Abstract
Objective:
To assess whether gene expression signatures associated with rheumatoid arthritis (RA) before pregnancy differ between women who improve or worsen during pregnancy, and determine whether these expression signatures are altered during pregnancy when RA improves or worsens.
Methods:
Clinical data and blood samples were collected before pregnancy (T0) and at the third trimester (T3) from 11 RA and 5 healthy women. RA disease activity was assessed using the Clinical Disease Activity Index (CDAI). At each time-point, RA-associated gene expression signatures were identified using differential expression analysis of RNA sequencing profiles between RA and healthy women.
Results:
Of the women with RA, 6 improved by T3 (RAimproved), 3 worsened (RAworsened) and 2 were excluded. At T0, mean CDAI scores were similar in both groups (RAimproved: 11.2±9.8; RAworsened: 13.8±6.7; Wilcoxon-rank test: p=0.6). In the RAimproved group, 89 genes were differentially expressed at T0 (q<0.05 and fold-change (FC)≥2) compared to healthy women. When RA improved at T3, 65 of 89 (73%) of these no longer displayed RA-associated expression. In the RAworsened group, a largely different RA gene expression signature (429 genes) was identified at T0. When RA disease activity worsened at T3, 207 of 429 (48%) lost their differential expression, while an additional 151 genes became newly differentially expressed.
Conclusion:
In our pilot dataset, pre-pregnancy RA expression signatures differed between women who subsequently improved or worsened during pregnancy, suggesting that inherent genomic differences perhaps influence how pregnancy impacts disease activity. Further, these RA signatures were altered during pregnancy, as disease activity changed.
Keywords: Rheumatoid arthritis, Gene expression signature, Pregnancy, RNA-seq
INTRODUCTION
Rheumatoid arthritis (RA) is a systemic inflammatory disease that leads to significant disability resulting from pain and swelling of inflamed joints and from joint destruction. To date, there is no cure. Pregnancy is known to have disease-modifying properties (1–4) on RA, with a significant proportion of women experiencing an improvement in disease activity during pregnancy, while in others, the disease may remain unchanged or may even worsen. Even though there are medications, including some traditional and biologic disease modifying anti-rheumatic drugs (DMARDs) that are considered safe for use in pregnancy (5, 6), many women with RA prefer to stop taking medications during pregnancy. However, because there are no known biomarkers at present to predict who will likely improve or worsen or whose RA will remain unchanged during pregnancy, these women are hesitant to plan a pregnancy because they do not know whether their disease will worsen if they stop taking medications in order to try to conceive.
Several case-control studies, based on gene expression data from microarrays (7–12) or RNA sequencing (RNA-seq), have been conducted to investigate gene expression signatures associated with RA (13). However, gene expression studies that have been conducted in the context of RA pregnancy did not examine RA-associated expression signatures in the non-pregnant state due to pre-pregnancy samples not being available (14–17). It is thus not known whether the pre-pregnancy RA expression signature can be used to predict whether RA will subsequently improve or worsen during pregnancy. Further, given that gene expression is a dynamic process, it is possible that the RA-associated gene expression signature may be altered during pregnancy. Genes modulating disease activity during pregnancy may show altered expression when disease activity changes over time, i.e. their expression may either no longer be associated with RA when disease activity is low or in remission during pregnancy, or additional genes may show RA-associated expression when RA worsens during pregnancy. However, the influence of pregnancy on the RA gene expression signature, if any, has not been investigated.
In the present study, we have used our unique prospective pilot pregnancy cohort of RA and healthy women that includes a pre-pregnancy time-point (18, 19) as a case-control dataset to examine gene expression signatures associated with RA at the pre-pregnancy baseline. We hypothesized that the baseline RA-associated gene expression signature among women who subsequently improved during pregnancy differs from that among women who worsened during pregnancy. We also evaluated a second hypothesis that the gene expression signature associated with RA at the pre-pregnancy baseline is altered during pregnancy, when disease activity improves or worsens.
SUBJECTS AND METHODS
Study subjects
Healthy women and women with RA of Danish descent who were planning a pregnancy were recruited and enrolled in our pregnancy cohort in Denmark and were prospectively followed, as previously described (18). A subset of 11 RA and 5 healthy women from this cohort, on whom we reported longitudinal changes in expression (19), was included in the present study. The women with RA fulfilled the 1987 revised American College of Rheumatology criteria for the disease (20). The study was approved by the Ethics Committee for Region Hovedstaden (Denmark), the Danish Data Protection Agency, and the Children’s Hospital Oakland Research Institute Institutional Review Board (IRB number: 2009–073). All subjects provided written informed consent prior to enrollment. Data and samples were collected as previously described (19).
Assessment of RA disease activity
RA disease activity was assessed using the Clinical Disease Activity Index (CDAI) (21), because it does not include acute phase reactants such as C-reactive protein (CRP) whose levels are known to fluctuate during pregnancy (22, 23); further, acute phase reactants do not contribute much additional information on top of what is provided by the CDAI (24). The change in CDAI (ΔCDAI) from before pregnancy (T0) to the third trimester (T3) was used to determine whether disease activity improved or worsened. Patients were categorized as having improved by T3 (RAimproved), if their ΔCDAI fit the criteria for a minimum clinically important difference (MCID) based on baseline (T0) disease activity; ΔCDAI values of 12, 6 and 1 were used as threshold when disease activity at T0 was high, moderate or low, respectively (25). Those women with an increase in CDAI from T0 to T3, satisfying the MCID criteria for worsening of disease activity, were included in the “worsened” subset, referred to as RAworsened.
RNA sequencing and Bioinformatic analyses
RNA extractions, processing and sequencing were performed as originally described (18). Pseudoalignment of the de-multiplexed raw sequence reads (FASTQ format) to the Ensembl reference human GRCh38 transcriptome assembly (release 98) and quantification of transcript abundances were performed using kallisto (v0.43.0) (26). BioMart annotations were used to combine transcript-level counts into gene-level estimates, i.e. counts were summed by Ensembl gene ID. Gene IDs that mapped to patches or alternate haplotypes rather than to the primary reference sequence were excluded to avoid duplication. Pseudogenes, genes without annotations as well as genes with very low read counts (<1 transcript per million) in at least 25% of all samples were filtered out. Any globin and rRNA transcripts still present were also filtered out. To adjust for variable sequencing depths across samples, the gene-level counts were normalized using the Trimmed Mean of M values (TMM) algorithm as implemented in the edgeR package (v3.26.8) (27, 28). To assess batch effects, normalized counts from pairs of technical replicates were plotted, and outliers were filtered out to achieve a Pearson correlation of at least 95% between replicates.
Statistical analyses
Case-control differential gene expression analysis
To identify gene expression signatures associated with RA at the T0 baseline, cross-sectional differential expression analysis was performed using edgeR (v3.26.8) (27), comparing normalized T0 gene-level counts between each RA subset (RAimproved or RAworsened) and healthy women. In each analysis, a negative binomial distribution was used to handle the over-dispersion in RNA-seq gene counts. Differential expression was tested using generalized linear model (GLM) likelihood ratio tests; differences between RA and healthy women were assessed using the contrast argument of the glmLRT function in edgeR. Correction for multiple testing was performed using the False Discovery Rate (FDR) method (29). A q-value threshold of 0.05, in combination with a fold-change (FC) of at least 2, was used to assess significance. To determine whether the pre-pregnancy expression signature changed when RA improved or worsened during pregnancy, the differential expression analysis was repeated using data from the same women (RAimproved or RAworsened vs. healthy) at the T3 time-point.
Functional analysis
Differentially expressed genes were analyzed for over-representation of Gene Ontology (GO) categories using a hypergeometric test implemented in the Web-based Gene Set Analysis Toolkit (WebGestalt) (30). A significance threshold of q<0.05 was used to define enrichment. Cystoscape (31) was used for functional annotations and visualization of protein interactions documented in the STRING database (32).
RESULTS
Study subjects
Of the 11 women with RA, 6 improved by T3 while 3 worsened, based on MCID thresholds. Two women were excluded because even though their disease activity improved during pregnancy, one was already in remission at T0, and the ΔCDAI value for the other (ΔCDAI=2.7) did not meet the MCID threshold of 6, for moderate baseline disease activity. The changes in disease activity scores from T0 to T3 were significantly correlated between the CDAI and the DAS28CRP3 (Pearson’s correlation=85%, p=0.004). The average age at conception was as follows: 28.9±6.0 years for RAimproved, 33.2±1.9 years for RAworsened, and 31.2±5.7 years for the healthy women. The women who improved had a shorter disease duration than those who worsened (RAimproved (mean±SD): 6.5±4.2 years and RAworsened: 8.9±1.1 years), although this difference was not statistically significant. Medications taken by the women with RA at each time-point are shown in Table 1. While mean disease activity (CDAI scores) at baseline did not differ significantly between the two RA subsets (RAimproved: 11.2±9.8 and RAworsened: 13.8±6.7; Wilcoxon-rank test: p=0.6), the mean values at T3 differed significantly (RAimproved: 2.2±1.3 and RAworsened: 31.7±15.1; Wilcoxon-rank test: p=0.02).
Table 1.
Medication use among the women with RA at each time-point
| Patient | Pre-pregnancy | 3rd trimester |
|---|---|---|
| Improved: | ||
| 1 | None | None |
| 2 | None | None |
| 3 | None | Prednisolone + Sulfasalazine |
| 4 | Sulfasalazine | Sulfasalazine |
| 5 | Sulfasalazine | Prednisolon |
| 6 | Prednisolone + Infliximab | Prednisolone + Sulfasalazine |
| Worsened: | ||
| 7 | Prednisolone + Sulfasalazine + Etanercept | Prednisolone + Sulfasalazine |
| 8 | Sulfasalazine + Adalimumab | Adalimumab |
| 9 | Infliximab | None |
The RA gene expression signature at the T0 (pre-pregnancy) baseline
RAimproved vs healthy women
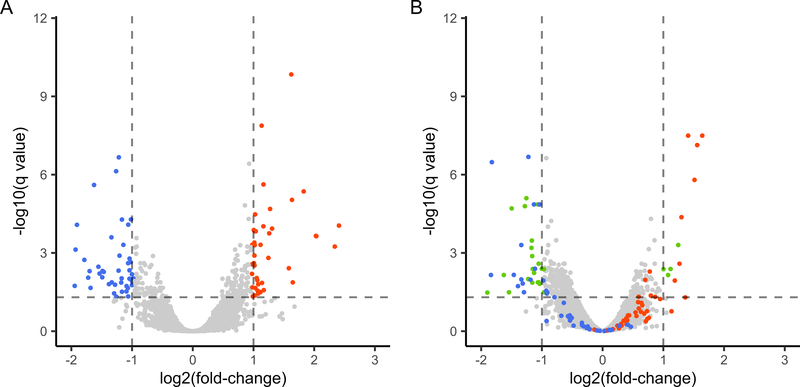
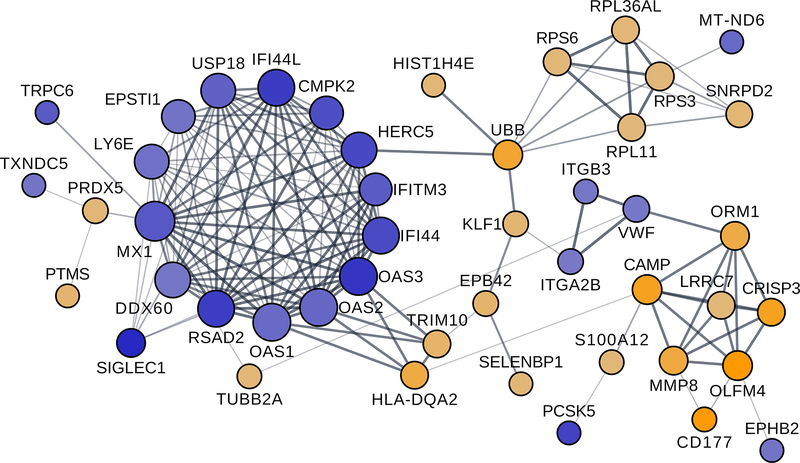
A total of 89 genes were differentially expressed (q<0.05; FC≥2) between the 6 RAimproved and 5 healthy women (Figure 1A and Supplementary Table S1). The genes that were over-expressed (n=44) in RA (e.g. C4BPA, CAMP, CD177, CRISP3, HLA-DQA2, MMP8, OLFM4, ORM1, S100A12) as well as those that were under-expressed (n=45) (e.g. CMPK2, HERC5, IFI44, IFI44L, IFITM3, IL1RL1, IL5RA, MX1, OAS1, OAS2, OAS3, SIGLEC1) were enriched in various immune-related gene ontology (GO) biological processes, as shown in Table 2, and in reactome pathways relating to interferon signaling (q=5.9E-06), antiviral mechanism by IFN-stimulated genes (q=7.0E-05), and p130Cas linkage to MAPK signaling for integrins (q=1.4E-02), among others. A large proportion (52%) of the 89 genes differentially expressed between RAimproved and healthy women at the T0 baseline encode proteins that are functionally related, as shown by protein networks, based on the STRING database (32), in Cytoscape (Figure 2); most of the under-expressed genes formed a tight cluster, distinct from the over-expressed genes.
Figure 1. RA-associated expression among RA women who improved during pregnancy.
Volcano plots showing differential expression between RA women who improved during pregnancy (RAimproved) and healthy women at two time points. (A) At pre-pregnancy (T0), 89 genes were differentially expressed (q<0.05; fold-change (FC)≥2) between RAimproved and healthy women, some being over-expressed in RA (orange dots) and others under-expressed (blue dots). (B) At the third trimester (T3), when RA improved, 65 of the 89 genes (73%) were no longer differentially expressed (orange and blue dots with −1≤log2(FC)≤1 and –log10(q-value)<1.3). Genes that became newly differentially expressed at T3 are shown as green dots.
Table 2.
Gene ontology (GO) biological processes enriched in genes with RA-associated expression before pregnancy among RAimproved women
| Gene Set | Description | Enrichment ratio | q value |
|---|---|---|---|
| GO:0032069 | Regulation of nuclease activity | 46.6 | 7.6E-04 |
| GO:0060337 | Type I interferon signaling pathway | 21.4 | 5.3E-05 |
| GO:1903901 | Negative regulation of viral life cycle | 20.8 | 3.1E-04 |
| GO:2001244 | Positive regulation of intrinsic apoptotic signaling pathway | 18.6 | 1.5E-02 |
| GO:0045087 | Innate immune response | 5.9 | 1.7E-06 |
| GO:0043312 | Neutrophil degranulation | 5.9 | 1.1E-03 |
| GO:0051707 | Response to other organism | 5.1 | 1.7E-05 |
| GO:0050878 | Regulation of body fluid levels | 4.8 | 2.2E-02 |
Figure 2. Protein network showing genes differentially expressed at the pre-pregnancy baseline within the same functional network.
A large proportion of the 89 genes differentially expressed between RAimproved women and healthy women at the pre-pregnancy baseline encode proteins that belong to a common functional network, based on protein interactions data from the STRING database. Most of the under-expressed genes (blue circles) formed a tight cluster, distinct from the over-expressed genes (orange circles).
RAworsened vs healthy women
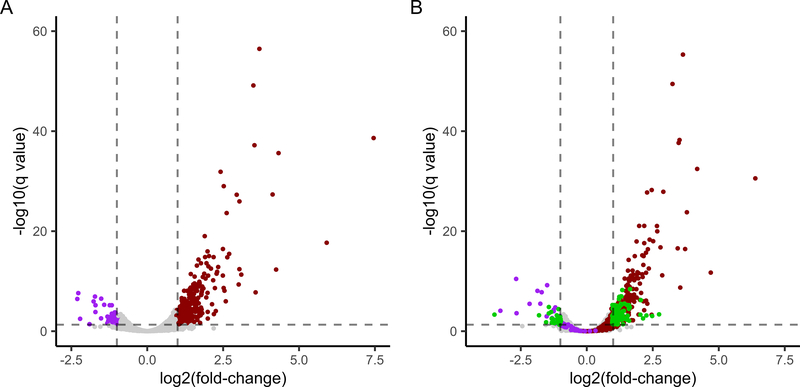
A total of 429 genes were differentially expressed (FC≥2; q<0.05) between RAworsened and healthy women at T0 (Figure 3A). This gene expression signature largely differed from the one identified above (RAimproved vs healthy); only 19 of the 429 genes overlapped with those differentially expressed between RAimproved and healthy women at T0, with the majority demonstrating similar expression patterns in both RA sub-groups compared to healthy women, i.e. over-expressed: OLFM4, UBB, ORM1, SEPTIN3, KRT1, TUBB2A or under-expressed: IL1RL1, IGLC3, IGLV2–14, PF4V1, FADS2, NKX3–1 (Supplementary Table S2). HLA-DRQA2, on the other hand, was 3-fold over-expressed among the RAimproved, and 3-fold under-expressed among the RAworsened women, compared to the healthy women.
Figure 3. RA-associated expression among RA women who worsened during pregnancy.
Volcano plots showing differential expression between RA women who worsened during pregnancy (RAworsened) and healthy women before and during pregnancy. (A) At pre-pregnancy (T0), 429 genes were differentially expressed (q<0.05; fold-change (FC)≥2) between RAworsened and healthy women, consisting of some that were over-expressed in RA (maroon dots) and some that were under-expressed (purple dots). (B) At the 3rd trimester (T3), when RA worsened, 207 of the 429 genes (48%) were no longer differentially expressed (maroon and purple dots with −1≤log2(FC)≤1 and –log10(q-value)<1.3). Numerous genes (green dots) became newly differentially expressed at T3 when disease activity worsened.
The RA gene expression signature is altered when RA improves or worsens during pregnancy
RAimproved vs healthy women at T3
When disease activity improved by T3, most of the baseline RA signature genes identified among RAimproved women (65 of 89, i.e. 73%) were no longer differentially expressed between the RAimproved and healthy women (Figure 1B and Supplementary Table S1). Of note, there were 24 genes that remained differentially expressed between RAimproved and healthy women at T3 (e.g. C4BPA, HLA-DQA2, IGLC3, IL5RA, MAOA, OLIG2, PTGDR2, TUBB2A), and an additional few (n=27) became newly differentially expressed (e.g. ADORA3, CYP27A1, DSC1, RAP1GAP, SCL29A1).
RAworsened vs healthy women at T3
When disease activity worsened by T3, 207 of the 429 genes (48%) differentially expressed at the T0 baseline lost their differential expression, while an additional 151 genes became newly differentially expressed (Figure 3B).
DISCUSSION
In the present study, our goal was to examine whether the pre-pregnancy RA gene expression signature differs between women who subsequently improved during pregnancy and those who worsened during pregnancy. We also examined whether the pre-pregnancy RA gene expression signature was altered in any way during pregnancy, when disease activity improved or worsened.
In our pilot dataset, even though mean disease activity was similar between the RAimproved and RAworsened groups at the pre-pregnancy (non-pregnant) baseline, there was very little overlap in the sets of genes showing RA-associated expression within each of the two groups. The few genes that overlapped between the two RA expression signatures included some that have previously been implicated in RA such as IL1RL1 (33), ORM1 (34), KRT1 (35), and HLA-DQA2 (36). Although HLA-DQA2 expression was associated with RA in both subsets, this gene demonstrated contrasting expression patterns in the two RA subsets; it was 3-fold over-expressed in the RAimproved group and 3-fold under-expressed in the RAworsened group, both compared to the same set of healthy women. While increased HLA-DQA2 expression has been reported in RA (37), a negative correlation has also been found between expression levels in synovial tissue fibroblast cells and Health Assessment Questionnaire (HAQ) scores (38). Nonetheless, the significance of these contrasting expression patterns in the two groups of women with RA is not entirely clear. The RA expression signature identified among the RAimproved women included many additional genes whose expression and/or methylation patterns have previously been associated with RA, such as S100A12 (39), CRISP3 (40), MMP8 (41), and CAMP (12). Of interest, the IFN-inducible genes IFI44, IFI44L, CMPK2, HERC5, MX1, SIGLEC1, OAS1, OAS2 and OAS3 were also part of the baseline RA expression signature among the RAimproved women. Compared to healthy women, these genes were under-expressed in this RA subset at baseline as we recently reported (19), in contrast to other studies of RA and other autoimmune conditions (9, 42, 43). While our pre-pregnancy RA signatures also included many genes that did not overlap with RA signatures from previous case-control studies, results were inconsistent even across those previous studies. This could be attributed to a number of factors including: the source of RNA [whole blood (our study) vs. peripheral blood mononuclear cells (PBMCs)] (9, 11, 14–17), synovial fibroblasts (13), or neutrophils (44); differences in gene expression technology used [RNA-seq (our study) vs. microarrays] (9, 11, 14–17); patient sample and sex ratio (only women, most of whom experienced improvement of RA disease activity during pregnancy (our study) vs. women and men) (9, 11, 13, 44).
In the present study, we also examined the influence of pregnancy on RA-associated gene expression signatures. This had not previously been reported since pre-pregnancy samples were not available in prior studies (14–17). We observed a dilution of the baseline RA signature during pregnancy, with the majority (73%) of signature genes showing similar expression in both RA and healthy women by the third trimester, when RA improved. These results are consistent with those of a previous study demonstrating minimal differences in PBMC expression profiles between RA and healthy women at the third trimester (15). While our study design does not allow us to determine whether the loss of association with RA at the 3rd trimester is specific to pregnancy or not, it is plausible that as the expression profiles of the women with RA undergo pregnancy-induced changes, they start to resemble those of healthy women, as we observed, and these changes are accompanied by an improvement of the disease. On the other hand, when RA worsened during pregnancy, many genes (n=151) demonstrated new expression patterns that became associated with RA, as would be expected when the “case” and “control” groups become phenotypically more different from each other.
In a previous study comparing the DAS28ESR and DAS28CRP, with and without patient global scores, during pregnancy, the DAS28CRP3 was found to perform better (45). However, since even CRP levels have been shown to fluctuate during pregnancy (22, 23), the DAS28CRP3 does not represent a gold standard for use in pregnancy. Other measures of disease activity such as the CDAI that do not include acute phase reactants have not been assessed for their performance during pregnancy. In the absence of a gold standard, we chose to use the CDAI to assess disease activity before and during pregnancy because acute phase reactants have been shown to add little information on top of the clinical variables already included in the CDAI (24). Additionally, the CDAI is more stringent than the DAS28 when assessing improvement of disease activity; it has been shown that patients can satisfy DAS28 remission criteria while still having active disease (46).
In our study, the availability of pre-pregnancy and pregnancy data from the same women who improved or worsened during pregnancy enabled us to investigate pre-pregnancy gene expression signatures between the two groups as well as the effect of pregnancy on those expression signatures. The use of RNA-seq technology to assess gene expression was another strength. Our study has some limitations. First, given that this is a pilot study, sample sizes were small. Nonetheless, patterns emerged that are supported by previous literature and thus further investigations in a larger sample are warranted. We did not examine proportions of cell types between disease states (RA vs healthy) and/or across time points because our goal was to identify overall systemic gene expression changes, resulting from altered expression of specific genes or from differences in cell proportions. Because we used total RNA from whole blood, expression profiles of neutrophils may have dominated the observed expression patterns. It is also possible that medications taken by the women with RA before pregnancy may have influenced the results. However, due to sample size limitations, we could not assess if this was the case, and we also were unable to adjust for dosage and/or specific medications.
In conclusion, we report here novel findings that women with RA who improved during pregnancy demonstrated differences in pre-pregnancy RA-associated gene expression compared to women who worsened. These differences in pre-pregnancy RA expression signatures suggest that inherent genomic differences between women with RA may influence how pregnancy alters disease activity. Our findings that RA-associated gene expression signatures are altered during pregnancy, when disease activity changes are also novel. Additional investigations in larger datasets are warranted to corroborate these preliminary findings and to identify novel drug targets and/or biomarkers of disease activity.
Supplementary Material
ACKNOWLEDGMENTS
We are immensely grateful to the study subjects for their participation in the study. We acknowledge our gratitude towards Dr. Hanne Kjærgaard for all her efforts in setting up the logistics for this study in Denmark; Dr. Kjærgaard passed away in 2013. We also thank the leadership team at the Juliane Marie Center in Denmark for their support. The Rheumatology departments at the following hospitals in Denmark facilitated collection of data and samples: Rigshospitalet (Glostrup), Odense Universitetshospital, Dansk Gigthospital (Sønderborg), Aarhus University Hospital (Skejby) and Regionshospitalet Viborg.
We thank all members of our project team for making this work possible: Anne-Grethe Rasmussen, Charlotte Schön Frengler, Dorte Heide, Randi Petersen, Tove Thorup Rasmussen, Lone Thomasen, Britta Hvidberg Nielsen, Teresa Rozenfeld, Kirsten Junker, Lis Kastberg Schubert, Lis Lund, Jette Barlach, Helle Bendtsen, Helle Andersen and Marjo Westerdahl for their contribution with data and sample collection; Rikke Godtkjær Andersen, Mie Rams Rasmussen, Katrine Elmgaard Jensen, Pia Pedersen, Stine Birkelund, Louise Mielke and Andreas Smed for management of data and samples. We also greatly appreciate valuable assistance provided by DANBIO personnel.
Dr. Damini Jawaheer accepts responsibility for the integrity and validity of the data collected and analyzed.
Funding: This work was supported in part by funds from: the National Institutes of Arthritis, Musculoskeletal and Skin Diseases (NIAMS), USA (Grants R21AR057931 and R01AR073111), Gigtforeningen, Denmark (Grant R87-A1477-B512), The Juliane Marie Center, Rigshospitalet (Denmark) and a private donor. These funders did not have any role in conducting this study or in interpretation and reporting of results.
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
REFERENCES
- 1.Hench PS. The ameliorating effect of pregnancy on chronic atrophic (infectious rheumatoid) arthritis, fibrositis, and intermittent hydrarthrosis. Mayo Clin Proc 1938;13:161–7. [Google Scholar]
- 2.Nelson JL, Ostensen M. Pregnancy and rheumatoid arthritis. Rheum Dis Clin North Am 1997;23:195–212. [DOI] [PubMed] [Google Scholar]
- 3.de Man YA, Dolhain RJ, van de Geijn FE, Willemsen SP, Hazes JM. Disease activity of rheumatoid arthritis during pregnancy: Results from a nationwide prospective study. Arthritis Rheum 2008;59:1241–8. [DOI] [PubMed] [Google Scholar]
- 4.Barrett JH, Brennan P, Fiddler M, Silman AJ. Does rheumatoid arthritis remit during pregnancy and relapse postpartum? Results from a nationwide study in the united kingdom performed prospectively from late pregnancy. Arthritis Rheum 1999;42:1219–27. [DOI] [PubMed] [Google Scholar]
- 5.Gotestam Skorpen C, Hoeltzenbein M, Tincani A, Fischer-Betz R, Elefant E, Chambers C, et al. The eular points to consider for use of antirheumatic drugs before pregnancy, and during pregnancy and lactation. Ann Rheum Dis 2016;75:795–810. [DOI] [PubMed] [Google Scholar]
- 6.Krause ML, Amin S, Makol A. Use of dmards and biologics during pregnancy and lactation in rheumatoid arthritis: What the rheumatologist needs to know. Ther Adv Musculoskelet Dis 2014;6:169–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huber R, Hummert C, Gausmann U, Pohlers D, Koczan D, Guthke R, et al. Identification of intra-group, inter-individual, and gene-specific variances in mrna expression profiles in the rheumatoid arthritis synovial membrane. Arthritis Res Ther 2008;10:R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ungethuem U, Haeupl T, Witt H, Koczan D, Krenn V, Huber H, et al. Molecular signatures and new candidates to target the pathogenesis of rheumatoid arthritis. Physiol Genomics 2010;42A:267–82. [DOI] [PubMed] [Google Scholar]
- 9.van der Pouw Kraan TC, Wijbrandts CA, van Baarsen LG, Voskuyl AE, Rustenburg F, Baggen JM, et al. Rheumatoid arthritis subtypes identified by genomic profiling of peripheral blood cells: Assignment of a type i interferon signature in a subpopulation of patients. Ann Rheum Dis 2007;66:1008–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bovin LF, Rieneck K, Workman C, Nielsen H, Sorensen SF, Skjodt H, et al. Blood cell gene expression profiling in rheumatoid arthritis. Discriminative genes and effect of rheumatoid factor. Immunol Lett 2004;93:217–26. [DOI] [PubMed] [Google Scholar]
- 11.Batliwalla FM, Baechler EC, Xiao X, Li W, Balasubramanian S, Khalili H, et al. Peripheral blood gene expression profiling in rheumatoid arthritis. Genes Immun 2005;6:388–97. [DOI] [PubMed] [Google Scholar]
- 12.Teixeira VH, Olaso R, Martin-Magniette ML, Lasbleiz S, Jacq L, Oliveira CR, et al. Transcriptome analysis describing new immunity and defense genes in peripheral blood mononuclear cells of rheumatoid arthritis patients. PLoS One 2009;4:e6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heruth DP, Gibson M, Grigoryev DN, Zhang LQ, Ye SQ. Rna-seq analysis of synovial fibroblasts brings new insights into rheumatoid arthritis. Cell Biosci 2012;2:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haupl T, Ostensen M, Grutzkau A, Burmester GR, Villiger PM. Interaction between rheumatoid arthritis and pregnancy: Correlation of molecular data with clinical disease activity measures. Rheumatology (Oxford) 2008;47 Suppl 3:iii19–22. [DOI] [PubMed] [Google Scholar]
- 15.Haupl T, Ostensen M, Grutzkau A, Radbruch A, Burmester GR, Villiger PM. Reactivation of rheumatoid arthritis after pregnancy: Increased phagocyte and recurring lymphocyte gene activity. Arthritis Rheum 2008;58:2981–92. [DOI] [PubMed] [Google Scholar]
- 16.Weix J, Forger F, Haupl T, Surbek D, Ostensen M, Villiger PM. Influence of pregnancy on the adipocytokine and peroxisome proliferator-activated receptor pathways in peripheral blood mononuclear cells from healthy donors and rheumatoid arthritis patients. Arthritis Rheum 2012;64:2095–103. [DOI] [PubMed] [Google Scholar]
- 17.Weix J, Haupl T, Raio L, Villiger PM, Forger F. The physiologic increase in expression of some type i ifn-inducible genes during pregnancy is not associated with improved disease activity in pregnant patients with rheumatoid arthritis. Transl Res 2013;161:505–12. [DOI] [PubMed] [Google Scholar]
- 18.Mittal A, Pachter L, Nelson JL, Kjaergaard H, Smed MK, Gildengorin VL, et al. Pregnancy-induced changes in systemic gene expression among healthy women and women with rheumatoid arthritis. PLoS One 2015;10:e0145204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goin DE, Smed MK, Pachter L, Purdom E, Nelson JL, Kjaergaard H, et al. Pregnancy-induced gene expression changes in vivo among women with rheumatoid arthritis: A pilot study. Arthritis Res Ther 2017;19:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The american rheumatism association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 1988;31:315–24. [DOI] [PubMed] [Google Scholar]
- 21.Mierau M, Schoels M, Gonda G, Fuchs J, Aletaha D, Smolen JS. Assessing remission in clinical practice. Rheumatology (Oxford) 2007;46:975–9. [DOI] [PubMed] [Google Scholar]
- 22.Skarzynska E, Zborowska H, Jakimiuk AJ, Karlinska M, Lisowska-Myjak B. Variations in serum concentrations of c-reactive protein, ceruloplasmin, lactoferrin and myeloperoxidase and their interactions during normal human pregnancy and postpartum period. J Trace Elem Med Biol 2018;46:83–7. [DOI] [PubMed] [Google Scholar]
- 23.Belo L, Santos-Silva A, Rocha S, Caslake M, Cooney J, Pereira-Leite L, et al. Fluctuations in c-reactive protein concentration and neutrophil activation during normal human pregnancy. Eur J Obstet Gynecol Reprod Biol 2005;123:46–51. [DOI] [PubMed] [Google Scholar]
- 24.Aletaha D, Nell VP, Stamm T, Uffmann M, Pflugbeil S, Machold K, et al. Acute phase reactants add little to composite disease activity indices for rheumatoid arthritis: Validation of a clinical activity score. Arthritis Res Ther 2005;7:R796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curtis JR, Yang S, Chen L, Pope JE, Keystone EC, Haraoui B, et al. Determining the minimally important difference in the clinical disease activity index for improvement and worsening in early rheumatoid arthritis patients. Arthritis Care Res (Hoboken) 2015;67:1345–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic rna-seq quantification. Nat Biotechnol 2016;34:525–7. [DOI] [PubMed] [Google Scholar]
- 27.Robinson MD, McCarthy DJ, Smyth GK. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of rna-seq data. Genome Biol 2010;11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B 1995;57:289–300. [Google Scholar]
- 30.Zhang B, Kirov S, Snoddy J. Webgestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic acids research 2005;33:W741–W8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, et al. String: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res 2005;33:D433–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leung BP, Xu D, Culshaw S, McInnes IB, Liew FY. A novel therapy of murine collagen-induced arthritis with soluble t1/st2. J Immunol 2004;173:145–50. [DOI] [PubMed] [Google Scholar]
- 34.Park YJ, Yoo SA, Hwang D, Cho CS, Kim WU. Identification of novel urinary biomarkers for assessing disease activity and prognosis of rheumatoid arthritis. Exp Mol Med 2016;48:e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharjee M, Balakrishnan L, Renuse S, Advani J, Goel R, Sathe G, et al. Synovial fluid proteome in rheumatoid arthritis. Clin Proteomics 2016;13:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andreasi RB, Khan MFJ, Galuppi E, Govoni M, Rubini M. Replication analysis of gene-gene interaction between hla-dqa2 and hla-dqb2 variants in italian rheumatoid arthritis patients. Annals of the Rheumatic Diseases 2017;76:207. [Google Scholar]
- 37.Niu X, Lu C, Xiao C, Ge N, Jiang M, Li L, et al. The crosstalk of pathways involved in immune response maybe the shared molecular basis of rheumatoid arthritis and type 2 diabetes. PLoS One 2015;10:e0134990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galligan CL, Baig E, Bykerk V, Keystone EC, Fish EN. Distinctive gene expression signatures in rheumatoid arthritis synovial tissue fibroblast cells: Correlates with disease activity. Genes Immun 2007;8:480–91. [DOI] [PubMed] [Google Scholar]
- 39.Nordal HH, Brun JG, Halse AK, Jonsson R, Fagerhol MK, Hammer HB. The neutrophil protein s100a12 is associated with a comprehensive ultrasonographic synovitis score in a longitudinal study of patients with rheumatoid arthritis treated with adalimumab. BMC Musculoskelet Disord 2014;15:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chapman EA, Lyon M, Simpson D, Mason D, Beynon RJ, Moots RJ, et al. Caught in a trap? Proteomic analysis of neutrophil extracellular traps in rheumatoid arthritis and systemic lupus erythematosus. Front Immunol 2019;10:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tchetverikov I, Ronday HK, Van El B, Kiers GH, Verzijl N, TeKoppele JM, et al. Mmp profile in paired serum and synovial fluid samples of patients with rheumatoid arthritis. Ann Rheum Dis 2004;63:881–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higgs BW, Liu Z, White B, Zhu W, White WI, Morehouse C, et al. Patients with systemic lupus erythematosus, myositis, rheumatoid arthritis and scleroderma share activation of a common type i interferon pathway. Ann Rheum Dis 2011;70:2029–36. [DOI] [PubMed] [Google Scholar]
- 43.Toro-Dominguez D, Carmona-Saez P, Alarcon-Riquelme ME. Shared signatures between rheumatoid arthritis, systemic lupus erythematosus and sjogren’s syndrome uncovered through gene expression meta-analysis. Arthritis Res Ther 2014;16:489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright HL, Thomas HB, Moots RJ, Edwards SW. Interferon gene expression signature in rheumatoid arthritis neutrophils correlates with a good response to tnfi therapy. Rheumatology (Oxford) 2015;54:188–93. [DOI] [PubMed] [Google Scholar]
- 45.de Man YA, Hazes JM, van de Geijn FE, Krommenhoek C, Dolhain RJ. Measuring disease activity and functionality during pregnancy in patients with rheumatoid arthritis. Arthritis Rheum 2007;57:716–22. [DOI] [PubMed] [Google Scholar]
- 46.Aletaha D, Smolen JS. Remission in rheumatoid arthritis: Missing objectives by using inadequate das28 targets. Nat Rev Rheumatol 2019;15:633–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.