Abstract
Human centromeres are composed of large tandem arrays of repetitive alpha satellite DNA, which are often sites of aberrant rearrangement in cancers ( Mitelman et al., 1997 ; Padilla- Nash et al., 2001 ). To date, annotation of the human centromere repetitive sequences remains incomplete, greatly hindering in-depth functional studies of these regions essential for chromosome segregation. In order to monitor sister chromatid exchange happening at the centromere (C-SCE) due to recombination and mutagenic events, I have applied the Chromosome-Orientation Fluorescence in situ Hybridization (CO-FISH) technique to centromeres (Cen-CO-FISH) in human cells. This hybridization-based method involves (1) the incorporation of nucleotide analogs through a single round of replication, (2) enzymatic digestion of the newly synthesized DNA strand and (3) subsequent hybridization of single-stranded probes, in absence of a denaturation step. The resulting signal allows to differentially label each sister chromatid based on the 5’-3’ directionality of the DNA and to score aberrant staining patterns indicative of C-SCE. The Cen-CO-FISH method applied to human centromeres revealed that human centromeres indeed undergo recombination in cycling cells resulting in C-SCE, and centromere instability is enhanced in cancer cell lines and primary cells undergoing senescence (Giunta and Funabiki, 2017). Here, I present the detailed protocol of the preparation, experimental procedure and data acquisition for the Cen-CO-FISH method in human cells. It also includes a conceptual overview of the technique, with examples of representative images and scoring guidelines. The Cen-CO-FISH represents a valuable tool to facilitate exploration of centromere repeats.
Keywords: Centromere, Fluorescence in situ hybridization , CO-FISH, Alpha satellite, Repetitive DNA, Genome stability, Recombination, Sister chromatid exchange
Background
The human genome project was marked completed in 2003, yet it omitted over 10% of the human repetitive DNA ( de Koning et al., 2011 ), including the centromere. The human centromere is a highly specialized genomic locus (Choo, 1997) playing a critical role during chromosome segregation where it serves as the site of kinetochore assembly to allow interaction with microtubules and sister chromatids separation during cell division (Cheeseman, 2014). Human centromeres are made of characteristic repetitive DNA sequences called alpha-satellites, whose linear assembly remains largely absent from the reference genomes. Here, I present the application of the Cen-CO-FISH technique to label human centromere and monitor recombination events resulting in crossover. Introduced by Bailey and colleagues over 20 years ago ( Bailey et al., 1996 ), the CO-FISH method has been widely applied to detect recombination, fragility, replication timing, fusion and inversions at telomeres repeats, as well as to monitor mitotic segregation patterns and non-random sister chromatid segregation ( Bailey et al., 2010 ). The application of this methodology to centromere, hereby called Cen-CO-FISH method, has revealed that the centromere-specific histone variant CENP-A, and CENP-A associated proteins CENP-C and CENP-T/W, work to prevent centromere instability and this functionality is compromised in cancer cell lines and in primary cells approaching replicative senescence that display higher number of C-SCE (Giunta and Funabiki, 2017). Cen-CO-FISH was used to assess centromere instability in cancer and during cellular senescence in human cells (Giunta and Funabiki, 2017) and it has been previously applied to study recombination ( Jaco et al., 2008 ; de La Fuente et al., 2015 ) and sister chromatid separation patterns in mouse cells ( Falconer et al., 2010 ). The wide application potentials of this methodology spans from quantitative detection of alpha satellite repeats, centromere recombination resulting in C-SCE, fragility, replication timing, fusion and inversions, as well as to monitor mitotic segregation patterns and non-random sister chromatids segregation ( Bailey et al., 2010 ; Yadlapalli and Yamashita, 2013). Cen-CO-FISH fills the gaps in the missing genetic information that have cast a shadow over the centromere and other repetitive regions, bringing new light into the possibilities for functional exploration of these important loci of our genome.
Materials and Reagents
6 or 10 cm Petri dish (Corning, Falcon®, catalog numbers: 353002 or 353003)
15 ml Falcon tube (Corning, Falcon®, catalog number: 352097)
Frosted slides (Superfrost Plus; Fisher Scientific, catalog number: 12-550-15)
Coverslips (24 x 60 mm) (Fisher Scientific, catalog number: 12-545-M)
Paper towel
Glass Pasteur pipette (Fisher Scientific, catalog number: 13-678-20A)
Gloves and lab coat
-
Human cells of interest and appropriate medium
Note: This protocol is for adherent cells, changes can be made for use for non-adherent cultures.
5’-Bromodeoxyuridine (BrdU) (MP Biomedicals, catalog number: 100166)Note: Prepare 10 mM stock solution in double distilled water (1,000x); make aliquots and store at -20 °C.
-
5’-Bromodeoxycytidine (BrdC) (Sigma-Aldrich, catalog number: B5002)
Note: Prepare 10 mM stock solution in double distilled water (1,000x); make aliquots and store at -20 °C.
Colcemid (Roche Diagnostics, catalog number: 10295892001; already diluted 10 μg/ml)–Store at 4 °C
Phosphate-buffered saline (PBS)
Trypsin-EDTA (Thermo Fisher Scientific, GibcoTM, catalog number: 25300)
Fetal bovine serum (Atlanta Biologicals)
Potassium chloride (KCl) (Fisher scientific, catalog number: P217-500)
-
RNase A (Sigma-Aldrich, catalog number: R5000)
Note: Prepare stock solution 50 mg/ml in 10 mM Tris-HCl pH 7.2. Aliquot and store at -20 °C
-
Hoechst 33258 (Thermo Fisher Scientific, InvitrogenTM, catalog number: H3569)
Note: Make a 10 μg/ml solution in double distilled water and store at 4 °C away from light.
Exonuclease III and buffer (Promega, catalog number: M1811)–Keep at -20 °C
DAPI (Sigma-Aldrich, catalog number: D9542)–0.5 mg/ml stock in water. Keep at 4 °C in the dark for one year
ProLong Gold Anti-fade Reagent (Thermo Fisher Scientific, InvitrogenTM, catalog number: P36934)
Nail varnish (Sally Hansen, Transparent Harderer)
Methanol (Fisher Scientific, catalog number: A452-4)
Glacial acetic acid (Fisher Scientific, catalog number: A38C-212)
Ethanol 100% (Decon, catalog number: 2716), 90% and 70%
Blocking reagent (Roche Diagnostics, catalog number: 11096176001)
Maleic acid (Sigma-Aldrich, catalog number: M0375)
Sodium chloride (NaCl) (Merck, catalog number: SX0420-5)
Sodium hydroxide (NaOH) (Fisher Scientific, catalog number: S318-500)
Tris-HCl pH 7.2 (Sigma-Aldrich, CAS number: 1185-53-1)
Formamide (Fisher Scientific, catalog number: BP228; use deionized for hybridization)
Bovine serum albumin (BSA)
Tween-20 (Hoefer, CAS number: GR128-500)
Sodium citrate (Sigma-Aldrich, CAS number: 6132-04-3)
-
Magnesium chloride (MgCl2) (Fisher Scientific, catalog number: AC41341-5000)
Manufacturer: Acros Organics, catalog number: 413410025.
Dithiothreitol (DTT) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0861)
Hypotonic solution (see Recipes)
Fixative solution (see Recipes)
Ethanol dilution (see Recipes)
Blocking solution (see Recipes)
Hybridization solution (see Recipes)
Hybridization wash #1 (see Recipes)
Hybridization wash #2 (see Recipes)
Peptide nucleic acid (PNA) probes (custom probes from PNABio) (see Recipes)
Sodium chloride and sodium citrate buffer (SSC, see Recipes)
Equipment
Pipettes (Gilson)
Centrifuge (Eppendorf, model: 5810 R)
Coplin Jars (Scienceware, Sigma-Aldrich, catalog number: S5641)
Heating block (VWR)
Water bath (Fisher Scientific, model: IsotempTM 210)
Stratalinker with 365-nm UV light blubs (Spectralinker XL-1000 1800 UV irradiator) (Spectronics Corporation, model: XL-1000)
Slides plastic tray–to fit into the Stratalinker
Hybridization chamber (see text for more details)
Orbital shaker
-
Imaging equipment:
DeltaVision Image Restoration microscope system (Applied Precision/GE Healthcare)
Olympus IX-70 microscope (Olympus, model: IX70)
100x/1.40 UPLSAPO objective lens
CoolSnap QE CCD camera (Photometrics)
Software
SoftWoRx (Sold by Applied Precision)
Metamorph 7.8 (Sold by Universal Imaging)
Prism 5 (Sold by GraphPad)
Procedure
-
Incorporation of BrdU:C and metaphase chromosome spread preparation
Cells are seeded into a 6 or 10 cm dish at least two days before harvesting, to be about 70% confluent at the time of harvesting. In a 10 cm dish, seed ~1 million cells.
-
Cells are then labeled for 16-20 h by incubating with 5’-Bromodeoxyuridine (BrdU) and 5’-Bromodeoxycytidine (BrdC). Thaw a 1,000x aliquot of BrdU and BrdC and mix 3:1 to obtain 7.5 mM BrdU + 2.5 mM BrdC. Add to the cells medium at a 1:1,000 dilution and incubate the cells overnight.
Note: Adjust the time of BrdU:C labeling according to whether your cells grow slower/faster to avoid double labeling or incompletely labeled cells. You are aiming for cells to incorporate BrdU and BrdC throughout S-phase and to prevent them to exit from mitosis after replication. Typically, HeLa cells are incubated with BrdU:C for 14-16 h before adding colcemid; to validate the time of BrdU:C incubation one can synchronize cells with thymidine at G1/S, release and monitor exit from mitosis by flow cytometry. Cells that exit mitosis 15 h post-thymidine release should be treated with BrdU:C for 16-17 h before addition of colcemid to ensure only cells that started labeling before S-phase are trapped by the colcemid block.
-
Add colcemid to a final concentration of 0.1 µg/ml (1:100 dilution of 10 µg/ml stock) to accumulate mitotic cells for about 2 h before harvesting.
Note: Avoid leaving cells into colcemid for over 3 h, because long exposure to colcemid will lead to very compacted chromosome morphology.
Harvest floating cells by saving the supernatant in a 15 ml Falcon tube, wash the dish once with PBS and collect the PBS wash as well into the same tube. Trypsinize for 5 min, block with medium containing fetal bovine serum, and collect the rest of the cells in the same tube.
Pellet cells for 5 min at 175 × g. Remove supernatant and resuspend cells in 10 ml of pre-warmed 75 mM KCl solution. Gently pipette up and down to resuspend the pellet.
Incubate the cells in the KCl hypotonic solution at 37 °C for 30 min (slowly invert the tube every 10 min to keep the cells suspended). Before pelleting the cells, adding 200 µl of fixative solution and mixing by slowly inverting the tubes twice helps the metaphases to remain intact during centrifugation, because cells are extremely fragile after the hypotonic treatment. Pellet the cells for 5 min at 175 × g.
Aspirate most supernatant, leaving about 200 µl of KCl to resuspend cells by tapping the tube or slowly pipetting up and down with a P1000.
-
Fix the resuspended pellet by adding 10 ml of freshly made ice-cold MeOH-acetic acid (3:1) fixative solution dropwise (prepare it 30 min before use, keep at -20 °C) while continuously vortexing the tube on a Vortex mixer at the lowest speed setting (#1).
Note: It is important to resuspend the pellet before addition of the fixative, and to add the fixative one drop at a time while continuously vortexing the cells at low speed for at least the first 2 ml. The remaining 8 ml can be added all at once.
Mix by inverting the tube a few times.
Cells can now be kept at 4 °C for several months, stored away from light.
Once you are ready to drop the cells, spin the tube down at 380 × g for 5 min. Aspirate the fixative leaving about 0.5-2 ml depending on how many cells are in each tube. Thoroughly resuspend cells in the remaining fixative.
If you are dropping onto Superfrost Plus slides, go directly to Step A13. If using regular slides, wash in soapy water and rinse 3 times with Milli-Q water before use. Keep slides in Milli-Q water in the fridge for an hour and dry well right before use.
-
Humidify the slide you are about to ‘drop’ on by hovering it over a hot wet towel placed on a heating block, until it steams up. You can also achieve this by ‘breathing’ onto the slide.
Note: Creating conditions of humidity, for instance by having wet paper towels onto an adjacent 90 °C block that continues to release vapor, and humidifying the air immediately adjacent to where slides are dropped, or using a hybridization hood with set humidity and temperature, helps with the quality of the spreads.
-
Using a glass Pasteur pipette, pipette up and down and aspirate the cell suspension. Drop several drops of cell suspension onto different places on the slide from about 20 cm away. You should aim to cover all parts of the slide with each drop.
Note on safety: Gloves and lab coat should be used to protect from possible splashes. The entire metaphase spread procedure can be performed in a laboratory chemical fume hood for additional protection.
-
Immediately after dropping, rest the slide for about 2-3 min onto a 42 °C block with a wet paper towel covering the block, cells side up.
Note: Factors that influence the spreading are: humidity, temperature, distance of dropping cells onto the slide and drying time.
Air-dry slides overnight at room temperature away from light. You can keep any cells left into the falcon tube stored in fresh fixative at 4 °C for up to one year.
-
Enzymatic digestion of BrdU:C labeled, newly synthesized strand
Dropped slides are inserted in a Coplin jar and rehydrated in PBS (pH 7.0-7.5) for 5 min (about 35 ml of solution is needed for each jar).
Treat slides with 0.5 mg/ml RNase A (in PBS, DNase free) for 10 min in a 37 °C water bath.
Stain slides with 1 µg/ml Hoechst 33258 (Sigma-Aldrich) diluted into 2x SSC for 15 min at room temperature with slight agitation.
Place slides in a shallow plastic tray and add just enough 2x SSC buffer to cover the slides. Expose the slides to 365 nm UV light at room temperature for 1,800 sec (30 min; equivalent to 5,400 J/m2) in a Stratalinker 1800.
Digest the BrdU:C labeled UV nicked DNA strand with 80 µl of 10 U/µl Exonuclease III (Promega) into buffer supplied by the manufacturer diluted to 1x (50 mM Tris-HCl, 5 mM MgCl2, 5 mM DTT, pH 8.0). Add the 80 µl solution onto a 24 x 60 coverslip that will make contact with the entire surface of the cells on the slide. Let it adhere to the slide, then place the slide cells side up for 10 min, away from light. Avoid air bubbles.
Remove the coverslips and repeat the Exonuclease III digestion step with fresh solution for another 10 min.
Wash slides in PBS for 5 min and dehydrate successively in 70%, 90%, 100% ethanol series for 2 min each.
Air dry slides and store at room temperature in the dark (can be left overnight).
-
Fluorescent in situ hybridization
Place 80 µl of hybridization mix onto coverslips and pick up the coverslip with the slide, then put the slide into the hybridization chamber (see Figures 1G-1I for instructions on how to make a hybridization chamber and for a visual illustration of these steps). Avoid air bubbles.
Hybridize with the Forward PNA probe at a 1:1,000 dilution (heated at 60 °C for 10 min right before use) in the dark for 2 h in the hybridization chamber.
Rinse in Wash #1 for 30 sec. Leave slides draining vertically for 10 sec.
-
Hybridize with the Reverse Complement PNA probe at a 1:1,000 dilution (heated at 60 °C for 10 min right before use) in the dark for 2 h.
Note: Inverting the order of the probes will not affect the hybridization or quality of the signal.
Wash slides in Wash #1 for 15 min 2 times on an orbital shaker.
Wash slides in Wash #2 for 5 min 3 times. To the second wash, add 1:500 DAPI from 0.5 mg/ml stock.
Dehydrate in 70%, 90%, 100% ethanol series for 3 min each.
Air dry slides for about 1 h.
Mount slides using Prolong Gold embedding medium (avoid bubbles) and seal using nail varnish. Slides are ready for imaging.
Slides can be stored up to 1 week at 4 °C or at -20 °C for longer storage.
Figure 1. Representative images of the Cen-CO-FISH procedure at different stages of the protocol.
A. Cells are harvested, hypotonically swollen and fixed overnight. Samples can be stored at 4 °C at this stage. Cells are then dropped onto glass slides to let the metaphases spread. Slides can be stored in the dark at room temperature (RT) before proceeding to Step B. B. Rehydrate the slides into PBS in a Coplin jar for 5 min. C. Remove PBS and add RNase A solution into the jar, incubate in a 37 °C water bath for 10 min. Remove RNase solution and incubate with Hoechst in 2x SSC for 15 min at RT. D. Place slides into plastic try and cover with just enough 2x SSC. E. Place tray into Stratalinker oven with 365 nm UV bulbs and expose for 30 min (1,800 sec). F. Prepare 80 μl of Exonuclease III solution for each slide and add the solution to a 24 x 60 coverslip that will make contact with the entire surface of the slide. G. Immediately after UV exposure, pick up the coverslip with Exonuclease III solution as shown. Invert the slide, adjust the coverslip to make sure it is central, the liquid is well distributed and there are no air bubbles. Incubate cells side up for 10 min at RT and then repeat Steps F-G one more time for an additional Exonuclease incubation. Wash in PBS and dehydrate in ethanol series. Store slides at RT in the dark (overnight). H. Prepare one of the probes (1:1,000-1:5,000) into hybridization solution, heat for 10 min at 60 °C before adding 80 μl onto the coverslips. Proceed as in Steps F-G shown. I. Place slides into a Hybridization chamber and incubate for 2 h at RT. To make a chamber, take a box, fill it with paper towels and wet throughout until all towels are humid. Do not put the slides directly on top of the wet paper. Use pipettes or other forms of support to raise the coverslips over the wet paper. Place slides cells side up and close the box away from light. Upon opening the box after 2 h, you should see a small amount of condensation on the lid. Wash with Hybridization buffer #1 and repeat Step H with the second, reverse complement probe (you can invert the order of the hybridization between forward and reverse complement probes, it should yield identical results). Wash in Hybridization buffer #1 and #2, including the DAPI step, dehydrate and air dry for at least one hour. Mount and seal before imaging.
Data analysis
Image acquisition and processing
Cells are imaged using a DeltaVision Image Restoration microscope system (Applied Precision/GE Healthcare), mounted on an Olympus IX-70 microscope and fitted with a 100x/1.40 UPLSAPO objective lens and a CoolSnap QE CCD camera (Photometrics).
15 or more metaphase spreads for each experiment are imaged to yield statistically significant and representative results. When imaging the cells, care should be taken to select the best spreads where chromosomes are nicely separated (Figure 2A) and approximately 46 chromosomes are present in the case of karyotypically stable diploid cells. Typical exposure times for the DAPI is 0.3 sec, for the TRITC channel (red probe) is 0.1 sec and for the FITC (green probe) is 0.5 sec.
The image is acquired as a z-stack containing about 15-30 0.2 μm sections. Acquired images are deconvolved using SoftWoRx (Applied Precision) and exported to Metamorph (Universal Imaging) for analysis as maximum projections, as shown in the examples in Figures 2A and 2B.
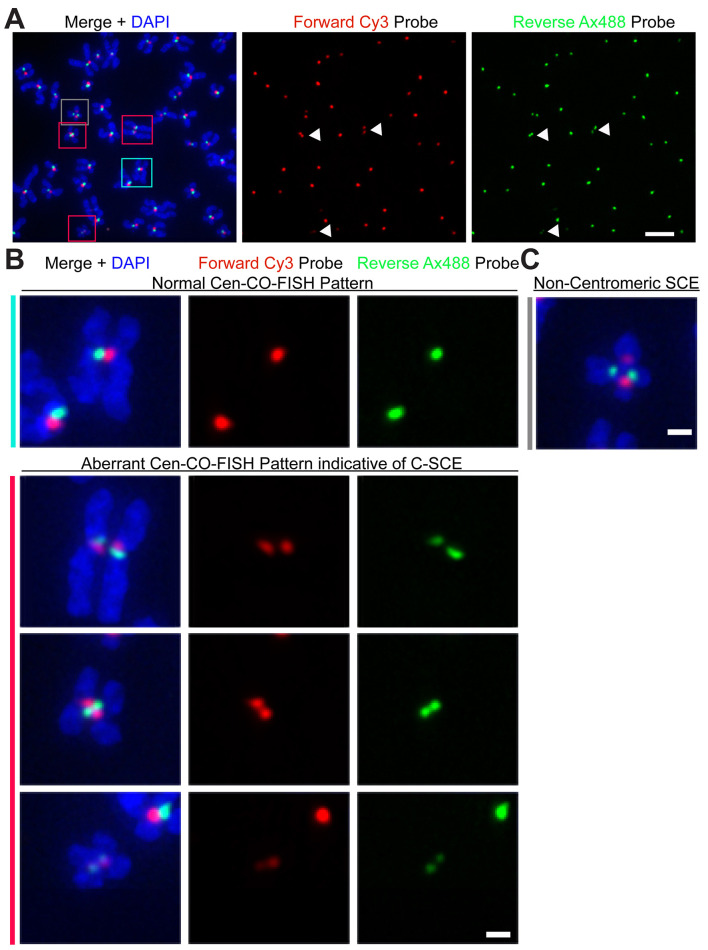
Figure 2. Detecting recombination events at human centromere using the Cen-CO-FISH method.
A. HeLa cells were stained with Cen-CO-FISH using forward (red) and reverse (green) probes raised against the CENP-B box sequence, present in all chromosomes in the metaphase spread shown in A. Individual channels allow the visual identification of aberrant Cen-CO-FISH staining patterns, often present as a ‘double dot’ for the red channel and the same for the green channel, as indicated by the white arrows. Color combined image merged with DAPI is also shown. Boxes are enlarged in Figure 2B. Scale bar = 5 µm. B. Enlarged boxes from Figure 2A, left panel, showing a representative image for a normal Cen-CO-FISH staining pattern (marked in teal), and three panels below showing aberrant patterns (marked in red) indicative of C-SCE. Scale bar = 1 µm. C. Examples of an aberrant staining pattern resulting from recombination outside the centromere sequence (marked in grey), thus excluded from quantification of centromere recombination and C-SCE. Scale bar = 1 µm.
Representative images and scoring
Following the Cen-CO-FISH protocol outlined, high-quality metaphase spreads are obtained stained with a specific centromere marker, as shown in Figure 2A. The forward probe, labeled in red, and the reverse strand probe, labeled in green, should not overlap but be adjacent to each other, labeling one individual sister chromatid of each chromosome (Figure 2B). Quantifications are performed by counting the number of aberrant Cen-CO-FISH patterns indicative of C-SCE in each metaphase spreads (Figure 2A, colored boxes). Examples of normal (top panel, teal box; Figure 2B) and aberrant Cen-CO-FISH pattern indicative of C-SCE are shown (bottom panels, red boxes; Figure 2B). In high chromosomal instability (CIN) lines or some cancer cell lines, a small number of aberrant patterns can be found resulting from fusion events occurring outside of the centromere region, as indicated by the presence of DAPI separating the two pairs of centromere signal, and should be excluded from the quantification of C-SCE (grey box; Figure 2C). The number of C-SCE can be presented as the percentage of aberrant centromeres over the total number of labeled centromere pairs for each metaphase and plotted in a scatter plot using Prism, as previously shown (Giunta and Funabiki, 2017).
Cen-CO-FISH is a reliable, reproducible and sensitive labeling technique to visualize repetitive genomic regions. The technique relies on two crucial steps: repetitiveness of the locus and incorporation of BrdU and BrdC into the newly synthesized strand. The probes used for the Cen-CO-FISH hybridization are fluorescently-labeled PNA 17 or 18-mer. Designing the probes to be aimed at sequences that are repeated enables the amplification of the fluorescence intensity. Centromere alpha satellite repeats spans from 0.5-5 Mb with sequences common to each human chromosomes ( Schueler et al., 2001 ; Fukagawa, 2004), yielding a robust Cen-CO-FISH signal for visualization, acquisition using conventional microscopy, and functional analyses. The reliance of the technique on labeling of the newly synthesized strand, which will subsequently be enzymatically digested, signify that this method can only be applied to cycling cells. However, this also represents an advantage for this method, where the yielded patterns are indicative of recombination events having happened during a single prior cell cycle. Thus, Cen-CO-FISH provides a bird’s-eye view on these dark genomic regions while giving specific temporal resolution and sequence orientation information to examine the behavior of the centromere repeats. Previous studies have reported the application of the Cen-CO-FISH technique to monitor centromere recombination in mouse cells ( Bailey et al., 1996 ; Jaco et al., 2008 ; Falconer et al., 2010 ) indicating that the technique can be successfully applied to tissue culture cells from different species using specifically designed probes.
Recipes
-
Hypotonic solution
75 mM KCl
Prewarm the required amount in a 37 °C water bath before each use
-
Fixative solution
3 parts methanol
1 part glacial acetic acid
Must be made fresh each time
Use ice-cold
-
Ethanol dilution
70% and 90% in double distilled water
-
Blocking solution
Dissolve the blocking reagent (Roche) into maleic acid buffer to make a 10% stock:
100 mM maleic acid
150 mM NaCl
Adjust pH to 7.5 with NaOH and store at 4 °C
Shake vigorously before each use
-
Hybridization solution
10 mM Tris-HCl pH 7.2
70% formamide
0.5% blocking reagent (from 10% stock)
-
Hybridization wash #1
10 mM Tris-HCl pH 7.2
70% formamide (Sigma-Aldrich)
0.1% BSA (dissolve in double distilled water before adding formamide)
-
Hybridization wash #2
0.1 M Tris-HCl pH 7.2
0.15 M NaCl
0.1% Tween-20
For the DAPI wash, add 1:500 DAPI using the 0.5 mg/ml stock
-
Peptide nucleic acid (PNA) probes (custom probes from PNABio)
Set #1 against the alpha satellite:
F3003 CENT-Cy3 (Cy3–OO–AAACTAGACAGAAGCATT)
Reverse complement CENT-RC-488 (Alexa488–O–AATGCTTCTGTCTAGTTT)
Set #2 against the CENP-B box:
F3002 CENP-B Cy3 (ATTCGTTGGAAACGGGA)
Reverse complement CENP-B-RC-488 (TCCCGTTTCCAACGAAT)
Notes:
Lyophilized probes were diluted to 50 μM in double distilled water based on manufacturer’s instructions. Aliquot and store at -80 °C to avoid multiple freeze-thawing.
For hybridization, thaw and spin the probes, and dilute into hybridization solution at a 1:1,000-1:5,000 dilution (adjust according to the signal obtained). Heat at 60 °C for 10 min before use.
-
Sodium chloride and sodium citrate buffer (SSC; 20x)
NaCl (3 M), sodium citrate (300 mM) dissolved in distilled water
Adjust pH to 7.0 and autoclave
Working dilution is 2x
Acknowledgments
I would like to thank H. Funabiki, F. Lottersberger, N. Bosco, and T. DeLange, as well as to K. Thomas, A. North and T. Tong at the Rockefeller University Bio-Imaging Resource Center (BIRC). I also want to thank Seneca Jason and A. Field for their support throughout this work. The work was funded by American-Italian Cancer Foundation fellowship and Women in Science Rockefeller fellowship. This protocol was adapted from procedures published in Giunta and Funabiki (2017) and Celli et al. (2006).
Competing interests statement: The author declares no competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Bailey S. M., Goodwin E. H., Meyne J. and Cornforth M. N.(1996). CO-FISH reveals inversions associated with isochromosome formation. Mutagenesis 11(2): 139-144. [DOI] [PubMed] [Google Scholar]
- 2. Bailey S. M., Williams E. S., Cornforth M. N. and Goodwin E. H.(2010). Chromosome Orientation fluorescence in situ hybridization or strand-specific FISH. Methods Mol Biol 659: 173-183. [DOI] [PubMed] [Google Scholar]
- 3. Celli G. B., Denchi E. L. and de Lange T.(2006). Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol 8(8): 885-890. [DOI] [PubMed] [Google Scholar]
- 4. Cheeseman I. M.(2014). The kinetochore. Cold Spring Harb Perspect Biol 6(7): a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choo K. H. A.(1997). The centromere. Oxford University Press, USA. [Google Scholar]
- 6. de Koning A. P., Gu W., Castoe T. A., Batzer M. A. and Pollock D. D.(2011). Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 7(12): e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de La Fuente R., Baumann C. and Viveiros M. M.(2015). ATRX contributes to epigenetic asymmetry and silencing of major satellite transcripts in the maternal genome of the mouse embryo. Development 142(10): 1806-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Falconer E., Chavez E. A., Henderson A., Poon S. S., McKinney S., Brown L., Huntsman D. G. and Lansdorp P. M.(2010). Identification of sister chromatids by DNA template strand sequences. Nature 463(7277): 93-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fukagawa T.(2004). Centromere DNA, proteins and kinetochore assembly in vertebrate cells. Chromosome Res 12(6): 557-567. [DOI] [PubMed] [Google Scholar]
- 10. Giunta S. and Funabiki H.(2017). Integrity of the human centromere DNA repeats is protected by CENP-A, CENP-C, and CENP-T. Proc Natl Acad Sci U S A 114(8): 1928-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jaco I., Canela A., Vera E. and Blasco M. A.(2008). Centromere mitotic recombination in mammalian cells. J Cell Biol 181(6): 885-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mitelman F., Mertens F. and Johansson B.(1997). A breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nat Genet 15 Spec No: 417-474. [DOI] [PubMed] [Google Scholar]
- 13. Padilla-Nash H. M., Heselmeyer-Haddad K., Wangsa D., Zhang H., Ghadimi B. M., Macville M., Augustus M., Schrock E., Hilgenfeld E. and Ried T.(2001). Jumping translocations are common in solid tumor cell lines and result in recurrent fusions of whole chromosome arms. Genes Chromosomes Cancer 30(4): 349-363. [DOI] [PubMed] [Google Scholar]
- 14. Schueler M. G., Higgins A. W., Rudd M. K., Gustashaw K. and Willard H. F.(2001). Genomic and genetic definition of a functional human centromere. Science 294(5540): 109-115. [DOI] [PubMed] [Google Scholar]
- 15. Yadlapalli S. and Yamashita Y. M.(2013). Chromosome-specific nonrandom sister chromatid segregation during stem-cell division. Nature 498(7453): 251-254. [DOI] [PMC free article] [PubMed] [Google Scholar]