Abstract
Manganese (Mn), an essential metal, can be toxic at elevated levels. In 2012, the first inherited cause of Mn excess was reported in patients with mutations in SLC30A10, a Mn efflux transporter. To explore the function of SLC30A10 in vitro, the current study used CRISPR/Cas9 gene editing to develop a stable SLC30A10 mutant Hep3B hepatoma cell line and collagenase perfusion in live mice to isolate primary hepatocytes deficient in Slc30a10. We also compared phenotypes of primary vs. non-primary cell lines to determine if they both serve as reliable in vitro models for the known physiological roles of SLC30A10. Mutant SLC30A10 Hep3B cells had increased Mn levels and decreased viability when exposed to excess Mn. Transport studies indicated a reduction of 54Mn import and export in mutant cells. While impaired 54Mn export was hypothesized given the essential role for SLC30A10 in cellular Mn export, impaired 54Mn import was unexpected. Whole genome sequencing did not identify any additional mutations in known Mn transporters in the mutant Hep3B mutant cell line. We then evaluated 54Mn transport in primary hepatocytes cultures isolated from genetically altered mice with varying liver Mn levels. Based on results from these experiments, we suggest that the effects of SLC30A10 deficiency on Mn homeostasis can be interrogated in vitro but only in specific types of cell lines.
Keywords: Manganese, SLC30A10, SLC39A14, liver, toxicity, Hep3B, CRISPR-Cas9, whole genome sequencing, primary hepatocytes
Introduction:
An essential trace metal, manganese (Mn) is important for human health (Aschner and Aschner 2005; Avila et al. 2013; Balachandran et al. 2020). It is required for growth and development, metabolism, and brain and immune function (Aschner and Aschner 2005). It also serves as a co-factor for enzymes such as the antioxidant protein superoxide dismutase 2. Mn is acquired from dietary sources such as green leafy vegetables, nuts, legumes, grains, and tea. Though dietary levels of essential trace metals such as Mn can vary substantially, our body regulates and maintains stable metal levels via control mechanisms such as absorption, excretion, compartmentalization, transport, and binding to cellular components (Jan et al. 2015). Alterations in these mechanisms can be detrimental to human health, leading to metal deficiency or toxicity, and Mn imbalance is no exception. Diseases of Mn imbalance are typically diseases of excess and toxicity. Mn toxicity can occur due to community (contaminated well water) and occupational exposures (welding and smelter work), environmental emissions (fossil fuels), iatrogenic exposure (total parenteral nutrition), and genetic deficiencies (mutations in Mn transport proteins SLC30A10 and SLC39A14). In addition, Mn deficiency is reported in patients and mice with mutations in the Mn transport protein SLC39A8 (Lin et al. 2017; Park et al. 2015).
Recently discovered inherited disorders of Mn imbalance in patients have extended our understanding of Mn transport and toxicity (Park et al. 2015; Quadri et al. 2012; Tuschl et al. 2012; Tuschl et al. 2016). Studies of these novel diseases have established a key set of genes that are essential for Mn transport and toxicity: SLC39A14, SLC30A10, and SLC39A8 (Jenkitkasemwong et al. 2018; Leyva-Illades et al. 2014; Lin et al. 2017). SLC39A14 and SLC30A10 are required for liver Mn import and export respectively whereas SLC39A8 is required for reclamation of Mn from bile into the liver. The interplay among these transporters and their tissue-specific role in Mn disposition is an active area of study. Recently, we and others have reported the in vivo role of Slc30a10 in Mn homeostasis using whole-body and tissue-specific gene knockouts in mice (Hutchens et al. 2017; Mercadante et al. 2019; Taylor et al. 2019; Xia et al. 2017). Currently, there is a lack of in vitro models to study the functional impact of SLC30A10 deficiency on Mn homeostasis. Most studies have used SLC30A10 overexpression in cell lines with no basal expression of SLC30A10. To address this, in the current study we establish and characterize in vitro models of SLC30A10 deficiency with the goal of using these models to better understand Mn transport and toxicity and its implication in human disease. We employed CRISPR/Cas9-dependent genome editing to develop a stable SLC30A10 mutant Hep3B hepatoma cell line and collagenase perfusion in live mice to generate primary hepatocytes deficient in Slc30a10. We also evaluated primary vs non-primary cell lines to determine if both can serve as reliable in vitro models for the known physiological roles of SLC30A10.
Materials and Methods:
Hep3B cell culture:
Hep3B, a human hepatocellular carcinoma (HCC) cell line with epithelial morphology, a modal number of 60, and subtetraploid mode of 82, was obtained from ATCC (HB-8064). Cells were cultured in Eagle's Minimum Essential Medium (MEM, Corning 10009CV) supplemented with 10% FBS (Gibco, 16000-044). Cells seeded on flasks or well-plates were maintained at 37 °C in a humidified incubator containing 5% CO2. Upon confluency, sub-culturing was carried out using 0.25% trypsin-EDTA (Gibco, 25200-056) with a subculture ratio of 1:4 to 1:6 twice per week. All experiments were carried out using cultures in the log phase of growth.
Generation of SLC30A10 mutant Hep3B cells:
CRISPR/Cas9-mediated biallelic SLC30A10 mutation was performed by GenScript (Piscataway, NJ). Briefly, the guide RNA (gRNA) targeting SLC30A10 exon 1 (forward TGGCGAGATGGGCCGCTACT; reverse CGGCGCAGTCCTGGAAGATGA) was designed and evaluated for on-target cleavage efficiency, then cloned into pGS-gRNA-CMV-SpCas9-puro2 vector. Hep3B cells were transfected with the gRNA-Cas9 construct using Lipofectamine 3000 (ThermoFisher, L3000-015). Transfected cells were enriched by puromycin selection. Isogenic clones were generated and confirmed to carry biallelic mutations at the targeted site by sequencing. To confirm SLC30A10 mutations, genomic DNA was extracted from cells using PureLink genomic DNA isolation kit (ThermoFisher, K1820-01), followed by PCR amplification of exon 1 (SLC30A10). The amplified DNA was then cloned and transformed into competent cells using TOPO® TA Cloning® kit (ThermoFisher, K4530-20). Cultures of transformants were used to isolate plasmid DNA followed by Sanger sequencing (Genewiz, MA). DNA sequence files were analyzed using Geneious (V10.2) software.
Growth analyses of Hep3B cell lines:
To determine population doubling time, parental (Hep3B) and SLC30A10 mutant Hep3B cells were seeded at 150,000 cells per T75 flasks and allowed to grow at 37 °C and 5% CO2 in a humidified incubator. Triplicate flasks were counted every 24 hours for up to 7 days using a hemocytometer. The total number of cells were plotted against days of incubation to determine the population doubling time.
RNA analyses:
Total RNA was isolated using TRIzol reagent (Invitrogen, 15596026) as previously described (Mercadante et al. 2019). One μg DNase-treated RNA was used for cDNA synthesis using High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, 4368813). qPCR was carried out on ViiA 7 Real-Time PCR System (Applied Biosystems) using PowerUp SYBR Green Master Mix (Applied Biosystems, A25742). RNA concentrations were determined using standard curve quantification approach followed by normalization using reference genes. Three separate reference genes (ACTB, GAPDH, HPRT1) were used to normalize the data for the gene of interest. Data showed similar trends with all three reference genes, therefore here we report normalized RNA data using HPRT1 as a reference gene. Primer sequences (Supplementary Table S3) (Quadri et al. 2012; Shimokawa et al. 2008; Steinmann et al. 2011; Wang and Seed 2003) used for qPCR analyses were tested for specificity using BLAST search and validated empirically by agarose gel electrophoresis and DNA sequencing of amplicons. Primer concentrations were optimized by testing a series of combinations of forward and reverse primer concentrations followed by melt curve analyses.
Metal analyses of Hep3B cell lines:
Cells were seeded in 100 mm tissue culture plates. At 80% confluency, cells were treated with MnCl2 for 24 hours. Post-treatment, culture media was removed and cells were washed with PBS, trypsinized, then pelleted. Cell pellets were washed twice with PBS containing 10 mM EDTA followed by a final PBS wash. Cells were then lysed using RIPA buffer on ice for 30 minutes with occasional shaking. Lysed cells were centrifuged and supernatants were used to determine total protein content by Lowry protein assay (BioRad, 5000112). Lysate were then digested using trace metal grade nitric acid (Fisher A509P212) and overnight incubation at 85 °C. Digested samples were analyzed by graphite furnace atomic absorption spectroscopy (PerkinElmer, AAnalyst 600) as previously described (Mercadante et al. 2019).
Cytotoxicity assays of Hep3B cells:
Cells were grown to 80% confluency then treated with MnCl2, CuCl2, or ZnSO4. Short-term and long-term cytotoxicity assessments were carried out for 24 hours and 7 days respectively. For long-term assessments, media containing metals were replenished on the fourth day after initiating metal treatment. Post-treatment, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT, Sigma 475989) solution (prepared in PBS) was added to the culture media at a final concentration of 0.5 mg/ml and incubated for 3 hours. Media with MTT reagent was aspirated and the formazan crystals were solubilized in DMSO. Quantity of formazan was measured by absorbance at 570 nm (SpectraMax, BioRad).
Transfection of SLC30A10 expression vector, fluorescent microscopy, and functional characterization:
A human SLC30A10-mRuby3-3xFLAG fusion construct was cloned into dual promoter PiggyBac (PB513B, System Bio) vector using In-Fusion HD EcoDry Cloning Kit (Takara Bio). Sequence fidelity of the vector (referred to as “PB-SLC30A10”) was confirmed by Sanger sequencing. In this vector, expression of SLC30A10 is driven by a CMV promoter and expression of GFP and a puromycin resistance gene by an EF1-alpha promoter. We attempted to achieve stable integration of the SLC30A10 expression vector into the genome of mutant Hep3B cells using PiggyBac Transposon System (System Bio) but were unable to isolate any stable integrants after puromycin selection. We were also unable to isolate stable integrants using transfection and puromycin selection in absence of transposase. For transient transfections, mutant cells were transfected with empty vector (“PB”) or PB-SLC30A10 Lipofectamine 3000 (Invitrogen). Transfection efficiency was checked by GFP fluorescence. Twenty-four hours after transfection, cells were treated with 2 mM MnCl2 for 24 hours followed by cytotoxicity assay (described above). For the fluorescent microscopy, 24 hours post-transfection, cells were stained with Hoechst 33342 nuclear stain (Invitrogen) and images were captured using a Nikon Ti-U Inverted Fluorescence Microscope and NIS-Elements F Package imaging software 3.22.00 Build 710 (Nikon Instruments Inc, USA). Images were processed using ImageJ software (NIH).
Generation, maintenance, and genotyping of animals:
Studies using animals were approved by the Institutional Animal Care and Use Committee at Brown University. Mice with whole body Slc30a10 deletion (Slc30a10KO/KO) and hepatocyte-specific Slc30a10 deletion (Slc30a10lox/lox Alb+) were generated and maintained as previously described (Mercadante et al. 2019). Mice carrying whole body Slc39a14 deletion were kindly provided by Dr. Knutson (University of Florida), who originally obtained them from Drs. Shintaro Hojyo and Toshiyuki Fukada (Tokushima Bunri University, Japan). Whole-body deletion for Slc39a14 was achieved by mating heterozygous mutants (Slc39a14+/KO). Genotypes of mice were confirmed by PCR analysis of gDNA as described earlier (Hojyo et al. 2011; Mercadante et al. 2019) or by commercial vendors (Transnetyx, TN). Animals were bred and maintained on a 12-hour light/dark cycles at the Center for Animal Resources and Education (CARE), Brown University. Animals were housed in ventilated cages and provided with standard rodent chow (LabDiet 5010, 120 ppm Mn) and water ad libitum. To delay the severity of phenotypes due to Slc30a10 deficiency, Slc30a10+/+ and Slc30a10KO/KO mice were raised on LabDiet 5010 (Envigo Inc) until P14 then switched onto Mn-deficient diet (1 ppm Mn, Envigo Inc) until hepatocyte isolation at 6 weeks of age.
Primary hepatocyte isolation:
Primary hepatocytes were isolated from five- to seven-week-old mice using a two-step collagenase digestion as previously described (Boylan et al. 2017; Cabral et al. 2018; Li et al. 2010; Mercadante et al. 2019). Briefly, the liver of an anesthetized mouse was perfused with 50 mL of pre-perfusion buffer (Ca2+/Mg2+-free HBSS, 5 mM HEPES, pH 7.4) followed by 50 mL of digestion buffer (Ca2+/Mg2+-free HBSS, 5 mM HEPES, 5 mM CaCl2, pH 7.6) containing collagenase-elastase (Worthington LK002067) and DNAse I (Worthington LK003172) at a flow rate of 5-7 mL/minute. Perfusion was considered effective when the liver was blanched immediately after perfusion and visibly digested towards the endpoint or the capsule started to separate from the surface. The liver was carefully removed and dispersed in ice-cold suspension buffer (HBSS, 5 mM HEPES, pH 7.4). Dispersed cells were filtered through a 70 μm membrane and centrifuged at 50 g, 4 °C for 4 minutes. Cells were resuspended and washed twice with suspension buffer to remove debris and non-parenchymal cells. The numbers of viable cells were determined using trypan blue dye and counting with a hemocytometer. Cells were plated in MEM-alpha (Gibco 12000022; with 10 μM proline, 10 μM pyruvate, 20 μM glutamate, 2 μM serine, 2 μM aspartate) containing FBS (10%) for 2 hours on Primaria plates (Corning 353847). After two hours, culture media was replaced with MEM-alpha (Gibco 12000022; with 10 μM proline, 10 μM pyruvate, 20 μM glutamate, 2 μM serine, 2 μM aspartate) containing BSA (0.5%) and allowed to grow overnight at 37 °C and 5% CO2 in a humidified incubator. Next day, cells were evaluated for morphological characteristics of hepatocytes (bi-nucleation) and contamination prior to their use for further experiments.
54Mn import assays:
Cells were seeded in 24-well plates and grown until confluent. Pulse phase was initiated by washing cells with PBS followed by addition of Opti-MEM reduced serum media (ThermoFisher 31985062) containing 0.5 μCi/mL 54Mn (Perkin Elmer). Cells were incubated at 37 °C for a specified period of time. Pulse media was removed and cells were washed with ice-cold PBS containing 0.5 mM EDTA. Cells were lysed using ice-cold RIPA buffer (25 mM Tris HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) on ice for 10 mins. Cell lysates were collected and counted for radioactivity by Triathler Gamma Counter with an external NaI well-type crystal detector (Hidex). Parallel incubations were carried out on ice on separate plates to account for non-specific binding at respective time points and subtracted from the counts obtained after 37 °C incubations. The subtracted counts were normalized to the total protein measured in cell lysates by Lowry-based protein assay (BioRad, 5000112).
54Mn export assays:
Cells were incubated with pulse media for 30 minutes at 37 °C as described above. The pulse media was removed and cells were washed with ice-cold PBS containing 0.5 mM EDTA. The chase phase was initiated by adding Opti-MEM without radiolabeled manganese. The chase media was collected and counted for radioactivity at respective time points. Cells were lysed using RIPA buffer on ice for 10 mins. Parallel incubations were carried out on ice to account for non-specific binding and subtracted from the counts obtained after 37 °C incubations at respective time points. The subtracted counts were normalized to the total protein measured in cell lysate using Lowry-based protein assay (BioRad, 5000112) and expressed relative to counts/μg protein at 0 minutes to account for differences in 54Mn import during the pulse phase.
Whole-genome sequencing (WGS):
WGS and bioinformatics analysis were performed by BGI Americas Corp. (Cambridge, MA). Genomic DNA was extracted using PureLink genomic DNA isolation kit (ThermoFisher, K1820-01); sample concentrations were determined by microplate reader (Qubit Fluorometer, Invitrogen) and integrity and purity by agarose gel electrophoresis. One μg DNA was randomly fragmented (Covaris, Woburn, MA). Magnetic beads were used to select fragments of average size 300-400 bp, followed by quantification using Qubit fluorometer. Fragments were subjected to end-repair, 3’ adenylation, and adaptor ligation at both ends, then amplified by ligation-mediated PCR. Products were heat denatured and circularized by the split oligo sequence, then converted into a library by modified rolling circle amplification. Sequence library was analyzed by Agilent Technologies 2100 Bioanalyzer and amplified to make DNA nanoballs, which were loaded into a patterned nanoarray. Paired-end reads were generated through BGISEQ-500 platform by employing combinatorial Probe-Anchor Synthesis. High-throughput sequencing was performed for each library to ensure that each sample met the average sequencing coverage requirement. Sequencing-derived raw image files were processed by BGISEQ-500 base-calling Software and the sequence data generated as paired-end reads and stored in FASTQ format. Raw data was then filtered and mapped to the human reference genome (GRCh37/HG19) using Burrows-Wheeler Aligner software (Li and Durbin 2010). To ensure accurate variant calling, recommended best practices were followed for variant analysis with the Genome Analysis Toolkit (GATK, https://www.broadinstitute.org/gatk/guide/best-practices). Local realignment around insertions and deletions (InDels) and base quality score recalibration were performed using GATK with duplicate reads removed by Picardtools (Broad Institute). The sequencing depth and coverage were calculated based on the alignments. Genomic variations, including single nucleotide polymorphisms (SNPs) and InDels, were detected by HaplotypeCaller software v3.3.0 from GATK. The variant quality score recalibration method was applied to obtain high-confidence variant calls. Copy number variants (CNVs) were identified using the CNVnator v0.2.7 read-depth algorithm. Structural variations (SV) were detected using Breakdancer or CREST. The SnpEff tool was applied to perform annotations for variants.
Statistics:
Statistical analyses were performed using GraphPad Prism 8 software (San Diego, CA). Data are presented as mean ± SEM. Each experiment was carried out independently at least three times. A p-value of less than 0.05 was considered statistically significant and represented as: p > 0.05 (ns), p ≤ 0.05 (*), p ≤ 0.01(**), p ≤ 0.001 (***), p ≤ 0.0001 (****). Data containing two groups were analyzed using an unpaired t-test whereas data with more than two groups were analyzed using either one-way or two-way ANOVA followed by Tukey’s post-hoc analysis. Line-of-best fit was determined using GraphPad Prism and the slopes of the lines were compared by linear regression analysis.
Results:
Development and characterization of SLC30A10-mutant Hep3B cell line:
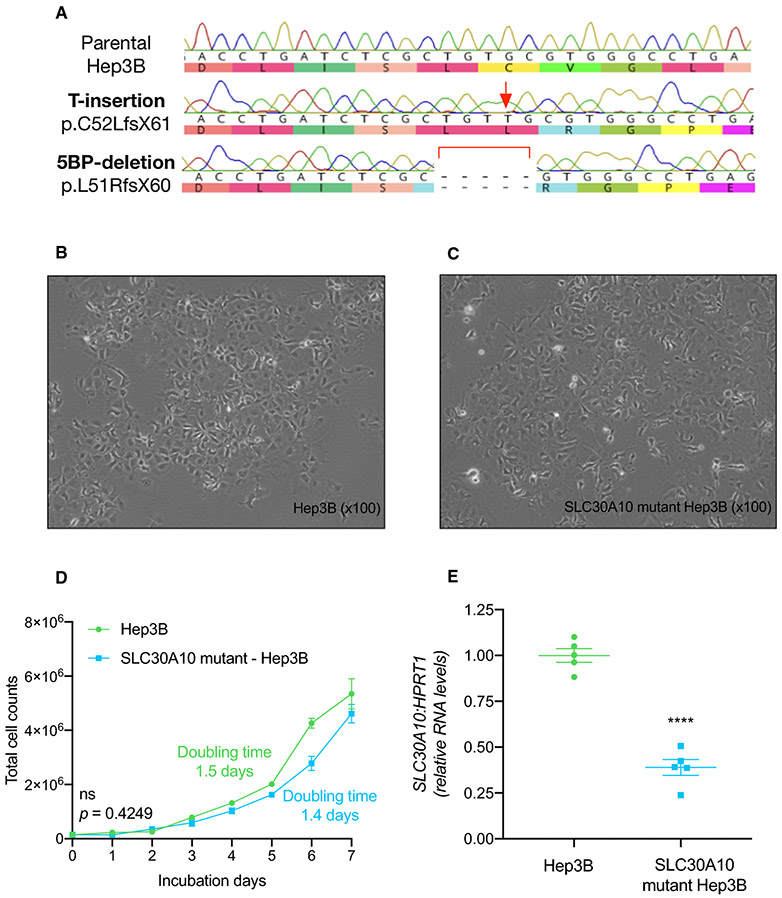
SLC30A10-mutant Hep3B cells were generated using CRISPR/Cas9 gene by targeting exon 1 of SLC30A10. We chose Hep3B cells due to its high basal expression of SLC30A10 when compared to HepG2, HEK293T, and HeLa cell lines (Supplementary Fig. S1). Sanger sequencing of genomic DNA from an isogenic clone revealed a T insertion and a 5-base-pair deletion in exon 1 of SLC30A10 (Fig. 1A). These frameshift mutations were predicted to result in pre-mature stop codons as p.C52LfsX61 and p.L51RfsX60 respectively. Morphologic examination by light microscopy showed epithelial, monolayer characteristics similar to parental Hep3B cells (Fig. 1B, C). Growth curve analysis indicated a similar growth rate for parental and mutant cell lines with population doubling times of 1.5 and 1.4 days respectively (Fig. 1D). QPCR showed a nearly 70% reduction in SLC30A10 expression in mutant cells compared to parental cells (Fig. 1E). SLC30A10 protein levels were not measured as the available commercial antibodies only detected SLC30A10 when overexpressed and our attempts to generate our own antibodies were not successful (data not shown). Overall, these results suggested that mutant cells exhibited similar growth rates and morphology yet lowered SLC30A10 expression when compared to parental cells.
Figure 1. Development and characterization of SLC30A10-mutant Hep3B cell line.
A) DNA chromatogram indicating mutant alleles harboring T insertion and 5-base-pair deletion in exon 1 of SLC30A10 gene. B, C) Representative light photomicrographs from parental and SLC30A10-mutant Hep3B cells indicating normal epithelial characteristics. Original magnification X100. D) Growth curve from mutant and parental Hep3B cell indicated non-significant population doubling time. E) Scatter dot plot showing normalized SLC30A10 expression indicating ~70% reduction in the mutant cell-lines. Data analyzed by two-tailed unpaired t-test. Data presented as p >0.05 (ns), p ≤0.05 (*), p ≤0.01(**), p ≤0.001 (***), p ≤0.0001 (****).
SLC30A10-mutant Hep3B cells are susceptible to Mn toxicity:
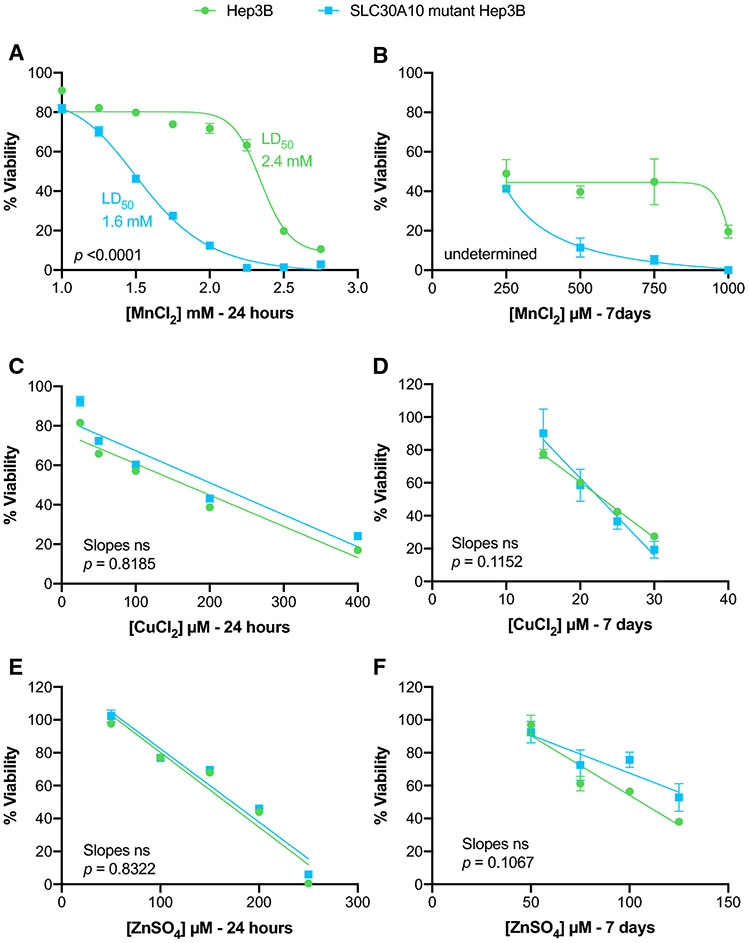
As SLC30A10 exports Mn (Leyva-Illades et al. 2014), we first tested mutant cell lines for susceptibility to metal toxicity. We hypothesized that if mutant cells have reduced SLC30A10 expression, exposure to Mn should result in Mn excess, toxicity, and loss of viability. Viability was assessed using the MTT cell proliferation assay in cells exposed to Mn, copper (Cu) or zinc (Zn) over 24 hours (short-term) or 7 days (long-term). Zinc was selected because SLC30A10 was previously believed to be a Zn transporter (hence named ZNT10) (Bosomworth et al. 2012)). Copper was chosen given that it is excreted through bile but not known to be transported by SLC30A10. LD50 values were lower in mutant than parental cells when exposed to Mn short-term (Fig. 2A). LD50 values were not determined for cells exposed to Mn long-term as the lowest concentration used resulted in more than 50% of cell death (Fig. 2B). Dose-dependent decreases in viability were similar for parental and mutant cells when exposed to Cu or Zn short- or long-term (Fig. 2C-F). Overall, mutant cells exhibited decreased viability when exposed to Mn but not Cu or Zn.
Figure 2. Cytotoxicity study to assess the mutant cell line.
Line graphs representing results from cell viability determined using MTT assay after exposure of parental and mutant cell lines to a series of concentrations of metals – A, B) MnCl2; C, D) CuCl2; and E, F) ZnSO4 for short- (24 hours) and long- (7 days) term exposure. Line-of-best fit was determined using GraphPad Prism and the slopes of the lines were compared by linear regression analysis. Difference in the LD50 values were compared with two-tailed unpaired t-test. Data presented as p >0.05 (ns), p ≤0.05 (*), p ≤0.01(**), p ≤0.001 (***), p ≤0.0001 (****).
Expression of human SLC30A10 attenuates sensitivity of mutant cells to Mn toxicity:
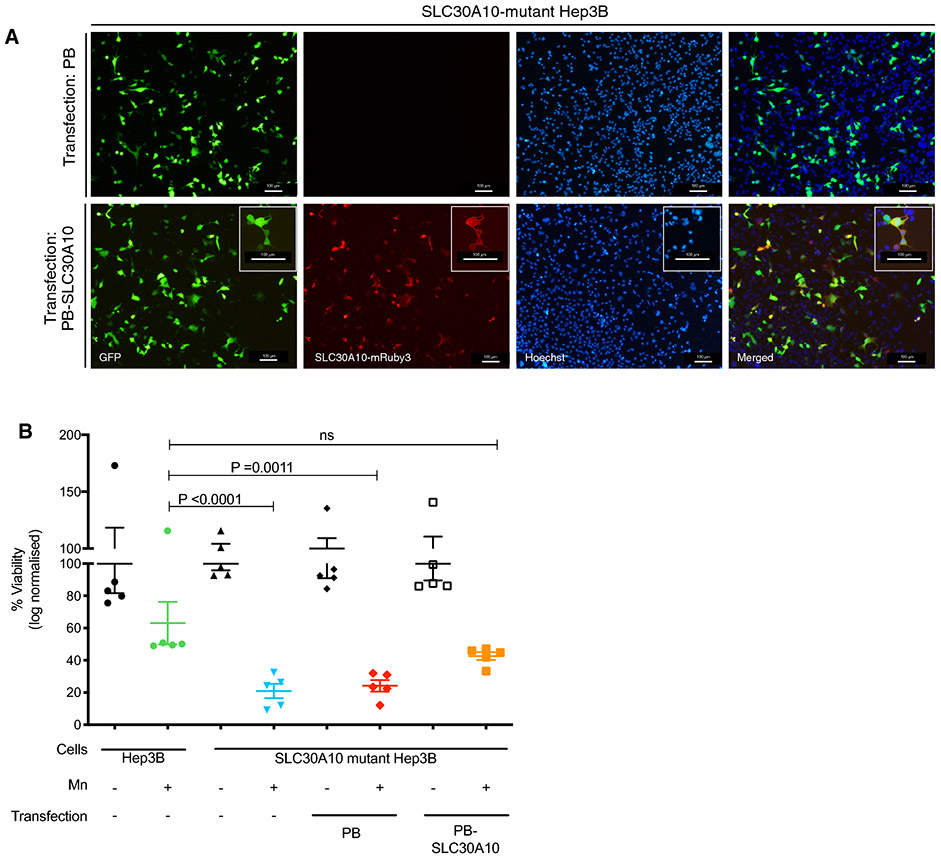
To further characterize the mutant cells, we carried out a rescue experiment in which the mutant cells were transfected with a plasmid expressing wild-type human SLC30A10. We observed about 40-50% transfection efficiency 24 hours after transfection as indicated by fluorescent microscopy (Fig 3A). As observed above (Fig 2A), cytotoxicity assays indicated parental cells treated with Mn were 60% viable while mutant cells treated with Mn showed 21% viability (p <0.0001, Fig 3B). When compared to Mn treatment of parental Hep3B cells, Mn exposure in untransfected mutant cells and mutant cells transfected with empty vector (PB) showed significant differences in cell viability (21%, p <0.0001 and 24%, p =0.0011 respectively), whereas mutant cells transfected with human SLC30A10 cDNA (PB-SLC30A10) did not (43%, p =0.7166). Such results indicate expression of wild-type SLC30A10 in mutant cells lead to partial attenuation of Mn toxicity in the Hep3B mutant cells.
Figure 3. Expression of SLC30A10 cDNA partially rescues Mn-induced cell death in mutant Hep3B cells.
A) Fluorescent micrographs showing expression of wild-type human SLC30A10 in the SLC30A10-mutant Hep3B cells (see methods section). B) Graph representing % viability (log normalized OD) of parental Hep3B cells, mutant cells, mutant cells transfected with empty vector (PB), and mutant cells transfected with human SLC30A10 expression vector (PB-SLC30A10). Data analyzed using one-way ANOVA followed by Tukey’s multiple comparison and presented as p >0.05 (ns), p ≤0.05 (*), p ≤0.01(**), p ≤0.001 (***), p ≤0.0001 (****)
Intracellular accumulation of Mn in SLC30A10 mutant cells upon Mn exposure:
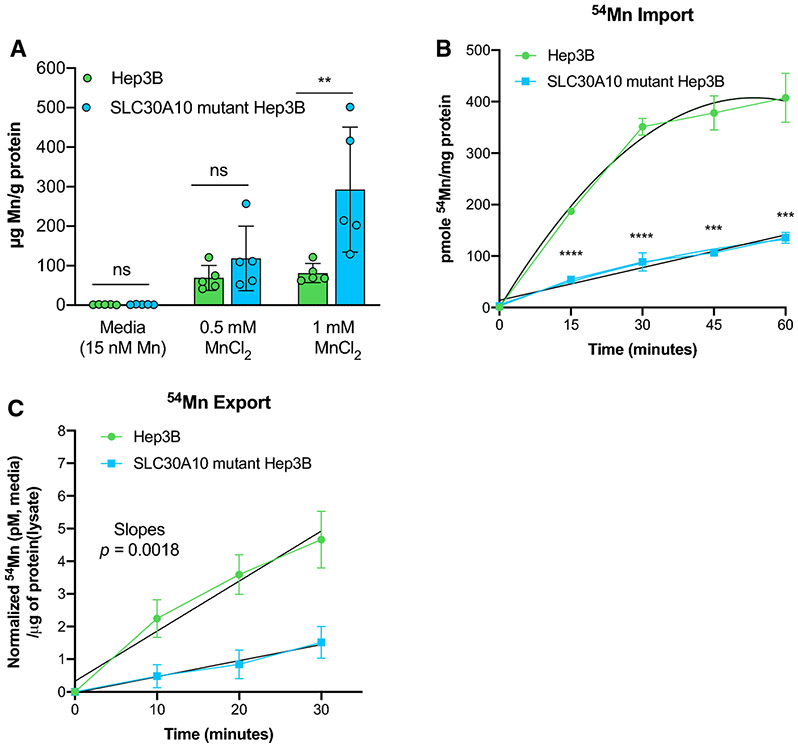
Susceptibility of mutant cells to Mn toxicity led us to investigate intracellular Mn levels post-sub-lethal Mn exposure. Mn levels were similar in parental and mutant cells when grown in basal media for 24 hours (Fig. 4A). At 0.5 mM MnCl2 exposure, there was a non-significant increase intracellular Mn in mutant cells (p=0.8971). At 1 mM MnCl2, mutant cells accumulated nearly 3.5 times more Mn compared to parental cells (p=0.0019). (We also measured Mn levels in the basal media and fetal bovine serum (data not shown). Mn levels in basal media were 15 nM and largely reflected the contribution of fetal bovine serum.)
Figure 4. Intracellular Mn and transport study in mutant cells.
A) Histogram representing intracellular manganese concentrations in mutant and parental cells determined using GF-AAS. Cells were incubated with MnCl2 for 24-hours followed by metal measurement. B) Line graph representing 54Mn import carried out by incubating cells with radio-labelled Mn showed impaired manganese import in mutant cells. C) Line graph representing 54Mn efflux from mutant and parental Hep3B cells using pulse-chase experiment. Cells were incubated with 54Mn for 30 mins (pulse) followed by chase period using replaced non-radioactive media. Data for Mn concentration was analyzed using 2way ANOVA followed by Tukey’s multiple comparison. Line-of-best fit was determined using GraphPad Prism and the slopes of the lines were compared by linear regression analysis. Data presented as p >0.05 (ns), p ≤0.05 (*), p ≤0.01(**), p ≤0.001 (***), p ≤0.0001 (****).
SLC30A10-mutant Hep3B cells exhibit altered 54Mn import and export:
Next, we evaluated Mn transport in mutant cells using 54Mn import and export assays. Import assays showed that mutant cells had impaired 54Mn uptake when compared to parental cells (Fig. 4B). Over the sixty-minute time course, mutant cells showed a linear relationship of 54Mn uptake, whereas parental cells had a non-linear relationship when the 54Mn content is normalized to μg protein in cell lysates were plotted against time (Fig. 4B). Similar results were observed when increasing amounts of non-radioactive Mn were added to pulse media; when cells were pulsed for longer times; or when calcium levels were altered in the media, given that Slc30a10-dependent Mn export is driven by active calcium-coupled exchange (Levy et al. 2019) (data not shown). Export assays indicated impaired 54Mn export in mutant cells when compared to parental cells, with both cell lines showing a linear relationship for 54Mn export over time (Fig. 4C, (Supplement Fig. S3A for data without normalization)). While impaired 54Mn export was hypothesized given that SLC30A10 is a Mn export protein, impaired 54Mn import was not expected.
Whole genomic sequencing reveal loss of variants in SLC30A10-mutant cells:
We next speculated that the 54Mn import defect in Hep3B mutant cells may reflect off-target effects of CRISPR genome editing or suppressor mutations leading to synthetic rescue of the mutant clone during the gene editing process. We performed whole genome sequencing (30x coverage) of parental and mutant cells (passage# 3) using a commercial service (BGI Inc.) to identify sequence differences between parental and mutant cell lines. Data analysis of whole genome sequencing did not reveal any mutations in the mutant cells for known and putative Mn transporters (SLC39A14 (ZIP14), SLC11A2 (DMT1), SLC39A8 (ZIP8), SLC40A1 (ferroportin), SLC30A10) except SLC30A10, however we did find multiple other mutations in other genes in mutant cells (Table S1). There were 1068 variants (59 InDels and 1009 SNPs) gained in the mutant cells. The biological significance of each of these mutations in metal homeostasis is unknown and out of scope of the current study. Targeted analysis also revealed a non-synonymous variation in exon 2 of SLC39A14 (Supplementary Fig. S2A), however this p.Leu33Pro (rs896378) variant was also present in parental Hep3B cells. Sequencing also revealed two synonymous variants, p.Leu65Leu (rs2293144) and p.His347His (rs6558052). We also found many variants (54 InDels and 844 SNPs) in the parental Hep3B hepatoma that were not reported in the human reference genome (GRCh37/HG19, Table S2) or COSMIC cell line database (Tate et al. 2019) for Hep3B cells.
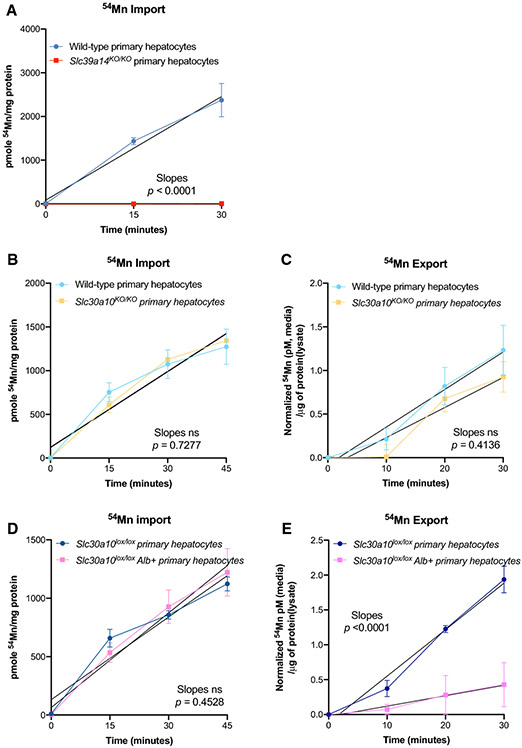
54Mn import and export assays confirm in vivo functions of SLC39A14 and SLC30A10 in primary hepatocyte cultures:
We considered that a cancer-derived cell line such as Hep3B may not exhibit the known physiologic role of SLC30A10 in cellular Mn export. To address this, we carried out 54Mn import and export assays on primary hepatocytes isolated from genetically altered mice. Primary hepatocytes isolated from mice deficient in Slc39a14, a protein essential for liver Mn import, showed blunted 54Mn import (Fig. 4A). 54Mn export was not analyzed because primary hepatocytes did not import sufficient 54Mn. We next focused on Slc30a10. (In an attempt to confirm membrane localization of Slc30a10 in primary hepatocytes, we performed confocal microscopy on primary hepatocytes isolated from mice expressing an Slc30a10-GFP fusion protein from the endogenous Slc30a10 gene (Mercadante et al. 2019), however confocal microscopy also revealed high levels of autofluorescence from primary hepatocytes not expressing the Slc30a10-GFP fusion (data not shown).) We employed mice with whole-body (Slc30a10KO/KO) and hepatocyte-specific Slc30a10 deficiency (Slc30a10lox/lox Alb+). We first tried to isolate primary hepatocytes from Slc30a10KO/KO mice raised on a standard rodent diet containing 120 ppm Mn however their blood vessels were too fragile to withstand cannulation and perfusion. To minimize the development of severe Mn excess in Slc30a10KO/KO mice, Slc30a10+/+ and Slc30a10KO/KO mice were switched onto a Mn-deficient diet (1 ppm Mn, Envigo Inc) until hepatocyte harvest at six weeks of age. This dietary approach decreased liver Mn levels in Slc30a10KO/KO mice by 5-fold but levels in mutant mice were still 13-fold higher than wild-type raised on the Mn-deficient diet (data not shown). Primary hepatocytes from Slc30a10+/+ and Slc30a10KO/KO mice raised on Mn-deficient diets showed linear relationships for 54Mn import over 45 minutes (Fig. 5B). 54Mn export was similar in primary hepatocytes from Slc30a10+/+ and Slc30a10KO/KO mice (Fig. 5C, (Supplement Fig. S3B for data without normalization)). Next, we performed transport studies in primary hepatocytes from Slc30a10lox/lox Alb+ mice, which develop mild Mn excess compared to Slc30a10KO/KO mice (Mercadante et al. 2019; Taylor et al. 2019). While 54Mn import was similar in Slc30a10lox/lox and Slc30a10lox/lox Alb+ hepatocytes (Fig. 5D), Slc30a10lox/lox Alb+ hepatocytes showed impaired 54Mn export compared to Slc30a10lox/lox hepatocytes (Fig. 5E, (Supplement Fig. S3C for data without normalization).
Figure 5. Transport study using primary hepatocytes cultures.
A) Line graph representing changes in 54Mn import in primary hepatocytes from wild-type and Slc39a14-deficient mice. B, C) Line graph representing 54Mn import (B) and export (C) using primary hepatocytes from wild-type and Slc30a10-deficient mice. D, E) Line graph representing 54Mn import (D) and export (E) using primary hepatocytes from Slc30a10lox/lox and Slc30a10lox/lox Alb mice. Line-of-best fit was determined using GraphPad Prism and the slopes of the lines were compared by linear regression analysis. Data presented as p >0.05 (ns), p ≤0.05 (*), p ≤0.01(**), p ≤0.001 (***), p ≤0.0001 (****).
Discussion:
Currently, there are few in vitro models of SLC30A10 deficiency. As SLC30A10 is essential for hepatobiliary Mn excretion, in this study we initially employed Hep3B, a hepatoma-derived cell line with high basal expression of SLC30A10. CRISPR-Cas9 gene editing was used to introduce SLC30A10 mutations into Hep3B cells, then parental and mutant cells were assessed for sensitivity to metal toxicity, intracellular Mn accumulation, and 54Mn transport. Though SLC30A10 expression in the mutant cell line was not completely abrogated, mutations in exon 1 similar to those present in our mutant cell line have shown reported in patients with Mn excess (Anagianni and Tuschl 2019).
The Hep3B parental cell line maintained viability for up to 24 hours in ~2 mM Mn. Although high levels of Mn are unlikely to represent physiological levels, these levels were required to impact viability in both parental and mutant cells. Such resilience to toxicity at high Mn levels was also observed in other cell lines (WIF-B, HepG2, AF5, HEK293T) which could withstand up to 1 mM Mn concentration for 16 hours (Thompson et al. 2018; Zogzas and Mukhopadhyay 2018). In contrast, compared to the parental cell line, our Hep3B mutant cell line was more sensitive to Mn toxicity and accumulated more Mn after exposure to Mn. In addition, complementation of mutant cells with wild-type SLC30A10 resulted in partial reversal of Mn toxicity (Fig 3B). (We speculate that partial attenuation may be explained by 50-60% transfection efficiency of Hep3B cells (Fig 3A).) These observations are consistent with the known role of SLC30A10 in Mn export. They are also consistent with published reports that SLC30A10 overexpression in the neuronal cell line AF5 rendered the cells less sensitive to Mn toxicity and that over-expression of wild-type SLC30A10 reduced Mn accumulation in HeLa cells (Carmona et al. 2019; Leyva-Illades et al. 2014). As excess Mn is known to disrupt the mitochondrial electron transport chain, we speculate that Mn toxicity of Hep3B mutant cell lines reflected mitochondrial dysfunction. However, the MTT assay we used to assess cell viability may not exclusively reflect mitochondrial toxicity. Nevertheless, in the current study our objective was to observe the sensitivity and the Hep3B mutant cell line to Mn, Cu, and Zn, not to establish the specific mechanism of Mn-induced of cytotoxicity.
While we did observe impaired 54Mn export in the Hep3B mutant cell line, we also observed an unexpected reduction of 54Mn import in mutant cells compared to parental cells. To our knowledge, this import defect does not reflect any known physiological role for SLC30A10 in metal import. As SLC30A10 is expressed at the apical surface of hepatocytes and intestinal enterocytes, SLC30A10 deficiency would result in Mn deficiency in vivo if SLC30A10 played a role in cellular Mn import. SLC30A10 deficiency in patients (Mukhtiar et al. 2016; Quadri et al. 2012; Stamelou et al. 2012; Tuschl et al. 2012), mice (Hutchens et al. 2017; Liu et al. 2017; Mercadante et al. 2019; Taylor et al. 2019), and zebrafish (Xia et al. 2017) results in Mn excess, not deficiency.
We considered that lack of cell polarization contributed to aberrant 54Mn import in Hep3B mutant cells. Unlike in HepG2 and WIF-B cells, polarization in Hep3B cells has not been reported in the literature. Primary hepatocytes cultured in the manner we employed in this study do not polarize either (Zeigerer et al. 2017). However, we did not observe impaired 54Mn import in Slc30a10KO/KO cells compared to Slc30a10+/+ cells or Slc30a10lox/lox Alb+ cells compared to Slc30a10lox/lox cells. Furthermore, we did observe minimal 54Mn import in Slc39a14KO/KO cells compared to Slc39a14+/+ cells, consistent with the known physiological role of SLC39A14 for cellular Mn import (Aydemir et al. 2017; Jenkitkasemwong et al. 2018; Xin et al. 2017). Given these data, we propose that lack of cell polarization is not primarily responsible for aberrant 54Mn import in Hep3B mutant cells.
CRISPR/Cas9 is extensively used for genome editing due to its efficiency and cost effectiveness however it can also result in unintended genomic site cleavage resulting in off-target effects (Wang et al. 2020; Zhang et al. 2015). In addition to off-target effects of CRISPR-Cas9 genome editing, we also considered that the import defect in mutant cells could reflect a suppressor mutation leading to synthetic rescue. WGS did not identify any sequence differences in SLC11A2, SLC39A14, SLC39A8, or SLC40A1 between parental and mutant cell lines. We did observe other sequence differences in multiple other genes between parental and mutant cell lines, but these may reflect accumulation of mutations in Hep3B, a cancer-derived cell line, that are unrelated to the Mn import defect. In view of these unsolicited sequence changes, we caution the use of CRISPR-Cas9 editing as a means of functional genomic screening of SLC30A10 to provide mechanistic understanding or preclinical toxicity testing.
Cultured cancer-derived or immortalized cell lines have been used for decades in biomedical research due to the fact that they are highly proliferative and relatively easy to manipulate pharmacologically and genetically. However, serial passaging of such cell lines may lead to drift in genotypic or phenotypic characteristics. (The Hep3B parental cell line was purchased from ATCC and subjected to only a few passages before CRISPR editing was initiated. Mutant cell lines were passaged only a few times before WGS was performed.) Cell passage-dependent drift has been reported in cell cultures (Briske-Anderson et al. 1997; Chang-Liu and Woloschak 1997; Esquenet et al. 1997; Hughes et al. 2007; O'Driscoll et al. 2006). WGS data derived from Hep3B cells revealed many variants gained in the parental Hep3B cells. Such mutations may represent genomic instability inherent in all cancer lines and/or accumulation of mutations after the Hep3B cell line was originally established. These changes may lead to the cell line not accurately representing the in vivo function of the original cell type. In contrast, primary cells isolated from animal/human tissue have a finite lifespan and limited proliferative capacity but are known to maintain relevant in vivo characteristics for a limited time (Alge et al. 2006; Pan et al. 2009).
To further explore the relevance of aberrant 54Mn import in Hep3B mutant cells, we analyzed primary hepatocytes isolated from Slc30a10-deficient mice. (As mentioned above, 54Mn import was assessed in Slc39a14+/+ and Slc39a14KO/KO cells and found to be minimal in the mutant cells.) We first tried to isolate primary hepatocytes from Slc30a10KO/KO mice raised on a standard rodent diet containing 120 ppm Mn, however blood vessels in these mice ruptured soon after perfusion was initiated; possibility due to severity of phenotypes due to Mn overload. We were able to isolate hepatocytes from Slc30a10KO/KO mice raised on Mn-deficient diets. (This dietary approach lowers liver Mn levels but does not completely normalize them to wild-type liver Mn levels (data not shown). We did not measure Mn levels in primary hepatocytes isolated from these mice.) Primary hepatocytes from Slc30a10+/+ and Slc30a10KO/KO mice raised on Mn-deficient diets exhibited no changes in 54Mn import or export. The similarity in 54Mn export between wild-type and mutant cells was not unexpected. In our characterization of Slc30a10KO/KO mice (Mercadante et al. 2019), we observed no difference in total bile Mn levels in Slc30a10+/+ and Slc30a10KO/KO mice despite severe Mn excess in the mutant mice. This was in contrast to a severe increase in bile Mn levels in wild-type mice raised on Mn-rich diets. We interpreted this as indicating that Slc30a10 is essential for biliary Mn export and that Slc30a10 deficiency leads to inappropriately low, but not absent, biliary Mn export—in conditions of Mn excess, Slc30a10-deficient hepatocytes do excrete Mn but at insufficient levels given their burden of Mn. While we did isolate hepatocytes from Slc30a10KO/KO mice raised on Mn-deficient diets, this dietary approach did not normalize liver Mn levels to wild-type levels. We propose that 54Mn export in primary Slc30a10KO/KO hepatocytes may be influenced by hepatocyte Mn content and may not solely reflect Slc30a10 deficiency.
To mitigate the potential effect of Mn excess on 54Mn transport studies in primary hepatocytes, we next employed hepatocytes from mice with liver-specific Slc30a10 deficiency (Slc30a10lox/lox Alb+). Notably, these mutant mice develop mild liver Mn excess yet have minimal total bile Mn levels and after 54Mn injection into portal vein export minimal 54Mn into bile (Mercadante et al. 2019). Transport studies indicated intact 54Mn import and impaired 54Mn export in Slc30a10lox/lox Alb+ hepatocytes relative to Slc30a10lox/lox hepatocytes. Overall, relative to Hep3B cells and primary Slc30a10KO/KO hepatocytes, primary Slc30a10lox/lox Alb+ hepatocytes exhibit Mn import and export most consistent with in vivo observations in Slc30a10-deficient mice.
In conclusion, based upon our results, we propose that the impacts of Slc30a10 deficiency on Mn transport can be recapitulated using primary hepatocytes from mice that lack severe Mn excess such as Slc30a10lox/lox Alb+ mice. However, cancer-derived cell lines or primary hepatocytes from mice with severe Mn excess, such Slc30a10KO/KO mice, should be used with caution.
Supplementary Material
Table S3: List of primer sequences used in the study. (Quadri et al. 2012; Shimokawa et al. 2008; Steinmann et al. 2011; Wang and Seed 2003)
Table S1. List of variants gained in SLC30A10-mutant Hep3B cells when compared to parental Hep3B cells.
Table S2. List of variants lost in parental Hep3B cells when compared to human reference genome (GRCh37/HG19).
Figure S1. Normalized SLC30A10 RNA levels in different cell lines. Hep3B hepatoma showed highest expression of SLC30A10 when compared to others.
Figure S2. Targeted sequence analysis revealed a presence of non-synonymous mutation in exon 2 of Mn importer SLC39A14 in mutant cells (S2A) and parental Hep3B cells (S2B).
Figure S3. Line graphs representing picomolar concentrations (not normalized) of 54Mn released in the media during the export assay. Export assays were conducted on A) cancer cell-lines, B) Slc30a10-deficient mice, C) Slc30a10lox/lox Alb mice.
Acknowledgments:
We acknowledge: Dr. Mitchell Knutson, University of Florida for providing Slc39a14 knockout mice who originally obtained from Drs Hojyo and Fukada; Dr. Christoph Schorl and the Genomics Core for assistance with RNA analyses (this facility has received partial support from the National Institutes of Health (NIGMS P30GM103410, NCRR P30RR031153, P20RR018728, and S10RR02763), National Science Foundation (EPSCoR 0554548), Lifespan Rhode Island Hospital, and the Division of Biology and Medicine, Brown University); Drs. Joseph Orchardo and David Murray for assistance with metal analysis; and Dr. Jennifer Sanders for assistance with primary hepatocyte isolation.
Funding:
Research was supported by National Institute of Health grants DK84122 and DK110049 (TBB).
Footnotes
Conflicts of interest/Competing interests: The authors declare no conflict of interest.
Ethics approval: Studies using animals were approved by the Institutional Animal Care and Use Committee at Brown University.
Consent to participate: N/A
Consent for publication: All authors agreed with the content and gave explicit consent to submit and publish this manuscript.
Availability of data and material: Authors are willing to share cell lines, datasets, and protocols employed. Further information and requests for resources should be directed to lead contact at thomas_bartnikas@brown.edu
Code availability: N/A
References:
- Alge CS, Hauck SM, Priglinger SG, Kampik A, Ueffing M (2006) Differential protein profiling of primary versus immortalized human RPE cells identifies expression patterns associated with cytoskeletal remodeling and cell survival J Proteome Res 5:862–878 doi: 10.1021/pr050420t [DOI] [PubMed] [Google Scholar]
- Anagianni S, Tuschl K (2019) Genetic Disorders of Manganese Metabolism Curr Neurol Neurosci Rep 19:33 doi: 10.1007/s11910-019-0942-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschner JL, Aschner M (2005) Nutritional aspects of manganese homeostasis Mol Aspects Med 26:353–362 doi: 10.1016/j.mam.2005.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila DS, Puntel RL, Aschner M (2013) Manganese in health and disease Met Ions Life Sci 13:199–227 doi: 10.1007/978-94-007-7500-8_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydemir TB et al. (2017) Metal Transporter Zip14 (Slc39a14) Deletion in Mice Increases Manganese Deposition and Produces Neurotoxic Signatures and Diminished Motor Activity J Neurosci 37:5996–6006 doi: 10.1523/JNEUROSCI.0285-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran RC et al. (2020) Brain manganese and the balance between essential roles and neurotoxicity J Biol Chem 295:6312–6329 doi: 10.1074/jbc.REV119.009453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosomworth HJ, Thornton JK, Coneyworth LJ, Ford D, Valentine RA (2012) Efflux function, tissue-specific expression and intracellular trafficking of the Zn transporter ZnT10 indicate roles in adult Zn homeostasis Metallomics 4:771–779 doi: 10.1039/c2mt20088k [DOI] [PubMed] [Google Scholar]
- Boylan JM, Francois-Vaughan H, Gruppuso PA, Sanders JA (2017) Engraftment and Repopulation Potential of Late Gestation Fetal Rat Hepatocytes Transplantation 101:2349–2359 doi: 10.1097/TP.0000000000001882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briske-Anderson MJ, Finley JW, Newman SM (1997) The influence of culture time and passage number on the morphological and physiological development of Caco-2 cells Proc Soc Exp Biol Med 214:248–257 doi: 10.3181/00379727-214-44093 [DOI] [PubMed] [Google Scholar]
- Cabral F, Miller CM, Kudrna KM, Hass BE, Daubendiek JG, Kellar BM, Harris EN (2018) Purification of Hepatocytes and Sinusoidal Endothelial Cells from Mouse Liver Perfusion Journal of visualized experiments : JoVE doi: 10.3791/56993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona A et al. (2019) SLC30A10 Mutation Involved in Parkinsonism Results in Manganese Accumulation within Nanovesicles of the Golgi Apparatus ACS Chem Neurosci 10:599–609 doi: 10.1021/acschemneuro.8b00451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang-Liu CM, Woloschak GE (1997) Effect of passage number on cellular response to DNA-damaging agents: cell survival and gene expression Cancer Lett 113:77–86 doi: 10.1016/s0304-3835(97)04599-0 [DOI] [PubMed] [Google Scholar]
- Esquenet M, Swinnen JV, Heyns W, Verhoeven G (1997) LNCaP prostatic adenocarcinoma cells derived from low and high passage numbers display divergent responses not only to androgens but also to retinoids J Steroid Biochem Mol Biol 62:391–399 doi: 10.1016/s0960-0760(97)00054-x [DOI] [PubMed] [Google Scholar]
- Hojyo S, Fukada T, Shimoda S, Ohashi W, Bin BH, Koseki H, Hirano T (2011) The zinc transporter SLC39A14/ZIP14 controls G-protein coupled receptor-mediated signaling required for systemic growth PLoS One 6:e18059 doi: 10.1371/journal.pone.0018059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes P, Marshall D, Reid Y, Parkes H, Gelber C (2007) The costs of using unauthenticated, over-passaged cell lines: how much more data do we need? Biotechniques 43:575, 577-578, 581-572 passim doi: 10.2144/000112598 [DOI] [PubMed] [Google Scholar]
- Hutchens S et al. (2017) Deficiency in the manganese efflux transporter SLC30A10 induces severe hypothyroidism in mice J Biol Chem 292:9760–9773 doi: 10.1074/jbc.M117.783605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan AT, Azam M, Siddiqui K, Ali A, Choi I, Haq QM (2015) Heavy Metals and Human Health: Mechanistic Insight into Toxicity and Counter Defense System of Antioxidants Int J Mol Sci 16:29592–29630 doi: 10.3390/ijms161226183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkitkasemwong S et al. (2018) SLC39A14 deficiency alters manganese homeostasis and excretion resulting in brain manganese accumulation and motor deficits in mice Proceedings of the National Academy of Sciences of the United States of America 115:E1769–E1778 doi: 10.1073/pnas.1720739115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Elkoshi N, Barber-Zucker S, Hoch E, Zarivach R, Hershfinkel M, Sekler I (2019) Zinc transporter 10 (ZnT10)-dependent extrusion of cellular Mn(2+) is driven by an active Ca(2+)-coupled exchange J Biol Chem 294:5879–5889 doi: 10.1074/jbc.RA118.006816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva-Illades D et al. (2014) SLC30A10 is a cell surface-localized manganese efflux transporter, and parkinsonism-causing mutations block its intracellular trafficking and efflux activity J Neurosci 34:14079–14095 doi: 10.1523/JNEUROSCI.2329-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform Bioinformatics 26:589–595 doi: 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WC, Ralphs KL, Tosh D (2010) Isolation and culture of adult mouse hepatocytes Methods Mol Biol 633:185–196 doi: 10.1007/978-1-59745-019-5_13 [DOI] [PubMed] [Google Scholar]
- Lin W et al. (2017) Hepatic metal ion transporter ZIP8 regulates manganese homeostasis and manganese-dependent enzyme activity J Clin Invest 127:2407–2417 doi: 10.1172/JCI90896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C et al. (2017) Hypothyroidism induced by loss of the manganese efflux transporter SLC30A10 may be explained by reduced thyroxine production J Biol Chem 292:16605–16615 doi: 10.1074/jbc.M117.804989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercadante CJ et al. (2019) Manganese transporter Slc30a10 controls physiological manganese excretion and toxicity J Clin Invest doi: 10.1172/JCI129710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtiar K, Ibrahim S, Tuschl K, Mills P (2016) Hypermanganesemia with Dystonia, Polycythemia and Cirrhosis (HMDPC) due to mutation in the SLC30A10 gene Brain Dev 38:862–865 doi: 10.1016/j.braindev.2016.04.005 [DOI] [PubMed] [Google Scholar]
- O'Driscoll L, Gammell P, McKiernan E, Ryan E, Jeppesen PB, Rani S, Clynes M (2006) Phenotypic and global gene expression profile changes between low passage and high passage MIN-6 cells J Endocrinol 191:665–676 doi: 10.1677/joe.1.06894 [DOI] [PubMed] [Google Scholar]
- Pan C, Kumar C, Bohl S, Klingmueller U, Mann M (2009) Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions Mol Cell Proteomics 8:443–450 doi: 10.1074/mcp.M800258-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH et al. (2015) SLC39A8 Deficiency: A Disorder of Manganese Transport and Glycosylation Am J Hum Genet 97:894–903 doi: 10.1016/j.ajhg.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadri M et al. (2012) Mutations in SLC30A10 cause parkinsonism and dystonia with hypermanganesemia, polycythemia, and chronic liver disease Am J Hum Genet 90:467–477 doi: 10.1016/j.ajhg.2012.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa T, Tostar U, Lauth M, Palaniswamy R, Kasper M, Toftgard R, Zaphiropoulos PG (2008) Novel human glioma-associated oncogene 1 (GLI1) splice variants reveal distinct mechanisms in the terminal transduction of the hedgehog signal J Biol Chem 283:14345–14354 doi: 10.1074/jbc.M800299200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamelou M, Tuschl K, Chong WK, Burroughs AK, Mills PB, Bhatia KP, Clayton PT (2012) Dystonia with brain manganese accumulation resulting from SLC30A10 mutations: a new treatable disorder Mov Disord 27:1317–1322 doi: 10.1002/mds.25138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann K, Richter AM, Dammann RH (2011) Epigenetic silencing of erythropoietin in human cancers Genes Cancer 2:65–73 doi: 10.1177/1947601911405043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JG et al. (2019) COSMIC: the Catalogue Of Somatic Mutations In Cancer Nucleic Acids Res 47:D941–D947 doi: 10.1093/nar/gky1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CA et al. (2019) SLC30A10 transporter in the digestive system regulates brain manganese under basal conditions while brain SLC30A10 protects against neurotoxicity J Biol Chem 294:1860–1876 doi: 10.1074/jbc.RA118.005628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson KJ, Hein J, Baez A, Sosa JC, Wessling-Resnick M (2018) Manganese transport and toxicity in polarized WIF-B hepatocytes Am J Physiol Gastrointest Liver Physiol 315:G351–G363 doi: 10.1152/ajpgi.00103.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuschl K et al. (2012) Syndrome of hepatic cirrhosis, dystonia, polycythemia, and hypermanganesemia caused by mutations in SLC30A10, a manganese transporter in man Am J Hum Genet 90:457–466 doi: 10.1016/j.ajhg.2012.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuschl K et al. (2016) Mutations in SLC39A14 disrupt manganese homeostasis and cause childhood-onset parkinsonism-dystonia Nat Commun 7:11601 doi: 10.1038/ncomms11601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Seed B (2003) A PCR primer bank for quantitative gene expression analysis Nucleic Acids Res 31:e154 doi: 10.1093/nar/gng154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y et al. (2020) Specificity profiling of CRISPR system reveals greatly enhanced off-target gene editing Sci Rep 10:2269 doi: 10.1038/s41598-020-58627-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z et al. (2017) Zebrafish slc30a10 deficiency revealed a novel compensatory mechanism of Atp2c1 in maintaining manganese homeostasis PLoS Genet 13:e1006892 doi: 10.1371/journal.pgen.1006892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Y et al. (2017) Manganese transporter Slc39a14 deficiency revealed its key role in maintaining manganese homeostasis in mice Cell Discov 3:17025 doi: 10.1038/celldisc.2017.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigerer A, Wuttke A, Marsico G, Seifert S, Kalaidzidis Y, Zerial M (2017) Functional properties of hepatocytes in vitro are correlated with cell polarity maintenance Exp Cell Res 350:242–252 doi: 10.1016/j.yexcr.2016.11.027 [DOI] [PubMed] [Google Scholar]
- Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH (2015) Off-target Effects in CRISPR/Cas9-mediated Genome Engineering Mol Ther Nucleic Acids 4:e264 doi: 10.1038/mtna.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogzas CE, Mukhopadhyay S (2018) Putative metal binding site in the transmembrane domain of the manganese transporter SLC30A10 is different from that of related zinc transporters Metallomics 10:1053–1064 doi: 10.1039/c8mt00115d [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3: List of primer sequences used in the study. (Quadri et al. 2012; Shimokawa et al. 2008; Steinmann et al. 2011; Wang and Seed 2003)
Table S1. List of variants gained in SLC30A10-mutant Hep3B cells when compared to parental Hep3B cells.
Table S2. List of variants lost in parental Hep3B cells when compared to human reference genome (GRCh37/HG19).
Figure S1. Normalized SLC30A10 RNA levels in different cell lines. Hep3B hepatoma showed highest expression of SLC30A10 when compared to others.
Figure S2. Targeted sequence analysis revealed a presence of non-synonymous mutation in exon 2 of Mn importer SLC39A14 in mutant cells (S2A) and parental Hep3B cells (S2B).
Figure S3. Line graphs representing picomolar concentrations (not normalized) of 54Mn released in the media during the export assay. Export assays were conducted on A) cancer cell-lines, B) Slc30a10-deficient mice, C) Slc30a10lox/lox Alb mice.