Abstract
Chemical and sedimentation procedures are used to purify virus particles. While these approaches are successful for wild-type viruses, they are often not feasible for purifying mutant viruses with assembly defects. We combined two published methods ( Atasheva et al., 2013 ; Moller- Tank et al., 2013 ), to generate a protocol that uses low-speed centrifugation to purify both wildtype and mutant enveloped virus particles at high yield with minimal handling steps. This protocol has successfully been used to purify alphavirus particles for imaging and structural studies ( Wang et al., 2015 ; Ramsey et al., 2017 ).
Keywords: Enveloped virus purification, Centrifugation, Assembly mutants
Background
Virus purification is traditionally based on chemical precipitation (e.g., PEG) or density gradient centrifugation. Centrifugation protocols involve pelleting particles at high speeds (> 100,000 × g) or through sedimentation matrices such as cesium, Nycodenz, Iodixanol, sucrose, glycerol, or tartrate. After sedimentation in the gradient, the purified virus sample usually requires additional steps to remove the gradient matrix, concentrate the purified particles, and buffer exchange into a stabilizing buffer for downstream applications. These approaches include dialysis, centrifugation through a centrifugal filter, or PEG precipitation. While these approaches will produce purified particles, there are several drawbacks: (1) overall yields can be low, (2) time required for the purification can extend to a week, (3) morphologically heterogeneous particles are not purified with equal efficiency, (4) particles can be damaged in the process, and (5) assembly mutants often do not survive the purification process making certain downstream analyses challenging.
The protocol described here uses a gentle approach to purify enveloped virus particles. We used minimal centrifugal force to reduce damage to particles which is observed as increased morphological heterogeneity in negative-stain transmission electron microscopy (TEM) or total loss of fragile particles by TEM or infectivity assays. In addition, we wanted to reduce the manipulation of purified virions post-purification. By merging two protocols from the literature ( Atasheva et al., 2013 ; Moller- Tank et al., 2013 ), we are able to purify different Alphaviruses (Sindbis [ Ramsey et al., 2017 ], Ross River [ Wang et al., 2015 ], Chikungunya [Mukhopadhyay and Wang, unpublished data] and assembly mutants of each) via low-speed centrifugation (LSC). No additional steps for gradient matrix removal, sample concentration, or buffer exchange are necessary. We are also able to use this protocol to purify viruses from different cell lines (mammalian and arthropod). These purified particles have been used for cryo-EM, mass spectrometry, and protease cleavage studies.
Materials and Reagents
-
Pipette tips
Note: Filter tips are not necessary unless you routinely use when working with your virus of choice.
150 mm cell culture dishes (Greiner Bio One International, catalog number: 639160, or equivalent)
Sterile polypropylene 50 ml conical tubes (Corning, catalog number: 430921, or equivalent)
Serological pipets, 5 and 10 ml (DWK Life Sciences, KIMBLE, catalog numbers: 56900-5110 and 56900-10110, or equivalent)
Kimwipes (KCWW, Kimberly-Clark, catalog number: 34155, or equivalent)
Cotton swabs (Ted Pella, catalog number: 80907, or equivalent)
pH test strips (Hach, catalog number: 2601300)
EM grids Formvar/Carbon 300 mesh (Ted Pella, catalog number: 01753-F)
BHK-21 cells (ATCC, catalog number: CCL-10)
C6/36 cells (ATCC, catalog number: CRL-1660)
1x sterile PBS (Corning, catalog number: 21-040-CM, or equivalent)
Coomassie blue
Page Ruler Prestained Protein Ladder (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 22616)
MEM media (Corning, catalog number: 15-010-CV, or equivalent)
100x L-glutamine (Corning, catalog number: 25-005-CV, or equivalent)
100x MEM nonessential amino acids (Corning, catalog number: 25-025-Cl, or equivalent)
100x antibiotic-antimycotic (Corning, catalog number: 30-004-Cl, or equivalent)
FBS (Corning, catalog number: 35-010-CV, or equivalent)
Sterile virus production serum free media (VP-SFM) (Thermo Fisher Scientific, GibcoTM, catalog number: 11681020, or equivalent)
HEPES (Fisher Scientific, catalog number: BP310-1, or equivalent)
Sodium chloride (NaCl) (Fisher Scientific, catalog number: BP358-10, or equivalent)
Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: EDS-100G, or equivalent)
Hydrochloric acid (HCl)
Uranyl acetate (Electron Microscopy Sciences, catalog number: 22400, or equivalent)
MEM complete media (see Recipes)
Supplemented VP-SFM (see Recipes)
HEPES-NaCl-EDTA (HNE) Resuspension buffer (see Recipes)
1% uranyl acetate (see Recipes)
Equipment
Pipettes
Tissue culture incubator, temperature and CO2 regulated (Thermo Fisher Scientific, Thermo ScientificTM, model: Forma 3130, or equivalent)
Refrigerated table-top centrifuge with fixed angle rotor (Eppendorf, model: 5804 R, with F34-6-38 rotor, or equivalent)
Biosafety cabinet (Thermo Fisher Scientific, Thermo ScientificTM, model: 1300 Series Class II, Type A2, catalog number: 1323TS, or equivalent)
Adjustable tilt rocker (Reliable Scientific, model: 55 rocking shaker)
Procedure
Seed 150 mm dishes with 8-9 x 106 BHK or C6/36 cells in 15 ml of complete MEM. For wild-type alphavirus purification, 2-3 150 mm dishes are routinely used in our lab.
Grow BHK cells at 37 °C in 5% CO2 and C6/36 cells at 28 °C in 5% CO2 for 2-4 days until cells are > 95% confluent.
Remove media using a serological pipette.
Infect cells with alphavirus stock at a multiplicity of infection (MOI) = 5 plaque-forming units per cell. Make up the final volume to 5 ml with Supplemented VP-SFM.
Rock for 1 h at speed control of 25 (on Reliable model 55 rocker) at room temperature so the virus has time to adhere and enter into cells. One-hour adsorption time is enough for both BHK and C6/36 cells.
Remove media containing the virus.
Add 5-10 ml PBS (equilibrated to room temperature) and tilt back and forth manually to wash the cells.
Pipette off PBS.
Repeat the PBS wash 2 more times. The goal is to remove as much of the MEM complete media as possible to avoid interference of residual FBS with the purification.
Add 15 ml of room temperature Supplemented VP-SFM to each 150 mm plate.
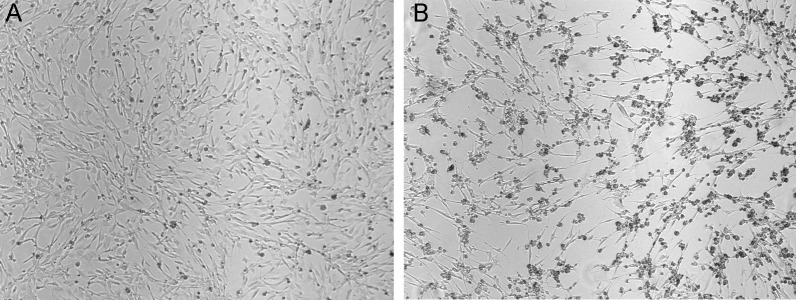
Incubate cells at an appropriate growth temperature. Media from BHK infected cells will be ready to harvest around 18-22 h post-infection. You should observe cytopathic effects and cell rounding, but minimal cell death (Figure 1). Prolonged incubation after the onset of the cytopathic effect will increase cell death and decrease virus yield. Infections in C6/36 cells incubate for 5 days but a cytopathic effect does not develop. After 2.5 days post-infection, add 5 ml of Supplemented VP-SFM media to the cells to supply additional nutrients. On day 5, collect the media and continue the next step.
Collect all the media in a 50 ml conical tube and clarify by centrifuging at 4,000 × g for 5 min, 4 °C. This step removes cell debris and dead cells. Transfer the supernatant into a new 50 ml conical tube.
At this step, the sample can be kept at 4 °C. In our lab, we do not store the sample for more than 2 days. Do not freeze the sample as there is no serum present and the enveloped virus particles will be damaged.
To purify the virus, centrifuge at 5,300 × g for 16 h at 15 °C. Use a fixed angle rotor in a refrigerated table-top centrifuge. Do not stop the centrifugation unless you are ready to continue with the subsequent steps immediately.
Stop the centrifuge and, as you remove the tubes from the rotor, circle the position where the pellets have been collected on the side of the tube. The pellets are translucent at best, and are usually formed where the side and cone of the 50 ml conical tube meet (Figure 2). It can help to mark the wall of the tube for the following steps.
Carefully decant the supernatant without disturbing the pellet.
Invert the conical tube onto a paper towel and let residual media drain down.
Using a Kimwipe and a cotton swab, wipe dry the sides and bottom of the conical tube being careful not to disturb the pellet.
Add 20-50 µl of the buffer of choice. PBS works well for biochemical assays and cryo-EM imaging but interferes with negative stain TEM. HNE buffer is good for both cryo-EM and negative stain TEM imaging. Volatile buffers such as ammonium acetate (100 mM, pH 7.5) are ideal for mass spectrometry applications.
Store the resuspended virus sample at 4 °C; do not freeze. Note that over time particles may precipitate out of solution. We have stored viruses for up to 1 year and have not noticed degradation by SDS-PAGE or TEM analysis.
Figure 1. Display of CPE in BHK cells.
Cells on the left (A) are uninfected and healthy whereas those on the right (B) have been infected with Sindbis virus for 20 h. Notice the cell rounding but yet minimal cell death. Images are at the same magnification (4,000x).
Figure 2. Position of pelleted viruses in centrifugation tubes after LSC.
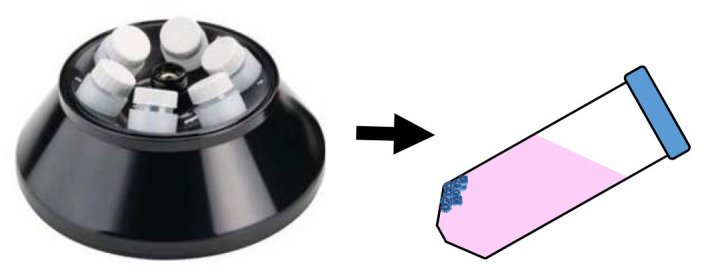
When using a fixed-angle rotor, the pelleted virus will appear as a clear, transparent pellet (blue dots) on the upper wall of the conical tube.
Data analysis
-
SDS-PAGE
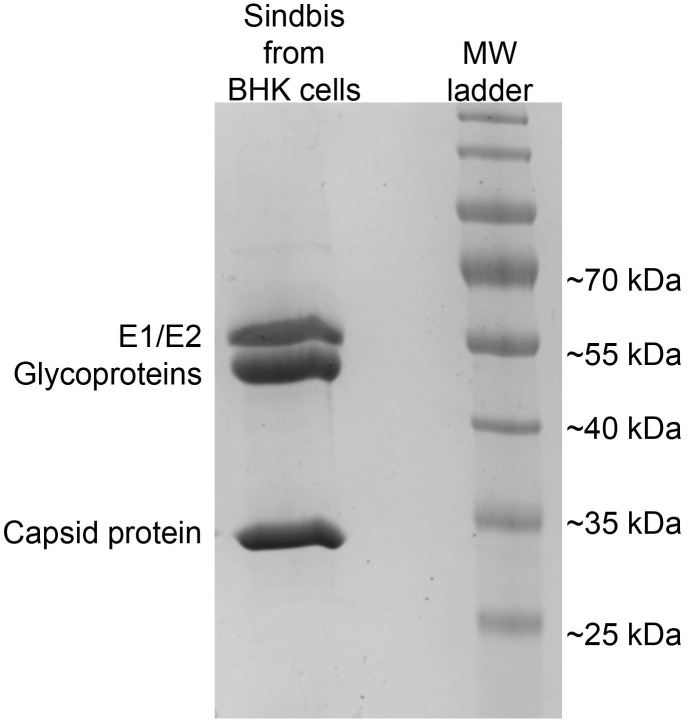
Particle purity can be assessed by SDS-PAGE (Figure 3). For best separation, we run a 10% gel prepared under standard conditions. Gels are electrophoresed at 200 V for 40-50 min and can be stained with Coomassie blue or other conventional stains.
-
TEM
Particle morphology is assessed by transmission electron microscopy (TEM) (Figure 4). Resuspended virus samples (3-5 µl) are applied to a carbon- and formvar-coated copper grid for 1 min and then blotted dry by holding a Kimwipe to the side of the grid. When blotting, place the paper at the edge of the grid in a perpendicular orientation. Thinner paper (or Kimwipes) wicks more completely than the thicker materials. Before the grid dries out (within seconds), add 5 µl of 1% uranyl acetate for 5 min. Uranyl acetate is removed by blotting with the side of the grid against a Kimwipe. Samples prepared this way can be imaged by any voltage TEM.
Figure 3. Purity of virions prepared by the LSC protocol shown by SDS-PAGE.
Sindbis virions from BHK-infected cells were purified using the LSC protocol. Three 150 mm dishes of cells were infected as described. LSC pellets were resuspended in 35 µl, and 2 µl of the resuspended pellet was loaded on the SDS-PAGE gel. The ladder is PageRuler Prestained Protein Ladder from Invitrogen. The bands seen on the gel correspond to the outer glycoproteins and the inner capsid protein and are labeled accordingly. Note, only viral proteins are observed.
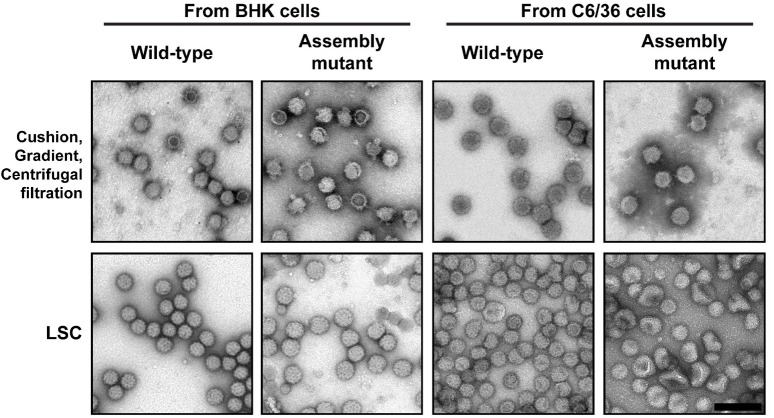
Figure 4. Purification of wild-type and assembly mutant virus.
Sindbis virions (wild-type and an assembly mutant) from both BHK-infected and C6/36-infected cells were purified using two protocols and then imaged by TEM. The top row shows the virion purification using three sequential steps: pelleting through a sucrose cushion, applying the pellet on a sucrose gradient, and buffer exchange and concentration of the viral band using centrifugal filtration. The bottom row shows the virus samples from the one step virion purification using the LSC protocol. In addition to having more particles on the grid, the LSC purified particles show fragile particles as indicated by the dent in the middle of the particle (arrow). These particles are most likely degraded or further damaged during the harsher purification methods. As a result, the diversity of assembly mutants is not represented in the upper panels. Scale bar (shown bottom right image) is 200 nm and is applicable to all EM images.
Notes
-
General notes
Each virus has a different infection MOI and harvest time. Additionally, different cell lines will also influence the optimal conditions for virus production. Performing a growth curve using your virus and host cell of interest is a good starting point for establishing purification harvest times.
After harvesting media (Step 12), if the media appears slightly acidic after harvesting the cells (Step 12) (observed by orange tint or measuring with pH paper), add sterile 1 M HEPES buffer, pH 7.5 to a final concentration of 5-10 mM to help to maintain the pH between 7-7.5.
When we want to use one purified virus preparation for multiple downstream analyses, we resuspend the low-speed centrifuged pellet in a larger volume (up to 500 µl) and then take portions of this sample for further applications.
In our laboratory, this protocol has been used for alphaviruses propagated in both BHK and C6/36 cells. We have not tried purifying viruses from other cell lines.
Recipes
-
MEM complete media
500 ml MEM
5 ml 100x L-glutamine
5 ml 100x nonessential amino acids
5 ml 100x antibiotic-antimycotic
50 ml FBS
Add all components to 500 ml bottle of MEM; the final volume is 565 ml
Store at 4 °C up to 6 months
-
Supplemented VP-SFM
1 L VP-SFM
10 ml 100x L-glutamine
10 ml 100x nonessential amino acids
10 ml 100x antibiotic-antimycotic
Store at 4 °C up to 6 months
Add all components to 1 L bottle of VP-SFM; the final volume is 1,030 ml
-
HEPES-NaCl-EDTA (HNE) Resuspension buffer (pH = 7.5)
20 mM HEPES
150 mM NaCl
0.1 mM EDTA
Adjust pH using HCl
Store at room temperature
-
1% uranyl acetate
Make a 10% solution (Vf = 1 ml) by adding 0.1 g uranyl acetate to 1 ml of MilliQ water. Invert to mix. The solution will be yellow
To make 1% solution (Vf = 1 ml), take 100 µl of 10% solution and add 900 µl of MilliQ water. Invert to mix
Store in the dark at room temperature
Note: Uranyl acetate waste should be disposed of according to the institutional policy.
Acknowledgments
This protocol was adapted from the previous publications Atasheva et al., 2013 and Moller- Tank et al., 2013 . It has been used in Wang et al., 2015 and Ramsey et al., 2017 . We acknowledge members of the Mukhopadhyay laboratory who shared purified alphavirus virions for our analyses. Funding for this work was from the National Science Foundation and Indiana University Office of the Vice Provost for Research. The authors declare no conflicts of interest or competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Atasheva S., Kim D. Y., Akhrymuk M., Morgan D. G., Frolova E. I. and Frolov I.(2013). Pseudoinfectious Venezuelan equine encephalitis virus: a new means of alphavirus attenuation. J Virol 87(4): 2023-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moller-Tank S., Kondratowicz A. S., Davey R. A., Rennert P. D. and Maury W.(2013). Role of the phosphatidylserine receptor TIM-1 in enveloped-virus entry. J Virol 87(15): 8327-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ramsey J., Renzi E. C., Arnold R. J., Trinidad J. C. and Mukhopadhyay S.(2017). Palmitoylation of Sindbis virus TF protein regulates its plasma membrane localization and subsequent incorporation into virions. J Virol 91(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang J. C., Chen C., Rayaprolu V., Mukhopadhyay S. and Zlotnick A.(2015). Self-assembly of an alphavirus core-like particle is distinguished by strong intersubunit association energy and structural defects. ACS Nano 9(9): 8898-8906. [DOI] [PMC free article] [PubMed] [Google Scholar]