Abstract
In recent years, deep learning techniques have been applied in musculoskeletal radiology to increase the diagnostic potential of acquired images. Generative adversarial networks (GANs), which are deep neural networks that can generate or transform images, have the potential to aid in faster imaging by generating images with a high level of realism across multiple contrast and modalities from existing imaging protocols. This review introduces the key architectures of GANs as well as their technical background and challenges. Key research trends are highlighted, including: (a) reconstruction of high-resolution MRI; (b) image synthesis with different modalities and contrasts; (c) image enhancement that efficiently preserves high-frequency information suitable for human interpretation; (d) pixel-level segmentation with annotation sharing between domains; and (e) applications to different musculoskeletal anatomies. In addition, an overview is provided of the key issues wherein clinical applicability is challenging to capture with conventional performance metrics and expert evaluation. When clinically validated, GANs have the potential to improve musculoskeletal imaging.
Keywords: Adults and Pediatrics, Computer Aided Diagnosis (CAD), Computer Applications-General (Informatics), Informatics, Skeletal-Appendicular, Skeletal-Axial, Soft Tissues/Skin
© RSNA, 2021
Summary
Generative adversarial networks have the potential to enhance musculoskeletal radiology.
Essentials
■ Generative adversarial network (GAN)–generated medical images are driving musculoskeletal radiology research forward by learning image representations of bones, joints, and soft tissues which have important diagnostic, prognostic, and therapeutic information.
■ GAN-based techniques have the potential to enhance all stages of the musculoskeletal radiology quantitative imaging chain by image synthesis, translation, enhancement, and segmentation as well as by assisting accurate image interpretation.
■ Despite substantial challenges with GANs, the flexible multiple modalities, contrast modeling, and high-quality information synthesis of GANs hold promise to facilitate deep learning for personalized and precision medicine.
Introduction
The implementation of artificial intelligence, a field in which models are created to interpret and learn from external data and then that learning is used to achieve assigned tasks, continues to transform the health care industry. Artificial intelligence is being developed or trialed for the management of medical data, accurate disease diagnostics, patient monitoring, and personalized treatment planning (1). Deep learning has become one of the most notable subfields of artificial intelligence owing to its ability to automatically represent large quantities of data using deep neural network layers, bypassing the handcrafted engineering steps required in machine learning pipeline designs (2,3).
Deep learning applications in musculoskeletal imaging are growing steadily owing to expanding computational power, relatively inexpensive storage devices, and availability of data. In musculoskeletal practice, breakthroughs in deep learning performance have occurred in three major areas: tissue segmentation (4), image reconstruction (5), and abnormality detection and classification (6,7). The emergence of deep learning applications in musculoskeletal imaging has been relatively slow, however, for several reasons: (a) musculoskeletal tissues—including small structural units or curved structures—are often assessed by three-dimensional (3D) volumetric imaging; (b) tiny structures with low tissue contrast require contrast enhancement, such as MRI or CT arthrography; (c) the range of abnormalities involved include a number of normal variations, aging-related findings, rare diseases, and subtle fractures; (d) the computational power required for an acceptable spatial resolution is high; (e) specific types of contrast agents are needed for the evaluation of various structures (eg, joints, spine); and (f) constructing datasets containing annotations based on high degree of expert consensus is relatively difficult owing to limited standardized reporting (8). Taken together, many factors must be considered when generating robust datasets for deep learning model development.
To overcome limitations in attaining these datasets, generative models that create synthetic, high-quality medical images have gained substantial attention owing to their ability to generate realistic data. This article focuses on generative adversarial networks (GANs) and their potential to overcome the clinical obstacles faced by artificial intelligence in the field of musculoskeletal imaging. An introduction to GANs and their applications in musculoskeletal imaging is presented herein, and the factors to be considered in their implementation to improve precision in medical procedures are outlined.
Overview of GANs
GANs have received considerable attention owing to their realistic data-generation capacities (9) (Fig 1). The main advantage of GANs is their ability to generate sharp, realistic images that appear to have been acquired normally and that provide insights into the natural features of the anatomic target of interest. This property has enabled GANs to perform successfully in image generation (10), image-to-image translation (11,12), image super-resolution (13), and 3D-object generation (14) tasks in the field of computer vision.
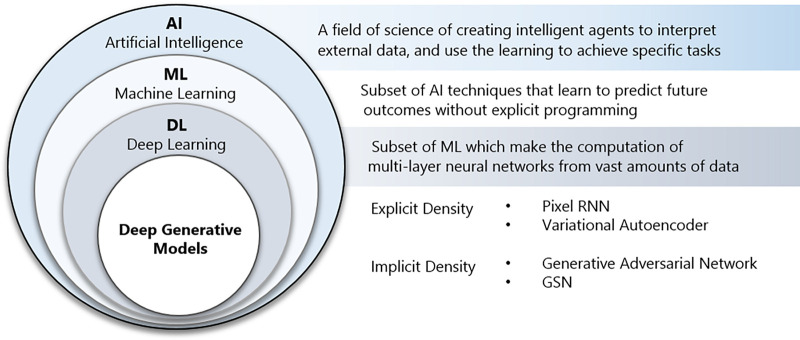
Figure 1:
An overview of the terms of artificial intelligence (AI), machine learning (ML), deep learning (DL), and generative adversarial network (GAN), as well as their nested relationships with each other. GSN = generative stochastic network, RNN = recurrent neural network.
GANs have provided ways to actively generate images that resemble those of the target anatomy through the joint optimization of two networks (Fig 2). GANs function by training two competing types of network—a generator and a discriminator. The generator creates a high-dimensional synthetic image from a low-dimensional input, such as arbitrary noise. Then, the generated image is fed into the discriminator, which attempts to distinguish the generated and real images. The generator’s loss function results in the model producing images that are increasingly indistinguishable from the original such that the discriminator cannot differentiate the generated and original images. During this process, the discriminator’s loss function penalizes misclassifications. While training, the two networks improve simultaneously, with the result that the generator network produces realistic and diverse outputs.
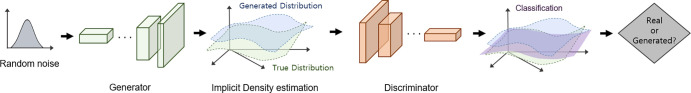
Figure 2:
Architecture of a generative adversarial network. The generator takes random inputs to generate new data, whereas the discriminator classifies the generated data and real data. The competitive training process of two deep neural networks results in new realistic data close to the true data distribution.
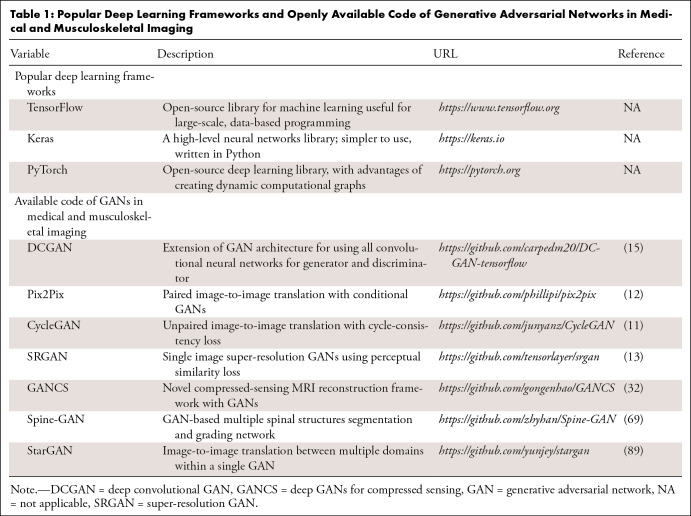
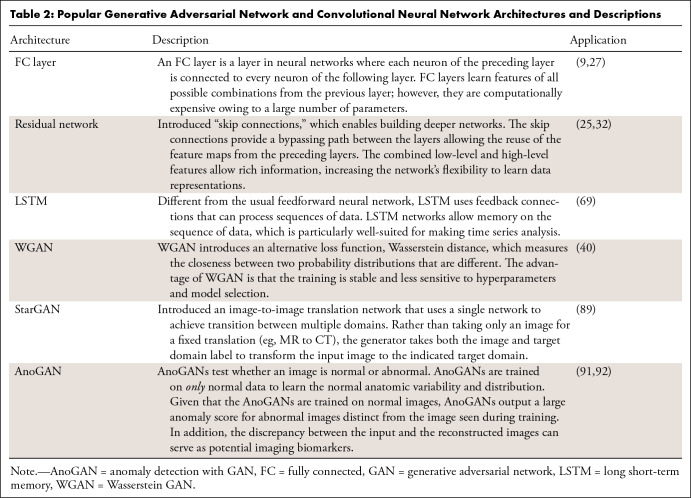
Musculoskeletal radiologists can leverage GANs by gaining an understanding of the practical applications and benchmark architectures to curate data accurately and make better clinical judgments of GAN outputs. To this end, we briefly introduce the representative variants of GANs that play a cardinal role in medical research. Table 1 contains popular deep learning frameworks for GAN implementation and a list of openly available GAN architectures with the corresponding code repositories. Table 2 provides additional descriptions of popular GAN methods and deep learning architectures to illustrate how the building blocks can be integrated for specific medical applications.
Table 1:
Popular Deep Learning Frameworks and Openly Available Code of Generative Adversarial Networks in Medical and Musculoskeletal Imaging
Table 2:
Popular Generative Adversarial Network and Convolutional Neural Network Architectures and Descriptions
Deep Convolutional GAN
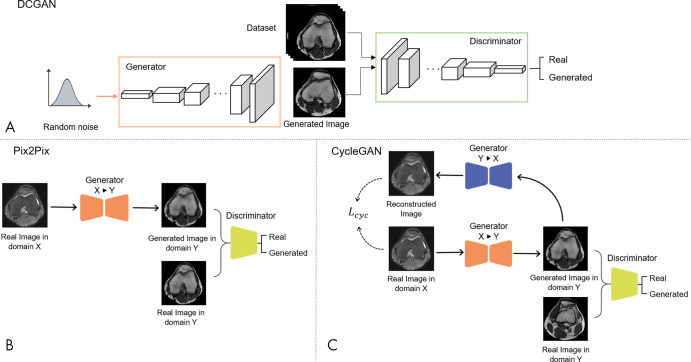
The most notable GAN design is the deep convolutional GAN (DCGAN) (Fig 3, A) (15). Fully connected layers, which are commonly used in GAN architectures, are not used in DCGANs. In addition, in DCGANs, the generator and the discriminator learn their own spatial downsampling and upsampling to follow an all-convolutional network architecture. Techniques such as batch normalization, rectified linear unit in the generator, and leaky rectified linear unit in the discriminator have been adapted to make DCGANs considerably more stable and easier to train, hence, actively researched and used for modern GAN architectures.
Figure 3:
Different generative adversarial network (GAN) framework. A, Architecture of deep convolutional GAN (DCGAN), where the generator and the discriminator consist of all-convolutional network architecture. B, Architecture of Pix2Pix framework, where paired training data are required in domain X and domain Y (supervised learning). C, Architecture of CycleGAN, where the model is trained in an unpaired fashion (unsupervised learning). Lcyc = cycle consistency loss, X = domain X, Y = domain Y.
Pix2Pix: A Conditional GAN
One successful GAN application has been in image-to-image translation (eg, filtering, image enhancement, colorization, denoising). Pix2Pix, a successful variant of the conditional GAN—a GAN that learns to generate images with prespecified conditions or characteristics—was proposed to achieve high-resolution image translation (12). The U-Net architecture (16) is used for the Pix2Pix generator, where the model attempts to generate corresponding target domain images from input images (Fig 3, B). Compared with the original GAN model, Pix2Pix requires rigidly aligned pairs of the desired outputs and the input counterparts (ie, MR and CT images of the same patient). However, paired images with almost-perfect spatial alignment are challenging to obtain in medical imaging and require additional image registration and processing.
CycleGAN
CycleGAN is one of the first models to gain extensive attention through image-to-image translation using unpaired images (11). Whereas Pix2Pix requires completely aligned image pairs to transform image from domain X to domain Y with a generator, GX→Y, CycleGAN introduces an additional generator, FY→X, to transform images from domain Y back to domain X (Fig 3, C). The idea behind CycleGAN is that when the input x is transferred through the entire transition cycle, X→Y→X, the output should be consistent with the input.
To achieve this, two cycles must be satisfied: the forward cycle, x → G (x) → F (G (x)) ≈ x, and the backward cycle, y → F (y) → G (F (y)) ≈ y. This performance is encouraged by minimizing the cycle consistency loss (Lcyc) that measures the discrepancy between the input image and the translated output of the cycle. Using T2-weighted and T1-weighted MRI translation as an example, the generator G is used on an input T2-weighted image to create a T1-weighted image from the input. The synthesized T1-weighted image is then converted back to a T2-weighted image using generator F. The discrepancy between the original and the translated T2-weighted image is minimized using a cycle consistency loss. The cycle consistency loss encourages generators to narrow the space of possible translations and thus synthesize images that are structurally consistent with the inputs. The transformed G (x) is fed into the discriminator DY to discriminate with real images y, whereas discriminator DX aims to discriminate translated images F (y) and real images x. In real clinical applications, most images are unpaired; thus, CycleGAN can be widely introduced into radiologic data science.
GANs in Musculoskeletal Imaging: State-of-the-Art Methods
MR Image Reconstruction
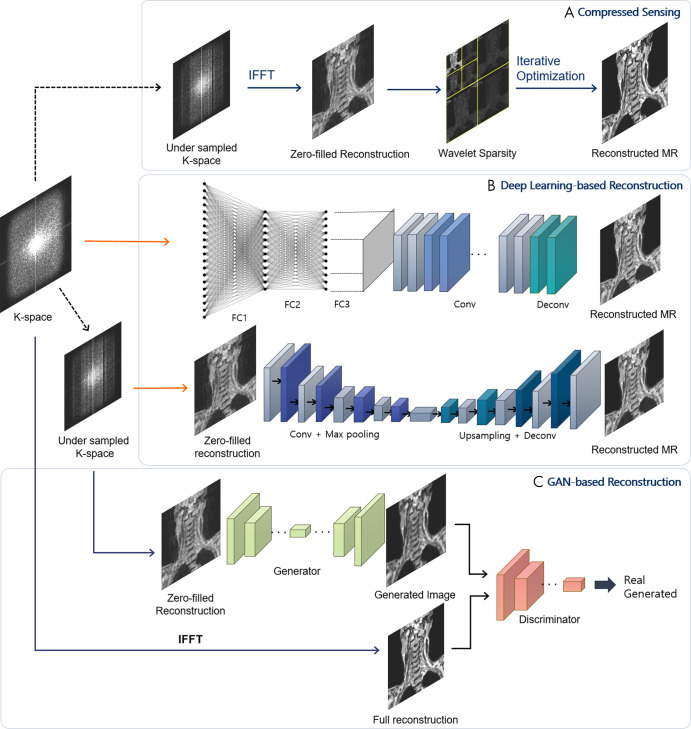
MRI offers excellent depiction of soft-tissue contrast and is a commonly applied, noninvasive, nonionizing radiation imaging modality for musculoskeletal tissues. It facilitates the assessment of a wide range of anatomic and pathologic features, including ligamentous injuries, tears in menisci and tendons, and occult bone injuries. Relatively long scan times can limit time-critical diagnosis and cause patient discomfort, however. Various acceleration techniques, including parallel MRI (17) and compressed-sensing MRI (18), have been applied to facilitate MR image acquisition. MR image reconstruction can be approached in a variety of ways, including compressed sensing, deep learning–based reconstruction, and GAN-based reconstruction (Fig 4).
Figure 4:
A schematic view of different architectures for MRI reconstruction. A, Compressed-sensing scheme. Starting from undersampled k-space data, the data are transformed to image and wavelet domains, and iterative optimization is applied to obtain full reconstruction. B, Deep learning–based reconstruction. Automated transform by manifold approximation (AUTOMAP) (27) uses fully connected (FC) layers with convolution layers to directly map k-space sensor data to image domain. Deep convolutional neural network models reconstruct MR images from zero-filled reconstructions. C, Generative adversarial network (GAN)–based reconstruction incorporates a discriminator to generate results closer to full reconstructions. IFFT = inverse fast-Fourier transform.
Earlier studies of compressed-sensing MRI reconstruction focused on using predefined sparsifying transforms, such as wavelet and total variation approaches, to produce a sparse representation in a given transform domain (19,20) (Fig 4, A). The sparse data enable the generation and removal of incoherent artifacts, and a nonlinear reconstruction method is applied to enforce image sparsity in the transform domain and data consistency with the acquired k-space data. Owing to the expense of nonlinear reconstruction and the unsatisfactory image reconstruction, however, dictionary learning frameworks using linear combinations of data-driven feature representations have been introduced (21). Dictionary frameworks still suffer from patch aggregation artifacts and large memory requirements to store shifted versions of the same features. Alternatively, convolutional sparse coding approaches that approximate the signal of interest by replacing regular dictionary dot products with convolutions have achieved fast and memory-efficient performance in compressed-sensing MRI tasks (22).
Deep learning approaches for reconstructing MR images (Fig 4, B) avoid complicated nonlinear optimization and parameter tuning. Initially, deep convolutional neural networks (CNNs) were used to learn the mappings between zero-filled and fully sampled MR images (23); this early research was followed by novel extensions using deep cascade of concatenated CNNs for MRI reconstruction (24) and residual networks to learn aliasing artifacts (25). Recent state-of-the-art deep learning algorithms have recast MRI reconstruction as a data-driven or model-driven task (26). In the data-driven approach, automated transform by manifold approximation (27) directly approximates the Fourier mapping between sensor-to-image domain using fully connected layers together with convolution layers. Model-driven methods, such as deep variational network (28) and alternating direction method of multiplier nets (29), incorporate deep learning into compressed-sensing reconstruction algorithms to learn the regularization parameters and functions, relaxing the hard constraint of the reconstruction algorithm and improving the reconstructed image quality.
GANs may become a feasible solution for high-quality sensor-to-image reconstruction, where GAN-based MRI reconstruction (Fig 4, C) mostly adopts additional loss functions to the deep de-aliasing GAN (30) architecture. Deep de-aliasing GANs add mean square error loss in both image and frequency domains and a perceptual loss to the original adversarial loss. The perceptual loss (defined as the distance of features extracted by pretrained VGG network [31] layers to learn the high-frequency pixel distributions of images) is added to achieve visually convincing anatomic details. Deep GANs for compressed sensing (32) incorporate residual networks and mixed L1 and L2 loss to remove high-frequency noise and artifacts while retaining details in knee images. In addition, MRI reconstruction based on a multifold chain of generators takes extra steps to refine the reconstructions, compared with a single-fold generator (33).
An additional potential advantage of GANs is their ability to restore severely ill-posed images of highly sparse “raw” sensor data. Deep CNNs are specially trained with a different perceptual loss to fit finer details that can cause artifacts to be intensified under noise (34). Since the additional adversarial loss in GANs trains the discriminator to distinguish better, fully sampled reconstructions and generated images successively, the generator may learn the refinements necessary to create artifact-free images. One day the advantages of GANs may outweigh those of deep CNNs in the reconstruction of highly sparse k-space data obtained from shorter scan times.
Image Enhancement
Denoising of CT images.— Low-dose CT is preferred over normal-dose CT because ionizing radiation doses have been associated with a small but increased risk of cancer (35); this increased risk increases the demand for low-dose CT. Images obtained with low-dose CT typically have highly nonlinear and nonuniformly distributed noise, however. CNN-based denoising methods have been investigated on models trained on paired datasets of normal-dose CT images and low-dose CT images (36,37). In these instances, low-dose CT images are generated by injecting artificial noise (eg, Poisson noise, Gaussian noise [38,39]) into the original normal-dose CT images acquired routinely. Since the networks typically use the pixel-wise loss for denoising, the drawback of this technique is that high-frequency image textures and structures are inevitably lost together with the noise. Thus, there is a need for other models for CT denoising.
Conditional GANs have been used with a paired dataset to generate images resembling normal-dose CT from low-dose CT inputs. Conditional GAN frameworks—with Wasserstein distance and perceptual loss (40), structural sensitivity loss in 3D space (41), and sharpness detection networks that enhance low-contrast regions (42)—have been investigated for mitigating CT noise. Stacked GAN (43) decomposes the CT enhancement into two subtasks. First, denoising GAN is applied to the noisy CT image; the second GAN enhances the resolution of the CT images for further lesion segmentation, which could include bone regions.
Conditional GAN-based methods are restricted in real clinical situations, however, as matched low-dose and normal-dose CT pairs typically are unavailable because of the unnecessary, extra radiation exposure to patients that would be required to obtain both sets of CT images. Artificial noise injection into images is oversimplified and cannot be generalized well for complicated, CT-specific noise distributions. In addition, development of independent algorithms for simulation of low-dose CT images is challenging owing to the need for statistical and domain knowledge such as x-ray photon statistics, electronic noise background, noise injection in sinogram domain, and multiple technical and/or scanner model-related parameters (39).
To resolve this unmatched pair challenge, an unsupervised denoising network can be learned using the CycleGAN framework. The forward and backward cycles of the two generators and discriminators learn the opposite direction mappings—one from low-dose to normal-dose images, the other from normal-dose to low-dose images. CycleGAN methods have been used to attenuate noise in the micro-CT tibia dataset (44) and with multiphase cardiac CT angiography (45) in the semisupervised and unsupervised settings. In general, both semisupervised and unsupervised CycleGAN approaches result in competitive denoising performance compared with the fully supervised deep learning methods with detailed texture and edge information preservation.
Super-resolution.— Super-resolution, a process of upscaling images to high spatial resolution from low spatial resolution, has suffered from limitations of the imaging equipment, the susceptibility of the medical images to patient movement (eg, voluntary or involuntary patient movement or pulsation of blood vessels), exposure to ionizing radiation, and long acquisition-time limitations. Super-resolution CNN-based methods (46,47) have outperformed classic super-resolution algorithms with respect to peak signal-to-noise ratio and structural similarity index. These methods have failed to produce consistent images that appear natural to the human eye, however.
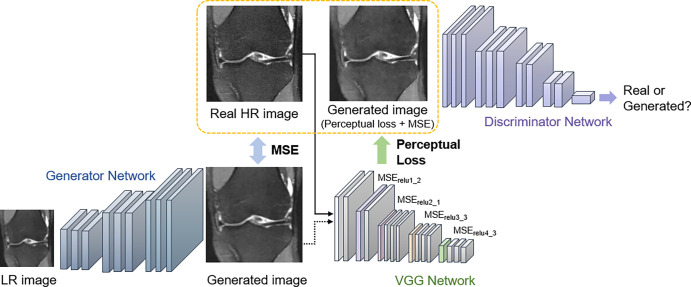
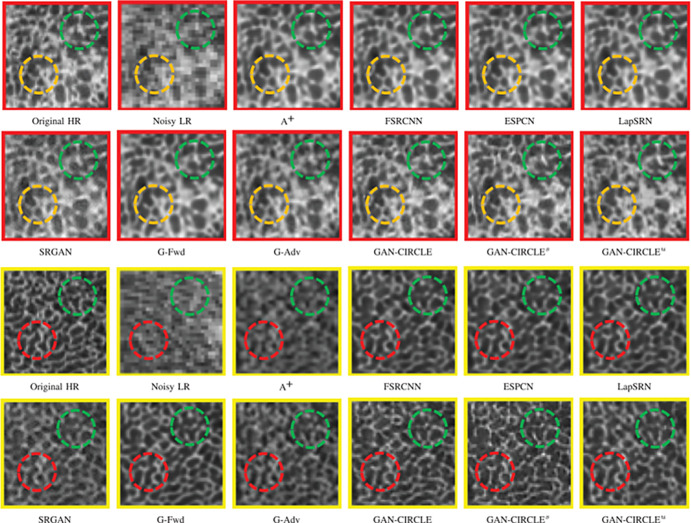
Super-resolution GAN (SRGAN) (13) approaches have recently attracted considerable attention, demonstrating remarkable performance compared with previous super-resolution algorithms. SRGAN incorporates perceptual loss to encourage the generator to learn the structural high-frequency details and pixel distributions of the high-resolution images (Fig 5). Compared with other types of SR methods, SRGAN achieves excellent performance in revealing visual information and recovering useful spatial and textural information. A 3D-SRGAN algorithm that generates thin-section interval CT images (virtual thin sections) from corresponding thick-interval images recovered better perceptual quality in high-intensity values such as bone boundaries (48). To reveal the microarchitecture and preserve anatomic information in CT bone imaging, a residual-based CycleGAN framework was used to generate high-resolution CT images by semisupervised and unsupervised learning (44). Notably, the proposed semisupervised CycleGAN-based enhancement was capable of capturing the fine structural textures with pixel sizes in the tibia bone dataset, showing competitive quantitative results with fully supervised super-resolution networks (Fig 6).
Figure 5:
The overall structure of super-resolution generative adversarial network. Perceptual loss is defined by the following: the generated image and the real high-resolution (HR) image are passed through a VGG-16 (or -19) network (31). The distance between specific rectified linear unit (relu) layers is compared to preserve high-frequency details between the images, and the adversarial loss further pushes the generated image to follow the natural image manifold. LR = low-resolution, MSE = mean squared error. Images used were obtained from the NYU Langone Health fastMRI Dataset (fastmri.med.nyu.edu) (5,95).
Figure 6:
Comparison of CT super-resolution and denoising case from micro-CT tibial bone images. The zoomed regions of interest of tibial bony structures are shown in boxes outlined in red or yellow. Generative adversarial network constrained by the identical, residual, and cycle learning ensemble (GAN-CIRCLE) recovers subtle structures and fine details better than other variations of the proposed networks, particularly in the regions marked by green, yellow, and red circles. The results of semisupervised GAN-CIRCLE (GAN-CIRCLEs) predict sharper images and richer anatomic textures compared with unsupervised GAN-CIRCLE (GAN-CIRCLEu). (Reprinted, with permission, from reference 44.) A+ = adjusted anchored neighborhood regression, ESPCN = efficient subpixel convolutional neural network, FSRCNN = fast super-resolution convolutional neural network, G-Adv = G-Adversarial, G-Fwd = G-Forward, HR = high-resolution, LapSRN = Laplacian pyramid super-resolution network, LR = low-resolution, SRGAN = super-resolution generative adversarial network.
Image Synthesis
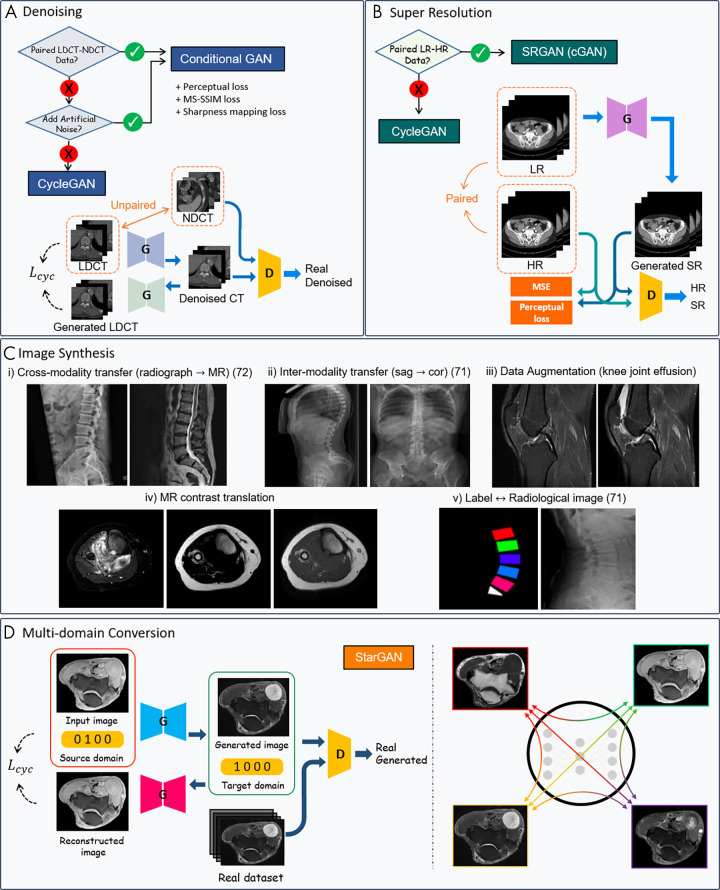
Data augmentation.— GANs can compensate for data scarcity or class imbalances by generating plausible samples to train a deep learning model. Compared with conventional data augmentation (eg, shifts, crops, horizontal or vertical flips, rotations), GAN-based data augmentation provides new images that resemble the original data to add to the training dataset (Fig 7). For example, one study used CycleGAN on normal thigh MR images to augment fatty infiltration, which facilitated the development of a segmentation network for thigh MRI with severe fatty infiltration as synthetic images were added to the training dataset (49). Additionally, the use of B-mode musculoskeletal US and synthetic segmentation masks trained with CycleGAN has been proposed to automate the generation and labeling of B-mode images (50). Very few GAN-based data augmentation procedures have been conducted in musculoskeletal imaging. Those conducted in other fields (eg, liver lesion synthesis from a limited CT dataset [51], body CT synthesis from digital body phantoms [52]), however, have demonstrated the possibility to capture the high degree of variability that might be applied to musculoskeletal disorders.
Figure 7:
Overview of generative adversarial network (GAN) applications in musculoskeletal imaging. Figures are directly cropped from corresponding studies: A, GAN schemes for low-dose CT denoising with and without paired images. B, GAN schemes for super-resolution (SR) with and without paired images. C, Examples of GAN-based image synthesis: (i) Synthetic spine MR image from radiographic images (72); (ii) sagittal (sag) to coronal (cor) radiograph translation (71); (iii) joint effusion synthesis from CycleGAN—knee MR images from Clinical Hospital Centre Rijeka, Croatia (85); (iv) multicontrast domain translation in knee MR images—synthetic T1-weighted MR image from T2 fat map and T2 water map using conditional GAN; and (v) synthetic planar radiographs created from labels (71). D, Unpaired multicontrast MR image synthesis with StarGAN, multiple domain mapping using a single generator (89). Lcyc = cycle consistency loss, D = discriminator, G = generator, HR = high-resolution, LDCT = low-dose CT, LR = low-resolution, MSE = mean squared error, MS-SSIM = multiscale structural similarity, NDCT = normal-dose CT, SRGAN = super-resolution GAN.
Cross-modality synthesis.— Cross-modality image synthesis is the process by which synthetic images are generated for one modality based on another (eg, generating synthetic CT images from MRI data). Despite requiring a large set of matched image pairs from two modalities, cross-modality image synthesis is an area of active research. Recently, image-to-image transfers using GANs have shown the potential to provide multiparametric functional and anatomic information without additional examination or diagnostic planning, thereby occupying an increasingly important role in personalized medicine. The conditional GAN frameworks are used for cases in which different image modalities can be perfectly registered with each other to ensure high fidelity. Moreover, CycleGAN networks can be used to manage cases in which perfect alignment between modalities is unfeasible.
The main role of GAN-based synthetic CT generation tasks is to overcome the shortcomings of MRI only–based treatment regarding missing electron density, PET/MRI attenuation correction, and difficulties of distinguishing between bones and air. Deep CNNs and conditional GANs have achieved moderate success with aligned MRI/CT images (53,54). Although the conditional GAN frameworks alleviate the problem of blurred structures of deep CNNs to some extent by including an adversarial loss, they remain limited by the pixel-wise loss component that requires an aligned MRI/CT dataset. CycleGAN, which does not require paired data, demonstrated effectiveness in qualitative analysis with fewer artifacts and blurring compared with paired data (55). However, finding the optimal translation on higher semantic levels and large variations (geometric, shape translations [56,57]) can be far more challenging. Customized loss functions, such as gradient consistency (58), mean p distance, and gradient difference loss (59), have been proposed as additional constraints to preserve boundaries of regions in the anatomic structures in synthetic CT volumes. Furthermore, in radiotherapeutic imaging, MRI-based dose calculation has been found to be feasible for the entire pelvis in patients with prostate cancer using the synthetic CT images generated by conditional GANs (60).
For multicontrast synthesis, CNN segmentation combined with GAN image translation has been explored to develop a generalized segmentation tool for multiple MRI contrasts from a single set of annotated data for training. Liu (61) used a CycleGAN to translate T1-weighted spoiled gradient-recalled echo knee image contrasts in the Segmentation of Knee Images 2010 image dataset (http://www.ski10.org) to proton density-weighted image and T2-weighted image knee MR image contrasts. The CycleGAN network incorporates a joint segmentation network to output translated proton density-weighted images and T2-weighted MR images, as well as segmentation masks. The proposed network achieved comparable segmentation performance on femoral and tibial cartilage, as well as on the femur and tibia compared with the supervised U-Net segmentation performance (61). The proposed generalized segmentation technique substantially reduces the annotation burden placed on musculoskeletal radiologists, and multiple structural information from various contrast images could be obtained from a single set of annotated data. Furthermore, recent work on the reconstruction of 3D CT in biplanar two-dimensional radiographs (62) shows the possibility of 3D cartilage visualization through two-dimensional MRI inputs, a promising strategy using 3D-GAN modeling.
GANs in Musculoskeletal Imaging: Specific Anatomies
Discriminative models, such as classification and regression models, have offered an impressive array of studies to identify pathologic abnormalities and facilitate automated quantitative analysis (63). CNN models for detecting fractures (64) and ligament injuries (65), automated severity grading of osteoarthritis (66), and assessment of bone age (67) have been developed to support high-performance diagnosis. Unlike discriminative models, the most notable studies of GANs for specific musculoskeletal anatomy mainly focused on image generation tasks in data augmentation, cross-modality synthesis, and segmentation networks.
Spine
Level-by-level description and identification of the degenerative disease of the vertebral bodies through daily musculoskeletal imaging is time-consuming. Therefore, automated localization and labeling of the spinal structures constitute the first step toward facilitating radiologic workflow efficiency. Complementary to deep learning approaches in medical image analysis of the spine (68), GAN-based approaches have been used to extract location information of vertebrae, disks, and spinal shape. Spine-GAN achieves segmentation of the neural foramen, intervertebral disks, and vertebrae concurrently by capturing the variability of spinal structures and spatial correlations using extensions such as atrous convolutions (ie, convolutions with holes) and long short-term memory module (69). The discriminator progressively strengthens the spatial consistency of the segmentation result by discriminating the segmentation output with the ground truth masks. In addition, a butterfly-shaped generator localizes the vertebrae in CT images, with the discriminator operating on the goodness of these predictions to encode a local vertebral spread (70).
In addition to vertebra labeling, GANs have exhibited the potential to become a feasible method for the generation of synthetic radiographs from labeled data describing the shape of the lumbar vertebrae (71). However, the networks show anatomic inaccuracies in generating the structures not depicted in the labeled data, such as the spinous processes and the distal aspect of the sacrum. The potential for super-resolution and image-to-image translation in conditional GANs delivers encouraging results in MRI contrast conversions such as T1-weighted to T2-weighted images and T2-weighted to short tau inversion-recovery images or turbo inversion recovery magnitude images (72); meanwhile, image translation from radiographs to MR images exhibits doubtful accuracy in regions not visible in radiographs, such as the thoracolumbar junction. Translations that lack a clear association between modality translations—a shortcoming for which current clinicians may distrust the synthesized output—require additional medical physics-based investigations.
Bone and Muscle
In musculoskeletal imaging, image synthesis is challenging because global fidelity, feature diversity, and high-frequency textures need to be preserved for precise delineation of muscles and bone structures. Since the pixel-wise loss and cycle consistency loss is not sufficient to maintain structure consistency for fine bone structures, different feature-preserving losses have been validated for skeletal assessment. For example, additional structural constraints, such as gradient consistency loss or correlation coefficient loss, forces the generator to create consistent boundaries (58,73). The advantages of GANs can also be maximized for bone lesions (74), fractures, and soft-tissue lesions to increase the incidences of rare diseases on radiographs or to overcome limited clinical education with an alternative to real images in the musculoskeletal domain for medical students and radiology trainees (75).
GANs for muscle imaging focus on the generation of synthetic images to overcome limited database and rare entities. Realistic and statistically cogent B-mode musculoskeletal US images can be translated from segmentation masks that encode the main features (two aponeuroses, fascicles, and pennation angle between 10° and 30°) to obtain large volumes of annotated data automatically (50). In addition, texture augmentation for fatty infiltration simulation has been achieved through CycleGAN (49). The recent GAN-based techniques may further facilitate artifact reduction and preservation of high resolution to create a large-scale standardized dataset for muscle imaging.
Implementations and Practical Considerations for GANs
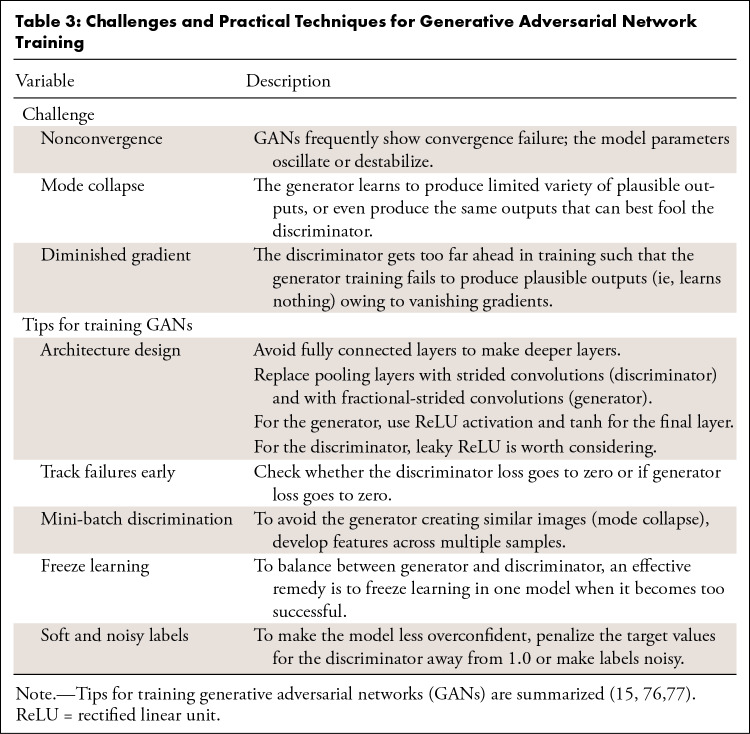
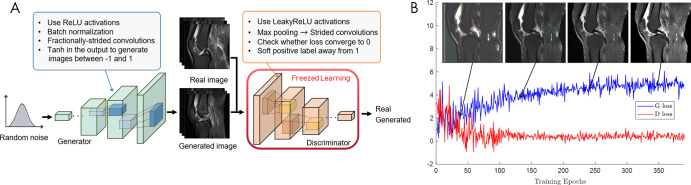
GANs appear to hold great promise across a wide range of applications. From a practical perspective, however, integration of GANs can be challenging in terms of training. We have reviewed different architectures and extensions, and Table 3 lists the challenges and recommendations detailed by researchers and studies for training GANs (15,76,77). Figure 8 shows an example of image translation progress and loss of generator and discriminator in the training process.
Table 3:
Challenges and Practical Techniques for Generative Adversarial Network Training
Figure 8:
Walk-through of generative adversarial network (GAN) training. A, Stability tips to improve GAN performance (Table 3). B, Example of image generation progress of GANs for joint effusion synthesis. The generated images at corresponding training stages are superimposed on the training loss graph. The generator and discriminator loss oscillates. Images from Clinical Hospital Centre Rijeka, Croatia (85). D loss = discriminator loss, G loss = generator loss, ReLU = rectified linear unit.
Image Quality Evaluation
No universally agreed-on performance metrics exist for evaluating the perceptual quality of GANs. Most CNN models incorporate similarity metrics for both loss functions in training and image distortion measurement in the test phase (eg, mean absolute error, peak signal-to-noise ratio, information fidelity criterion, structural similarity index). However, high similarity of distortion measures does not indicate high perceptual quality of the synthesized images (78), which increases the need for the development of a new measurement that satisfies clinical applicability. Human judgment-based evaluation is a popular qualitative approach in which such measurements are computed through real versus synthesized questionnaires or diagnostic quality scoring (44,79). One way to alleviate the problems of expensive and time-consuming human evaluations is to use downstream tasks such as classification, segmentation, or a referenceless blur factor (80) as a validation tool for quality assessment of the generated images. In addition, many quantitative methods have been designed as an alternative to human assessors; for these, the inception score (76) and the Fréchet inception distance (81) have been widely used owing to reasonably accurate estimates of the quality and diversity of generated images afforded by those measures. The adequacy of these evaluation metrics remains unexplored in the medical domain, however, and needs further study.
Artifact Introduction
One issue that arises when using GANs is that generated images may contain various subtle artifacts that may be difficult to detect. In the process of GAN-based image synthesis, the generator can start creating high-frequency patterns owing to its objective of producing finer details to create perceptually convincing translations that represent the natural image manifold (13,32). Additionally, checkerboard artifacts may be introduced when the generator produces images with deconvolution (82). Quantifying the hallucination risk of GANs is challenging and requires future research. As such, GAN-induced high-frequency textures, especially on images that were not seen during the training phase, must be assessed by an expert radiologist to prevent clinical judgment on erroneous output. In the case of MRI reconstruction, the breaking point of k-space undersampling rates at which GANs can start to create high-frequency noise and hallucinations requires study (32).
Anatomic Distortion
For GANs that incorporate VGG-based perceptual loss, distortion of anatomic features has been detected in prior studies, possibly owing to the fact that VGG networks are pretrained on natural images, which have different structural features than medical images (41). Similarly, as a result of the ill-posed problem in unsupervised learning of CycleGAN, finding the optimal translation for geometric and structural levels is far more challenging compared with transferring local textures (56,57,83). For musculoskeletal image translation with large anatomic variations, we anticipate that other new research areas will expand to assess the preservation of anatomic structures, particularly those related to loss functions.
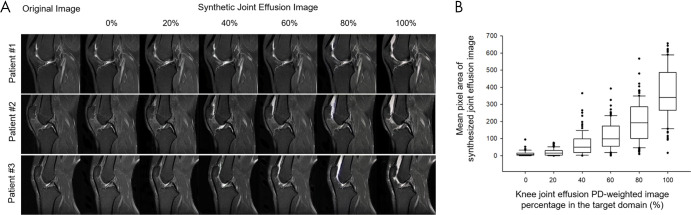
In addition, since CycleGAN is trained to implicitly approximate the underlying structure of the distinct anatomic structures, the performance relies heavily on the distribution of the target structure. Thus, in an extremely biased target domain composition (eg, all tumor or all healthy), the CycleGAN adds or removes tumor features in the target images from a balanced source domain distribution (84). To demonstrate the effect of target domain distribution on CycleGAN translations, we previously conducted a study with a publicly available dataset (Clinical Hospital Centre Rijeka, Croatia [85]) to synthesize joint effusion on sagittal proton density-weighted knee examinations without pathologic findings. The comparison in Figure 9 illustrates the bias impact of the changing ratio of joint effusion images (from 0% to 100%) in the target domain. Joint effusion increases from left to right, and when trained on 100% effusion the translated images display a large amount of joint effusion. This emphasizes the need to address the possible effects of distribution mismatching on GAN performances through careful curation of data by health care professionals.
Figure 9:
A, Joint effusion synthesis from healthy class through CycleGAN domain translation while the ratio of joint effusion samples in the target domain is increased. The distribution of joint effusion samples is increased from 0% to 100%, in increments of 20%. The images of the source domain (knee MRI without effusion) are shown in the left-most column, with the translated image in the target domain shown in the right-most column. The magnitude of the joint effusion increases from left to right. B, The graph (box plots) for the mean pixel area of synthesized joint effusion images with different ratios of joint effusion samples in the target domain. The horizontal line in each box represents the median (50th percentile) of the calculated strain ratios, and the top and bottom of each box represents the 25th and 75th percentiles, respectively. Images from Clinical Hospital Centre Rijeka, Croatia (85). GAN = generative adversarial network, PD = proton-density.
GAN Application in Musculoskeletal Imaging: Quantitative Precision Image Analysis
Deep learning–based quantitative methods have proven highly efficient in assisting with musculoskeletal clinical assessments, assessing objective imaging features (eg, osteoarthritis, joint degeneration [66,86]). Using both high-resolution visual features and accurate musculoskeletal structure segmentation, GAN-based applications can become an essential quantitative imaging method for the precision treatment of various musculoskeletal conditions.
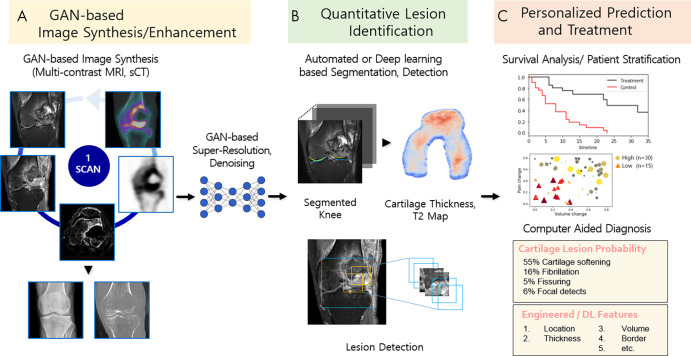
In recent years, several GAN-based techniques have been developed to perform image generation between multiple modalities. Synthetic MRI techniques—for instance, the reconstruction of various MR image contrasts from a single scan—had been studied actively before GANs were proposed (87,88). The motivations for multidomain translation based on the StarGAN architecture (89), however, arose from its capacity and efficiency to learn all domains with one single generator. Implementing StarGAN for multiple domain translation may lead to the generation of images of different imaging protocols, institutions, vendors, and patient populations; it may even benefit clinicians who want to monitor individual treatment responses in order to supplement precision treatments or personalized plans. Multidomain image translation combined with GAN-based image enhancement (eg, super-resolution, denoising) is a highly valuable image processing technique and is anticipated to improve the quantitative and personalized assessment of musculoskeletal disorders (Fig 10, A).
Figure 10:
Schematic of future applications of generative adversarial networks (GANs) to musculoskeletal imaging chain and clinical management. A, Flexible multiple domain translation tasks using GANs will create various synthetic radiologic images (eg, MRI contrast or modality conversion). GAN-based super-resolution and denoising methods offer the potential to enhance image quality using lower radiation dose and reduced scan and reconstruction time. B, Machine learning methods (eg, GAN-based segmentation or detection models) are applied for tissue and lesion identification to extract quantitative information from images. C, Use of computer-aided diagnosis systems to evaluate the information to make personalized predictions of disease diagnosis, treatment planning, and survival analysis. DL = deep learning, sCT = synthetic CT.
In addition to the promise of multimodality image synthesis, we believe there is substantial promise for GANs to provide high value in automated quantitative segmentation. Through domain adaptations between different tissue contrasts or modalities, GAN-driven segmentation facilitates the fully automated segmentation process by generalizing a single set of annotated data to different domains (61), thereby resolving the problem of annotation scarcity in musculoskeletal imaging. Moreover, semantic segmentation using GANs (69,90) corrects the higher-level inconsistencies between the segmentation model prediction and the ground truth masks, which is critical to obtain accurate cartilage and bone segmentation (Fig 10, B).
In addition to the implementation of GANs for image acquisition, the increasing use of GANs as models for identifying potential imaging biomarkers has encouraged progress in personalized precision medicine. Trained on only data containing normal anatomy, anomaly detection with GANs (AnoGANs) learn the anatomic variability of healthy distributions. When tested on unseen cases or anomalous images, AnoGANs output a large anomaly score for query images distinct from the images seen during training, thereby serving as potential imaging biomarker candidates (91). AnoGANs can be trained to detect metastatic bone tumors (92) and may have the potential to capture cartilage injuries, ligamentous or meniscal tears, bone marrow abnormalities, and marrow-replacing lesions. In addition, GANs have exhibited capabilities in modeling disease progression (93), which can be applied to the prediction of osteoarthritis progression, treatment response, and surgical repair outcome. In this regard, a variety of GAN techniques may eventually allow image diagnosis of the subtle progressions of skeletal tumors and tumorlike lesions (94), supplemented by computer-aided systems as an adjunct instrument for individualized diagnosis and treatment guidance (Fig 10, C).
Conclusion
GANs have the potential to be the next step in artificial intelligence–powered radiology owing to their capacity to go beyond recognition and classification toward the generation of high-resolution, synthetic images. Although GANs have a promising future in musculoskeletal research settings, their clinical application requires careful curation of the dataset so as to include—by high-quality annotations—cases relevant to the clinical question. It will be the role of the radiologist to understand the inherent properties of the given data within the GAN procedure and to collaborate with clinicians and data scientists in relevant applications. Because the use of GAN-based tools has the potential to benefit every step in the overall quantitative imaging chain and assist accurate image interpretation, it will be imperative for future musculoskeletal radiologists to adopt these powerful tools appropriately. From flexible manipulation of multiple radiologic images to the accurate and precise imaging for quantitative assessment, increased incorporation of GANs will provide cost-effective, reproducible, and efficient approaches for patient monitoring and precision treatment.
Supported by a National Research Foundation (NRF) grant funded by the Korean government, Ministry of Science and ICT (MSIP, 2018R1A2B6009076).
Disclosures of Conflicts of Interest: Y.S. Activities related to the present article: institution received National Research Foundation grant funded by the Korean government, Ministry of Science and ICT (MSIP 2018R1A2B6009076). Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. J.Y. disclosed no relevant relationships. Y.H.L. Activities related to the present article: institution received National Research Foundation grant funded by the Korean government, Ministry of Science and ICT (MSIP 2018R1A2B6009076). Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships.
Abbreviations:
- AnoGAN
- anomaly detection with GAN
- CNN
- convolutional neural network
- DCGAN
- deep convolutional GAN
- GAN
- generative adversarial network
- SRGAN
- super-resolution GAN
- 3D
- three dimensional
References
- 1.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med 2019;25(1):44–56. [DOI] [PubMed] [Google Scholar]
- 2.Lee JG, Jun S, Cho YW, et al. Deep learning in medical imaging: general overview. Korean J Radiol 2017;18(4):570–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521(7553):436–444. [DOI] [PubMed] [Google Scholar]
- 4.Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R. Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med 2018;79(4):2379–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zbontar J, Knoll F, Sriram A, et al. fastMRI: An Open Dataset and Benchmarks for Accelerated MRI. arXiv:1811.08839 [preprint]. https://arxiv.org/abs/1811.08839. Posted November 21, 2018. Accessed February 7, 2021.
- 6.Liu F, Zhou Z, Samsonov A, et al. Deep learning approach for evaluating knee MR images: achieving high diagnostic performance for cartilage lesion detection. Radiology 2018;289(1):160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Schacky CE, Sohn JH, Liu F, et al. Development and Validation of a Multitask Deep Learning Model for Severity Grading of Hip Osteoarthritis Features on Radiographs. Radiology 2020;295(1):136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gholamrezanezhad A, Kessler M, Hayeri SM. The need for standardization of musculoskeletal practice reporting: learning from ACR BI-RADS, liver imaging–reporting and data system, and prostate imaging–reporting and data system. J Am Coll Radiol 2017;14(12):1585–1587. [DOI] [PubMed] [Google Scholar]
- 9.Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial nets. Adv Neural Inf Process Syst 2014;2672–2680. [Google Scholar]
- 10.Wang TC, Liu MY, Zhu JY, Tao A, Kautz J, Catanzaro B. High-Resolution Image Synthesis and Semantic Manipulation with Conditional GANs. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, June 18–23, 2018. Piscataway, NJ: IEEE, 2018; 8798–8807. [Google Scholar]
- 11.Zhu JY, Park T, Isola P, Efros AA. Unpaired Image-to-Image Translation Using Cycle-Consistent Adversarial Networks. In: 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, October 22–29, 2017. Piscataway, NJ: IEEE, 2017; 2223–2232. [Google Scholar]
- 12.Isola P, Zhu JY, Zhou T, Efros AA. Image-to-Image Translation with Conditional Adversarial Networks. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, July 21–26, 2017. Piscataway, NJ: IEEE, 2017; 1125–1134. [Google Scholar]
- 13.Ledig C, Theis L, Huszár F, et al. Photo-Realistic Single Image Super-Resolution Using a Generative Adversarial Network. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, July 21–26, 2017. Piscataway, NJ: IEEE, 2017; 4681–4690. [Google Scholar]
- 14.Gadelha M, Maji S, Wang R. 3D Shape Induction from 2D Views of Multiple Objects. In: 2017 International Conference on 3D Vision (3DV), Qingdao, China, October 10–12, 2017. Piscataway, NJ: IEEE, 2017; 402–411. [Google Scholar]
- 15.Radford A, Metz L, Chintala S. Unsupervised Representation Learning with Deep Convolutional Generative Adversarial Networks. arXiv:1511.06434 [preprint]. https://arxiv.org/abs/1511.06434. Posted November 19, 2015. Accessed February 7, 2021.
- 16.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells W, Frangi A, eds. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science, vol 9351. Cham, Switzerland: Springer, 2015; 234–241. [Google Scholar]
- 17.Heidemann RM, Özsarlak O, Parizel PM, et al. A brief review of parallel magnetic resonance imaging. Eur Radiol 2003;13(10):2323–2337. [DOI] [PubMed] [Google Scholar]
- 18.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med 2007;58(6):1182–1195. [DOI] [PubMed] [Google Scholar]
- 19.Tang J, Nett BE, Chen GH. Performance comparison between total variation (TV)-based compressed sensing and statistical iterative reconstruction algorithms. Phys Med Biol 2009;54(19):5781–5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lustig M, Donoho DL, Santos JM, Pauly JM. Compressed sensing MRI. IEEE Signal Process Mag 2008;25(2):72–82. [Google Scholar]
- 21.Ravishankar S, Bresler Y. MR image reconstruction from highly undersampled k-space data by dictionary learning. IEEE Trans Med Imaging 2011;30(5):1028–1041. [DOI] [PubMed] [Google Scholar]
- 22.Quan TM, Jeong WK. Compressed Sensing Reconstruction of Dynamic Contrast Enhanced MRI Using GPU-Accelerated Convolutional Sparse Coding. In: 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, April 13–16, 2016. Piscataway, NJ: IEEE, 2016; 518–521. [Google Scholar]
- 23.Wang S, Su Z, Ying L, et al. Accelerating Magnetic Resonance Imaging Via Deep Learning. In: 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, April 13–16, 2016. Piscataway, NJ: IEEE, 2016; 514–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlemper J, Caballero J, Hajnal JV, Price AN, Rueckert D. A deep cascade of convolutional neural networks for dynamic MR image reconstruction. IEEE Trans Med Imaging 2018;37(2):491–503. [DOI] [PubMed] [Google Scholar]
- 25.Lee D, Yoo J, Ye JC. Deep Residual Learning for Compressed Sensing MRI. In: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, VIC, Australia, April 18–21, 2017. Piscataway, NJ: IEEE, 2017; 15–18. [Google Scholar]
- 26.Liang D, Cheng J, Ke Z, Ying L. Deep MRI Reconstruction: Unrolled Optimization Algorithms Meet Neural Networks. arXiv:1907.11711 [preprint]. https://arxiv.org/abs/1907.11711. Posted July 26, 2019. Accessed February 7, 2021.
- 27.Zhu B, Liu JZ, Cauley SF, Rosen BR, Rosen MS. Image reconstruction by domain-transform manifold learning. Nature 2018;555(7697):487–492. [DOI] [PubMed] [Google Scholar]
- 28.Hammernik K, Klatzer T, Kobler E, et al. Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med 2018;79(6):3055–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun J, Li H, Xu Z. Deep ADMM-Net for compressive sensing MRI. Adv Neural Inf Process Syst 2016;10–18. [Google Scholar]
- 30.Yang G, Yu S, Dong H, et al. DAGAN: Deep de-aliasing generative adversarial networks for fast compressed sensing MRI reconstruction. IEEE Trans Med Imaging 2018;37(6):1310–1321. [DOI] [PubMed] [Google Scholar]
- 31.Simonyan K, Zisserman A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv:1409.1556 [preprint]. https://arxiv.org/abs/1409.1556. Posted September 4, 2014. Accessed February 7, 2021.
- 32.Mardani M, Gong E, Cheng JY, et al. Deep generative adversarial neural networks for compressive sensing MRI. IEEE Trans Med Imaging 2019;38(1):167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quan TM, Nguyen-Duc T, Jeong WK. Compressed sensing MRI reconstruction using a generative adversarial network with a cyclic loss. IEEE Trans Med Imaging 2018;37(6):1488–1497. [DOI] [PubMed] [Google Scholar]
- 34.Deng M, Goy A, Li S, Arthur K, Barbastathis G. Probing shallower: perceptual loss trained Phase Extraction Neural Network (PLT-PhENN) for artifact-free reconstruction at low photon budget. Opt Express 2020;28(2):2511–2535. [DOI] [PubMed] [Google Scholar]
- 35.Hendee WR, O’Connor MK. Radiation risks of medical imaging: separating fact from fantasy. Radiology 2012;264(2):312–321. [DOI] [PubMed] [Google Scholar]
- 36.Chen H, Zhang Y, Zhang W, et al. Low-dose CT via convolutional neural network. Biomed Opt Express 2017;8(2):679–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang E, Min J, Ye JC. A deep convolutional neural network using directional wavelets for low-dose X-ray CT reconstruction. Med Phys 2017;44(10):e360–e375. [DOI] [PubMed] [Google Scholar]
- 38.Chen H, Zhang Y, Kalra MK, et al. Low-dose CT with a residual encoder-decoder convolutional neural network. IEEE Trans Med Imaging 2017;36(12):2524–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leuschner J, Schmidt M, Baguer DO, Maaß P. The LoDoPaB-CT Dataset: A Benchmark Dataset for Low-Dose CT Reconstruction Methods. arXiv:1910.01113 [preprint]. https://arxiv.org/abs/1910.01113. Posted October 1, 2019. Accessed February 7, 2021.
- 40.Yang Q, Yan P, Zhang Y, et al. Low-dose CT image denoising using a generative adversarial network with Wasserstein distance and perceptual loss. IEEE Trans Med Imaging 2018;37(6):1348–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.You C, Yang Q, Gjesteby L, et al. Structurally-sensitive multi-scale deep neural network for low-dose CT denoising. IEEE Access 2018;6(41839):41855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi X, Babyn P. Sharpness-aware low-dose CT denoising using conditional generative adversarial network. J Digit Imaging 2018;31(5):655–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang Y, Cai J, Lu L, et al. CT Image Enhancement Using Stacked Generative Adversarial Networks and Transfer Learning for Lesion Segmentation Improvement. In: Shi Y, Suk HI, Liu M, eds. Machine Learning in Medical Imaging. MLMI 2018. Lecture Notes in Computer Science, vol 11046. Cham, Switzerland: Springer, 2018; 46–54. [Google Scholar]
- 44.You C, Li G, Zhang Y, et al. CT Super-Resolution GAN Constrained by the Identical, Residual, and Cycle Learning Ensemble (GAN-CIRCLE). IEEE Trans Med Imaging 2020;39(1):188–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang E, Koo HJ, Yang DH, Seo JB, Ye JC. Cycle-consistent adversarial denoising network for multiphase coronary CT angiography. Med Phys 2019;46(2):550–562. [DOI] [PubMed] [Google Scholar]
- 46.Chaudhari AS, Fang Z, Kogan F, et al. Super-resolution musculoskeletal MRI using deep learning. Magn Reson Med 2018;80(5):2139–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong C, Loy CC, He K, Tang X. Image super-resolution using deep convolutional networks. IEEE Trans Pattern Anal Mach Intell 2016;38(2):295–307. [DOI] [PubMed] [Google Scholar]
- 48.Kudo A, Kitamura Y, Li Y, Iizuka S, Simo-Serra E. Virtual Thin Slice: 3D Conditional GAN-based Super-Resolution for CT Slice Interval. In: Knoll F, Maier A, Rueckert D, Ye J, eds. Machine Learning for Medical Image Reconstruction. MLMIR 2019. Lecture Notes in Computer Science, vol 11905. Cham, Switzerland: Springer, 2019; 91–100. [Google Scholar]
- 49.Gadermayr M, Li K, Müller M, et al. Domain-specific data augmentation for segmenting MR images of fatty infiltrated human thighs with neural networks. J Magn Reson Imaging 2019;49(6):1676–1683. [DOI] [PubMed] [Google Scholar]
- 50.Cronin NJ, Finni T, Seynnes O. Using deep learning to generate synthetic B-mode musculoskeletal ultrasound images. Comput Methods Programs Biomed 2020;196105583. [DOI] [PubMed] [Google Scholar]
- 51.Frid-Adar M, Diamant I, Klang E, Amitai M, Goldberger J, Greenspan H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018;321(321):331. [Google Scholar]
- 52.Russ T, Goerttler S, Schnurr AK, et al. Synthesis of CT images from digital body phantoms using CycleGAN. Int J Comput Assis Radiol Surg 2019;14(10):1741–1750. [DOI] [PubMed] [Google Scholar]
- 53.Liu F, Jang H, Kijowski R, Bradshaw T, McMillan AB. Deep learning MR imaging–based attenuation correction for PET/MR imaging. Radiology 2018;286(2):676–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nie D, Trullo R, Lian J, et al. Medical Image Synthesis with Context-Aware Generative Adversarial Networks. In: Descoteaux M, Maier-Hein L, Franz A, Jannin P, Collins D, Duchesne S, eds. Medical Image Computing and Computer Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science, vol 10435. Cham, Switzerland: Springer, 2017; 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolterink JM, Dinkla AM, Savenije MH, Seevinck PR, van den Berg CA, Išgum I. Deep MR to CT. Synthesis Using Unpaired Data. In: Tsaftaris S, Gooya A, Frangi A, Prince J, eds. Simulation and Synthesis in Medical Imaging. SASHIMI 2017. Lecture Notes in Computer Science, vol 10557. Cham, Switzerland: Springer, 2017; 14–23. [Google Scholar]
- 56.Wu W, Cao K, Li C, Qian C, Loy CC. Transgaga: Geometry-Aware Unsupervised Image-to-Image Translation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, June 15–20, 2019. Piscataway, NJ: IEEE, 2019; 8012–8021. [Google Scholar]
- 57.Gokaslan A, Ramanujan V, Ritchie D. Improving shape deformation in unsupervised image-to-image translation. In: Kim K, Tompkin J, eds. Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, September 8–14, 2018. Cham, Switzerland: Springer, 2018; 649–665. [Google Scholar]
- 58.Hiasa Y, Otake Y, Takao M, et al. Cross-Modality Image Synthesis from Unpaired Data Using CycleGAN. In: Gooya A, Goksel O, Oguz I, Burgos N, eds. Simulation and Synthesis in Medical Imaging. SASHIMI 2018. Lecture Notes in Computer Science, vol 11037. Cham, Switzerland: Springer, 2018; 31–41. [Google Scholar]
- 59.Lei Y, Harms J, Wang T, et al. MRI-only based synthetic CT generation using dense cycle consistent generative adversarial networks. Med Phys 2019;46(8):3565–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maspero M, Savenije MHF, Dinkla AM, et al. Dose evaluation of fast synthetic-CT generation using a generative adversarial network for general pelvis MR-only radiotherapy. Phys Med Biol 2018;63(18):185001. [DOI] [PubMed] [Google Scholar]
- 61.Liu F. SUSAN: segment unannotated image structure using adversarial network. Magn Reson Med 2019;81(5):3330–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ying X, Guo H, Ma K, Wu J, Weng Z, Zheng Y. X2CT-GAN: Reconstructing CT from Biplanar X-Rays with Generative Adversarial Networks. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, June 15–20, 2019. Piscataway, NJ: IEEE, 2019; 10619–10628. [Google Scholar]
- 63.Chea P, Mandell JC. Current applications and future directions of deep learning in musculoskeletal radiology. Skeletal Radiol 2020;49(2):183–197. [DOI] [PubMed] [Google Scholar]
- 64.Cheng CT, Ho TY, Lee TY, et al. Application of a deep learning algorithm for detection and visualization of hip fractures on plain pelvic radiographs. Eur Radiol 2019;29(10):5469–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang PD, Wong TT, Rasiej MJ. Deep learning for detection of complete anterior cruciate ligament tear. J Digit Imaging 2019;32(6):980–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tiulpin A, Thevenot J, Rahtu E, Lehenkari P, Saarakkala S. Automatic knee osteoarthritis diagnosis from plain radiographs: A deep learning-based approach. Sci Rep 2018;8(1):1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee H, Tajmir S, Lee J, et al. Fully automated deep learning system for bone age assessment. J Digit Imaging 2017;30(4):427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Forsberg D, Sjöblom E, Sunshine JL. Detection and labeling of vertebrae in MR images using deep learning with clinical annotations as training data. J Digit Imaging 2017;30(4):406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Han Z, Wei B, Mercado A, Leung S, Li S. Spine-GAN: Semantic segmentation of multiple spinal structures. Med Image Anal 2018;50(23):35. [DOI] [PubMed] [Google Scholar]
- 70.Sekuboyina A, Rempfler M, Kukačka J, et al. Btrfly Net: Vertebrae Labelling with Energy-Based Adversarial Learning of Local Spine Prior. In: Frangi A, Schnabel J, Davatzikos C, Alberola-López C, Fichtinger G, eds. Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science, vol 11073. Cham, Switzerland: Springer, 2018; 649–657. [Google Scholar]
- 71.Galbusera F, Niemeyer F, Seyfried M, et al. Exploring the potential of generative adversarial networks for synthesizing radiological images of the spine to be used in in silico trials. Front Bioeng Biotechnol 2018;653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Galbusera F, Bassani T, Casaroli G, et al. Generative models: an upcoming innovation in musculoskeletal radiology? A preliminary test in spine imaging. Eur Radiol Exp 2018;2(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ge Y, Xue Z, Cao T, Liao S. Unpaired whole-body MR to CT synthesis with correlation coefficient constrained adversarial learning. In: Angelini ED, Landman BA, eds. Proceedings of SPIE: Medical Imaging 2019—Image Processing. Vol 10949. Bellingham, Wash: International Society for Optics and Photonics, 2019. [Google Scholar]
- 74.Gupta A, Venkatesh S, Chopra S, Ledig C. Generative image translation for data augmentation of bone lesion pathology. arXiv:1902.02248 [preprint]. https://arxiv.org/abs/1902.02248. Posted February 6, 2019. Accessed February 7, 2021.
- 75.Finlayson SG, Lee H, Kohane IS, Oakden-Rayner L. Towards generative adversarial networks as a new paradigm for radiology education. arXiv:1812.01547 [preprint]. https://arxiv.org/abs/1812.01547. Posted December 4, 2018. Accessed February 7, 2021.
- 76.Salimans T, Goodfellow I, Zaremba W, Cheung V, Radford A, Chen X. Improved techniques for training gans. Adv Neural Inf Process Syst 2016;2234–2242. [Google Scholar]
- 77.Pfau D, Vinyals O. Connecting generative adversarial networks and actor-critic methods. arXiv:1610.01945 [preprint]. https://arxiv.org/abs/1610.01945. Posted October 6, 2016. Accessed February 7, 2021.
- 78.Blau Y, Michaeli T. The Perception-Distortion Tradeoff. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, June 18–22, 2018. Piscataway, NJ: IEEE, 2018; 6228–6237. [Google Scholar]
- 79.Chuquicusma MJ, Hussein S, Burt J, Bagci U. How to Fool Radiologists with Generative Adversarial Networks? A Visual Turing Test for Lung Cancer Diagnosis. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, April 4–7, 2018. Piscataway, NJ: IEEE, 2018; 240–244. [Google Scholar]
- 80.Chaudhari AS, Stevens KJ, Wood JP, et al. Utility of deep learning super-resolution in the context of osteoarthritis MRI biomarkers. J Magn Reson Imaging 2020;51(3):768–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heusel M, Ramsauer H, Unterthiner T, Nessler B, Hochreiter S. Gans trained by a two time-scale update rule converge to a local nash equilibrium. Adv Neural Inf Process Syst 2017;6626–6637. [Google Scholar]
- 82.Odena A, Dumoulin V, Olah C. Deconvolution and checkerboard artifacts. Distill 2016;1(10):e3. [Google Scholar]
- 83.Fu H, Gong M, Wang C, Batmanghelich K, Zhang K, Tao D. Geometry-Consistent Generative Adversarial Networks for One-Sided Unsupervised Domain Mapping. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, June 15–20, 2019. Piscataway, NJ: IEEE, 2019; 2427–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cohen JP, Luck M, Honari S. Distribution Matching Losses Can Hallucinate Features in Medical Image Translation. In: Frangi A, Schnabel J, Davatzikos C, Alberola-López C, Fichtinger G, eds. Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science, vol 11070. Cham, Switzerland: Springer, 2018; 529–536. [Google Scholar]
- 85.Štajduhar I, Mamula M, Miletić D, Ünal G. Semi-automated detection of anterior cruciate ligament injury from MRI. Comput Methods Programs Biomed 2017;140(151):164. [DOI] [PubMed] [Google Scholar]
- 86.Zhou Z, Zhao G, Kijowski R, Liu F. Deep convolutional neural network for segmentation of knee joint anatomy. Magn Reson Med 2018;80(6):2759–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yi J, Lee YH, Song HT, Suh JS. Clinical feasibility of synthetic magnetic resonance imaging in the diagnosis of internal derangements of the knee. Korean J Radiol 2018;19(2):311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tanenbaum LN, Tsiouris AJ, Johnson AN, et al. Synthetic MRI for clinical neuroimaging: results of the Magnetic Resonance Image Compilation (MAGiC) prospective, multicenter, multireader trial. AJNR Am J Neuroradiol 2017;38(6):1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Choi Y, Choi M, Kim M, Ha JW, Kim S, Choo J. StarGAN: Unified Generative Adversarial Networks for Multi-domain Image-to-Image Translation. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, June 18–23, 2018. Piscataway, NJ: IEEE, 2018; 8789–8797. [Google Scholar]
- 90.Xue Y, Xu T, Zhang H, Long LR, Huang XJN. SegAN: Adversarial Network with Multi-scale L1 Loss for Medical Image Segmentation. Neuroinformatics 2018;16(3-4):383–392. [DOI] [PubMed] [Google Scholar]
- 91.Schlegl T, Seeböck P, Waldstein SM, Schmidt-Erfurth U, Langs G. Unsupervised Anomaly Detection with Generative Adversarial Networks to Guide Marker Discovery. In: Niethammer M, Styner M, Aylward S, et al., eds. Information Processing in Medical Imaging. IPMI 2017. Lecture Notes in Computer Science, vol 10265. Cham, Switzerland: Springer, 2017; 146–157. [Google Scholar]
- 92.Watanabe H, Togo R, Ogawa T, Haseyama M. Bone Metastatic Tumor Detection Based on AnoGAN Using CT Images. In: 2019 IEEE 1st Global Conference on Life Sciences and Technologies (LifeTech), Osaka, Japan, March 12–14, 2019. Piscataway, NJ: IEEE, 2019; 235–236. [Google Scholar]
- 93.Bowles C, Gunn R, Hammers A, Rueckert D. Modelling the progression of Alzheimer’s disease in MRI using generative adversarial networks. In: Angelini ED, Landman BA, eds. Proceedings of SPIE: Medical Imaging 2018—Image Processing. Vol 10574. Bellingham, Wash: International Society for Optics and Photonics, 2018. [Google Scholar]
- 94.Miller TT. Bone tumors and tumorlike conditions: analysis with conventional radiography. Radiology 2008;246(3):662–674. [DOI] [PubMed] [Google Scholar]
- 95.Knoll F, Zbontar J, Sriram A, et al. fastmri: A publicly available raw k-space and dicom dataset of knee images for accelerated mr image reconstruction using machine learning. Radiol Artif Intell 2020;2(1):e190007. [DOI] [PMC free article] [PubMed] [Google Scholar]