Abstract
Purpose
Electrocardiogram (ECG) is one of the most essential tools for detecting heart problems. Till today most of the ECG records are available in paper form. It can be challenging and time-consuming to manually assess the ECG paper records. Hence, automated diagnosis and analysis are possible if we digitize such paper ECG records.
Methods
The proposed work aims to convert ECG paper records into a 1-D signal and generate an accurate diagnosis of heart-related problems using deep learning. Camera-captured ECG images or scanned ECG paper records are used for the proposed work. Effective pre-processing techniques are used for the removal of shadow from the images. A deep learning model is used to get a threshold value that separates ECG signal from its background and after applying various image processing techniques threshold ECG image gets converted into digital ECG. These digitized 1-D ECG signals are then passed to another deep learning model for the automated diagnosis of heart diseases into different classes such as ST-segment elevation myocardial infarction (STEMI), Left Bundle Branch Block (LBBB), Right Bundle Branch Block (RBBB), and T-wave abnormality.
Results
The accuracy of deep learning-based binarization is 97%. Further deep learning-based diagnosis approach of such digitized paper ECG records was having an accuracy of 94.4%.
Conclusions
The digitized ECG signals can be useful to various research organizations because the trends in heart problems can be determined and diagnosed from preserved paper ECG records. This approach can be easily implemented in areas where such expertise is not available.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40846-021-00632-0.
Keywords: Paper ECG, Digitization, Deep learning, Diagnosis
Introduction
Digital technologies have revolutionalized signal analysis and automated diagnosis. It is essential to convert biomedical signals, such as electro-myogram (EMG), electro-cardiograms (ECG), and electro-encephalograms (EEG) into digital form, for such computerized investigations. Digitized signals have advantages, such as security, easy to store, transmit, and retrieve. In this manuscript, we focus on ECG digitization and automated diagnosis using it. A doctor checks a patient’s ECG data to determine the heart-related problems of the patient. The data can be extracted from these records, and a doctor can diagnose whether the person has a heart problem or had faced any complications in the past. All these things can be automated when all the ECG signals are in digital format. The ECG signal is a plot of voltage on the Y-axis against time on the X-axis. Once the ECG machine is on, it begins to record the activity of the heart, and data is presented as a zigzag graph that the doctors evaluate. If we digitize ECG paper records then mathematical operations can be done using them. We can pre-filter, remove the excess noise, and feed it to the featured deep learning network. Nowadays, digital ECG recorders are available, wherein the data is stored on a compact disk (CD) as a picture file but they are extremely expensive. Hence, the traditional ECG recorders that give a print on the graph paper are widely used. ECG monitors are used too which do not store the data. We aim to make ECG diagnosis available to common people so that just by clicking the photo of a paper ECG record on their smartphone, the patient should able to get a diagnosis report without personally going to the doctor.
Several studies and research work have been reported in the literature for ECG digitization and conversion of ECG paper records to 1-D signals. Binarization from degraded document images is studied by Biswas et al. [1]. A fine image is produced in the end by blurring the degraded images using a gaussian filter. Another technique of binarization of degraded images through contrasting, was carried out by Su et al. [2]. A contrast map was first constructed and then binarized. This binary image was then combined with Canny’s edge map to identify the text stroke edge pixels. Sauvola et al. [3] worked on image binarization by considering the page as a collection of subcomponents such as text, background, and picture. Two algorithms were applied to determine a local threshold for each pixel. Swamy et al. [4] proposed an algorithm for the existing ECG paper trace to digital time series with adaptive and image processing techniques. Also, the proposed technique was enhanced to calculate the heart rate from the obtained time series with an accuracy of 95%.
The importance of digitalization was studied and Mallawaarachchi et al. [5] proposed several tools for ECG extraction while maintaining a minimum user involvement requirement. The proposed method was tested on data of 550 trace snippets and comparative analysis showed an average accuracy of 96%. Waits et al. [6] mainly focused on the problems caused by the degraded ECG records. They developed a method called perceptual spectral centroid for solving this problem by correctly predicting the ECG signal with good accuracy. Mitra et al. [7] studied the automated data extraction system for converting ECG paper records to digital time databases. They performed the Fourier transform of the generated database and saw its response properties for every ECG signal. The process for binarizing and enhancing degraded documents was done by Gatos et al. [8]. It could deal with degradations that occur due to shadows, non-uniform illumination, low contrast, large signal-dependent noise, smear, and strain.
The degradation of ECG records and inefficiency in storing previous records was studied by Rupali et al. [9]. The authors have used the entropy-based bit plane slicing (EBPS) algorithm for extracting digital ECG records from the degraded ECG paper. The proposed method could also help in retrospective cardiovascular analysis. Jayaraman et al. [10] designed a technique for ECG morphology interpretation and arrhythmia detection based on time series. In this method, binarization of images was done through image processing and data acquisition. A conversion of ECG signal from ECG strips or papers was done by Kumar et al. [11]. The ECG strips are scanned and then using MATLAB, the data was obtained for the ECG taken from Indian patients, with an accuracy of 99%.
Damodaran et al. [12] suggested the extraction of ECG morphological features from ECG paper. The algorithm evaluated 25 patient’s ECG data from 12-lead ECG equipment and further enhanced the accuracy of the heart rate signal. The authors achieved an accuracy of 99%. A method based on K-means was proposed by Shi et al. [13] to extract ECG data from paper recordings of 105 patients. The recordings had different degradations and the precision rate of the approach was 99%. Chebil et al. [14] proposed some improvements to the existing digitization process by selecting appropriate image resolution during scanning and using neighborhood and median approach for the extraction and digitization of the ECG waveform. Badilini et al. [15] described an image processing engine that first detects the underlying grid and then extrapolates the ECG waveforms using a technique based on active contour modeling. Ravichandran et al. [16] designed a MATLAB-based tool to convert ECG information from paper charts into digital ECG signals. The conversion was performed by detecting the graphical grid on ECG charts using grayscale thresholding. After that, the ECG signal was digitized based on its contour and a template-based optical character recognition was used to extract patient demographic information from the paper records.
Sao et al. [17] analyzed and classified an ECG signal using various artificial neural network (ANN) algorithms. They compared all the algorithms with one another to classify 12 Lead ECGs according to the disease. Lyon et al. [18] reviewed the different computational methods used for ECG analysis, mainly focusing on machine learning and 3D computer simulations, their accuracy, clinical implications, and contributions to medical advances. A survey on the current state-of-the-art methods of ECG-based automated abnormalities heartbeat classification was done by Luz et al. [19]. The ECG signal pre-processing, the heartbeat segmentation techniques, the feature description methods, and the learning algorithms used were compared in the literature.
A Deep Neural Network (DNN) for the classification of the heartbeat was done by Sannino et al. [20] using Tensor Flow and Google deep learning library. Deep learning techniques have been implemented in the diagnosis of cardiac arrhythmia using ECG signals with minimal possible data pre-processing by Swapna et al. [21]. An end-to-end approach for addressing QRS complex detection and measurement of Electrocardiograph (ECG) paper using convolutional neural networks (CNNs) was done by Yu et al. [22]. An algorithm to extract ECG signals automatically from scanned 12 lead ECG paper recordings by operations including edge detection, image binarization, and skew correction was designed by Sun et al. [23].
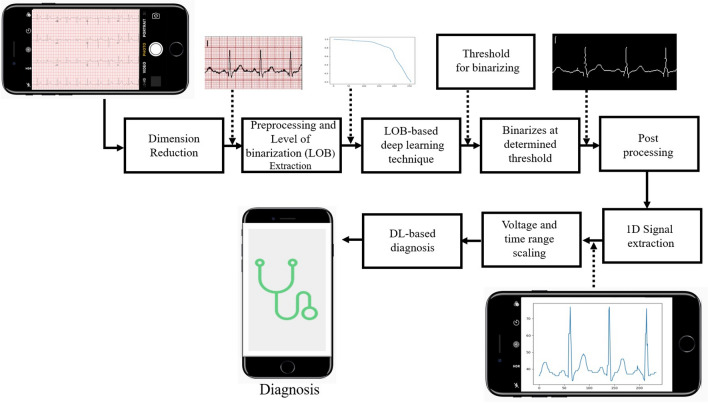
As shown in Fig. 1, first we convert 12 lead paper ECG record, which is generally available in hospitals to a single lead. This single-lead ECG signal goes through a pre-processing phase for image enhancement. In pre-processing, shadow removal is also done for camera-captured images. Then by brute force technique, we check the threshold value and plot a characteristic curve (graphical plot of level of binarization (LOB) and normalized sum (NS)) for each grey value from 0 to 255. Then the signal is fed to the ML model that was trained using the characteristic curves and corresponding threshold values to predict the threshold values automatically. On getting the threshold value from the DL model, a binarized image of a single lead paper ECG record is obtained where only the ECG signal is preserved and the background grid is removed. This image further goes through the post-processing phase, where the ECG lead names (e.g. v1, avf, etc.) are removed and the broken ECG signal is converted into a continuous ECG signal if required. Dilation and skeletonization are done later to ensure a single pixel value for the ECG signal in each column. Later, the post-processed binary image is vertically scanned to convert a 2-D image into a 1-D ECG signal. This 1-D signal goes as an input to the deep learning diagnostic model which predicts the patient’s heart-related problems and gives analysis similar to a cardiologist.
Fig. 1.
Flow diagram for automated binarization and diagnosis using deep learning from 12-lead paper ECG records. The 12 lead was converted to a single lead, pre-processed for shadow removal, and the Level of Binarization (LOB) characteristic curve is extracted. Then the DL model is used, and the background is removed. The signal is post-processed to remove the labels. The 1-D signal is then extracted using vertical scanning. A deep learning-based diagnosis was done at the end
Methods
Image Aquisition
The ECG records were available in paper form. The first step is converting it into an image by scanning or through camera capture. We had received data from Saidhan Hospital and STEMI Global. The database contains images of ECG captured from the camera (Samsung Galaxy S7) and also 12 lead ECG data records in the form of pdf files (Model: ECG600G).
Pre-processing
The 12 lead ECG data records are available in the form of pdf files. These pdf files had to be converted into an image format for further processing like converting 12 lead ECG records into single leads. 12 lead ECG was available in the form of pdf. But to process it we would be needing it in an image form (JPEG or PNG). This was done using the pdf2image python library.
After obtaining the images of 12 lead ECG, 12 different images were obtained with a single lead in each image. There were some files with continuous 12-leads that were manually converted which was done by writing a semi-manual algorithm using the OpenCV library. Here, the left and right mouse clicks were used to draw horizontal and vertical lines on the 12 lead ECG image to get a grid in which each box of the grid contains a single lead.
These boxes were saved as separate images to get 12 different images from one single 12 lead ECG image. (Code is given in the Supplementary Information.) For improving the computation complexities and accuracy, it was important to study every lead of the ECG. Rectangular boxes were located by shape detection and appropriately cropped to get 12 different image files. (The code for automated single lead extraction is also given in Supplementary Information.)
Binary Image Extraction
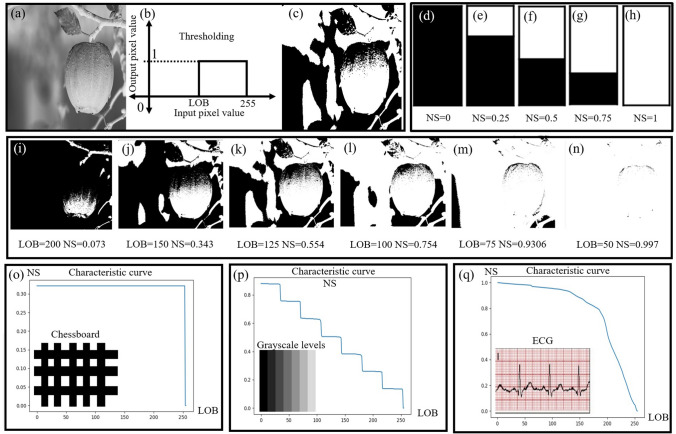
We find all the possible threshold values for the ECG signal by the brute force method. Possible threshold values range from 0 to 255. By applying these threshold values, we can obtain four types of images (viz. fully black images, fully white, images with the grid and signal, images without the grid, and only the signal). We only wanted to extract the signal without the grid. The process was repeated manually for all images where a 1-D characteristic curve and a single threshold value were generated to generate the training set for the deep learning-based model. Once trained, this model provided an automated threshold for any given input image. Once we got the desired image with only the ECG signal we noted the value of LOB for the image. In the case of thresholding, we have taken the data of level of binarization at every threshold i.e. from 0 to 255 and this was repeated for 66 images. The global thresholding concept of the LOB characteristic curve used for image Binarization is shown in the Fig. 2. Binarization of a sample grayscale image is shown in Fig. 2a–c. Figure 2d–h shows the concept of the normalized sum which is zero for a completely black image and one for a completely white image. From Fig. 2i–n, we can observe that as the value of LOB decreases, normalized sum (NS) gets incremented as the number of white pixels increases. At LOB = 50, white pixels occupy majority place and we get the value of NS as 0.997. At LOB = 200, the picture is mostly black and hence, NS = 0.073. Figure 2o shows a chessboard image with its corresponding characteristic curve, which shows the step transition from black to white. Similarly, different gray level image (as shown in Fig. 2q) has a characteristic curve with eight steps. The characteristic curve is also plotted for a single-lead ECG paper record showing two slopes, one for ECG signal and other for background grid. This curve is used as input to the deep learning model for automated threshold calculation.
Fig. 2.
Concept of LOB characteristic curve a–c thresholding of sample grayscale image for a particular level of Binarization (LOB) from 0 to 255, d–h normalized sum (NS) varies from 0 to 1 for complete black to complete white image, i–n binarized images at different threshold values (LOB) and NS, o Chessboard image (as shown in the snippet) and its corresponding characteristics curve showing the step transition from black to white, p image with different Grayscale levels and its corresponding characteristics curve, q characteristics curve of single-lead ECG (shown in the snippet) showing two slopes, one for ECG signal and other for background grid
Deep Learning-based Binarization
After extracting a LOB characteristic curve in the previous step, we made a deep learning model to predict threshold values for automated binarization. In this case, we have used a deep learning model to predict the threshold which uses the level of binarization. For thresholding using deep learning, we have prepared a dataset containing all (0 to 255) thresholding values during the Level of binarization value, with respect to a single image. The delta of the characteristic curve was calculated to avoid overfitting during the DL training phase. We subtracted the current value from its next value to find the delta of the characteristic curve. Then we took the inverse of the data so that we can get the required features. Then we applied the sequential model and the layer used was the Dense Layer, and the activation function used was ReLU. We had one input layer, two hidden layers, and one output layer. Then we applied EarlyStopping, ModelCheckpoint, and ReduceLROnPlateau function. EarlyStopping function is used when the learning rate of the model doesn’t change. To save the best models, the ModelCheckpoint function was used so that we can use them for prediction. ReduceLROnPlateau decreases the learning rate (LR) when the accuracy of the model doesn’t increase. Later the best model was chosen. Then we applied the model and got the threshold predicted values from the DL model. The entire data was exported into an excel sheet.
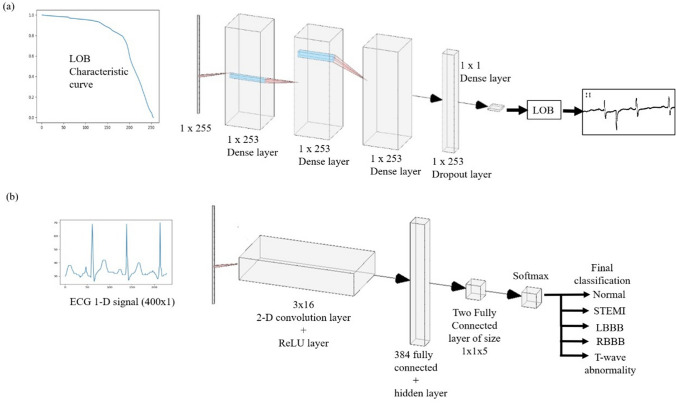
Figure 3a shows the deep Learning model for determining the correct threshold value for the input ECG report for binarization. The image was first binarized and then during vertical scanning, it was converted into a 1D array. The width of the image acts as the size of the input 1D array. In this, we first passed the characteristic curve as an input to the Dense layer of size . This is passed through two Dense hidden layers of size . After this, we use the dropout layer of size again which is passed through a Dense model of dimension . After this, we get the number called LOB which is the threshold value. This is the value at which we should binarize the image for getting the correct binarized output.
Fig. 3.
a Deep learning Model for determining threshold value. The input is LOB characteristic curve of size is . The 1-D signal is passed through multiple Dense layers. The output is then passed through a dropout layer. The data is then passed through the Dense model of dimension and we get the predicted output threshold value. b Deep learning Model for diagnosis. The input signal of the size is passed to the convolution layer along with the ReLU layer of dimension . After that, it is passed through a series of 384, and two fully connected layers along with hidden layers. Then it is passed through the Softmax layer which further diagnoses the ECG report
Post-processing
Once we have the binary signal, we stepped into the discontinuity detection process to check if there was any broken signal while binarizing. Now, a signal being an analog quantity, it would always show a smooth change and not a sudden change. Hence, to re-construct the broken signal, we have used a 1-D signal reconstruction algorithm. This algorithm helped join the broken parts from ECG and avoid sudden change. In the first, dilation and skeletonization are performed. The next step was the lead name removal, which was present on every lead image. We performed vertical scanning of every column of the image array, starting from the bottom of the columns to the top. During the scan, we knew that if we were going to get the first white pixel from the bottom, it would be the ECG lead, so we ignore that because we want to preserve the ECG signal. All the white pixels above the lead pixels of that column will just be the lead name character white pixels, which need to be removed. So, we convert the character pixels to black. These lead names act as a disturbance to the image, and hence it needs to be removed. We removed all the names, characters, or printed values so that only the signal remains and rest all unwanted data gets filtered out. To remove shadow impressions, we split the image into RGB values then we applied dilation which resulted in the reduction of the black shadow. Further, we applied median blur followed by the normalization to remove the salt pepper noise. The above procedures were implemented for all three RGB values. In this 1-D signal reconstruction algorithm, the broken binary signal obtained was made continuous, and then the lead name removal was done. For the first subpart, OpenCV operations were applied to the broken signal. This signal was then dilated, which increased the thickness of the ECG signal. This filled the broken gaps. Skeletonization was then applied to this thick signal to reduce the thickness again. So, by dilating and later skeletonizing, the broken signals were made continuous. Finally, we merged the result of all three parts to produce our final result without shadow impressions.
Signal Extraction
The objective was that we need to find the value of ECG in terms of voltage and time. We know that the dimension of a single box in hard-copy ECG paper is 1 cm 1 cm. We also know that an image is an array of pixels but in an image, the dimension of a single box in terms of pixel value will not be the same as 1 cm 1 cm. So the approach is that we are calculating the single box size of the graph in terms of the pixel, then we are calculating the different peak values of ECG in terms of pixel and remapping them back to the voltage. After post-processing, the 2-D image is vertically scanned for the identification of ECG signal pixels, and the identified signal pixels are then stored as an array. For obtaining time and voltage values, we performed a different type of detection where the red squares were preserved, and the ECG signal was removed. From those red squares, each red square in the time domain was 0.2 seconds, and in the voltage domain, it was 0.5 mV. So basically, from the domains, we got the scale for X-axis and Y-axis. And then, these scales were converted into pixels. So whatever X and Y coordinate values we were getting from pixels, we converted it into time and voltage. From time and voltage, we fetched a corresponding 1-D signal with the determined time and voltage values.
Deep Learning Model for Diagnosis
The most significant waves seen in a normal ECG are P wave, QRS complex, and a T wave. Different types of abnormalities such as ST-segment elevation myocardial infarctio (STEMI), Left Bundle Branch Block (LBBB), Right Bundle Branch Block (RBBB), and T-wave abnormality can be found out from the ECG records of a patient. STEMI is a type of myocardial infarction. A part of the heart muscle becomes dead due to the obstruction of blood supply in that area. LBBB is an ECG abnormality in the conduction system of the heart where the left ventricle contracts later than the right ventricle. In the case of RBBB, the right bundle branch is not directly activated by impulses whereas the left ventricle functions normally. Sinus rhythm is necessary for the conduction of electrical impulses within the heart. A strangely shaped T wave may signify disruption in re-polarization of the heartbeat and possibly be identified as sinus rhythm T-wave abnormality. Deep learning-based ECG diagnosis algorithm classifies the given ECG images into five different classes (normal, STEMI, LBBB, RBBB, and T-wave abnormality).
The deep learning model for the diagnosis using digitized ECG for heart abnormalities consists of various layers. For the deep learning model, we have used 400 points sample. The input is a 400 1 matrix 1-D signal which is passed to a 2-D convolutional layer of size is 3 16. After getting the binary image without any noise and lead character, we perform vertical scanning of the image array starting from the left-bottom of the image. When we get the first white pixel in a column, we know it is an ECG signal and store its corresponding value on the y-axis. Iterate it for the remaining 399 columns. This is how we get the 400 point training data and then we also store the corresponding disease value as label data. The extra space is filled by padding the same numbers. We then apply a ReLU layer to it. Two fully connected layers act as a hidden layer after applying the ReLU layer. There is a 5 neuron output layer that is given to the softmax and classification layer for classifying the abnormalities into 5 different classes.
Figure 3b shows this model, in which we provide the binarized ECG image as an input to the model, of dimension . This is further passed through a 2-Dimensional convolution network along with the ReLU layer which is of dimension 3x16. After this, we use a series of 384 and 2 fully connected layers, which also have hidden layers with them. Finally, we use the Softmax layer which gives the diagnosed output. We classify them under the following categories: Normal, Stemi, LBBB, RBBB, and T-wave abnormality.
In this model, we have taken input (1-D ECG signal) of size (400, 1, 1). Further, we have passed this input in our model. The model comprises a convolutional layer with filter size = 3, number of filters = 16, followed by ReLu layer. We have used 3 fully connected layers followed by the SoftMax layer for classification into different labeled data. The input format of the dataset (400, 1, 1) allowed us to use the 2D convolution layer. If we set the ’Padding’ option to ’same’, then the size of the padding for the output during the training will be same as the input. The software adds the same amount of padding to the top and bottom, and to the left and right. If the padding that must be added vertically has an odd value, then the software adds extra padding to the bottom. If the padding that must be added horizontally has an odd value, then the software adds extra padding to the right.
Result
We have analyzed 3200 ECG samples (scanned + camera capture + machine-generated). The 12 lead ECG data records are available in the form of pdf files. These pdf files had to be converted into an image format for further processing like converting 12 lead ECG records into single leads. We find all the possible threshold values for the ECG signal by the brute force method. Possible threshold values range from 0 to 255. By applying these threshold values, we can obtain four types of images (viz. fully black images, fully white, images with the grid and signal, images without the grid, and only the signal). We only wanted to extract the signal without the grid. The process was repeated manually for all images where a 1-D characteristic curve and a single threshold value were generated to generate the training set for the deep learning-based model. For the thresholding using machine learning, we have prepared a dataset containing 255 values of Level of binarization value with respect to a single image. This was repeated for 66 images. Results show that the characteristic curve of the ECG waveform is a superior feature that helped to improve the accuracy of binarization and diagnosis. The characteristic curve of ECG shows two slopes, one for the background grid and the other for the corresponding ECG signal. Post LOB binarization, we found out that it was necessary to remove the characters, apply skeletonization to reduce the width of the ECG signal after binarization. Once the images were skeletonized, it was easy to detect signals using vertical scanning. The only issue with the vertical scanning is when the R waveform is too steep, as seen in Fig. 5b. In vertical scanning, we scan every column of the image, starting from the bottom of the column. As we go up, the first white pixel that we encounter in that column, we consider that pixel as an ECG lead pixel. Any pixel which comes after that in the same column is just considered as noise, so we remove it by converting that white pixel into a black pixel same as the background. But as the R waveform is too steep, more than one white pixel can be present in the same column, which will be then considered as a noise and then get removed. This might pose an issue for the R waveform which may lead to the discontinuity of the ECG lead. Again, if the characters like ECG lead names are overlapping the ECG signal, there is a probability that ECG damages the signal. Hence, the discontinuous ECG signal needs to be post-processed in order to correct the waveform using discontinuity detection.
Fig. 5.
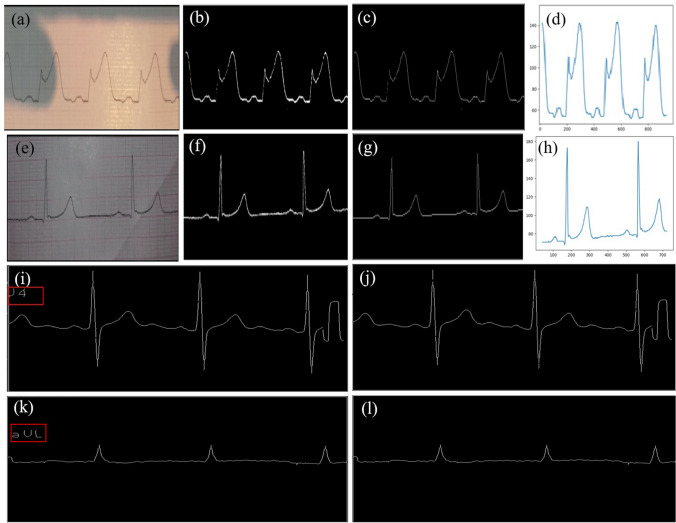
a The camera captured paper ECG record with shadow. Results after binarization (b), skeletonization and character removal (c) and 1-D signal extraction (d) from images with shadow. f–h extraction of 1-D signal from the image with non-uniform illumination shown in (e). Binarized ECG image with undesired lead names (shown in i and k, red box), removal of lead names j, l that avoids distortion in signal extraction
The accuracy achieved for threshold prediction using the LOB characteristic curve and deep learning (DL) was around 97% and RMS error of 0.034%. Root mean squared error is the square root of the mean of the square of all of the errors. The use of RMSE is very common, and it is considered an excellent general-purpose error metric for numerical predictions. The trained DL was designed using a dense network; it had a dropout layer, which also has a single value at the output. This output value is then used for thresholding the images.
We have also found that a shadow removal algorithm is very beneficial for all the images clicked via smartphones. Furthermore, uniform luminance is necessary, which can be achieved by luminance correction algorithms. Therefore, these two steps helped in thresholding, and they should not be a part of deep learning because even with LOB-based deep learning, it does not follow the pre-processing steps, and we face errors. Hence, shadow removal and uniform luminance correction are mandatory for any ECG digitization. Also, the character removal helped us to decode the signal correctly, and that should be done in post-processing. For character removal, OCR and subtraction of optical characters can be carried out.
After 1-D extraction using vertical scanning from binarized images, the signal is given to another DL model that converted the digitized signal. The DL model diagnosed four different diseases, namely, LBBB, RBBB, T-wave abnormality, and STEMI. We had passed almost 300 images. Only for T-wave abnormality due to data images constraint, we could pass only 145 images. We also found that it was effortless to detect the T-wave abnormality. The normal ECGs and LBBB had a lot of overlapping between them, close to almost 3 to 4%. STEMI and RBBB generally do not get misclassified with each other. The worst misclassification was faced by class 3, which is LBBB, which was classified as STEMI, normal, or even in some cases RBBB.
Hence, we think that extracting the 1-D ECG signal from paper records opens a new pathway for designing various digitizing algorithms and automated diagnostics. It could be the most beneficial for the people in rural areas where there is a lack of such digital ECG equipment and experienced doctors for the diagnosis of heart-related problems. The smartphone-based application for automated Binarization and diagnosis from paper ECG records will be really helpful in rural areas. In addition, in the coronavirus disease pandemic, COVID-19 infected patients are given a medicine named hydroxychloroquine (HCQ). The person’s blood flow rate and muscle activity can be determined by the heart rate of the person. Mostly HCQ or similar medicines can dilute the blood and dissolve the blood clots. If the heart cannot pump the diluted blood, the oxygen level drops down and can be fatal. It will be tough to determine the cause of death unless we have the ECG records of the patient.
Hence, developing such an image processing and deep learning-based app could be conducive to the social community. It would be an excellent researching opportunity for upcoming new researchers to automatically diagnose these signals, which we found challenging in the current manuscript.
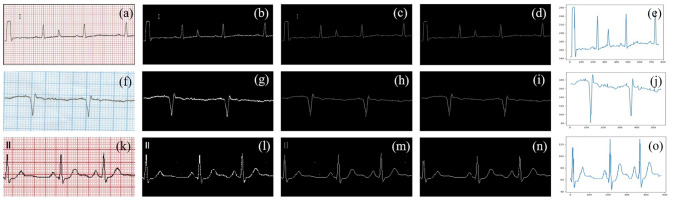
As seen from Fig. 4, we extracted a 1-D signal from the different colored input ECG paper records. The complete process of the signal extraction is shown in this figure. Figure. 4 shows different backgrounds (red graph (a), blue graph (f), software-generated background (k) does not affect our digitization process. Figure 4b, g, l show the process of thresholding on those images. The next step involves the process of skeletonizing. Figure 4c, h, m shows the output of this process. Further, we do the character removal process as shown in Fig. 4d, i, n. Finally, the 1-d signal is extracted by doing vertical scanning of those images shown in Fig. 4e, j and o.
Fig. 4.
1-D ECG signal extraction from an input image of ECG paper record irrespective of the background (textbfa), (f), (k) ECG signal recorded on different background color grids. b, g, l binarized images after thresholding. Images after dilation and skeletonizing (c), (h), (m) Images after character removal (d), (i), (n). Final 1-D signal extraction using vertical scanning (e), (j), (o)
As seen from Fig. 5 the camera captured paper ECG record comes along with the shadow. To convert the shadow image into uniform illumination we applied the shadow removal algorithm. Binarization was achieved by applying the corresponding threshold value as shown in Fig. 5b and h. Then we applied the dilation along with the skeletonization followed by character removal in ash shown in Fig. 5c and g. Now, the resulting image was pure ECG line. Hence, we applied vertical traversing to extract the 1D signal, as shown in Fig. 5d and h. The Binarized ECG image as shown in Fig. 5i and k with the undesired lead name (highlighted within the red box) should be removed to avoid distortion in signal extraction. Hence, we applied a letter removal algorithm, after which Fig. 5j and l has been generated.
Discussion
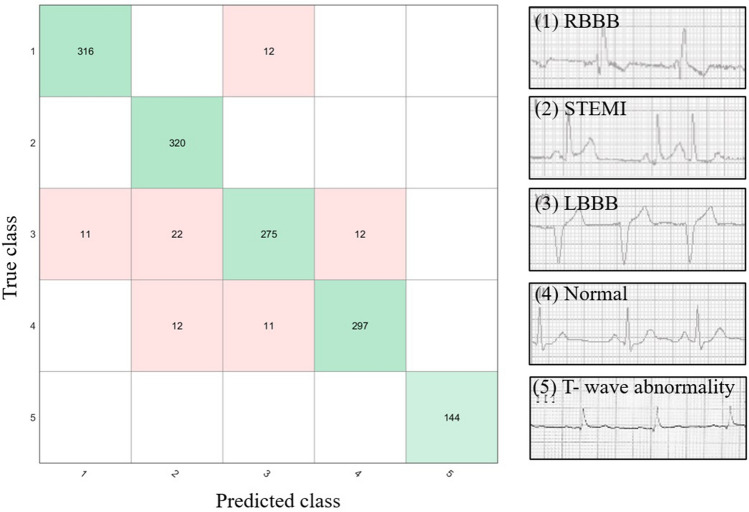
A confusion matrix for the deep learning model designed for the diagnosis of heart abnormalities into five classes as shown in Fig. 6. The proposed model correctly classified all T-wave abnormalities. Out of a total of 328 signals tested for classification of RBBB, only 12 were wrongly classified. In the case of STEMI classification, all 320 signals were correctly labeled.
Fig. 6.
The confusion matrix for the deep learning model is used for diagnosis of the heart abnormalities
As shown in Table 1, we have analyzed 3200 ECG samples (scanned + camera capture + machine-generated) which are very high as compared to the other literature that has been reported. Although the accuracy reported by Shi et al. [13] is 99% but the number of samples used by them is less as compared to our method. Our proposed method achieved an accuracy of 97% with the least root mean square (RMS) error of 0.034%.
Table 1.
Comparison table with same number of images and same dataset
| Method | Accuracy(%) | RMS error (%) | Number of Images in dataset |
|---|---|---|---|
| Contrast enhancement filtering | 99 | 1.1 | 20 |
| EBPS | 98.3 | 1.87 | 20 |
| LOB (proposed) | 99.46 | 0.9 | 20 |
As shown in Table 2, we have analyzed 3200 ECG samples (scanned + camera capture + machine-generated) which are very high as compared to the other literature that has been reported. Mallawaarachchi et al. [5] used 550 samples for testing and the accuracy achieved was 96%. Although the accuracy as reported by Damodaran et al. [12] and Shi et al. [13] is 99% but the number of samples that were used by them was less as compared to our method. Our proposed method had the accuracy to be 97% with the least root mean square (RMS) error of 0.034%. Badilini et al. [15] and Ravichandran et al. [16] had the RMS error to be 16.8% and 12% respectively which was high as compared to that of the proposed method. There is a scope of improvement as we have classified the abnormalities into 5 different classes. However, more than 12 types of diseases are known that can be detected using the ECG waveforms.
Table 2.
Comparison with existing methods
Conclusion
The manuscript presented the automated digitization of ECG paper records and automated diagnosis of heart-related abnormalities. Our approach can be easily implemented in rural areas where such expertise is not available. Our system mainly classifies the ECG signals into 5 different heart diseases viz. RBBB, LBBB, STEMI, T-wave abnormality, and normal ECG. The input image is first pre-processed by applying the very efficient shadow removal algorithm. The threshold of this image is then calculated by a 3 layer dense DL model with 97% accuracy. The post-processing of this image includes character removal and then converting it into a 1-D signal, which is fed to the DL model and an accuracy of 94.4% is achieved to classify the ECG into one of the diseases. Automated diagnosis can further be useful for various other research work.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Saidhan hospital and STEMI global for providing us with ECG dataset and their help in diagnostics.
Funding
No funding was involved in the present work.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with animals or humans performed by any of the authors. All the necessary permissions were obtained from the Institute Ethical Committee and concerned authorities.
Informed Consent
No informed consent was required as the studies does not involve any human participant.
References
- 1.Biswas, B., Bhattacharya, U., & Chaudhuri, B. B. (2014). In: Proceedings of the 22nd International Conference on Pattern Recognition, pp. 3008–3013.
- 2.Su B, Lu S, Tan CL. IEEE Transactions on Image Processing. 2012;22(4):1408. doi: 10.1109/TIP.2012.2231089. [DOI] [PubMed] [Google Scholar]
- 3.Sauvola J, Pietikäinen M. Pattern Recognition. 2000;33(2):225. doi: 10.1016/S0031-3203(99)00055-2. [DOI] [Google Scholar]
- 4.Swamy, P., Jayaraman, S., & Chandra, M. G. (2010). In: Proceedings of the International Conference on Bioinformatics and Biomedical Technology, pp. 400–403.
- 5.Mallawaarachchi, S., Perera, M. P. N., & Nanayakkara, N. D. (2014). In: Proceedings of the IEEE Conference on Biomedical Engineering and Sciences (IECBES), pp. 868–873.
- 6.Waits GS, Soliman EZ. Journal of Electrocardiology. 2017;50(1):123. doi: 10.1016/j.jelectrocard.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Mitra S, Mitra M, Chaudhuri BB. Computers in Biology and Medicine. 2004;34(7):551. doi: 10.1016/j.compbiomed.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Gatos B, Pratikakis I, Perantonis SJ. Pattern Recognition. 2006;39(3):317. doi: 10.1016/j.patcog.2005.09.010. [DOI] [Google Scholar]
- 9.Patil R, Karandikar R. Journal of Electrocardiology. 2018;51(4):707. doi: 10.1016/j.jelectrocard.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Jayaraman, S., Swamy, P., Damodaran, V., & Venkatesh, N. (2012). In: Proceedings of the Advances in Electrocardiograms-Methods and Analysis, pp. 127–140.
- 11.Kumar, V., Sharma, J., Ayub, S., & Saini, J. (2012). In: Proceedings of the Fourth International Conference on Computational Intelligence and Communication Networks, pp. 317–321.
- 12.Damodaran, V., Jayaraman, S., & Poonguzhali, S. (2011). In: Proceedings of the Defense Science Research Conference and Expo (DSR), pp. 1–4.
- 13.Shi, G., Zheng, G., & Dai, M. (2011). In: Proceedings of the 2011 Computing in Cardiology, pp. 797–800.
- 14.Chebil, J., Al-Nabulsi, J., & Al-Maitah, M. (2008). In: Proceedings of the 2008 International Conference on Computer and Communication Engineering, pp. 1308–1312.
- 15.Badilini F, Erdem T, Zareba W, Moss AJ. Journal of Electrocardiology. 2005;38(4):310. doi: 10.1016/j.jelectrocard.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Ravichandran L, Harless C, Shah AJ, Wick CA, Mcclellan JH, Tridandapani S. IEEE journal of translational engineering in health and medicine. 2013;1:1800107. doi: 10.1109/JTEHM.2013.2262024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sao, P., Hegadi, R., & Karmakar, S. (2015). In: Proceedings of the International Journal of Science and Research, National Conference on Knowledge, Innovation in Technology and Engineering, pp. 82–86.
- 18.Lyon A, Mincholé A, Martínez JP, Laguna P, Rodriguez B. Journal of the Royal Society Interface. 2018;15(138):20170821. doi: 10.1098/rsif.2017.0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luz EJS, Schwartz WR, Cámara-Chávez G, Menotti D. Computer Methods and Programs in Biomedicine. 2016;127:144. doi: 10.1016/j.cmpb.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Sannino G, De Pietro G. Future Generation Computer Systems. 2018;86:446. doi: 10.1016/j.future.2018.03.057. [DOI] [Google Scholar]
- 21.Swapna G, Soman K, Vinayakumar R. Procedia Computer Science. 2018;132:1192. doi: 10.1016/j.procs.2018.05.034. [DOI] [Google Scholar]
- 22.Yu, R., Gao, Y., Duan, X., Zhu, T., Wang, Z., & Jiao, B. (2018). In: Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 4636–4639. [DOI] [PubMed]
- 23.Sun, X., Li, Q., Wang, K., He, R., & Zhang, H. (2019). In: Proceedings of the Computing in Cardiology (CinC), p. 1.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.