0.18 | 0.19 | 1.24 | 1.48 | |
| Adj p | 0.653 | 0.900 | 0.822 | 0.975 | 0.111 | 0.366 | 0.983 | 0.001 | 0.328 | 0.996 | 0.002 | 1.000 | 0.999 | 0.719 | 0.333 | ||
| Horse | Diff | − 3.10 | 2.28 | − 1.74 | 1.37 | − 0.39 | − 2.04 | 3.59 | − 0.24 | 1.49 | 5.53 | − 8.36 | − 0.63 | 0.95 | 3.18 | 0.58 | |
| Adj p | 0.280 | 0.059 | 0.693 | 0.637 | 0.972 | 0.679 | 0.733 | 1.000 | 0.839 | 0.006 | 0.001 | 0.949 | 0.678 | 0.054 | 0.932 |
Significant p-values (Tukey HSD correction, p<0.05) and the matching mean differences in gene expression to humans are indicated in bold.
Table 3.
Mahalanobis distances of the four model species to humans for all genes combined under healthy, transiently and constantly inflamed conditions as well as for the different functional gene groups: tenogenic markers, collagens, MMPS and inflammatory mediators.
| Mouse | Rat | Sheep | Horse | |
|---|---|---|---|---|
| Mahalanobis distances all genes per condition | ||||
| Healthy | 20.01 | 27.44 | 24.78 | 23.05 |
| Transient Inflammation | 13.49 | 18.66 | 10.62 | 9.59 |
| Constant Inflammation | 10.50 | 17.45 | 8.94 | 12.97 |
| Mahalanobis distances all conditions per group of genes | ||||
| SCX, TNC, TNMD | 5.45 | 12.21 | 10.54 | 6.88 |
| COL1, COL3, COL5 | 8.28 | 13.73 | 12.88 | 8.45 |
| MMP1, MMP3, MMP13 | 8.53 | 11.27 | 8.60 | 4.65 |
| COX2, IL6, | 15.82 | 14.25 | 18.51 | 17.77 |
Figure 4.
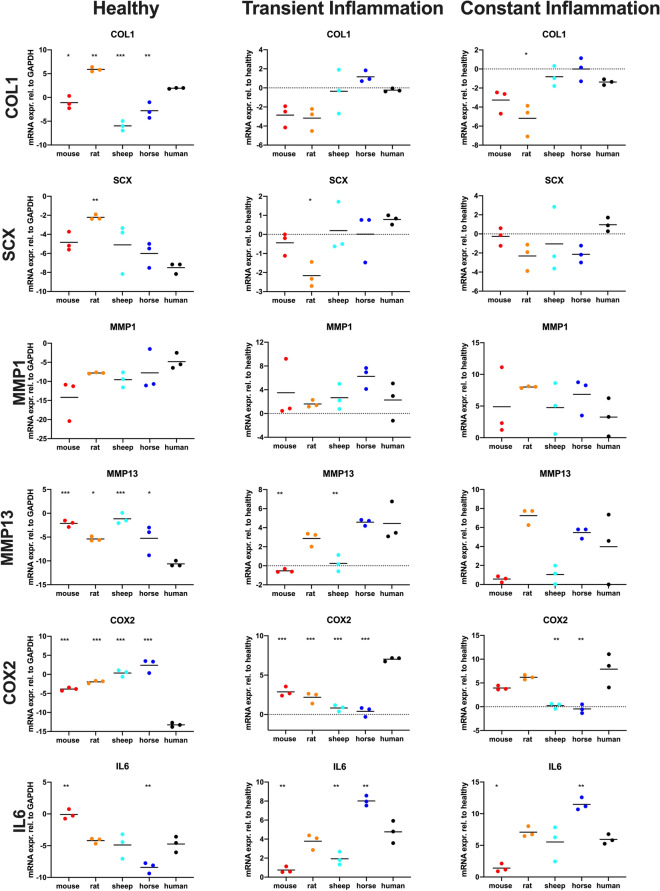
Scatter dot plots showing COL1, SCX, MMP1, MMP13, COX2 and IL6 gene expression (presented as log2) of healthy tenocytes and tenocytes exposed to inflammatory stimuli for 24 h (transient inflammation) or continuously (constant inflammation) in different species (the black lines indicate the respective means). Gene expression in the inflammatory conditions is shown relative to the healthy tenocytes. Each dot represents a different biological replicate. Differences were evaluated using ANOVA with Tukey HSD test, *p < 0.05; **p < 0.01; ***p < 0.001.
Figure 5.
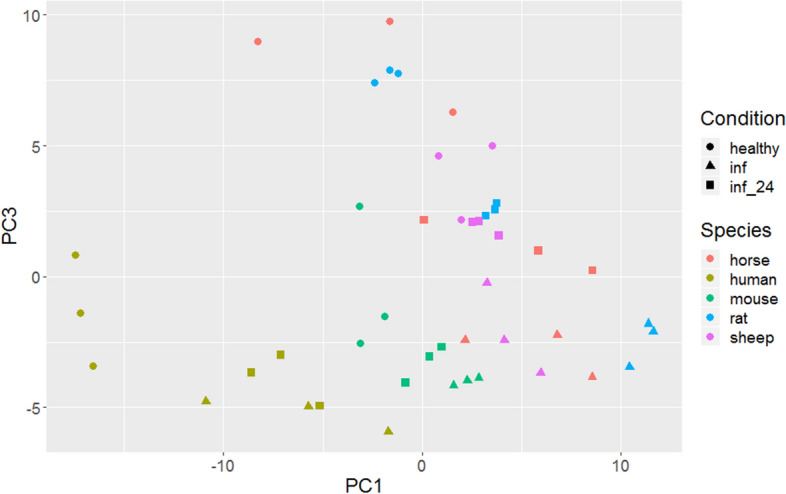
Plot of PC1 (explaining 36% of the variance) vs. PC3 (explaining 14% of the variance) of a Principal Component Analysis of gene expression values. Species are colour coded, conditions (healthy, transiently inflamed, and continuously inflamed) are differentiated by symbols. Each dot represents a different biological replicate.
To condense the information from the univariate ANOVA results to overall measures of similarity between the different animals and humans we calculated the multivariate Mahalanobis distances of the four species to humans. The Mahalanobis distance is a non-dimensional measure of dissimilarity, where between group distances are weighted by the inverses of within group variability, much like the test statistic of a t-test for one variable. Similar to its use in graphically detecting outliers in multiple dimensions, we use it to show multivariate dissimilarity in gene expression among species. In supplementary Fig. 1, we summarize conditions within a gene. From Fig. 4, it is evident that Cox2 is rather variable among species (especially human healthy cells differ from all animals, least so from mice) while showing relatively little variation within species under all conditions. Hence the Mahalanobis distances to humans are comparatively large with mice being closest to humans (suppl. Figure 1). The second immune gene, Il6, shows also relatively little within species variation (Fig. 4), but comparatively less between species variation; especially rats and sheep are similar to humans, while mice and horses are slightly further away (Fig. 4, suppl. Figure 1). The Mahalanobis distances (Table 3) of the immune genes are relatively large compared to the other gene combinations due to low within species variability. The overall distance of the immune genes is a compromise between the two genes, such that rats are slightly more similar to humans than mice, with sheep and horses further away. When different functional groups of genes are analysed, variable Mahalanobis distances of the four model species from humans are found (Table 3). While mice and horses are closest to humans in their tendon marker and collagen expression, horses appear to be by far the best model for MMPs (Table 3). For the overall gene expression of healthy tenocytes, Mahalanobis distances from human tenocytes are large and similar among species, with mouse tendon cells appearing least and rat tenocytes most distant from human. For transient inflammation, Mahalanobis distances to humans are generally lower, but the spread of differences widens, with horses, sheep and mice relatively close and rats clearly furthest. For constant inflammation, the pattern is qualitatively similar to that under transient inflammation. Overall, the pattern of multivariate dissimilarity of species varies widely and unpredictably among species pairs with no single species most similar to humans.
Principal Component Analysis (PCA) is an exploratory technique for reducing the complexity of data. We used expression data from all genes and plot the results for the different species x condition combinations (Fig. 5, suppl. Figure 2). The plot of PC1 vs. PC3 is easier to interpret than of other combinations of PCs. (Fig. 5). With the exception of humans and mice, species are generally not well-separated. Within all species, the different conditions are spread along an oblique line with healthy to the upper left and inflamed to the lower right. Within mice, healthy is also separated from inflamed, while in sheep the pattern is less clear. Pairwise plots of other PCs show similar but less clear patterns (suppl. Figure 2).
Discussion
Choice of the most appropriate animal model is the most essential and challenging element of animal-based research, and also an important aspect of the 3Rs (i.e., replace, reduce, refine) to ensure the best use of animals59–61. Unfortunately, the choice of animal models frequently is based more on convention, financial and practical considerations, such as housing and husbandry requirements or the availability of reagents and biochemical tests, than compelling scientific evidence of the fit to human diseases and clinical contexts4,5,9,62–64. The lack of formal requirements for animal models is due to the traditional assumption that genetic homology derived from a common evolutionary origin also implies functional similarities of gene regulation, signalling pathways and developmental systems between species (the “unity in diversity” concept)7. Species may, however, differ in critical aspects and rarely have assumed similarities been empirically demonstrated7. The diversity of human patients and symptoms is thus unlikely fully represented in highly inbred rodents65,66. Even humanized models, which have contributed significantly to research by facilitating functional studies in vivo, cannot replicate the complexity of human disease67.
Both the European Medicines Agency (EMA) and the USA Federal Food and Drug Administration require the use of fit-for-purpose animal models to evaluate efficacy, durability, dose–response, degradation and safety of new therapeutics for market approval. Recently, these regulatory authorities published guidelines identifying requirements to demonstrate the relevance of animal models for investigational new product testing by cross-species comparison of the structural homology of the target, its distribution, signal transduction pathways and pharmacodynamics68,69. Furthermore, several voluntary initiatives have established criteria to encourage the evidence-based selection of animal models for stroke and schizophrenia11,70,71. To this end, both the model species and disease-induction protocols, need to be validated by comparing the animal model with the gold standard or the target species11. As no gold standard for tendon research is available, this study compared tenocyte morphology, proliferation rate, wound healing speed, and gene expression of two small animals (mouse and rat) and two large animals (sheep and horse) to human under healthy as well as “diseased” (transiently and continuously inflamed) conditions to determine similarities and differences among species. It can serve as the foundation for a rational, evidence-based choice of optimal animal models for specific aspects of human tendinopathy.
Tendon injury induces a local inflammatory response, which initiates the healing process. Tendon healing occurs in three chronologic phases: inflammation (0–7 days), proliferation (1–6 weeks), and remodeling (6 weeks – 6 months). While these stages overlap, they are characterized by temporally and functionally distinct cytokine profiles and cellular processes72. The initial inflammatory phase is characterized by influx of inflammatory cells, which release chemotactic and proinflammatory cytokines and growth factors that lead to recruitment and proliferation of macrophages and resident tendon fibroblasts44,72–83. In addition, tenocytes produce also several endogenous cytokines and growth factors which contribute to the healing process in an auto‐ and paracrine manner44,75. During the proliferative stage tenocytes proliferate and produce an immature neomatrix with a predominance of type III rather than type I collagen44,72,80–82,84. Lastly, in the course of the remodeling phase, the cellularity decreases, matrix synthesis is reduced and collagen fibrils and tenocytes align linearly with the direction of tension44,72–79,81–83. However, in both man and horse suffering from naturally occurring tendon disease, the normal architecture, composition and function of the tendon are never completely restored, predisposing them to recurring injury and tendinopathy44,72–79,81–83.
Given the importance of inflammation in tendon injury and repair, with pro-inflammatory cytokines acting as a regulatory link between several catabolic and anabolic systems and as a double-edged sword both promoting and impeding tendon repair44,83,85–88, this study focused on the comparative response to inflammation.
We used IL-1β and TNF-α, two hallmark cytokines of inflammation in tendons, which are associated with tendon injury and tendinopathy in vivo and in vitro, to induce disease-relevant inflammation28,30,33,45,89–97. IL-1 activates the NF-κB pathway in tenocytes, induces the production of inflammatory mediators including COX2 and IL6, and matrix remodelling factors such as MMP1, MMP3 and MMP1328,30,89–91. It can even cause loss of the tenocyte phenotype, which is associated with decreased expression of tendon-related genes, e.g. COL1, SCX and TNMD28,30,89–91. Similarly TNF-α can strongly activate tenocytes, stimulating them to produce more cytokines, including IL-1β, TNF-α, IL- 628,30,92–94,96. Accordingly, we used tendon-specific markers (SCX, TNC, TNMD, COL1, COL3, COL5), matrix remodelling proteinases (MMP1, MMP3, MMP13) and inflammatory factors (COX2, IL6, NF-κB, p53) in addition to proliferation and wound healing speed as read-outs to evaluate the response to IL-1β/ TNF-α induced inflammation.
Interestingly, while mouse and rat tenocytes most closely matched human tenocytes’ low proliferation capacity and minimal effect of inflammation on proliferation, the human wound closure speed was best approximated by rats and horses. Tenocyte migration to the injured tissue and proliferation are essential processes in tendon healing98,99. Accordingly, inflammatory stimulation, e.g. with IL-1β, has been shown to increase tenocyte migration and proliferation, the capacity for which decreases with age80,83,87,88,100–106. In this study, we observed a decrease in tenocyte migration and proliferation following inflammatory stimulation in all species (statistically significant for sheep tenocyte proliferation as well as rat and mouse tendon cell migration under constant inflammation) except rats (non-significant trend toward increased proliferation), which may be due to our use of tenocytes from individuals in disease-relevant age groups.
The overall gene expression of human tenocytes was most similar to murine under healthy, equine under transient and ovine under constant inflammatory conditions. The species difference between human and the four animal models was particularly evident in the expression of the main tendon matrix component COL1. Healthy tenocytes of all four model species exhibited significant differences to human in their expression of COL1. Col1 typically amounts to appr. 95% of total tendon collagen or 50–80% of tendon dry weight107, but cytokines, such as IL-1β and TNF-α, suppress COL1 synthesis, which leads to reduced stiffness28,44,84,108. In this study the decrease in COL1 synthesis following inflammatory stimulation could be observed in all species and was most pronounced in rat and mice, least in sheep and most similar to humans in horses.
The expression of the transcription factor SCX, a specific marker of the tendon/ligament lineage109, while low in all species under healthy conditions, only increased in humans upon constant inflammation. SCX is a transcription factor that regulates tendon genes, including Col1 and Tnmd, and is required for normal tendon development110,111 and adult tendon repair in mice112,113. An increase in its expression is likely to result in changes in the expression of its downstream genes and to be beneficial to tendon healing post injury113. The essential contribution of SCX was also shown in SCX-null mice, which fail to convert from producing primarily COL3 to synthesizing mainly COL1 during tendon repair, supporting the hypothesis that the transcriptional control of collagen type I is mediated by SCX113. Overall for the six tenogenic factors, rat tenocytes showed the largest difference to humans in the Mahalanobis distance, while tendon cells from mice and horses most closely equaled humans, indicating that these species might be most suitable for studies evaluating ECM production and tendon healing.
For matrix remodelling proteinases, the species differences were most prevalent for healthy tenocytes: tendon cells of all model species differed from humans for MMP13 and all but horses for MMP3. MMPs are key players in physiological and pathological tendon ECM remodeling, contributing to the degradation of tendon ECM and hence the loss of the biomechanical resistance and durability of tendon44,114–116. An increase in MMP expression has also been implicated in the pathogenesis of tendinopathy44,116. MMP13 specifically was upregulated in rotator cuff tendon tears and flexor tendon injury117–119. In this study, inflammatory stimulation increased MMP13 expression in tenocytes of all species, only minimally in mice and horses but 4–eightfold in rats, sheep and humans. In contrast, all species showed similarly increased MMP1 expression following inflammatory stimulation; no significant differences were observed in MMP1 expression in any species in any condition compared to humans. This corresponds well with other studies showing upregulation of MMP1 in ruptured tendons suggesting a high level of collagen degradation by this enzyme120. In total, for the functional group of ECM remodelling genes, horses again provided the best and rats the worst match to humans as shown in the Mahalanobis distance analysis.
For the expression of inflammatory mediators, the Mahalanobis distances of all species were larger than for the other functional gene groups. Although the immunophysiology of larger animal species has traditionally been presumed to be closer to humans than rodents47,121, rat tenocytes most closely approximated human tendon cells in this category. Additionally, in healthy condition, mice presented the lowest distance from all animals, rising again the question if larger animals truly are more similar to human. Remarkably, healthy and transiently inflamed tenocytes of all four model species, as well as constantly inflamed ovine and equine tenocytes, showed significant differences to human in their expression of COX2. Following inflammatory stimulation, COX2 was only significantly upregulated in humans, mice and rats. Upregulation of COX2 plays an important, multifaceted role in the inflammatory cascade in injured tendons through the synthesis of prostaglandins125. COX2 is essential in the early injury response as evidenced by impaired tendon repair following administration of selective COX2 inhibitors in the early repair phase122. The lacking upregulation of sheep and horses therefore invites further investigation into the early tendon healing response in the different species in vivo.
Correspondingly, IL6, a cytokine with strong association with inflammation in tendon disease58,123,124, displayed significantly different expression in transiently inflamed tenocytes of all species except rats. Statistically significant differences in IL6 expression compared to human were also evident under constant inflammatory conditions for mice and horses. IL6 plays an essential role in tendon healing as repair processes in IL6 knock-out mice are impaired38. It tenocytes in two ways: i) IL6 stimulates tenocyte proliferation and survival and ii) it inhibits their tenogenic differentiation via the Janus tyrosine kinases/Stat3 signaling pathway44,125.
Cell properties may be influenced not only by species and interdonor differences but also by cell isolation and processing methods126,127. In the present study, two isolation methods, enzymatic digestion and cell migration out of tendon explants, have been used depending on the available sample size. Enzymatic digestion was used for smaller sample sizes as higher cell yields are achieved with this method, while the explant technique is less invasive and requires less manipulation and labour. As both methods were used for all species and alterations in experimental conditions have been shown to be of minor importance to cell behaviour compared to cell source and interdonor variability126, the isolation method is unlikely to have significantly influenced the species-specific gene expression profiles observed in this study.
In summary, the results of our study show that all four model species approximate some aspects of the behaviour of human tenocytes well and others poorly. No animal model sufficiently emulates human tenocytes’ cellular and molecular features and response to inflammation to be considered the gold-standard for tendon research. Translational medicine will need to continue to rely on a fit-for-purpose selection of animal models to approximate the human condition, based on the essential characteristics that must be mimicked for a particular research question19. Peculiarities, strengths, and weaknesses of the model species need to be accounted for in the study design, analysis and interpretation19,128,129. Data from multiple animal models should be combined to optimize translational predictive validity.
Materials and methods
Tenocytes of four mammalian species (mouse, rat, sheep, horse) were compared with human tenocytes (n = 3 donors, i.e., biological replicates, per species). All methods and experimental protocols in this study were carried out in accordance and compliance with relevant institutional and national guidelines and regulations.
Tenocyte isolation from animals
All animals were euthanized for reasons unrelated to this study. Based on the "Good Scientific Practice. Ethics in Science und Research" regulation implemented at the University of Veterinary Medicine Vienna, the Institutional Ethics Committee (“Ethics and Animal Welfare Committee”) of the University of Veterinary Medicine Vienna does not require approval of in vitro cell culture studies, if the cells were isolated from tissue, which was obtained either solely for diagnostic or therapeutic purposes or in the course of institutionally and nationally approved experiments.
Species-specific, energy-storing, weight-bearing tendons were harvested from skeletally mature animals immediately following euthanasia: Achilles tendons from sheep (Merino-cross breed, female, aged 2–5 years), rats (Fischer344 breed, female, aged 3–4 months) and mice (c57bl/6 breed, female, aged 8–12 weeks); superficial digital flexor tendons from the front limb of horses (7–15 years, geldings). Under sterile conditions, the paratenon was removed and the tendons were sectioned into small pieces (< 0.5 × 0.5 × 0.5 cm). Isolation of cells was performed either by enzymatic digestion using 3 mg/ml collagenase type II (Gibco Life technologies, Vienna, Austria) for 6–8 h or migration from explants (explants were removed after 7–10 days) or a combination of both. Cells were expanded until 80—90% confluency before passaging.
Human tenocytes
Human tenocytes obtained with ethical approval and informed consent from the Achilles tendon of three male human donors (aged 60–90 years) in accordance with relevant guidelines and regulations (Declaration of Helsinki) were purchased in cryopreserved condition in passage two from two different providers (Pelo Biotech GmbH, Germany and Zen-Bio, North Carolina, USA with review of the protocols and consent forms by an independent review board (Institutional Review Board, Pearl Pathways, LLC) which is accredited by the Association for the Accreditation of Human Research Protection Program Inc.).
Cell culture
The culture medium was identical for all species: minimal essential medium (α-MEM, Sigma-Aldrich, Vienna, Austria) supplemented with 10% fetal bovine serum (FBS-12A, Capricon, Ebsdorfergrund, Germany), 1% L-Glutamine (L-Alanyl L-Glutamine 200 Mm, Biochrom), 100 units mL-1penicillin and 0.1 mg mL-1streptomycin (P/S, Sigma-Aldrich, Vienna, Austria).
Cells were cultured at 37 °C, 5% CO2 until the desired passage and number of cells was obtained. Experiments were performed with cells either in passage 3 or 4.
Morphology
Cells were imaged both at low and high confluency using the EVOS FL Auto imaging system in phase contrast with a 40 × and 400 × objective (ThermoFisher Scientific, AMEP4680). Cell phenotypes and cell sheet patterns were characterised for all species and compared to human cells. Tenocyte dimensions (length and width) were measured for each of the five species.
Inflammatory stimulation
Gene expression, proliferation and migration of tenocytes of all five species were compared under standard culture conditions (healthy control) as well as under transient (24 h) and constant exposure to inflammatory stimuli (10 ng/ml IL1β (Immuno Tools, Friesoythe, Germany) and 10 ng/ml TNFα (Immuno Tools, Friesoythe, Germany)) as previously described28,89,90,93,115,129. Successful induction of inflammation was confirmed by upregulation of inflammatory markers (COX2, IL6, see results). After 24-h exposure to inflammation, the transient inflammation group received fresh culture medium, while for the constant inflammation group fresh medium was again supplemented with inflammatory factors (Fig. 2).
Proliferation assay
Tenocytes were plated in 96-well plates (3000 cells/well in technical triplicates) and cultured under control (healthy), transient and constant inflammatory conditions. The cell number per plate was quantified via DNA fluorescence using the CyQuant assay (Invitrogen) according to the manufacturer’s recommendations on day 0, 1, 2, and 3 (Fig. 2). As cell proliferation sets in after a lag time of about 24 h and relative proliferation rates decrease steadily, we used log(cell nr) as the target variable and log(time in hours minus 23) as regression variable for the parametric statistical analysis.
Wound healing (scratch) assay
Migration of tenocytes was evaluated in a wound healing model using a magnetic scratch device to create standardized cell-free gaps of 1.5 mm width in confluent sheets of tenocytes130. Cells were seeded in 12-well plates (100,000 cells/well in technical triplicates) and left to adhere overnight. Inflammatory stimuli were added to the transient and constant inflammation groups and scratches were created 48 h after seeding under control (healthy), transient and constant inflammatory conditions (Fig. 2). The cell-free area was imaged at 24 h intervals (0, 24, 48, 72, 96 h, Fig. 2) in phase contrast using the EVOS FL Auto imaging system with a 4 × fluorite objective using coordinate recovery function. The gap size was measured using the MRI Wound healing Tool (http://dev.mri.cnrs.fr/projects/imagej-macros/wiki/Wound_Healing_Tool) in ImageJ (https://imagej.nih.gov/ij/, version 2.0.0-rc-43/1.50e). As gap closure approached 100% in the fastest group, healthy rat tenocytes, at 48 h, this time point was chosen as cut-off for slope calculations and comparison of conditions. For the parametric analysis, we used the untransformed gap area [mm2] as target variable and the untransformed time between 0 and 48 h (before the gap closed in any of the samples) as regression variable.
Quantitative PCR
Tenocytes were seeded in 12-well plates (100,000 cells/well in technical triplicates) and cultured under control (healthy), transient and constant inflammatory conditions. Cells were harvested for RNA isolation using RNA isolation reagent (Trizol, ThermoFisher Scientific, MA, USA) 48 h after initiation of inflammation, as previously described131. The 48 h time point was chosen as it allows assessment of the response to inflammation as well as to removal of inflammatory stimuli.
Briefly, a solution of Trizol and Chloroform (Sigma-Aldrich) in a ratio of 5 to 1 was used. Total RNA was recovered by the addition of isopropyl alcohol (Sigma-Aldrich) and glycerol (Thermo Scientific). The mixture was incubated on ice and centrifuged for 45 min at 13,000 rpm. The total RNA pellet was washed with 75% ethanol and solubilized in RNase-free water. Genomic DNA was removed by a DNA removal kit (Life Technologies, Carlsbad, California, USA). Two nanograms of RNA from each sample was used for the qPCR reaction (qPCR One-Step Eva Green kit, Bio&Sell, Feucht, Germany).
We measured gene expression of tendon markers (TNC, TNDM, SCX), collagens (COL1, COL3, COL5), matrix-metalloproteinases (MMP1, MMP3, MMP13), inflammatory factors (IL6, COX2, NFkB, p53), a marker for aberrant tenocyte differentiation (ALP) and for focal adhesion and migration (FAK) in the four model species and humans under healthy, transiently and constantly inflamed conditions. All primers were designed using the Primer3 software. Primer sequences are shown in supplementary table 2. The transcript level for the 15 genes of interest was normalized to the transcript level of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and presented as ratio to GAPDH58. The ratio between COL 1 and COL 3 was also evaluated for further matrix remodelling characterization, with a higher COL1:COL3 ratio indicating a stronger tenogenic phenotype132,133. For the parametric analysis, we used the log2 transformed ratios of the target gene to GAPDH as target variable.
Statistical analysis
For statistical analyses, the R statistical programming language134 and GraphPad (version 8.4.2) were used. Target and regression variables (where appropriate) are given in the respective subsections. Data are presented descriptively as mean and standard deviation. Generally, linear models (analyses of variance and covariances, ANCOVA) were used, e.g., for wound healing the untransformed area was the target variable, time a regression variable, and species and condition factors; as interactions of two explanatory variables time*species, time*condition, condition*species, and biological replicate nested within species were included; furthermore, the three-way interaction time*species*condition was also included. Note that all terms with time are to be interpreted as slopes or differences in slopes. The Tukey’s HSD (honestly significant difference) test was used to account for multiple testing, where appropriate. Confidence intervals of parameter estimates were calculated.
Note that many different target variables are available, i.e., data are multidimensional. With qPCR alone, 15 genes of interest were measured under three conditions. For each gene separately, an ANOVA with species and condition was calculated. Furthermore, we condensed information by calculating the multivariate (Mahalanobis) distance of the log2-transformed mRNA concentrations, for the three conditions of each gene, and report the distance of each of the four mammalian species from the human values. For a single condition, all 15 genes of interest could be used for calculating the multivariate distance. Additionally, we grouped genes into classes, e.g., all collagens or all matrix-metalloproteinases and calculated multivariate distances for the classes separately, this time jointly for the different conditions. We also calculated a principal component analysis (PCA) of the log2-transformed qPCR data for all gene, treatment, and species combinations together. The proportions of the variance explained by the different PC's are reported and the rotated data for the different treatment and species combinations are shown in graphs for the most important components.
Supplementary Information
Acknowledgements
The authors acknowledge Sinan Gültekin for his technical and John Breteler for his graphical support. This research was supported by the Austrian Research Promotion agency (grant number 856858) and the University of Veterinary Medicine Vienna tandem PhD programme.
Author contributions
G.O.: study conception and design, data acquisition and interpretation, manuscript preparation. M.F.: study conception and design, data acquisition and interpretation, manuscript preparation. C.V.: data analysis and interpretation, manuscript preparation. I. R.: data acquisition, figure preparation. F. J.: study conception and design, data analysis and interpretation, manuscript preparation all authors reviewed the manuscript.
Funding
Open access funding provided by University of Veterinary Medicine Vienna.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Gil Lola Oreff and Michele Fenu.
Change history
11/6/2021
The original online version of this Article was revised: In the original version of this Article the Funding section was omitted. The correct Funding section now reads: “Open access funding provided by University of Veterinary Medicine Vienna.”
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-91914-9.
References
- 1.Wendler A, Wehling M. The translatability of animal models for clinical development: biomarkers and disease models. Curr. Opin. Pharmacol. 2010;10:601–606. doi: 10.1016/j.coph.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Wendler A, Wehling M. Translatability score revisited: differentiation for distinct disease areas. J. Transl. Med. 2017;15:226. doi: 10.1186/s12967-017-1329-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henderson, V. C., Kimmelman, J., Fergusson, D., Grimshaw, J. M. & Hackam, D. G. Threats to validity in the design and conduct of preclinical efficacy studies: a systematic review of guidelines for in vivo animal experiments. PLoS Med10, e1001489 (2013). [DOI] [PMC free article] [PubMed]
- 4.Pandora Pound, M. R.-H. Is it possible to overcome issues of external validity in preclinical animal research? Why most animal models are bound to fail. Journal of Translational Medicine16, 203 (2018). [DOI] [PMC free article] [PubMed]
- 5.van der Worp, H. B. et al. Can Animal models of disease reliably inform human studies? PLoS Med7, e1000245 (2010). [DOI] [PMC free article] [PubMed]
- 6.Bottagisio M, Lovati AB. A review on animal models and treatments for the reconstruction of Achilles and flexor tendons. J. Mater. Sci. Mater. Med. 2017;28:45. doi: 10.1007/s10856-017-5858-y. [DOI] [PubMed] [Google Scholar]
- 7.Wall RJ, Shani M. Are animal models as good as we think? THE. 2008;69:2–9. doi: 10.1016/j.theriogenology.2007.09.030. [DOI] [PubMed] [Google Scholar]
- 8.Harrison RK. Phase II and phase III failures: 2013–2015. Nat. Rev. Drug Discovery. 2016;15:817–818. doi: 10.1038/nrd.2016.184. [DOI] [PubMed] [Google Scholar]
- 9.Kimmelman, J., Mogil, J. S. & Dirnagl, U. Distinguishing between exploratory and confirmatory preclinical research will improve translation. Plos Biol12, e1001863 (2014). [DOI] [PMC free article] [PubMed]
- 10.Ireson CR, Alavijeh MS, Palmer AM, Fowler ER, Jones HJ. The role of mouse tumour models in the discovery and development of anticancer drugs. Br. J. Cancer. 2019;121:101–108. doi: 10.1038/s41416-019-0495-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varga OE, Hansen AK, Sandøe P, Olsson IAS. Validating animal models for preclinical research: a scientific and ethical discussion. Altern. Lab. Anim. 2019;38:245–248. doi: 10.1177/026119291003800309. [DOI] [PubMed] [Google Scholar]
- 12.Sams-Dodd F. Strategies to optimize the validity of disease models in the drug discovery process. Drug Discovery Today. 2006;11:355–363. doi: 10.1016/j.drudis.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Denayer T, Stöhr T, Van Roy M. Animal models in translational medicine: validation and prediction. New Horizons in Translational. Med. 2014;2:5–11. [Google Scholar]
- 14.Abraham, A. C. et al. Targeting the NF-κB signaling pathway in chronic tendon disease. Sci. Transl. Med.11, eaav4319 (2019). [DOI] [PMC free article] [PubMed]
- 15.Docheva D, Müller SA, Majewski M, Evans CH. Biologics for tendon repair. Adv. Drug Deliv. Rev. 2015;84:222–239. doi: 10.1016/j.addr.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walden G, et al. A clinical, biological, and biomaterials perspective into tendon injuries and regeneration. Tissue Eng. Part B Rev. 2017;23:44–58. doi: 10.1089/ten.teb.2016.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abat, F. et al. Current trends in tendinopathy: consensus of the ESSKA basic science committee. Part I: biology, biomechanics, anatomy and an exercise-based approach. J. Exp. Orthopaed.4, 1–11 (2017). [DOI] [PMC free article] [PubMed]
- 18.Crowe LAN, et al. S100A8 & S100A9: Alarmin mediated inflammation in tendinopathy. Sci. Rep. 2019;9:1528. doi: 10.1038/s41598-018-37684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hast MW, Zuskov A, Soslowsky LJ. The role of animal models in tendon research. Bone Joint Res. 2014;3:193–202. doi: 10.1302/2046-3758.36.2000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huisman E, Thornton G, Roberts C, Scott A. Identification of biomarkers for early tendon degeneration using an in-vivo rabbit model. Br. J. Sports Med. 2013;47(e2):53–e2. [Google Scholar]
- 21.Kaux, J. F., Forthomme, B., Le Goff, C. & Crielaard, J. M. Current opinions on tendinopathy. J. Sports Sci. Med. (2011). [PMC free article] [PubMed]
- 22.Riley GP. Gene expression and matrix turnover in overused and damaged tendons. Scand. J. Med. Sci. Sports. 2005;15:241–251. doi: 10.1111/j.1600-0838.2005.00456.x. [DOI] [PubMed] [Google Scholar]
- 23.Bruns J, Kampen J, Kahrs J, Plitz W. Achilles tendon rupture: experimental results on spontaneous repair in a sheep-model. Knee Surg. Sports Traumatol. Arthrosc. 2000;8:364–369. doi: 10.1007/s001670000149. [DOI] [PubMed] [Google Scholar]
- 24.Gajhede-Knudsen M, Ekstrand J, Magnusson H, Maffulli N. Recurrence of Achilles tendon injuries in elite male football players is more common after early return to play: an 11-year follow-up of the UEFA Champions League injury study. Br. J. Sports Med. 2013;47:763–768. doi: 10.1136/bjsports-2013-092271. [DOI] [PubMed] [Google Scholar]
- 25.Barboni, B. et al. Indirect co-culture with tendons or tenocytes can program amniotic epithelial cells towards stepwise tenogenic differentiation. PLoS ONE7, e30974 (2012). [DOI] [PMC free article] [PubMed]
- 26.Butler DL, et al. Functional tissue engineering for tendon repair: a multidisciplinary strategy using mesenchymal stem cells, bioscaffolds, and mechanical stimulation. J. Orthop. Res. 2008;26:1–9. doi: 10.1002/jor.20456. [DOI] [PubMed] [Google Scholar]
- 27.Dakin SG, et al. Chronic inflammation is a feature of Achilles tendinopathy and rupture. Br. J. Sports Med. 2018;52:359–367. doi: 10.1136/bjsports-2017-098161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.John T, et al. Effect of pro-inflammatory and immunoregulatory cytokines on human tenocytes. J. Orthop. Res. 2010;28:1071–1077. doi: 10.1002/jor.21079. [DOI] [PubMed] [Google Scholar]
- 29.Dakin, S. G. et al. Inflammation activation and resolution in human tendon disease. Science Translational Medicine7, 311ra173–311ra173 (2015). [DOI] [PMC free article] [PubMed]
- 30.Tang C, et al. The roles of inflammatory mediators and immunocytes in tendinopathy. J. Orthopaedic Transl. 2018;14:23–33. doi: 10.1016/j.jot.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tarafder S, et al. Tendon stem/progenitor cells regulate inflammation in tendon healing via JNK and STAT3 signaling. FASEB J. 2017;31:3991–3998. doi: 10.1096/fj.201700071R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rees JD, Stride M, Scott A. Tendons - time to revisit inflammation. Br. J. Sports Med. 2013;48:1553–1557. doi: 10.1136/bjsports-2012-091957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Millar NL, Murrell GAC, McInnes IB. Inflammatory mechanisms in tendinopathy – towards translation. Nat. Rev. Rheumatol. 2017;13:110–122. doi: 10.1038/nrrheum.2016.213. [DOI] [PubMed] [Google Scholar]
- 34.Thorpe CT, Clegg PD, Birch HL. A review of tendon injury: why is the equine superficial digital flexor tendon most at risk? Equine Vet. J. 2010;42:174–180. doi: 10.2746/042516409X480395. [DOI] [PubMed] [Google Scholar]
- 35.Barsby T, Bavin EP, Guest DJ. Three-dimensional culture and transforming growth factor beta3 synergistically promote tenogenic differentiation of equine embryo-derived stem cells. Tissue Eng Part A. 2014;20:2604–2613. doi: 10.1089/ten.tea.2013.0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beredjiklian PK, et al. Regenerative versus reparative healing in tendon: a study of biomechanical and histological properties in fetal sheep. Ann. Biomed. Eng. 2003;31:1143–1152. doi: 10.1114/1.1616931. [DOI] [PubMed] [Google Scholar]
- 37.Chhabra A, et al. GDF-5 deficiency in mice delays Achilles tendon healing. J. Orthop. Res. 2003;21:826–835. doi: 10.1016/S0736-0266(03)00049-4. [DOI] [PubMed] [Google Scholar]
- 38.Lin TW, Cardenas L, Glaser DL, Soslowsky LJ. Tendon healing in interleukin-4 and interleukin-6 knockout mice. J. Biomech. 2006;39:61–69. doi: 10.1016/j.jbiomech.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 39.Lui PPY, Maffulli N, Rolf C, Smith RKW. What are the validated animal models for tendinopathy? Scand. J. Med. Sci. Sports. 2011;21:3–17. doi: 10.1111/j.1600-0838.2010.01164.x. [DOI] [PubMed] [Google Scholar]
- 40.Warden SJ. Animal models for the study of tendinopathy. Br. J. Sports Med. 2007;41:232–240. doi: 10.1136/bjsm.2006.032342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carpenter JE, Hankenson KD. Animal models of tendon and ligament injuries for tissue engineering applications. Biomaterials. 2004;25:1715–1722. doi: 10.1016/s0142-9612(03)00507-6. [DOI] [PubMed] [Google Scholar]
- 42.Lake SP, Ansorge HL, Soslowsky LJ. Animal models of tendinopathy. Disabil. Rehabil. 2008;30:1530–1541. doi: 10.1080/09638280701785460. [DOI] [PubMed] [Google Scholar]
- 43.Cadby JA, et al. Further characterisation of an experimental model of tendinopathy in the horse. Equine Vet. J. 2013;45:642–648. doi: 10.1111/evj.12035. [DOI] [PubMed] [Google Scholar]
- 44.Schulze-Tanzil G, et al. The role of pro-inflammatory and immunoregulatory cytokines in tendon healing and rupture: new insights. Scand. J. Med. Sci. Sports. 2011;21:337–351. doi: 10.1111/j.1600-0838.2010.01265.x. [DOI] [PubMed] [Google Scholar]
- 45.Millar NL, et al. Inflammation is present in early human tendinopathy. Am. J. Sports Med. 2010;38:2085–2091. doi: 10.1177/0363546510372613. [DOI] [PubMed] [Google Scholar]
- 46.Dakin, S. G. et al. Macrophage sub-populations and the lipoxin A4 receptor implicate active inflammation during equine tendon repair. PLoS ONE7, e32333 (2012). [DOI] [PMC free article] [PubMed]
- 47.Cibelli J, et al. Strategies for improving animal models for regenerative medicine. Cell Stem Cell. 2013;12:271–274. doi: 10.1016/j.stem.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Virchenko O, Fahlgren A, Rundgren M, Aspenberg P. Early Achilles tendon healing in sheep. Arch. Orthop. Trauma Surg. 2008;128:1001–1006. doi: 10.1007/s00402-008-0691-x. [DOI] [PubMed] [Google Scholar]
- 49.Barboni B, et al. Achilles Tendon Regeneration Can Be Improved by Amniotic Epithelial Cell Allotransplantation. Cell transplant. 2012;21:2377–2395. doi: 10.3727/096368912X638892. [DOI] [PubMed] [Google Scholar]
- 50.Huri G, et al. A novel repair method for the treatment of acute Achilles tendon rupture with minimally invasive approach using button implant: a biomechanical study. Foot Ankle Surg. 2013;19:261–266. doi: 10.1016/j.fas.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 51.Dudhia J, et al. Aging enhances a mechanically-induced reduction in tendon strength by an active process involving matrix metalloproteinase activity. Aging Cell. 2007;6:547–556. doi: 10.1111/j.1474-9726.2007.00307.x. [DOI] [PubMed] [Google Scholar]
- 52.Dakin SG, Dudhia J, Smith RKW. Resolving an inflammatory concept: The importance of inflammation and resolution in tendinopathy. Vet. Immunol. Immunopathol. 2014;158:121–127. doi: 10.1016/j.vetimm.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsang, A. S. et al. Effects of tendon injury on uninjured regional tendons in the distal limb: An in-vivo study using an ovine tendinopathy model. PLoS ONE14, e0215830 (2019). [DOI] [PMC free article] [PubMed]
- 54.Khan MR, et al. Evaluation of the effects of synovial multipotent cells on deep digital flexor tendon repair in a large animal model of intra-synovial tendinopathy. J. Orthop. Res. 2019;38:128–138. doi: 10.1002/jor.24423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith MM, et al. Modulation of aggrecan and ADAMTS expression in ovine tendinopathy induced by altered strain. Arthritis Rheum. 2008;58:1055–1066. doi: 10.1002/art.23388. [DOI] [PubMed] [Google Scholar]
- 56.Biasutti, S. et al. Spatiotemporal variations in gene expression, histology and biomechanics in an ovine model of tendinopathy. PLoS ONE12, e0185282 (2017). [DOI] [PMC free article] [PubMed]
- 57.Jacobsen, E. et al. Focal Experimental Injury Leads to Widespread Gene Expression and Histologic Changes in Equine Flexor Tendons. PLoS ONE10, e0122220 (2015). [DOI] [PMC free article] [PubMed]
- 58.Legerlotz K, Jones ER, Screen HRC, Riley GP. Increased expression of IL-6 family members in tendon pathology. Rheumatology. 2012;51:1161–1165. doi: 10.1093/rheumatology/kes002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ferreira, G. S. et al. A standardised framework to identify optimal animal models for efficacy assessment in drug development. PLoS ONE14, e0218014 (2019). [DOI] [PMC free article] [PubMed]
- 60.Wood MW, Hart LA. Selecting appropriate animal models and strains: making the best use of research, information and outreach. AATEX. 2007;14:303–306. [Google Scholar]
- 61.Swearengen JR. Choosing the right animal model for infectious disease research. Anim. Models Exp. Med. 2018;1:100–108. doi: 10.1002/ame2.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh VP, et al. Critical evaluation of challenges and future use of animals in experimentation for biomedical research. Int. J. Immunopathol. Pharmacol. 2016;29:551–561. doi: 10.1177/0394632016671728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Vries, R. B. M., Buma, P., Leenaars, M., Ritskes-Hoitinga, M. & Gordijn, B. Reducing the number of laboratory animals used in tissue engineering research by restricting the variety of animal models. articular cartilage tissue engineering as a case study. Tissue Eng. Part B Rev.18, 427–435 (2012). [DOI] [PubMed]
- 64.de Vries RBM, et al. The usefulness of systematic reviews of animal experiments for the design of preclinical and clinical studies. ILAR J. 2014;55:427–437. doi: 10.1093/ilar/ilu043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bolker J. There's more to life than rats and flies. Nature. 2012;491:31–33. doi: 10.1038/491031a. [DOI] [PubMed] [Google Scholar]
- 66.Mestas J, Hughes CCW. Of mice and not men: differences between mouse and human immunology. J. Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 67.Laudanski K, et al. Potential pitfalls of the humanized mice in modeling sepsis. Int. J. Inflamm. 2018;2018:1–9. doi: 10.1155/2018/6563454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.US Food and Drug Administration - Center for Drug Evaluation and Research. Product development under the animal rule: guidance for industry. (2015).
- 69.European Medicines Agency. Guideline on strategies to identify and mitigate risks for first-in-human and early clinical trials with investigational medicinal products. (2017). [DOI] [PMC free article] [PubMed]
- 70.Regenberg A, et al. The role of animal models in evaluating reasonable safety and efficacy for human trials of cell-based interventions for neurologic conditions. J. Cereb. Blood Flow Metab. 2008;29:1–9. doi: 10.1038/jcbfm.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Markou A, Chiamulera C, Geyer MA, Tricklebank M, Steckler T. Removing obstacles in neuroscience drug discovery: the future path for animal models. Neuropsychopharmacol. 2008;34:74–89. doi: 10.1038/npp.2008.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leong NL, et al. Tendon and ligament healing and current approaches to tendon and ligament regeneration. J. Orthop. Res. 2020;38:7–12. doi: 10.1002/jor.24475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang J, et al. gene expression of transforming growth factor beta-1 in rabbit zone II flexor tendon wound healing: evidence for dual mechanisms of repair. Plast. Reconstr. Surg. 1997;100:937–944. doi: 10.1097/00006534-199709001-00016. [DOI] [PubMed] [Google Scholar]
- 74.Chan BP, et al. Effects of basic fibroblast growth factor (bFGF) on early stages of tendon healing: a rat patellar tendon model. Acta Orthop. Scand. 2000;71:513–518. doi: 10.1080/000164700317381234. [DOI] [PubMed] [Google Scholar]
- 75.Chbinou N, Frenette J. Insulin-dependent diabetes impairs the inflammatory response and delays angiogenesis following Achilles tendon injury. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004;286:R952–7. doi: 10.1152/ajpregu.00536.2003. [DOI] [PubMed] [Google Scholar]
- 76.Sharma P, Maffulli N. Basic biology of tendon injury and healing. The Surgeon. 2005;3:309–316. doi: 10.1016/s1479-666x(05)80109-x. [DOI] [PubMed] [Google Scholar]
- 77.Sharma P, Maffulli N. Biology of tendon injury: healing, modeling and remodeling. J. Musculoskelet. Neuronal Interact. 2006;6:181–190. [PubMed] [Google Scholar]
- 78.Tohyama, H., Yasuda, K., Uchida, H. & Nishihira, J. The responses of extrinsic fibroblasts infiltrating the devitalised patellar tendon to IL-1β are different from those of normal tendon fibroblasts. J. Bone Joint Surg. Br. 89-B, 1261–1267 (2007). [DOI] [PubMed]
- 79.Chen CH, et al. Tendon healing in vivo: gene expression and production of multiple growth factors in early tendon healing period. YJHSU. 2008;33:1834–1842. doi: 10.1016/j.jhsa.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 80.Chang HN, et al. The effect of aging on migration, proliferation, and collagen expression of tenocytes in response to ciprofloxacin. J. Orthop. Res. 2012;30:764–768. doi: 10.1002/jor.21576. [DOI] [PubMed] [Google Scholar]
- 81.Loiselle AE, et al. Remodeling of murine intrasynovial tendon adhesions following injury: MMP and neotendon gene expression. J. Orthop. Res. 2009;27:833–840. doi: 10.1002/jor.20769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thomopoulos S, Parks WC, Rifkin DB, Derwin KA. Mechanisms of tendon injury and repair. J. Orthop. Res. 2015;33:832–839. doi: 10.1002/jor.22806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paterson YZ, et al. Genome-wide transcriptome analysis reveals equine embryonic stem cell-derived tenocytes resemble fetal, not adult tenocytes. Stem Cell Res Ther. 2020;11:53. doi: 10.1186/s13287-020-01692-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maffulli N, Ewen SWB, Waterston SW, Reaper J, Barrass V. Tenocytes from ruptured and tendinopathic achilles tendons produce greater quantities of type iii collagen than tenocytes from normal achilles tendons. Am. J. Sports Med. 2016 doi: 10.1177/03635465000280040901. [DOI] [PubMed] [Google Scholar]
- 85.Eming SA, Hammerschmidt M, Krieg T, Roers A. Interrelation of immunity and tissue repair or regeneration. Semin. Cell Dev. Biol. 2009;20:517–527. doi: 10.1016/j.semcdb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 86.Manning CN, et al. The early inflammatory response after flexor tendon healing: a gene expression and histological analysis. J. Orthop. Res. 2014;32:645–652. doi: 10.1002/jor.22575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang Y, et al. Aspirin inhibits inflammation and scar formation in the injury tendon healing through regulating JNK/STAT-3 signalling pathway. Cell Prolif. 2019;52:1397. doi: 10.1111/cpr.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Adam, B. et al. Oral Ibuprofen Interferes with Cellular Healing Responses in a Murine Model of Achilles Tendinopathy. J Musculoskelet Disord Treat4, (2018). [DOI] [PMC free article] [PubMed]
- 89.Zhang K, Asai S, Yu B, Enomoto-Iwamoto M. IL-1β irreversibly inhibits tenogenic differentiation and alters metabolism in injured tendon-derived progenitor cells in vitro. Biochem. Biophys. Res. Commun. 2015;463:667–672. doi: 10.1016/j.bbrc.2015.05.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tsuzaki M, et al. IL-1β induces COX2, MMP-1, -3 and -13, ADAMTS-4, IL-1β and IL-6 in human tendon cells. J. Orthop. Res. 2006;21:256–264. doi: 10.1016/S0736-0266(02)00141-9. [DOI] [PubMed] [Google Scholar]
- 91.Yang G, Im H-J, Wang JH-C. Repetitive mechanical stretching modulates IL-1beta induced COX-2, MMP-1 expression, and PGE2 production in human patellar tendon fibroblasts. Gene. 2005;363:166–172. doi: 10.1016/j.gene.2005.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Machner A, et al. Higher susceptibility to Fas ligand induced apoptosis and altered modulation of cell death by tumor necrosis factor-α in periarticular tenocytes from patients with knee joint osteoarthritis. Arthritis Res. Ther. 2003;5:R253. doi: 10.1186/ar789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hosaka Y, Kirisawa R, Ueda H, Yamaguchi M, Takehana K. Differences in tumor necrosis factor (TNF)alpha and TNF receptor-1-mediated intracellular signaling factors in normal, inflamed and scar-formed horse tendons. J. Vet. Med. Sci. Jpn. Soc. Vet. Sci. 2005;67:985–991. doi: 10.1292/jvms.67.985. [DOI] [PubMed] [Google Scholar]
- 94.D'Addona A, Maffulli N, Formisano S, Rosa D. Inflammation in tendinopathy. The Surgeon. 2017;15:297–302. doi: 10.1016/j.surge.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 95.Stolk M, et al. New insights into tenocyte-immune cell interplay in an in vitro model of inflammation. Sci. Rep. 2017;7:9801. doi: 10.1038/s41598-017-09875-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gaida JE, et al. Evidence of the TNF-α system in the human Achilles tendon: expression of TNF-α and TNF receptor at both protein and mRNA levels in the tenocytes. Cells Tissues Organs. 2012;196:339–352. doi: 10.1159/000335475. [DOI] [PubMed] [Google Scholar]
- 97.Kimmerling KA, McQuilling JP, Staples MC, Mowry KC. Tenocyte cell density, migration, and extracellular matrix deposition with amniotic suspension allograft. J. Orthop. Res. 2019;37:412–420. doi: 10.1002/jor.24173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tsai W-C, et al. Effects of celecoxib on migration, proliferation and collagen expression of tendon cells. Connect. Tissue Res. 2007;48:46–51. doi: 10.1080/03008200601071295. [DOI] [PubMed] [Google Scholar]
- 99.Larson BJ, Longaker MT, Lorenz HP. Scarless fetal wound healing: a basic science review. Plast. Reconstr. Surg. 2010;126:1172–1180. doi: 10.1097/PRS.0b013e3181eae781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tsai W-C, et al. Decreased proliferation of aging tenocytes is associated with down-regulation of cellular senescence-inhibited gene and up-regulation of p27. J. Orthop. Res. 2011;29:1598–1603. doi: 10.1002/jor.21418. [DOI] [PubMed] [Google Scholar]
- 101.Klatte-Schulz F, et al. Influence of age on the cell biological characteristics and the stimulation potential of male human tenocyte-like cells. Eur. Cell. Mater. 2012;24:74–89. doi: 10.22203/ecm.v024a06. [DOI] [PubMed] [Google Scholar]
- 102.Lee YW, et al. Effects of redox modulation on cell proliferation, viability, and migration in cultured rat and human tendon progenitor cells. Oxid. Med. Cell. Longev. 2017;2017:8785042. doi: 10.1155/2017/8785042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jackson JE, Kopecki Z, Anderson PJ, Cowin AJ. In vitro analysis of the effect of Flightless I on murine tenocyte cellular functions. J. Orthop. Surg. Res. 2020;15:1–14. doi: 10.1186/s13018-020-01692-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lui PPY, Wong CM. Biology of tendon stem cells and tendon in aging. Front. Genet. 2020;10:2716. doi: 10.3389/fgene.2019.01338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kohler J, et al. Uncovering the cellular and molecular changes in tendon stem/progenitor cells attributed to tendon aging and degeneration. Aging Cell. 2013;12:988–999. doi: 10.1111/acel.12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jiang D, et al. Effect of young extrinsic environment stimulated by hypoxia on the function of aged tendon stem cell. Cell Biochem. Biophys. 2014;70:967–973. doi: 10.1007/s12013-014-0004-7. [DOI] [PubMed] [Google Scholar]
- 107.Riley GP, et al. Tendon degeneration and chronic shoulder pain: changes in the collagen composition of the human rotator cuff tendons in rotator cuff tendinitis. Ann. Rheum. Dis. 1994;53:359–366. doi: 10.1136/ard.53.6.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Qi J, et al. Interleukin-1beta increases elasticity of human bioartificial tendons. Tissue Eng. 2006;12:2913–2925. doi: 10.1089/ten.2006.12.2913. [DOI] [PubMed] [Google Scholar]
- 109.Liu Y, Suen C-W, Zhang J-F, Li G. Current concepts on tenogenic differentiation and clinical applications. J. Orthopaed. Transl. 2017;9:28–42. doi: 10.1016/j.jot.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shukunami C, et al. Scleraxis is a transcriptional activator that regulates the expression of Tenomodulin, a marker of mature tenocytes and ligamentocytes. Sci. Rep. 2018;8:3155. doi: 10.1038/s41598-018-21194-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Léjard V, et al. Scleraxis and NFATc regulate the expression of the Pro-α1(I) collagen gene in tendon fibroblasts. J. Biol. Chem. 2007;282:17665–17675. doi: 10.1074/jbc.M610113200. [DOI] [PubMed] [Google Scholar]
- 112.Sakabe T, et al. Transcription factor scleraxis vitally contributes to progenitor lineage direction in wound healing of adult tendon in mice. J. Biol. Chem. 2018;293:5766–5780. doi: 10.1074/jbc.RA118.001987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McClellan A, et al. A novel mechanism for the protection of embryonic stem cell derived tenocytes from inflammatory cytokine interleukin 1 beta. Sci. Rep. 2019;9:53. doi: 10.1038/s41598-019-39370-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Archambault JM, Elfervig-Wall MK, Tsuzaki M, Herzog W, Banes AJ. Rabbit tendon cells produce MMP-3 in response to fluid flow without significant calcium transients. J. Biomech. 2002;35:303–309. doi: 10.1016/s0021-9290(01)00217-2. [DOI] [PubMed] [Google Scholar]
- 115.Archambault J, Tsuzaki M, Herzog W, Banes AJ. Stretch and interleukin-1beta induce matrix metalloproteinases in rabbit tendon cells in vitro. J. Orthop. Res. 2002;20:36–39. doi: 10.1016/S0736-0266(01)00075-4. [DOI] [PubMed] [Google Scholar]
- 116.Arnoczky SP, Lavagnino M, Egerbacher M, Caballero O, Gardner K. Matrix metalloproteinase inhibitors prevent a decrease in the mechanical properties of stress-deprived tendons: an in vitro experimental study. Am. J. Sports Med. 2007;35:763–769. doi: 10.1177/0363546506296043. [DOI] [PubMed] [Google Scholar]
- 117.Berglund M, Hart DA, Wiig M. The inflammatory response and hyaluronan synthases in the rabbit flexor tendon and tendon sheath following injury. J. Hand Surg. Eur. 2007;32:581–587. doi: 10.1016/J.JHSE.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 118.Lo IKY, Marchuk LL, Hollinshead R, Hart DA, Frank CB. Matrix metalloproteinase and tissue inhibitor of matrix metalloproteinase mrna levels are specifically altered in torn rotator cuff tendons. Am. J. Sports Med. 2017;32:1223–1229. doi: 10.1177/0363546503262200. [DOI] [PubMed] [Google Scholar]
- 119.Sun HB, et al. Coordinate regulation of IL-1β and MMP-13 in rat tendons following subrupture fatigue damage. Clin. Orthop. Relat. Res. 2008;466:1555–1561. doi: 10.1007/s11999-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jones GC, et al. Expression profiling of metalloproteinases and tissue inhibitors of metalloproteinases in normal and degenerate human achilles tendon. Arthritis Rheum. 2006;54:832–842. doi: 10.1002/art.21672. [DOI] [PubMed] [Google Scholar]
- 121.Seok J, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. U.S.A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sauerschnig M, et al. Effect of COX-2 inhibition on tendon-to-bone healing and PGE2 concentration after anterior cruciate ligament reconstruction. Eur. J. Med. Res. 2018;23:276. doi: 10.1186/s40001-017-0297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Morita W, Dakin SG, Snelling SJB, Carr AJ. Cytokines in tendon disease: a systematic review. Bone Joint Res. 2017;6:656–664. doi: 10.1302/2046-3758.612.BJR-2017-0112.R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Radovanović G, Wolfarth B, Legerlotz K. Interleukin-6 levels drop after a 12 week long physiotherapeutic intervention in patients with Achilles tendinopathy—a pilot study. Transl. Sports Med. 2019;2:233–239. [Google Scholar]
- 125.Chen S, et al. Interleukin-6 promotes proliferation but inhibits tenogenic differentiation via the janus kinase/signal transducers and activators of transcription 3 (JAK/STAT3) pathway in tendon-derived stem cells. Med. Sci. Monit. 2018;24:1567–1573. doi: 10.12659/MSM.908802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gittel C, et al. Isolation of equine multipotent mesenchymal stromal cells by enzymatic tissue digestion or explant technique: comparison of cellular properties. BMC Vet. Res. 2013;9:221. doi: 10.1186/1746-6148-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wagenhäuser MU, et al. Collagen type I and decorin expression in tenocytes depend on the cell isolation method. BMC Musculoskelet. Disord. 2012;13:140. doi: 10.1186/1471-2474-13-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wehling M. Assessing the translatability of drug projects: what needs to be scored to predict success? Nat. Rev. Drug Discovery. 2009;8:541–546. doi: 10.1038/nrd2898. [DOI] [PubMed] [Google Scholar]
- 129.Davidson MK, Lindsey JR, Davis JK. Requirements and selection of an animal model. Isr. J. Med. Sci. 1987;23:551–555. [PubMed] [Google Scholar]
- 130.Fenu, M. Evaluation and quantification of novel scratch assay to mimic wound healing model. (2018).
- 131.Haltmayer, E. et al. Co-culture of osteochondral explants and synovial membrane as in vitro model for osteoarthritis. PLoS ONE14, e0214709 (2019). [DOI] [PMC free article] [PubMed]
- 132.Yao L, Bestwick CS, Bestwick LA, Maffulli N, Aspden RM. Phenotypic drift in human tenocyte culture. Tissue Eng. 2006;12:1843–1849. doi: 10.1089/ten.2006.12.1843. [DOI] [PubMed] [Google Scholar]
- 133.Costa-Almeida R, Calejo I, Reis RL, Gomes ME. Crosstalk between adipose stem cells and tendon cells reveals a temporal regulation of tenogenesis by matrix deposition and remodeling. J. Cell. Physiol. 2017;233:5383–5395. doi: 10.1002/jcp.26363. [DOI] [PubMed] [Google Scholar]
- 134.R Core Team. R: A language and environment for statistical computing. (R Foundation for Statistical Computing, 2017).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.