Abstract
Single-molecule fluorescence detection of protein and other biomolecules requires a polyethylene glycol (PEG)-passivated surface. Individual channels on a PEG-passivated slide are typically used only a few times, limiting the number of experiments per slide. Here, we report several strategies for regenerating PEG surfaces for multiple rounds of experiments. First, we show regeneration of DNA- or RNA-tethered surfaces by washing out the bound protein by 0.1% sodium dodecyl sulfate, which is significantly more effective than 6 M urea, 6 M GdmCl, or 100 μM proteinase K. Strikingly, 10 consecutive experiments in five different systems produced indistinguishable results both in molecule count and protein activity. Second, duplexed DNA unwound by helicase or denatured by 50 mM NaOH was reannealed with a complementary strand to regenerate the duplexed substrate with an exceptionally high recovery rate. Third, the biotin-PEG layer was regenerated by using 7 M NaOH to strip off NeutrAvidin, which can be reapplied for additional experiments. We demonstrate five cycles of regenerating antibody immobilized surface by which three different protein activity was measured. Altogether, our methods represent reliable and reproducible yet simple and rapid strategies that will enhance the efficiency of single-molecule experiments.
Significance
Single-molecule fluorescence detection has become a widely used technology in many laboratories. For single-molecule imaging that involves proteins, both unlabeled and labeled, it is critical to passivate the surface to minimize nonspecific adsorption of the protein. Polyethylene glycol (PEG) is a polymer brush layer that can be used for such a passivation strategy. The PEG surface, despite the long labor required for preparation, is typically only used once or twice to ensure clean and accurate molecular detection. Here, we present three chemical solutions that can be used to regenerate PEG surface in a highly reliable manner. 0.1% sodium dodecyl sulfate, 50 mM NaOH, and 7 M NaOH were used to regenerate PEG surface to three distinct levels with highly reproducible results.
Introduction
Over the past two decades, single-molecule fluorescence techniques have been greatly advanced and extensively applied to study complex biological systems (1, 2, 3, 4, 5). In particular, single-molecule fluorescence resonance energy transfer (smFRET) has been widely employed to study mechanisms underlying many biological reactions and provide real-time kinetics without needing to synchronize the reactions (6, 7, 8). One field of view (25 × 75 μm2) measures fluorescence signals from several hundred single molecules that are immobilized on a quartz surface and imaged by total internal reflection microscopy (9,10). It is essential to passivate the imaging surface to exclude nonspecific adsorption of molecules such as proteins (7,11). Two primary passivation methods include bovine serum albumin (BSA)-biotin and polyethylene glycol (PEG) treatment. BSA-biotin is sufficient for measuring conformational dynamics of nucleic acid without proteins (6,12). This noncovalent blocking may not withstand harsh chemical treatments such as sodium dodecyl sulfate (SDS), urea, or GdmCl, which are occasionally used during smFRET measurements (13,14). By contrast, PEG treatment passivates the surface by a covalent linkage of the PEG polymer to aminosilane, which efficiently prevents nonspecific protein binding (6,9,15). It was shown that Tween 20 treatment of a PEG surface can further improve the passivation strength by 5- to 10-fold (16,17).
Recently, more protocols have been developed for PEG surface passivation, with every procedure requiring multiple steps of chemical treatments (18). The level of surface passivation is critical because improperly passivated surfaces lead to excessive nonspecific binding (6,7,15). Two rounds of PEGylation can remarkably improve the passivation quality (15). Thus, carefully prepared PEG slides can be used multiple times without issues arising from nonspecific binding (16). Nevertheless, one slide channel is typically used only once or a few times because of the irreversible nature of many experiments such as unwinding reactions or concerns about remaining molecules that may perturb subsequent experiments. The question is if used slides can be washed completely and fully restored to previous surface conditions.
Here, we present several regeneration strategies that enable nearly complete recovery of the standard PEG surface (19, 20, 21, 22). The first procedure is to wash out proteins bound to DNA or RNA by applying 0.1% SDS. The wash time is <1 min, and recovery is extremely high, as evidenced by the maintenance of DNA or RNA molecule count and the protein binding activity achieved in 10 consecutive runs of experiments. The second method involves denaturing duplexed DNA with 50 mM NaOH and reannealing with a complementary strand to either recover the same duplex or convert to a different substrate. The result displayed nearly 10 rounds of perfectly recovered substrate in terms of molecule count and protein activity measured before and after the regeneration. Finally, 7 M NaOH was applied to break the biotin-NeutrAvidin linkage, and the PEG-biotin surface was fully regenerated by coating with fresh NeutrAvidin five times in a row. Three applications of this process to different biotin-tagged antibody or DNA/RNA systems all resulted in highly successful regeneration. Importantly, the three strategies can be used in succession according to the experimental demand. Taken together, our regeneration methods are time-saving, cost-effective, and simple yet highly reliable and reproducible.
Materials and methods
Preparation of DNA constructs for fluorescence resonance energy transfer measurement
The high-performance-liquid-chromatography-purified biotinylated and nonbiotinylated oligonucleotides (from Integrated DNA Technologies, Coralville, IA) labeled with cyanine 3 (Cy3) or cyanine 5 (Cy5) were mixed at a 1:1.2 ratio (nonbiotinylated strand in excess) to make a partial duplex (10 μM) for fluorescence resonance energy transfer (FRET) measurements. They were annealed in T50 buffer (10 mM Tris-HCl (pH 7.5) and 50 mM NaCl) by heating up to 95°C for 2 min, then gradually cooling at the rate of 2°C/min down to 40°C followed by 5°C/min cooling to reach 4°C (thermal cycler).
Preparation of PEG-passivated slides and sample chamber assembly
The preparation of PEG slides involves a series of steps that have been well documented previously (19,23,24). Briefly, the slides are soaked in methanol and acetone overnight, rinsed with water thoroughly, boiled in microwave for 5–10 min in water, sonicated in 5% Alconox (New York, NY) for 20–30 min. We scrub the slides with gloved hands, rinse, and sonicate for 20–30 min in water and burn with a strong propane torch for at least 30 s to remove all sources of fluorescence. Meanwhile, coverslips are cleaned in 5% Alconox by scrubbing with a finger and rinsing with water. Then, the slides and coverslips are sonicated for 20–30 min in water and etched in 1 M potassium hydroxide for 30–45 min followed by rinsing with water. The slides are burned for 2–3 min, and coverslips are quickly sterilized by passing through the flame four to five times. The fully dried slides and coverslips are coated with aminosilane for 30–45 min followed by a rinse with water. The slides and coverslips are treated with a mixture of 98% mPEG (m-PEG-5000; Laysan Bio, Arab, AL) and 2% biotin-PEG (biotin-PEG-5000; Laysan Bio) and incubated overnight (at least 4 h). The slides and coverslips are then washed with water and dried completely using nitrogen gas or air and stored at −20°C for future experiments. Finally, the flow channels are created by applying thinly cut double-sided tape slices on the slide and putting the coverslip on top, making a sandwich between the slide and the coverslip. The edges between the slide and coverslip are sealed by epoxy.
smFRET measurements
All smFRET experiments are conducted at room temperature (∼23 ± 2°C) using a custom-built prism-type total internal reflection inverted fluorescence microscope (Olympus IX 71, Tokyo, Japan), as described earlier (6,19). Annealed partial duplex DNA stock labeled with biotin, Cy3, and Cy5 is diluted to 15–20 pM and flowed to the flow chamber. The DNA is immobilized on the PEG- passivated surface via a biotin-NeutrAvidin (50 μg/ml) linkage, and unbound excess molecules are washed out. All smFRET measurements are performed in an imaging buffer containing 10 mM Tris-HCl (pH 7.5) and respective salts of different experiments, with 10% glycerol with an oxygen scavenging system (10 mM trolox, 0.5% glucose, 1 mg/ml glucose oxidase, and 4 μg/ml catalase) to avoid blinking and to improve dye stability.
Data acquisition and analysis
The sample chamber is imaged by the prism-type total internal reflection equipped with a solid-state 532- and 634-nm diode laser (Compass 315M; Coherent, Santa Clara, CA) to excite Cy3 and Cy5, respectively (6). The fluorescence of Cy3 and Cy5 is simultaneously collected using water immersion objective (60× NA; Olympus). To separate the donor and acceptor signals, a dichroic mirror (cutoff = 630 nm) is used, and the Cy3 and Cy5 images are projected on EMCCD camera (iXon 897; Andor Technology, Belfast, UK) side by side. The real-time videos of the fluorescence single molecules are recorded with 100-ms frame integration time unless otherwise stated. One field of view captures 200–400 molecules on average.
The recorded data are processed using an Interactive Data Language program and analyzed by a MATLAB script (The MathWorks, Natick, MA). More than 4000 molecules (20 frames of 20 short videos) collected from different imaging surfaces generate an FRET histogram via an automated MATLAB script (The MathWorks). The donor leakage was corrected based on the FRET-value of the donor-only molecules. Further, to exclude the donor-only contribution to the histogram at the low-FRET region, Cy3 and Cy5 molecules were excited sequentially (10 frames for Cy3, 1 frame dark, and 10 frames for Cy5) by using the green and red lasers, respectively. Each FRET histogram is normalized and fitted to a Gaussian distribution in Origin using an unrestrained peak center position.
0.1% SDS addition to protein-DNA-RNA complexes
A 10% stock of SDS was prepared in water. To recover free DNA-RNA from bound protein (i.e., to wash off the bound protein from the nucleic acid), we apply 0.1% SDS either in 100 mM NaCl or 1–5 mM MgCl2 salt containing buffer to maintain the salt concentration of each complex. To wash off the unbound protein from the slide, the sample chamber is washed two to three times with a blank buffer (T50) followed by the corresponding reaction buffer required for the experiment. It is important to avoid KCl during the SDS wash because it can result in precipitation of the SDS-potassium complex, which reduces the SDS effect in denaturing protein. Below is a list of different protein-DNA-RNA complexes to which 0.1% SDS was used multiple times for regeneration. We note that in some cases, if 0.1% SDS is inefficient to regenerate, then 0.5% SDS was applied.
POT1 binding to telomere G-quadruplex
The sequence of the partial duplex DNA used in this experiment is 5′-TGGCGACGGCAGCGAGGCTTAGGGTTAGGGTTAGGGTTAGGG/Cy3/-3′+5′-/Cy5/GCCTCGCTGCCGTCGCCA/biotin/-3′. Recombinant human POT1 protein was expressed in a baculovirus/insect cell system and was purified as previously described (25). For every experimental replicate, 25 nM POT1 is added to the immobilized telomere G-quadruplex (G4) in the sample chamber. POT1 binding kinetics are calculated from the dwell time between the moment of flow to the moment of the first FRET decline, which represents the POT1 binding.
RecA filament formation along poly T40 tail
The sequence of the partial duplex DNA used in this experiment is 5′-TGGCGACGGCAGCGAGGC(dT)40/Cy3/-3′+5′-/Cy5/GCCTCGCTGCCGTCGCCA/biotin/-3′. In every experimental replicate, RecA (1 μM; from New England Biolabs (NEB), Ipswich, MA) and 1 mM ATP in imaging buffer (50 mM KCl and 3 mM MgCl2) is added to the immobilized partial duplex for RecA assembly. The kinetic rate of RecA filament formation is calculated by fitting the histogram of FRET histogram to a Gaussian distribution and calculating the degree of formation (transition from DNA-only to RecA-bound FRET peak) at different times after RecA addition.
FUS association with poly U50 tail
The sequence of the partial duplex DNA used in this experiment is 5′-/Cy3/(drU)50rGrCrCrUrCrGrCrUrGrCrCrGrUrCrGrCrCrA-3′+5′-/biotin/rUrGrGrCrGrArCrGrGrCrArGrCrGrArGrGrC/Cy5/-3′. FUS expression plasmid is transformed into BL21 (DE3) competent cells (NEB) and purified as described previously (26). In every repeat of the experiment, 5 nM FUS is added to the immobilized partial duplex in the sample chamber. Subsequently, higher concentration of FUS (500 nM) is added to test the dynamic FRET fluctuation behavior of multimer FUS as previously observed (26).
DHX36 engaging with DNA/RNA G4
The sequence of the partial duplex DNA used in this experiment is 5′-TGGCGACGGCAGCGAGGCTTGGGTGGGTAGGGTGGG(dT)9/Cy3/-3′+5′ /Cy5/GCCTCGCTGCCGTCGCCA/biotin/-3′. DHX36 protein is expressed in BL21 (DE3) cells and purified as previously described (27). In each experimental replicate, 10 nM DHX36 in the imaging buffer (50 mM KCl and 3 mM MgCl2) is added to the immobilized DNA/RNA G4 with T9/U9 tail in the sample chamber. The dynamic traces are recorded after washing the free protein.
PcrA translocation on T40 tail
The sequence of the partial duplex DNA used in this experiment is 5′-/Cy3/(dT)40GCCTCGCTGCCGTCGCCA-3′+5′-/biotin/TGGCGACGGCAGCGAGGC/Cy5/-3′. PcrA is purified from Bacillus stearothermophilus as described previously (28). 100 pM PcrA in imaging buffer (10 mM KCl and 5 mM MgCl2) is applied to the immobilized partial duplex immobilized in the sample chamber. Data are recorded with 50-ms frame integration time, and more than 1,000 cycles of repetitive FRET fluctuations are used to calculate the dwell time using MATLAB code (The MathWorks).
For each of the above protein-DNA-RNA systems, the same experiments were repeated up to 10 times. The slide then was stored in 4°C (to be used within 1–2 days) or −20°C (to be used after 2–3 days or more) for further use. The sample chamber was washed with experimental buffer before and after the storage. In some instances, the number of molecules decreased because of buffer contamination, such as RNase. In these cases, we found we could reapply the FRET construct into the same channel and recover good molecule density for single-molecule measurement.
Proteinase K, 6 M urea, and 6 M GdmCl compared with 0.1% SDS
8 M urea and 8 M GdmCl stocks are each prepared in water and filtered through a 0.22-μm membrane. As in the case of SDS, 100 μM proteinase K (NEB), 6 M urea, or 6 M GdmCl with corresponding buffer conditions is applied to the protein-DNA-RNA complexes described above to recover free DNA-RNA. After each round of incubation, the surface is washed thoroughly with the corresponding imaging buffer for further measurements. If the protein is not completely removed from the DNA-RNA, then 0.1% SDS can be applied for complete removal of protein for further experiment.
Unwinding and reannealing
Rep-X helicase is applied to unwind a partial duplex that contains 18-bp duplex and a poly-T15 tail at the 3′ end for helicase loading. The number of FRET spots are counted before and after applying Rep-X with ATP. Short videos (2 s) are recorded at different imaging areas to calculate the unwinding rate. The number of fluorescence spots plotted against time produces a curve that is fitted to a first exponential decay function. After unwinding, the biotinylated strand remains immobilized on the surface. For reannealing, 5 nM of the complementary single-stranded DNA (ssDNA) is applied in 5-mM-MgCl2-containing buffer and incubated for 5 min. The excess free ssDNA is washed out using T50, and another round of unwinding experiment is performed. The same reannealing and unwinding process is repeated 10 times. G4-containing substrate tends to nonspecifically stick to the surface. In this case, the surface can be further blocked by applying biotin (1 μM), BSA (0.4 mg/ml), and yeast transfer RNA (t-RNA) (0.2 mg/ml), which greatly reduces the nonspecific binding.
Breaking the biotin-NeutrAvidin linkage
To break the immobilized biotin-NeutrAvidin linkage, 7 M NaOH is applied to the flow chamber. After 2 min of incubation, the chamber is washed three times with T50, and another layer of NeutrAvidin is applied to coat the PEG-biotin surface. The same process can be repeated multiple times. Because 7 M NaOH is a strong reagent, it may reduce the surface passivation quality after several applications; hence, the surface can be further blocked by biotin (1 μM), BSA (0.4 mg/ml), and yeast t-RNA (0.2 mg/ml), as stated above.
Results and discussion
Removal of protein by 0.1% SDS to regenerate DNA/RNA-tethered PEG surface
One of the most common smFRET and PIFE (29,30) experimental setups involves tethering fluorescently labeled and biotinylated DNA or RNA on a PEG-passivated surface and applying protein to detect binding and activity. This experimental setup is ideal for measuring binding and unbinding rates and protein-induced conformational changes within the nucleic acid substrates. When the experiment requires multiple runs, such as titrating different protein concentrations or changing buffer conditions, it is typical to use a new channel rather than reusing the same channel. This is done to ensure that each experiment is performed in the same condition and to prevent contamination by leftover protein. However, the protocol to generate slides is time intensive and involves expensive reagents. Therefore, it would be highly beneficial to regenerate surfaces for multiple uses by removing all the protein and restoring the clean, unbound substrate tethered to the surface. A complete regeneration would be expected to preserve the number of substrate molecules tethered to surface, the FRET-values of the free and bound substrate, and protein binding kinetics and activity.
To test the regeneration of substrate-tethered surfaces, we conducted five sets of experiments on DNA or RNA binding proteins previously studied in our laboratory. Each experiment entails a unique structure of nucleic acid labeled with Cy3 and Cy5 designed for FRET measurements. Each FRET-labeled substrate is immobilized to the PEG-biotin-coated quartz surface via biotin-NeutrAvidin linkage (Fig. 1, A–C). In each case, FRET histograms were made in the absence and presence of a protein by collecting FRET-values from more than 4000 molecules in 20 different imaging areas. Each protein-nucleic-acid system was treated with 0.1% SDS to wash the protein and tested for recovery.
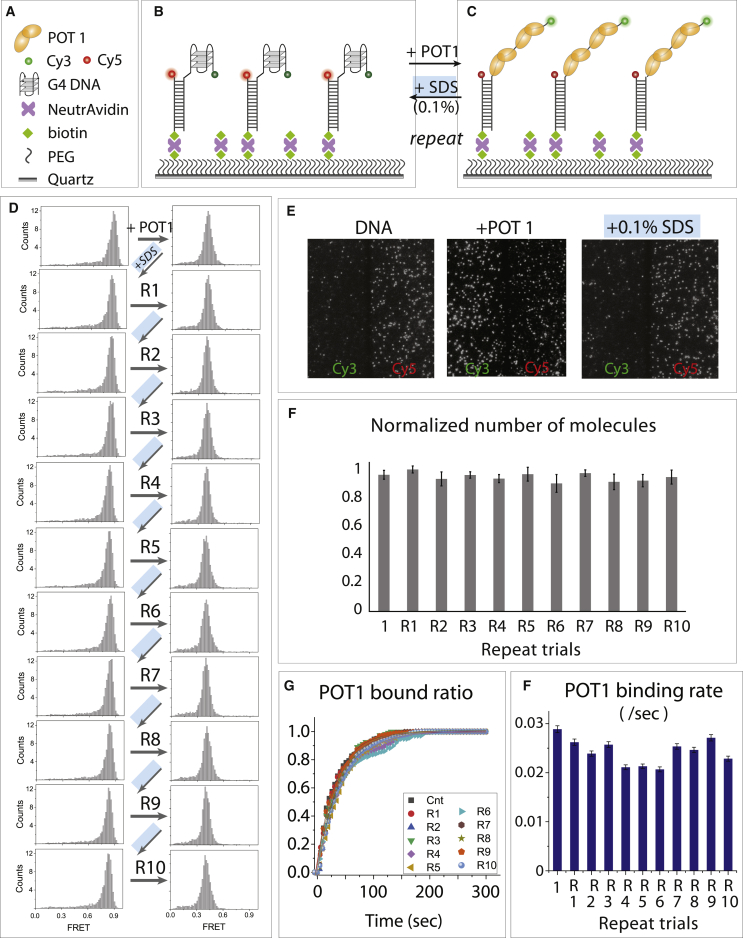
Figure 1.
Multiple rounds of the regenerating telomeric DNA-bound PEG surface by 0.1% SDS. (A–C) smFRET schematics that show telomeric DNA designed for smFRET experiment and POT1 binding, which induces FRET decrease. (D) The FRET histogram of G-quadruplex (G4) before and after POT1 binding followed by 0.1% SDS wash repeated 10 times, as denoted by R1 to R10. (E) Representative fields of view before and after POT1 binding followed by 0.1% SDS wash. (F) The number of molecules counted during each repeat of trials and normalized with control. Error bar represents the standard deviation (SD) from molecule counts of 20 different field of view. (G) Single-exponential fitting of POT1 bound fraction. (H) The bar graph of POT1 binding rate taken at R1–R10 trials and errors are reported from curve fitting. To see this figure in color, go online.
POT1 binding to telomeric G4
Telomeric DNA consists of a long duplex followed by a 3′ single-stranded overhang composed of tandem TTAGGG repeats (31). POT1 is a member of the shelterin complex, which binds exclusively to the telomeric overhang and disrupts the G4 structure (22,32). Accordingly, the unbound telomeric G4 DNA construct exhibits high FRET because of the folded G4 bringing the two dyes into close proximity. Upon addition of POT1, a shift to low FRET indicates the disruption of the G4 structure (Fig. 1, B–D). The POT1 bound state is extremely stable, remaining in low FRET even after multiple cycles of buffer wash. When 0.1% SDS is added, however, the POT1-bound low-FRET peak completely shifts back to the high-FRET peak, corresponding to the unbound folded G4 state. We repeated the process of POT1 addition followed by washing with 0.1% SDS 10 times. In every repeat, the FRET histograms of the POT1-bound state (low FRET) and free G4 state (high FRET) were indistinguishable from previous runs, indicating the high degree of reproducibility enabled by regeneration (Fig. 1 D). The representative field of view recorded before and after addition of POT1 and SDS, respectively, showed similar density of DNA molecules and intensity level after the first round (Fig. 1 E) as well as each repeat of trials. The number of molecules counted in every repeat remained approximately same, confirming that the integrity of the surface is maintained for up to 10 experiments (Fig. 1 F). For each experimental repeat, we also measured the binding rate of POT1 to G4 by performing real-time flow experiments (Fig. S1; (22,33,34)). The binding rates calculated from the exponential fitting of the bound fraction decay curve was highly similar in all cases, further confirming the high reproducibility over multiple slide uses (Fig. 1, G and H).
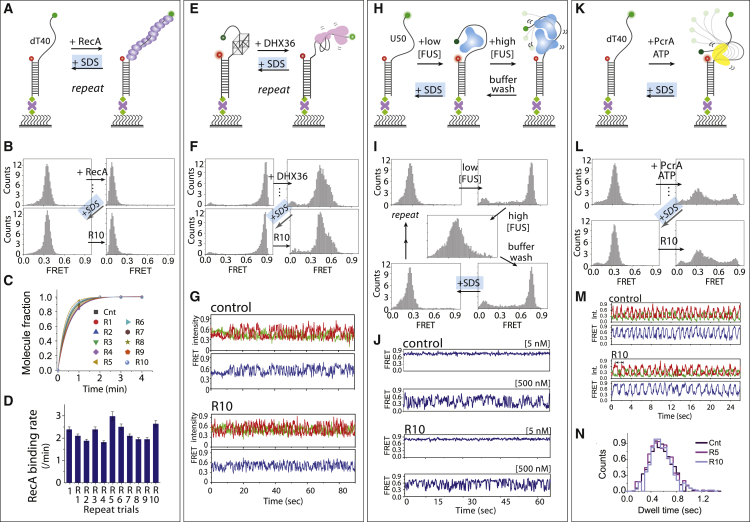
RecA filament formation on poly-T DNA
RecA is an Escherichia coli protein that forms a helical filament on ssDNA to catalyze homologous recombination (35). The 3′ poly-T40 tail DNA exhibits 0.3 FRET in the free state and shifts to 0.1 FRET upon addition of RecA, consistent with filament formation (Fig. 2 A). When 0.1% SDS is added, there is an immediate disappearance of the 0.1 FRET and concomitant appearance of 0.3 FRET, signifying the complete disassembly of the RecA filament. Repeating this experiment 10 times yielded identical histograms (Fig. 2 B). Using real-time smFRET traces, RecA assembly kinetics were shown to be nearly identical for each repeat of the experiment, signifying the highly reproducible surface regeneration (Fig. 2, C and D).
Figure 2.
Regeneration of various DNA/RNA surface by 0.1% SDS treatment. (A, E, H, and K) Schematic smFRET experiments including (A) 3′ poly T40 tail and RecA binding, (E) G4 with T15 tail and DHX36 binding, (H) 5′ poly U50 tail and FUS engagement, and (K) 5′ poly T40 and PcrA binding and translocation. (B, F, I, and L) The FRET histogram of control and 10th repeats of respective protein binding. (C and D) Single-exponential fitting of RecA-bound fraction and corresponding rate of 10 repeat trials (errors are reported from curve fitting). (G, J, and M) Representative smFRET traces of (G) DHX36 bound to G4, (J) FUS bound to poly U50, (M) PcrA bound to poly T50, and (N) PcrA-induced translocation repeat dwell time. To see this figure in color, go online.
DHX36 mediated unfolding of G4 DNA-RNA
DHX36 specifically binds and resolves the parallel conformation of G4-DNA-RNA with a single-stranded tail (14,27). The FRET-value shifts from 0.9 to 0.5 upon DHX36 binding to G4-DNA. Again, 0.1% SDS was sufficient to remove the bound protein and revert the histogram back to 0.9. Reapplication of DHX36 followed by an SDS wash was repeated 10 times with consistent results (Fig. 2, E and F; Fig. S2). As reported previously, DHX36 binding to the parallel G4-DNA displayed repetitive cycles of unfolding and refolding (36), and similar FRET fluctuations were observed in all 10 rounds of experiments (Fig. 2 G). We also performed the same experiment on G4-RNA, which also showed effective surface regeneration with 0.1% SDS (Fig. S3), demonstrating that regeneration works for both DNA and RNA substrates equally well.
FUS binding and dynamics on poly-U RNA
FUS is a nuclear RNA binding protein that interacts with single-stranded RNA (ssRNA) as we reported previously (26,37). As before, FUS was applied to the poly-U50 tail containing partial duplex, and surface regeneration by 0.1% SDS was tested (Fig. 2 H). Consistent with previous results (26), free U50 displays a low-FRET peak (∼0.2) that shifts to high FRET (∼0.8) upon the addition of 5 nM FUS (Fig. 2 I). Addition of a higher FUS concentration (500 nM) produces a broad mid-FRET peak (∼0.5), whereas a buffer wash restores the high-FRET peak. We found that the addition of 0.1% SDS to FUS-bound RNA restores a low-FRET peak (∼0.2), corresponding to the free RNA construct (Fig. 2 I). The same process repeated 10 times produced a nearly identical result. Furthermore, the steady high-FRET and dynamic FRET fluctuations that result from 5 to 500 nM FUS, respectively, were highly reproducible throughout the 10 rounds (Fig. 2 J; (26)).
PcrA induced repetitive looping of poly-T40
PcrA is a 3′-5′ helicase that exhibits repetitive looping activity of a 5′ ssDNA tail (28). As seen in above examples, the FRET histogram that shifted upon PcrA binding was completely reverted back to the DNA-only state by 0.1% SDS treatment (Fig. 2, K and L). 10 rounds of successive experiments produced highly similar FRET fluctuations were consistent with the previous finding (Fig. 2 M; (28)). The dwell time analysis conducted by collecting time intervals between FRET fluctuations showed extremely similar values across all 10 experiments, confirming the highly reproducible surface regeneration (Fig. 2 N).
Although we only tested up to 10 cycles of slide regeneration, we expect that further repetition would be possible. Furthermore, we found that the same slide could be used after overnight storage at 4 or −20°C with identical results (see Materials and methods for details).
0.1% SDS is more potent in regeneration than other protein denaturants
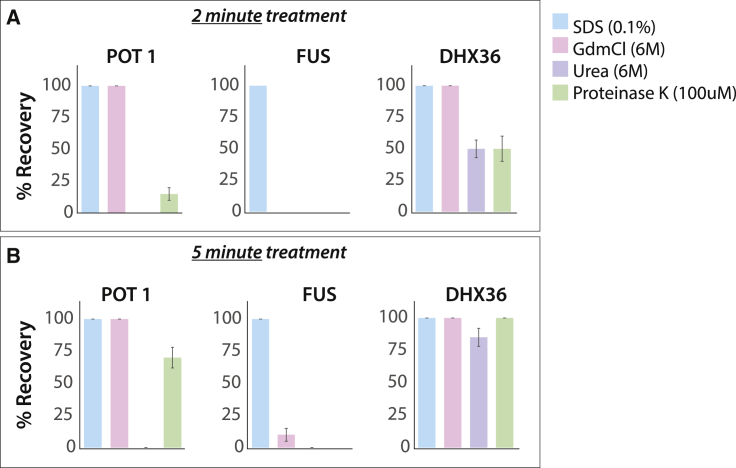
Next, we asked if other protein denaturing reagents such as urea, GdmCl, or proteinase K can be also used for regenerating the DNA/RNA-immobilized PEG surface. First, the POT1-bound telomeric overhang was challenged by addition of 6 M urea followed by a 10-min incubation. The resulting FRET histogram remained unchanged, indicating that 6 M urea is unable to dissociate POT1 (Fig. S4). By contrast, 6 M GdmCl was effective in removing POT1 from telomeric ssDNA (Fig. S4). Interestingly, proteinase K (100 μM) showed a slow disengagement of POT1, taking 15 min for a complete reversal of the FRET peak (Fig. S4). In the case of DHX36-bound DNA-RNA-G4, 6 M urea was slow (∼10 min to complete), whereas 6 M GdmCl was fast (∼2 min) in removing DHX36 (Fig. S5). Unexpectedly, proteinase K (100 μM) displayed a slow reversal in FRET peak, but the addition of new DHX36 was unable to reproduce protein activity, suggesting an incomplete digestion of DHX36 by proteinase K that disabled further binding (Fig. S5). For FUS-bound poly-U50 tail, 6 M urea and proteinase K (100 μM) was not effective in dislodging FUS even after incubation for 20 min (Fig. S6). Interestingly, 6 M GdmCl showed ∼40% recovery after 20 min (Fig. S6). Taken together, 0.1% SDS is significantly more effective and consistent in clearing bound protein than other reagents (Fig. 3).
Figure 3.
SDS is far more efficient in regenerating the substrate bound surface than other denaturants. The percentage of recovery was obtained from shifted FRET peak (area under the curve) from protein bound state to free DNA/RNA only state (Fig. S4–S6) at different denaturant after incubation of (A) 2 min and (B) 5 min. Error bars represent the SD from three different trials. To see this figure in color, go online.
Duplex substrate regeneration after unwinding or melting
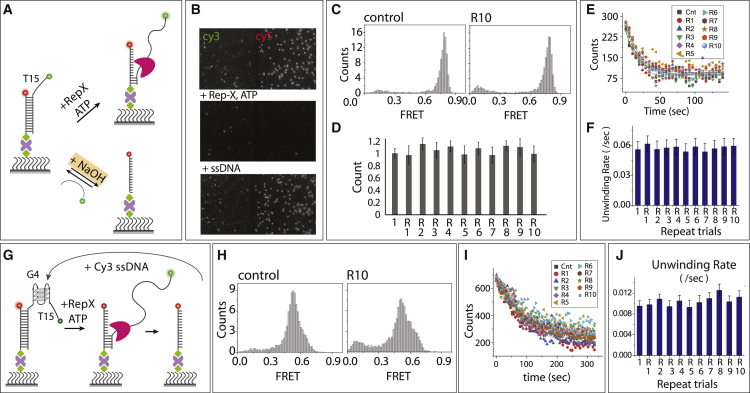
We recently reported that of superfamily 1 helicase, Rep unwinds tightly folded G4 DNA in vitro and in E. coli (19). DNA or RNA unwinding can be studied using a similar dual-labeled smFRET configuration as described above. However, in this case, the nonbiotinylated strand is lost upon complete unwinding of the duplex (19), limiting the use of one channel for one unwinding experiment. Here, we show that the duplex substrate can be recovered on the surface by simply reannealing with the complementary ssDNA. We prepared a partial duplex with the 3′-T15 tail to which superhelicase Rep-X (38) was added with ATP (Fig. 4 A). After complete unwinding that removed the Cy3 strand, we applied 5 nM of complementary ssDNA in 5-mM-MgCl2-containing buffer to promote annealing. After a 5-min incubation, we cleared the excess Cy3 strand by buffer wash and repeated the Rep-X unwinding assay. Based on the molecule count, reannealing mostly restored all duplex substrates lost because of the unwinding (Fig. 4 B). The FRET histograms taken at each cycle showed a highly reproducible result (Fig. 4 C). Likewise, the rate of unwinding deduced by counting the number of molecules over time was similar in each trial (Fig. 4, D–F). Next, we performed the same unwinding and reannealing experiments for G4-containing partial duplex and observed a high level of reproducibility as well (Fig. 4, G–J). We note that G4 containing DNA and RNA has a higher tendency to adsorb to the NeutrAvidin-coated surface, necessitating additional surface blocking with biotin, BSA, and yeast t-RNA.
Figure 4.
Duplex recovery by denaturation and reannealing. (A) Schematic model of Rep-X-induced unwinding of partial duplex and reannealing by ssDNA. Low concentration of NaOH (∼50 mM) denatures the DNA duplex, which can be recovered by reannealing. (B) Representative fields of view before and after unwinding followed by reannealing. (C) The FRET histogram of control and after 10th reannealing. (D) The number of molecules counted during each repeat of reannealing and normalized with control. Error bar represents the SD from molecule counts of 20 different field of view. (E) Number of molecules counted for each repeated trial during unwinding of partial duplex and unwinding rate obtained from the single-exponential fitting (F). (G) Schematic model of Rep-X-induced G4 unwinding and reannealing by G4-ssDNA. (H) The FRET histogram of control and after 10th reannealing. (I) Number of molecules counted over time of G4 unwinding and the calculated unwinding rate (J). (F and J) Errors are reported from curve fitting. To see this figure in color, go online.
In addition to helicase unwinding, we can also denature the surface bound duplex by treating with 50 mM NaOH, leaving behind the biotinylated, surface-tethered Cy5 strand. Complementary ssDNA can then be annealed to the tethered ssDNA (Fig. 4 A). This approach allows the sequence of the Cy3 strand to be changed readily as long as the duplex region is maintained. We tested reannealing with a G4 DNA strand followed by DHX36 binding. The result shows that the T15 tail was completely exchanged by the G4 strand, evidenced by the expected FRET peak changes in each condition (Fig. S7).
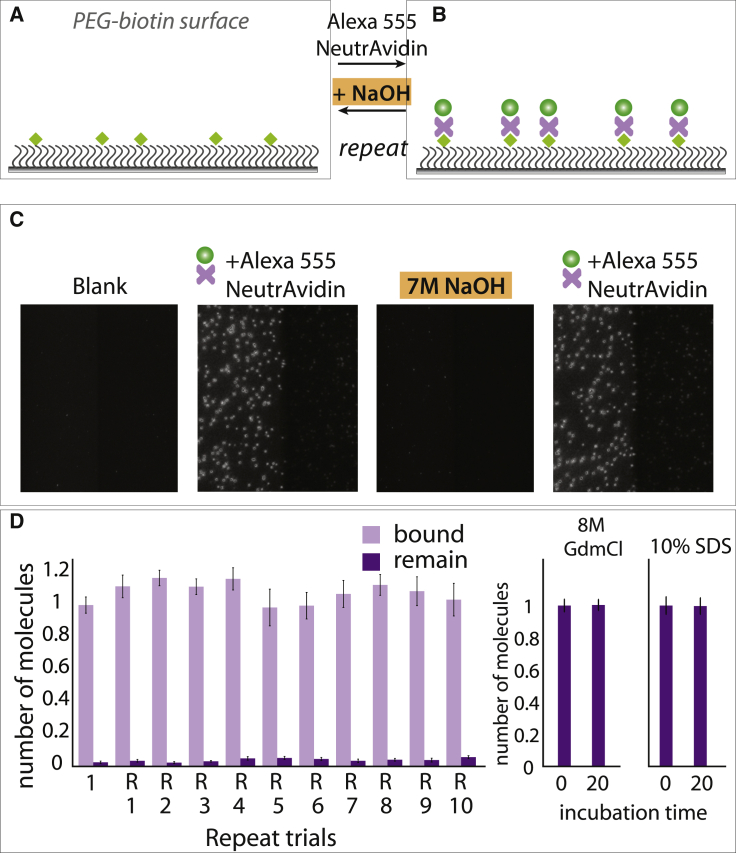
Breaking the biotin-streptavidin linkage with 7 M NaOH
Next, we asked if the PEG-biotin surface can be regenerated by stripping off the NeutrAvidin layer, which requires breaking of biotin-NeutrAvidin linkage, the strongest known noncovalent interaction in nature with Kd of 10−14 M (39). If this is successful, one can reuse the same surface for entirely different experiments. We found that 7 M NaOH treatment can break the biotin-NeutrAvidin linkage (Fig. 5, A and B). To visualize this process, Alexa-Fluor-555-labeled streptavidin (Thermo Fisher Scientific, Waltham, MA) was applied to the PEG-biotin surface, and binding was probed by the appearance of the fluorescence signal. Incubation of 7 M NaOH for 2 min followed by a buffer wash resulted in disappearance of fluorescence signal. When the same concentration of Alexa-Fluor-555-labeled streptavidin (Thermo Fisher Scientific) was added to the same channel, a similar density of fluorescence was observed. The surface was regenerated successfully up to 10 times (Fig. 5 C). The count of fluorescent molecules further confirmed the reproducible binding of streptavidin rather than chemical damage of Alexa-Fluor-555 dye (Thermo Fisher Scientific) in each round of 7 M NaOH treatment (Fig. 5 D). By contrast, 8 M GdmCl and 10% SDS were insufficient to remove Alexa-Fluor-555-labeled streptavidin (Thermo Fisher Scientific) from the biotin even after 20 min of incubation (Fig. 5 D).
Figure 5.
Breaking the NeutrAvidin-streptavidin linkage by 7 M NaOH. (A and B) Schematic diagram of PEG-passivated slide coated with Alexa-Fluor-555-labeled streptavidin (Thermo Fisher Scientific) and detachment by 7 M NaOH treatment. (C) Representative fields of view before and after treatment of Alexa-Fluor-555-labeled streptavidin (Thermo Fisher Scientific) and 7 M NaOH, respectively. (D) Molecule count during each repeat of binding and unbinding trials (left side). Shown is the molecule count of Alexa-Fluor-555-streptavidin (Thermo Fisher Scientific) bound on surface before and after 8 M GdmCl and 10% SDS treatment. Error bar represents the SD from molecule counts of 20 different field of view. To see this figure in color, go online.
Using the 7 M NaOH regeneration strategy combined with 0.1% SDS, we conducted a series of five different experiments involving DNA, RNA, and proteins, all in the same channel. In addition to the reproducibility, these experiments reveal the compatibility of using multiple reagents in the same channel depending on the experimental needs (Fig. S8).
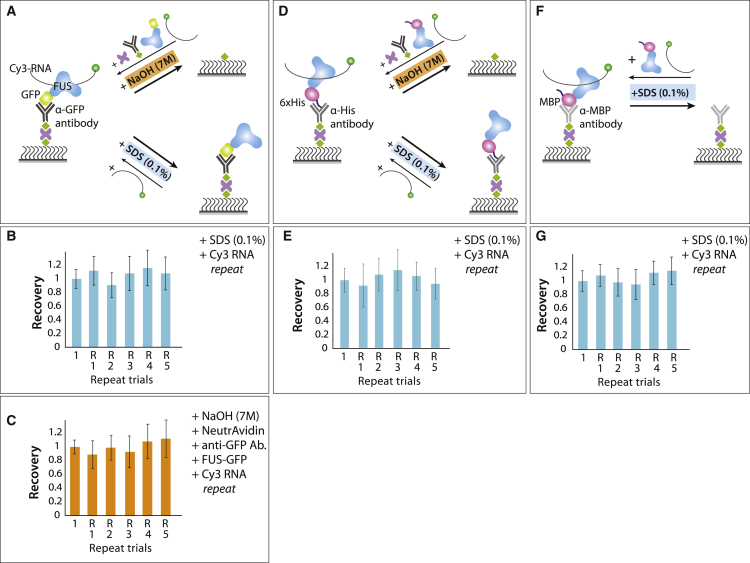
Antibody-bound surface regeneration for single-molecule pull-down
We tested if 7 M NaOH treatment can be employed for single-molecule pull-down experiments (8). First, the biotin-conjugated anti-GFP antibody was applied to the NeutrAvidin-coated surface. Next, GFP-tagged FUS was applied. Cy3-labeled poly-U50 ssRNA was added to probe the FUS-RNA interaction (Fig. 6 A; (26)). Then, 7 M NaOH was applied and incubated for 2 min and washed out. The same procedure repeated five times produced nearly complete recovery (Fig. 6 C). We tested two additional cases including anti-histidine and anti-maltose-binding-protein (MBP), and both showed reproducible recovery of the molecule count. We found that the strong 7 M NaOH reagent reduces the surface passivating effect when used more than five times but that blocking the surface with BSA and yeast t-RNA significantly reduced nonspecific binding (Fig. S9). Interestingly, when we applied 0.1% SDS to the FUS-bound, Cy3-labeled ssRNA, the ssRNA disappeared, indicated by the loss of fluorescent molecules (Fig. 6 A). When Cy3-RNA was reapplied, the RNA engaged with FUS, suggesting that 0.1% SDS was not harsh enough to denature FUS but is sufficient to release the bound ssRNA (Fig. 6 B). This also indicates that 0.1% SDS does not disrupt the interaction between GFP and anti-GFP. The same binding/unbinding process was tested on histidine-tagged FUS, which was immobilized on the surface through a biotinylated anti-His antibody (Fig. 6 D). Similarly, FUS-bound ssRNA disappeared after the addition of 0.1% SDS and reengaged when freshly applied. Hence, the histidine to anti-His antibody interaction also remains unaffected by 0.1% SDS. The number of molecules counted in each trial showed a high recovery rate (Fig. 6 E). The same test applied to MBP-FUS and the anti-MBP antibody displayed similar binding efficiency to Cy3-ssRNA. Upon 0.1% SDS wash, Cy3-ssRNA did not bind, as indicated by the absence of fluorescence spots. When the MBP-FUS was added again, the Cy3-ssRNA signal appeared to the same level (Fig. 6 F). This indicates that the anti-MBP-MBP interaction was disrupted by 0.1% SDS. Repeated runs of this experiment resulted in highly reproducible outcomes (Fig. 6 G).
Figure 6.
Antibody-bound surface regeneration. (A) Schematic diagram of Cy3-U50 engaged with FUS immobilized to the surface via (A) anti-GFP antibody, (D) anti-histidine antibody, and (F) anti-MBP antibody and regeneration strategies by NaOH and SDS. (B, E, and G) Normalized molecule count during each repeat of experiments with 0.1% SDS and (C) 7 M NaOH treatment. All error bars represent the SD from molecule counts of 20 different field of view. To see this figure in color, go online.
Conclusion
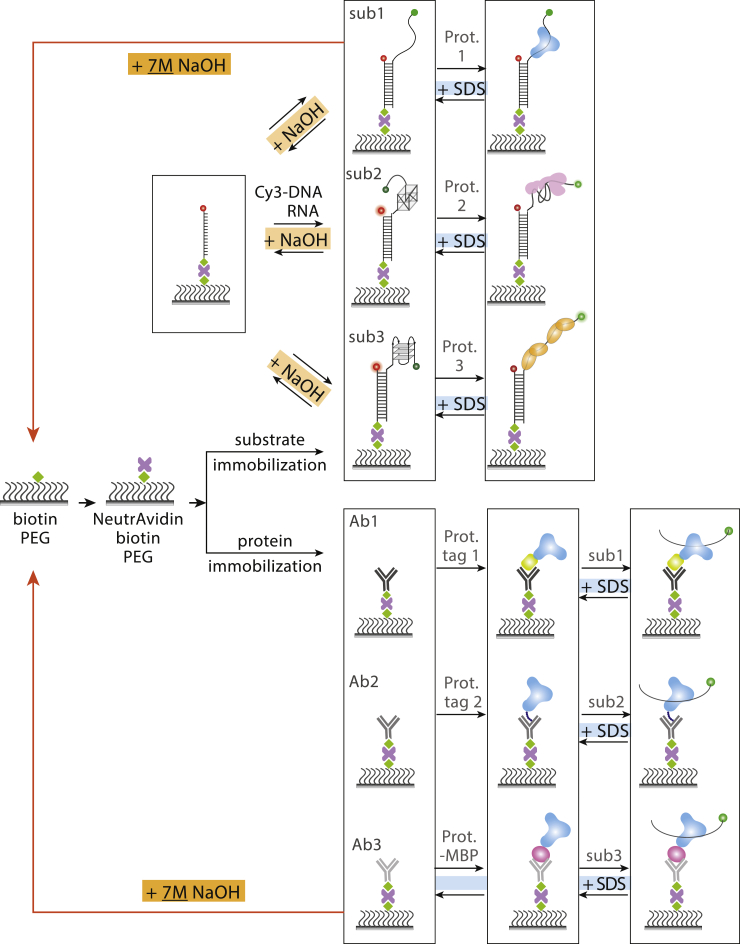
For single-molecule experiments, PEG slides offer more advantages than noncovalently linked passivation surfaces (6,15). However, making a high-quality PEG surface requires a time-consuming step-by-step process that takes 7–8 h on average. In general, one channel within a PEG slide is used only once to avoid potential complications that arise from remaining molecules that may affect the next round of measurements. Often, single-molecule experiments require multiple runs of experiments such as protein or ATP titrations or buffer condition changes (19). In this work, we demonstrate that a single-PEG-passivated channel can be used multiple times either to repeat the same measurements or to conduct different types of experiments (Fig. 7). We tested five different DNA-RNA-binding proteins and three types of antibodies for which 0.1% SDS showed significantly higher regeneration capability than other denaturants (Fig. 7). The slide channel integrity was preserved throughout many repeats of experiments and even after overnight storage in 4 or −20°C. The regeneration was demonstrated not only by the preserved molecule count but also by the retained protein activity such as ATP-dependent motion of helicases such as Rep-X and PcrA or the rate of protein binding in the case of POT1 and RecA. The integrity of the NeutrAvidin-coated surface arises from the strong bond between PEG-biotin-NeutrAvidin, which can withstand even 10% SDS treatment. Helicase-induced unwinding and loss of a nonbiotin strand can be recovered back to the duplex by in situ annealing with a complementary strand. 50 mM NaOH is sufficient to denature a duplex into a single strand, which can be reannealed with the same strand or made into a new substrate with the same in situ annealing protocol. Addition of 7 M NaOH was harsh enough to break the biotin-NeutrAvidin linkage, which can essentially generate a new PEG-biotin slide surface (Fig. 7). Importantly, the various chemical reagents used above can be employed in succession within one slide based on the specific experimental demand (Fig. 7). The three detergents at particular concentrations we used here, including 0.1% SDS, 50 mM NaOH, and 7 M NaOH, were achieved and optimized carefully by trial and error. For example, we found SDS to be superior to urea, GdmCl, and proteinase K by trying them all out in varying concentrations. Taken together, these strategies will enhance efficiency and increase cost-effectiveness without compromising the data quality for researchers who rely on the PEG-biotin slide for single-molecule fluorescence imaging.
Figure 7.
Summary. Shown are the PEG slide regeneration strategies that can be applied in succession depending on the experimental demands. Importantly, 7 M NaOH treatment can regenerate the original PEG-biotin surface for a completely new experiment. To see this figure in color, go online.
Author contributions
T.P. designed and performed all the experiments in consultation with T.H. and S.M. T.P. analyzed all the data. T.P., S.M. wrote the manuscript, and T.H. edited the manuscript.
Acknowledgments
We thank Dr. Patricia L. Opresko, Dr. Momčilo Gavrilov, Dr. Michael Banco, Dr. Adrian R. Ferré-D’Amaré, and Kevin Rhine for providing the POT1, Rep-X, PcrA, DHX36, and FUS proteins, respectively. We thank Dr. Laura Ganser for editing and Bersabel Wondimagegnhu for careful proofreading of the manuscript and Dr. Amarshi Mukherjee and Dr. Manindra Bera for helpful discussions.This work was supported by National Institute of Health General Medicine (1R0S1GM115631-01A1 to T.P. and S.M. and 1R01-CA 207342-01A1 and 1RF1 NS113636-01 to S.M.).
Editor: Mark C. Williams.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.02.031.
Supporting material
References
- 1.Neuman K.C., Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim H., Ha T. Single-molecule nanometry for biological physics. Rep. Prog. Phys. 2013;76:016601. doi: 10.1088/0034-4885/76/1/016601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karunatilaka K.S., Rueda D. Single-molecule fluorescence studies of RNA: a decade’s progress. Chem. Phys. Lett. 2009;476:1–10. doi: 10.1016/j.cplett.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornish P.V., Ha T. A survey of single-molecule techniques in chemical biology. ACS Chem. Biol. 2007;2:53–61. doi: 10.1021/cb600342a. [DOI] [PubMed] [Google Scholar]
- 5.Zhuang X. Single-molecule RNA science. Annu. Rev. Biophys. Biomol. Struct. 2005;34:399–414. doi: 10.1146/annurev.biophys.34.040204.144641. [DOI] [PubMed] [Google Scholar]
- 6.Roy R., Hohng S., Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamichhane R., Solem A., Rueda D. Single-molecule FRET of protein-nucleic acid and protein-protein complexes: surface passivation and immobilization. Methods. 2010;52:192–200. doi: 10.1016/j.ymeth.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain A., Liu R., Ha T. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 10.Zhuang X., Bartley L.E., Chu S. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 11.Rasnik I., McKinney S.A., Ha T. Surfaces and orientations: much to FRET about? Acc. Chem. Res. 2005;38:542–548. doi: 10.1021/ar040138c. [DOI] [PubMed] [Google Scholar]
- 12.Di Fiori N., Meller A. Automated system for single molecule fluorescence measurements of surface-immobilized biomolecules. J. Vis. Exp. 2009:1542. doi: 10.3791/1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuertes G., Banterle N., Lemke E.A. Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements. Proc. Natl. Acad. Sci. USA. 2017;114:E6342–E6351. doi: 10.1073/pnas.1704692114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tippana R., Chen M.C., Myong S. RNA G-quadruplex is resolved by repetitive and ATP-dependent mechanism of DHX36. Nat. Commun. 2019;10:1855. doi: 10.1038/s41467-019-09802-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandradoss S.D., Haagsma A.C., Joo C. Surface passivation for single-molecule protein studies. J. Vis. Exp. 2014:e50549. doi: 10.3791/50549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hua B., Han K.Y., Ha T. An improved surface passivation method for single-molecule studies. Nat. Methods. 2014;11:1233–1236. doi: 10.1038/nmeth.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan H., Xia Y., Wang W. A simple procedure to improve the surface passivation for single molecule fluorescence studies. Phys. Biol. 2015;12:045006. doi: 10.1088/1478-3975/12/4/045006. [DOI] [PubMed] [Google Scholar]
- 18.Selvin P.R., Ha T. Cold Spring Harbor Laboratory Press; New York: 2008. Single-Molecule Techniques: A Laboratory Manual. [Google Scholar]
- 19.Paul T., Voter A.F., Myong S. E. coli Rep helicase and RecA recombinase unwind G4 DNA and are important for resistance to G4-stabilizing ligands. Nucleic Acids Res. 2020;48:6640–6653. doi: 10.1093/nar/gkaa442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paul T., Mishra P.P. Direct observation of spatial configuration and structural stability of locked Y-shaped DNA structure. RSC Advances. 2016;6:103270–103274. [Google Scholar]
- 21.Paul T., Bera S.C., Mishra P.P. Direct observation of breathing dynamics at the mismatch induced DNA bubble with nanometre accuracy: a smFRET study. Nanoscale. 2017;9:5835–5842. doi: 10.1039/c6nr09348e. [DOI] [PubMed] [Google Scholar]
- 22.Lee H.-T., Sanford S., Myong S. Position-dependent effect of guanine base damage and mutations on telomeric G-quadruplex and telomerase extension. Biochemistry. 2020;59:2627–2639. doi: 10.1021/acs.biochem.0c00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul T., Bera S.C., Mishra P.P. Single-molecule FRET studies of the hybridization mechanism during noncovalent adsorption and desorption of DNA on graphene oxide. J. Phys. Chem. B. 2016;120:11628–11636. doi: 10.1021/acs.jpcb.6b06017. [DOI] [PubMed] [Google Scholar]
- 24.Bera S.C., Paul T., Mishra P.P. Direct observation of the external force mediated conformational dynamics of an IHF bound Holliday junction. Faraday Discuss. 2018;207:251–265. doi: 10.1039/c7fd00184c. [DOI] [PubMed] [Google Scholar]
- 25.Sowd G., Lei M., Opresko P.L. Mechanism and substrate specificity of telomeric protein POT1 stimulation of the Werner syndrome helicase. Nucleic Acids Res. 2008;36:4242–4256. doi: 10.1093/nar/gkn385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niaki A.G., Sarkar J., Myong S. Loss of dynamic RNA interaction and aberrant phase separation induced by two distinct types of ALS/FTD-linked FUS mutations. Mol. Cell. 2020;77:82–94.e4. doi: 10.1016/j.molcel.2019.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen M.C., Tippana R., Ferré-D’Amaré A.R. Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature. 2018;558:465–469. doi: 10.1038/s41586-018-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J., Myong S., Ha T. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang H., Kim H., Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc. Natl. Acad. Sci. USA. 2011;108:7414–7418. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang H., Myong S. Protein induced fluorescence enhancement (PIFE) for probing protein-nucleic acid interactions. Chem. Soc. Rev. 2014;43:1221–1229. doi: 10.1039/c3cs60201j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moye A.L., Porter K.C., Bryan T.M. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat. Commun. 2015;6:7643. doi: 10.1038/ncomms8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 33.Hwang H., Buncher N., Myong S. POT1-TPP1 regulates telomeric overhang structural dynamics. Structure. 2012;20:1872–1880. doi: 10.1016/j.str.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang H., Kreig A., Myong S. Telomeric overhang length determines structural dynamics and accessibility to telomerase and ALT-associated proteins. Structure. 2014;22:842–853. doi: 10.1016/j.str.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joo C., McKinney S.A., Ha T. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 36.Tippana R., Hwang H., Myong S. Single-molecule imaging reveals a common mechanism shared by G-quadruplex-resolving helicases. Proc. Natl. Acad. Sci. USA. 2016;113:8448–8453. doi: 10.1073/pnas.1603724113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rhine K., Makurath M.A., Myong S. ALS/FTLD-linked mutations in FUS glycine residues cause accelerated gelation and reduced interactions with wild-type FUS. Mol. Cell. 2020;80:666–681.e8. doi: 10.1016/j.molcel.2020.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arslan S., Khafizov R., Ha T. Protein structure. Engineering of a superhelicase through conformational control. Science. 2015;348:344–347. doi: 10.1126/science.aaa0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chivers C.E., Koner A.L., Howarth M. How the biotin-streptavidin interaction was made even stronger: investigation via crystallography and a chimaeric tetramer. Biochem. J. 2011;435:55–63. doi: 10.1042/BJ20101593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.