Summary
Gene therapy is becoming an increasingly valuable tool to treat many genetic diseases with no or limited treatment options. This is the case for hundreds of monogenic metabolic disorders of hepatic origin, for which liver transplantation remains the only cure. Furthermore, the liver contains 10–15% of the body’s total blood volume, making it ideal for use as a factory to secrete proteins into the circulation. In recent decades, an expanding toolbox has become available for liver-directed gene delivery. Although viral vectors have long been the preferred approach to target hepatocytes, an increasing number of non-viral vectors are emerging as highly efficient vehicles for the delivery of genetic material. Herein, we review advances in gene delivery vectors targeting the liver and more specifically hepatocytes, covering strategies based on gene addition and gene editing, as well as the exciting results obtained with the use of RNA as a therapeutic molecule. Moreover, we will briefly summarise some of the limitations of current liver-directed gene therapy approaches and potential ways of overcoming them.
Keywords: Non-viral vectors, viral vectors, gene addition, gene silencing, gene editing, hepatocytes, immune response, efficacy, toxicity
Abbreviations: AAT, α1-antitrypsin; AHP, acute hepatic porphyrias; AIP, acute intermittent porphyria; AAV, adeno-associated virus; Ad, adenovirus; ASGCT, American Society of Gene and Cell Therapy; ALAS1, aminolevulic synthase 1; APCs, antigen-presenting cells; ASOs, antisense oligonucleotides; apoB/E, apolipoprotein B/E; ASGPR, asialoglycoprotein receptor; CRISPR, clustered regularly interspaced short palindromic repeats; CN, Crigel-Najjar; CRISPR/Cas9, CRISPR associated protein 9; CBS, cystathionine β-synthase; dCas9, dead Cas9; DSBs, double-strand breaks; ERT, enzyme replacement therapy; FH, familial hypercholesterolemia; FSP27, fat-specific protein 27; GalNAc, N-acetyl-D-galactosamine; GT, gene therapy; GSD1a, glycogen storage disorder 1a; GO, glycolate oxidase; GUSB, β-glucuronidase; HDAd, helper-dependent adenovirus; HemA/B, haemophilia A/B; HT, hereditary tyrosinemia; HDR, homology-directed repair; IDS, iduronate 2-sulfatase; IDUA, α-L-iduronidase; IMLD, inherited metabolic liver diseases; ITR, inverted terminal repetition; LDH, lactate dehydrogenase; LV, lentivirus; LNP, Lipid nanoparticles; lncRNA, long non-coding RNA; LTR, long terminal repeat; LDLR, low-density lipoprotein receptor; MMA, methylmalonic acidemia; miRNAs, microRNAs; MPR, metabolic pathway reprograming; MPS, mucopolysaccharidosis; MPS type I, MPSI; MPS type VII, MPSVII; NASH, non-alcoholic steatohepatitis; NHEJ, non-homologous end joining; NHPs, non-human primates; OLT, orthotopic liver transplantation; OTC, ornithine transcarbamylase; PKU, phenylketonuria; PB, piggyBac; PEG, polyethylene glycol; PEI, polyethyleneimine; PH1, Primary hyperoxaluria type 1; PFIC3, progressive familial cholestasis type 3; PA, propionic acidemia; PCSK9, proprotein convertase subtilisin/kexin type 9; RV, retrovirus; SB, Sleeping Beauty; siRNA, small-interfering RNA; S/MAR, scaffold matrix attachment regions; STK25, serine/threonine protein kinase 25; SRT, substrate reduction therapy; TALEN, transcription activator-like effector nucleases; TTR, transthyretin; UCD, urea cycle disorders; VLDLR, very-low-density lipoprotein receptor; WD, Wilson’s disease; ZFN, zinc finger nucleases
Key points.
-
•
The liver is a very attractive organ for gene therapy because of its role in many essential metabolic functions, the natural hepatic tropism of many gene therapy vectors, and its capacity to act as a protein factory for distribution to the whole body.
-
•
In this rapidly advancing field, an extended toolbox has become available for liver-directed gene delivery, including non-viral and viral vectors with high transduction capacity and specificity for hepatocytes.
-
•
The use of RNA-mediated gene silencing and AAV-mediated gene delivery is transforming the potential therapeutic options for patients with inherited metabolic liver diseases.
-
•
The combination of genome editing with viral and non-viral vectors is a powerful approach that may potentially represent a curative solution to many inherited disorders.
-
•
The management of liver toxicity caused by the induction of an inflammatory response to gene transfer vectors is essential.
-
•
New strategies to circumvent the inhibitory action of humoral and cellular immune responses against gene therapy vectors are under development, and their clinical testing will occur in the near future.
Introduction
According to the latest definition by the American Society of Gene and Cell Therapy (ASGCT) in 2019, gene therapy (GT) is “the introduction, removal, or change in the content of a person's genetic code with the goal of treating or curing a disease".1 This definition includes the standard GT approaches based on gene addition or supplementation, but also gene editing strategies to modify, to repair or to introduce DNA sequences into the cellular genome, as well as procedures that involve gene silencing using RNA interference or targeted nucleases (Fig. 1).1
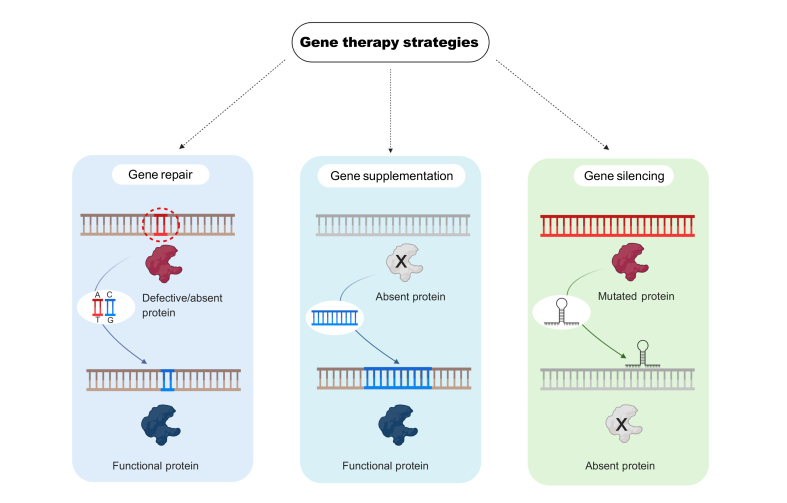
Fig. 1.
Gene therapy strategies currently being used for the treatment of liver diseases can be divided into 3 main categories.
Gene repair involves the correction of an existing mutation to restore the expression of the correct version of the protein. Gene supplementation (or addition) is simply the delivery of the correct version of the gene expressing the missing or defective protein. Gene silencing requires the elimination or reduction of the expression of a protein to reduce toxic metabolites.
GT began in the 1990s as a promising therapeutic alternative for many genetic disorders, including inherited monogenic disorders.[2], [3], [4] However, GT suffered major setbacks, including the unfortunate death of a young patient in a clinical trial evaluating the therapeutic efficacy of an adenoviral vector to treat ornithine transcarbamylase (OTC) deficiency and the development of leukaemia in some patients with severe combined immunodeficiency disorder treated with haematopoietic stem cells modified using retroviral vectors.4,5 Since then, the field has gone through a period of intense research on vector development, new GT therapeutic modalities and preclinical safety studies that have ultimately led to successful clinical applications. In the past 5 years several GT products with very different characteristics have been approved: antisense oligonucleotides (ASOs: Mipomersen, Nusinersen, Etaplirsen, Golodirsen), RNA interference (Patisiran, Givosiran, Lumasiran), lentiviral-transduced cells (Autologous CD34+ cells transduced with a lentiviral vector containing the human ADA gene, Axicabtagene Ciloleucel, Tisagenlecleucel, brexucabtagene autoleucel) or recombinant adeno-associated viral (AAV) vectors (Alipogene tiparvovec, voretigene neparvovec-rzyl, Onasemnogene abeparvovec).[6], [7], [8] The most recent progress in GT for hepatic indications will be reviewed here.
GT can be divided into 2 modalities according to the administration procedure: ex vivo and in vivo. In ex vivo GT, the patient's cells are extracted, cultured, amplified, and modified with the vector of choice prior to being reinfused back into the patient. In in vivo GT, the therapeutic agent is administered directly to the patient using different routes of administration that may be systemic or local (Fig. 2). In vivo GT is more applicable to the treatment of hepatic disorders due to the limited ability to manipulate and expand hepatocytes ex vivo.
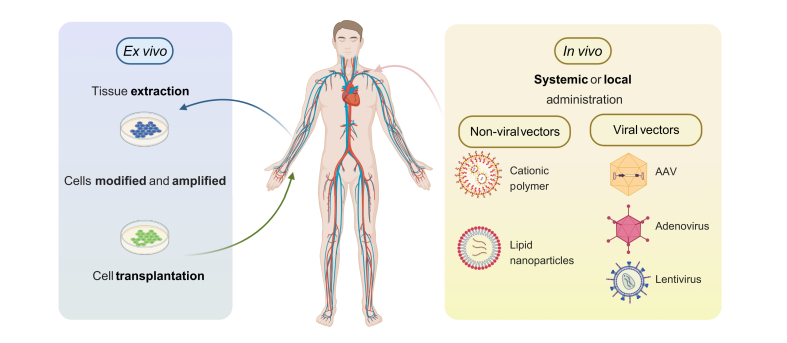
Fig. 2.
Gene therapy strategies.
Ex vivo gene therapy starts with an extraction of cells that are transduced with the vector carrying the therapeutic gene and then reintroduced into the body. In vivo gene therapy is based on the direct administration of the gene delivery vector or genetic material to the organism and can utilise different types of vectors, including non-viral and viral vectors. AAV, adeno-associated virus.
The liver is a critical organ for most metabolic pathways and thus the source of many inherited metabolic liver diseases (IMLDs). A common feature of all these disorders is a deficiency in the synthesis or functionality of proteins involved in biochemical pathways essential for metabolism. Conventional enzyme replacement therapy (ERT) is only available for a limited number of IMLDs and the only curative treatment option is orthotopic liver transplantation (OLT), which is limited by organ donor availability and involves lifelong immunosuppression. The fact that OLT can be curative supports the hypothesis that restoration of the expression of a defective protein in the liver can lead to a resolution of the disease. Thus, liver-directed GT and gene editing strategies have emerged as promising alternatives to OLT for IMLDs.[9], [10], [11] Furthermore, substrate reduction therapy (SRT) or metabolic pathway reprograming (MPR) are attractive strategies for several metabolic disorders associated with the accumulation of toxic metabolic products.12,13 Taking advantage of small-interfering RNA (siRNA) technology or target-specific gene editing with the goal of inhibiting key metabolic enzymes, new molecular-based approaches such as genetically based next-generation SRT or MPR are under development.12,13
Importantly, the liver is a protein factory, secreting the majority of the most abundant proteins present in the circulation. Because of this property, the liver represents a potential bioreactor for the production of recombinant proteins.14 This is the case in the GT-based treatment of inherited bleeding disorders like haemophilia A (HemA) or B (HemB), but also ERT-treatable lysosomal storage diseases.15,16
Owing to the large size and negative charge of genetic material, carrier vehicles are required to achieve delivery into the interior of target cells. There are 2 large families of vectors depending on their origin: viral and non-viral. In this review, we profile the latest advances in the development of approaches for liver-directed GT and some of their clinical applications.
Non-viral vectors
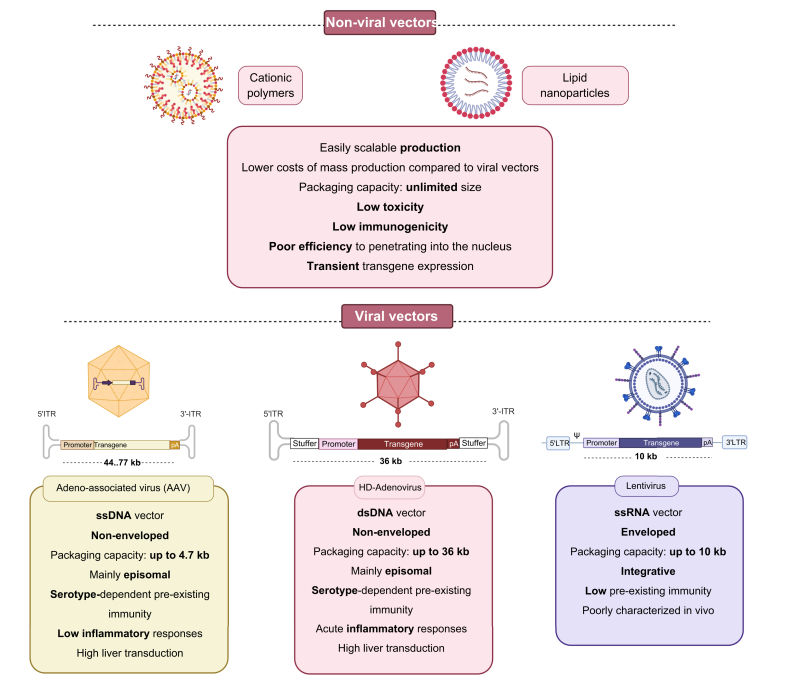
Most non-viral vectors are composed of polymeric or lipid particles that package and protect genetic material in their interior to facilitate entry into the cell. The use of non-viral vectors offers several advantages over the use of viral vectors, such as their easily scalable production, a long shelf life, a theoretically unlimited size of the genetic material payload, and a better safety profile. The limitations of non-viral vectors are their poor efficiency at penetrating into the nucleus and their limited ability to achieve long-lasting transgene expression (Fig. 3).
Fig. 3.
Non-viral and viral vectors used most frequently in liver-directed gene therapy.
Each possesses varying characteristics, benefits, and limitations, which are essential for their selection in a wide variety of gene therapy applications.
Administration of naked genetic material
The simplest form of non-viral vector administration is via naked genetic material. However, this strategy is impaired by very low efficacy cell entry. In the case of hepatocytes, several methods can be used to improve this low efficacy of transfer. One of the most straightforward methods is hydrodynamic injection, which is based on the rapid injection of a relatively high volume of a solution that induces very high venous pressure in the liver and facilitates the entry of genetic material into the hepatocytes.17,18 This technique is commonly used for proof-of-concept studies in mice.19 Although hydrodynamic injections are difficult to translate to humans, intravascular hydrodynamic procedures with partial catheterisation have shown some success in large animals, such as pigs, sheep, and non-human primates (NHPs).19,20
The introduction of naked DNA into the cell can be enhanced by several other technological methods, such as electroporation or ultrasound. The use of DNA by electroporation in the liver of rats was shown for the first time in 1996.21 Recently, liver electroporation has been used for the expression of α1-antitrypsin (AAT) in AAT-deficient mice, resulting in a reduction of pulmonary emphysema.22 Transcutaneous ultrasound has been proven to be efficient for the delivery of genetic material to the liver of mice and pigs.23,24 So far, the use of these approaches in liver-directed GT is still restricted to experimental animal studies.
However, one of the main limitations of the administration of naked DNA, as with other approaches based on episomal delivery, is the rapid loss of transgene expression due to the loss of the genetic material in dividing cells or epigenetic silencing. To overcome this limitation, different strategies have been developed such as the introduction of stabilisation sequences, the use of transposons and more recently the use of gene editing endonucleases (Fig. 4). These strategies have also been used in combination with several other delivery methods explained below.
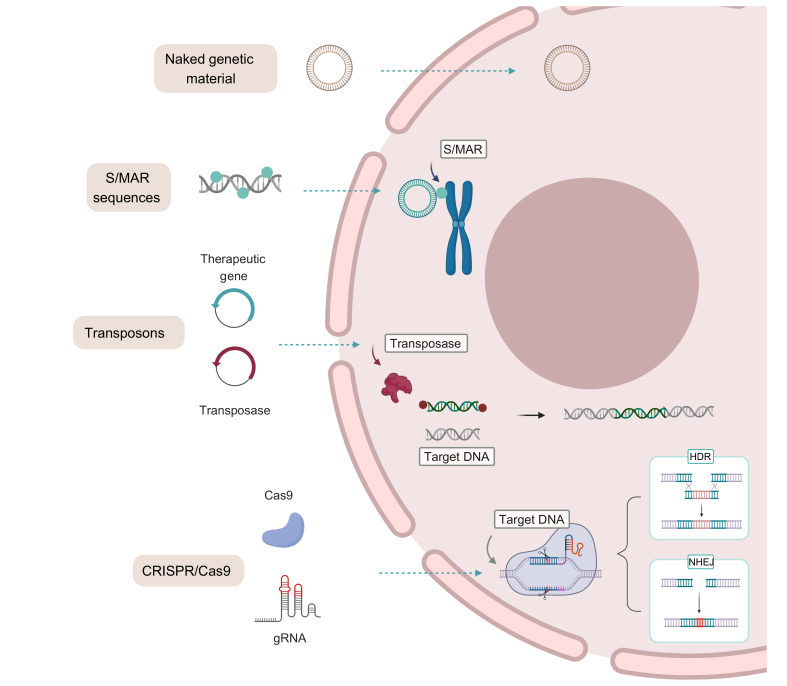
Fig. 4.
Examples of several strategies that have advanced the field of gene therapy.
Naked plasmid DNA, plasmid DNA containing S/MAR sequences, transposons, and gene editing nucleases (e.g. CRISPR/Cas9). dsDNA, double-stranded DNA; S/MAR, scaffold matrix attachment regions; ssDNA, single-stranded DNA; ssRNA, single-stranded RNA.
The use of stabilisation sequences is a potential workaround that has been explored. Scaffold matrix attachment regions (S/MAR) are genomic DNA sequences that bind chromatin to the nuclear matrix during interphase and are involved in DNA replication and transcription.25 DNA vectors containing a S/MAR sequence can provide persistent mitotic stability in dividing cells and avoid epigenetic silencing allowing for sustained transgene expression. However, despite the initial success of this strategy to achieve sustained transgene expression in the liver of mice and pigs,14,26 S/MAR has not yet reached clinical trials.
For permanent modifications to the cellular genome, 2 different strategies have been developed: transposons and gene editing nucleases. Discovered by Barbara McClintock in the 1940s,27 transposons are DNA sequences that jump from one location in the genome to another. Transposon sequences are flanked by terminal inverted repeats (TIRs) that are recognised and cleaved by a transposase enzyme allowing their reinsertion into other locations. Thus, transposon systems have been modified to integrate therapeutic sequences into the host genome.28 Currently, the most promising transposons for GT applications are derived from Sleeping Beauty (SB) or piggyBac (PB) systems.[29], [30], [31], [32] The SB transposon system has been shown to be efficient in treating murine models of HemA, HemB, AAT deficiency, β-glucuronidase (GUSB) deficiency (mucopolysaccharidosis (MPS) type VII, MPSVII), α-L-iduronidase (IDUA) deficiency (MPS type I, MPSI), and hereditary tyrosinemia (HT).29,30 Recently, the delivery of SB transposons by hydrodynamic injection into the liver of dogs resulted in high levels of IDUA and GUSB activity in the circulation. PB transposon-mediated delivery of factor VIII- or factor IX-encoding complementary DNAs in murine models of HemA and HemB resulted in the stable expression of circulating coagulation factors.31,32 Although of therapeutic interest, off-target integration of transposons should be carefully evaluated due to the potential risk of insertional mutagenesis.
Gene editing tools have greatly evolved in recent decades, creating new opportunities for the treatment of many genetic disorders. Three major platforms hold the most promise: zinc finger nucleases (ZFN), transcription activator-like effector nucleases (TALEN) and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (CRISPR/Cas9).33 Two clinical trials are currently ongoing that use ZFN gene editing for the treatment of MPSI and MPSII based on the integration of the therapeutic gene into the albumin locus of hepatocytes.34,35 However, due to its simplicity, high efficiency, and easy customisation, CRISPR/Cas9 is the gene editing tool of choice nowadays.[36], [37], [38], [39] Several clinical trials using CRISPR/Cas9 are ongoing or about to be initiated but most of them are based on ex vivo editing for the treatment of cancer, and a very small number target genetic diseases. Nevertheless, at the preclinical stage, the use of CRISPR/Cas9 targeting the liver is being applied to HemA and HemB via targeted therapeutic gene integration, and to glycogen storage disorder 1a (GSD1a) and AAT deficiency by precise correction of gene mutations.38 Gene editing can also be used as a genetic MPR or SRT strategy, as has been shown for the treatment of familiar hypercholesterolemia (FH), primary hyperoxaluria type 1 (PH1), and tyrosinemia via the elimination of proprotein convertase subtilisin/kexin type 9 (PCSK9), glycolate oxidase (GO)/lactate dehydrogenase (LDH) expression, and hydroxyphenylpyruvate dioxygenase (HPD), respectively.[39], [40], [41]
Chemically modified siRNA and antisense oligonucleotides
As we have described, some IMLDs lead to the accumulation of toxic metabolites. Patients affected by these disorders can benefit from SRT or MPR, for which siRNA or ASOs can serve as tools. Conjugates of N-acetyl-D-galactosamine (GalNAc) and siRNAs have been used for in vivo targeting of hepatocytes thanks to the specific binding to the asialoglycoprotein receptor (ASGPR).42,43 One of the most advanced therapies is the use of GalNac-siRNA targeting aminolevulic synthase 1 (ALAS1) mRNA for the treatment of acute hepatic porphyrias (AHPs).44 ALAS1 is the first enzyme in the heme synthesis pathway and its inhibition reduces the accumulation of toxic metabolites and the number of acute porphyria attacks. This same strategy has been used for the treatment of patients with PH1, reducing oxalate levels in urine by 75%. The products named Givosiran and Lumasiran have recently been approved by the FDA.44,45 GalNAc-modified siRNAs are also being developed for the treatment of acquired or hereditary hyperlipidaemias, chronic HBV infection, non-alcoholic steatohepatitis (NASH), GSD1a, and hereditary haemochromatosis.42,43
A chemically modified ASO has already been approved by the FDA for the treatment of homozygous FH (Mipomersen). Mipormersen targets apolipoprotein B (APOB) mRNA and reduces circulating cholesterol levels.46 GalNAc-modified ASOs are under development for the treatment of NASH by targeting the expression of serine/threonine protein kinase 25 (STK25) or fat-specific protein 27 (FSP27). They are also being developed for the inhibition of HBV replication.47 Interestingly, ASOs have been shown to be very promising tools for the modulation of the expression of long non-coding RNAs (lncRNAs) which have been shown to play important roles in different liver pathologies.48
Polycationic and lipid nanoparticles
Two major classes of non-viral vectors that have shown high efficiency in transferring genetic material to the liver are cationic polymers (polycations) and liposomal formulations.
Cationic polymers form nanoparticles with nucleic acids through electrostatic interactions, enabling the transport of nucleic acids into the cell. One of the most frequently used is polyethylenimine (PEI), which can mediate high transduction efficiencies.49 Furthermore, the use of galactosylated PEI targeting the ASGPR proved to be very efficient in transducing human hepatic cell lines and the livers of mice and rats. PEI nanoparticles and derivatives have been used experimentally for the delivery of different drugs, including siRNA and microRNAs (miRNAs), for the treatment of liver malignancies, but no clinical studies have yet been performed.50
Lipid nanoparticles (LNPs) have very similar composition to cell membranes. They are formed by amphiphilic lipids that when dispersed in an aqueous environment spontaneously form spherical structures with a hydrophilic interior. LNPs are a suitable carrier for nucleic acid delivery because of their excellent biocompatibility, biodegradability, low toxicity and immunogenicity, structural flexibility, and ease of large-scale preparation. The use of LNPs has experienced a significant resurgence in GT as vehicles for siRNA and mRNAs.51
However, the major challenge when developing LNP-based gene therapies is finding effective, tissue-specific delivery strategies.52 Traditionally, targeting is achieved by physically or chemically conjugating ligands for specific receptors onto the nanoparticle surface. A targeting approach was developed by binding a multivalent GalNAc-cluster to the LNP. These nanoparticles have been shown to deliver mRNA molecules to hepatocytes with high efficiency, correcting mouse models of genetic diseases such as methylmalonic acidemia (MMA), PH1, GSD1a, citrin deficiency, acute intermittent porphyria (AIP), maple syrup urine disease, arginase deficiency, OTC deficiency, and progressive familial cholestasis type 3 (PFIC3).[53], [54], [55], [56], [57], [58], [59] The duration of action of a single dose of mRNA-LNP lasted between 2–3 weeks, requiring repeat administration to prolong a curative effect. These encouraging results supported the initiation of a phase I/II trial for the treatment of MMA (Table 1).
Table 1.
Liver-targeted gene therapy clinical trials.
| Indication | Sponsor | Therapeutic agent/route of administration | CT Phase | CT number |
|---|---|---|---|---|
| Haemophilia B | Avigen/CHOP | AAV2-F9WT/Hepatic artery | I/II | 00076557 |
| SJCRH/UCL | AAV8sc-F9WT/iv infusion | I | 00979238 | |
| uniQure | AAV5-coF9WT/iv infusion | I/II | 02396342 | |
| Spark Therapeutics | AAV-SPARK100-F9Padua/iv infusion | I/II | 0284096 | |
| Shire Pharmaceuticals | AAV8sc-F9Padua/iv infusion | I/II | 01687608 | |
| uniQure | AAV5-coF9Padua/iv infusion | IIb | 03489291 | |
| Freeline | AAVS3-coF9Padua/iv infusion | I/II | 03369444 | |
| Dimension Therapeutics | AAVrh10-coF9WT/iv infusion | I/II | 02618915 | |
| Haemophilia A | Biomarin Pharmaceutical | AAV5-coBDDF8/iv infusion | I/II | 02576795 |
| Spark Therapeutics | AAVLK03-coBDDF8/iv infusion | I/II | 03003533 | |
| Pfizer | AAV6-coBDDF8/iv infusion | I/II | 03061201 | |
| UCL/SJCRH | AAV8-coF8/iv infusion | I | 030018130 | |
| Bayern | AAVhu37-coBDDF8/iv infusion | I/II | 03588299 | |
| Baxalta/Shire | AAV8-coBDDF8/iv infusion | I | 03370172 | |
| OTC deficiency | University of Pennsylvania | Adeno-OTC/Hepatic artery | I | NCT00004498 |
| NICHD | Adeno-OTC/Intrahepatic injection | I | NCT00004386 | |
| Ultragenix | AAV8-OTC (DTX301)/iv infusion | I/II | NCT02991144 | |
| Phenylketonuria | Homology Medicines | AAVHSC15-PAH (HMI-102)/iv infusion | I/II | NCT03952156 |
| Biomarin Pharmaceutical | AAV5-PAH (BMN 307)/iv infusion | I/II | NCT04480567 | |
| Acute intermittent porphyria | DIGNA/Uniqure | AAV5 PBGD/iv infusion | I | NCT02082860 |
| Alnylam Pharmaceuticals | siRNA ALAS1 (Givosiran)/subcutaneous | I/II | NCT02452372 | |
| Methylmalonic acidemia | MODERNA | NLP-RNA MMA (mRNA3927)/iv | I/II | NCT03810690 |
| LogicBio Therapeutics | AAVLK03-MMA integrative/iv infusion | I/II | NCT04581785 | |
| Familial hypercholesterolemia | NCRR/UPENN | autologous hepatocytes/retrovirus LDL iv | I/II | NCT00004809 |
| Alnylam Pharmaceuticals | siRNA-PCSSC/subcutaneous | I/II | NCT02314442 | |
| REGENX BIO/UPENN | AAV8-hLDLR/iv infusion | I/II | NCT02651675 | |
| Fabry | Freeline Therapeutics | AAV3S-αGLA/iv infusion | I/II | NCT04040049 |
| Sangamo therapeutics | AAV6-αGLA/iv infusion | I/II | NCT04046224 | |
| 4D Molecular Therapeutics | AAV4D-C102-αGLA/iv infusion | I/II | NCT04519749 | |
| Sangamo Therapeutics | AAV6-ZFN IDUA/iv infusion | I/II | NCT02702115 | |
| Sangamo Therapeutics | AAV6-ZFN IDS/iv infusion | I/II | NCT03041324 | |
| MPS type VI | Telethon Ins | AAV8-hARSB/iv infusion | I/II | NCT03173521 |
| Audentes Therapeutics | AAV8-hGAA/iv infusion | I/II | NCT04174105 | |
| Spark Therapeutics | AAVx-hGAA/iv infusion | I/II | NCT04093349 | |
| Asklepios Biopharceuticals | AAV8-hGAA/iv infusion | I/II | NCT03533673 | |
| Gangliosidosis GM1 | Lysogene | AAVrh10-GLB1/iv infusion | I/II | NCT04273269 |
| NHGRI | AAV9-GLB1/iv infusion | I/II | NCT03952637 | |
| Danon disease | Rocket Pharmaceuticals | AAV9-LAMP2B/iv infusion | I/II | NCT03882437 |
| GSD1a Von Gierke |
Ultragenix Pharmaceuticals | AAV8-G6PC/iv infusion | I/II | NCT03517085 |
| Wilson’s disease | Vivet Therapeutics | AAVx-ATP7B/iv infusion | I/II | NCT03520751 |
| Crigler-Najjar | Genethon-Selecta Bio | AAV8-UGT1a/iv infusion | I/II | NCT03466463 |
| Audentes | AAV8-UGT1a/iv infusion | I/II | NCT03223194 | |
| Primary hyperoxaluria type 1 | Alnylam Pharmaceuticals | siRNA HAO (Lumasiran)/subcutaneous | III | NCT04125472 |
AAV, adeno-associated virus; CHOP, Children's Hospital of Philadelphia; NHGRI, National Human Genome Research Institute; NICHD, National Institute of Child Health and Human Development; SJCRH, St. Jude Children's Research Hospital; UCL, University College London; UPENN, University of Pennsylvania.
Source: Clinicaltrials.gov. January 2021. (x: nondisclosed information).
An interesting case of liver targeting using LNPs is Onpattro, an LNP composed of an ionizable cationic lipid, a phospholipid, cholesterol, and a polyethylene glycol (PEG)-lipid carrying an siRNA targeting transthyretin (TTR), for the treatment of TTR-mediated amyloidosis.60 This LNP is coated by the host apolipoprotein E (ApoE) that triggers its transport into hepatocytes via low-density lipoprotein receptor (LDLR)-mediated endocytosis. The treatment suppresses the deposition of amyloid fibrils and was approved as the first siRNA drug (patisiran) by the FDA in 2018 for the treatment of TTR-type familial amyloid polyneuropathy.61 A similar strategy is under clinical development for the treatment of elevated LDL-cholesterol and hypercholesterolemia using siRNA-LNPs targeting PCSK9 and ApoB, respectively.62 siRNA-containing LNPs are also being explored for the treatment of hepatocellular carcinoma (HCC) and chronic viral hepatitis infections. siRNAs targeting the oncogene MYC and polo-like kinase-1 (PLK1) have been tested in phase I/II clinical trials and though well tolerated have achieved limited antitumor effects.63 Recently, a phase II clinical trial has been completed based on the inhibition of HBV replication using LNPs containing 3 chemically modified siRNAs that inhibit HBV antigen expression and replication in preclinical HBV infection models.64
Additionally, LNPs are now being used for the delivery of gene editing molecules such as ZFNs and Cas9 mRNA together with a single guide (sg)RNA. mRNA encoding ZFNs formulated into LNP have enabled >90% knockout of gene expression in mice by targeting the TTR or PCSK9 gene for the treatment of TTR amyloidosis and FH, respectively.65 More importantly, LNPs have been used for the delivery of ZFNs together with a viral vector carrying a promoterless DNA sequence capable of homologous recombination that can result in high levels of the integrated sequence.34,35 This strategy is being explored, as previously mentioned, for the treatment of MPS I and II (Table 1).
Viral vectors
To generate a viral vector for GT, the viral genes necessary for replication and those that cause pathogenicity are normally removed from the viral genome and replaced by the genetic sequence to be delivered. Maintaining the replicative capability of the virus is advantageous for some applications, such as for oncolytic or tumour cell killing viruses. Viral vectors that have been used for liver-targeted GT mainly include vectors based on AAVs, adenoviruses (Ads), retroviruses (RVs) and lentiviruses (LVs), as different vectors serve for different applications (Fig. 3).[9], [10], [11]
Vectors based on AAVs
The AAV genome is a single-stranded DNA of 4.7 kb that contains 2 genes, rep (encoding the proteins involved in replication) and cap (encoding the viral capsid). Flanking the ends of the genome are inverted terminal repeats (ITRs) folded in a fork-shaped secondary structure that contain the replication initiation and termination sequences and the packaging signal. AAVs have some characteristics that make them ideal vectors for GT: replication deficiency, no human pathology associated with infection, and the requirement of co-infection with another virus such as adenovirus to complete their life cycle. In the absence of co-infection by another virus, the AAV genome is integrated into chromosome 19, in the so-called AAV locus, where it remains dormant.66
To produce recombinant AAV (rAAV) vectors, the rep and cap genes are removed from the viral genome and replaced with therapeutic sequence with only the ITRs maintained.66,67 During the production process of rAAV, both rep and cap genes are provided in trans. The absence of rep results in the virus losing its ability to integrate, and once inside the nucleus the viral genome remains mostly episomal, which reduces the risk of insertional mutagenesis. Furthermore, using different cap genes, either naturally occurring or artificially designed, can result in AAV capsids with diverse properties in terms of their seroreactivity (serotypes) and tropism for different tissue or cell types. Of particular importance, in animal models most AAVs barely induce any immune response, which allows the infection to go unnoticed and the transduced cells to persevere unscathed, maintaining a durable expression of the therapeutic transgene. rAAVs can efficiently transduce quiescent cells such as hepatocytes.68 However, in patients the administration of rAAV has been shown to activate T cell immune responses against the capsid protein that require the use of steroids to avoid the elimination of transduced hepatocytes.69
AAV vectors of nearly all serotypes efficiently accumulate in the liver following intravenous administration due to their natural hepatic tropism.68 However, the extent of vector particle accumulation does not necessarily correlate with the level of transgene expression.70 Moreover, hepatotropism of each serotype differs depending on the species and the transduction efficacy observed in mouse models does not always translate to other species. In fact, studies performed in NHP and mice with humanised livers have revealed important differences in the transduction efficiency of different AAV serotypes between human/NHP and murine hepatocytes.71 To identify new capsids with increased capacity to transduce the human liver, different strategies to engineer the viral capsid have been explored. The simplest involves attaching ligands known to bind to specific receptors present on the surface of hepatocytes to the viral capsid.71,72 Chemical coupling of GalNAc ligands onto lysine residues present on the surface of the AAV capsid has been shown to enhance AAV binding to hepatocytes.72
AAV capsid variants can also be generated through capsid libraries containing random modifications that are subjected to high throughput in vitro or in vivo selection screens.73,74 For this method, capsid libraries can be generated by the insertion of random peptides in specific domains of the capsid or by capsid shuffling. Using “humanised” mouse models is the preferred choice for optimising AAV vectors for human liver targeting, albeit without the ability to study the human host responses to the AAV. This screening system led to the creation of AAV-LK03,75 a variant with improved in vivo tropism for human hepatocytes, which is currently being tested in a clinical trial for HemA (Table 1). The AAV-LK03 capsid protein has 98.9% sequence identity with the naturally occurring AAV3B, which was initially discarded for further development due to the low transduction efficiency in mice.76 Recently, several groups have been working on developing a new AAV capsid based on the AAV3B serotype.[77], [78], [79] One result of this endeavour has been the identification of AAVS3, a serotype now in phase I/II clinical trials for HemB and Fabry disease.
Because of their many advantages, AAV vectors have emerged as the leading candidates for liver-targeted GT. The clear front runners in the clinical application of AAV gene delivery are therapies for HemA and HemB, in which the liver is used as a factory to produce the missing coagulation factor. Since the first clinical trial using a AAV serotype 2 vector to express coagulation factor IX for HemB, several trials have been performed on different naturally occurring or synthetic serotypes, including AAV5, AAV8, AAVrh10, AAVS3, AAV-Spark100, and AAV-Spark200 for HemA and HemB, with varying results (Table 1).[79], [80], [81], [82], [83], [84] While sustained protein expression was achieved in some, in others clinical development was discontinued due to safety issues or insufficient efficacy, highlighting the importance of a proper selection of the serotype and expression cassettes. Anyway, these studies showed that long-term liver transduction using AAV is possible and have paved the way for the use of rAAVs in the treatment of IMLD.[9], [10], [11],66,67
The first IMLD experimentally treated with an AAV vector was FH via infusion of an AAV vector, carrying the very low-density lipoprotein receptor (VLDLR) gene, into the portal circulation of FH mice, which resulted in a 40% reduction in serum cholesterol and triglyceride. Importantly, the reduction was stable for the duration of the study. Since then, the number of studies reporting therapeutic efficacy in IMLD animal models has increased exponentially, with proof-of-concept studies for AIP, PH1, Wilson’s disease (WD), MMA, propionic acidemia (PA), PKU, HT1, PFIC3, Crigler-Najjar (CN), GSD1a, and urea cycle disorders (UCDs). Albeit using different AAV serotypes, promoters and delivery routes, in general, all studies showed long-term therapeutic efficacy in the absence of major safety issues. [9], [10], [11],66,67
The first clinical trial using AAV for an IMLD was for the treatment of AIP. Phase I data showed an excellent safety profile, but only moderate therapeutic effect.86 Nowadays, different clinical trials for the treatment of IMLD are ongoing or about to start, including for PKU, OTC deficiency, CN, FH, Fabry, MPS type VI, Pompe, GSD1a, and WD (Table 1).
One of the limitations of AAV vectors is related to their episomal nature, which – while reducing their mutagenic potential – results in the loss of the vector genomes and concomitant loss of the therapeutic effect when administered to a growing or regenerating liver.87 This is of particular importance for the treatment of the paediatric population. In order to avoid the disappearance of therapeutic vector genomes, several strategies are under development, such as the use of rAAV genomes containing homology arms for a safe integration in the cellular genome, the selection of AAVs with integration capacity, transposon-carrying AAVs, or the AAV-mediated delivery of gene editing systems.
The simplest strategy has been AAVs that induce spontaneous targeted integration in the albumin locus of a promoterless sequence carrying the therapeutic gene flanked by albumin homology arms.88,89 Using this approach, the therapeutic gene is placed upstream of the albumin stop codon resulting in the production of an mRNA that is translated into a chimera of albumin and the therapeutic protein. This strategy has been proven to be efficient in animal models of haemophilia, CN, AAT deficiency and MMA,[90], [91], [92] as well as reaching clinical trials to treat patients with MMA. However, only ~1% of the hepatocytes are initially edited, thus this strategy is valid only when low levels of transduced hepatocytes are sufficient or when the edited hepatocytes have a selective advantage and can repopulate the liver. Unfortunately, this is not the case for the majority of IMLDs. This low efficiency of recombination has been improved by co-administration of an AAV-carrying specific CRISPR-Cas9 which introduces double-strand breaks in the albumin locus.92
Alternatively, a group of new AAVs isolated from human CD34+ haematopoietic stem cells (named AAVHSCs) have recently been described; they are highly efficient at mediating homologous recombination in the absence of any associated nuclease. It has been shown that AAVHSC-mediated gene insertion can correct the disease phenotype in a PKU mouse model.93 A phase I/II clinical trial has recently been initiated using AAVHSC15 in patients with late-onset PKU (Table 1).
The use of AAV vectors to deliver gene editing nucleases and donor templates for gene correction by homology-directed repair (HDR) is a field of intense interest.94 HDR-based correction of point mutations has been achieved in a humanised mouse model of OTC deficiency.95 AAV-mediated gene editing can also be utilised to reduce the expression of specific proteins, e.g. in MPR or SRT strategies. This approach has been successfully applied in animal models of FH and PH1 by suppressing the expression of PCSK9 and HO-1 proteins, respectively.40,96 More recently, AAVs carrying sequences for transposon systems have been explored as a strategy for the permanent correction of the transduced cell. The use of the AAV-PB transposon system has been shown to induce sustained gene expression and therapeutic efficacy in mouse models of PFIC3 and UCDs.97,98 The AAV-PB system has also been used successfully for insulin gene delivery in a mouse model of diabetes, resulting in normoglycaemia and glucose tolerance.99
Adenoviral vectors
Adenoviruses (Ads) are more complex than AAVs, with a 36 kb linear double-stranded DNA genome encoding various structural and non-structural genes. They are non-enveloped viruses and the serotypes most frequently used are human serotypes 2 and 5. However, in recent years Ads isolated from different species have been used to bypass the pre-existing humoral immune response against these serotypes. In immune-competent individuals, human Ad cause only a mild and self-limited disease. Ad possesses very attractive characteristics for liver gene delivery, such as marked hepatotropism, high levels of gene expression in a wide variety of cells, easy and routine production of high functional titres, large packaging capacity, and low genotoxicity.100 The first Ad-based recombinant viruses lacked replication genes but maintained a large part of the genome, allowing for exogenous genetic material of up to 8.5 kb to be introduced. These so-called first-generation Ad vectors are highly efficient in transducing hepatocytes; however, the manipulation to prevent replication of the virus failed to eliminate the expression of a number of structural proteins that caused an important inflammatory and immune reaction.100 This immune response resulted in the elimination of infected cells in a relatively short time, and therefore only achieved a transient therapeutic effect. First-generation Ad vectors have been shown to be effective for the development of vaccines and antitumoral treatments, including hepatic tumours, but not for the treatment of inherited diseases. In order to achieve long-term expression, third-generation Ad vectors, or helper-dependent adenovirus (HDAd) were developed.101,102 In HDAd, all viral sequences are removed except for the terminal and the packaging signal sequences, allowing delivery of large sequences (up to 36 kb) and providing long-term transgene expression, which has been demonstrated in mice and NHPs. HDAd expressing LDLR for the treatment of FH has been shown to improve lipid profile and reduced aortic atherosclerosis in rodents and NHPs.103 The therapeutic efficacy of HDAd has also been clearly demonstrated in animal models of PH1, CN, AIP, GSD1a, and OTC.[100], [101], [102]
Despite these attractive features, there are still several hurdles arising mainly from the immunogenicity of Ad vectors, which strongly limits their efficient and safe application in clinical trials.104,105 Furthermore, in the case of HDAd vectors, the lack of an appropriate clinical grade production platform remains a major limitation to clinical use.106
To address these obstacles, a variety of strategies to evade host immune responses have been developed that can be classified into 2 broad categories: host treatment with immunosuppressive drugs or modification of the capsid.107 The latter can be achieved in several ways, including chemical and physical modifications of the Ad capsid with polymers, Ad capsid-display of immuno-evasive proteins, and genetic modification of the Ad capsid. Among these various strategies, there has been a great deal of interest in the coating of Ad with non-immunogenic polymers like PEG, which has been shown to significantly enhance liver transduction and reduce the production of inflammatory cytokines and neutralising antibodies (NAbs).107
As with AAV, the Ad genome remains episomal after infecting the cell. To establish a permanent Ad-mediated genetic change, Ad vectors are being used for the delivery of CRISPR/Cas9 editing systems or transposon elements to the liver. A 2-vector system has been explored to deliver CRISPR/Cas9 and a therapeutic DNA sequence targeting integration into a safe harbour locus of the genome for the treatment of HemB or AAT deficiency.108,109 In both cases, high rates of HDR were observed with long-term expression and without apparent long-term damage to the mouse livers. Co-transduction of HDAd with SB transposons in a canine HemB model resulted in the stable expression of factor IX for nearly 1,000 days.110
Retroviral and lentiviral vectors
Retroviruses (RV) and lentiviruses (LV) are enveloped single-stranded RNA viruses that produce their own reverse transcriptase (RT) resulting in a double-stranded DNA provirus that integrates into the cell genome. The viral genome contains the env, gag, and pol genes flanked by long terminal repeats (LTRs), which carry enhancer/promoter elements that are required for integration.11,111 The env and gag genes encode structural viral proteins while pol encodes non-structural proteins including the RT. Lentiviruses have 2 additional genes, tat and rev, that encode regulatory proteins.111
For vector generation, the LTRs are maintained, and the viral genes are eliminated and replaced by the therapeutic sequence, with a capacity of 8–9 kb. RVs are unable to transduce non-dividing cells, which explains why this vector is more often considered for ex vivo GT. LVs, in contrast, are able to transduce dividing and mitotically quiescent primary cells, including differentiated hepatocytes.111
RV and LV vectors are mainly used for the correction of haematological disorders. However, the use of LV-transduced bone marrow cells for the treatment of lysosomal storage disorders, such as adrenoleukodystrophy, has shown spectacular results.112
Early experimental approaches for the genetic treatment of liver diseases using these vectors were based on the transduction of primary hepatocytes and reimplantation after genetic modification into the liver or spleen. Using this strategy, persistent transgene expression was obtained in mice, rats, dogs, and NHPs.[113], [114], [115], [116], [117], [118], [119] Furthermore, long-term disease improvement was achieved in FH, HT, CN and haemophilia animal models, leading to the first human trials.[115], [116], [117], [118] In patients, RV-based GT required ex vivo culturing of hepatocytes obtained from a liver biopsy that are transduced with the recombinant RV vector and subsequently infused directly into the liver. This therapeutic strategy was used in patients with homozygous FH achieving only a mild improvement and only in some of the patients. The limited efficacy was due to a very low percentage of hepatocyte engraftment (Table 1).119 Furthermore, this approach requires the use of invasive procedures to isolate and reinfuse the transduced hepatocytes.
Transduction efficiency has been improved with the use of LV vectors achieving in vitro transduction in close to 90% of cells.120,121 Hepatocyte transplantation, however, remains relatively inefficient and variable, likely due to poor engraftment and limited persistence of engrafted hepatocytes if there is not a proliferative advantage. Recently, liver-directed ex vivo GT using a LV vector to integrate a corrected Fah gene was shown to cure liver disease in a pig model of HT1 thanks to the selective advantage of modified hepatocytes.122
In transitioning from ex vivo approaches to systemic administration of RV and LV vectors for the treatment of liver diseases, initial studies combined the administration of RV vectors with partial hepatectomy, hepatoxic drugs or hepatocyte growth factor, to induce hepatocyte division and facilitate viral integration.[123], [124], [125], [126], [127], [128] Long-term transgene expression was hampered by the induction of adaptive immune responses against the transgene, necessitating immunosuppressive treatment.129,130
The capacity of systemic administration of recombinant LV vectors to cure disease has been shown in mouse models of CN, MMA, and GSD1a.[131], [132], [133] The risk of insertional mutagenesis initially associated with LV has diminished with the development of self-inactivating (SIN) vectors, in which the LTR enhancer/promoter elements have been deleted. The administration of SIN-LV vectors to adult mice and dogs resulted in stable liver expression of factor IX, induction of tolerance against the recombinant protein and no evidence of genotoxicity.134 However, mild acute toxicity and low efficacy were observed due to collateral transduction of resident antigen-presenting cells (APCs) in the liver (Kupffer cells and hepatic stellate cells). Strategies are currently being investigated to prevent vector uptake by APCs in order to improve hepatocyte transduction and reduce systemic toxicity. The incorporation of the human phagocytosis inhibitor CD47 into LV particles resulted in a significant increase in transgenic expression and a reduction in toxicity in monkeys.135 In the coming years, there may be an increase in the use of LVs for the treatment of liver diseases through direct administration, as new LV pseudotypes with high capacity to transduce hepatocytes (but avoid other cell types) are being developed.
Clinical experience with systemic administration of RV and LV for liver diseases is scarce. In 2003, 13 patients with HemA received a recombinant RV expressing coagulation factor VII that was well tolerated, but no significant clinical benefits were observed.136 Two clinical trials are posted in clinical trials.gov for the treatment of HemA and HemB with a LV vector, but they are currently not recruiting (NCT03217032 and NCT03961243).
Relevant aspects to consider, current limitations and possible future directions in liver-targeted gene therapy
Given the increasing number of publications showing the therapeutic efficacy of liver-targeted gene delivery, there is a strong need to focus more effort on overcoming the challenges inherent in translating these approaches to the clinic. Among the main challenges are the innate and adaptive immune responses to the GT product, the potential hepatic toxicity, and the limitations of large-scale and clinical grade vector production.7,10,11,137,138 In the case of viral vectors for liver hepatic delivery, clinical experience with Ad has not as of yet expanded beyond cancer treatment, since its application to IMLD is hampered by inflammatory responses and low production yields, while the use of RV or LV vectors for liver targeting is still scarce; thus, herein, we have mainly focused on experience with AAV vectors.
The administration of GT vectors does not go unnoticed by the immune system. Both non-viral and viral vectors induce the activation of both innate and adaptive immune responses that can jeopardise the therapeutic effect. Moreover, the induction of inflammatory responses by the administration of recombinant vectors to a diseased liver might result in the aggravation of the liver pathology.10,137 Furthermore, sustained transgene expression has been shown to be limited by the activation of the adaptive immune response that results in the elimination of transduced hepatocytes. To prevent the disappearance of the therapeutic genetic material, a recurrent and straightforward option already successfully applied in both preclinical and clinical trials is the administration of immunosuppressive treatments.137,139,140 However, all carry the associated risk of leaving the patient transiently immunocompromised and thus exposed to infection, and depending on the characteristics of the disease, this is a downside to be carefully evaluated. Several alternatives being explored are the use of polymer-coated vectors or the modification of the vector surface to prevent uptake by APCs and reduce the activation of the immune response and increase cell transduction. Additionally, these immune responses can be mitigated by modifying the CpG sequence content of the recombinant genetic material in order to reduce Toll-like receptor 9 pathway activation.141
Furthermore, very recently, concern has been raised about hepatoxicity associated with systemic administration of rAAV due to the death of 3 young patients suffering a fatal neuromuscular disorder that received a high vector dose. The 3 patients were older and heavier than the other treated patients, and more importantly they had evidence of pre-existing hepatobiliary disease. Thus, special attention should be given when treating patients with pre-existing hepatic conditions, which is common for patients with IMLD.142 However, it is noteworthy that clinical trials using AAV for AIP, OTC deficiency, or GSD1a have not reported major adverse events.85
An additional very important immune-related limitation is the presence of pre-existing humoral antibodies and memory T cells against the viral vectors which can completely block liver transduction. A large percentage of the population has NAbs against AAV or Ad vectors.140 In addition, when a patient has been treated with a subtherapeutic vector dose or the therapeutic effect has waned, they cannot be effectively treated again due to the presence of NAbs. This is particularly relevant when the target population is paediatric, since the effectiveness of treatment may be diluted over time due to liver growth. The development of strategies to overcome this problem is the focus of many research groups. Some very simple strategies are the use of alternative serotypes without cross-reactivity, the development of less immunogenic serotypes, or the chemical modification of the capsid. As an alternative, the physical removal of NAbs by plasmapheresis or immunoadsorption have been successfully applied in animal models.143 Recently, an interesting strategy has been reported to eliminate NAbs based on the use of bacterial proteases capable of degrading human IgGs. This strategy, used to prevent antibody-mediated kidney rejection after transplantation, has been shown to allow vector redosing in mice and NHPs.144
An important aspect that should be carefully evaluated is the oncogenic potential of GT. In the case of AAV vectors, after alarming results obtained in some animal models of oncogenic integration and some reports indicating an association of the integration of the WT AAV genome with the development of HCC in humans,145 it is important to acknowledge that the analysis of AAV genome integration in the liver of rAAV-treated patients has shown a safe profile throughout long-term follow-up (12-15 years post-vector administration) showing no evidence of sustained hepatic toxicity or development of HCC.146,147 However, regardless of the vector used for the delivery of the genetic material, all patients should be closely followed to monitor potential oncogenic integration. Furthermore, specific integration in safe harbours might represent a safer strategy that also enables long-term expression.
Conclusion
The liver, due to its central role in metabolism and its role as a protein factory, is the target organ for the treatment of many inherited and acquired metabolic disorders and represents a very attractive platform for the production of circulating therapeutic proteins. Several liver-directed GT approaches using non-viral and viral vectors have been shown to provide long-lasting therapeutic effects in clinically relevant animal models; however, only a fraction of them have reached the clinic (Table 1). Nevertheless, promising results from ongoing liver-targeted clinical trials and the marketing authorisation of a number of GT products encourage optimism about the future of GT for liver diseases.
Although many challenges remain, ongoing efforts and progress will allow many more currently untreatable genetic disorders to be added to the list of treatable and even curable diseases without the need for OLT in the near future.
Financial support
Grant support: RTI2018-101936-B-I00 to G.G-A. from Agencia Estatal de Investigación and fondos FEDER.
Authors’ contributions
SM: performed the systemic review of the literature, drafted the manuscript, and prepared the figures; NDW: performed the systemic review of the literature and drafted the manuscript; NZ: performed the systemic review of the literature, drafted and revised the manuscript; RA: supervised, drafted and revised the manuscript. GGA: supervised and revised the manuscript.
Conflict of interest
NDW and GGA are Vivet Therapeutics employees and hold stock of the company.
Please refer to the accompanying ICMJE disclosure forms for further details.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2021.100300.
Contributor Information
Rafael Aldabe, Email: raldabe@unav.es.
Gloria Gonzalez-Aseguinolaza, Email: ggasegui@unav.es.
Supplementary data
The following is the supplementary data to this article:
References
- 1.American Society of Gene & Cell Therapy . 2019. Gene and cell therapy FAQs.https://www.asgct.org/education/more-resources/gene-and-cell-therapy-faqs [Google Scholar]
- 2.Blaese R.M., Culver K.W., Miller A.D., Carter C.S., Fleisher T., Clerici M. T lymphocyte-directed gene therapy for ADA-SCID: initial trial results after 4 years. Science. 1995;270:475–480. doi: 10.1126/science.270.5235.475. [DOI] [PubMed] [Google Scholar]
- 3.Bordignon C., Notarangelo L.D., Nobili N., Ferrari G., Casorati G., Panina P. Gene therapy in peripheral blood lymphocytes and bone marrow for ADA- immunodeficient patients. Science. 1995;270:470–475. doi: 10.1126/science.270.5235.470. [DOI] [PubMed] [Google Scholar]
- 4.Raper S.E., Chirmule N., Lee F.S., Wivel N.A., Bagg A., Gao G.P. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol Genet Metab. 2003;80:148–158. doi: 10.1016/j.ymgme.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 5.Hacein-Bey-Abina S., von Kalle C., Schmidt M., Le Deist F., Wulffraat N., McIntyre E. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 6.Ginn S.L., Amaya A.K., Alexander I.E., Edelstein M., Abedi M.R. Gene therapy clinical trials worldwide to 2017: an update. J Gene Med. 2018;20 doi: 10.1002/jgm.3015. [DOI] [PubMed] [Google Scholar]
- 7.Shahryari A., Saghaeian Jazi M., Mohammadi S., Razavi Nikoo H., Nazari Z., Hosseini E.S. Development and clinical translation of approved gene therapy products for genetic disorders. Front Genet. 2019;10:868. doi: 10.3389/fgene.2019.00868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bulaklak K., Gersbach C.A. The once and future gene therapy. Nat Commun. 2020;11:5820. doi: 10.1038/s41467-020-19505-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cozmescu A.C., Counsell J., Gissen P. Gene therapies targeting the liver. J Hepatol. 2021;74:235–236. doi: 10.1016/j.jhep.2020.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Baruteau J., Waddington S.N., Alexander I.E., Gissen P. Gene therapy for monogenic liver diseases: clinical successes, current challenges and future prospects. J Inherit Metab Dis. 2017;40:497–517. doi: 10.1007/s10545-017-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zabaleta N., Hommel M., Salas D., Gonzalez-Aseguinolaza G. Genetic-based approaches to inherited metabolic liver diseases. Hum Gene Ther. 2019;30:1190–1203. doi: 10.1089/hum.2019.140. [DOI] [PubMed] [Google Scholar]
- 12.Coutinho M.F., Santos J.I., Matos L., Alves S. Genetic substrate reduction therapy: a promising approach for lysosomal storage disorders. Diseases. 2016;4:33–48. doi: 10.3390/diseases4040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pankowicz F.P., Jarrett K.E., Lagor W.R., Bissig K.D. CRISPR/Cas9: at the cutting edge of hepatology. Gut. 2017;66:1329–1340. doi: 10.1136/gutjnl-2016-313565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quiviger M., Giannakopoulos A., Verhenne S., Marie C., Stavrou E.F., Vanhoorelbeke K. Improved molecular platform for the gene therapy of rare diseases by liver protein secretion. Eur J Med Genet. 2018;61:723–728. doi: 10.1016/j.ejmg.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Arruda V.R., Doshi B.S. Gene therapy for hemophilia: facts and quandaries in the 21st century. Mediterr J Hematol Infect Dis. 2020;12 doi: 10.4084/MJHID.2020.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leal A.F., Espejo-Mojica A.J., Sánchez O.F., Ramírez C.M., Reyes L.H., Cruz J.C. Lysosomal storage diseases: current therapies and future alternatives. J Mol Med. 2020;98:931–946. doi: 10.1007/s00109-020-01935-6. [DOI] [PubMed] [Google Scholar]
- 17.Song Y.K., Liu F., Zhang G., Liu D. Hydrodynamics-based transfection: simple and efficient method for introducing and expressing transgenes in animals by intravenous injection of DNA. Methods Enzymol. 2002;346:92–105. doi: 10.1016/s0076-6879(02)46050-8. [DOI] [PubMed] [Google Scholar]
- 18.Zhang G., Budker V., Wolff J.A. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 1999;10:1735–1737. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- 19.Sandra L., Herrero M.J., Aliño S.F. Translational advances of hydrofection by hydrodynamic injection. Genes. 2018;9:136. doi: 10.3390/genes9030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokoo T., Kamimura K., Abe H., Kobayashi Y., Kanefuji T., Ogawa K. Liver-targeted hydrodynamic gene therapy: recent advances in the technique. World J Gastroenterol. 2016;22:8862–8868. doi: 10.3748/wjg.v22.i40.8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heller R., Jaroszeski M., Atkin A., Moradpour D., Gilbert R., Wands J. In vivo gene electroinjection and expression in rat liver. FEBS Lett. 1996;389:225–228. doi: 10.1016/0014-5793(96)00590-x. [DOI] [PubMed] [Google Scholar]
- 22.Sutter M.A., Cremona T.P., Nita I., Cavarra E., Lungarella G., Lewis E.C. In vivo electroporation-mediated, intrahepatic Alpha1 antitrypsin gene transfer reduces pulmonary emphysema in pallid mice. Pharmaceutics. 2020;12:793–807. doi: 10.3390/pharmaceutics12090793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Endo-Takahashi Y., Negishi Y. Microbubbles and nanobubbles with ultrasound for systemic gene delivery. Pharmaceutics. 2020;12:964–978. doi: 10.3390/pharmaceutics12100964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noble-Vranish M.L., Song S., Morrison K.P., Tran D.M., Sun R.R., Loeb K.R. Ultrasound-mediated gene therapy in swine livers using single-element, multi-lensed, high-intensity ultrasound transducers. Mol Ther Methods Clin Dev. 2018;10:179–188. doi: 10.1016/j.omtm.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heng H.H.Q. Chromatin loops are selectively anchored using scaffold/matrix-attachment regions. J Cell Sci. 2004;117:999–1008. doi: 10.1242/jcs.00976. [DOI] [PubMed] [Google Scholar]
- 26.Giannakopoulos A., Quiviger M., Stavrou E., Verras M., Marie C., Scherman D. Efficient episomal gene transfer to human hepatic cells using the pFAR4-S/MAR vector. Mol Biol Rep. 2019;46:3203–3211. doi: 10.1007/s11033-019-04777-9. [DOI] [PubMed] [Google Scholar]
- 27.McClintock B. The stability of broken ends of chromosomes in zea mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishihara H. Evolution of transposable elements and evolution of eukaryote genomes mediated by transposable elements. Genes Genet Syst. 2019;94:231. doi: 10.1266/ggs.94.231. [DOI] [PubMed] [Google Scholar]
- 29.Podetz-Pedersen K.M., Olson E.R., Somia N.V., Russell S.J., McIvor R.S. A broad range of dose optima achieve high-level, long-term gene expression after hydrodynamic delivery of sleeping beauty transposons using hyperactive SB100x transposase. Mol Ther Nucleic Acids. 2016;5:e279. doi: 10.1038/mtna.2015.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hackett P.B., Jr., Aronovich E.L., Hunter D., Urness M., Bell J.B., Kass S.J. Efficacy and safety of Sleeping Beauty transposon-mediated gene transfer in preclinical animal studies. Curr Gene Ther. 2011;11:341–349. doi: 10.2174/156652311797415827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakanishi H., Higuchi Y., Kawakami S., Yamashita F., Hashida M. piggyBack transposon-mediated long-term gene expression in mice. Mol Ther. 2010;18:707–714. doi: 10.1038/mt.2009.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Matteo M., Samara-Kuko E., Ward N.J., Waddington S.N., McVey J.H., Chuah M.K. Hyperactive piggyBac transposons for sustained and robust liver-targeted gene therapy. Mol Ther. 2014;22:1614–1624. doi: 10.1038/mt.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruiz de Galarreta M., Lujambio A. Therapeutic editing of hepatocyte genome in vivo. J Hepatol. 2017;67:818–828. doi: 10.1016/j.jhep.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 34.Laoharawee K., DeKelver R.C., Podetz-Pedersen K.M., Rohde M., Sproul S., Nguyen H.O. Dose-dependent prevention of metabolic and neurologic disease in murine MPS II by ZFN-mediated in vivo genome editing. Mol Ther. 2018;26:1127–1136. doi: 10.1016/j.ymthe.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ou L., DeKelver R.C., Rohde M., Tom S., Radeke R., St Martin S.J. ZFN-mediated in vivo genome editing corrects murine hurler syndrome. Mol Ther. 2019;27:178–187. doi: 10.1016/j.ymthe.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alves-Bezerra M., Furey N., Johnson C.G., Bissig K.D. Using CRISPR/Cas9 to model human liver disease. JHEP Rep. 2019;1:392–402. doi: 10.1016/j.jhepr.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uddin F., Rudin C.M., Sen T. CRISPR gene therapy: applications, limitations, and implications for the future. Front Oncol. 2020;10:1387–1404. doi: 10.3389/fonc.2020.01387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu T., Li L., Liu Y.C., Cao W., Chen J.S., Hu S. CRISPR/Cas9-related technologies in liver diseases: from feasibility to future diversity. Int J Biol Sci. 2020;16:2283–2295. doi: 10.7150/ijbs.33481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gong J., Wang H.X., Lao Y.H., Hu H., Vatan N., Guo J. A versatile nonviral delivery system for multiplex gene-editing in the liver. Adv Mater. 2020;32 doi: 10.1002/adma.202003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zabaleta N., Barberia M., Martin-Higueras C., Zapata-Linares N., Betancor I., Rodriguez S. CRISPR/Cas9-mediated glycolate oxidase disruption is an efficacious and safe treatment for primary hyperoxaluria type I. Nat Commun. 2018;9:5454–5463. doi: 10.1038/s41467-018-07827-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pankowicz F.P., Barzi M., Legras X., Hubert L., Mi T., Tomolonis J.A. Reprogramming metabolic pathways in vivo with CRISPR/Cas9 genome editing to treat hereditary tyrosinaemia. Nat Commun. 2016;7:12642. doi: 10.1038/ncomms12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang Y. Preclinical and clinical advances of GalNAc-decorated nucleic acid therapeutics. Mol Ther Nucleic Acids. 2017;6:116–132. doi: 10.1016/j.omtn.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Debacker A.J., Voutila J., Catley M., Blakey D., Habib N. Delivery of oligonucleotides to the liver with GalNAc: from research to registered therapeutic drug. Mol Ther. 2020;28:1759–1771. doi: 10.1016/j.ymthe.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balwani M., Sardh E., Ventura P., Peiró P.A., Rees D.C., Stölzel U. Phase 3 trial of RNAi therapeutic givosiran for acute intermittent porphyria. N Engl J Med. 2020;382:2289–2301. doi: 10.1056/NEJMoa1913147. [DOI] [PubMed] [Google Scholar]
- 45.Liebow A., Li X., Racie T., Hettinger J., Bettencourt B.R., Najafian N. An investigational RNAi therapeutic targeting glycolate oxidase reduces oxalate production in models of primary hyperoxaluria. J Am Soc Nephrol. 2017;28:494–503. doi: 10.1681/ASN.2016030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levin A.A. Treating disease at the RNA level with oligonucleotides. N Engl J Med. 2019;380:57–70. doi: 10.1056/NEJMra1705346. [DOI] [PubMed] [Google Scholar]
- 47.Alkhouri N., Reddy G.K., Lawitz E. Oligonucleotide-based therapeutics: an emerging strategy for the treatment of chronic liver diseases. Hepatology. 2020 doi: 10.1002/hep.31569. [DOI] [PubMed] [Google Scholar]
- 48.Mahpour A., Mullen A.C. Our emerging understanding of the roles of long non-coding RNAs in normal liver function, disease, and malignancy. JHEP Rep. 2020;3:100177. doi: 10.1016/j.jhepr.2020.100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lungwitz U., Breunig M., Blunk T., Göpferich A. Polyethylenimine-based non-viral gene delivery systems. Eur J Pharm Biopharm. 2005;60:247–266. doi: 10.1016/j.ejpb.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 50.Xue L., Yan Y., Kos P., Chen X., Siegwart D.J. PEI fluorination reduces toxicity and promotes liver-targeted siRNA delivery. Drug Deliv Transl Res. 2021;11:255–260. doi: 10.1007/s13346-020-00790-9. [DOI] [PubMed] [Google Scholar]
- 51.Witzigmann D., Kulkarni J.A., Leung J., Chen S., Cullis P.R., van der Meel R. Lipid nanoparticle technology for therapeutic gene regulation in the liver. Adv Drug Deliv Rev. 2020;159:344–363. doi: 10.1016/j.addr.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 53.Wei G., Cao J., Huang P., An P., Badlani D., Vaid K. Synthetic human ABCB4 mRNA therapy rescues severe liver disease phenotype in a BALB/c.Abcb4-/- mouse model of PFIC3. J Hepatol. 2020;20 doi: 10.1016/j.jhep.2020.12.010. 33849-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang L., Park J.S., Yin L., Laureano R., Jacquinet E., Yang J. Dual mRNA therapy restores metabolic function in long-term studies in mice with propionic acidemia. Nat Commun. 2020;11:5339. doi: 10.1038/s41467-020-19156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Truong B., Allegri G., Liu X.B., Burke K.E., Zhu X., Cederbaum S.D. Lipid nanoparticle-targeted mRNA therapy as a treatment for the inherited metabolic liver disorder arginase deficiency. Proc Natl Acad Sci U S A. 2019;116:21150–21159. doi: 10.1073/pnas.1906182116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao J., An D., Galduroz M., Zhuo J., Liang S., Eybye M. mRNA therapy improves metabolic and behavioral abnormalities in a murine model of citrin deficiency. Mol Ther. 2019;27(7):1242–1251. doi: 10.1016/j.ymthe.2019.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.An D., Schneller J.L., Frassetto A., Liang S., Zhu X., Park J.S. Systemic messenger RNA therapy as a treatment for methylmalonic acidemia. Cell Rep. 2017;21:3548–3558. doi: 10.1016/j.celrep.2017.11.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang L., Berraondo P., Jericó D., Guey L.T., Sampedro A., Frassetto A. Systemic messenger RNA as an etiological treatment for acute intermittent porphyria. Nat Med. 2018;24:1899–1909. doi: 10.1038/s41591-018-0199-z. [DOI] [PubMed] [Google Scholar]
- 59.Prieve M.G., Harvie P., Monahan S.D., Roy D., Li A.G., Blevins T.L. Targeted mRNA therapy for ornithine transcarbamylase deficiency. Mol Ther. 2018;26:801–813. doi: 10.1016/j.ymthe.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Akinc A., Maier M.A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat Nanotechnol. 2019;14:1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 61.Luigetti M., Servidei S. Patisiran in hereditary transthyretin-mediated amyloidosis. Lancet Neurol. 2021;20:21–23. doi: 10.1016/S1474-4422(20)30397-5. [DOI] [PubMed] [Google Scholar]
- 62.Ray K.K., Wright R.S., Kallend D., Koenig W., Leiter L.A., Raal F.J. Two phase 3 trials of inclisiran in patients with elevated LDL cholesterol. N Engl J Med. 2020;382:1507–1519. doi: 10.1056/NEJMoa1912387. [DOI] [PubMed] [Google Scholar]
- 63.El Dika I., Lim H.Y., Yong W.P., Lin C.C., Yoon J.H., Modiano M. An open-label, multicenter, phase I, dose escalation study with phase II expansion cohort to determine the safety, pharmacokinetics, and preliminary antitumor activity of intravenous TKM-080301 in subjects with advanced hepatocellular carcinoma. Oncologist. 2019;24 doi: 10.1634/theoncologist.2018-0838. 747-e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alexopoulou A., Vasilieva L., Karayiannis P. New approaches to the treatment of chronic hepatitis B. J Clin Med. 2020;9:3187. doi: 10.3390/jcm9103187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Conway A., Mendel M., Kim K., McGovern K., Boyko A., Zhang L. Non-viral delivery of zinc finger nuclease mRNA enables highly efficient in vivo genome editing of multiple therapeutic gene targets. Mol Ther. 2019;27:866–877. doi: 10.1016/j.ymthe.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Daya S., Berns K.I. Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev. 2008;21:583–593. doi: 10.1128/CMR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hastie E., Samulski J.R. Recombinant adeno-associated virus vectors in the treatment of rare diseases. Expert Opin Orphan Drugs Inform Healthc. 2015:675–689. doi: 10.1517/21678707.2015.1039511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Palaschak B., Herzog R.W., Markusic D.M. AAV-mediated gene delivery to the liver: overview of current technologies and methods. Methods Mol Biol. 2019;1950:333–360. doi: 10.1007/978-1-4939-9139-6_20. [DOI] [PubMed] [Google Scholar]
- 69.Peyvandi F., Garagiola I. Clinical advances in gene therapy updates on clinical trials of gene therapy in haemophilia. Haemophilia. 2019;25:738–746. doi: 10.1111/hae.13816. [DOI] [PubMed] [Google Scholar]
- 70.Vercauteren K., Hoffman B.E., Zolotukhin I., Keeler G.D., Xiao J.W., Basner-Tschakarjan E. Superior in vivo transduction of human hepatocytes using engineered AAV3 capsid. Mol Ther. 2016;24:1042–1049. doi: 10.1038/mt.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Büning H., Srivastava A. Capsid modifications for targeting and improving the efficacy of AAV vectors. Mol Ther Methods Clin Dev. 2019;12:248–265. doi: 10.1016/j.omtm.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mével M., Bouzelha M., Leray A., Pacouret S., Guilbaud M., Penaud-Budloo M. Chemical modification of the adeno-associated virus capsid to improve gene delivery. Chem Sci. 2020;11:1122. doi: 10.1039/c9sc04189c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Westhaus A., Cabanes-Creus M., Rybicki A., Baltazar G., Navarro R.G., Zhu E. High-throughput in vitro, ex vivo, and in vivo screen of adeno-associated virus vectors based on physical and functional transduction. Hum Gene Ther. 2020;31:575–589. doi: 10.1089/hum.2019.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koerber J.T., Jang J.H., Schaffer D.V. DNA shuffling of adeno-associated virus yields functionally diverse viral progeny. Mol Ther. 2008;16:1703–1709. doi: 10.1038/mt.2008.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lisowski L., Dane A.P., Chu K., Zhang Y., Cunningham S.C., Wilson E.M. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506:382–386. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Paulk N.K., Pekrun K., Zhu E., Nygaard S., Li B., Xu J. Bioengineered AAV capsids with combined high human liver transduction in vivo and unique humoral seroreactivity. Mol Ther. 2018;26:289–303. doi: 10.1016/j.ymthe.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li S., Ling C., Zhong L., Li M., Su Q., He R. Efficient and targeted transduction of nonhuman primate liver with systemically delivered optimized AAV3B vectors. Mol Ther. 2015;23:1867–1876. doi: 10.1038/mt.2015.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ling C., Yin Z., Li J., Zhang D., Aslanidi G., Srivastava A. Strategies to generate high-titer, high-potency recombinant AAV3 serotype vectors. Mol Ther Methods Clin Dev. 2016;3:16029. doi: 10.1038/mtm.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Biswas M., Marsic D., Li N., Zou C., Gonzalez-Aseguinolaza G., Zolotukhin I. Engineering and in vitro selection of a novel AAV3B variant with high hepatocyte tropism and reduced seroreactivity. Mol Ther Methods Clin Dev. 2020;19:347–361. doi: 10.1016/j.omtm.2020.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rangarajan S., Walsh L., Lester W., Perry D., Madan B., Laffan M. AAV5-Factor VIII gene transfer in severe hemophilia A. N Engl J Med. 2017;377:2519–2530. doi: 10.1056/NEJMoa1708483. [DOI] [PubMed] [Google Scholar]
- 81.Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 82.Nathwani A.C., Tuddenham E.G., Rangarajan S., Rosales C., McIntosh J., Linch D.C. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.High K.A., George L.A., Eyster M.E., Sullivan S.K., Ragni M.V. A phase 1/2 trial of investigational Spk-8011 in hemophilia a demonstrates durable expression and prevention of bleeds. Blood. 2018;132(S1):487. [Google Scholar]
- 84.George L.A., Sullivan S.K., Giermasz A., Rasko J.E.J., Samelson-Jones B.J., Ducore J. Hemophilia B gene therapy with a high specific- activity factor IX variant. N Engl J Med. 2017;377:2215–2227. doi: 10.1056/NEJMoa1708538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pasi K.J., Rangarajan S., Mitchell N., Lester W., Symington E., Madan B. Multiyear follow-up of AAV5-hFVIII-SQ gene therapy for hemophilia A. N Engl J Med. 2020;382:29–40. doi: 10.1056/NEJMoa1908490. [DOI] [PubMed] [Google Scholar]
- 86.D'Avola D., López-Franco E., Sangro B., Pañeda A., Grossios N., Gil-Farina I. Phase I open label liver-directed gene therapy clinical trial for acute intermittent porphyria. J Hepatol. 2016;65:776–783. doi: 10.1016/j.jhep.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 87.Nakai H., Yant S.R., Storm T.A., Fuess S., Meuse L., Kay M.A. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J Virol. 2001;75:6969–6976. doi: 10.1128/JVI.75.15.6969-6976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Borel F., Tang Q., Gernoux G., Greer C., Wang Z., Barzel A. Survival advantage of both human hepatocyte xenografts and genome-edited hepatocytes for treatment of α-1 antitrypsin deficiency. Mol Ther. 2017;25:2477–2489. doi: 10.1016/j.ymthe.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barzel A., Paulk N.K., Shi Y., Huang Y., Chu K., Zhang F. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature. 2015;517:360–364. doi: 10.1038/nature13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Porro F., Bortolussi G., Barzel A., De Caneva A., Iaconcig A., Vodret S. Promoterless gene targeting without nucleases rescues lethality of a Crigler-Najjar syndrome mouse model. EMBO Mol Med. 2017;9:1346–1355. doi: 10.15252/emmm.201707601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chandler R.J., Venturoni L.E., Liao J., Hubbard B.T., Schneller J.L., Hoffmann V. Promoterless, nuclease-free genome editing confers a growth advantage for corrected hepatocytes in mice with methylmalonic acidemia. Hepatology. 2020 doi: 10.1002/hep.31570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.De Caneva A., Porro F., Bortolussi G., Sola R., Lisjak M., Barzel A. Coupling AAV-mediated promoterless gene targeting to SaCas9 nuclease to efficiently correct liver metabolic diseases. JCI Insight. 2019;5 doi: 10.1172/jci.insight.128863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chatterjee S., Sivanandam V., Wong K.K., Jr. Adeno-associated virus and hematopoietic stem cells: the potential of adeno-associated virus hematopoietic stem cells in genetic medicines. Hum Gene Ther. 2020;31:542–552. doi: 10.1089/hum.2020.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Conboy I., Murthy N., Etienne J., Robinson Z. Making gene editing a therapeutic reality. F1000Res. 2018 Dec 21;7 doi: 10.12688/f1000research.16106.1. F1000 Faculty Rev-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ginn S.L., Amaya A.K., Liao S.H.Y., Zhu E., Cunningham S.C., Lee M. Efficient in vivo editing of OTC-deficient patient-derived primary human hepatocytes. JHEP Rep. 2019;2:100065. doi: 10.1016/j.jhepr.2019.100065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Siew S.M., Cunningham S.C., Zhu E., Tay S.S., Venuti E., Bolitho C. Prevention of cholestatic liver disease and reduced tumorigenicity in a murine model of PFIC type 3 using hybrid AAV-piggyBac gene therapy. Hepatology. 2019;70:2047–2061. doi: 10.1002/hep.30773. [DOI] [PubMed] [Google Scholar]
- 98.Cunningham S.C., Siew S.M., Hallwirth C.V., Bolitho C., Garg G., Michael I.P. Modeling correction of severe urea cycle defects in the growing murine liver using a hybrid recombinant adeno-associated virus/piggyback transposase gene delivery system. Hepatology. 2015;62:417–428. doi: 10.1002/hep.27842. [DOI] [PubMed] [Google Scholar]
- 99.La Q.T., Ren B., Logan G.J., Cunningham S.C., Khandekar N., Nassif N.T. Use of a hybrid adeno-associated viral vector transposon system to deliver the insulin gene to diabetic NOD mice. Cells. 2020;9:2227. doi: 10.3390/cells9102227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gao J., Mese K., Bunz O., Ehrhardt A. State-of-the-art human adenovirus vectorology for therapeutic approaches. FEBS Lett. 2019;593:3609–3622. doi: 10.1002/1873-3468.13691. [DOI] [PubMed] [Google Scholar]
- 101.Ricobaraza A., Gonzalez-Aparicio M., Mora-Jimenez L., Lumbreras S., Hernandez-Alcoceba R. High-capacity adenoviral vectors: expanding the scope of gene therapy. Int J Mol Sci. 2020;21:3643. doi: 10.3390/ijms21103643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Castello R., Borzone R., D’Aria S., Annunziata P., Piccolo P., Brunetti-Pierri N. Helper-dependent adenoviral vectors for liver-directed gene therapy of primary hyperoxaluria type 1. Gene Ther. 2016;23:129–134. doi: 10.1038/gt.2015.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Leggiero E., Labruna G., Iaffaldano L., Lombardo B., Greco A., Fiorenza D. Helper-dependent adenovirus-mediated gene transfer of a secreted LDL receptor/transferrin chimeric protein reduces aortic atherosclerosis in LDL receptor-deficient mice. Gene Ther. 2019;26:121–130. doi: 10.1038/s41434-019-0061-z. [DOI] [PubMed] [Google Scholar]
- 104.Somanathan S., Calcedo R., Wilson J.M. Adenovirus-antibody complexes contributed to lethal systemic inflammation in a gene therapy trial. Mol Ther. 2020;28:784–793. doi: 10.1016/j.ymthe.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shirley J.L., de Jong Y.P., Terhorst C., Herzog R.W. Immune responses to viral gene therapy vectors. Mol Ther. 2020;28:709–722. doi: 10.1016/j.ymthe.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu J., Seol D.W. Helper virus-free gutless adenovirus (HF-GLAd): a new platform for gene therapy. BMB Rep. 2020;53:565–575. doi: 10.5483/BMBRep.2020.53.11.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sun Y., Lv X., Ding P., Wang L., Sun Y., Li S. Exploring the functions of polymers in adenovirus-mediated gene delivery: evading immune response and redirecting tropism. Acta Biomate. 2019;97:93–104. doi: 10.1016/j.actbio.2019.06.059. [DOI] [PubMed] [Google Scholar]
- 108.Palmer D.J., Turner D.L., Ng P. A single "All-in-One" helper-dependent adenovirus to deliver donor DNA and CRISPR/Cas9 for efficient homology-directed repair. Mol Ther Methods Clin Dev. 2020;17:441–447. doi: 10.1016/j.omtm.2020.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Palmer D.J., Turner D.L., Ng P. Bi-allelic homology-directed repair with helper dependent adenoviruses. Mol Ther Methods Clin Dev. 2019;15:285–293. doi: 10.1016/j.omtm.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hausl M.A., Zhang W., Müther N., Rauschhuber C., Franck H.G., Merricks E.P. Hyperactive sleeping beauty transposase enables persistent phenotypic correction in mice and a canine model for hemophilia B. Mol Ther. 2010;18:1896–1906. doi: 10.1038/mt.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ferry N., Pichard V., Sébastien Bony D.A., Nguyen T.H. Retroviral vector-mediated gene therapy for metabolic diseases: an update. Curr Pharm Des. 2011;17:2516–2527. doi: 10.2174/138161211797247587. [DOI] [PubMed] [Google Scholar]
- 112.Poletti V., Biffi A. Gene-based approaches to inherited neurometabolic diseases. Hum Gene Ther. 2019;30:1222–1235. doi: 10.1089/hum.2019.190. [DOI] [PubMed] [Google Scholar]
- 113.Kay M.A., Baley P., Rothenberg S., Leland F., Fleming L., Ponder K.P. Expression of human alpha 1-antitrypsin in dogs after autologous transplantation of retroviral transduced hepatocytes. Proc Natl Acad Sci U S A. 1992;89:89–93. doi: 10.1073/pnas.89.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Grossman M., Raper S.E., Wilson J.M. Transplantation of genetically modified autologous hepatocytes into nonhuman primates: feasibility and short-term toxicity. Hum Gene Ther. 1992;3:501–510. doi: 10.1089/hum.1992.3.5-501. [DOI] [PubMed] [Google Scholar]
- 115.Overturf K., Al-Dhalimy M., Tanguay R., Brantly M., Ou C.N., Finegold M. Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat Genet. 1996;12:266–273. doi: 10.1038/ng0396-266. [DOI] [PubMed] [Google Scholar]
- 116.Raper S.E., Grossman M., Rader D.J., Thoene J.G., Clark B.J., 3rd, Kolansky D.M. Safety and feasibility of liver-directed ex vivo gene therapy for homozygous familial hypercholesterolemia. Ann Surg. 1996;223:116–126. doi: 10.1097/00000658-199602000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tada K., Roy-Chowdhury N., Prasad V., Kim B.H., Manchikalapudi P., Fox I.J. Long-term amelioration of bilirubin glucuronidation defect in Gunn rats by transplanting genetically modified immortalized autologous hepatocytes. Cell Transpl. 1998;7:607–616. doi: 10.1177/096368979800700611. [DOI] [PubMed] [Google Scholar]
- 118.Andreoletti M., Loux N., Vons C., Nguyen T.H., Lorand I., Mahieu D. Engraftment of autologous retrovirally transduced hepatocytes after intraportal transplantation into nonhuman primates: implication for ex gene therapy. Hum Gene Ther. 2001;12:169–179. doi: 10.1089/104303401750061230. [DOI] [PubMed] [Google Scholar]
- 119.Grossman M., Rader D.J., Muller D.W., Kolansky D.M., Kozarsky K., Clark B.J., 3rd A pilot study of ex vivo gene therapy for homozygous familial hypercholesterolaemia. Nat Med. 1995;1:1148–1154. doi: 10.1038/nm1195-1148. [DOI] [PubMed] [Google Scholar]
- 120.Oertel M., Rosencrantz R., Chen Y.Q., Thota P.N., Sandhu J.S., Dabeva M.D. Repopulation of rat liver by fetal hepatoblasts and adult hepatocytes transduced ex vivo with lentiviral vectors. Hepatology. 2003 May;37(5):994–1005. doi: 10.1053/jhep.2003.50183. [DOI] [PubMed] [Google Scholar]
- 121.Bierwolf J., Volz T., Lütgehetmann M., Allweiss L., Riecken K., Warlich M. Primary human hepatocytes repopulate livers of mice after in vitro culturing and lentiviral-mediated gene transfer. Tissue Eng Part A. 2016;22:742–753. doi: 10.1089/ten.tea.2015.0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hickey R.D., Nicolas C.T., Allen K., Mao S., Elgilani F., Glorioso J. Autologous gene and cell therapy provides safe and long-term curative therapy in A large pig model of hereditary tyrosinemia type 1. Cell Transpl. 2019;28:79–88. doi: 10.1177/0963689718814188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kay M.A., Rothenberg S., Landen C.N., Bellinger D.A., Leland F., Toman C. In vivo gene therapy of hemophilia B: sustained partial correction in factor IX-deficient dogs. Science. 1993;262:117–119. doi: 10.1126/science.8211118. [DOI] [PubMed] [Google Scholar]
- 124.Ferry N., Duplessis O., Houssin D., Danos O., Heard J.M. Retroviral-mediated gene transfer into hepatocytes in vivo. Proc Natl Acad Sci U S A. 1991;88:8377–8381. doi: 10.1073/pnas.88.19.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Borgnon J., Djamouri F., Lorand I., Rico V.D., Loux N., Pages J.C. Follistatin allows efficient retroviral-mediated gene transfer into rat liver. Biochem Biophys Res Commun. 2005;328:937–943. doi: 10.1016/j.bbrc.2005.01.049. [DOI] [PubMed] [Google Scholar]
- 126.Minami H., Tada K., Chowdhury N.R., Chowdhury J.R., Onji M. Enhancement of retrovirus-mediated gene transfer to rat liver in vivo by infusion of hepatocyte growth factor and triiodothyronine. J Hepatol. 2000;33:183–188. doi: 10.1016/s0168-8278(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 127.Oh S.H., Kim S.H., Kim H.W., Kim Y.J. An efficient retrovirus-mediated transduction of human blood coagulation factor VIII cDNA in regenerating rat liver. Ann Hematol. 1999;78:213–218. doi: 10.1007/s002770050504. [DOI] [PubMed] [Google Scholar]
- 128.Forbes S.J., Themis M., Alison M.R., Sarosi I., Coutelle C., Hodgson H.J. Synergistic growth factors enhance rat liver proliferation and enable retroviral gene transfer via a peripheral vein. Gastroenterology. 2000;118:591–598. doi: 10.1016/s0016-5085(00)70266-6. [DOI] [PubMed] [Google Scholar]
- 129.Puppi J., Guillonneau C., Pichard V., Bellodi-Privato M., Cuturi M.C., Anegon I. Long term transgene expression by hepatocytes transduced with retroviral vectors requires induction of immune tolerance to the transgene. J Hepatol. 2004;41:222–228. doi: 10.1016/j.jhep.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 130.Aubert D., Ménoret S., Chiari E., Pichard V., Durand S., Tesson L. Cytotoxic immune response blunts long-term transgene expression after efficient retroviral-mediated hepatic gene transfer in rat. Mol Ther. 2002;5:388–396. doi: 10.1006/mthe.2002.0561. [DOI] [PubMed] [Google Scholar]
- 131.Tada K., Chowdhury N.R., Neufeld D., Bosma P.J., Heard M., Prasad V.R. Long-term reduction of serum bilirubin levels in Gunn rats by retroviral gene transfer in vivo. Liver Transpl Surg. 1998;4:78–88. doi: 10.1002/lt.500040111. [DOI] [PubMed] [Google Scholar]
- 132.Chandler R.J., Venditti C.P. Gene therapy for methylmalonic acidemia: past, present, and future. Hum Gene Ther. 2019;30:1236–1244. doi: 10.1089/hum.2019.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Clar J., Mutel E., Gri B., Creneguy A., Stefanutti A., Gaillard S. Hepatic lentiviral gene transfer prevents the long-term onset of hepatic tumours of glycogen storage disease type 1a in mice. Hum Mol Genet. 2015;24:2287–2296. doi: 10.1093/hmg/ddu746. [DOI] [PubMed] [Google Scholar]
- 134.Cantore A., Ranzani M., Bartholomae C.C., Volpin M., Valle P.D., Sanvito F. Liver-directed lentiviral gene therapy in a dog model of hemophilia B. Sci Transl Med. 2015;7:277ra28. doi: 10.1126/scitranslmed.aaa1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Milani M., Annoni A., Moalli F., Liu T., Cesana D., Calabria A. Phagocytosis-shielded lentiviral vectors improve liver gene therapy in nonhuman primates. Sci Transl Med. 2019;22 doi: 10.1126/scitranslmed.aav7325. 11(493) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Powell J.S., Ragni M.V., White G.C., 2nd, Lusher J.M., Hillman-Wiseman C., Moon T.E. Phase 1 trial of FVIII gene transfer for severe hemophilia A using a retroviral construct administered by peripheral intravenous infusion. Blood. 2003;102:2038–2045. doi: 10.1182/blood-2003-01-0167. [DOI] [PubMed] [Google Scholar]
- 137.Muhuri M., Maeda Y., Ma H., Ram S., Fitzgerald K.A., Tai P.W. Overcoming innate immune barriers that impede AAV gene therapy vectors. J Clin Invest. 2021;131 doi: 10.1172/JCI143780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kerpel-Fronius S., Baroutsou V., Becker S., Carlesi R., Collia L., Franke-Bray B. Development and use of gene therapy orphan drugs-ethical needs for a broader cooperation between the pharmaceutical industry and society. Front Med. 2020;7:608249. doi: 10.3389/fmed.2020.608249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chand D., Mohr F., McMillan H., Tukov F.F., Montgomery K., Kleyn A. Hepatotoxicity following administration of onasemnogene abeparvovec (AVXS-101) for the treatment of spinal muscular atrophy. J Hepatol. 2020 doi: 10.1016/j.jhep.2020.11.001. S0168-8278(20)33748-X. [DOI] [PubMed] [Google Scholar]
- 140.Verdera H.C., Kuranda K., Mingozzi F. AAV vector immunogenicity in humans: a long journey to successful gene transfer. Mol Ther. 2020;28:723–746. doi: 10.1016/j.ymthe.2019.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Konkle B.A., Walsh C., Escobar M.A., Josephson N.C., Young G., von Drygalski A. BAX 335 hemophilia B gene therapy clinical trial results - potential impact of CpG sequences on gene expression. Blood. 2020:2019004625. doi: 10.1182/blood.2019004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Buscara L., Gross D.A., Daniele N. Of rAAV and men: from genetic neuromuscular disorder efficacy and toxicity preclinical studies to clinical trials and back. J Pers Med. 2020;10:258. doi: 10.3390/jpm10040258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Salas D., Kwikkers K.L., Zabaleta N., Bazo A., Petry H., van Deventer S.J. Immunoadsorption enables successful rAAV5-mediated repeated hepatic gene delivery in nonhuman primates. Blood Adv. 2019;3:2632–2641. doi: 10.1182/bloodadvances.2019000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Leborgne C., Barbon E., Alexander J.M., Hanby H., Delignat S., Cohen D.M. IgG-cleaving endopeptidase enables in vivo gene therapy in the presence of anti-AAV neutralizing antibodies. Nat Med. 2020;26:1096–1101. doi: 10.1038/s41591-020-0911-7. [DOI] [PubMed] [Google Scholar]
- 145.La Bella T., Imbeaud S., Peneau C., Mami I., Datta S., Bayard Q. Adeno-associated virus in the liver: natural history and consequences in tumour development. Gut. 2020;69:737–747. doi: 10.1136/gutjnl-2019-318281. [DOI] [PubMed] [Google Scholar]
- 146.Gil-Farina I., Fronza R., Kaeppel C., Lopez-Franco E., Ferreira V., D'Avola D. Recombinant AAV integration is not associated with hepatic genotoxicity in nonhuman primates and patients. Mol Ther. 2016;24:1100–1105. doi: 10.1038/mt.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.George L.A., Ragni M.V., Rasko J.E.J., Raffini L.J., Samelson-Jones B.J., Ozelo M. Long-term follow-up of the first in human intravascular delivery of AAV for gene transfer: AAV2-hFIX16 for severe hemophilia B. Mol Ther. 2020;28:2073–2082. doi: 10.1016/j.ymthe.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.