Abstract
Therapeutic regimens for the COVID‐19 pandemics remain unmet. In this line, repurposing of existing drugs against known or predicted SARS‐CoV‐2 protein actions have been advanced, while natural products have also been tested. Here, we propose that p‐cymene, a natural monoterpene, can act as a potential novel agent for the treatment of SARS‐CoV‐2‐induced COVID‐19 and other RNA‐virus‐induced diseases (influenza, rabies, Ebola). We show by extensive molecular simulations that SARS‐CoV‐2 C‐terminal structured domain contains a nuclear localization signal (NLS), like SARS‐CoV, on which p‐cymene binds with low micromolar affinity, impairing nuclear translocation of this protein and inhibiting viral replication, as verified by preliminary in vitro experiments. A similar mechanism may occur in other RNA‐viruses (influenza, rabies and Ebola), also verified in vitro for influenza, by interaction of p‐cymene with viral nucleoproteins, and structural modification of their NLS site, weakening its interaction with importin A. This common mechanism of action renders therefore p‐cymene as a possible antiviral, alone, or in combination with other agents, in a broad spectrum of RNA viruses, from SARS‐CoV‐2 to influenza A infections.
Keywords: Ebola, importin A, influenza A, nucleocapsid protein, nucleoprotein, p‐cymene, rabies, SARS‐CoV‐2
Left panel: Docking of p‐cymene within the NLS sequence of N protein, shown in yellow. The ligand interacts preferentially with Arginine 262 and Threonine 265, in the center of the NLS sequence (aa 258–268), therefore impairing the interaction of N with importin‐α. Right panel: p‐cymene inhibits (black curve) and prevents (red curve) SARS‐CoV‐2 replication, in a dose‐dependent manner.

Abbreviations
- CPE
cytopathic effect
- DMEM
Dulbecco's modified essential medium
- GRAS
generally recognized as safe
- MD
molecular dynamics
- MDCK
Madin‐Darby Canine Kidney cells
- MEM
minimal essential medium
- MSM
Markov state modeling
- N
SARS‐CoV‐2 nucleocapsid protein
- N‐CTD
SARS‐CoV‐2 N protein C‐terminal domain
- NES
nuclear export signal
- NLS
nuclear localization signal
- N‐NTD
SARS‐CoV‐2 N protein N‐terminal domain
- NP
nucleoprotein
- PDB
protein database
- Rg
radii of gyration
- RMSD
root mean square deviation
- RT‐PCR
real‐time quantitative PCR
- Vero cells
Verda reno cells
Significance Statement.
p‐cymene inhibits SARS‐CoV‐2 and impairs Influenza H1N1 viral replication, in vitro, at non‐toxic concentrations. This action is exerted by p‐cymene interaction with SARS‐CoV‐2 nucleocapsid and influenza nucleoprotein, as revealed by extensive in silico molecular simulation and molecular dynamics. We propose p‐cymene as a possible antiviral agent, alone or as an adjuvant therapeutic, in cases of COVID‐19, or other diseases caused by RNA‐viruses.
1. INTRODUCTION
An unforeseen International effort led to the identification of the cause of the novel COVID‐19 disease (the SARS‐CoV‐2 virus) and the detailed analysis of its genome, 1 a few months since its outbreak. Sustained academic and industrial effort led to the production of successful vaccines, 2 , 3 , 4 or vaccine candidates, 5 , 6 based on different technologies (see Ref. [7] for an exhaustive analysis of technologies and report of all candidate vaccines). However, an efficient dedicated therapy has not yet been advanced. Actual therapeutic approaches comprise antibodies or antibody cocktails against the spike protein of the virus, employed for host cell entry. 8 , 9 In addition, a number of alternative viral targets are actually explored as potential drug candidates. 10 , 11 , 12 , 13 Indeed, the viral genome, one of the longest found in RNA viruses, codes for a number of distinct proteins, comprising a small (pp1a) and large (pp1ab) polyprotein (gene ORF1ab), while other genes (S, E, M, and N) encode the structural proteins Spike, Envelope, Membrane, and Nucleocapsid, respectively. Ten open reading frames (ORF1–ORF10) encode accessory proteins with speculated functionality. 14 A number of these proteins have been investigated as potential drug targets. Finally, the in‐depth analysis of viral biology and its interaction with cellular mechanisms led to repurposing of a number of drugs, with the notable recent paradigm of colchicine. 15 , 16
The nucleocapsid (N) protein is one of the most important SARS‐CoV‐2 proteins. 17 A detailed analysis of this protein has been recently published. 18 The authors report that N‐protein plays an important role in viral replication, assembly, and immune regulation. Although the full crystal structure of the protein has not yet been resolved, they report that SARS‐CoV‐2 N protein possesses a very high homology with the SARS‐CoV N protein. Both contain two structural domains (N‐Terminal Domain, NTD and C‐Terminal Domain, CTD), linked by a non‐structured protein part. Previous studies revealed that coronavirus N‐NTD is the RNA‐binding domain, while N‐CTD is the dimerization domain, that also has a certain RNA‐binding activity. This plurality of N actions makes it a good candidate for therapy. It is to note that, although N is an internal viral protein, the presence of IgA, IgM, and IgG antibodies against N antigen in the sera of COVID‐19 infected patients has been reported by immunoblot assays, indicating its importance in host immunity. 17 Interestingly, previous reports (see Ref. [19] and references therein) revealed the existence of nuclear localization signals (NLS), especially in the CTD part, as well as nuclear export signals (NES) in SARS‐CoV virus N, suggesting that this RNA‐binding protein may serve as a dynamic shuttle between the cytoplasm, nucleus, and nucleolus.
Among the multitude of products tested against the COVID‐19 disease, a number of natural products have been assayed (critically reviewed in Ref. [20] and references therein), targeting mainly the viral proteases in vitro. However, no natural product has been proposed to target the N protein. Recently, we have reported that a combination of three aromatic plants (Thymbra capitata (L.) Cav., Origanum dictamnus L., Salvia fruticosa Mill., 21 and references therein) is efficient against upper respiratory tract viral infections. Further in vitro studies revealed that a primary target of this preparation is the viral nucleoprotein (NP), inhibiting its nuclear translocation, 22 and impairing viral protein transcription, while a proof‐of‐concept clinical trial suggests a possible action of the preparation in mild, ambulatory, COVID‐19 positive patients (Lionis et al. 23 ). Based on these data, we have analyzed, in silico, the different constituents of our preparation, against the SARS‐CoV‐2 N protein and other viral NPs (influenza, rabies, Ebola). Here, we report that p‐cymene, a natural monoterpene, at non‐toxic concentrations, impairs viral replication. In silico molecular simulation and molecular dynamics (MD) studies suggest that p‐cymene can bind to the CTD part of SARS‐CoV‐2 N protein, as well as to influenza, rabies, Ebola NPs. We further report that p‐cymene impairs viral infection in vitro. Therefore, p‐cymene might represent a valid therapy, alone or in combination with other drugs, in cases of COVID‐19, or other diseases caused by RNA‐viruses.
2. MATERIAL AND METHODS
2.1. Docking simulations
The sequence of nucleocapsid proteins from SARS‐CoV‐2, influenza, Ebola and rabies viruses, in fasta format, was retrieved from the NCBI protein database (https://www.ncbi.nlm.nih.gov/protein/) and introduced to the GalaxyWeb server (http://galaxy.seoklab.org/), routine TMB. 24 , 25 , 26 , 27 For SARS‐CoV‐2 CTD, we used a model based on the PDB 6WJI crystal, 28 while for influenza, rabies and Ebola NP, we have relied on the models from the GalaxyWeb, as these proteins have not yet been crystalized. p‐cymene mol2 file format was prepared by the OpenBabel program (http://openbabel.org). 29
Each protein (in pdb format) and ligand (in mol2) files were uploaded to the GalaxyWeb server and a fully flexible docking (involving the receptor and the ligand) was performed, followed by optimization and subsequent refinement, through the GalaxyRefine algorithm. 24 , 27 The best solution (denoted as “Model 1”) was retained. The corresponding pdb file (containing the protein–ligand complex) was retrieved. The 3D structures of the ligated and unligated N protein were compared and the root mean square deviation (RMSD) was calculated by UCSF Chimera 1.11.2 program. 30
The sequence of human coronavirus 229E nucleocapsid protein mRNA (GenBank: J04419.1), part of the whole SARS‐CoV‐2 mRNA, was used as a template, in order to simulate the interaction of SARS‐CoV‐2 N‐protein‐viral mRNA. It was introduced to the RNAfold web server (http://rna.tbi.univie.ac.at/cgi‐bin/RNAWebSuite/RNAfold.cgi) to predict the secondary structure of the single‐stranded RNA sequence, by using minimum free energy prediction. 31 The resulting mRNA secondary structure was then introduced to the RNAComposer server (http://rnacomposer.cs.put.poznan.pl/), 32 , 33 a fully automated modeling service, which returned a single 3D‐RNA structure model of the viral mRNA.
For protein–protein and protein–RNA interactions, we have used the HEX 8.0.0 program (http://hex.loria.fr/). 34
2.2. Molecular dynamics
The crystal structure of the SARS‐CoV‐2 nucleocapsid protein (C‐terminal dimerization domain, residues 249–389, PDB entry: 6WJI 28 ) and human importin‐α (residues 71–197) containing the interaction site with the NLS sequence of cargo proteins 35 were used as initial coordinates to build MD models. A nucleocapsid–importin complex was produced based on docking calculations with the HEX 34 program.
The Amber03 36 protein force field was used for the residues and ions. The Amber03 parameters for p‐cymene were derived based on the ACPYPE algorithm. 37
Classical MD production trajectories were run with the leap‐frog integrator in GROMACS 2020 38 for the nucleocapsid–importin complex with and without p‐cymene. Seven replicas (trajectories) were run per sample (with or without p‐cymene) starting from seven different structures extracted at the equilibration process. The equations of motion were integrated for 2.0 μs per replica, in a total simulation time of 28 μs. The classical MD trajectories were analyzed by Markov state modeling (MSM). 39 , 40 , 41 The classical MD runs were followed by the multiple walkers (MW) variant of the metadynamics method 42 again with seven replicas per sample for 0.7 μs each replica, with selected nucleocapsid‐importing distances as collective variables to be biased in the enhanced sampling.
Α detailed description of the MD/enhanced sampling implementation and data analysis are presented in Supporting Information, Material and Methods.
2.3. Biochemical and cellular methods
2.3.1. Cell culture, virus infections, and cell proliferation analysis
SARS‐CoV‐2 (isolate 30–287) was obtained through culture in Vero E6 cells (ATCC® CRL‐1586), from an infected patient, in Alexandroupolis, Greece. Virus stock was prepared by infecting fully confluent Vero E6 cells in DMEM, 10% fetal bovine serum, with antibiotics, at 37°C, 5% CO2. Four days after inoculation, the supernatant was frozen at −80°C until use. Infections were carried out in 96‐well plates, using SARS‐CoV‐2 (m.o.i. of 0.1) on Vero E6 cells. Cells were treated with different concentrations of p‐cymene, in a volume of 15 μl, per 150 μl of medium, for 48 h. Cell morphology was observed with phase contrast, in an inverted microscope, to record CPE. Culture supernatants were analyzed using real‐time RT‐PCR.
Madin‐Darby Canine Kidney cells (MDCK; ATCC® CCL34™) cells were grown according to standard conditions, as described in detail in. 43 Human influenza virus strain A FM/1/47 (H1N1) was obtained from ATCC. Influenza virus strain was studied in MDCK cells in serum‐free MEM, containing 1 μg/ml trypsin TPCK‐treated (Sciex), 2 mM L‐glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin. Evaluation of virus titers and viral growth curves were carried, according to standard protocols. 44 In the present study, we have probed different concentrations of p‐cymene (up to 2 mg/ml).
Analysis of the effects of p‐cymene on the cell viability was performed in MDCK and Vero E6 cells using the tetrazolium MTT assay for 24, 48, and 72 h respectively. 43
2.3.2. Titration assay
Titration of SARS‐CoV‐2 infected cells was carried in 96‐well plates, using Vero E6 cells and TCID50 was calculated according to the method of Reed and Muench 45 and by sigmoidal fitting. Plates were incubated at 37°C for 4 days, and the cytopathic effect (CPE) was scored by observation under an inverted phase‐contrast microscope.
The antiviral activity of p‐cymene on Influenza A virus was assayed in plaque‐reduction assay (expressed in the number of forming plaques), with Ribavirin as a positive control (25 μg/ml). After 3–4 days and upon plaque formation, cells were fixed with paraformaldehyde and cell monolayers were stained with crystal violet. Following staining the plaques that developed were counted, and the viral titer was evaluated by the formula: X = (# of plaques)/(d × V) where X = the titer, d = dilution factor and V = the volume of diluted virus instilled on the cells, as detailed previously. 22
2.3.3. Real‐time quantitative PCR
To determine the SARS‐CoV‐2 viral load, RNA was extracted from 96‐well supernatants (100 μl) using NucleoSpin Dx Virus according to the manufacturer (Macherey Nagel). Multi‐target real‐time RT‐PCR was performed using COVID‐19 SARS‐Cov‐2 Real‐TM according to the manufacturer (Sacace Biotechnologies).
For influenza experiments, MDCK cells (2 × 105) were plated in 12‐well plates and after 24‐h infected with influenza A FM/1/47/H1N1 or HRV14 for 2 h at 37°C. Subsequently, cells were incubated in the absence (control) or the presence of p‐cymene for 12 h. Total RNA was extracted with NucleoSpin RNA kit (Macherey‐Nagel). The in vitro quantification of influenza A genome was performed using the Techne qPCR human influenza A (M1) genome kit (TechneTM), following the manufacturer's instructions, in a Stratagene Mx300 P qPCR System. Relative expression levels of target genes were calculated from Ct values.
2.3.4. Immunofluorescence analysis
Madin‐Darby Canine Kidney cells were plated on glass coverslips in 24‐well plates. After 24 h, they were infected with influenza A FM/1/47/H1N1 for 1 h at 37°C. MEM supplemented or not with p‐cymene was added, and cells were fixed 6 h after the infection with 4% PFA and permeabilized with 0.1% TritonX‐100, for 10 min. Cells were stained with a mouse monoclonal anti‐NP antibody (Santa Cruz), followed by a secondary FITC‐labeled goat anti‐mouse antibody (Santa Cruz). The nuclei were stained with DAPI. Fluorescent images were acquired with an epifluorescent Leica DMIRE2 microscope equipped with a Leica DFC300FX digital camera.
2.3.5. Immunoblot assays
For the analysis of whole‐cell extracts, cells were collected and pelleted at 2500 g for 10 min. The M‐PER Mammalian Protein Extraction Reagent (Cat. No 78503; Thermo Scientific) along with protease inhibitors (Cat. No 78415; Thermo Scientific) was used. Samples were boiled in SDS gel‐loading buffer, separated by electrophoresis and transferred on to PVDF membrane. Membrane was blocked with TBST buffer with 5% (w/vol) dried non‐fat milk and incubated with primary antibody of NP (sc‐80481; Santa Cruz). Following incubation with the primary antibody, membrane was incubated with secondary antibody (goat Anti‐mouse IgG Antibody, Peroxidase Conjugated, H+L [AP124P] Sigma). The membrane was developed using Luminata Forte Western HRP Substrate (Cat. No WBLUF0100; Millipore) by the ChemiDoc™ MP System (Cat. No 170‐8280; Bio‐Rad).
2.4. Statistical analysis
Statistical analysis was performed in GraphPad Prisma V6.05 (GraphPad Software Inc.) and Origin 2018 (OriginLab Co.), with the tests described in Section 3. A threshold of p < .05 was retained for significance.
2.5. Nomenclature of targets and ligands
Key protein targets in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY, 46 and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20. 47 The ligand (p‐cymene) is hyperlinked to the PubChem database (https://pubchem.ncbi.nlm.nih.gov/).
3. RESULTS
3.1. In silico simulation reveals that p‐cymene interacts with the Importin‐binding domain of SARS‐CoV‐2 N protein and impairs its association with the IMPα‐IMPβ‐RanGDP complex
Nucleocapsid SARS‐CoV‐2 is a large protein of 419 amino acids (NCBI Reference Sequence: YP_009724397.2). Until today, the crystal structure of the complete protein has not been resolved. By contrast, a number of crystals (PDB codes 6M3M, 6VYO, 6YI3) of the N‐, RNA‐binding, terminal part, and the C‐ dimerization part of the protein (PDB code 6WJI) have been reported (Figure S1). Previous reports suggest that SARS‐CoV N protein (90% homolog to SARS‐CoV‐2) may quickly shuttle between the nucleus and the cytoplasm, a process regulated by N protein phosphorylation, 48 while the transfer of the viral RNA to the nucleus has also been suggested. 19 Therefore, we have scanned the SARS‐CoV‐2 N protein for the existence of the prototype NLSα sequence PRQKRTATKAY. 35 It was identified in position 258–268 of the N protein, within its structured C‐terminal part (Figure 1A). Simulation of N binding with the complex IMPα‐IMPβ‐RanGDP and viral RNA (see Ref. [35] for methodological details) (Figure 1B) confirmed the high‐affinity binding of Importin complex to N (ΔG −1000.5 kcal/mol for importins and −418.7 kcal/mol for RNA, localized at the structured N‐terminal part of the protein, Figure 1B).
FIGURE 1.
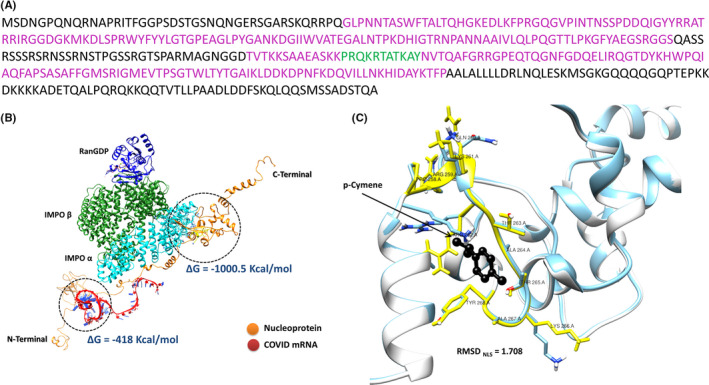
(A) Sequence of the SARS‐CoV‐2 N protein. In purple are shown the crystalized parts of the protein, while in green is presented the nuclear localization signal (NLS) sequence. (B) In silico simulation of the binding of the IMPα‐IMPβ‐RanGDP complex and viral RNA with SARS‐CoV‐2 N protein. ΔG for the interaction of N protein with the IMPα‐IMPβ‐RanGDP complex and with viral RNA are also shown. (C) Docking of p‐cymene within the NLS sequence of N protein shown in yellow. As shown the ligand interacts preferentially with Arginine 262 and Threonine 265 at the center of the NLS sequence (aa 258–268), therefore impairing the interaction of N with importins
Molecular docking simulation of the p‐cymene binding on SARS‐CoV‐2 N protein revealed a high‐affinity association of the ligand in the structured C‐terminal part of the molecule (ΔG = −5.6 kcal/mol, corresponding to an apparent affinity of 113 μΜ). p‐Cymene binds within the NLS sequence of the protein (Figure 1C) and interacts with amino acids Arg262 and Thr265. This interaction induces a significant conformational change of the NLS region of the N protein (RMSD = 1.708 Å), resulting in a re‐orientation of amino acids Arg262 (RMSD = 2.801 Å), Lys266 (RMSD = 2.759 Å), and Glu260 (RMSD = 2.650 Å) of the NLSα sequence (Figure 1C). These changes impair the interaction of N with the IMPα‐IMPβ‐RanGDP complex. As expected, viral RNA‐binding interaction with N protein remains unaltered (ΔG = −418.7 kcal/mol in the absence and −453.4 kcal/mol in the presence of p‐cymene).
Molecular dynamics simulations of N protein complex with Importin A, in the absence or the presence of p‐cymene (Figure 2), revealed that p‐cymene is able to destabilize the nucleocapsid–importin complex. The Free Energy Surfaces for the dissociation of the nucleocapsid–Importin complex in the absence or the presence of p‐cymene respectively are reported herein. Two main minima are observed (M1, M2). The transition from M1 to M2 and beyond (larger distances and complex radii of Gyration) is happening on a shallower energy surface in the presence of p‐cymene, compared with its absence, and a reduced energy barrier, suggestive of a weaker association of Importin A with N protein in the former case, verifying molecular docking data. The structures of the two complexes, at minima M1 and M2, are also given for reference. The MSM analysis of the classical MD trajectories has indicated several important residues of the nucleocapsid protein that are involved in the dynamics of its interaction with importins (see Supporting Information). Interestingly enough, ten out of the most important residues of the SARS‐CoV‐2 N protein (83, 193, 256, 276, 280, 361, 369, 370, 374, 387) are associated with the formation of salt bridges between nucleocapsid and Importin‐α in the absence of p‐cymene. To the contrary, only five out of the most important residues of the nucleocapsid protein (193, 259, 276, 280, 361) are associated with the formation of salt bridges between nucleocapsid and Importin‐α in the presence of p‐cymene. This definitely indicates a weakening of the nucleocapsid–Importin‐A association in the presence of p‐cymene.
FIGURE 2.
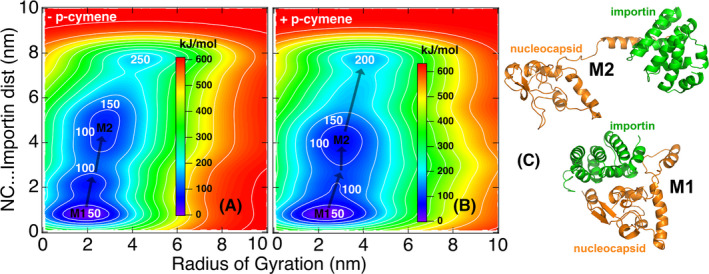
Free energy surfaces for the dissociation of the nucleocapsid–Importin α complex in the absence (A) and the presence of p‐cymene (B). (C) The associated structures at the M1–M2 minima of the complex
Previous reports suggest that SARS‐CoV‐2 protein dimerizes through an interaction of its N‐CTD η1 domain. 49 p‐Cymene is predicted to bind at this region (amino acids Arg262 and Thr265), impairing the antiparallel dimer formation of the N‐CTD protein (Figure S2).
3.2. p‐Cymene is predicted to bind to the NLS‐domain of a number of other viral NP proteins and impairs importin binding
A number of RNA viruses contain NPs, which may shuttle between the cytoplasm to the nucleus. This transfer depends also on the binding of NPs to host importins and the subsequent nuclear internalization, through nuclear pores. Here, we confirmed the presence of a NLS for Importin α (NLSα, Figure 3A) in a number of RNA viruses (Influenza A, Rabies, Ebola), employing the aforementioned methodology. p‐Cymene docking simulation on these NPs revealed an interaction within their NLS region. In detail, p‐cymene interacted with Influenza A NP at the region 169–177, with Rabies NP at the regions 241–250, 312–315, and 322–325 and with Ebola NP at the regions 162–163, 204–211, and 299–300 (Figure 3B–D). Interestingly, binding of p‐cymene to the specific proteins inhibits (Influenza H1N1 and Ebola) or impairs significantly (Rabies) the interaction of NP with Importin α (Figure 3E).
FIGURE 3.
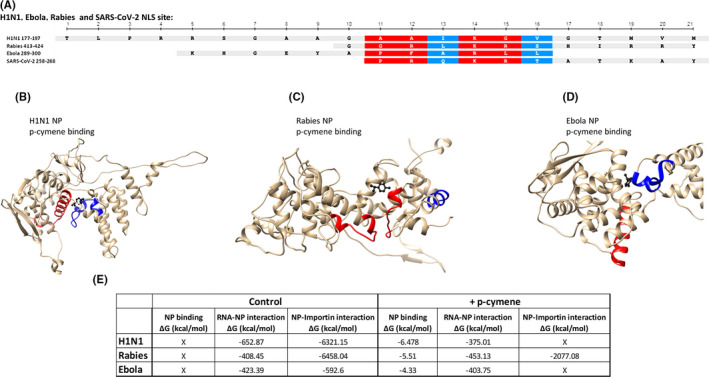
(A) Comparison of nuclear localization signal (NLS) sequences in Influenza H1N1, Ebola, Rabies, and SARS‐CoV‐2 viruses. (B–D) Simulation binding of p‐cymene (red color) on Influenza H1N1, Rabies, and Ebola nucleoproteins (NPs). The NLS sequence of each protein is presented in blue color. (E) Table summarizing the RNA and Importin binding to Influenza H1N1, Rabies, and Ebola NPs in the absence or the presence of p‐cymene
p‐Cymene interacted with Influenza A (H1N1) NP through binding with Thr177, the first of amino acid of NLS of this NP (Figure S3). This binding induced a significant conformational change of the NLS region of the protein (1.252 Å). In addition, as shown in Figure S4A, the orientation of Arg180 and Arg181, necessary for importin recognition, is significantly modified after p‐cymene binding (RMSDARG180 = 2.969 Å and RMSDARG181 = 1.831 Å), by adopting a reversion of their orientation axis, hiding the NLS sequence from the importin complex. A similar change of the orientation of NLSα amino acids occured in rabies NP (RMSD of the NLS region after p‐cymene binding = 2.090 Å). In addition, significant conformational changes of the amino acids Arg418 and Arg423 (RMSD changes after p‐cymene binding = 2.043 and 3.576 Å respectively, Figure S4B) occured after p‐cymene binding. These conformational changes resulted in a significant modification of the NLSα region, partially impairing the binding of Importin A and subsequently the efficient nuclear translocation of the NP. Finally, p‐cymene binds to the Ebola NP (Figure 2D; Figure S4C). In particular, p‐cymene binded to Leu299 and Leu300, two amino acids of the NLS of this NP. The RMSD of Ebola NP NLSα increases by 1.041 Å after p‐cymene binding due mainly to Lys281 and Arginine 298 conformational changes (t RMSDLYS281 = 1.591 Å and RMSDARG298 = 1.609 Å, after p‐cymene binding, respectively). These key amino acids for importin recognition rotated by almost 180°, and impair the recognition of the NLS sequence from the importin complex.
3.3. In vitro validation of the inhibitory effect of p‐cymene on SARS‐CoV‐2 and Influenza viruses
3.3.1. SARS‐CoV‐2
Treatment of SARS‐CoV‐2 infected Vero cells with variable concentrations of p‐cymene resulted in a significant decrease of plaque formation (Figure 4A) and viral titer in the culture supernatants (up to 90%, verified by Q‐PCR), at concentrations ranging from 0.0125 to 200 μg/ml, after 2 days of incubation (Figure 4). Up to 95% reduced titer was also observed during pre‐treatment of Vero cells, by Q‐PCR, prior to infection with SARS‐CoV‐2 (Figure 4B). The IC50 during co‐treatment was 74.5 μg/ml, while pre‐treatment IC50 was reduced to 57 μg/ml, as calculated by sigmoidal fitting of data. Importantly, p‐cymene at concentrations <100 μg/ml does not impair cell viability (Figure S5).
FIGURE 4.
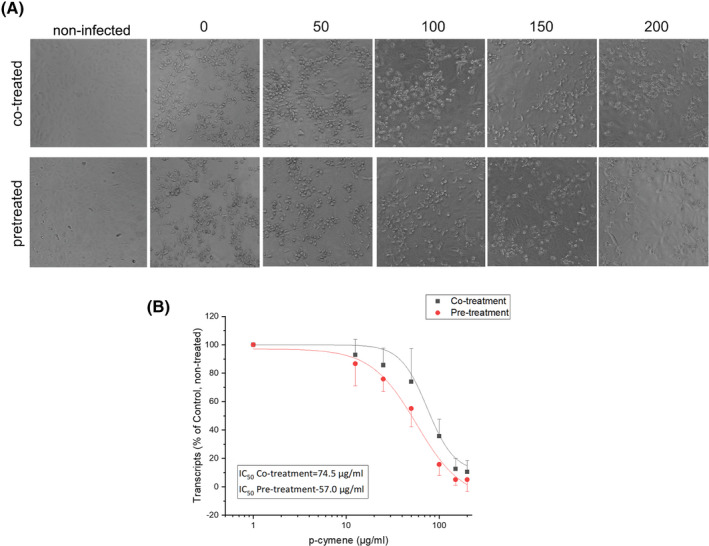
(A) Phase‐contrast photographs of cells infected with SARS‐CoV‐2 and co‐treated or pre‐treated for 2 h with the indicated concentrations of p‐cymene. (B) The inhibition of SARS‐CoV‐2 RNA in the supernatant of cell cultures is presented (mean ± SD) of cells co‐treated or pre‐treated for 2 h with the indicated concentrations of p‐cymene together with a sigmoidal fit of data. The obtained IC50s are also presented
3.3.2. Influenza A (H1N1)
Incubation of MDCK cells with variable concentrations of p‐cymene for 72 h revealed a modest effect of plaque formation, much lower than that of ribavirin (25 μg/ml), used as a positive control (Figure 5A). However, p‐cymene (20 μg/ml, as higher concentrations induced a decreased cell viability, Figure S5) significantly decreased the production of progeny virus (Figure 5B), and also decreased the genomic expression of viral proteins, as shown in Figure 5C for M1 matrix protein genome, assayed by Q‐PCR and the expression of influenza NP protein levels (Figure 5D). Finally, in view of our in silico data on the impairment of Importin A binding and the subsequent reduction of viral RNA transport to the nucleus, we assayed by immunofluorescence the cellular distribution of influenza NP. As shown in Figure 5E, after a 6 h incubation of infected cells with 20 μg/ml of p‐cymene, a reduced number of NP‐positive stained cells and a decrease of nuclear translocation of NP were found.
FIGURE 5.
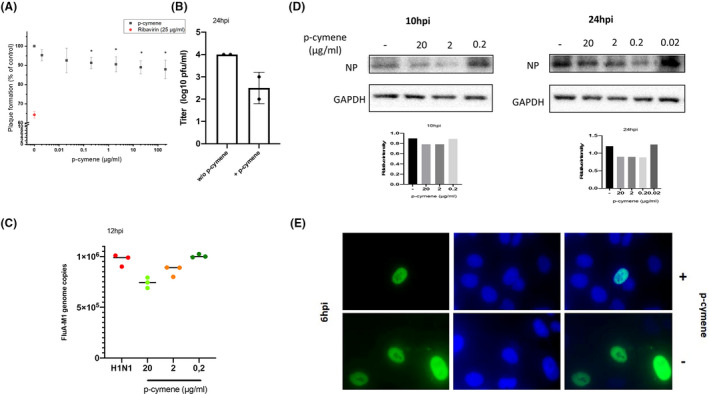
(A) Plaque reduction assay of Madin‐Darby Canine Kidney (MDCK) cells infected with Influenza H1N1 virus (0.05 PFU/cell) and incubated with variable concentrations of p‐cymene for 72 h. Figure shows mean ± SD of two experiments in triplicates. Ribavirin (red dot) at 25 μg/ml is presented as a positive control. *p < .05 as compared to non‐treated cells. (B) p‐cymene reduces the production of progeny virus, as assayed by quantitative PCR of the supernatants of MDCK cells were pretreated with p‐cymene (20 μg/ml) and infected with influenza A/H1N1 virus (0.1 PFU per cell) for 24 h. (C) Quantification by qRT‐PCR of FluA M1 genome copies in MDCK cells infected with FluA/H1N1 at MOI 0.5 PFU/cell. Data shown are means ± SD of two independent experiments. (D) Immunoblot analysis of influenza nucleoprotein (NP) in total MDCK cell lysates incubated for 10 and 24 h post‐infection with variable concentrations of p‐cymene. (E) The role of p‐cymene in the NP distribution. MDCK cells were infected with FluA/H1N1 (MOI 5) in the absence or presence of p‐cymene. Six hours after the infection, cells were fixed and subsequently stained using influenza A anti‐NP primary antibody followed by FITC secondary antibody (green). DAPI was used to visualize the nucleus area (blue)
4. DISCUSSION
COVID‐19 pandemics imposed a number of, not yet resolved, problems to the scientific community. In spite of the combined world‐wide scientific effort and the analysis of SARS‐CoV‐2 virus, 1 neither a prophylactic or therapeutic regimen has been proposed, with the exception of monoclonal antibodies 8 , 9 and dexamethasone, 50 , 51 which target hospitalized patients, in intensive care units. In this respect, the search of small, non‐expensive molecules, efficient in non‐critically‐ill patients, remains unmet. In this line, repurposing existing drugs against known or predicted SARS‐CoV‐2 protein actions have been documented (reviewed and discussed in Refs [52, 53, 54]), while natural products have also been tested (reviewed in Ref. [20]). Finally, quercetin has been proposed as an alternative for dexamethasone. 55
Here, we propose that p‐cymene can act as a potential novel agent for the treatment of RNA viruses‐induced diseases (influenza, rabies, Ebola), including SARS‐CoV‐2‐induced COVID‐19. In addition, our preliminary in vitro studies showed that the effect of p‐cymene is exerted at the low micromolar range, an effect compatible with our simulation data and the reported change in Gibbs free energy (ΔG −5.6 kcal/mol for p‐cymene interaction with SARS‐CoV‐2 N protein, and −6.5, −5.5, and −4.3 kcal/mol for influenza A, rabies, and Ebola NPs respectively). In addition, the decrease of the transcripts and the protein content of key viral proteins provides further evidence for the validity of the proposed mechanism. We further suggest as a possible mechanism of action its interaction with SARS‐CoV‐2 N‐protein and viral NPs, impairing their cytoplasmic‐nuclear shuttle, supported by extensive molecular docking and MD studies.
Our results on SARS‐CoV‐2 were performed in the African green monkey kidney Vero E6 cell line. These cells are the standard cell line used for SARS‐CoV and SARS‐CoV‐2 antiviral screening, as they are highly permissive to virus infections and lack interferon production allowing for CPE observation. However, resent studies 56 , 57 , 58 , 59 suggest that both infection and therapeutic responses might be cell line‐dependent. Indeed, in an exhaustive study 59 it was reported that only few human cell lines (CaCo‐2, Huh7, 293T, and Calu‐3), derived from colorectal adenocarcinoma, hepatocellular carcinoma, embryonic kidney, and lung adenocarcinoma respectively, were susceptible to SARS‐CoV‐2 infection, together with a number of other animal cell lines. Interestingly, the susceptibility for SARS‐CoV‐2 infection was not observed in other cell lines from the same anatomical site. As pulmonary infections are one of the primary sites of SARS‐CoV‐2 symptom manifestation, Calu‐3 cells were also extensively used for drug screening, 56 , 57 , 58 with the authors suggesting that evaluation of novel antiviral candidate drugs should be performed in different cell lines to exclude potential cell‐dependent artefacts. Interestingly, endosomal acidification was required for SARS‐CoV‐2 entry to maintain the low pH necessary for endosomal cysteine protease activity, required for priming Spike for membrane fusion in Vero and Huh7.5, while the plasma membrane‐associated serine protease TMPRSS2 is necessary for the viral glycoprotein entry priming in lung epithelial cells. 60 Although, here, we have not assayed the response of other cell lines to p‐cymene, the proposed mechanism of action (binding to the SARS‐CoV‐2 N protein, modifying its trafficking and action) is beyond differences in viral entry mechanisms. However, a thorough determination of p‐cymene effect in a number of SARS‐CoV‐2 human and animal cell lines is required, before its proposed use as a possible drug.
In contrast to SARS‐CoV‐2 infected cells, the effect of p‐cymene did not induce a complete elimination of influenza viral infection, at non‐toxic concentrations, suggesting that, this agent should be used in combination with other anti‐viral compounds. A previous study 22 revealed that a combination of plant essential oils, in which p‐cymene is a major constituent, 61 is by far more potent in inhibiting Influenza A (as well as Influenza B and rhinovirus RSV14) strains. Interestingly, molecular docking of all major constituents on Influenza A NP revealed that the majority of constituents bind at the same groove as p‐cymene (Figure S6), suggesting an additive action of the different compounds.
p‐Cymene is a naturally occurring organic compound that is classified as a hydrocarbon, related to a monoterpene. p‐Cymene is an isoprenoid lipid molecule with one aromatic ring. It exists as a solid and is considered to be practically insoluble in water, with a molecular weight of 134.222 g/mol, water solubility 23.4 mg/L and boiling point 177.1°C. It is a constituent of the essential oils of more than 100 plant species, such a T. vulgaris and O. vulgare subsp glandulosum. 62 , 63 According to the Code of Federal Regulations of the U.S. Food and Drug Administration (21CFR172.515) it is considered to be “Generally Recognized As Safe” and the EU (REACH, document Proposal for Harmonized classification and Labeling based on Regulation EC No 1272/2008, Annex VI, Part 2, Substance Name: 1‐isopropyl‐4‐methylbenzene; ρ‐cymene). A search in Pub‐Chem revealed that the in vitro IC50 of the agent is >1 mg/L tested in human monocytes, 64 while its LD50 is 1125 mg/kg IP and 1695 mg/kg PO (https://pubchem.ncbi.nlm.nih.gov/compound/7463#section=Toxicity), much higher than the concentration impairing viral proliferation VERO E6 (50–70 μg/ml) or MDCK cells (20 μg/ml), reported here. In humans, p‐cymene, at doses >3 g/kg induced only mild gastrointestinal symptoms (nausea, vomiting) or headache. The properties of p‐cymene in humans have been recently reviewed. 65 According to the authors (and references therein), p‐cymene possesses antibacterial, antifungal, and antiparasitic activities, combined with anti‐inflammatory actions. However, as the authors discuss, p‐cymene is not the main constituent in natural extracts, responsible for these actions, but it might act through synergism, antagonism, or additive effects. Here, we extend these actions, by reporting a proper antiviral effect, and suggest, by in silico studies, a novel mechanism of action through binding to NPs, including the nucleocapsid (N) protein of SARS‐CoV‐2, impairing the protein cytoplasmic‐nuclear shuttle. It derives that p‐cymene can be used as a safe agent in cases of COVID‐19 positive patients, at the concentrations revealed from our study.
SARS‐CoV‐2 N protein is mainly cytoplasmic, with very rare reports suggesting a possible nuclear translocation. This 46‐kDa protein presents a ~90% homology to the SARS‐CoV N, and a major factor conferring the enhanced pathogenicity of SARS‐CoV‐2. 66 N represents the most abundant SARS‐CoV‐2 protein in the infected cell, 67 , 68 , 69 while anti‐N SARS‐CoV‐2 antibodies are the main humoral response of COVID‐19 patients. 70 The N protein is the central component of SARS‐CoV‐2 virions. It forms the viral ribonucleoprotein (RNP) complex through binding to viral RNA; it has a primary role in viral mRNA transcription and replication, cytoskeleton organization, and immune regulation. In addition, it functions as a suppressor of cellular RNA transcription (Ref. [18] and references herein). Interestingly, SARS‐CoV‐2 has specific nuclear localization (NLS), as also reported here, and NES, 71 suggesting a possible cytoplasmic‐nuclear shuttle. However, in our analysis only one NLSα is found, contrasting the three NLSα signals reported previously. 71 SARS‐CoV‐2 N protein is a major target of phosphorylation by host cell protein kinases, with 22 putative phosphorylation sites. 68 Simulations performed in this work show a possible association of SARS‐CoV‐2 N‐protein with importin α, a process inhibited by p‐cymene. In support of such a mechanism, previous work has revealed that nuclear SARS‐CoV N protein (90% homolog to SARS‐CoV‐2) shuttles between the nucleus and the cytoplasm, a process regulated by N protein phosphorylation by several protein kinases, including glycogen synthase kinase‐3, protein kinase A, casein kinase II, and cyclin‐dependent kinase. 48 The authors suggest that nuclear localization of SARS‐CoV N protein is very short, and that the phosphorylated protein is very quickly translocated to the cytoplasm after being bound to 14‐3‐3 protein isoforms. Interestingly, a very recent report has shown a similar interaction of phosphorylated SARS‐CoV‐2 N protein with all 14‐3‐3 isoforms, 72 leading to a similar quick nucleo‐cytoplasmic shuttling of SARS‐CoV‐2 protein. In this respect, our data suggesting an interaction of p‐cymene with SARS‐CoV‐2 N protein, impairing its interaction with Importin α and subsequently inhibiting its cytoplasmic‐nuclear shuttle, might represent a valid pharmaceutic intervention in the course of COVID‐19 diseased patients.
Interestingly, p‐cymene binds also in the vicinity of the NLSα sequence of NPs of other RNA viruses (influenza, rabies, and Ebola), as revealed by simulation, modifying significantly the 3‐D conformation of the NLS site, and inhibiting its interaction with importin α, a mechanism known for influenza virus and suggested for Ebola, through an alternative O‐glycosylation and O‐phosphorylation, 73 a valid mechanism in a number of other proteins. 74 Finally, we suggest a similar mechanism for the rabies virus, although this should be confirmed by in vitro experiments. Therefore, this common mechanism of action renders p‐cymene as a possible antiviral agent, in a broad spectrum of RNA viruses from influenza A to COVID‐19, as verified by our in vitro studies of cells infected with influenza or SARS‐CoV‐2.
Is therefore p‐cymene a potential anti‐viral compound? p‐Cymene's rapid absorption, leading to a time of maximum concentration of 0.33 h, 75 is in favor of its use. However, p‐cymene is also eliminated very rapidly (T 1/2 0.44 h) from the plasma, although plasma concentration versus time profile of p‐cymene shows a double‐peak which might be due to distribution, reabsorption, or enterohepatic circulation. 75 Finally, taking into consideration the half‐life of p‐cymene, assuming an absorption of 50% after an oral dose, the effective concentration of 20–50 μg/ml in the interstitial fluid, a dose of 12 mg of p‐cymene should be administered every 6 h for a 70 kg adult, in order to maintain an effective concentration. This dose is far below that reported to induce any side effect in animals or in humans (https://pubchem.ncbi.nlm.nih.gov/compound/7463#section=Toxicity).
In conclusion, data presented in this study suggest that p‐cymene, a non‐toxic natural compound, may be used alone or as a companion therapeutic for the control of RNA viral diseases, including COVID‐19 and influenza. However, the in silico identified mechanism of action should be further verified by additional in vitro experiments.
DISCLOSURE
AP, MT, CL, MK, SP, GS, and EC are inventors in a PCT patent application (priority # GR‐is 20200100068, GB‐2002141.6, PCT/EP2021/053389), related to the subject of this work. MK, GS, CL, SP, and EC are shareholders of the University of Crete spin‐off company Nature Crete Pharmaceuticals PC.
AUTHOR CONTRIBUTION
Castanas, Sourvinos, Pirintsos, and Karakasiliotis participated in research design. Panagiotopoulos, Tseliou, Karakasiliotis, Kotzambasi, Daskalakis, and Kesesidis, Kampa conducted experiments. Karakasiliotis, Daskalakis, Notas, Sourvinos, and Castanas performed data analysis. Karakasiliotis, Daskalakis, Lionis, Kampa, Pirintsos, Sourvinos, and Castanas wrote or contributed in the writing of the manuscript.
ETHICS
Ethics issues are not applicable to this work.
Supporting information
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge PRACE for awarding us access to Joliot‐Curie at GENI@CEA (Irene), France, through the “PRACE support to mitigate impact of COVID‐19 pandemic” call and the project “Epitope vaccines based on the dynamics of mutated SARS‐CoV‐2 proteins at all atom resolution.” We also acknowledge Greece and the European Union (European Social Fund‐ESF) for funding through the Operational Program «Human Resources Development, Education and Lifelong Learning» in the context of the project “Strengthening Human Resources Research Potential via Doctorate Research” (MIS‐5000432), implemented by the State Scholarships Foundation (ΙΚΥ)» to AAP (PhD scholarship) and a Hellenic Foundation for Research and Innovation (H.F.R.I.) Grant to MK (# 3725). The financial support of Galenica SA is also acknowledged. All data are included in the main and the Supplemental part of this work.
Panagiotopoulos A, Tseliou M, Karakasiliotis I, et al. p‐cymene impairs SARS‐CoV‐2 and Influenza A (H1N1) viral replication: In silico predicted interaction with SARS‐CoV‐2 nucleocapsid protein and H1N1 nucleoprotein. Pharmacol Res Perspect. 2021;9:e00798. 10.1002/prp2.798
DATA AVAILABILITY STATEMENT
All data are included in the text, figures, and supporting information provided.
REFERENCES
- 1. O'Leary VB, Ovsepian SV. Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). Trends Genet. 2020;36(11):892‐893. 10.1016/j.tig.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Voysey M, Clemens SAC, Madhi SA, et al. Safety and efficacy of the ChAdOx1 nCoV‐19 vaccine (AZD1222) against SARS‐CoV‐2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397(10269):99‐111. 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baden LR, El Sahly HM, Essink B, et al. Efficacy and safety of the mRNA‐1273 SARS‐CoV‐2 vaccine. N Engl J Med. 2021;384(5):403‐416. 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid‐19 vaccine. N Engl J Med. 2020;383(27):2603‐2615. 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sadoff J, Le Gars M, Shukarev G, et al. Interim results of a phase 1–2a trial of Ad26.COV2.S Covid‐19 vaccine. N Engl J Med. 2021;1‐2. 10.1056/NEJMoa2034201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Logunov DY, Dolzhikova IV, Shcheblyakov DV, et al. Safety and efficacy of an rAd26 and rAd5 vector‐based heterologous prime‐boost COVID‐19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397(10275):671‐681. 10.1016/S0140-6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dai L, Gao GF. Viral targets for vaccines against COVID‐19. Nat Rev Immunol. 2021;21(2):73‐82. 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hansen J, Baum A, Pascal KE, et al. Studies in humanized mice and convalescent humans yield a SARS‐CoV‐2 antibody cocktail. Science. 2020;369(6506):1010‐1014. 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baum A, Ajithdoss D, Copin R, et al. REGN‐COV2 antibodies prevent and treat SARS‐CoV‐2 infection in rhesus macaques and hamsters. Science. 2020;370(6520):1110‐1115. 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nie X, Qian L, Sun R, et al. Multi‐organ proteomic landscape of COVID‐19 autopsies. Cell. 2021;184(3):775‐791.e14. 10.1016/j.cell.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schneider WM, Luna JM, Hoffmann HH, et al. Genome‐scale identification of SARS‐CoV‐2 and pan‐coronavirus host factor networks. Cell. 2021;184(1):120‐132.e14. 10.1016/j.cell.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network‐based drug repurposing for novel coronavirus 2019‐nCoV/SARS‐CoV‐2. Cell Discov. 2020;6:14. 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fernandes MS, da Silva FS, Freitas A, de Melo EB, Trossini GHG, Paula FR. Insights on 3D structures of potential drug‐targeting proteins of SARS‐CoV‐2: application of cavity search and molecular docking. Mol Inform. 2021;40(2):e2000096. 10.1002/minf.202000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565‐574. 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tardif J, Bouabdallaoui N, L’Allier P, et al. Efficacy of colchicine in non‐hospitalized patients with COVID‐19. MedRxiv. 2021. 10.1101/2021.01.26.21250494. [DOI] [Google Scholar]
- 16. Deftereos SG, Giannopoulos G, Vrachatis DA, et al. Effect of colchicine vs standard care on cardiac and inflammatory biomarkers and clinical outcomes in patients hospitalized with coronavirus disease 2019. JAMA Netw Open. 2020;3(6):e2013136. 10.1001/jamanetworkopen.2020.13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zeng W, Liu G, Ma H, et al. Biochemical characterization of SARS‐CoV‐2 nucleocapsid protein. Biochem Biophys Res Commun. 2020;527(3):618‐623. 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peng Y, Du N, Lei Y, et al. Structures of the SARS‐CoV‐2 nucleocapsid and their perspectives for drug design. EMBO J. 2020;39(20):e105938. 10.15252/embj.2020105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Surjit M, Lal S. The nucleocapsid protein of the SARS coronavirus: structure, function and therapeutic potential. In: Lal S, ed. Molecular Biology of the SARS‐Coronavirus. Berlin, Germany: Springer; 2010:129‐151. [Google Scholar]
- 20. Benarba B, Pandiella A. Medicinal plants as sources of active molecules against COVID‐19. Front Pharmacol. 2020;11:1189. 10.3389/fphar.2020.01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pirintsos S, Bariotakis M, Kampa M, Sourvinos G, Lionis C, Castanas E. The therapeutic potential of the essential oil of Thymbra capitata (L.) Cav., Origanum dictamnus L., Salvia fruticosa Mill. and a case of plant‐based pharmaceutical development. Front Pharmacol. 2020;11:522213. 10.3389/fphar.2020.522213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tseliou M, Pirintsos SA, Lionis C, Castanas E, Sourvinos G. Antiviral effect of an essential oil combination derived from three aromatic plants (Coridothymus capitatus (L.) Rchb. f., Origanum dictamnus L. and Salvia fruticosa Mill.) against viruses causing infections of the upper respiratory tract. J Herb Med. 2019;17–18:100288. 10.1016/j.hermed.2019.100288. [DOI] [Google Scholar]
- 23. Lionis C, Karakasiliotis I, Petelos E, et al. A mixture of essential oils from three Cretan Aromatic Plants (thyme, Greek sage and Cretan dittany, CAPeo) inhibits SASR‐CoV‐2 proliferation: in vitro evidence and a Proof‐of‐Concept intervention study in mild ambulatory COVID‐19‐positive patients. MedRxiv. 2021. 10.1101/2021.01.11.20248947. [DOI] [Google Scholar]
- 24. Lee GR, Heo L, Seok C. Effective protein model structure refinement by loop modeling and overall relaxation. Proteins. 2016;84(Suppl 1):293‐301. 10.1002/prot.24858. [DOI] [PubMed] [Google Scholar]
- 25. Ko J, Park H, Heo L, Seok C. GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res. 2012;40(W1):W294‐W297. 10.1093/nar/gks493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shin W, Lee G, Heo L, Lee H, Seok C. Prediction of protein structure and interaction by GALAXY protein modeling programs. Bio Design. 2014;2(1):1‐11. [Google Scholar]
- 27. Heo L, Park H, GalaxyRefine SC. Protein structure refinement driven by side‐chain repacking. Nucleic Acids Res. 2013;41(W1):W384‐W388. 10.1093/nar/gkt458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Minasov G, Shuvalova L, Wiersum G, Satchell K. 2.05 angstrom resolution crystal structure of C‐terminal dimerization domain of nucleocapsid phosphoprotein from SARS‐CoV‐2. 2020. www.rcsb.org/structure/6WJI#entity‐1. Accessed April 22, 2020.
- 29. O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J Cheminform. 2011;3(1):33. 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605‐1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 31. Hofacker IL, Stadler PF. Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics. 2006;22(10):1172‐1176. 10.1093/bioinformatics/btl023. [DOI] [PubMed] [Google Scholar]
- 32. Popenda M, Szachniuk M, Antczak M, et al. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012;40(14):e112. 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Antczak M, Popenda M, Zok T, et al. New functionality of RNAComposer: an application to shape the axis of miR160 precursor structure. Acta Biochim Pol. 2016;63(4):737‐744. 10.18388/abp.2016_1329. [DOI] [PubMed] [Google Scholar]
- 34. Ritchie DW. Evaluation of protein docking predictions using Hex 3.1 in CAPRI rounds 1 and 2. Proteins. 2003;52(1):98‐106. 10.1002/prot.10379. [DOI] [PubMed] [Google Scholar]
- 35. Panagiotopoulos AA, Polioudaki C, Ntallis SG, et al. The sequence [EKRKI(E/R)(K/L/R/S/T)] is a nuclear localization signal for importin 7 binding (NLS7). Biochim Biophys Acta Gen Subj. 2021;1865(5):129851. 10.1016/j.bbagen.2021.129851. [DOI] [PubMed] [Google Scholar]
- 36. Duan Y, Wu C, Chowdhury S, et al. A point‐charge force field for molecular mechanics simulations of proteins based on condensed‐phase quantum mechanical calculations. J Comput Chem. 2003;24(16):1999‐2012. 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 37. Sousa da Silva AW, Vranken WF. ACPYPE—AnteChamber PYthon Parser interfacE. BMC Res Notes. 2012;5(1):367. 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berendsen H, van der Spoel D, van Drunen R. GROMACS: a message‐passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91:43‐56. [Google Scholar]
- 39. Chodera JD, Noe F. Markov state models of biomolecular conformational dynamics. Curr Opin Struct Biol. 2014;25:135‐144. 10.1016/j.sbi.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Prinz JH, Wu H, Sarich M, et al. Markov models of molecular kinetics: generation and validation. J Chem Phys. 2011;134(17):174105. 10.1063/1.3565032. [DOI] [PubMed] [Google Scholar]
- 41. Pande VS, Beauchamp K, Bowman GR. Everything you wanted to know about Markov State Models but were afraid to ask. Methods. 2010;52(1):99‐105. 10.1016/j.ymeth.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Raiteri P, Laio A, Gervasio FL, Micheletti C, Parrinello M. Efficient reconstruction of complex free energy landscapes by multiple walkers metadynamics. J Phys Chem B. 2006;110(8):3533‐3539. 10.1021/jp054359r. [DOI] [PubMed] [Google Scholar]
- 43. Glatthaar‐Saalmuller B, Fal AM, Schonknecht K, Conrad F, Sievers H, Saalmuller A. Antiviral activity of an aqueous extract derived from Aloe arborescens Mill. against a broad panel of viruses causing infections of the upper respiratory tract. Phytomedicine. 2015;22(10):911‐920. 10.1016/j.phymed.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thulasi Raman SN, Latreille E, Gao J, et al. Dysregulation of Ephrin receptor and PPAR signaling pathways in neural progenitor cells infected by Zika virus. Emerg Microbes Infect. 2020;9(1):2046‐2060. 10.1080/22221751.2020.1818631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Epidemiol. 1938;27(3):493‐497. 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 46. Harding SD, Sharman JL, Faccenda E, et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res. 2018;46(D1):D1091‐D1106. 10.1093/nar/gkx1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Alexander SPH, Kelly E, Mathie A, et al. THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: introduction and other protein targets. Br J Pharmacol. 2019;176(Suppl 1):S1‐S20. 10.1111/bph.14747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Surjit M, Kumar R, Mishra RN, Reddy MK, Chow VT, Lal SK. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14–3‐3‐mediated translocation. J Virol. 2005;79(17):11476‐11486. 10.1128/JVI.79.17.11476-11486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou R, Zeng R, von Brunn A, Lei J. Structural characterization of the C‐terminal domain of SARS‐CoV‐2 nucleocapsid protein. Mol Biomed. 2020;1:2. 10.1186/s43556-020-00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Horby P, Lim WS, et al. Dexamethasone in hospitalized patients with Covid‐19—preliminary report. N Engl J Med. 2020. 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cain DW, Cidlowski JA. After 62 years of regulating immunity, dexamethasone meets COVID‐19. Nat Rev Immunol. 2020;20(10):587‐588. 10.1038/s41577-020-00421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cadegiani FA. Repurposing existing drugs for COVID‐19: an endocrinology perspective. BMC Endocr Disord. 2020;20(1):149. 10.1186/s12902-020-00626-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Khan Z, Ghafoor D, Khan A, et al. Diagnostic approaches and potential therapeutic options for coronavirus disease (COVID‐19). New Microbes New Infect. 2020:100770. 10.1016/j.nmni.2020.100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Asselah T, Durantel D, Pasmant E, Lau G, Schinazi RF. COVID‐19: Discovery, diagnostics and drug development. J Hepatol. 2020. 10.1016/j.jhep.2020.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pawar A, Pal A. Molecular and functional resemblance of dexamethasone and quercetin: a paradigm worth exploring in dexamethasone‐nonresponsive COVID‐19 patients. Phytother Res. 2020. 10.1002/ptr.6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Holwerda M, V'Kovski P, Wider M, Thiel V, Dijkman R. Identification of an antiviral compound from the pandemic response box that efficiently inhibits SARS‐CoV‐2 infection in vitro. Microorganisms. 2020;8(12). 10.3390/microorganisms8121872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dittmar M, Lee JS, Whig K, et al. Drug repurposing screens reveal cell‐type‐specific entry pathways and FDA‐approved drugs active against SARS‐Cov‐2. Cell Rep. 2021;35(1):108959. 10.1016/j.celrep.2021.108959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sheahan TP, Sims AC, Zhou S, et al. An orally bioavailable broad‐spectrum antiviral inhibits SARS‐CoV‐2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med. 2020;12(541):eabb5883. 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chu H, Chan JF, Yuen TT, et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS‐CoV‐2 and SARS‐CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID‐19: an observational study. Lancet Microbe. 2020;1(1):e14‐e23. 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280.e8. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Duijker G, Bertsias A, Symvoulakis EK, et al. Reporting effectiveness of an extract of three traditional Cretan herbs on upper respiratory tract infection: results from a double‐blind randomized controlled trial. J Ethnopharmacol. 2015;163:157‐166. 10.1016/j.jep.2015.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mechergui K, Jaouadi W, Coelho JP, Khouja ML. Effect of harvest year on production, chemical composition and antioxidant activities of essential oil of oregano (Origanum vulgare subsp glandulosum (Desf.) Ietswaart) growing in North Africa. Ind Crop Prod. 2016;90:32‐37. 10.1016/j.indcrop.2016.06.011. [DOI] [Google Scholar]
- 63. Jordan MJ, Martinez RM, Goodner KL, Baldwin EA, Sotomayor JA. Seasonal variation of Thymus hyemalis Lange and Spanish Thymus vulgaris L. essential oils composition. Ind Crop Prod. 2006;24(3):253‐263. 10.1016/j.indcrop.2006.06.011. [DOI] [Google Scholar]
- 64. Kavoosi G, Teixeira da Silva JA. Inhibitory effects of Zataria multiflora essential oil and its main components on nitric oxide and hydrogen peroxide production in glucose‐stimulated human monocyte. Food Chem Toxicol. 2012;50(9):3079‐3085. 10.1016/j.fct.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 65. Marchese A, Arciola CR, Barbieri R, et al. Update on monoterpenes as antimicrobial agents: a particular focus on p‐cymene. Materials. 2017;10(8):947. 10.3390/ma10080947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gussow AB, Auslander N, Faure G, Wolf YI, Zhang F, Koonin EV. Genomic determinants of pathogenicity in SARS‐CoV‐2 and other human coronaviruses. Proc Natl Acad Sci U S A. 2020;117(26):15193‐15199. 10.1073/pnas.2008176117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bar‐On YM, Flamholz A, Phillips R, Milo R. SARS‐CoV‐2 (COVID‐19) by the numbers. Elife. 2020;9. 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bouhaddou M, Memon D, Meyer B, et al. The global phosphorylation landscape of SARS‐CoV‐2 infection. Cell. 2020;182(3):685‐712.e19. 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bojkova D, Klann K, Koch B, et al. Proteomics of SARS‐CoV‐2‐infected host cells reveals therapy targets. Nature. 2020;583(7816):469‐472. 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Smits VAJ, Hernandez‐Carralero E, Paz‐Cabrera MC, et al. The nucleocapsid protein triggers the main humoral immune response in COVID‐19 patients. Biochem Biophys Res Commun. 2021;543:45‐49. 10.1016/j.bbrc.2021.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kumar A, Parveen A, Kumar N, et al. Characterization of nucleocapsid (N) protein from novel coronavirus SARS‐CoV‐2. 2020. 10.20944/preprints202005.0413.v1. [DOI]
- 72. Tugaeva KV, Hawkins DEDP, Smith JLR, et al. The mechanism of SARS‐CoV‐2 nucleocapsid protein recognition by the human 14‐3‐3 proteins. J Mol Biol. 2021;433(8):166875. 10.1016/j.jmb.2021.166875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Watanabe S, Noda T, Kawaoka Y. Functional mapping of the nucleoprotein of Ebola virus. J Virol. 2006;80(8):3743‐3751. 10.1128/JVI.80.8.3743-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kamemura K, Hart G. Dynamic interplay between O‐glycosylation and O‐phosphorylation of nucleocytoplasmic proteins: a new paradigm for metabolic control of signal transduction and transcription. Prog Nucleic Acid Res Mol Biol. 2003;73:107‐136. 10.1016/s0079-6603(03)01004-3. [DOI] [PubMed] [Google Scholar]
- 75. Hu X, Chu Y, Ma G, et al. Simultaneous determination of ascaridole, p‐cymene and alpha‐terpinene in rat plasma after oral administration of Chenopodium ambrosioides L. by GC‐MS. Biomed Chromatogr. 2015;29(11):1682‐1686. 10.1002/bmc.3479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
All data are included in the text, figures, and supporting information provided.


