Summary
Peptide mobility shift assays provide a sensitive measure of kinase enzymatic activity and can be used to evaluate kinase inhibitors. Herein, we describe a protocol adapted for rapid assessment of doublecortin-like kinase inhibitors. Advantages include rapid iterations of therapeutic compound assessment and the ability to characterize kinase mutations, such as drug-resistant mutants for biological rescue experiments, on kinase activity.
For complete details on the use and execution of this protocol, please refer to Liu et al. (2020).
Subject areas: cancer, high-throughput screening, molecular/chemical probes, protein biochemistry
Graphical abstract

Highlights
-
•
A peptide substrate mobility shift assay (MSA) measures DCLK1 kinase activity
-
•
The MSA platform can generate enzyme kinetics data
-
•
The MSA enables sensitive rank ordering of DCLK1 kinase inhibitors
-
•
The assay is compatible with a 384-well format
Peptide mobility shift assays provide a sensitive measure of kinase enzymatic activity and can be used to evaluate kinase inhibitors. Herein, we describe a protocol adapted for rapid assessment of doublecortin-like kinase inhibitors. Advantages include rapid iterations of therapeutic compound assessment and the ability to characterize kinase mutations, such as drug-resistant mutants for biological rescue experiments, on kinase activity.
Before you begin
DCLK1 kinase activity supports tumor cell growth and is a cancer target. Here we report an assay for rapid evaluation of the potency of DCLK1 kinase inhibitors. The assay utilizes purified DCLK1 kinase and a peptide substrate. Kinase activity can be measured by taking advantage of a mobility shift in the mobility of the peptide substrate, measured by microcapillary electrophoresis, after phosphorylation. An automated platform enables kinetic studies and high-throughput measurements from dense plate formats.
Preparation of buffers and reagents
Timing: 4 h
-
1.
Prepare cell culture media, protein purification buffers, and assay buffer as described in materials and equipment.
-
2.
Obtain the DCLK1 expression construct and BL21 DE3 cells.
-
3.
Bacteria are cultured using standard procedures (Elbing and Brent, 2019a, 2019b).
Production of recombinant DCLK1 protein
Timing: 4 days
-
4.Expression DCLK1 kinase domain in E.coli.
-
a.Transform DCLK1 kinase domain DNA construct in E.coli BL21 DE3 cells.
-
b.Grow cells to OD 600 of 0.6 at 37°C, then reduce the temperature to 18°C.
-
c.Induce protein expression with 0.6 mM isopropyl β-D-1 thiogalactopyranoside (IPTG) and allow culture to continue for ∼10 h at 18°C.
-
a.
-
5.DCLK1 protein purification
-
a.Harvest cells by centrifugation at 17,000 g for 30 min at 4°C.
-
b.Suspend cell pellet in Lysis buffer with protease inhibitors (1 mM Benzamidine and 1 mM phenylmethylsulfonyl fluoride (PMSF).
 CRITICAL: Protease inhibitors should be added from stocks right before suspending cells.
CRITICAL: Protease inhibitors should be added from stocks right before suspending cells. -
c.Lyse cells by passing 3 times through a homogenizer (EmulsiFlex-C3, pressure: 8000–10000 psi) while keeping on ice.
-
d.Centrifuge lysate at 20K for 1 h and filter the supernatant through a 0.2 micron membrane.
-
e.Capture protein using Nickle-NTA resin and elute with 300 mM imidazole elution solution using standard methods (Nallamsetty et al., 2005; Nallamsetty and Waugh, 2007; Tropea et al., 2007)
-
f.Buffer exchange and further purify by passing over a Superdex S200 column using standard methods (Nallamsetty et al., 2005; Nallamsetty and Waugh, 2007; Tropea et al., 2007).
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. coli BL21 | New England Biolabs | Cat #C2527I |
| E. coli DH5α | New England Biolabs | Cat #C2988J |
| Chemicals, peptides, and recombinant proteins (it is useful to spell out the names of the chemicals) | ||
| LB (lysogeny broth) | RPI (Mount Prospect, IL) | Cat #L24040 |
| ATP stock 10 mM (liquid) (adenosine triphosphate) | Promega (Madison, WI) | Cat #V915 |
| HEPES, Free Acid (solid, >99%) | RPI (Mount Prospect, IL) | Cat #H75030 |
| Imidazole (solid, >99%) | Sigma (St. Louis, MO) | Cat #792527 |
| NaCl (solid, >99%) (sodium chloride) | RPI (Mount Prospect, IL) | Cat #S23020 |
| Glycerol (liquid) | RPI (Mount Prospect, IL) | Cat #G22020 |
| Brij-35 (liquid, 30%) | Sigma (St. Louis, MO) | Cat #B4184 |
| CR-3 (liquid, 200 ×) | PerkinElmer (Houston, TX) | Cat #760050 |
| CR-8 (liquid, 2%) | PerkinElmer (Houston, TX) | Cat #760278 |
| EDTA (solid, >99%) (ethylenediaminetetraacetic acid) | Sigma (St. Louis, MO) | Cat #EDS-1KG |
| Tween-20 (liquid) | Sigma (St. Louis, MO) | Cat #P1379 |
| DTT (solid, ≥97%) (1,4-dithiothreitol) | Sigma (St. Louis, MO) | Cat #10197777001 |
| IPTG (solid, ≥99%) (isopropyl β-D-1-thiogalactopyranoside) | Sigma (St. Louis, MO) | Cat #11411446001 |
| Protease Inhibitor Cocktail | Sigma (St. Louis, MO) | Cat #P8465 |
| DCLK1-IN-1 | Gray Lab, Dana-Farber Cancer Institute | (Ferguson et al., 2020a; Ferguson et al., 2020b) |
| FL-Peptide 12: 5-FAM-KKLRRTLSVA-COOH 1.5 mM | Caliper Life Sciences | Cat #760356 |
| Benzamidine | Sigma (St. Louis, MO) | Cat # 12072 |
| PMSF | Tocris | Cat #4486 |
| DMSO (dimethyl sulfoxide) | Sigma (St. Louis, MO) | Cat # D2650 |
| MgCl2 (magnesium chloride) | Sigma (St. Louis, MO) | Cat # 208337 |
| DCLK1 kinase domain DNA construct | Dr. Ana Clara Redondo, University of Oxford | N/A |
| Software and algorithms | ||
| Prism 8 | GraphPad | N/A |
| Other | ||
| Greiner LUMITRAC™ 200 384-well plates | Millipore Sigma | Cat #M6936-40EA |
| Nickle-NTA resin | Bio-Rad | Cat #732-4612 |
| Superdex S200 column | Cytiva | Cat #17104302 |
Materials and equipment
| Perkin Elmer LabChip® EZ reader and parameters | |
|---|---|
| Model | #122919 |
| Pressure | −1.8 PSI |
| Downstream Voltage | −500 V |
| Upstream Voltage | −2250 V |
| Buffer Sip | 35 s |
| Sample/Marker sip Time | 0.2 s |
| Final Delay | 80 s |
| Peak Order | product first |
| Cycles | 60 for enzyme titration; 30 for ATP titration; 1 or 2 for IC50 |
LB (Luria-Bertani)
| Reagent | Final concentration | Amount |
|---|---|---|
| LB | 1× | 25 g |
| ddH2O | n/a | to 1 L |
| Total | n/a | 1 L |
It is good for 1 month at 25°C.
HEPES buffer (pH 7.3 or 7.5)
| Reagent | Final concentration | Amount |
|---|---|---|
| Hepes | 1 M | 238.3 g |
| ddH2O | n/a | to 1 L |
| Total | n/a | 1 L |
It is good for 6 months at 25°C.
pH adjust this solution using NaOH
NaCl buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl | 5 M | 292.2 g |
| ddH2O | n/a | to 1 L |
| Total | n/a | 1 L |
It is good for 6 months at 25°C.
MgCl2 buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| MgCl2 | 1 M | 95.2 g |
| ddH2O | n/a | to 1 L |
| Total | n/a | 1 L |
It is good for 6 months at 25°C.
Imidazole buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Imidazole | 2 M | 136.2 g |
| ddH2O | n/a | to 1 L |
| Total | n/a | 1 L |
It is good for 6 months at 25°C.
EDTA buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| EDTA | 1 M | 292.2 g |
| ddH2O | n/a | to 1 L |
| Total | n/a | 1 L |
It is good for 6 months at 25°C.
DTT buffer 10 mL
| Reagent | Final concentration | Amount |
|---|---|---|
| DTT | 1 M | 1.5 g |
| ddH2O | n/a | to 10 mL |
| Total | n/a | 10 mL |
It is good for 6 months at −20°C.
Benzamidine stock solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Benzamidine | 500 mM | 873.15 mg |
| ddH2O | n/a | to 10 mL |
| Total | n/a | 10 mL |
It is good for 6 months at 4°C.
PMSF stock solution
| Reagent | Final concentration | Amount |
|---|---|---|
| PMSF | 100 mM | 50 mg |
| DMSO | n/a | to 2.87 mL |
| Total | n/a | 2.87 mL |
It is good for 6 months at −20°C.
Lysis buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Hepes pH 7.3 (1 M) | 50 mM | 50 mL |
| NaCl (5 M) | 350 mM | 70 mL |
| Imidazole (2 M) | 20 mM | 10 mL |
| Glycerol | 5% | 5 mL |
| Benzamidine (500 mM) | 1 mM | 2 mL |
| PMSF (100 mM) | 1 mM | 10 mL |
| ddH2O | n/a | 853 mL |
| Total | n/a | 1 L |
It is good for 6 months at 4°C.
Note: The buffer should be filtered.
Imidazole elution solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Imidazole (2 M) | 300 nM | 1.5 mL |
| Lysis Buffer | n/a | 998.5 mL |
| Total | n/a | 1 L |
It is good for 6 months at 4°C.
Gel filtration buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Hepes pH 7.3 (1 M) | 10 mM | 10 mL |
| NaCl (5 M) | 700 mM | 140 mL |
| MgCl2 (1 M) | 1 mM | 1 mL |
| Glycerol | 5% | 5 mL |
| ddH2O | n/a | 844 mL |
| Total | n/a | 1 L |
It is good for 6 months at 4°C.
Note: The buffer should be filtered.
Kinase buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Hepes pH 7.5 (1 M) | 100 mM | 100 mL |
| Brij-35 (30%) | 0.003% | 100 μL |
| Tween-20 | 0.004% | 40 μL |
| MgCl2 (1M) | 10 mM | 10 mL |
| DTT (add before use) | 2 mM | 2 mL |
| ddH2O | n/a | 887.86 mL |
| Total | n/a | 1 L |
It is good for 6 months at 4°C.
Note: The buffer should be filtered.
Separation buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Hepes pH 7.3 (1 M) | 100 mM | 100 mL |
| Brij-35 (30%) | 0.015% | 500 μL |
| EDTA (1M) | 1 mM | 2 mL |
| CR-3 | 0.1% | 25 mL |
| DMSO | 5% | 50 mL |
| CR-8 (add before use) | 0.5% | 5 mL |
| ddH2O | n/a | 817.4 mL |
| Total | n/a | 1 L |
It is good for 6 months at 4°C.
Note: The buffer should be filtered.
Kinase stop buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Separation buffer | 1× | 961 mL |
| EDTA (1 M) | 40 mM | 39 mL |
| Total | n/a | 1 L |
It is good for 6 months at 4°C.
2× peptide mix
| Reagent | Final concentration | Amount |
|---|---|---|
| Peptide 12 | 2.0 μM | 13.3 μL |
| ATP | 200 μM | 20 μL |
| Reaction buffer | n/a | 9.967 mL |
| Total | n/a | 10 mL |
It is good for 1 day at 4°C.
Trough buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 2× Peptide Mix | 1× | 0.5 mL |
| Reaction buffer | n/a | 0.5 mL |
| Total | n/a | 1 mL |
It is good for 1 day at 4°C.
Step-by-step method details
Run a real-time reaction to establish DCLK1 activity
Timing: 6 h
-
1.Prepare buffers, assay chip and the EZ Reader instrument.
-
a.Add DTT to 1× Kinase Buffer to a final working concentration of 2 μM.
-
b.Remove the chip from the container and place the chip in holder filled with ultrapure deionized water. Dump out the EDTA solution from the chip wells. Rinse with water 3 times and with Reaction Buffer 3 times.
-
c.Dry the top and bottom surfaces of the chip using vacuum suction. This step is very important. Leaving any water on the surfaces will result in equipment failure.
-
d.Make 2× Peptide Mix in Reaction Buffer. 1× Peptide Mix: 1 μM of peptide 12 and 100 μM of ATP.
-
e.Make 1× Trough Buffer. Dilute 0.5 mL of Peptide Mix with 0.5 mL of Reaction Buffer. Add 450 mL to each side of EZ Reader Trough.
-
f.Refer to the EZ Reader user manual for additional details.
-
a.
-
2.Perform the reaction in 384-well plates in a total volume of 80 μL. The reaction comprises recombinant DCLK1, ATP and one FAM-labeled peptide substrate (peptide 12 for DCLK1) in Reaction Buffer.
-
a.Prepare 2× DCLK1 solution. Typically, the highest final enzyme concentration will be 10 nM, therefore 2× is 20 nM. Make 400 μL of 2× Reaction Buffer with DCLK1 enzyme included. Use this to create 1:1 dilutions in 40 μL volumes. A typical dilution experiment will involve 6 total concentrations (A1-A6) with duplicates (E1-E6).
-
b.Prepare the instrument to receive the plate. Select wells, set voltages and pressure, and choose a number of cycles that gives a one-hour time course.
-
c.Add 40 μL of 2× peptide mix (peptide 12) to wells to initiate reaction. The final concentrations of DCLK1 are 10 nM, 5 nM, 2.5 nM, 1.25 nM, 0.625 nM and 0.3125 nM. Peptide 12 (substrate) is 1 μM and ATP is 200 μM.
-
a.
-
3.
Plot the results to determine a DCLK1 concentration that converts 30% of substrate to product in 1 h. Other time points could be used, but 1 h is generally convenient. Also, the reaction rate should be linear (Figure 1).
CRITICAL: Establishing an acceptable DCLK1 concentration is crucial to subsequent steps. Repeat this step multiple times to confirm it is reproducible in your personal workflow. You may need to titrate the ATP concentration to confirm you are working at the Michaelis constant, denoted by Km, for ATP. To determine the Km, we performed a dose response experiment starting at 150 μM and diluting 1:2. Km, the substrate concentration that gives the half maximal velocity, is calculated using GraphPad Prism (Motulsky, 2016). We chose concentration of DCLK1 that gave ∼30% conversion at 1 h from step 3. Run the kinetics reaction and determine the Km for ATP. Also note that for screening we work in small volumes to conserve sample. This involves addition of 20 μL of peptide mix to 20 μL of reaction buffer. Because of this small volume, it is best to keep time courses relatively short to avoid changes in the assay volume due to evaporation. For DCLK1, we use a 1-h kinase-substrate reaction. The stability of enzyme at room temperature should also be considered. Adjust the reaction velocity and reaction time based on protein stability.
Figure 1.
Concentration-dependent conversion rates of DCLK1
Run a one-time reaction to measure IC50 of DCLK1 inhibitors
Timing: 3 h
Setup the DCLK1-DCLK1 substrate reaction with potential DCLK1 inhibitors in 384 plates.
-
4.Set up the reaction plates.
-
a.Our typical plate layout provides for an 8-point dose curve from 432 nM to 0.25 nM. (Figure 2).
-
b.Prepare 2× Enzyme (DCLK1) Mix. Use the DCLK1 concentration that can convert 30% of substrate to product in one hour as described above. Make 2× DCLK1 in Reaction Buffer. For example, if the reaction concentration is 5 nM, the 2× Enzyme Mix contains 10 nM of DCLK1 in Reaction Buffer.
-
c.Add 20 μL of 2× Enzyme Mix to appropriate wells on screening plate. The “no enzyme” control will contain only Reaction Buffer.
-
d.Add Inhibitors, positive controls and negative controls. To determine the IC50 of inhibitors, do an inhibitor dose titration. Figure 2 gives an example of 8-point dose response in triplicate using a 384-well plate to test 14 inhibitors. The positive control and DMSO control use only one concentration instead of a dose range (Figure 2).
-
e.Allow to reach room temperature.
-
a.
-
5.Prepare 2× Peptide Mix.
-
a.Make 2× Peptide Mix. 1× Peptide Mix: Peptide 12 (DCLK1) at 1 μM; ATP at 100 μM
- Stock solution: Peptide 12 at 3.0 mM; ATP at 100 mM
-
a.
-
6.Start the enzyme-substrate reaction.
-
a.After 1 h incubation from step 1, add 20 μL of 2× Peptide Mix into each well and start the enzyme-substrate reaction.
-
b.Quench the reaction with 40 μL of 1× Separation Buffer that includes 40 mM EDTA.
-
a.
Pause point: Quenched reactions can theoretically be stored at 4°C, but we found this occasionally degraded the peptide fluorescence signal.
- 7.
Figure 2.
Layout of the reaction plate
The reaction is based on 8-point dose response design in triplicate. The positive control is a known DCLK1 inhibitor at a concentration that completely inhibits DCLK1 activity. The negative control is no enzyme. 14 inhibitors per plate can be tested in this format.
Figure 3.
Post-run analysis setup
Expected outcomes
Substrate peptide mobility conversion rates should be around 35% at baseline and less if inhibition is present (Figure 4). We also expect the conversion rates should be below 5% in the absence of DCLK1 enzyme or in the presence of the positive control (a known DCLK1 inhibitor). The layout of Figure 4 is the same as that of Figure 2.
Figure 4.
Peptide conversion rate display in the EZ Reader software
LabChip EZ Reader software can also compute the inhibition rates (%) using the formula:
Inhibition (%) = ((Conversion_DMSO – Conversion_DCLK1_inhibitors)/Conversion_DMSO) ∗ 100%. (Figure 5).
Figure 5.
Inhibition rates of reaction calculated in the EZ Reader control software
The inhibition rates are relative to DMSO controls
Quantification and statistical analysis
Inhibition or conversion data are exported to GraphPad (Prism 8.4.3) for IC50 analysis. A layout of the sipper map is provided to assist with analysis (Figure 6).
- 1.
-
2.Analyze the data.
-
a.Click Analyze and then choose Nonlinear regression.
-
b.On the Nonlinear regression window, choose the "Dose-response - Inhibition".
-
c.Then choose "log(inhibitor) vs. response -- Variable slope (four parameters)".
-
d.Click OK (Figure 9).
-
e.View the results (Figures 10 and 11).
-
a.
Figure 6.
Sipper map for 12-sipper chips
Figure 7.
Graph type selection in GraphPad
Figure 8.
Data input structure for GraphPad
Figure 9.
Model selection menu in GraphPad
Figure 10.
Model fit parameters in GraphPad
Figure 11.
The IC50 curves of DCLK1 inhibitors
The IC50 curves were generated using GraphPad Prism
Limitations
As with many biochemical assays, reproducibility is contingent on maintaining consistency of reagents, particularly the enzyme, from experiment to experiment. To accomplish this, it is recommended that users avoid freeze thaw cycles of enzyme and ATP. In other words, we recommend aliquoting reagent pools into fractions for individual use and freezing so that each experiment starts from fresh aliquots; any excess of thawed aliquots be discarded (not refrozen and used later).
Troubleshooting
Problem 1
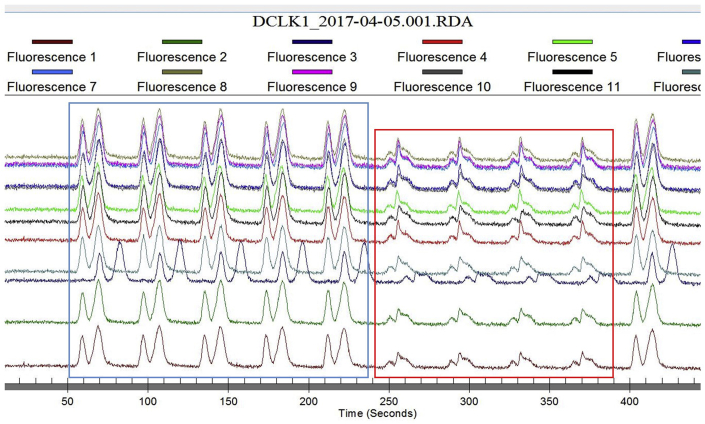
The product and substrate peaks traces from the EZ reader should be sharp and well separated (Figure 12 left). However, for a variety of reasons peak quality can degrade (Figure 12 right – red box). This may be instrument-related, but can also be related to assay conditions.
Figure 12.
Normal (blue) and abnormal (red) product/substrate peaks from EZ reader
Potential solution
a. In one case we noted that quenching the reaction with 40 mM EDTA caused degradation of peaks. One solution is to lower the concentration of EDTA.
b. If the problem persists, the reaction can be terminated by heating to greater than 60°C.
Problem 2
DCLK1 protein has low activity.
Potential solution
ATP may have expired. Also, the enzyme may need to be activated by incubating with using ATP so that it can autophosphorylate.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ken Westover (Kenneth.westover@utsouthwestern.edu).
Materials availability
All materials described are commercially available at the time of publication.
Data and code availability
Not applicable.
Acknowledgments
This work was supported by grants from the American Cancer Society award 132205-RSG-18-039-01-DMC (K.D.W.), Cancer Prevention and Research Institute of Texas RP170373 (K.D.W.), and Welch Foundation Grant I1829 (K.D.W.).
Author contributions
K.D.W. designed this study, and Y.L. performed the experiments and data analysis.
Declaration of interests
K.D.W. has received consulting fees from Sanofi Oncology and is a member of the SAB for Vibliome Therapeutics. K.D.W. has received research funding from Revolution Medicines. K.D.W. declares that none of these relationships are directly or indirectly related to the content of this manuscript.
Contributor Information
Yan Liu, Email: yan3.liu@utsouthwestern.edu.
Kenneth D. Westover, Email: kenneth.westover@utsouthwestern.edu.
References
- Elbing K.L., Brent R. Growth of E. coli in liquid medium. Curr. Protoc. Mol. Biol. 2019;125:e81. doi: 10.1002/cpmb.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbing K.L., Brent R. Recipes and tools for culture of Escherichia coli. Curr. Protoc. Mol. Biol. 2019;125:e83. doi: 10.1002/cpmb.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson F.M., Liu Y., Harshbarger W., Huang L., Wang J., Deng X., Capuzzi S.J., Muratov E.N., Tropsha A., Muthuswamy S. Synthesis and structure-activity relationships of DCLK1 kinase inhibitors based on a 5,11-Dihydro-6H-benzo[e]pyrimido[5,4-b][1,4]diazepin-6-one scaffold. J. Med. Chem. 2020;63:7817–7826. doi: 10.1021/acs.jmedchem.0c00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson F.M., Nabet B., Raghavan S., Liu Y., Leggett A.L., Kuljanin M., Kalekar R.L., Yang A., He S., Wang J. Discovery of a selective inhibitor of doublecortin like kinase 1. Nat. Chem. Biol. 2020;16:635–643. doi: 10.1038/s41589-020-0506-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ferguson F.M., Li L., Kuljanin M., Mills C.E., Subramanian K., Harshbarger W., Gondi S., Wang J., Sorger P.K. Chemical biology toolkit for DCLK1 reveals connection to RNA processing. Cell Chem. Biol. 2020;27:1229–1240 e1224. doi: 10.1016/j.chembiol.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motulsky H. GraphPad Software, LLC; 2016. GraphPad Curve Fitting Guide. [Google Scholar]
- Nallamsetty S., Austin B.P., Penrose K.J., Waugh D.S. Gateway vectors for the production of combinatorially-tagged His6-MBP fusion proteins in the cytoplasm and periplasm of Escherichia coli. Protein Sci. 2005;14:2964–2971. doi: 10.1110/ps.051718605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallamsetty S., Waugh D.S. A generic protocol for the expression and purification of recombinant proteins in Escherichia coli using a combinatorial His6-maltose binding protein fusion tag. Nat. Protoc. 2007;2:383–391. doi: 10.1038/nprot.2007.50. [DOI] [PubMed] [Google Scholar]
- Tropea J.E., Cherry S., Nallamsetty S., Bignon C., Waugh D.S. A generic method for the production of recombinant proteins in Escherichia coli using a dual hexahistidine-maltose-binding protein affinity tag. Methods Mol. Biol. 2007;363:1–19. doi: 10.1007/978-1-59745-209-0_1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.