Abstract
Mammalian milk including microRNAs (miRNAs) as a novel class of noncoding RNAs, that can be transferred to infants and it plays on a critical role in biological functions such as immune regulation and development. However, the origin and functional importance of milk-derived miRNAs are still undetermined. This study applied RNA sequencing to explore the featured profiles of miRNA expression in colostrum and mature milk-originated exosomes from human, bovine, and caprine milk. These dietary exosome-derived miRNAs are highly conserved in human, bovine and caprine milk. Interestingly, abundant miRNAs expressed in human milk are similarly conserved across species. In addition, we confirmed that immune-related miRNAs (miR-30a-5p, miR-22-3p, and miR-26a) are commonly observed in the colostrum and mature milk of cows and caprines as well as humans. Our results provide new insights and resources for investigating the functionality of immune-associated miRNAs and evaluating physiological and biological condition in human, bovine and caprine milk as biomarkers.
Keywords: Colostrum, Mature milk, Dietary exosome, MicroRNA, Mammals
INTRODUCTION
Milk is a major nutritional source for infants, and breastfeeding is one of the most valuable participants to infant health [1]. In addition, breastfeeding reduces the risk of acute infant-related diseases by providing an infant with a developmental program and protection against infection [2–4]. The components of breast milk can contribute the immune response of newborns, and they influence on the development of infant and children’s immune systems. However, the mechanisms of immune modulation by breast milk are still unclear and might predicted from the role of biological macromolecules existed in breast milk including antibodies, growth factors, and nucleic acids [5]. An additional unique bioactive component of breast milk recently discovered is microRNA (miRNA) [6,7].
Since 1993 discovered on their importance, it has been well-established that miRNAs composed with 18–22 nucleotides in length as a small class of noncoding RNAs (ncRNAs), principally contribute as post-transcriptional regulators of target gene expression by RNA interference [8]. In particular, they have served as key players of gene regulation at the post-transcriptional level in humans, animals and plants [9–11]. In addition, a variety of miRNAs are present in body fluidic system contacting tears, saliva, plasma, and urine as well as commonly in breast milk [12]. Exosomes are membrane-bound extracellular vesicles (EVs) with a diameter of 40 to 100 nm that are released from the cells into the extracellular space after being formed within multivesicular bodies in the endosome system [13]. The exosomal cargo is genetic material including DNA, mRNA, miRNA, proteins and lipids [14]. In particular, they are abundant in various type of RNA including coding RNA and ncRNA [8]. Interestingly, previous studies indicated that exosomes are rich in breast milk and contains miRNAs related to the immune system as well as the nutrition of newborns [8,15,16]. Exosomes are continuously secreted into mammary fluidic system (e.g., mature milk and colostrum), and they act a key player for intracellular communication by carrying their contents (e.g., miRNA) to target cells [17,18]. Initially, existence of milk-derived exosomes were reported from human breast milk [19] and later in milk and colostrum from animals such as cows [20], caprines [21], and pigs [15]. Previous studies have characterized and investigated on a number of miRNAs that originated from milk exosomes of various mammals [6,8,22]. However, less information is available on the identification of miRNAs in exosomes derived from milk components present in various mammals. Identification of miRNAs in exosomes isolated from colostrum and mature milk from various mammals can be useful for exploring biological functionality related to the newborn’s immune system. The purpose of this study was to identify profiling of miRNAs and characterize their genetic diversities in exosomes derived from the colostrum and mature milk of three different species of mammals.
MATERIALS AND METHODS
Milk samples
Bovine colostrum and mature milk samples were collected from 6 healthy Holstein cows (for colostrum, it was taken from the cows within 3 days postpartum, n = 3; for mature milk, it was taken from cows at 30 days postpartum, n = 3). In the same manner, caprine colostrum and mature milk samples were also obtained from the Saanen breed for colostrum (n = 3) and mature milk (n = 3). Moreover, human breast milk samples were collected from two volunteers (Jeonju, Jeollabuk-do, Korea). Human breast colostrum and mature milk samples were collected from healthy mothers during the first 3 days and 30 days after delivery, respectively. Each milk sample was collected, and then the collected samples totaling 50–100 mL were put into storage bags. All milk samples were frozen immediately after collection and were kept at −80°C until use. All procedures in this study were approved by the Investigational Review Board (IRB) of Jeonju University (jjIRB-2020-1205).
Isolation of dietary exosome from the milk samples
Isolation of exosomes from the milk was performed using established methods in previous studies with minor modifications [23]. Briefly, the milk samples were centrifuged at 1,500×g and 4°C for 30 min to remove fat, cells, and debris. The supernatant was defatted at 16,500×g for 1 h at 4°C to remove the casein. The casein-removed supernatant (whey) was then passed through 0.45- and 0.22-μm filters to remove residual cell debris. The clear supernatant was carefully collected to avoid contamination of the pelletized exosomes. The pelletized exosomes were resuspended in phosphate-buffered saline (PBS), followed by ultracentrifugation for washing (100,000×g, 4°C, 90 min). After washing twice with additional ultracentrifugation (135,000×g, 4°C, 90 min), the exosomes were reconstituted in PBS.
Extraction of miRNA and small RNA-sequencing
Extraction of miRNA from the milk exosomes was performed using established methods in previous studies with minor modifications [24]. Briefly, miRNA was extracted from milk exosomes using combined phenol and column methods: phenol methods using Qiazol® (Qiagen, Hilden, Germany) and column methods using the miRNeasy® Mini kit (miRNeasy I, Qiagen). All methods were employed according to the manufacturer’s instruction. The miRNAs extracted were re-suspended with 50 µL of nuclease-free water. The integrity of the miRNAs as well as quality and quantity were determined on an Agilent 2100 Bioanalyzer using an RNA 6000 Pico Kit (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s manual. The sequencing service and library preparation were provided by Macrogen, Korea. All libraries were sequenced using the HiSeq platform (Illumina).
Bioinformatics analysis
Low quality reads were removed from the raw data, high quality reads were then subjected to sRNAbench for adapter sequence trimming and length distribution analysis read. The sequences with read length larger than 15 nt were aligned against human, bovine, and caprine miRNA database (miRBase, release ver. 22) [25] with the default parameters to identify known miRNAs using sRNAbench and did not allow any discrepancies. Each library was processed separately, and the expression level of miRNA was estimated by sRNAbench, which normalized the number of reads for each miRNA RPM. The miRNA unique sequences were applied to investigate the level of the miRNA expression based on the normalized read count.
Statistical analysis
All data are expressed as the means ± SD. Statistical analysis was performed by one-way analysis of variance (ANOVA). A value of p < 0.05 was considered statistically significant.
RESULTS AND DISCUSSION
Comparison of miRNAs in exosomes from human breast colostrum and mature milk
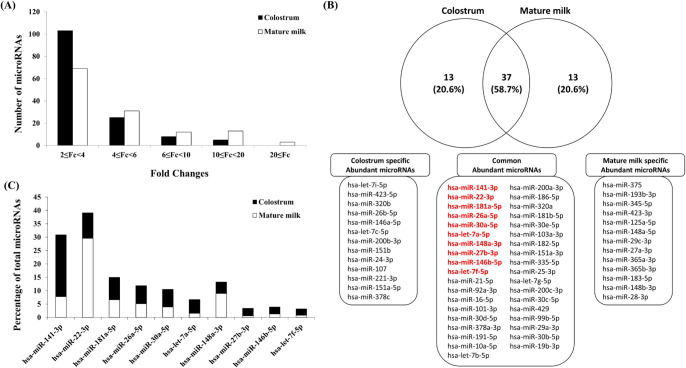
Initially, human breast milk was profiled for miRNA using a small RNA sequencing method to compare colostrum collected within 3 days after delivery and mature milk collected 30 days after delivery. The comparison of expression between the exosomes of human colostrum and mature milk was performed mainly on read count, and similar to cow milk, it was confirmed that the majority of the increased miRNAs had 2–4 times increased expression (Fig. 1A). In particular, it was confirmed that there were many miRNAs with increased expression of 2 to 4 times or less in the exosomes of human colostrum and that there were many miRNAs with increased expression of 4 times or more in the exosomes of mature human milk. The top 50 miRNAs most abundant in each exosome of human colostrum and mature milk are plotted in Fig. 1B. As a result, 37 out of 50 (58.7% of each sample) were found in common, among which the top 6 miRNAs in exosomes of human colostrum were included. When comparing the ratios of the top 10 common miRNAs in exosomes of human colostrum and mature milk, it was confirmed that they existed at different ratios (Fig. 1C). In the case of exosomes of human colostrum, it was confirmed that hsa-miR-22-3p was the most abundant, and in the case of exosomes of mature milk, it was confirmed that hsa-miR-141-3p was the most abundant. The identified miRNAs existed abundantly in human milk previous studies is consistent with results from previous studies [26,27]. Of the unique miRNAs most highly expressed in several studies [26,27], 9 miRNAs (except hsa-miR-27b-3p) were also found in the top 10 group in our study. This small but notable difference can be attributed to different ranking approaches, but it also implies a human-specific difference in the relative abundance of human milk exosome miRNAs. Additionally, based on results described previously [26–29], immune-related miRNAs were compared with those identified in our study. Among the top 50 miRNAs, 29 (58%) and 25 (50%) immune-related miRNAs were present in colostrum and mature milk, respectively. In particular, there were 23 (62.16%) immune-related miRNAs in abundant miRNAs common to both colostrum and mature milk. These characterized miRNAs reinforce the idea that immune-associated miRNAs are abundant in breast milk exosomes, suggesting that more of these miRNAs could be detected.
Fig. 1. MicroRNAs expressed in exosomes derived from human breast colostrum and mature milk.
(A) Expression patterns of miRNAs expressed in human colostrum and mature milk. (B) Venn diagram depicting miRNAs expressed in human colostrum and mature milk. (C) Abundance profile of miRNAs in percentage (%) for each miRNA of colostrum and mature milk in the top 10 miRNAs read (indicated with red) by next generation sequencing (NGS).
Comparison of miRNAs in exosomes from bovine colostrum and mature milk
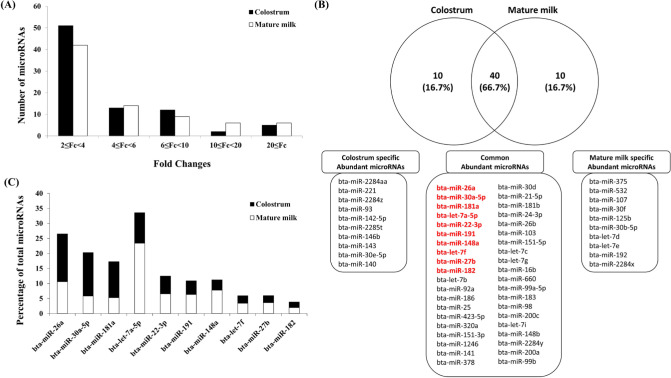
After collecting the colostrum and mature milk, it was quickly frozen at −20°C and quickly transferred to the laboratory, and then exosomes were isolated and stored at −80°C. The miRNAs were isolated from exosomes isolated from bovine colostrum and mature milk. In the comparison of expression, miRNAs with 2- to 4-fold increased expression were the most abundant (Fig. 2A). In the exosomes of bovine colostrum, bta-miR-221 showed up to 48-fold expression compared to bovine mature milk, and in the exosomes of bovine mature milk, bta-miR-375 showed up to a 23-fold increase in expression. These miRNAs are thought to be some of the miRNAs showing functional differences between bovine colostrum and mature milk. The top 50 miRNAs, most abundantly present in bovine colostrum and mature milk exosomes, respectively, are plotted in Fig. 2B using a Venn diagram. When comparing the top miRNAs in the bovine colostrum and mature milk exosomes, there were around 40 miRNAs (66.7% of each sample) in common, among which all of the top 10 miRNAs in the colostrum were included. In mature milk exosomes, 9 of them were included in the top 10. Colostrum-specific and mature milk-specific miRNAs were present in each of the 10 samples, and the corresponding lists are shown in Fig. 1B. The proportions in each sample of the top 10 miRNAs were different (Fig. 2C), indicating that there is potential for functional differences between bovine colostrum and mature milk. The overall ratio of the top 10 miRNAs was approximately 73% in bovine colostrum and approximately 75% in mature milk. There were 4 immune-related miRNAs (bta-miR-26a, bta-181a, bta-let-7a-5p, bta-miR-191, and bta-miR-27b) in the top 10 miRNAs among the common abundant miRNAs in colostrum and mature milk-derived exosomes [15,30]. In addition, among the many miRNAs common to colostrum and mature exosomes, bta-miR-26a, the most abundant miRNA in colostrum, and bta-let-7a-5p, the most abundant miRNA in mature milk, are immune-related miRNAs. Among the common top 10 miRNAs, 7 miRNAs (bta-miR-26a, bta-miR-181a, bta-let-7a-5p, bta-miR-191, bta-miR-148a, bta-let-7f, and bta-miR-27b) were abundant in milk, as shown in previous studies [15,30,31]. The shared 7 miRNAs might be associated with fundamental functions of bovine milk.
Fig. 2. MicroRNA expressed in exosomes from bovine colostrum and mature milk.
(A) Expression patterns of miRNAs expressed in bovine colostrum and mature milk. (B) Venn diagram depicting miRNAs expressed in bovine colostrum and mature milk. (C) Abundance profile of miRNAs in percentage (%) for each miRNA of colostrum and mature milk in the top 10 miRNAs read (indicated with red) by next generation sequencing (NGS).
Comparison of miRNAs in exosomes from caprine colostrum and mature milk
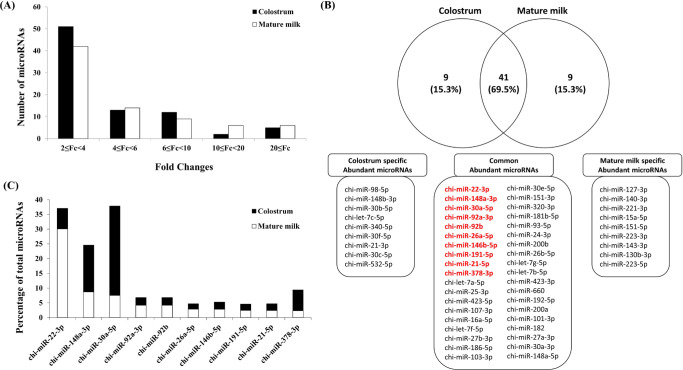
Caprine milk was also profiled for miRNA using a small RNA sequencing method to compare colostrum collected within 3 days after delivery and mature milk collected 1 month after delivery. As a result of comparing the miRNA expression of caprine colostrum and mature milk exosomes, miRNAs with increased expression 2 to 4 times similar to cow and human milk exosomes accounted for the majority (Fig. 3A). The top 50 miRNAs, which are most abundant in caprine colostrum and mature milk exosomes, are plotted in a Venn diagram (Fig. 3B). As a result, among the top 50 miRNAs, 41 (69.5% of each sample) were common, and among them, all of the top 10 miRNAs in the exosomes of caprine mature milk were included. When comparing the ratio of each sample of the top 10 miRNAs, it was confirmed that 3 (chi-miR-148a-3p, chi-miR-30a-5p and chi-miR-378-3p) of the top 10 miRNAs were more abundant in colostrum, and the remaining 7 were more abundant in mature milk (Fig. 3C). Of the top 10 miRNAs most highly expressed in caprine milk in previous studies [5,32], six miRNAs (chi-miR-21-5p, chi-miR-92a-3p, chi-miR-92b, chi-miR148a-3p, chi-miR-146b-5p, and chi-miR-30a-5p) were included in the top 10 groups in our study. In addition, miR-148a is reported to be one of the most highly expressed miRNAs in caprine milk before and after pasteurization [5], and this study also found high levels of miR-148a. Interestingly, three of the four miRNAs (chi-miR-27b, chi-miR-103, and chi-miR-200a) reported to be involved in milk fat synthesis during caprine lactation were included in colostrum and mature milk [33]. In addition, two of the five miRNAs (chi-miR-146 and chi-miR-223) reported as immune-related miRNAs in caprine milk were identified in this study [34].
Fig. 3. MicroRNA expressed in exosomes from caprine colostrum and mature milk.
(A) Expression patterns of miRNAs expressed in caprine colostrum and mature milk. (B) Venn diagram depicting miRNAs expressed in human colostrum and mature milk. (C) Abundance profile of miRNAs in percentage (%) for each miRNA of colostrum and mature milk in the top 10 miRNAs read (indicated with red) by next generation sequencing (NGS).
Comparative analysis of miRNAs from human, bovine and caprine mature and colostrum milk-derived exosomes
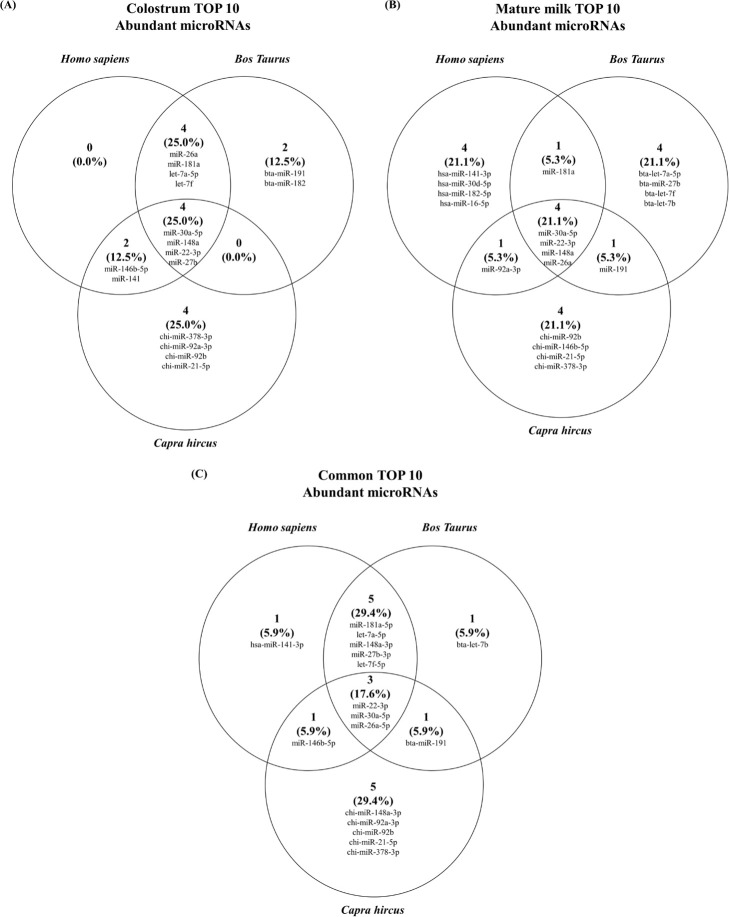
The top 10 miRNAs abundant in milk exosome-derived miRNAs for each species were investigated (Fig. 4). First, in colostrum exosome-derived miRNAs, a total of 4 miRNAs were common in humans, bovines and caprines (Fig. 4A). In addition, humans and bovines had a total of 8 miRNAs in common, and humans and caprines had 6 miRNAs in common. However, in the case of bovines and caprines, only four miRNAs existed in common with humans. In the case of mature milk, a total of 4 miRNAs were common in humans, bovines and caprines, similar to the colostrum-derived miRNAs (Fig. 4B). Humans and bovines, humans and caprines, and bovines and caprines each had 5 common miRNAs. Next, in miRNAs derived from both colostrum and mature milk, a total of 3 miRNAs existed in common from humans, bovines, and caprines. Interestingly, we found that milk-derived exosomes of bovines and humans have the most miRNAs (8 miRNAs) in common. These results propose that miRNAs in human and bovine milk are more similar than those in caprine milk.
Fig. 4. Comparative analysis of exosome-derived microRNA in human, bovine and caprine colostrum and mature milk.
(A) Venn diagram depicting the top 10 miRNAs expressed in human, bovine and caprine colostrum and mature milk. (B) Venn diagram depicting the top 10 miRNAs expressed in mature milk of humans, bovines and caprines. (C) Venn diagram depicting the top 10 miRNAs expressed in common in colostrum and mature milk from humans, bovines and caprines.
Bovine and caprine milk are most widely applied as dietary dairy products, and it is important to determine their levels of miRNA expression, as it is a major component of infant formula [5]. Milk exosomal miRNAs are transferred from humans to humans as well as from livestock milk to influence on the expression of essential genes in target cells and tissues [35,36]. We showed in this study that highly expressed milk-derived miRNAs are primarily immune-related miRNAs [5,15,26–30,32]. In human colostrum, 6 miRNAs (miR-26a, miR-181a, let-7a-5p, miR-30a-5p, miR-22-3p, and miR-141) of the top 10 miRNAs are related to immunity, and 2 of them are common to cow and caprine colostrum. In human mature milk, 5 miRNAs (miR-141-3p, miR-181a, miR-30a-5p, miR-22-3p, and miR-26a) in the top 10 were related to immunity, and 3 of them were common to cow and caprine milk. Interestingly, let-7a-5p, a miRNA that was relatively less expressed in human milk, was more abundant in cows and there was more miR-146b-5p in caprines. Among the miRNAs that are commonly expressed in human colostrum and mature milk, 7 miRNAs (including miR-22-3p, miR-30a-5p, and miR-26a-5p) of the top 10 miRNAs were associated with immunity, and 3 of them were identified as common between cattle and caprines. Interestingly, we indicated that the miRNAs that are highly expressed in human milk are similarly conserved across species. Milk is an abundant source of exosomal miRNAs that are evolutionarily conserved among mammals. Therefore, functional studies of other species of highly conserved exosomal miRNAs can be used to a variety of mammalian milk samples.
Taken together, we found that miR-30a-5p, miR-22-3p, and miR-26a are highly conserved in colostrum and mature milk in cows, caprines, and humans, suggesting their possible importance in neonatal growth. Our results provide new insights and resources for discovering the noble functionality of immune-associated miRNAs from dietary exosome [37] and for developing infant formulas taking into account the differences on miRNA expression in human, bovine and caprine milk as well as evaluating biological features as diagnostic biomarkers.
Acknowledgements
Not applicable.
Competing interests
No potential conflict of interest relevant to this article was reported.
Funding sources
This research was supported by the High Value-Added Food Technology Development Program of the Korean Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry, and Fisheries (iPET), the Ministry for Food, Agriculture, Forestry, and Fisheries of the Korea (318090-03-1-WT011) and the National Research Foundation of Korea Grant funded by the Korean government (NRF-2018R1D1A3B07050304).
Availability of data and material
Upon reasonable request, the datasets of this study can be available from the corresponding author.
Authors’ contributions
Conceptualization: Yun B, Kim Y, Park DJ, Oh S.
Data curation: Yun B, Kim Y, Park DJ, Oh S.
Formal analysis: Yun B, Kim Y, Park DJ, Oh S.
Methodology: Yun B, Kim Y, Park DJ, Oh S.
Software: Yun B, Kim Y, Park DJ, Oh S.
Validation: Yun B, Kim Y, Park DJ, Oh S.
Investigation: Yun B, Kim Y, Park DJ, Oh S.
Writing - original draft: Yun B, Kim Y, Park DJ, Oh S.
Writing - review & editing: Yun B, Kim Y, Park DJ, Oh S.
Ethics approval and consent to participate
All procedures in this study were approved by the Investigational Review Board (IRB) of Jeonju University (jjIRB-210215-HB-2021-0212).
REFERENCES
- 1.Hoddinott P, Tappin D, Wright C. Breast feeding. BMJ. 2008;336:881–7. doi: 10.1136/bmj.39521.566296.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alsaweed M, Lai CT, Hartmann PE, Geddes DT, Kakulas F. Human milk miRNAs primarily originate from the mammary gland resulting in unique miRNA profiles of fractionated milk. Sci Rep. 2016;6:1–13. doi: 10.1038/srep20680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassiotou F, Geddes DT. Programming of appetite control during breastfeeding as a preventative strategy against the obesity epidemic. J Hum Lact. 2014;30:136–42. doi: 10.1177/0890334414526950. [DOI] [PubMed] [Google Scholar]
- 4.Newburg DS, Walker WA. Protection of the neonate by the innate immune system of developing gut and of human milk. Pediatr Res. 2007;61:2–8. doi: 10.1203/01.pdr.0000250274.68571.18. [DOI] [PubMed] [Google Scholar]
- 5.Golan-Gerstl R, Elbaum Shiff Y, Moshayoff V, Schecter D, Leshkowitz D, Reif S. Characterization and biological function of milk-derived miRNAs. Mol Nutr Food Res. 2017;61:1700009. doi: 10.1002/mnfr.201700009. [DOI] [PubMed] [Google Scholar]
- 6.Hata T, Murakami K, Nakatani H, Yamamoto Y, Matsuda T, Aoki N. Isolation of bovine milk-derived microvesicles carrying mRNAs and microRNAs. Biochem Biophys Res Commun. 2010;396:528–33. doi: 10.1016/j.bbrc.2010.04.135. [DOI] [PubMed] [Google Scholar]
- 7.Izumi H, Kosaka N, Shimizu T, Sekine K, Ochiya T, Takase M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J Dairy Sci. 2012;95:4831–41. doi: 10.3168/jds.2012-5489. [DOI] [PubMed] [Google Scholar]
- 8.Özdemir S. Identification and comparison of exosomal microRNAs in the milk and colostrum of two different cow breeds. Gene. 2020;743:144609. doi: 10.1016/j.gene.2020.144609. [DOI] [PubMed] [Google Scholar]
- 9.Alsaweed M, Hartmann PE, Geddes DT, Kakulas F. MicroRNAs in breastmilk and the lactating breast: potential immunoprotectors and developmental regulators for the infant and the mother. Int J Environ Res Public Health. 2015;12:13981–4020. doi: 10.3390/ijerph121113981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Lee RC, Feinbaum RL, Ambros V, The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 12.Alsaweed M, Hepworth AR, Lefèvre C, Hartmann PE, Geddes DT, Hassiotou F. Human milk microRNA and total RNA differ depending on milk fractionation. J Cell Biochem. 2015;116:2397–407. doi: 10.1002/jcb.25207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng L, Sun X, Scicluna BJ, Coleman BM, Hill AF. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 2014;86:433–44. doi: 10.1038/ki.2013.502. [DOI] [PubMed] [Google Scholar]
- 14.Li SP, Lin ZX, Jiang XY, Yu XY. Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta Pharmacol Sin. 2018;39:542–51. doi: 10.1038/aps.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen T, Xi QY, Ye RS, Cheng X, Qi QE, Wang SB, et al. Exploration of microRNAs in porcine milk exosomes. BMC Genomics. 2014;15:100. doi: 10.1186/1471-2164-15-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shandilya S, Rani P, Onteru SK, Singh D. Small interfering RNA in milk exosomes is resistant to digestion and crosses the intestinal barrier in vitro. J Agric Food Chem. 2017;65:9506–13. doi: 10.1021/acs.jafc.7b03123. [DOI] [PubMed] [Google Scholar]
- 17.Pua HH, Happ HC, Gray CJ, Mar DJ, Chiou NT, Hesse LE, et al. Increased hematopoietic extracellular RNAs and vesicles in the lung during allergic airway responses. Cell Rep. 2019;26:933–44. doi: 10.1016/j.celrep.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu H, Ling M, Xue J, Dai X, Sun Q, Chen C, et al. Exosomal microRNA-21 derived from bronchial epithelial cells is involved in aberrant epithelium-fibroblast cross-talk in COPD induced by cigarette smoking. Theranostics. 2018;8:5419–33. doi: 10.7150/thno.27876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Admyre C, Johansson SM, Qazi KR, Filén JJ, Lahesmaa R, Norman M, et al. Exosomes with immune modulatory features are present in human breast milk. J Immunol. 2007;179:1969–78. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- 20.Reinhardt TA, Lippolis JD, Nonnecke BJ, Sacco RE. Bovine milk exosome proteome. J Proteomics. 2012;75:1486–92. doi: 10.1016/j.jprot.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 21.Li Z, Lan X, Guo W, Sun J, Huang Y, Wang J, et al. Comparative transcriptome profiling of dairy goat microRNAs from dry period and peak lactation mammary gland tissues. PLOS ONE. 2012;7:e52388. doi: 10.1371/journal.pone.0052388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Herwijnen MJC, Driedonks TAP, Snoek BL, Kroon AMT, Kleinjan M, Jorritsma R, et al. Abundantly present miRNAs in milk-derived extracellular vesicles are conserved between mammals. Front Nutr. 2018;5:81. doi: 10.3389/fnut.2018.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maburutse BE, Park MR, Oh S, Kim Y. Evaluation and characterization of milk-derived microvescicle isolated from bovine colostrum. Korean J Food Sci Anim Resour. 2017;37:654–62. doi: 10.5851/kosfa.2017.37.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh S, Park MR, Son SJ, Kim Y. Comparison of total RNA isolation methods for analysis of immune-related microRNAs in market milks. Korean J Food Sci Anim Resour. 2015;35:459–65. doi: 10.5851/kosfa.2015.35.4.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao Y, Du X, Li J, Lönnerdal B. Human milk exosomes and their microRNAs survive digestion in vitro and are taken up by human intestinal cells. Mol Nutr Food Res. 2017;61:1700082. doi: 10.1002/mnfr.201700082. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q, et al. Immune-related microRNAs are abundant in breast milk exosomes. Int J Biol Sci. 2012;8:118–23. doi: 10.7150/ijbs.8.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:343–51. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 29.Sonkoly E, Ståhle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18:131–40. doi: 10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Quan SY, Nan XM, Wang K, Zhao YG, Jiang LS, Yao JH, et al. Replacement of forage fiber with non-forage fiber sources in dairy cow diets changes milk extracellular vesicle-miRNA expression. Food Funct. 2020;11:2154–62. doi: 10.1039/C9FO03097B. [DOI] [PubMed] [Google Scholar]
- 31.Kosaka N, Izumi H, Sekine K, Ochiya T. MicroRNA as a new immune-regulatory agent in breast milk. Silence. 2010;1:7. doi: 10.1186/1758-907X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou J, An X, Song Y, Cao B, Yang H, Zhang Z, et al. Detection and comparison of microRNAs in the caprine mammary gland tissues of colostrum and common milk stages. BMC Genetics. 2017;18:1–8. doi: 10.1186/s12863-017-0498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin X, Luo J, Zhang L, Zhu J. MicroRNAs synergistically regulate milk fat synthesis in mammary gland epithelial cells of dairy goats. Gene Expr J Liv Res. 2013;16:1–13. doi: 10.3727/105221613X13776146743262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Y. Expressional analysis of immune-related miRNAs in breast milk. Genet Mol Res. 2015;14:11371–6. doi: 10.4238/2015.September.25.4. [DOI] [PubMed] [Google Scholar]
- 35.Baier SR, Nguyen C, Xie F, Wood JR, Zempleni J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J Nutr. 2014;144:1495–500. doi: 10.3945/jn.114.196436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen T, Xie MY, Sun JJ, Ye RS, Cheng X, Sun RP, et al. Porcine milk-derived exosomes promote proliferation of intestinal epithelial cells. Sci Rep. 2016;6:33862. doi: 10.1038/srep33862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yun B, Maburutse BE, Kang M, Park MR, Park DJ, Kim Y, et al. Short communication: dietary bovine milk-derived exosomes improve bone health in an osteoporosis-induced mouse model. J Dairy Sci. 2020;103:7752–60. doi: 10.3168/jds.2019-17501. [DOI] [PubMed] [Google Scholar]