Abstract
Purpose
To develop an unsupervised deep learning model on MR images of normal brain anatomy to automatically detect deviations indicative of pathologic states on abnormal MR images.
Materials and Methods
In this retrospective study, spatial autoencoders with skip-connections (which can learn to compress and reconstruct data) were leveraged to learn the normal variability of the brain from MR scans of healthy individuals. A total of 100 normal, in-house MR scans were used for training. Subsequently, as the model was unable to reconstruct anomalies well, this characteristic was exploited for detecting and delineating various diseases by computing the difference between the input data and their reconstruction. The unsupervised model was compared with a supervised U-Net– and threshold-based classifier trained on data from 50 patients with multiple sclerosis (in-house dataset) and 50 patients from The Cancer Imaging Archive. Both the unsupervised and supervised U-Net models were tested on five different datasets containing MR images of microangiopathy, glioblastoma, and multiple sclerosis. Precision-recall statistics and derivations thereof (mean area under the precision-recall curve, Dice score) were used to quantify lesion detection and segmentation performance.
Results
The unsupervised approach outperformed the naive thresholding approach in lesion detection (mean F1 scores ranging from 17% to 62% vs 6.4% to 15% across the five different datasets) and performed similarly to the supervised U-Net (20%–64%) across a variety of pathologic conditions. This outperformance was mostly driven by improved precision compared with the thresholding approach (mean precisions, 15%–59% vs 3.4%–10%). The model was also developed to create an anomaly heatmap display.
Conclusion
The unsupervised deep learning model was able to automatically detect anomalies on brain MR images with high performance.
Supplemental material is available for this article.
Keywords: Brain/Brain Stem Computer Aided Diagnosis (CAD), Convolutional Neural Network (CNN), Experimental Investigations, Head/Neck, MR-Imaging, Quantification, Segmentation, Stacked Auto-Encoders, Technology Assessment, Tissue Characterization
© RSNA, 2021
Summary
An autoencoder was trained on healthy brain anatomy on MR images and then used to successfully detect anomalies on test datasets containing images of microangiopathy, glioblastoma, and multiple sclerosis.
Key Results
■ An unsupervised deep learning model trained on healthy brain scans was able to detect a wide range of anomalies on independent test datasets, and performance was comparable with that of a supervised U-Net.
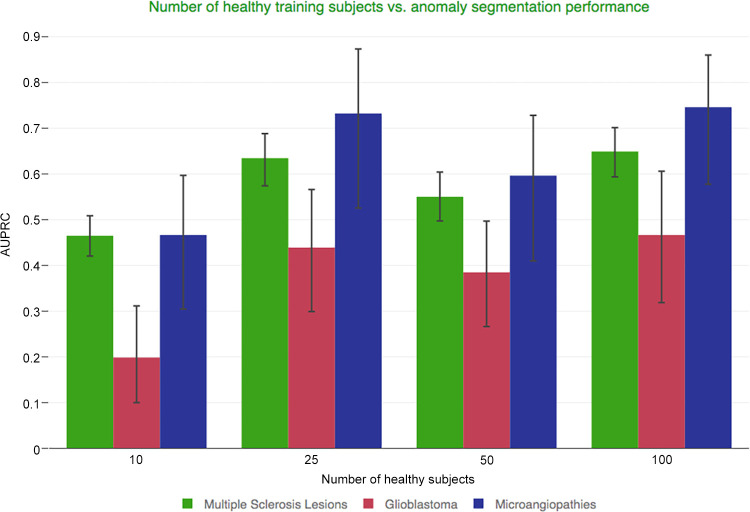
■ As opposed to supervised strategies, with the unsupervised strategy, even small numbers of healthy training patients yielded acceptable model performance (mean area under the precision-recall curve for training with 25 vs 100 patients: multiple sclerosis, 0.64 vs 0.65; glioblastoma, 0.44 vs 0.47; and microangiopathy, 0.73 vs 0.75).
Introduction
MRI of the brain is of fundamental importance for the diagnosis and treatment of neurologic diseases. Reading and interpreting MR studies is an intricate process, which can be prone to errors. Bruno et al (1) reported that in up to 5%–10% of images, a relevant pathologic condition is missed. To support radiologists in the clinic, outstanding levels of performance have been achieved with recent supervised deep learning methods. However, their training requires vast amounts of labeled data, which are tedious to produce. Additionally, these approaches suffer from limited generalizability because, in general, training data rarely comprise the gamut of all possible pathologic appearances. Obtaining enough training data at all for less common diseases can be particularly challenging.
Given these constraints, solving the problem of limited data by approaching it in a way similar to the way in which humans are trained to recognize normal anatomy first constitutes an attractive alternative approach. In Schlegl et al (2), such an attempt was made for modeling normality in retinal optical coherence tomographic data with generative adversarial networks; however, this was done at the level of small image patches, which, together with the unstable training and costly inference of the method, renders the approach impractical for high-resolution MR images. In contrast, autoencoders can model normality through a reconstruction objective and thus do not suffer from training instabilities and can, more importantly, work with high-resolution data, such as data from full MR images. In the medical field, a few attempts involving autoencoders for unsupervised anomaly detection have been made. Vaidhya et al (3) used unsupervised three-dimensional (3D) stacked denoising autoencoders for patch-based glioma detection and segmentation on brain MR images; however, this was only used as a pretraining step for a supervised model. In another recent work (4), constrained adversarial autoencoders were proposed to detect and delineate anomalies on brain MR images; however, this was only applied at low resolution in such a way that the method was not able to deal with small lesions like those in multiple sclerosis. Baur et al (5) proposed the use of spatial autoencoders to facilitate the detection of small lesions, but the method was constrained to few consecutive axial sections.
Here, we propose a method for unsupervised anomaly detection on whole-brain MR images by modeling the normal variability of brain anatomy in which we leverage spatial autoencoders with skip-connections (6). In contrast to previous works, which either operate on small image patches and are thus computationally expensive or operate on low-resolution data and thus risk losing small anomalous structures, in this work, the use of spatial autoencoders allowed us to build a model that can capture “global” normal anatomic appearance at high resolution of entire brain volumes. The specific design enhances reconstruction quality such that not only large pathologic lesions but also small multiple sclerosis lesions can be detected. We evaluated our method on in-house and public MR images with three different pathologic conditions and report both qualitative and quantitative brain lesion detection and delineation results.
Materials and Methods
Patient Datasets
This retrospective study was approved by the local institutional review board, and informed consent was waived. Two different models were trained in this study: an unsupervised model was trained on normal MR images, and a supervised U-Net was trained on MR images of multiple sclerosis, glioblastoma, and microangiopathy pathologic characteristics. Development and testing of these two models were done using a total of six different in-house or publicly available MR datasets: 109 individuals with normal MR images (in-house), 99 patients with multiple sclerosis (in-house), 20 patients with glioblastoma (in-house), 10 patients with microangiopathy (in-house), 51 patients from the White Matter Hyperintensity segmentation challenge (7), and 70 patients from The Cancer Imaging Archive (TCIA) (8).
All in-house images were acquired between 2015 and 2018. The MR images from the healthy individuals (n = 109) were used for training and validating the unsupervised model. These MR images consisted of 3D T2 fluid-attenuated inversion recovery (FLAIR) images with no detected abnormalities. These individuals were scanned for various clinical reasons (most often headache) and were randomly selected from our database. A total of 100 patients were used for model training, and nine patients were used for model validation. In-house MR images from patients with multiple sclerosis, glioblastoma, and microangiopathy were randomly selected from the in-house databases. These MR images were used in the training of the supervised U-Net, as well as in the validation and testing of both the unsupervised model and supervised U-Net (dataset splits are shown in Table 1). Last, images from both the White Matter Hyperintensity segmentation challenge dataset and TCIA low-grade glioma and glioblastoma datasets were randomly selected for inclusion.
Table 1:
Patient and Scanner Information
Table 1 lists all datasets as well as sequence details. Table E1 (supplement) lists the patient identifiers of the public datasets. Patient data from the multiple sclerosis cohorts (9,10) and data for patients with glioblastomas (11) were previously reported as being used for the development (9) or evaluation (11) of segmentation tools as well as for the development of a classifier for the detection of multiple sclerosis (10).
MRI Parameters for the In-House Datasets
Images from our in-house, local database were acquired during clinical routine with 3-T MRI scanners from Philips (Achieva, Achieva dStream) or Siemens (Verio). All images were 3D FLAIR acquisitions, although there were imaging parameters that varied between scanners, as listed in Table 1.
Image Preprocessing
All images were rigidly co-registered to the SRI24 atlas (12) to harmonize spatial orientation, skull-stripped with ROBEX (RObust Brain Extraction) (13), and denoised using CurvatureFlow (14). To increase numeric stability, every scan was further normalized into the range [0,1] based on its respective 98th percentile.
Image Assessment and Segmentation
Ground truth segmentations for local images were checked or generated by B.W. (neuroradiologist with 8 years of training) using the freely available software ITK-Snap (https://www.itksnap.org/pmwiki/pmwiki.php). Images were segmented as 3D volumes using a thresholding-based, supervised approach implemented in ITK-Snap. All segmentations were validated in all three orientations (axial, sagittal, coronal) for spatial coherence.
Unsupervised Training with Normal MR Images for Healthy Brain Anatomy
An overview of our framework is given in Figure 1. A total of 100 MR images of patients without pathologic brain conditions (training data) served to train a specifically designed autoencoder (see Fig 1, C for the underlying architecture). In the training stage (Fig 1, A), this neural network was first presented only with FLAIR images of healthy human brains, which allowed the network to capture the underlying distribution of normal brain anatomy. The training process was performed by iteratively updating the learnable network parameters using the backpropagation algorithm (stochastic gradient descent) to minimize the L2 distance between input samples and their reconstructions.
Figure 1:
Overview of the model framework: the proposed anomaly-detection concept at a glance. An autoencoder is optimized to compress and reconstruct MR images of healthy brain anatomy (training phase, A). A simple subtraction (during the inference phase, B) of the reconstructed image from the corresponding input data reveals lesions in the brain. C, Underlying neural network architecture.
Subtraction Images after Processing and Binarization
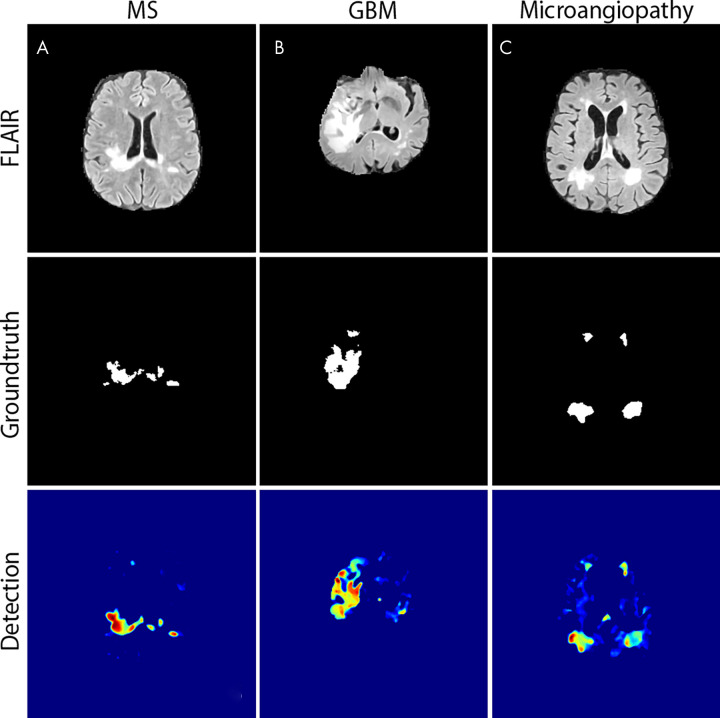
During testing, we obtained subtraction images by computing the difference between an input and its reconstruction (Fig 1, B). For this assessment, the images were fed through the network, and subtraction images were computed from the original and respective reconstructions. Each subtraction image was displayed as a heatmap (Fig 2), in which high residuals (setting themselves apart from the blue-colored nonanomalous pixels) depict anomalous structures, such as multiple sclerosis lesions, tumors, or ischemic white matter lesions. To remove filamentary residuals, we applied a 3D median filter with kernel size of 5 × 5 × 5. The resulting image was plotted as a heatmap (Fig 2) or was thresholded to obtain a binary segmentation. To automatically choose a cutoff for binarization, we leveraged another 18 annotated MR images for validation. These comprised three patients with multiple sclerosis, glioblastoma, and microangiopathy, as well as nine healthy control subjects (Table 1). Inference was performed on all sections containing brain tissue, and a cutoff that yielded the highest Dice score on those samples was determined. After thresholding, a connected component analysis was performed to discard any lesion candidates smaller than 2 voxels in any direction.
Figure 2:
Example of heatmaps. Examples of A, multiple sclerosis (MS) lesions, B, a glioblastoma (GBM), and C, microangiopathy. The first row shows the respective axial fluid-attenuated inversion recovery (FLAIR) section, the middle row shows the ground truth segmentation, and the bottom row shows the result of our anomaly detection displayed as a heatmap.
Comparison Methods
We compared our method with a supervised deep learning model, U-Net (15), which directly generates probability maps for lesion segmentations. Unlike the autoencoder, the U-Net was trained on an independent cohort of 100 additional samples with segmentation ground truth, comprising 50 patients with multiple sclerosis from our local database and 50 patients with glioma from the TCIA (see Table 1). The network structure followed the original description by Ronneberger et al (15) but was adjusted to handle data at a 256 × 256 resolution. We used a cutoff detection probability of 0.5 to binarize the predictions of the U-Net and to obtain state-of-the-art performance. In parallel, we evaluated a naive thresholding-based classifier, which only requires data for determining the threshold rather than also requiring training data. For this purpose, we calculated a cutoff maximizing the Dice score on the same 18 validation patients as outlined above for the autoencoder. For a fair comparison, the same pre- and postprocessing was applied to all three methods.
Ablation Studies
Autoencoders were trained with either 10, 25, 50, or 100 MR images from healthy individuals and then applied on the testing set to assess the impact on the delineation of the anomalies.
We additionally modeled normal brain anatomy at three different resolutions to investigate the impact of image resolution on the segmentation performance regarding different pathologic conditions.
Statistical Analysis
An anomaly was considered as detected if its ground truth–connected component overlapped with a 3D conglomeration of binarized residuals and if the latter was less than twice the size of ground truth–connected component in any dimension. To quantify the delineation performance, we computed the precision and recall at continuous cutoffs є [0,1] to obtain the precision-recall curve. The larger the area under the precision-recall curve (AUPRC), the better the performance. Additionally, we calculated the precision, recall, and F1 score (which is the harmonic mean of precision and recall; for anomaly detection) as well the Dice score (for anomaly delineation) on the predefined cutoff. Although the F1 score and Dice score are mathematically equivalent, we draw a distinction to differentiate between detection and delineation performance. Statistical analyses were implemented in Python using the scientific Scikit-learn and SciPy modules.
Results
Anomaly Detection Performance
On our in-house dataset and the public TCIA dataset, our unsupervised approach showed a higher F1 score than both the supervised U-Net classifier and the naive, threshold-based classifier for anomaly detection (Table 2). Although our approach substantially outperformed the threshold-based approach (with mean F1 scores ranging from 17% to 62% vs 6.4% to 15%), the comparison with a U-Net favored our approach for some tasks (the glioblastoma or microangiopathy datasets) and favored the U-Net for other tasks, although the differences were generally small (with mean F1 scores for the U-Net ranging from 20% to 64%). On closer inspection (Table 2), our method produced fewer false-positive results than both the threshold-based classifier and the U-Net (higher precision) while maintaining better detection performance than the threshold-based classifier (higher recall). With the exception of our in-house microangiopathy dataset and the public TCIA dataset, the U-Net identified most of the lesions. On the two external datasets, all methods had lower performance.
Table 2:
Lesion Detection Rates using the F1 Score, Recall, and Precision
Anomaly Delineation Performance
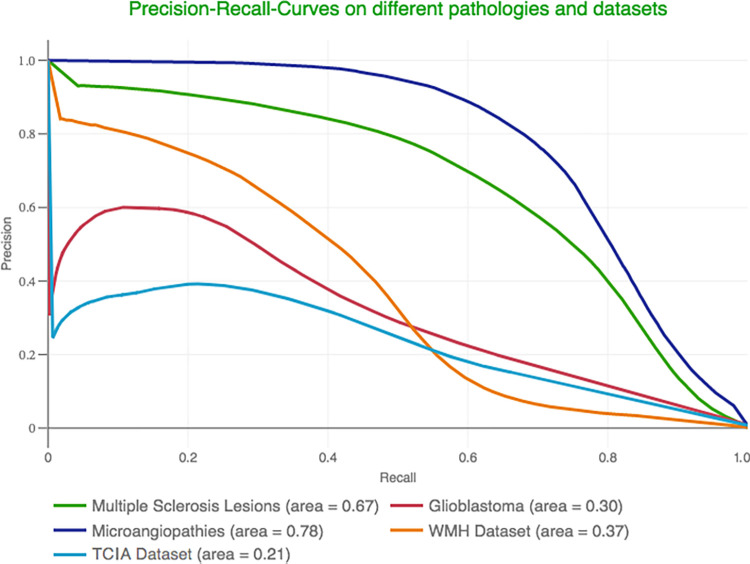
For rating the quality of anomaly delineations (ie, the precise coverage of every anomalous voxel), we report the AUPRC (Fig 3) over all testing patients combined. Additionally, we computed the Dice score using a cutoff determined with a held-out validation set of 18 patients (Fig 4). In this context, the trained network was able to reach Dice scores of 0.39 (AUPRC, 0.30) and 0.73 (AUPRC, 0.79) for glioblastoma and microangiopathy, respectively. Multiple sclerosis lesions, which can be particularly small, were delineated with a Dice score of 0.65 and an AUPRC of 0.67. On the two external datasets, the Dice scores were 0.45 (AUPRC, 0.37) for the White Matter Hyperintensity data and 0.35 (AUPRC, 0.21) for the TCIA data.
Figure 3:
Precision–recall curves for anomaly delineation. The models’ performance in segmenting multiple sclerosis lesions, glioblastomas, microangiopathies, and pathologic conditions from two other public datasets quantified with a precision–recall curve. TCIA = glioma dataset from The Cancer Imaging Archive, WMH = dataset from the White Matter Hyperintensity segmentation challenge.
Figure 4:
Patient-wise Dice scores. The models’ patient-wise segmentation performance on multiple sclerosis lesions, glioblastomas, microangiopathies, and pathologic conditions in two other public datasets, measured with the help of the Dice score. Data are portrayed using box plots. TCIA = glioma dataset from The Cancer Imaging Archive, WMH = dataset from the White Matter Hyperintensity segmentation challenge.
Minimum Number of Healthy MR Images Required for Optimal Model Performance
Figure 5 and Table E2 (supplement) show the patient-wise AUPRC statistics with bootstrapped CIs (alpha = .05) for different training set sizes on our in-house datasets. Increasing the number of training patients from 10 to 25 improved the AUPRC for all three tasks to values comparable with those using the entire training dataset of 100 patients.
Figure 5:
Ablation study using a variable number of healthy individuals’ MR images. The number of healthy training participants and their impact on the model’s anomaly delineation performance, quantified with the help of the area under the precision-recall curve (AUPRC) that is computed (AUPRC per patient).
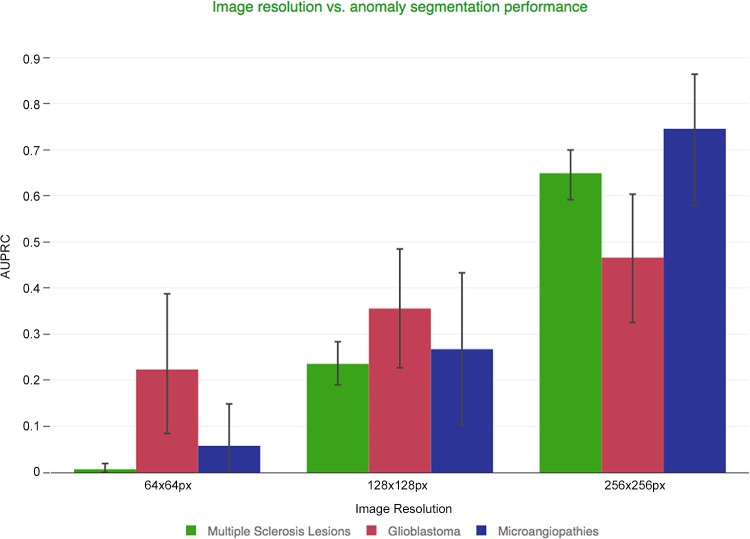
Influence of Image Resolution on Delineation Performance
Image resolution is a crucial parameter to consider when trade-offs must be made to accommodate model complexity and the variability of the brain. For small, hyperintense lesions, the best results were obtained at the native (highest) resolution. In contrast, large, space-occupying anomalies, such as glioblastomas, benefit less from a higher resolution (Fig 6).
Figure 6:
Ablation study on autoencoder performance at different image resolutions. Image resolution and its impact on the model’s anomaly-delineation performance, quantified with the help of the area under the precision–recall curve (AUPRC) (AUPRC per patient).
Discussion
In this study, we propose an inverse solution to unsupervised anomaly detection. Instead of focusing on training a network to identify pathologic conditions, we trained an unsupervised network on normal brain MR images to enable detection of arbitrary pathologic characteristics as deviations from the norm. We demonstrate the applicability of this concept to detect morphologically diverse pathologic characteristics on brain MR images.
For the in-house microangiopathy and multiple sclerosis datasets, which have good contrast between homogeneous, hyperintense lesions and the surrounding normal anatomy, the proposed anomaly detection approach showed good performance; however, there was a noticeable drop in performance for glioblastoma detection and delineation. The displacement of the brain in patients with gliomas poses a problem for the network in terms of detecting and delineating gliomas as well as the other hyperintense lesions. The reconstruction is often imperfect aside from the glioma itself, leading to a higher number of false-positive results in those regions. Baur et al (16) have recently proposed a scale-space approach to anomaly segmentation, which, by aggregating information from different spatial resolutions, might improve detection and segmentation of such space-occupying lesions. Furthermore, we observe that domain shifting is challenging for both supervised models and our model: when analyzing the highly diversified external datasets, both models had lower performance than they demonstrated when analyzing the in-house datasets of multiple sclerosis and microangiopathy lesions. More diverse training sets, as well as image augmentation techniques, could improve the domain adaptation. In particular, combining our unsupervised detection approach with unsupervised domain-adaptation strategies (17) offers exciting research opportunities. Although our approach outperforms a thresholding-based approach in terms of its F1 scores, which balance precision and recall (Table 1), results were mixed when comparing our approach to a U-Net. More details on technical considerations and thorough investigations of conceptual and architectural choices can be found in our comparative study of different recent unsupervised deep learning–based approaches proposed for anomaly detection in brain MRI (18).
Investigating the number of samples necessary to learn normal brain anatomy from MR images, we found that use of as few as 25 samples yields models with acceptable performance. A more elaborate investigation of the sample size can be found in a comparative study by Baur et al (18), which corroborates these findings across different autoencoder-based methods. The small sample size used in our study is in stark contrast to supervised deep learning algorithms, which often rely on very large numbers to achieve good performance. Recently, Chilamkurthy et al (19) published an algorithm, which they trained on over 250 000 CT scans, for detecting and classifying pathologic conditions on head CT images. As expected, training the model with higher-resolution images improved the detection of small pathologic findings such as multiple sclerosis lesions. The ability of our approach to analyze high-resolution MR images (1 mm, isotropic) was an advantage. Investigating model uncertainty by, for example, using dropout during inference (20) might help to further understand how to improve model architecture and training. Last, fusing supervised and unsupervised approaches for anomaly detection or segmentation holds promise for further improving performance (21), especially in view of our results, which favor our approach for some datasets and favor the U-Net for others.
There were limitations to our work. Although our algorithm detects anomalies on MR images, it does not classify them. Nonetheless, architectures in which specific (supervised) classification algorithms are triggered on detections by our unsupervised anomaly detection algorithm are quite conceivable. Furthermore, the medical literature on errors in radiology clearly shows that this first step (detection of pathologic conditions) is when most mistakes occur and is consequently the point at which computer-aided detection systems offer the most clinical use. Only a single modality (FLAIR) was analyzed in this study. How well our work generalizes to different levels of contrast (eg, contrast-enhanced T1 images or CT scans) necessitates future studies. Furthermore, domain adaptation (see discussion above) remains a challenge.
In summary, we demonstrated an algorithm trained on normal brain anatomy from MR images and exploited this trained model to detect a wide range of anomalies on an independent test dataset. This model can be trained on a small number of nonlabeled samples, circumventing typical obstacles encountered in training supervised deep learning models. Generalizing this model to other modalities and parts of the body highlights the potential of such unsupervised anomaly detection models to meaningfully support the diagnostic workflow in radiology.
C.B. and B.W. contributed equally to this work.
Supported by the Zentrum Digitalisierung Bayern. B.W. is supported by the Deutsche Forschungsgemeinschaft SFB-824.
Disclosures of Conflicts of Interest: C.B. disclosed no relevant relationships. B.W. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: institution received grants from DFG, ZDB. Other relationships: disclosed no relevant relationships. M.M. Activities related to the present article: institution received grants from Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) priority program SPP2177, Radiomics: Next Generation of Biomedical Imaging) – project number 428223038; consulting fee from Jahr. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. C.Z. disclosed no relevant relationships. N.N. Activities related to the present article: consulting fee from Herr. Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. S.A. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: author has grant from German Academic Exchange Service (DAAD). Other relationships: disclosed no relevant relationships.
Abbreviations:
- AUPRC
- area under the precision-recall curve
- FLAIR
- fluid-attenuated inversion recovery
- TCIA
- The Cancer Imaging Archive
- 3D
- three dimensional
References
- 1.Bruno MA, Walker EA, Abujudeh HH. Understanding and confronting our mistakes: the epidemiology of error in radiology and strategies for error reduction. RadioGraphics 2015;35(6):1668–1676. [DOI] [PubMed] [Google Scholar]
- 2.Schlegl T, Seeböck P, Waldstein SM, Schmidt-Erfurth U, Langs G. Unsupervised anomaly detection with generative adversarial networks to guide marker discovery. In: Niethammer M, Styner M, Aylward S, et al., eds. Information processing in medical imaging. IPMI 2017. Vol 10265, Lecture Notes in Computer Science. Cham, Switzerland: Springer, 2017; 146–157. [Google Scholar]
- 3.Vaidhya K, Thirunavukkarasu S, Alex V, Krishnamurthi G. Multi-modal brain tumor segmentation using stacked denoising autoencoders. In: Crimi A, Menze B, Maier O, Reyes M, Handels H, eds. Brainlesion: glioma, multiple sclerosis, stroke and traumatic brain injuries. BrainLes 2015. Vol 9556, Lecture Notes in Computer Science. Cham, Switzerland: Springer, 2015; 181–194. [Google Scholar]
- 4.Chen X, Konukoglu E. Unsupervised detection of lesions in brain MRI using constrained adversarial auto-encoders. ArXiv 1806.04972 [preprint] https://arxiv.org/abs/1806.04972. Posted June 13, 2018. Accessed January 26, 2021.
- 5.Baur C, Wiestler B, Albarqouni S, Navab N. Deep autoencoding models for unsupervised anomaly segmentation in brain MR images. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T, eds. Brainlesion: glioma, multiple sclerosis, stroke and traumatic brain injuries. BrainLes 2018. Vol 11383, Lecture Notes in Computer Science. Cham, Switzerland: Springer, 2018; 161–169. [Google Scholar]
- 6.Baur C, Wiestler B, Albarqouni S, Navab N. Bayesian skip-autoencoders for unsupervised hyperintense anomaly detection in high resolution brain MRI. In: Proceedings of the 17th International Symposium on Biomedical Imaging (ISBI). Piscataway, NJ: Institute of Electrical and Electronics Engineers, 2020; 1905–1909. [Google Scholar]
- 7.Kuijf HJ, Biesbroek JM, De Bresser J, et al. Standardized assessment of automatic segmentation of white matter hyperintensities and results of the WMH segmentation challenge. IEEE Trans Med Imaging 2019;38(11):2556–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bakas S, Akbari H, Sotiras A, et al. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data 2017;4(1):170117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt P, Gaser C, Arsic M, et al. An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. Neuroimage 2012;59(4):3774–3783. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Alberts E, Pongratz V, et al. Predicting conversion from clinically isolated syndrome to multiple sclerosis-An imaging-based machine learning approach. Neuroimage Clin 2019;21101593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber T, Alber G, Bette S, et al. Progressive disease in glioblastoma: benefits and limitations of semi-automated volumetry. PLoS One 2017;12(2):e0173112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohlfing T, Zahr NM, Sullivan EV, Pfefferbaum A. The SRI24 multichannel atlas of normal adult human brain structure. Hum Brain Mapp 2010;31(5):798–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iglesias JE, Liu CY, Thompson PM, Tu Z. Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans Med Imaging 2011;30(9):1617–1634. [DOI] [PubMed] [Google Scholar]
- 14.Sethian JA. Level set methods and fast marching methods. CIT J Comput Inf Technol 2003;11(1):1–2. [Google Scholar]
- 15.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A, eds. Medical image computing and computer-assisted intervention. MICCAI 2015. Vol 9351, Lecture Notes in Computer Science. Cham, Switzerland: Springer, 2015; 234–241. [Google Scholar]
- 16.Baur C, Wiestler B, Albarqouni S, Navab N. Scale-space autoencoders for unsupervised anomaly segmentation in brain MRI. ArXiv 2006.12852 [preprint] https://arxiv.org/abs/2006.12852. Posted June 23, 2020. Accessed January 26, 2021. [DOI] [PubMed]
- 17.Li H, Loehr T, Wiestler B, Zhang J, Menze B. e-UDA: efficient unsupervised domain adaptation for cross-site medical image segmentation. ArXiv 2001.09313 [preprint] https://arxiv.org/abs/2001.09313. Posted January 25, 2020. Accessed January 26, 2021.
- 18.Baur C, Denner S, Wiestler B, Albarqouni S, Navab N. Autoencoders for unsupervised anomaly segmentation in brain MR images: a comparative study. ArXiv 2004.03271 [preprint] https://arxiv.org/abs/2004.03271. Posted April 7, 2020. Accessed January 26, 2021. [DOI] [PubMed]
- 19.Chilamkurthy S, Ghosh R, Tanamala S, et al. Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. Lancet 2018;392(10162):2388–2396. [DOI] [PubMed] [Google Scholar]
- 20.Gal Y, Ghahramani Z. Dropout as a Bayesian approximation: representing model uncertainty in deep learning. In: Proceedings of The 33rd International Conference on Machine Learning. ML Research Press, 2016; 1050–1059. [Google Scholar]
- 21.Baur C, Wiestler B, Albarqouni S, Navab N. Fusing unsupervised and supervised deep learning for white matter lesion segmentation. In: Proceedings of The 2nd International Conference on Medical Imaging with Deep Learning. ML Research Press, 2019; 63–72. [Google Scholar]