Abstract
Traditionally, cell analysis has focused on using molecular biomarkers for basic research, cell preparation, and clinical diagnostics; however, new microtechnologies are enabling evaluation of the mechanical properties of cells at throughputs that make them amenable to widespread use. We review the current understanding of how the mechanical characteristics of cells relate to underlying molecular and architectural changes, describe how these changes evolve with cell-state and disease processes, and propose promising biomedical applications that will be facilitated by the increased throughput of mechanical testing: from diagnosing cancer and monitoring immune states to preparing cells for regenerative medicine. We provide background about techniques that laid the groundwork for the quantitative understanding of cell mechanics and discuss current efforts to develop robust techniques for rapid analysis that aim to implement mechanophenotyping as a routine tool in biomedicine. Looking forward, we describe additional milestones that will facilitate broad adoption, as well as new directions not only in mechanically assessing cells but also in perturbing them to passively engineer cell state.
Keywords: single-cell analysis, inertial microfluidics, mechanical phenotyping, cell sorting, cancer, mechanophenotyping, enrichment
INTRODUCTION
Understanding the mechanical properties of single cells is integral to understanding general cell and tissue behavior (1), and this understanding is becoming increasingly important for translational applications as well. Not to be confused with the forces exerted by cells, the mechanical properties of cells (or mechanical biomarkers) describe the deformability, or the resistance to deformation, of a cell in response to an applied load. Deformability (or effective stiffness) is akin to other characteristic properties, such as gene and protein expression, which are used to phenotype cell populations, but deformability differs in that it is an integrative characteristic of many molecular changes. Recent findings have shown that mechanical phenotyping is an exciting, alternative means for diagnosing cancer, detecting rare cells, and predicting phenotype. Beyond simple characterization, cellular mechanical biomarkers may also be used to enrich specific cell types for both basic science investigations and regenerative medicine therapies. This article begins with a brief overview of the cellular components contributing to whole-cell mechanical properties and follows with a discussion of how these properties are relevant to clinical diagnostics and therapies. Traditional techniques for testing single cells, as well as the most recent high-throughput approaches, are described. The review concludes with a vision of where the field of mechanical phenotyping is headed.
MECHANICAL PROPERTIES OF SINGLE CELLS
Molecular and Architectural Contributions
The mechanical behavior of individual cells is inextricably linked to their intracellular components, with particular importance historically given to the cytoskeleton. The phenomena of mechanosensing and mechanotransduction have been hypothesized to occur via connections among membrane proteins, the cytoskeleton, and the nucleus (2, 3). These links in the chain are integrally important in elucidating how stresses and strains propagate throughout a cell. Similarly, understanding the mechanical properties associated with each component and the interconnections among components can elucidate the overall mechanical properties of the cell.
Cytoskeleton.
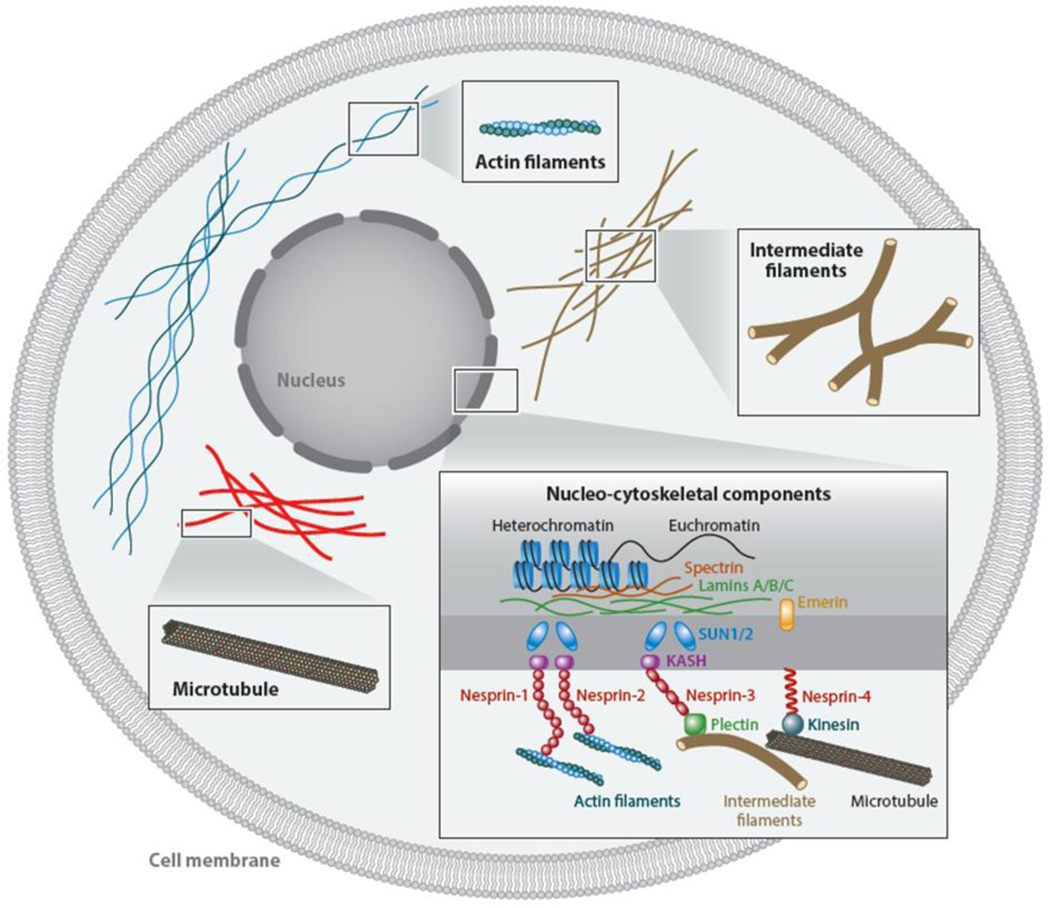
The main components of the cytoskeleton include microfilaments, intermediate filaments, and microtubules (Figure 1). Microfilaments are linear polymers of actin subunits that can assemble into helical filaments (F-actin). They resist tension in the cell and can polymerize or depolymerize within minutes, which facilitates efficient cell motility. Microfilaments also maintain the positioning of organelles in the cell and provide overall resistance to deformation from external stimuli. Intermediate filaments serve largely to provide structure in cells and have been classified into six types according to similarities in amino acid sequences and protein structures. They form an elaborate network in the cytoplasm, extending from the nucleus, whose lamina largely comprises intermediate filaments (lamins), to the plasma membrane. Intermediate filaments connect with other cytoskeletal elements, organelles, and the cell membrane to form a fully connected network. They also resist tension and are thought to contribute significantly at larger deformations, otherwise remaining mechanically passive (4). That said, recent work has indicated that keratins (5) and vimentin (6), two types of intermediate filaments, can contribute significantly to small-deformation responses as well. Microtubules are polymers of tubulin that serve to transport intracellular components throughout the cell during normal biological processes while also contributing to overall cellular structure and behavior. Microtubules resist compression, as opposed to microfilaments and intermediate filaments. The entire cytoskeleton acts in concert to balance tensile and compressive loads, and it is this combined network that contributes to a cell’s mechanical phenotype (7–9).
Figure 1.
Schematic of intracellular components contributing to cellular mechanical phenotypes. Major cytoskeletal elements include microfilaments, intermediate filaments, and microtubules. Mechanical and biological characteristics intersect via nucleocytoskeletal structures. Local and whole-cell deformations interact with the major elements to varying degrees, being highly dependent on cell type and morphology.
In addition to the three main cytoskeletal elements, there are many other proteins critical to the overall functionality of the mechanical network. Of particular interest are nucleocytoskeletal proteins that link the nucleus to the cytoskeleton. Intensive study is looking at the role key molecules play in mechanosensing---such as lamin A, B1, B2, and C; spectrin; kinesin; plectin; emerin; SUN proteins, and KASH proteins (i.e., nesprins 1, 2, 3, and 4)---and to a lesser extent, how these molecules affect the mechanical phenotype of a cell [reviewed in (2, 3)].
Besides the relative content of each cytoskeletal protein, their organization has a significant role in how a cell generates or resists forces (10–12). Likewise, cellular interactions with the microenvironment can dramatically influence measured mechanical properties. When spread on a surface, the cytoskeletal network forms a web of filamentous structures that changes over time as the cell moves. In general, the mechanical properties of adhered cells in this morphology are less compliant than in rounded cells (13–15). However, this relationship may not extend fully to the suspended state unless the structural elements contributing to the exhibited phenotype remain consistent in both states (16).
The cytoskeleton makes both passive and active contributions to cellular mechanical properties. In an equilibrium state, when the cell is not dividing or moving, a portion of the cytoskeletal structure will remain largely static, and, hence, will impart a general mechanical phenotype to the cell. This passive contribution is often representative of the cell and can be measured using a number of techniques (described in the section entitled Techniques for Measuring the Mechanical Properties of Single Cells). However, the cytoskeleton is never completely static. Portions will naturally depolymerize and polymerize in response to both intracellular and extracellular stimuli. These biological or mechanical signals can induce structural changes, which if extensive enough, may significantly alter the overall mechanical characteristics of the cell. This is especially noticeable during the cell cycle, when extensive cytoskeletal remodeling occurs to allow for cell division (17). During cell division, the generation of an internal osmotic pressure also creates a prestress on the cortical actomyosin network, leading to increases in measured stiffness (18). Major rearrangements in cytoskeletal structure that correspond to cell-extracellular matrix and cell-cell contacts will also result in significant changes in mechanical properties, which occur in concert with the expression of differentiation-specific markers (19). Since many common biochemical techniques to assay cytoskeletal architecture and activity are destructive and cannot be used with living cells, mechanical phenotyping provides an attractive alternative for determining a cell’s state based on these properties.
Cell membrane.
The cell membrane is an integral contributor to cellular mechanical behavior due to its numerous connections to the underlying cytoskeleton. As a viscoelastic covering, it effectively distributes external forces acting on the cell to the intracellular tensile and compressive elements. Alone, the cell membrane has relatively weak mechanical properties, as demonstrated by its high extensibility when tethers are pulled away from its surface (20). As a mechanical component, the cell membrane is very important at the nanoscale, where deformations do not penetrate to the underlying cytoskeletal elements or organelles. However, the membrane is part of the whole and is connected to the rest of the intracellular environment. The phenomena of mechanosensing and mechanotransduction involve the propagation of stresses and strains from the cell membrane (localized at, for example, integrins or cadherins) through the cytoskeletal network to the nucleus (21). Since the cell membrane is necessarily the origin of these extracellular stimuli, it is important for understanding the mechanics of a cell. For smaller deformations induced by fluid flow or magnetic particles, the viscoelastic properties of the membrane can contribute to measured behaviors. This is apparent when cells are placed in suspension, a state that is highly relevant to high-throughput flow-based mechanical approaches to assessment, where the viscoelasticity of the cell membrane contributes to the shear-induced distribution of cells within microfluidic channels (22).
Nucleus and other organelles.
Although the nucleus and organelles are thought to serve primarily biochemical functions, their presence can also influence both local and whole-cell mechanical property measurements. The relative contributions of organelles depend, in large part, on the physical size of each component. For example, cells with a high nuclear-cytoplasmic ratio will be dominated by the mechanical characteristics of the nucleus, the stiffest organelle. Interestingly, pluripotent stem cells are often characterized as having large nuclei relative to their cytoplasm, suggesting that their whole-cell properties are more similar to their nuclear properties than to differentiated, somatic cell types (23, 24). The morphology of a cell can also influence how much the nucleus contributes to its mechanical properties. High-resolution elasticity maps of spread or flattened cells show significantly different mechanical properties for measurements made over the nucleus versus over cytoplasmic and cytoskeletal areas (25). As mentioned above, the nucleus has been hypothesized to serve as the ultimate mechanosensory unit in the cell, where propagated forces or strains, or both, effect changes that alter gene transcription (2, 3). This potential role has spurred extensive investigation into not only whole-cell mechanical properties but also into nuclear mechanical properties as a means for characterizing cell type (26). Nuclear lamins have been widely implicated in the mechanics of the nucleus (2, 3, 27); however, chromatin architecture and compaction also appear to play a significant part in large nuclear deformations. Interestingly, both a reduction in nuclear lamins A and C and in the amount of compact heterochromatin are characteristic of a pluripotent state.
Association with Tissue of Origin
A rough correlation has been observed between cellular or nuclear elasticity and tissue microelasticity, suggesting that the mechanical phenotype of cells is strongly influenced by the compliance of surrounding structures (26). Tissue-specific interactions likely impart a mechanical phenotype to cells that is related to their location in the body, giving rise to the characteristic values (that is, the mechanical biomarkers) reported by investigators studying individual cell types, which is discussed in the next sections. It should be kept in mind that cross-study comparisons are difficult to make due to the lack of standardization among testing approaches and research groups. As such, we emphasize broad trends among the cell types that have been studied and a few special cases where multiple cell types have been evaluated in self-contained studies.
Somatic cells.
A structure--function relationship exists at the level of the cell, which necessitates that mechanical properties be well suited to the microenvironment imparted by the surrounding tissue. Empirical evidence suggests that this cell--tissue accommodation can result in mechanical characteristics that can be used to identify cell types, and even subpopulations of cell types, in specific tissues (14, 26, 28–30).
Somatic cells span a large range of elastic properties, from very soft, neuronal cell types to much stiffer, bone and muscle cell types [reviewed in (14, 29)]. Compliant, or soft, cell types, such as neurons and myeloid and lymphoid cells, have Young’s moduli in the range of 0.1–0.2 kilopascals (kPa) (31, 32). Less compliant, or stiff, cell types, such as osteoblasts and cardiomyocytes, can range between 2 and 10 kPa, with contracted muscle cells having reported moduli as high as 100 kPa (33). As mentioned, cross-study comparisons are complicated by a lack of standardization of experimental protocols and techniques. Therefore, studies that include cells from multiple tissues in their experimental design are often the best means to observe relative differences in mechanical properties. Darling et al. (28) have shown that superficial-zone chondrocytes are approximately twice as stiff as middle- or deep-zone chondrocytes (1.2 kPa versus 0.6 kPa, respectively). In a different study ((14), they also reported values for spread and spherical human osteoblasts (6.5 kPa for spread cells; 2.6 kPa for spherical), chondrocytes (1.8 kPa; 1.4 kPa), adipocytes (0.9 kPa, spherical only), adipose-derived stem cells (2.5 kPa; 2.6 kPa) and bone-marrow-derived stem cells (3.2 kPa; 2.5 kPa). Azeloglu et al. (34) characterized the mechanical properties of both nuclear and cytoplasmic regions for alveolar type I cells (2.5 kPa for nuclear; 2.5 kPa for cytoplasmic) and type II cells (3.1 kPa; 4.7 kPa) and lung fibroblasts (3.3 kPa; 6.0 kPa). The reason each of these cells exhibits a characteristic mechanical phenotype is largely associated with the biological role they fill. However, researchers can use these properties as a means to further identify a specific cell type, potentially for sorting or enrichment purposes. Red blood cells (RBCs) are a special case of somatic cell that has been extensively studied to determine mechanical phenotypes associated with health and disease (35–37), with applications focused primarily on disease diagnosis rather than on cell purification.
Stem cells.
During the past decade researchers have focused intensively on how stem cells sense and respond to the mechanical properties of the materials around them (38). In the last few years, attention has also focused on the mechanical properties of stem cells themselves, particularly during differentiation. During this process, major changes occur in gene expression and protein abundance, resulting in similarly drastic changes in cytoskeletal structure and architecture. In fact, biological and mechanical factors often interact during differentiation, with the mechanical cues serving as a driving factor in some cases (39, 40). Evaluating the mechanical properties of stem cells before, during, and after differentiation holds clues to what factors are most important in controlling this unique behavior.
The mechanical biomarkers associated with stem cells hold great promise for sorting-based enrichment for cellular therapies. Traditional biochemical approaches have been limited by the lack of a universal surface marker for mesenchymal stem cells (41). Recently, Gonzaléz-Cruz et al. (15) showed that the elastic and viscoelastic properties of mesenchymal stromal and stem cells indicated not only the population’s differentiation potential for a given lineage but also its ultimate synthetic capabilities. Likewise, Ekpenyong et al. (42) showed that cellular mechanical biomarkers define the function of myeloid precursor cells in blood, allowing them to be used as a differentiation marker that can be targeted for novel therapies. Interestingly, Bongiorno et al. (43) showed that cellular mechanical biomarkers were more reliable indicators of osteogenesis in differentiating stem cells than traditional markers such as bone sialoprotein and osteocalcin. These combined results suggest that the mechanical properties of stem cells are an excellent target for enrichment (or depletion) in regenerative medicine therapies.
Effects of Disease
One of the primary applications of the mechanical phenotyping of cells is to identify unhealthy cells. Whether as a means to better understand disease or simply as a diagnostic approach, testing the mechanical properties of single cells can provide clear indications of dysfunction. Even within a healthy individual, mechanical changes can be observed regularly in cells such as neutrophils, which modulate their cytoskeleton to allow for easier migration through the endothelium and interstitial spaces (42). Monitoring the mechanical properties of these and other immune cells may provide another means of detecting whether a patient can effectively fight infection.
Cancer is a prominent target of investigation. Researchers have repeatedly observed that malignant cells exhibit a more compliant phenotype than their healthy counterparts [extensively reviewed by Suresh (44)], presumably because compliance provides an advantage for invading surrounding tissues. More specifically, as cells become malignant, mechanical changes have been attributed to an increasingly disorganized cytoskeleton and less pronounced cortical actin (45, 46). For a selection of cancers, Cross et al. (47) showed that malignant cells were more than 70% softer than cells from normal tissue. Reinforcing these results, Remmerbach et al. (48) found that oral cancer cells (squamous cell carcinoma) were 3.5 times more compliant than healthy cells.
Other diseases can also exhibit cellular changes that are reflected in measurements of mechanical properties. Sometimes these changes are related directly to intracellular processes (e.g., abnormal expression of genes or proteins in cancer cells), but others are influenced by external factors. Malaria is one disease that has been extensively studied for its effects on the mechanical properties of cells. When RBCs are infected with the parasite Plasmodium falciparum, they lose their deformability, with cell stiffness increasing by more than 10-fold (49). This mechanical change increases the risk of occlusions in the spleen and peripheral capillaries (50), lowering oxygen concentrations in downstream tissues and eventually leading to necrosis. RBC deformability is also affected in diseases such as sickle cell, sepsis, and diabetes (51, 52). Osteoarthritis is another representative disease that changes the mechanical phenotype of affected cells. Chondrocytes in articular cartilage exhibit a less compliant mechanical phenotype in diseased tissue compared with healthy tissue, which is likely induced as a compensatory mechanism for the higher strains existing in the degraded cartilage (53).
The aging process, although not a disease, also induces changes in the mechanical properties of individual cells, which may contribute, in part, to the degradation of cellular function (54, 55). For most cell types, age causes an increase in cell stiffness along with an inability to fully recover from large deformations (56). This cytoskeletal dysfunction leads to poor mechanosensing and mechanotransduction, affecting multiple signaling cascades that impact normal cell function. In some cases, the decreased elasticity of cells may be caused by changes to the surrounding matrix (i.e., stiffer matrix equals stiffer cells). For other cases, such as cells of the immune system, intracellular signals are the more likely cause of mechanical changes. Regardless of the reason, determining the extent of these changes and how they affect the overall health of the organism is an important area of study.
Modulation by Exogenous Chemical Treatment
Cellular mechanical properties can be dramatically altered using exogenous chemicals that affect components of the cytoskeleton or nucleus. Traditionally, researchers have used these chemicals to modify the architecture of the cytoskeleton, resulting in a modified mechanical phenotype. Concurrent biological effects also occur that are typically detrimental to cell viability. Many chemotherapeutic drugs act on this premise. For example, paclitaxel permanently stabilizes the microtubule network, effectively preventing mitosis and inhibiting metastasis (44). Other drugs, such as dexamethasone and daunorubicin, produce an overall stiffening of whole-cell properties in leukemic cells (32). Studies of cell mechanics often use exogenous chemicals, such as cytochalasin B and D, latrunculin A, or jasplakinolide, to disrupt the organization of actin filaments, resulting in decreased elasticity (7). Blebbistatin has been used to inhibit nonmuscle myosin II, preventing active contraction of the cytoskeleton and causing a concurrent softening of the whole cell (57). Modifying the mechanical properties of the nucleus, e.g., by inhibiting histone deacetylases (HDACs) with trichostatin A and expanding the amount of open euchromatin, can decrease the stiffness of the organelle by 50%, significantly changing how forces and strains are propagated through the cell (58). Interestingly, chemicals that specifically target microtubules, such as paclitaxel and colchicine, have largely been found to yield no measurable effect on whole-cell mechanical properties (7). Monitoring the effects of treatment with exogenous chemicals is straightforward by using conventional or high-throughput mechanical-testing techniques. Although these drugs may be crude in that there are secondary effects, and the mechanical phenotype arises from the sum of multiple cellular changes, these drugs will continue to be a primary area of interest in helping to uncover proteins or other molecular determinants of mechanical phenotype.
POTENTIAL APPLICATIONS OF MECHANICAL PHENOTYPING
Cancer Diagnostics
Architectural features of cells have long been recognized as being useful for identifying and staging malignancy and analyzing these features in cells from body fluids and biopsies is routine in cytology and pathology labs. High-throughput approaches to mechanical phenotyping, which are also sensitive to cellular architecture, are ideally suited to making an impact in the cytopathology lab through quantitative and automated diagnosis of these samples.
There is a need to apply new technologies to improve sensitivity, reduce processing time and cost, and provide quantitative and comparable test results. Traditionally, features of the cytoplasm and, in particular, nuclear architecture---such as chromatin condensation, nuclear envelope shape, metaphase nuclei, and the nuclear-cytoplasmic ratio (Figure 2a)---are used to arrive at a diagnosis of potential malignancy, which is important to direct subsequent clinical care for the patient (59, 60). As an example, the specificity of pleural effusion cytology routinely tops 98%, but the sensitivity of the technique for malignancy ranges widely, between 40% and 90% (61). Therefore, many malignant samples are missed or require additional procedures to accurately assess the origin of the effusion and, therefore, the clinical treatment. Processing time and cost are also issues because the conventional preparation of samples of body fluids for cytological analysis involves many processing steps, the use of staining reagents, and prescreening reads by cytotechnicians prior to formal readings by the cytopathologist. Mechanical phenotyping without the need for labeling can potentially assay the same architectural features of cells quickly and without requiring significant time from a technician.
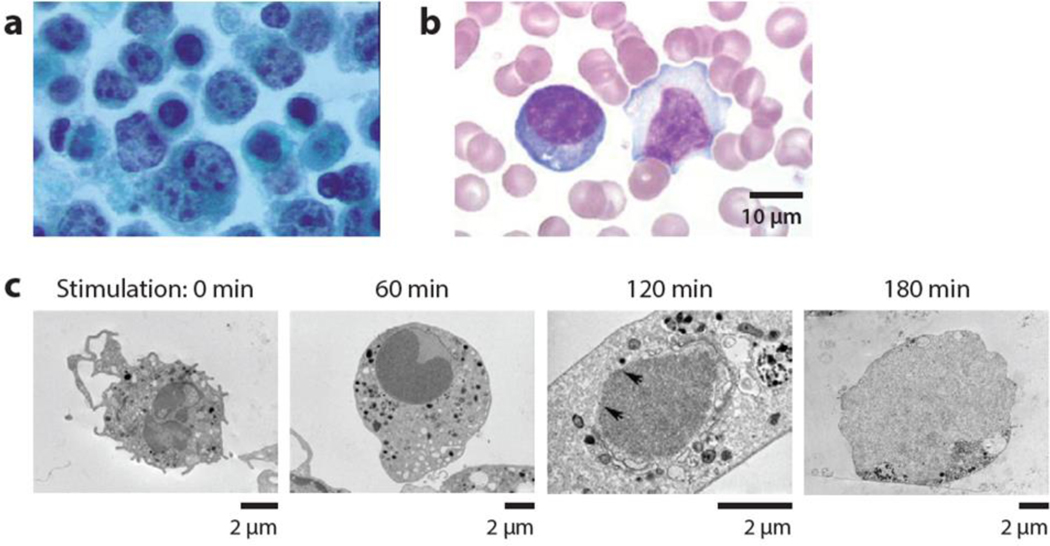
Figure 2.
Changes occurring in cellular structure with disease. (a) Lymphoma cells present with abnormally shaped nuclei, with overall light staining of chromatin and dense staining of punctate chromatin. Figure adapted from Reference 60 with permission. (b) Reactive lymphocytes with expanded basophilic cytoplasm and irregular nuclear shape. In contrast, resting lymphocytes have reduced cytoplasm and regular nuclear contours (http:www.wikidoc.orgindex.phpReactive_lymphocyte). (c) Neutrophils undergoing a process of NETosis, a form of programmed cell death in which the nuclear membrane disassembles and chromatin is released. The nuclear envelope dissolves and chromatin mixes with granules during a 3-hour period (67).
Quantitative analyses of mechanical phenotypes, by augmenting qualitative cytomorphological analyses, are also expected to yield important benefits (62). Quantitative metrics allow for standardization and enhance the communication of results between clinical sites, thus leading to diagnostic tests that have been honed for specific purposes (e.g., high-sensitivity measurements when the risk of missing a diagnosis would have important consequences, and high-specificity measurements for population studies). Currently, cytopathology results are rarely imaged and digitally archived because of the expense of storing large numbers of high-resolution image files. However, quantitative values of the architectural features of cells that have been assayed mechanically can be stored in an information-rich format that is readily interpretable and easily shared, thus facilitating productive interactions among clinicians.
Approaches using mechanical phenotyping have just begun to be applied to address clinical problems in the analysis of body fluids. Cells from pleural effusions (63) and exfoliated cells from the oral cavity (48) have been analyzed using a variety of mechanophenotyping approaches. In a study of more than 100 patients, Tse et al. (63) showed the ability to quantitatively identify malignant pleural effusions with high accuracy, including discriminating leukemias from inflammatory processes. Importantly, it was the distribution of hundreds to thousands of measurements of single-cell deformability and size that allowed distinctions to be made between the malignant or inflammatory origins of the effusions. The deformability-cytometry approach employed in this example, along with other possibilities described in the section entitled High-Throughput Approaches to Mechanical Assessment, are expected to address the problems of analyzing such heterogeneous samples. These technologies will lead to a flurry of new activity to enable body fluids to be analyzed to identify malignancy, potentially more accurately, with flexible quantitative measures and at lower cost.
Analysis of Immune Status
Like cancer cells in body fluids, leukocytes are freely available in blood and easily accessed. Analyses of leukocyte size and granularity are routinely conducted as part of normal clinical care using hematology analyzers to determine subpopulations of granulocytes, monocytes, and lymphocytes. In many illnesses the relative number of these cell subtypes varies and can be diagnostically useful. In addition to variations in the relative abundance of subtypes of leukocytes, processes of cytoskeletal or nuclear reorganization occur for each subtype during a variety of disease processes (http:www.wikidoc.orgindex.phpReactive_lymphocyte).
When lymphocytes are activated, e.g., as a result of infection with the Epstein--Barr virus, gross changes occur in nuclear condensation and cell size (Figure 2b). Measurements of deformability may be used diagnostically to quickly rule in or rule out viral infections that lead to large quantities of circulating reactive or activated lymphocytes.
Activated lymphocytes also are involved in the rejection of transplanted organs or graft-versus-host disease. Currently, the gold standard tests used to determine transplant rejection following presentation with nonspecific symptoms, such as fever, involve biopsy of the transplanted organ to identify the subpopulations of invading leukocytes (64), an invasive, risky, and expensive procedure. Although further work is needed in vivo, approaches using mechanical phenotyping have shown promise in identifying activated lymphocytes in vitro. Lectin- and anti-CD3-activated peripheral blood mononuclear cells have been observed to exhibit substantial increases in deformability and a larger spread in deformability (65). Screening peripheral blood for populations of cells with these telltale signs may be used to indicate that a rejection process is brewing and help guide doses of immunomodulatory drugs.
When neutrophils are stimulated and undergo a process termed NETosis (66, 67), drastic changes in nuclear architecture occur during a period of a few hours (Figure 2c). Chromatin decondenses, the nuclear envelope dissolves, and chromatin mixes with granules before the cell bursts, releasing chromatin NETs (neutrophil extracellular traps) that act to trap and kill bacteria (68). Clearly, these gross architectural changes are expected to lead to corresponding changes in mechanical properties. With recent evidence linking NET formation to coagulopathy and organ damage accompanying sepsis (69), a way to rapidly measure the mechanical properties of a population of neutrophils may prove useful in diagnosing sepsis and suggesting earlier treatments to avoid subsequent organ damage. Such a measure would report on the current status of the patient, given the short period of time during which neutrophils undergo NETosis, and so should also be beneficial for postdiagnosis monitoring.
Drug Screening
Drugs that affect cytoskeletal or nuclear architecture are often employed in treating cancer or other disorders, and assays of cell mechanics have the potential to screen for drug sensitivity. Drugs such as estramustine, colchicine, paclitaxel, eribulin, and discodermolide act by modifying cytoskeletal function, specifically microtubule dynamics, in cells that are actively dividing (70, 71). Cells that are sensitive to these anticancer drugs are expected to display discernible differences in mechanical properties before and after treatment, but cells that are resistant to these drugs would have little variation in their mechanical properties pre- and posttreatment. Further, performing mechanical measurements on single cells would also yield information about heterogeneity within a population and lead to identification of rare, resistant cells, which can be masked when using screening methods that act on bulk-lysed populations. Thus, a simple technique for measuring the viscoelastic properties of hundreds of thousands of cells may have clinical implications in screening cells from cancer biopsies for resistance and sensitivity to chemotherapeutics, thus yielding a new approach to personalized medicine (72). Importantly, end-point measures of cell death are not ideal for biopsied primary cells, which may not survive well in culture conditions, thus earlier measures of cell response to drugs (e.g., mechanical changes) may be better suited.
Using cell mechanics as a proxy for cytoskeletal changes may also enable screening libraries of drugs to identify drugs which affect the cytoskeleton or nuclear architecture. Similar to current high-throughput screening, cells in standard well-plates could be exposed to various drug libraries at a range of concentrations. These cells would then be measured using one of the techniques described in the section entitled High-Throughput Approaches to Mechanical Assessment to obtain a data set of the mechanical parameters of single cells. Notably, this screening process would be label-free, requiring no immunolabeling or complex interrogation of cell behavior (such as a motility assay).
Cell Separation
In general, the diagnostic applications of a mechanical biomarker rely solely on measuring the mechanical properties of a cell; however, potential therapeutic applications or downstream molecular diagnostics could benefit from mechanics-based approaches to separation.
Blood.
In blood, the separation of diseased RBCs or pathologically activated white blood cells (WBCs) may have therapeutic benefit (73). Using mechanics as a label-free biomarker has intrinsic advantages for such an application, in which unadulterated, healthy blood cells are reintroduced to a patient. Such a dialysis-like therapeutic (74) could potentially be applied to remove more deformable, activated immune cells to reduce organ failure in sepsis patients, malignant bone marrow cells for autologous transplantation of the remaining nonmalignant cells, or stiffer malaria-infected cells, or sickled RBCs during a sickle-cell crisis. Recently, the purification of malaria-infected cells from confounding host leukocyte cells and DNA has been argued to be important to enable whole-genome sequencing and identification of drug resistance in parasites (75).
Regenerative medicine.
The implantation of differentiated cells derived from pluripotent stem cells is showing promise for treating major health problems, such as heart disease, vision loss, immune-system disorders, and neurological diseases. Despite these and other exciting developments, there remain several concerns about safety, practical manufacturing issues, and process-control issues that limit the widespread deployment of stem-cell-based therapies. The presence of residual naive, partially differentiated, or partially reprogrammed cells, which pose a high risk of teratoma formation, is an ongoing concern that can be mitigated only by requiring robust quality control and separation procedures (76, 77). The development of a tumor in even a single patient among a thousand receiving treatment would be devastating to the public’s confidence in regenerative medicine.
In addition, economically viable therapies will require automated and scalable cell-separation and purification processes that minimize exposure to exogenous agents, which each require separate quality-control processes, according to US Food and Drug Administration guidelines. New high-throughput techniques that identify and sort cells based on their mechanical phenotype could be compatible with scalable production or quality controls to remove residual teratoma-forming pluripotent stem cells that are significantly more deformable (78), or to select subpopulations of mesenchymal stem cells that are more therapeutically active (15, 79).
Molecular underpinnings of whole-cell mechanics.
Just as fluorescence-activated cell sorting (FACS) enabled the identification of molecular signatures accompanying subsets of leukocytes based on their immunophenotypes, sorting based on mechanophenotypes could provide unique understanding of the molecular underpinnings of cell mechanics. Such tools could enable new insights into which genes are responsible for controlling cellular architecture, and in heterogeneous populations of diseased cells, help isolate genetic mutations or epigenetic changes underlying uniquely stiff or compliant phenotypes. One can envision basic studies that may more definitively answer key questions in the field, such as: Are the most compliant or stiff cancer cells in a population the most invasive? Is the most deformable population of activated lymphocytes the most productive in secreting cytokines, antibodies, or cell-killing enzymes? Do stem cells with the highest level of deformability correspond to those that are most pluripotent? Is deformability inextricably linked to differentiation potential or vice versa? Answering such questions requires the use of robust methods to sort large populations of cells based on their mechanophenotype.
TECHNIQUES FOR MEASURING THE MECHANICAL PROPERTIES OF SINGLE CELLS
Researchers have investigated cellular mechanical properties for decades, and, in some cases, centuries. A variety of tools has been used to examine the deformation response of materials at the microscopic level (80). The optimal choice of technique depends largely on the goal of the experiment. However, it should be kept in mind that each has its strengths and weaknesses, and some may introduce bias due to the nature of the test. For example, micropipette aspiration (MA) and atomic force microscopy (AFM) can both be used to determine the Young’s modulus of a cell, but the former typically tests cells in suspension whereas the latter typically tests cells adhered to a surface. Even if cells in both cases exhibit a rounded morphology, the measured properties will be influenced significantly by underlying cytoskeletal differences [i.e., surface-adhered cells tested by AFM appear less viscous than those in suspension tested by MA (28)]. Likewise, a method that uses a very small probe for indentation will acquire highly localized measures of mechanical properties. In comparison, a method that deforms the entire cell will produce a different set of mechanical properties. The choice of method should be balanced against knowledge of a cell’s architecture. In the preceding example, localized measures can be highly variable if made across an entire cell due to contributions from cytoskeletal structures and underlying organelles, such as the nucleus (25). Additionally, the testing parameters used in an experiment can result in similar biases, with applied strains, frequencies, rates, and durations all potentially influencing the measured properties. In general, maintaining a consistent testing regimen will provide the best environment for making comparisons among cell types, regardless of the testing approach used. This is true for both conventional and high-throughput applications.
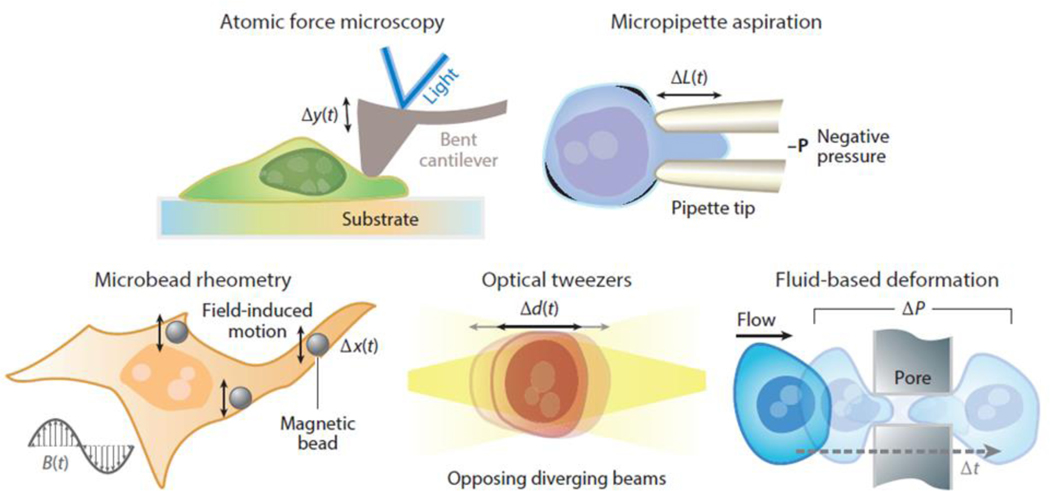
This section introduces the most common techniques used for measuring the mechanical properties of cells, but it is not intended as an exhaustive review. Emphasis is given to AFM, MA, microbead rheometry, optical tweezers and traps (OTs), and fluid-based deformation cytometry because these are the most common approaches used for the mechanical characterization of single cells (see Table 1 and Figure 3), with each spawning higher-throughput possibilities. Select studies are included to illustrate the application of these methods both in their infancy as well as more recently, with particular emphasis placed on features that are relevant for high-throughput mechanical assessment.
Table 1.
Summary of conventional approaches to testing single cells
| Technique | Cell restrictions | Mechanical properties | Typical applied force range | High-throughput |
|---|---|---|---|---|
| Atomic force microscopy | Adherent cells | Elastic and viscoelastic properties of a local region or a whole cell | pN--μN | Potentially |
| Micropipette aspiration | Nonadherent or detached, adherent cells | Elastic and viscoelastic properties of a local region or a whole cell | pN--nN | Potentially |
| Magnetic tweezers | Adherent cells | Elastic and viscoelastic properties of a local region | pN | No |
| Optical tweezers and traps | Adherent or nonadherent cells | Membrane elasticity, whole-cell deformability | fN--pN | Potentially |
| Fluid-based deformation cytometry | Nonadherent or detached, adherent cells | Whole-cell deformability | pN | Yes |
Figure 3.
Traditional techniques that have been applied to measure mechanical properties of cells.
Atomic- Force Microscopy
Monitoring the deformation of a cell in response to physical compression or indentation is one of the most common approaches to assay the mechanical properties of a single cell. Indentation tests encompass a broad spectrum of mechanical characterizations, from simple elastic indentation to time-dependent viscoelastic relaxation to dynamic frequency-dependent rheology. Cell poking, cytoindentation, and AFM all work on the similar premise of applying a force and monitoring the deformation response of a cell. From these data, elastic and viscoelastic properties can be extracted.
Kenneth S. Cole in 1932 (81) was perhaps the first scientist to use cantilever-beam theory to probe the mechanical properties of a cell. He compressed a sea urchin egg with a tiny beam and measured the internal pressure as 40 dyn/cm2 or 4 Pa. Earlier studies of the cell membrane were reviewed nearly 80 years ago by Harvey & Danielli (82). A more sophisticated approach, cell poking, was developed in the 1980s by McConnaughey & Peterson (83), which allowed for quantification of the elastic properties of the whole cell. This technique is essentially the same as modern AFM-based tests, but researchers now have greater accuracy and sensitivity in the measurements they can make.
AFM was invented by Gerd Binnig in 1986 as an extremely high resolution scanning-probe microscope (84). It was applied to the study of single-cell mechanics by Radmacher et al. (85) in the early 1990s and has continued to grow in use ever since. AFM, when used for force spectroscopy rather than surface imaging, applies classic beam theory to determine the moduli of tested materials. The vertical bending of a cantilever is measured with a laser, and the result is used to calculate an applied force at the tip, which is typically shaped as a sphere, cone, or pyramid. Although the selection of cantilever stiffness and probe geometry are highly dependent on the design of the experiment, for whole-cell indentations it is recommended to use a very soft cantilever (stiffness, k, approximately 0.01–0.06 N/m) and a spherical tip (approximately 5 μm). Ideally, AFM-based tests should produce indentations and cantilever deflections that are optimal for evaluating the mechanical properties of whole cells (13–15, 28, 86–92). Empirically, indentations should be less than 10% of the cell diameter or height, a common mathematical model constraint, and cantilever deflections should be reliably above noise levels in a fluid environment (> 5–10 nm). A spherical tip provides more conformal contact and lower local strains than sharp tips, and, if appropriately sized, provides a more accurate bulk measurement of the mechanical property of the cell, lessening the influence of individual intracellular components (93).
The mathematical models used in conjunction with AFM, or any mechanical test, are approximations of the physical behavior of the cell. As such, it is difficult to definitively state that one model is more appropriate than another. The most prevalent approach is to use the Hertz model with indentation data to determine Young’s modulus. The choice of model, as well as the testing parameters mentioned above, can dramatically influence the reported properties of a cell and should be carefully considered when making comparisons across studies.
Although simple indentation tests are the most common approach to characterization, living cells exhibit a viscoelastic phenotype, so alternative tests are necessary to fully describe their deformation behavior. Time-dependent tests, such as stress relaxation (28) or creep indentation (94), have been used extensively to characterize viscoelastic characteristics. In these experiments, either strain or stress is held constant while the other parameter is observed over time. So, for example, in stress-relaxation experiments, a constant indentation is made in the cell while the force is recorded for 0.5–1 min. The temporal drop in force describes the viscous character of the cell. Another common method for characterizing viscoelastic properties is to apply a dynamic loading profile with the AFM tip and monitor the frequency-dependent response (9, 95). Results from this microrheological test describe the storage and loss moduli, which represent the elastic and viscous characteristics of the cell, respectively.
AFM is an excellent technique for precisely measuring the mechanical properties of individual cells in a highly controlled environment and will continue to be a critical tool for validating the mechanical properties of cells that have been evaluated by other, less established approaches.
Micropipette Aspiration
A common alternative to indentation-type mechanical measurements of single cells is micropipette aspiration, also known as MA. This approach involves applying suction pressure to a cell while monitoring the extension of the membrane (and sometimes nucleus) into the bore of a micropipette. The first MA device was developed by Mitchison & Swann in 1954 (96) to investigate the surface tension of the membrane of a sea urchin egg cell. This technique has also been used for decades to study RBCs (36), and in more recent years has been expanded to investigate many other cell types, including neutrophils, endothelial cells, and chondrocytes (97). Similarly to AFM, MA can be used to evaluate the viscoelastic properties of cells, both in local regions or as a whole-cell measurement. The choice of micropipette bore size is important and figures into the mathematical equations used to calculate mechanical properties from experimental data. Vacuum pressure is also critically important, since it controls how much of the cell is drawn into the micropipette. Small extensions (requiring approximately 1 Pa) primarily measure properties of the cell membrane, whereas large extensions (requiring approximately 1000 Pa) measure cytoskeletal and cytoplasmic properties. Cell nuclei, both alone and in living cells, have also been studied extensively using MA (24, 26, 98).
As opposed to AFM, the principles and experimental approach of MA is much more conducive to high-throughput applications. This is in large part due to its compatibility with microfluidic designs, which can more easily replicate the cell-extension process in narrow channels. Furthermore, measuring cells in a suspended state allows for a more symmetric geometry, at least with respect to the whole-cell shape, which can lessen variability in the viscoelastic properties being measured (99). Due to the nature of its design, MA is less versatile in the types of tests it can be used for, being limited primarily to creep experiments.
Microbead Rheometry
Rheological testing involves applying physical perturbations at different frequencies while monitoring the resultant deformation. For microscopic samples, the use of microbead rheometry or magnetic tweezers is an attractive option for doing this because the test can evaluate elastic and viscoelastic properties in highly localized regions. Practically, this is accomplished by placing magnetic microbeads on or in a cell and monitoring their motion (21, 100, 101). Although a passive response can be of interest, most measurements are made in the presence of a magnetic field, which can be varied in intensity and frequency (102). This includes the specialized technique called magnetic twisting cytometry, which rotates a bead rather than translating it. External perturbation of the microbeads allows researchers to more fully understand the dynamic responses of individual, cellular components, such as the cytoskeleton, cytosol, and cell membrane.
The advantage of microbead rheometry over other techniques is that it can provide a very detailed description of the mechanical properties of a cell by interrogating multiple locations---corresponding to multiple localized beads---at one time. This is not feasible with AFM or MA. Likewise, microbead rheometry can assess intracellular mechanics more easily than other approaches. The major drawback is in the magnitude of forces that can be applied (piconewtons), which limits it to measuring local biological features. Whole-cell mechanical properties could be determined by taking many local measurements throughout the cell and combining them in a way that reflects an overall phenotype.
Developing microbead rheometry into a high-throughput application may be difficult, especially for intracellular measurements. To accurately capture the movement of microbeads on and in a cell, the testing environment has to be highly controlled and a high-resolution imaging system must be used. Although the magnetic field aspect of the technique could allow for concurrent testing of many cells, monitoring the movement of beads in all the cells with sufficient resolution would be extremely challenging. It is possible that some form of microbead rheometry could be incorporated into a continuous system, coupling the dynamic, bead--cell response in a magnetic field to its position in a static flow field in a microfluidic setup. However, recording the mechanical properties of a single cell in this way would be impractical.
Optical Tweezers and Traps
Light-based cell-manipulation approaches, including optical tweezers and traps (known as OTs), are attractive testing methods that do not require mechanical contact with the cells under examination. OTs use a highly focused laser beam to create a 3D light gradient that exerts attractive and repulsive forces on a bead or cell, relying on a dielectric contrast with the surrounding solution (103, 104). Conventional OTs are limited by the size of the particle that can be manipulated using a single trap. However, many studies have focused instead on trapping beads, rather than the cell itself, and using them to probe the properties of the membrane (105). In this way, tethers can be drawn away from the cell to measure the tensile properties of the membrane and underlying cytoskeleton (20). Variations on the traditional single-spot OTs include the diode laser bar trap, which can exert the same focusing forces but over a much larger area (an approximately 100 μm line) (106).
One of the limitations of using OTs for measuring the properties of single cells is the relatively small-magnitude force that can be applied, typically less than a couple hundred piconewtons. Although this works well for investigating molecular bonds, it is not strong enough to deform most cell types sufficiently to obtain information about whole-cell properties. However, the optical stretcher is a related OT device that can apply upward of a nanonewton of force, which is sufficient to deform a whole nucleated cell (103, 107). The device works by placing cells between two, nonfocused laser beams, which act to stretch the cell in a nondestructive manner. Even greater deformation is possible by using more powerful lasers, but care has to be taken to minimize the effect of localized heating on a cell’s viability and mechanical properties (108).
Similar to MA, OTs can easily measure the mechanical properties of cells in suspension, which facilitates high-throughput mechanical assessment. They can also be integrated into microfluidic systems, either as a means to sort cells among channels (109, 110) or as the main, mechanical characterization technique (106, 111). These approaches are discussed in more detail in the in the section entitled High-Throughput Approaches to Mechanical Assessment.
Fluid-Based Deformation Cytometry
Because the deformation of a cell in a flow field depends on its mechanical characteristics, fluid-based assessments offer one of the most likely means of translating the mechanical testing of a single cell to a high-throughput format (51). Discussion here is limited to initial applications of fluid-based deformation for measuring cellular mechanical properties. More detailed descriptions of the high-throughput applications are provided in the following section.
Early approaches applying this technique used cone viscometers or parallel-plate flow channels to precisely apply a known shear force to a cell (37). By tethering the cell to the channel wall, uniaxial stretching could be applied to measure the elasticity of the cell membrane. When used in combination with finite-element modeling, measurements of mechanical properties have similar precision to techniques such as AFM, MA, and OTs (112).
Passing cells through microchannels, and monitoring the deformation behavior of each, is one of the more feasible options for achieving high-throughput biomechanical, flow cytometry (51, 113). In its simplest form, this approach uses a precisely sized channel to constrict and deform the cells passing through, using changes in cell geometry and dwell time to extrapolate viscoelastic parameters (114, 115). Fluid-based deformation techniques are currently limited in the types of mechanical properties that they can measure as well as in the accuracy of those measurements. Techniques such as AFM are needed to provide reliable measurements of the properties of single cells, at least for validation and verification of fluid-based tests. Improvements are being made so that both elastic and viscoelastic properties can be reliably measured, although it is difficult to separate the two responses in this type of testing environment.
HIGH-THROUGHPUT APPROACHES to MECHANICAL ASSESSMENT
For many of the envisioned applications discussed above it is necessary to process thousands to millions of cells within minutes to hours. For example, a diagnostic sample may contain a large background of normal cells within which diseased cells represent a small minority (1% or less). In such a case, sufficient throughput is required to process upward of thousands of cells in minutes to obtain statistically accurate results concerning the target cells of interest. A short processing time is valuable to lessen the physical changes that can occur as cells sit in buffer or media prior to processing. Even higher throughputs are needed to prepare therapeutic batches of cells containing hundreds of millions of cells. This section highlights the emerging tools that are addressing this throughput challenge and opening up mechanical-phenotyping approaches to new biomedical applications (Figure 4). These techniques are classified based on the physical origin of the stress field leading to measured strains.
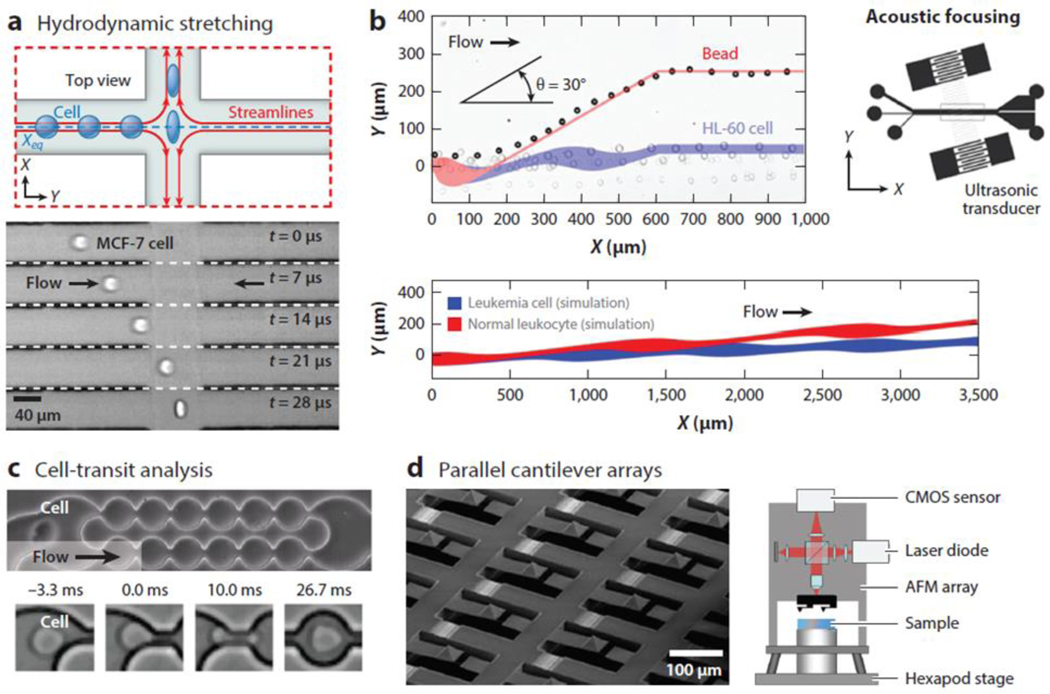
Figure 4.
Emerging platforms for high-throughput mechanophenotyping. (a) Approaches based on hydrodynamic stretching deform cells in microfluidic channels using, for example, an extensional flow; these approaches use high-speed imaging to measure a cell’s properties. (b) Approaches using acoustic-based separation apply differing forces depending on a cell’s compressibility, leading to deformability-based separation. Experimental results demonstrate separation of rigid beads from leukemia cells, and simulations suggest the ability to separate leukemic cells from healthy leukocytes based on their compressibility differences. An angled ultrasonic transducer is used in this work. (c) Cell-transit analyzers measure cell deformation or transit-time information as cells pass through constrictions. (d) Atomic force microscopy (AFM) arrays aim to parallelize physical measurements using unique optical setups to measure cantilever deflection. Abbreviation: CMOS, complementary metal oxide semiconductor.
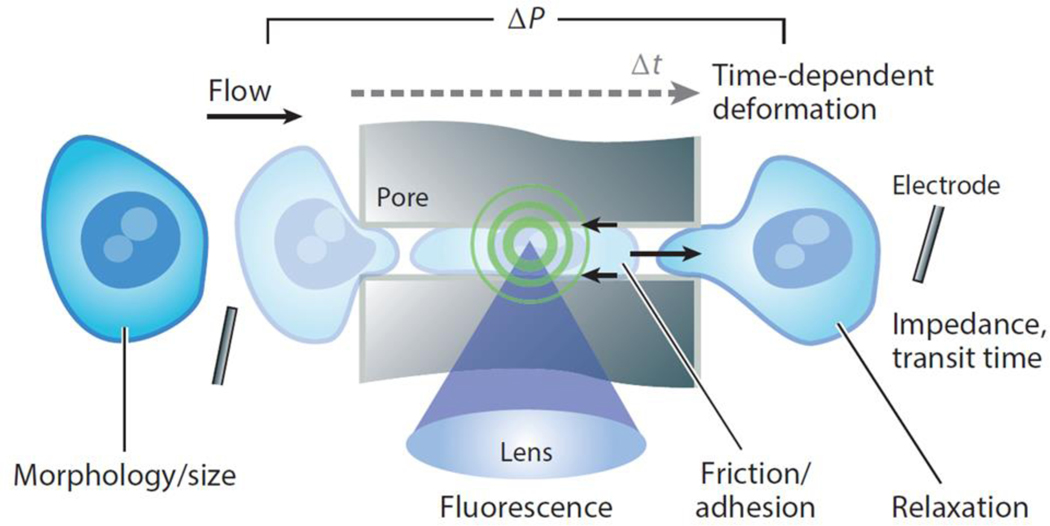
Transit Through Constrictions
Perhaps the simplest method of assaying the mechanical properties of cells in a high-throughput fashion is by measuring changes in cellular shape and transit times as cells pass through constrictions (Figure 4c). There is a long history of such approaches: almost 30 years ago Koutsouris et al. (116) measured electrically the time it took cells to transit through pores to characterize subpopulations of RBCs. During the past decades, microfluidic technology has been introduced and has increased the precision and throughput of these measurements. It has also become apparent that cell size and adhesive properties are convolved with cell deformability such that more precise time-dependent readouts are required to better separate these properties (117–119). The varied implementations of measurements of cell deformation and transit through a constriction differ mainly in their readout methods, which are based on impedance or conductance through a pore (120, 121), high-speed imaging (122–126), or on precision sensing of mass within a resonant cantilever (118). Recent efforts have also aimed at decoupling cell-deformation time during entry into the constriction, which requires the entire cell to change shape, from transit time following deformation, which may depend more on elastic restoring forces and surface friction within the channel (118). Separating these effects, or measuring transit times along surfaces with different coatings, is likely to increase the information obtained from these approaches. Interestingly, by combining passage through constrictions with microfluidic flow, deformability-based separations have also been achieved (127, 128), which will be advantageous in preparative applications.
Approaches to measuring cell transit times have sufficient throughput to measure subpopulations within a reasonable period. Analyses of 1 cell/s (118) to approximately 100 cells/s (121) have been demonstrated using significant automation. The main disadvantage of cell-transit approaches is that the constriction size needs to be well matched to the size of the cell of interest because cells that are too large or too small will either clog the channels or pass through without providing a unique signal. Therefore, these techniques have mainly been used on monodisperse cell populations (e.g., purified RBCs) and are less well suited for use with heterogeneously sized cells from samples of body fluid or cell cultures without prior purification into homogeneous size groupings.
Hydrodynamic Approaches
Hydrodynamic approaches separate cells and measure their mechanical properties by using intrinsic fluid-dynamic stresses that are tuned by the design of the microfluidic channels. The two main classes of hydrodynamic approaches rely on (a) deformability-induced lift to cause the lateral migration of cells in a continuous flow and, therefore, achieve deformability-based separation and (b) hydrodynamic cell stretching and imaging in extensional microfluidic flows in which strain in a controlled hydrodynamic stress field is measured by high-speed microscopy (Figure 4a). Lift on deformable cells in flow has been recognized since the 1930s as an explanation for the Fåhræus--Lindqvist effect, in which the viscosity of a blood solution depends on the diameter of the tube. Briefly, a cell in a shear flow will continuously deform and rotate (i.e., undergo a tank treading motion) in a way that depends on the local shear rate and the cell’s mechanical properties. This deformed shape, which is conserved as the cell travels downstream, is a dominant factor leading to a lift force directed toward regions of lower shear (129). This lift force can be directly used and balanced against other hydrodynamic lift forces in the dilute limit to separate cells based on deformability (130). At higher cellular-volume fractions in blood (approximately 40–50%) deformable RBCs migrate preferentially to the center of the channel and exclude, for example, stiffer malaria-infected RBCs or WBCs through a process called margination. Margination has been used to enrich WBCs from blood (131) and concentrate RBCs with malarial parasites (132). Note that only small deformations in cells are observed in these continuous shear-based separation processes.
Hydrodynamic stretching, in contrast, is able to deform cells at high rates at high levels of strain (> 50%). In this class of approaches, cells are centered through a focusing method before being introduced into a fluid junction with an extensional flow field (Figure 4a) (65). Variations of this approach use either inertial focusing (133) or viscoelastic focusing (134) to center cells prior to stretching to increase the uniformity of the stress field. In addition to an extensional flow junction with two opposing inlets and two outlets, a pinching flow has also been demonstrated that contains three inlets and a single outlet, a design that allows the same cells to be stretched in two different stress fields to increase information content (i.e., to assay responses to small and large strains) (135, 136).
Hydrodynamic approaches yield the highest throughputs of any current technique (up to 20,000 cells/s) (135); however, because the stress field acting on a cell in a microflow depends on cell size and shape, the measurements are sensitive to these parameters. New approaches to modeling or calibrating a particular flow field are needed to better decouple contributions made by the size of the cell from the mechanical properties measured (137). Another area needing additional improvement is image analysis following high-speed imaging. Often this is done subsequent to the experiment and requires 10−−15 min of high-end computation time. New hardware-based image-analysis implementations promise to eliminate this bottleneck (138, 139).
Acoustic Approaches
Standing acoustic waves have only recently been coupled to a microfluidic cavity to yield forces on cells that depend on the relative compressibility and density of the cell compared with the surrounding fluid, as well as on cell size. In some initial work implementing this concept, the trajectories of cells were followed after switching on an acoustic field, and compressibility parameters were extracted by assuming previously measured cell densities and sizes. By following the trajectories of cells in this standing acoustic wave, cancer cell lines were determined to have a compressibility of > 4.0 × 10−10 Pa−1, and nonmalignant cells had a compressibility of < 4.0 × 10−10 Pa−1 (140). In this approach, large numbers of cells could not be evaluated rapidly because the cells’ paths were measured in a noncontinuous fashion, but high-throughput and continuous-flow separation based on compressibility is possible using acoustic fields (Figure 4b) (141). This has been shown by separating HL-60 leukemic cells from like-sized (15 μm) rigid beads. Simulations also suggest that this approach has the ability to separate malignant cells from healthy cells (Figure 4b).
Importantly, the approach was able to process approximately 300 cells/s, suggesting that it could be applicable for rapid analysis of cells. Further work using acoustic fields will need to explore the limits of differences in compressibility that are measurable, and the relationship of compressibility to traditionally measured effective elasticity. Also, approaches to deconvolve differences in density and size on a cell-by-cell basis will be required because these also affect the acoustic contrast factor and subsequent force.
Automated Atomic Force Microscopy
As discussed earlier, AFM has been extensively used to characterize the mechanical properties of cells. Given this interest, it is not surprising that the automation and parallelization of AFM instrumentation has been proposed to achieve higher-throughput measurements of cell mechanics. Yuan and coworkers (142) have developed an automated system that includes feeding back information from the analysis of microscopic images to enable robotic positioning and indenting. This level of automation still requires several seconds per measurement due to the nature of the indentation process, suggesting that parallelization is necessary to achieve more practical measurement throughput. An arrayed implementation has also been designed for the purpose of measuring cells that are prepatterned on a substrate (Figure 4d) (143). In this approach, a 4 ×17 cantilever-probe array was fabricated to register with a patterned array of cells. To parallelize the accurate position sensing of the different probe tips, instead of measuring laser deflection, an interferometric readout scheme was implemented using a standard CMOS (complementary metal oxide semiconductor) camera. Although this approach can increase throughput many-fold, there remain several challenges to ensuring registration between the array and the surface so that similar indentations are made over all of the cells in the array, with initial cell positioning being critical to achieve maximum throughput. Perhaps because of these remaining problems, significant additional developments adapting AFM as a high-throughput approach have not been pursued.
Osmotic Approaches
Another method of applying stress to a cell---although focused on the cell membrane---is through osmotic shock. Historical techniques have used such an approach to characterize membrane integrity and cortical cytoskeletal strength, especially in RBCs (144). These early osmotic fragility tests characterized the quantity of cells lysed at a given time using bulk absorption measurements of hemoglobin or measures of intact pellet size after centrifugation. More recently, individual cells undergoing lysis have been tracked using video microscopy to study osmotic fragility in RBCs in a static setting (145). The approaches taken to increase throughput of osmotic fragility tests make use of microfluidic devices that create osmotic shock by either electroporation (146) or exposure to a hypotonic solution (147). Following membrane disruption or hypotonic exposure, RBCs gradually lyse and lose contrast as the index of refraction of the internal cytoplasm begins to match that of the external fluid, and this can be imaged with a high-speed camera. Single cells can be tracked using some approaches, and the lysis time can be determined quantitatively. Because of the serial and continuous nature of the assay, thousands of cells can be analyzed in a reasonable period at rates of approximately 1 cell/s (146). A unique feature of osmotic approaches is that the mechanism of colloid--osmotic lysis focuses forces on the cell membrane and, therefore, should be most sensitive to membrane properties; however, these approaches would be less well suited to the analysis of nucleated cells.
Optical Approaches
Forces applied by OTs have also been adapted for higher-throughput measurements. Practically, these approaches all stretch cells directly with optical forces rather than with pairs of beads attached to cells because such bead-attached cells are a statistical minority in a population. The optical stretcher is the first approach to make a significant advance (148), achieving throughputs of approximately 1 cell/min, which is mainly limited by the need to wait for a sufficient time to image small, creeping cellular deformations. Smaller deformations are inherent to the technique, given the relatively weak optical forces applied and the fundamental trade-off between increased optical forces and sample heating. Note that the longer timescales of observation also bring some advantages in terms of extracting viscoelastic and plastic responses from the time-dependent deformation and relaxation curves (85). Increased-throughput optical stretching has been demonstrated (up to 1 RBC/s) using a diode laser bar to continuously stretch cells while they travel in the flow direction (149). Using a carrier frequency to modulate stretching with this approach, Plasmodium-infected RBCs were effectively discriminated from healthy RBCs at even higher rates (> 20 RBCs/s) (150). Further work is required to know whether the increased throughput could also carry over to the analysis of stiffer nucleated cells measured with the optical stretcher.
FUTURE DIRECTIONS IN THE FIELD
Miniaturization and microfluidic technology have had significant impacts on the field of cell mechanics by allowing improved interfacing and control at the cellular length scale. As discussed in this review, these tools are now providing the foundation to enable mechanical phenotyping to be translated for use in both clinical and research applications. Because each technology measures separate aspects of the mechanical properties of a cell, it will be important to determine which approach to measurement provides the optimal discrimination capabilities and cost advantages for each application. One area of future work will be to combine multiple measurements of the mechanical and physical properties of cells together to achieve a more accurate and information-rich picture of the cellular physome. Dudani et al. (135) combined two stretching measurements at low and high strains to achieve a more information-rich output. Analysis of the time-dependence of deformation, along with a cell’s size, morphology, and strain relaxation in flow through systems, should be easy to implement in most of the techniques described above (Figure 5). If trends follow those of immunofluorescence, the addition of parameters inevitability will lead to discoveries of new cell subpopulations.
Figure 5.
Emerging tools are expected to combine multiple parameters, including traditional immunofluorescence markers, with biomechanical and physical markers (e.g., morphological, electrical, adhesive, and viscoelastic). An imagined future device for high-throughput assessment of multiparameter data is shown.
In addition, since traditional physical measures of deformability---such as compressibility, elastic modulus, and stiffness---are not familiar to clinicians and cell biologists, in order to hasten the adoption of new techniques it will be necessary to link these integrative characteristics of a cell’s state with quantities that are more familiar, such as molecular abundance (e.g., through simultaneous immunofluorescence). Combining capabilities for measuring deformability and immunofluorescence will be easiest to implement in the continuous-flow microfluidic techniques, such as the analysis of cell transit through constrictions, and hydrodynamic-stretching approaches. As these systems come online, it will be exciting to understand how molecular properties may or may not correlate with mechanics at the single-cell level.
Finally, since the majority of high-throughput techniques that are used to measure mechanical properties apply significant stress and strain to cells, mechanical stimulation could also be used in the future to deliver genes or to direct differentiation. Applying high levels of stress to the cell membrane as the cell transits through constrictions is known to create pores, and these can facilitate molecular delivery (151, 152). Both the mechanical stiffness of the substrate (38) and force (153) are also known to contribute to cell-fate decisions, suggesting that the high-throughput application of known forces could be used in this realm to modulate differentiation states in therapeutic cell populations while limiting the need for exogenous compounds.
It certainly looks like we will continue squeezing cells for information (and perhaps to deliver information!) in the foreseeable future.
ACKNOWLEDGMENTS
This work was supported by awards from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (grant R01 AR063642 to E.M.D.), the National Science Foundation (CAREER Award CBET1253189 to E.M.D.; CAREER Award CBET1150588 to D.D.), and the David and Lucile Packard Foundation (Packard Fellowship to D.D.). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Science Foundation.
Footnotes
DISCLOSURE STATEMENT
EMD is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review. DD has financial interests in CytoVale Inc. which is commercializing high-throughput mechanical phenotyping technology.
LITERATURE CITED
- 1.Janmey PA, McCulloch CA. (2007) Cell mechanics: integrating cell responses to mechanical stimuli. Annual review of biomedical engineering. 9:1–34. PubMed PMID: 17461730. [DOI] [PubMed] [Google Scholar]
- 2.Isermann P, Lammerding J. (2013) Nuclear mechanics and mechanotransduction in health and disease. Current biology : CB. 23(24):R1113–21. PubMed PMID: 24355792; PMCID: 3883624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swift J, Discher DE. (2014) The nuclear lamina is mechano-responsive to ECM elasticity in mature tissue. J Cell Sci. 127(Pt 14):3005–15. PubMed PMID: 24963133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin Z, Kreplak L, Buehler MJ. (2009) Hierarchical structure controls nanomechanical properties of vimentin intermediate filaments. PloS one. 4(10):e7294. PubMed PMID: 19806221; PMCID: 2752800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seltmann K, Fritsch AW, Kas JA, Magin TM. (2013) Keratins significantly contribute to cell stiffness and impact invasive behavior. Proceedings of the National Academy of Sciences of the United States of America. 110(46):18507–12. PubMed PMID: 24167274; PMCID: 3832002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo M, Ehrlicher AJ, Mahammad S, Fabich H, Jensen MH, Moore JR, et al. (2013) The role of vimentin intermediate filaments in cortical and cytoplasmic mechanics. Biophysical journal. 105(7):1562–8. PubMed PMID: 24094397; PMCID: 3791300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rotsch C, Radmacher M. (2000) Drug-induced changes of cytoskeletal structure and mechanics in fibroblasts: an atomic force microscopy study. Biophysical journal. 78(1):520–35. PubMed PMID: 10620315; PMCID: 1300659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heidemann SR, Kaech S, Buxbaum RE, Matus A. (1999) Direct observations of the mechanical behaviors of the cytoskeleton in living fibroblasts. The Journal of cell biology. 145(1):109–22. PubMed PMID: 10189372; PMCID: 2148213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahaffy RE, Shih CK, MacKintosh FC, Kas J. (2000) Scanning probe-based frequency-dependent microrheology of polymer gels and biological cells. Physical review letters. 85(4):880–3. PubMed PMID: 10991422. [DOI] [PubMed] [Google Scholar]
- 10.Elson EL. (1988) Cellular mechanics as an indicator of cytoskeletal structure and function. Annual review of biophysics and biophysical chemistry. 17:397–430. PubMed PMID: 3293593. [DOI] [PubMed] [Google Scholar]
- 11.Janmey PA. (1998) The cytoskeleton and cell signaling: component localization and mechanical coupling. Physiological reviews. 78(3):763–81. PubMed PMID: 9674694. [DOI] [PubMed] [Google Scholar]
- 12.Stossel TP. (1984) Contribution of actin to the structure of the cytoplasmic matrix. The Journal of cell biology. 99(1 Pt 2):15s–21s. PubMed PMID: 6086665; PMCID: 2275585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darling EM, Zauscher S, Block JA, Guilak F. (2007) A thin-layer model for viscoelastic, stress-relaxation testing of cells using atomic force microscopy: do cell properties reflect metastatic potential? Biophysical journal. 92(5):1784–91. Epub 2006/12/13. PubMed PMID: 17158567; PMCID: 1796808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darling EM, Topel M, Zauscher S, Vail TP, Guilak F. (2008) Viscoelastic properties of human mesenchymally-derived stem cells and primary osteoblasts, chondrocytes, and adipocytes. Journal of biomechanics. 41(2):454–64. Epub 2007/09/11. PubMed PMID: 17825308; PMCID: 2897251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez-Cruz RD, Fonseca VC, Darling EM. (2012) Cellular mechanical properties reflect the differentiation potential of adipose-derived mesenchymal stem cells. Proceedings of the National Academy of Sciences of the United States of America. 109(24):E1523–9. PubMed PMID: 22615348; PMCID: 3386052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maloney JM, Nikova D, Lautenschlager F, Clarke E, Langer R, Guck J, et al. (2010) Mesenchymal stem cell mechanics from the attached to the suspended state. Biophysical journal. 99(8):2479–87. Epub 2010/10/21. PubMed PMID: 20959088; PMCID: 2955350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMahon HT, Gallop JL. (2005) Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 438(7068):590–6. PubMed PMID: 16319878. [DOI] [PubMed] [Google Scholar]
- 18.Stewart MP, Helenius J, Toyoda Y, Ramanathan SP, Muller DJ, Hyman AA. (2011) Hydrostatic pressure and the actomyosin cortex drive mitotic cell rounding. Nature. 469(7329):226–30. PubMed PMID: 21196934. [DOI] [PubMed] [Google Scholar]
- 19.Mundel P, Reiser J, Zuniga Mejia Borja A, Pavenstadt H, Davidson GR, Kriz W, et al. (1997) Rearrangements of the cytoskeleton and cell contacts induce process formation during differentiation of conditionally immortalized mouse podocyte cell lines. Exp Cell Res. 236(1):248–58. PubMed PMID: 9344605. [DOI] [PubMed] [Google Scholar]
- 20.Titushkin I, Cho M. (2006) Distinct membrane mechanical properties of human mesenchymal stem cells determined using laser optical tweezers. Biophysical journal. 90(7):2582–91. PubMed PMID: 16399828; PMCID: 1403190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang N, Butler JP, Ingber DE. (1993) Mechanotransduction across the cell surface and through the cytoskeleton. Science. 260(5111):1124–7. PubMed PMID: 7684161. [DOI] [PubMed] [Google Scholar]
- 22.Hur SC, Henderson-MacLennan NK, McCabe ERB, Di Carlo D. (2011) Deformability-based cell classification and enrichment using inertial microfluidics. Lab on a chip. 11(5):912–20. PubMed PMID: WOS:000287409600019. [DOI] [PubMed] [Google Scholar]
- 23.Ribeiro AS, Dahl KN. (2010) The nucleus as a central structure in defining the mechanical properties of stem cells. Conference proceedings : Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE Engineering in Medicine and Biology Society Conference. 2010:831–4. PubMed PMID: 21096312. [DOI] [PubMed] [Google Scholar]
- 24.Chalut KJ, Hopfler M, Lautenschlager F, Boyde L, Chan CJ, Ekpenyong A, et al. (2012) Chromatin decondensation and nuclear softening accompany Nanog downregulation in embryonic stem cells. Biophysical journal. 103(10):2060–70. PubMed PMID: 23200040; PMCID: 3512036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darling EM. (2011) Force scanning: a rapid, high-resolution approach for spatial mechanical property mapping. Nanotechnology. 22(17):175707. PubMed PMID: 21411911; PMCID: 3150532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, et al. (2013) Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 341(6149):1240104. PubMed PMID: 23990565; PMCID: 3976548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart CL, Kozlov S, Fong LG, Young SG. (2007) Mouse models of the laminopathies. Exp Cell Res. 313(10):2144–56. PubMed PMID: 17493612; PMCID: 1949387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darling EM, Zauscher S, Guilak F. (2006) Viscoelastic properties of zonal articular chondrocytes measured by atomic force microscopy. Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society. 14(6):571–9. Epub 2006/02/16. PubMed PMID: 16478668. [DOI] [PubMed] [Google Scholar]
- 29.Kuznetsova TG, Starodubtseva MN, Yegorenkov NI, Chizhik SA, Zhdanov RI. (2007) Atomic force microscopy probing of cell elasticity. Micron. 38(8):824–33. PubMed PMID: 17709250. [DOI] [PubMed] [Google Scholar]
- 30.Kanthilal M, Darling EM. (2014) Characterization of mechanical and regenerative properties of human, adipose stromal cells. Cellular and molecular bioengineering. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benzina O, Szabo V, Lucas O, Saab MB, Cloitre T, Scamps F, et al. (2013) Changes induced by peripheral nerve injury in the morphology and nanomechanics of sensory neurons. Journal of biomedical optics. 18(10):106014. PubMed PMID: 24165740. [DOI] [PubMed] [Google Scholar]
- 32.Rosenbluth MJ, Lam WA, Fletcher DA. (2006) Force microscopy of nonadherent cells: a comparison of leukemia cell deformability. Biophysical journal. 90(8):2994–3003. PubMed PMID: 16443660; PMCID: 1414579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X, Sun Z, Foskett A, Trzeciakowski JP, Meininger GA, Muthuchamy M. (2010) Cardiomyocyte contractile status is associated with differences in fibronectin and integrin interactions. Am J Physiol-Heart C. 298(6):H2071–H81. PubMed PMID: WOS:000277863100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Azeloglu EU, Bhattacharya J, Costa KD. (2008) Atomic force microscope elastography reveals phenotypic differences in alveolar cell stiffness. Journal of applied physiology. 105(2):652–61. PubMed PMID: 18535125; PMCID: 2519939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennett V. (1985) The membrane skeleton of human erythrocytes and its implications for more complex cells. Annual review of biochemistry. 54:273–304. PubMed PMID: 3161450. [DOI] [PubMed] [Google Scholar]
- 36.Rand RP, Burton AC. (1964) Mechanical Properties of the Red Cell Membrane. I. Membrane Stiffness and Intracellular Pressure. Biophysical journal. 4:115–35. PubMed PMID: 14130437; PMCID: 1367460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hochmuth RM, Mohandas N, Blackshe Pl. (1973) Measurement of Elastic-Modulus for Red-Cell Membrane Using a Fluid Mechanical Technique. Biophysical journal. 13(8):747–62. PubMed PMID: WOS:A1973Q356300002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engler AJ, Sen S, Sweeney HL, Discher DE. (2006) Matrix elasticity directs stem cell lineage specification. Cell. 126(4):677–89. PubMed PMID: 16923388. [DOI] [PubMed] [Google Scholar]
- 39.Nobusue H, Onishi N, Shimizu T, Sugihara E, Oki Y, Sumikawa Y, et al. (2014) Regulation of MKL1 via actin cytoskeleton dynamics drives adipocyte differentiation. Nature communications. 5:3368. PubMed PMID: 24569594. [DOI] [PubMed] [Google Scholar]
- 40.Labriola NR, Darling EM. (2015) Temporal heterogeneity in single-cell gene expression and mechanical properties during adipogenic differentiation. Journal of biomechanics. 48(6):1058–66. Epub 2015/02/17. PubMed PMID: 25683518; PMCID: PMC4380682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuan RS, Boland G, Tuli R. (2003) Adult mesenchymal stem cells and cell-based tissue engineering. Arthritis Res Ther. 5(1):32–45. PubMed PMID: 12716446; PMCID: 154434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ekpenyong AE, Whyte G, Chalut K, Pagliara S, Lautenschlager F, Fiddler C, et al. (2012) Viscoelastic properties of differentiating blood cells are fate- and function-dependent. PloS one. 7(9):e45237. PubMed PMID: 23028868; PMCID: 3459925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bongiorno T, Kazlow J, Mezencev R, Griffiths S, Olivares-Navarrete R, McDonald JF, et al. (2014) Mechanical stiffness as an improved single-cell indicator of osteoblastic human mesenchymal stem cell differentiation. Journal of biomechanics. 47(9):2197–204. PubMed PMID: 24296276; PMCID: 4024369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suresh S. (2007) Biomechanics and biophysics of cancer cells. Acta biomaterialia. 3(4):413–38. PubMed PMID: 17540628; PMCID: 2917191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar S, Weaver VM. (2009) Mechanics, malignancy, and metastasis: the force journey of a tumor cell. Cancer metastasis reviews. 28(1–2):113–27. PubMed PMID: 19153673; PMCID: 2658728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swaminathan V, Mythreye K, O’Brien ET, Berchuck A, Blobe GC, Superfine R. (2011) Mechanical stiffness grades metastatic potential in patient tumor cells and in cancer cell lines. Cancer research. 71(15):5075–80. PubMed PMID: 21642375; PMCID: 3220953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cross SE, Jin YS, Rao J, Gimzewski JK. (2007) Nanomechanical analysis of cells from cancer patients. Nature nanotechnology. 2(12):780–3. PubMed PMID: 18654431. [DOI] [PubMed] [Google Scholar]
- 48.Remmerbach TW, Wottawah F, Dietrich J, Lincoln B, Wittekind C, Guck J. (2009) Oral cancer diagnosis by mechanical phenotyping. Cancer research. 69(5):1728–32. PubMed PMID: 19223529. [DOI] [PubMed] [Google Scholar]
- 49.Fedosov DA, Lei H, Caswell B, Suresh S, Karniadakis GE. (2011) Multiscale modeling of red blood cell mechanics and blood flow in malaria. PLoS computational biology. 7(12):e1002270. PubMed PMID: 22144878; PMCID: 3228770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cranston HA, Boylan CW, Carroll GL, Sutera SP, Williamson JR, Gluzman IY, et al. (1984) Plasmodium falciparum maturation abolishes physiologic red cell deformability. Science. 223(4634):400–3. PubMed PMID: 6362007. [DOI] [PubMed] [Google Scholar]
- 51.Zheng Y, Nguyen J, Wei Y, Sun Y. (2013) Recent advances in microfluidic techniques for single-cell biophysical characterization. Lab on a chip. 13(13):2464–83. PubMed PMID: 23681312. [DOI] [PubMed] [Google Scholar]
- 52.Kirschenbaum LA, Aziz M, Astiz ME, Saha DC, Rackow EC. (2000) Influence of rheologic changes and platelet-neutrophil interactions on cell filtration in sepsis. American journal of respiratory and critical care medicine. 161(5):1602–7. PubMed PMID: 10806162. [DOI] [PubMed] [Google Scholar]
- 53.Trickey WR, Lee GM, Guilak F. (2000) Viscoelastic properties of chondrocytes from normal and osteoarthritic human cartilage. Journal of orthopaedic research : official publication of the Orthopaedic Research Society. 18(6):891–8. PubMed PMID: 11192248. [DOI] [PubMed] [Google Scholar]
- 54.Chahine NO, Blanchette C, Thomas CB, Lu J, Haudenschild D, Loots GG. (2013) Effect of age and cytoskeletal elements on the indentation-dependent mechanical properties of chondrocytes. PloS one. 8(4):e61651. PubMed PMID: 23613892; PMCID: 3628340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xue F, Lennon AB, McKayed KK, Campbell VA, Prendergast PJ. (2013) Effect of membrane stiffness and cytoskeletal element density on mechanical stimuli within cells: an analysis of the consequences of ageing in cells. Computer methods in biomechanics and biomedical engineering. PubMed PMID: 23947334. [DOI] [PubMed] [Google Scholar]
- 56.Starodubtseva MN. (2011) Mechanical properties of cells and ageing. Ageing research reviews. 10(1):16–25. PubMed PMID: 19897057. [DOI] [PubMed] [Google Scholar]
- 57.Martens JC, Radmacher M. (2008) Softening of the actin cytoskeleton by inhibition of myosin II. Pflugers Archiv : European journal of physiology. 456(1):95–100. PubMed PMID: 18231808. [DOI] [PubMed] [Google Scholar]
- 58.Krause M, Te Riet J, Wolf K. (2013) Probing the compressibility of tumor cell nuclei by combined atomic force-confocal microscopy. Physical biology. 10(6):065002. PubMed PMID: 24304807. [DOI] [PubMed] [Google Scholar]
- 59.Hong S, Krafft AE. (2001) Primary effusion lymphoma with herpesvirus 8 DNA in patients coinfected with HIV and hepatitis C virus: a report of 2 cases. The AIDS reader. 11(8):418–22. PubMed PMID: 11570267. [PubMed] [Google Scholar]
- 60.Mach AJ, Adeyiga OB, Di Carlo D. (2013) Microfluidic sample preparation for diagnostic cytopathology. Lab on a chip. 13(6):1011–26. PubMed PMID: 23380972; PMCID: 4041400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sahn SA. (2008) The value of pleural fluid analysis. The American journal of the medical sciences. 335(1):7–15. PubMed PMID: 18195577. [DOI] [PubMed] [Google Scholar]
- 62.Renshaw AA. (2007) Reporting risk of malignancy/dysplasia in cytology: a potential way to improve communication, if not reputation. Cancer. 111(6):465–6. PubMed PMID: 17924580. [DOI] [PubMed] [Google Scholar]
- 63.Tse HT, Gossett DR, Moon YS, Masaeli M, Sohsman M, Ying Y, et al. (2013) Quantitative diagnosis of malignant pleural effusions by single-cell mechanophenotyping. Science translational medicine. 5(212):212ra163. PubMed PMID: 24259051. [DOI] [PubMed] [Google Scholar]
- 64.De Vlaminck I, Valantine HA, Snyder TM, Strehl C, Cohen G, Luikart H, et al. (2014) Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Science translational medicine. 6(241):241ra77. PubMed PMID: 24944192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gossett DR, Tse HT, Lee SA, Ying Y, Lindgren AG, Yang OO, et al. (2012) Hydrodynamic stretching of single cells for large population mechanical phenotyping. Proceedings of the National Academy of Sciences of the United States of America. 109(20):7630–5. PubMed PMID: 22547795; PMCID: 3356639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brinkmann V, Zychlinsky A. (2007) Beneficial suicide: why neutrophils die to make NETs. Nature reviews Microbiology. 5(8):577–82. PubMed PMID: 17632569. [DOI] [PubMed] [Google Scholar]
- 67.Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, et al. (2007) Novel cell death program leads to neutrophil extracellular traps. The Journal of cell biology. 176(2):231–41. PubMed PMID: 17210947; PMCID: 2063942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. (2004) Neutrophil extracellular traps kill bacteria. Science. 303(5663):1532–5. PubMed PMID: 15001782. [DOI] [PubMed] [Google Scholar]
- 69.Camicia G, Pozner R, de Larranaga G. (2014) Neutrophil Extracellular Traps in Sepsis. Shock. PubMed PMID: 25004062. [DOI] [PubMed] [Google Scholar]
- 70.Jordan MA, Wilson L. (1998) Microtubules and actin filaments: dynamic targets for cancer chemotherapy. Curr Opin Cell Biol. 10(1):123–30. PubMed PMID: 9484604. [DOI] [PubMed] [Google Scholar]
- 71.Kavallaris M. (2010) Microtubules and resistance to tubulin-binding agents. Nature reviews Cancer. 10(3):194–204. PubMed PMID: 20147901. [DOI] [PubMed] [Google Scholar]
- 72.Schilsky RL. (2010) Personalized medicine in oncology: the future is now. Nature reviews Drug discovery. 9(5):363–6. PubMed PMID: 20431568. [DOI] [PubMed] [Google Scholar]
- 73.DARPA. Biological Technologies Office. Available from: http://www.darpa.mil/Our_Work/BTO/Programs/DialysisLikeTherapeutics(DLT).aspx. [Google Scholar]
- 74.Kang JH, Super M, Yung CW, Cooper RM, Domansky K, Graveline AR, et al. (2014) An extracorporeal blood-cleansing device for sepsis therapy. Nat Med. 20(10):1211–6. Epub 2014/09/14. PubMed PMID: 25216635. [DOI] [PubMed] [Google Scholar]
- 75.Auburn S, Marfurt J, Maslen G, Campino S, Ruano Rubio V, Manske M, et al. (2013) Effective preparation of Plasmodium vivax field isolates for high-throughput whole genome sequencing. PloS one. 8(1):e53160. PubMed PMID: 23308154; PMCID: 3537768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lowry WE, Quan WL. (2010) Roadblocks en route to the clinical application of induced pluripotent stem cells. J Cell Sci. 123(Pt 5):643–51. PubMed PMID: 20164303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Werbowetski-Ogilvie TE, Bosse M, Stewart M, Schnerch A, Ramos-Mejia V, Rouleau A, et al. (2009) Characterization of human embryonic stem cells with features of neoplastic progression. Nat Biotechnol. 27(1):91–7. PubMed PMID: 19122652. [DOI] [PubMed] [Google Scholar]
- 78.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. (2007) Physical plasticity of the nucleus in stem cell differentiation. Proceedings of the National Academy of Sciences of the United States of America. 104(40):15619–24. PubMed PMID: 17893336; PMCID: 2000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee WC, Shi H, Poon Z, Nyan LM, Kaushik T, Shivashankar GV, et al. (2014) Multivariate biophysical markers predictive of mesenchymal stromal cell multipotency. Proceedings of the National Academy of Sciences of the United States of America. 111(42):E4409–18. Epub 2014/10/10. PubMed PMID: 25298531; PMCID: PMC4210311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bao G, Suresh S. (2003) Cell and molecular mechanics of biological materials. Nature materials. 2(11):715–25. PubMed PMID: 14593396. [DOI] [PubMed] [Google Scholar]
- 81.Cole KS. (1932) Surface forces of the Arbacia egg. J Cell Compar Physl. 1(1):1–9. PubMed PMID: WOS:000200175800001. [Google Scholar]
- 82.Harvey EN, Danielli JF. (1938) Properties of the cell surface. Biol Rev Camb Philos. 13(4):319–41. PubMed PMID: WOS:000188136400001. [Google Scholar]
- 83.McConnaughey WB, Petersen NO. (1980) Cell poker: an apparatus for stress-strain measurements on living cells. The Review of scientific instruments. 51(5):575–80. PubMed PMID: 6988931. [DOI] [PubMed] [Google Scholar]
- 84.Binnig G, Quate CF, Gerber C. (1986) Atomic force microscope. Physical review letters. 56(9):930–3. PubMed PMID: 10033323. [DOI] [PubMed] [Google Scholar]
- 85.Radmacher M, Tillamnn RW, Fritz M, Gaub HE. (1992) From molecules to cells: imaging soft samples with the atomic force microscope. Science. 257(5078):1900–5. PubMed PMID: 1411505. [DOI] [PubMed] [Google Scholar]
- 86.Shin D, Athanasiou K. (1999) Cytoindentation for obtaining cell biomechanical properties. Journal of orthopaedic research : official publication of the Orthopaedic Research Society. 17(6):880–90. PubMed PMID: 10632455. [DOI] [PubMed] [Google Scholar]
- 87.Darling EM, Guilak F. (2008) A neural network model for cell classification based on single-cell biomechanical properties. Tissue engineering Part A. 14(9):1507–15. Epub 2008/07/16. PubMed PMID: 18620486; PMCID: 2748927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Darling EM, Pritchett PE, Evans BA, Superfine R, Zauscher S, Guilak F. (2009) Mechanical properties and gene expression of chondrocytes on micropatterned substrates following dedifferentiation in monolayer. Cellular and molecular bioengineering. 2(3):395–404. Epub 2010/07/14. PubMed PMID: 20625462; PMCID: 2898162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gilchrist CL, Darling EM, Chen J, Setton LA. (2011) Extracellular matrix ligand and stiffness modulate immature nucleus pulposus cell-cell interactions. PloS one. 6(11):e27170. PubMed PMID: 22087260; PMCID: 3210142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gonzalez-Cruz RD, Darling EM. (2013) Adipose-derived stem cell fate is predicted by cellular mechanical properties. Adipocyte. 2(2):87–91. PubMed PMID: 23805404; PMCID: 3661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yim EK, Darling EM, Kulangara K, Guilak F, Leong KW. (2010) Nanotopography-induced changes in focal adhesions, cytoskeletal organization, and mechanical properties of human mesenchymal stem cells. Biomaterials. 31(6):1299–306. PubMed PMID: 19879643; PMCID: 2813896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jaasma MJ, Jackson WM, Keaveny TM. (2006) Measurement and characterization of whole-cell mechanical behavior. Annals of biomedical engineering. 34(5):748–58. PubMed PMID: 16604292. [DOI] [PubMed] [Google Scholar]
- 93.Harris AR, Charras GT. (2011) Experimental validation of atomic force microscopy-based cell elasticity measurements. Nanotechnology. 22(34):345102. PubMed PMID: 21795774. [DOI] [PubMed] [Google Scholar]
- 94.Wu HW, Kuhn T, Moy VT. (1998) Mechanical properties of L929 cells measured by atomic force microscopy: effects of anticytoskeletal drugs and membrane crosslinking. Scanning. 20(5):389–97. PubMed PMID: 9737018. [DOI] [PubMed] [Google Scholar]
- 95.Radmacher M, Fritz M, Kacher CM, Cleveland JP, Hansma PK. (1996) Measuring the viscoelastic properties of human platelets with the atomic force microscope. Biophysical journal. 70(1):556–67. PubMed PMID: 8770233; PMCID: 1224955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mitchison JM, Swann MM. (1954) The Mechanical Properties of the Cell Surface .2. The Unfertilized Sea-Urchin Egg. J Exp Biol. 31(3):461–72. PubMed PMID: WOS:A1954UF63300011. [Google Scholar]
- 97.Hochmuth RM. (2000) Micropipette aspiration of living cells. Journal of biomechanics. 33(1):15–22. PubMed PMID: 10609514. [DOI] [PubMed] [Google Scholar]
- 98.Rowat AC, Foster LJ, Nielsen MM, Weiss M, Ipsen JH. (2005) Characterization of the elastic properties of the nuclear envelope. Journal of the Royal Society, Interface / the Royal Society. 2(2):63–9. PubMed PMID: 16849165; PMCID: 1578255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wottawah F, Schinkinger S, Lincoln B, Ananthakrishnan R, Romeyke M, Guck J, et al. (2005) Optical rheology of biological cells. Physical review letters. 94(9):098103. PubMed PMID: 15784006. [DOI] [PubMed] [Google Scholar]
- 100.Crick FHC, Hughes AFW. (1950) The Physical Properties of Cytoplasm - a Study by Means of the Magnetic Particle Method .1. Experimental. Exp Cell Res. 1(1):37–80. PubMed PMID: WOS:A1950UL49200003. [Google Scholar]
- 101.Valberg PA. (1984) Magnetometry of ingested particles in pulmonary macrophages. Science. 224(4648):513–6. PubMed PMID: 6710153. [DOI] [PubMed] [Google Scholar]
- 102.Hoffman BD, Massiera G, Van Citters KM, Crocker JC. (2006) The consensus mechanics of cultured mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 103(27):10259–64. PubMed PMID: 16793927; PMCID: 1502445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ashkin A. (1970) Acceleration and Trapping of Particles by Radiation Pressure. Physical review letters. 24(4):156-&. PubMed PMID: WOS:A1970F157100011. [Google Scholar]
- 104.Ashkin A. (1997) Optical trapping and manipulation of neutral particles using lasers. Proceedings of the National Academy of Sciences of the United States of America. 94(10):4853–60. PubMed PMID: 9144154; PMCID: 24595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang H, Liu KK. (2008) Optical tweezers for single cells. Journal of the Royal Society, Interface / the Royal Society. 5(24):671–90. PubMed PMID: 18381254; PMCID: 2408388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Applegate R Jr., Squier J, Vestad T, Oakey J, Marr D. (2004) Optical trapping, manipulation, and sorting of cells and colloids in microfluidic systems with diode laser bars. Optics express. 12(19):4390–8. PubMed PMID: 19483988. [DOI] [PubMed] [Google Scholar]
- 107.Guck J, Ananthakrishnan R, Mahmood H, Moon TJ, Cunningham CC, Kas J. (2001) The optical stretcher: a novel laser tool to micromanipulate cells. Biophysical journal. 81(2):767–84. PubMed PMID: 11463624; PMCID: 1301552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chan CJ, Whyte G, Boyde L, Salbreux G, Guck J. (2014) Impact of heating on passive and active biomechanics of suspended cells. Interface focus. 4(2):20130069. PubMed PMID: 24748957; PMCID: 3982451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Buican TN, Smyth MJ, Crissman HA, Salzman GC, Stewart CC, Martin JC. (1987) Automated single-cell manipulation and sorting by light trapping. Applied optics. 26(24):5311–6. PubMed PMID: 20523522. [DOI] [PubMed] [Google Scholar]
- 110.Enger J, Goksor M, Ramser K, Hagberg P, Hanstorp D. (2004) Optical tweezers applied to a microfluidic system. Lab on a chip. 4(3):196–200. PubMed PMID: 15159778. [DOI] [PubMed] [Google Scholar]
- 111.Lincoln B, Wottawah F, Schinkinger S, Ebert S, Guck J. (2007) High-throughput rheological measurements with an optical stretcher. Methods in cell biology. 83:397–423. PubMed PMID: 17613318. [DOI] [PubMed] [Google Scholar]
- 112.Qiu J, Baik AD, Lu XL, Hillman EM, Zhuang Z, Dong C, et al. (2014) A noninvasive approach to determine viscoelastic properties of an individual adherent cell under fluid flow. Journal of biomechanics. 47(6):1537–41. PubMed PMID: 24581798; PMCID: 3974127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vanapalli SA, Duits MH, Mugele F. (2009) Microfluidics as a functional tool for cell mechanics. Biomicrofluidics. 3(1):12006. PubMed PMID: 19693387; PMCID: 2717604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gifford SC, Frank MG, Derganc J, Gabel C, Austin RH, Yoshida T, et al. (2003) Parallel microchannel-based measurements of individual erythrocyte areas and volumes. Biophysical journal. 84(1):623–33. PubMed PMID: 12524315; PMCID: 1302643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Korin N, Bransky A, Dinnar U. (2007) Theoretical model and experimental study of red blood cell (RBC) deformation in microchannels. Journal of biomechanics. 40(9):2088–95. PubMed PMID: 17188279. [DOI] [PubMed] [Google Scholar]
- 116.Koutsouris D, Guillet R, Lelievre JC, Guillemin MT, Bertholom P, Beuzard Y, et al. (1988) Determination of erythrocyte transit times through micropores. I--Basic operational principles. Biorheology. 25(5):763–72. PubMed PMID: 3252926. [DOI] [PubMed] [Google Scholar]
- 117.Adamo A, Sharei A, Adamo L, Lee B, Mao S, Jensen KF. (2012) Microfluidics-based assessment of cell deformability. Analytical chemistry. 84(15):6438–43. PubMed PMID: 22746217; PMCID: 3418411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Byun S, Son S, Amodei D, Cermak N, Shaw J, Kang JH, et al. (2013) Characterizing deformability and surface friction of cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 110(19):7580–5. PubMed PMID: 23610435; PMCID: 3651488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen J, Zheng Y, Tan QY, Shojaei-Baghini E, Zhang YL, Li J, et al. (2011) Classification of cell types using a microfluidic device for mechanical and electrical measurement on single cells. Lab on a chip. 11(18):3174–81. PubMed PMID: WOS:000294263400019. [DOI] [PubMed] [Google Scholar]
- 120.Adamo A, Arione A, Sharei A, Jensen KF. (2013) Flow-Through Comb Electroporation Device for Delivery of Macromolecules. Analytical chemistry. 85(3):1637–41. PubMed PMID: WOS:000314676100056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zheng Y, Shojaei-Baghini E, Azad A, Wang C, Sun Y. (2012) High-throughput biophysical measurement of human red blood cells. Lab on a chip. 12(14):2560–7. PubMed PMID: 22581052. [DOI] [PubMed] [Google Scholar]
- 122.Abkarian M, Faivre M, Stone HA. (2006) High-speed microfluidic differential manometer for cellular-scale hydrodynamics. Proceedings of the National Academy of Sciences of the United States of America. 103(3):538–42. PubMed PMID: 16407104; PMCID: 1334647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bow H, Pivkin IV, Diez-Silva M, Goldfless SJ, Dao M, Niles JC, et al. (2011) A microfabricated deformability-based flow cytometer with application to malaria. Lab on a chip. 11(6):1065–73. PubMed PMID: WOS:000287867100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Otto O, Rosendahl P, Mietke A, Golfier S, Herold C, Klaue D, et al. (2015) Real-time deformability cytometry: on-the-fly cell mechanical phenotyping. Nature methods. 12(3):199–202, 4 p following Epub 2015/02/03. PubMed PMID: 25643151. [DOI] [PubMed] [Google Scholar]
- 125.Rosenbluth MJ, Lam WA, Fletcher DA. (2008) Analyzing cell mechanics in hematologic diseases with microfluidic biophysical flow cytometry. Lab on a chip. 8(7):1062–70. PubMed PMID: 18584080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rowat AC, Jaalouk DE, Zwerger M, Ung WL, Eydelnant IA, Olins DE, et al. (2013) Nuclear Envelope Composition Determines the Ability of Neutrophil-type Cells to Passage through Micron-scale Constrictions. Journal of Biological Chemistry. 288(12):8610–8. PubMed PMID: WOS:000316564500061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lin BK, McFaul SM, Jin C, Black PC, Ma HS. (2013) Highly selective biomechanical separation of cancer cells from leukocytes using microfluidic ratchets and hydrodynamic concentrator. Biomicrofluidics. 7(3). PubMed PMID: WOS:000321145800015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang G, Mao W, Byler R, Patel K, Henegar C, Alexeev A, et al. (2013) Stiffness dependent separation of cells in a microfluidic device. PloS one. 8(10):e75901. PubMed PMID: 24146787; PMCID: 3797716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Amini H, Lee W, Di Carlo D. (2014) Inertial microfluidic physics. Lab on a chip. 14(15):2739–61. PubMed PMID: 24914632. [DOI] [PubMed] [Google Scholar]
- 130.Hur SC, Henderson-MacLennan NK, McCabe ERB, Di Carlo D. (2011) Deformability-based cell classification and enrichment using inertial microfluidics. Lab on a chip. 11(5):912–20. PubMed PMID: WOS:000287409600019. [DOI] [PubMed] [Google Scholar]
- 131.Shevkoplyas SS, Yoshida T, Munn LL, Bitensky MW. (2005) Biomimetic autoseparation of leukocytes from whole blood in a microfluidic device. Analytical chemistry. 77(3):933–7. PubMed PMID: WOS:000226759000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hou HW, Bhagat AA, Chong AG, Mao P, Tan KS, Han J, et al. (2010) Deformability based cell margination--a simple microfluidic design for malaria-infected erythrocyte separation. Lab on a chip. 10(19):2605–13. PubMed PMID: 20689864. [DOI] [PubMed] [Google Scholar]
- 133.Di Carlo D, Irimia D, Tompkins RG, Toner M. (2007) Continuous inertial focusing, ordering, and separation of particles in microchannels. Proceedings of the National Academy of Sciences of the United States of America. 104(48):18892–7. PubMed PMID: 18025477; PMCID: 2141878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cha S, Shin T, Lee SS, Shim W, Lee G, Lee SJ, et al. (2012) Cell stretching measurement utilizing viscoelastic particle focusing. Analytical chemistry. 84(23):10471–7. PubMed PMID: 23163397. [DOI] [PubMed] [Google Scholar]
- 135.Dudani JS, Gossett DR, Tse HT, Di Carlo D. (2013) Pinched-flow hydrodynamic stretching of single-cells. Lab on a chip. 13(18):3728–34. PubMed PMID: 23884381. [DOI] [PubMed] [Google Scholar]
- 136.Zheng Y, Chen J, Cui T, Shehata N, Wang C, Sun Y. (2014) Characterization of red blood cell deformability change during blood storage. Lab on a chip. 14(3):577–83. PubMed PMID: 24296983. [DOI] [PubMed] [Google Scholar]
- 137.Henon Y, Sheard GJ, Fouras A. (2014) Erythrocyte deformation in a microfluidic cross-slot channel. RSC Advances. 4(68):36079–88. [Google Scholar]
- 138.Lee D, Meng P, Jacobsen M, Tse H, Di Carlo D, Kastner R. A hardware accelerated approach for imaging flow cytometry. Field Programmable Logic and Applications (FPL), 2013 23rd International Conference on; Sept. 2–4, 2013: IEEE; 2013. [Google Scholar]
- 139.Tse HTK, Meng PF, Gossett DR, Irturk A, Kastner R, Di Carlo D. (2011) Strategies for Implementing Hardware-Assisted High-Throughput Cellular Image Analysis. Jala-J Lab Autom. 16(6):422–30. PubMed PMID: WOS:000297189800004. [DOI] [PubMed] [Google Scholar]
- 140.Hartono D, Liu Y, Tan PL, Then XY, Yung LY, Lim KM. (2011) On-chip measurements of cell compressibility via acoustic radiation. Lab on a chip. 11(23):4072–80. PubMed PMID: 22020269. [DOI] [PubMed] [Google Scholar]
- 141.Ding X, Peng Z, Lin SC, Geri M, Li S, Li P, et al. (2014) Cell separation using tilted-angle standing surface acoustic waves. Proceedings of the National Academy of Sciences of the United States of America. PubMed PMID: 25157150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang Z, Liu L, Wang Y, Xi N, Dong Z, Li M, et al. (2012) A fully automated system for measuring cellular mechanical properties. Journal of laboratory automation. 17(6):443–8. PubMed PMID: 23015516. [DOI] [PubMed] [Google Scholar]
- 143.Favre M, Polesel-Maris J, Overstolz T, Niedermann P, Dasen S, Gruener G, et al. (2011) Parallel AFM imaging and force spectroscopy using two-dimensional probe arrays for applications in cell biology. Journal of molecular recognition : JMR. 24(3):446–52. PubMed PMID: 21504022. [DOI] [PubMed] [Google Scholar]
- 144.Hunter FT. (1940) A Photoelectric Method for the Quantitative Determination of Erythrocyte Fragility. The Journal of clinical investigation. 19(5):691–4. PubMed PMID: 16694786; PMCID: 435004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ionescu-Zanetti C, Wang LP, Di Carlo D, Hung P, Di Blas A, Hughey R, et al. (2005) Alkaline hemolysis fragility is dependent on cell shape: Results from a morphology tracker. Cytom Part A. 65A(2):116–23. PubMed PMID: WOS:000229353200003. [DOI] [PubMed] [Google Scholar]
- 146.Bao N, Kodippili GC, Giger KM, Fowler VM, Low PS, Lu C. (2011) Single-cell electrical lysis of erythrocytes detects deficiencies in the cytoskeletal protein network. Lab on a chip. 11(18):3053–6. PubMed PMID: 21785802; PMCID: 3286654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhan Y, Loufakis DN, Bao N, Lu C. (2012) Characterizing osmotic lysis kinetics under microfluidic hydrodynamic focusing for erythrocyte fragility studies. Lab on a chip. 12(23):5063–8. PubMed PMID: 23047457. [DOI] [PubMed] [Google Scholar]
- 148.Guck J, Schinkinger S, Lincoln B, Wottawah F, Ebert S, Romeyke M, et al. (2005) Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophysical journal. 88(5):3689–98. PubMed PMID: WOS:000228688800062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sraj I, Eggleton CD, Jimenez R, Hoover E, Squier J, Chichester J, et al. (2010) Cell deformation cytometry using diode-bar optical stretchers. Journal of biomedical optics. 15(4). PubMed PMID: WOS:000281335400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sawetzki T, Eggleton CD, Desai SA, Marr DWM. (2013) Viscoelasticity as a Biomarker for High-Throughput Flow Cytometry. Biophysical journal. 105(10):2281–8. PubMed PMID: WOS:000327285100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hallow DM, Seeger RA, Kamaev PP, Prado GR, LaPlaca MC, Prausnitz MR. (2008) Shear-induced intracellular loading of cells with molecules by controlled microfluidics. Biotechnol Bioeng. 99(4):846–54. PubMed PMID: WOS:000253277300011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sharei A, Zoldan J, Adamo A, Sim WY, Cho N, Jackson E, et al. (2013) A vector-free microfluidic platform for intracellular delivery. Proceedings of the National Academy of Sciences of the United States of America. 110(6):2082–7. PubMed PMID: 23341631; PMCID: 3568376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Heisenberg CP, Bellaiche Y. (2013) Forces in tissue morphogenesis and patterning. Cell. 153(5):948–62. PubMed PMID: 23706734. [DOI] [PubMed] [Google Scholar]