Abstract
Background
Consanguineous families have a relatively high prevalence of genetic disorders caused by bi-allelic mutations in recessive genes. This study aims to evaluate the effectiveness and efficiency of a consanguinity-based exome sequencing approach to capturing genetic mutations in inherited retinal dystrophy families with consanguineous marriages.
Methods
Ten unrelated consanguineous families with a proband affected by inherited retinal dystrophy were recruited in this study. All participants underwent comprehensive ophthalmic examinations. Whole exome sequencing was performed, followed by a homozygote-prior strategy to rapidly filter disease-causing mutations. Bioinformatic prediction of pathogenicity, Sanger sequencing and co-segregation analysis were carried out for further validation.
Results
In ten consanguineous families, a total of 10 homozygous mutations in 8 IRD genes were identified, including 2 novel mutations, c.1654_1655delAG (p. R552Afs*5) in gene FAM161A in a patient diagnosed with retinitis pigmentosa, and c.830T > C (p.L277P) in gene CEP78 in a patient diagnosed with cone and rod dystrophy.
Conclusion
The genetic etiology in consanguineous families with IRD were successfully identified using consanguinity-based analysis of exome sequencing data, suggesting that this approach could provide complementary insights into genetic diagnoses in consanguineous families with variant genetic disorders.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13023-021-01902-5.
Keywords: Inherited retinal dystrophy, Consanguinity, Exome sequencing, Genetic mutation
Background
Inherited retinal dystrophy (IRD) is a group of severe and irreversible vision-threatening disorders, affecting approximately 2 million people worldwide [1]. It is characterized by degeneration and dysfunction of specialized and light-sensitive neurons in the retina, such as cone and/or rod photoreceptor cells [2–4]. IRD is comprised of a wide spectrum of diseases with enormous genetic heterogeneity [1, 5–7]. The most common form of IRD is retinitis pigmentosa (RP), which is already known to be attributed to the primary loss of rods, followed by secondary loss of cones. RP initially affects night and peripheral vision, but eventually, patients will experience central vision impairment [8, 9]. Other diseases in this group include cone and rod dystrophy (CORD), Leber congenital amaurosis (LCA), Bietti crystalline dystrophy (BCD) and syndromic diseases such as Usher syndrome and Bardet-Biedl syndrome [10–13]. Currently, more than 250 disease-causing genes have been identified in patients with IRD. Transmission of these genes has been shown to resemble Mendelian traits, including autosomal recessive (AR), autosomal dominant (AD), and X-linked (XL) modes of inheritance. Although great efforts have been made to dissect the genetic basis of IRD, the genetic causes of IRD in a large proportion of patients remain unknown [14–16].
Many communities around the world have a long tradition of marrying to relatives. The highest prevalence of inbreeding was observed in North and sub-Saharan Africa, the Middle East, and West, Central, and South Asia [17]. In the rural areas of China, consanguineous marriages such as cousin couples are relatively common, especially among minority ethnic groups. According to data from a population-based survey in the 1990s, the average consanguinity marriage rate among Chinese Han population was 1.4% and it was significantly higher in rural areas than in urban areas, while among Yi population which is one of the major ethnic minority of China, the rate was as high as 14.6% [18]. In the recent decade, nationwide quantitative information on consanguinity is not available, but some population surveys conducted in ethnic minority settlements indicate that the situation of consanguineous marriage has not changed much.
Consanguinity increases the likelihood of pathogenic mutations in a homoallelic state. A higher prevalence of rare hereditary diseases caused by homozygous mutations in AR genes has been observed among members of consanguineous families compared to those of non-consanguineous families [19, 20]. Several studies have also reported genetic causes of IRDs among consanguineous families worldwide, especially in the areas where consanguinity rate is particularly high [21–24].
However, few studies were performed among patients with IRD in Chinese consanguineous families. Our in-house data show that about 11% of IRD probands have consanguineous parents. Therefore, there is a need for genetic screening and counseling among with IRD patients in Chinese consanguineous families.
To date, next-generation sequencing (NGS) techniques have been widely applied to the research of IRDs, among which whole exome sequencing (WES) has demonstrated the greatest reliability and efficiency in testing genetic mutations [25–27]. In this study, deep sequencing of all causative genes of IRD was performed on ten probands, and WES data were analyzed with a consanguinity-based analysis strategy. This study not only broadened the genetic mutation spectrum but also enriched knowledge on the genotype–phenotype correlations of IRDs, suggesting that molecular genetic testing using a consanguinity-based approach is of great value for the diagnosis of IRDs.
Methods
Subjects and clinical evaluations
This study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of Beijing Tongren Hospital. Informed consent was acquired from all participating subjects. Ten probands, together with one affected sibling and 23 unaffected family members were recruited from ten unrelated consanguineous families. Medical history was obtained and comprehensive ophthalmic examinations, including fundus photography, optical coherence tomography (OCT), visual field testing, and electroretinogram (ERG), were performed. Peripheral blood samples from all participants were collected.
Exome sequencing and homozygote strategy
Exome sequencing and subsequent procedures, including quality control and variation annotation, were performed as previously described [28–31]. Homozygous mutant alleles in all 281 known IRD genes were filtered first. Known IRD genes were gathered based on RetNet [32] (https://sph.uth.edu/RETNET/), OMIM (https://www.omim.org/) and numerous scientific literatures. After the implementation of the homozygote-prior filtering strategy, the remaining variants were confirmed by Sanger sequencing. Co-segregation analysis was performed in all available subjects from the ten families registered in this study.
Assessment of pathogenicity
Variants with a minor allele frequency (MAF) of > 0.5% for recessive genes and > 0.01% for dominant genes in the 1000 Genomes Project (1000G), Exome Aggregation Consortium (ExAC) and Genome Aggregation Database (gnomAD) were excluded. The pathogenicity of each candidate variant was predicted by online analysis programs. Missense variants were evaluated with Mutation Taster (http://www.mutationtaster.org/), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), SIFT (http://provean.jcvi.org/protein) and CADD (https://cadd.gs.washington.edu/). Predicted crystal structures of the wild-type proteins and mutants were obtained using SWISS-MODEL (http://swissmodel.expasy.org) and demonstrated by PyMol software. Finally, the pathogenicity of variants was categorized as pathogenic, likely-pathogenic, uncertain significance, benign and likely benign according to ACMG (American College of Medical Genetics and Genomics) classification guidelines [33].
Results
Clinical features
In the current study, six probands were diagnosed with RP, while the other four probands were non-RP and their diagnoses included CORD, BCD, and LCA (Table 1).
Table 1.
Clinical features of 10 probands
| ID | Sex | Diagnosis | Initial sympton | NYS | Age of onset (y) | Age at visit (y) | BCVA (decimal) OD/OS | ERG (scotopic/photopic) | VF |
|---|---|---|---|---|---|---|---|---|---|
| F1-III:4 | M | RP, HM | Night blindness | N | 39 | 46 | 0.4/0.6 | NA | Constricted |
| F2-III:3 | M | RP, MC | Night blindness. Central vision impairment | N | < 1 | 27 | 0.1/0.1 | Both severely reduced | NA |
| F3-III:5 | F | RP | Night blindness | N | 20 | 51 | LP | NA | NA |
| F4-III:4 | F | RP | Night blindness | N | 20 | 60 | HM/HM | Both severely reduced | NA |
| F5-III:1 | M | RP | Night blindness | N | 14 | 34 | 0.5/0.6 | Both severely reduced | Constricted |
| F6-III:1 | F | RP | Night blindness | N | 6 | 23 | 0.8/0.6 | Severely reduced/slightly reduced | Constricted |
| F7-III:4 | M | CORD, POAG | Central vision impairment | N | 40 | 45 | CF at 1 m/0.02 | Both severely reduced | Central scotoma |
| F8-III:5 | F | CORD | Central vision impairment | N | 32 | 43 | CF at 40 cm/0.01 | Both severely reduced, greater loss in cone | Central scotoma |
| F9-III:2 | M | BCD | Night blindness | N | 43 | 44 | 0.6/0.8 | Severely reduced/slightly reduced | Constricted |
| F10-III:1 | F | LCA | Vision impairment | Y | < 1 | 29 | HM/HM | Extinguished | NA |
NYS, Nystagmus; M, male; F, female; RP, retinitis pigmentosa; HM, high myopia; MC, coloboma-like lesions in macula; CORD, cone and rod dystrophy; POAG, primary open-angle glaucoma; BCD, Bietti crystalline dystrophy; BCVA, best corrected visual acuity; OD, right eye; OS, left eye; VF, visual field; ERGs, Electroretinograms; NA, not available
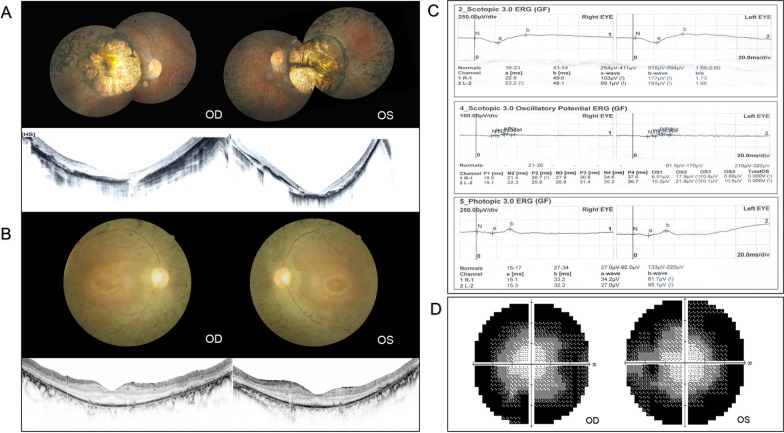
Among the six RP probands, four presented as classic RP (F3-III:5, F4-III:4, F5-III:1 and F6-III:1). They complained of night blindness as the initial symptom, with progressive constriction of visual field presenting in the following years. Typical RP manifestations were observed in proband F5-III:1 (Fig. 1A). Atypical RP was observed in proband F1-III:4 and F2-III:3. F1-III:4 was diagnosed with RP and high myopia, and manifested typical RP fundus combined with a large myopic conus and a tigroid fundus change. F2-III:3 had atypical RP with involvement of the macula. Fundus photography demonstrated bilateral prominent coloboma-like lesions in the macula. OCT showed a severely thinned macula structure and lack of full-thickness in the inner and outer segments of photoreceptors (Fig. 1).
Fig. 1.
Clinical manifestations of typical and atypical RP patients. A Fundus photos of F2-III:3 presented bilateral prominent macular atrophy, with chorioretinal attenuation and extensive bone spicule pigmentation. OCT showed severe thinning of macular structure along with loss of photoreceptors. B–D Clinical manifestations of F5-III:1. B Fundus photos showed typical RP presentations, including waxy pallor disc, attenuated retinal vasculature, and mid-peripheral bone-spicule pigmentation. OCT illustrated severe loss of photoreceptors with preservation in the fovea. C Full-field electroretinogram (ERG) demonstrated a reduced rod and cone response amplitude. D Constricted visual field
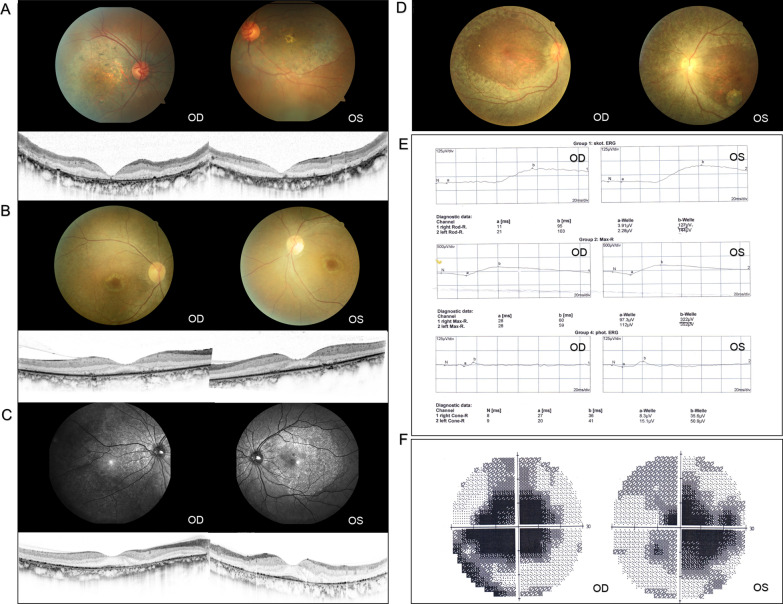
Out of four probands with non-RP, F7-III:4 and F8-III:5 were diagnosed with CORD. They suffered a decrease of central vision in their 3rd or 4th decade, and BCVA had declined to 0.02 and 0.01 (decimal) respectively at their visit time. F7-III:4 was diagnosed with CORD combined with primary open-angle glaucoma (POAG). Fundus photography revealed an enlargement of the optic cup with a cup to disc ratio (C/D) as 0.4, a decrease of neuroretinal rim width in the inferior and superior, and a distinct appearance of maculopathy, including pigment degeneration and deposition over the macular region. OCT showed the absence of foveal photoreceptors and retinal pigment epithelium (RPE). In addition, the visual field test revealed a serious impairment of the central visual field. The full-field ERG showed that the amplitude of cones became evidently flat while that of rods declined slightly (Fig. 2A, D, E). HRT F8-III:5 was diagnosed with CORD and presented loss of foveal photoreceptor with a blurred outer layer structure (Fig. 2B), and ERG showed a greater loss of cone responses compared with rod responses.
Fig. 2.
Clinical manifestations of non-RP patients. A Fundus photos of F7-III:4 diagnosed with CORD revealed an enlargement of the optic cup with a C/D of 0.4, decrease of neuroretinal rim width in the inferior and superior, attenuated retinal vasculature, and retinal pigment epithelium (RPE) changes over the macular region. OCT showed the absence of foveal photoreceptors and RPE. B Fundus and OCT of F8-III:5 diagnosed with CORD presented Bull’s eye maculopathy with loss of foveal photoreceptor and a blurred outer layer structure. C Fundus and OCT images of F9-III:2 diagnosed with BCD showed bilateral multiple crystalline deposits and RPE dystrophy with loss of photoreceptors. D Fundus images of F10-III:1 diagnosed with LCA exhibited show a waxy pallor optic disc, attenuated retinal vasculature, and extensive chorioretinal atrophy. E Full-field ERG of F7-III:4 manifested a significantly decreased amplitude of cones while the response of rods declined slightly in both eyes. F Visual field test of F7-III:4 revealed a serious impairment of the central visual field
F9-III:2 and F10-III:1 were diagnosed as BCD and LCA respectively, and typical disease manifestations such as multiple crystalline deposits in BCD and extensive chorioretinal atrophy in LCA were observed (Fig. 2).
Molecular diagnosis
A gene list comprised of 281 known IRD-causing genes was screened and evaluated for implicated mutations by whole exome sequencing, with an overall average coverage of over 150 X (Additional file 1: Table S1).
Through this homozygote-prior strategy, the disease-causing mutations in all 10 families were identified in a highly efficient manner, with a total of 10 homozygous mutations in 8 IRD genes determined, including 8 previously reported mutations, and 2 novel mutations involving genes FAM161A in patient F1-III:4 with RP and CEP78 in F8-III:5 with CORD (Table 2, Fig. 3). The identified mutations segregated with phenotype within all pedigrees.
Table 2.
Genetic mutations identified in this study
| ID | Gene | Mutation | Protein change | Transcript | MuationTaster | Poly Phen-2 | Sift | CADD | gnomAD | References |
|---|---|---|---|---|---|---|---|---|---|---|
| F1-III:4 | FAM161A | c.1654_1655delAG | p.R552Afs*5 | NM_032180.3 | D | / | / | / | 0 | Novel |
| F2-III:3 | RDH12 | c.437 T > A | p.V146D | NM_152443.3 | D | D | D | 27 | 0.00001193 | Gong et al. [34] |
| F3-III:5 | USH2A | c.9469C > T | p.Q3157X | NM_206933.4 | D | / | / | 37 | 0.00001631 | Jiang et al. [35] |
| F4-III:4 | EYS | c.7228 + 1G > A | splicing | NM_001142800.2 | D | / | / | 25.9 | 0.00002783 | Gu et al. [36] |
| F5-III:1 | EYS | c.6174 T > G | p.Y2058X | NM_001142800.2 | D | / | / | 37 | 0 | ClinVar submitted |
| F6-III:1 | USH2A | c.9570 + 1G > A | splicing | NM_206933.4 | D | / | / | 24.6 | 0.0000408 | Xu et al. [37] |
| F7-III:4 | CRB1 | c.3991C > T | p.R1331C | NM_201253.3 | D | D | D | 32 | 0.00001989 | Tsang et al. [38] |
| F8-III:5 | CEP78 | c.830 T > C | p.L277P | NM_032171.3 | P | D | D | 23.2 | 0.00003244 | Novel |
| F9-III:2 | CYP4V2 | c.1091-2A > G | splicing | NM_207352.4 | D | / | / | 33 | 0.00004771 | Yin et al. [39] |
| F10-III:1 | IQCB1 | c.445_448delCTCT | p.L149Sfs*30 | NM_001023570.4 | D | / | / | / | 0 | Otto et al. [40] |
Homo, homozygous; Mutation Taster-D, disease causing; P, polymorphism; Polyphen-2-D, probably damaging; Sift-D, damaging
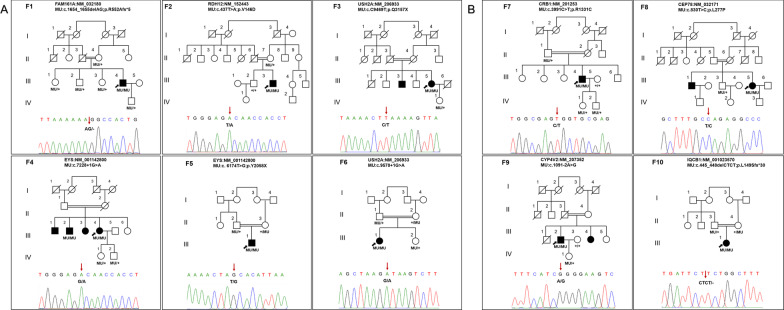
Fig. 3.
Pedigrees and segregation data. Pedigrees and segregation data of families diagnosed with RP and non-RP are illustrated in A and B, respectively. Squares and circles represent males and females. Filled symbols represent the affected patients. Arrows indicate the probands. MU: mutation. + : wildtype
The deletion of nucleotides in FAM161, c.1654_1655delAG (p. R552Afs*5) resulted in frameshift mutations and affected the normal RNA maturation process, leading to a production of truncated protein. This frameshift mutation had a severe impact on protein function and was absent in the gnomAD database, and therefore, by definition in ACMG guidelines, was a pathogenic mutation.
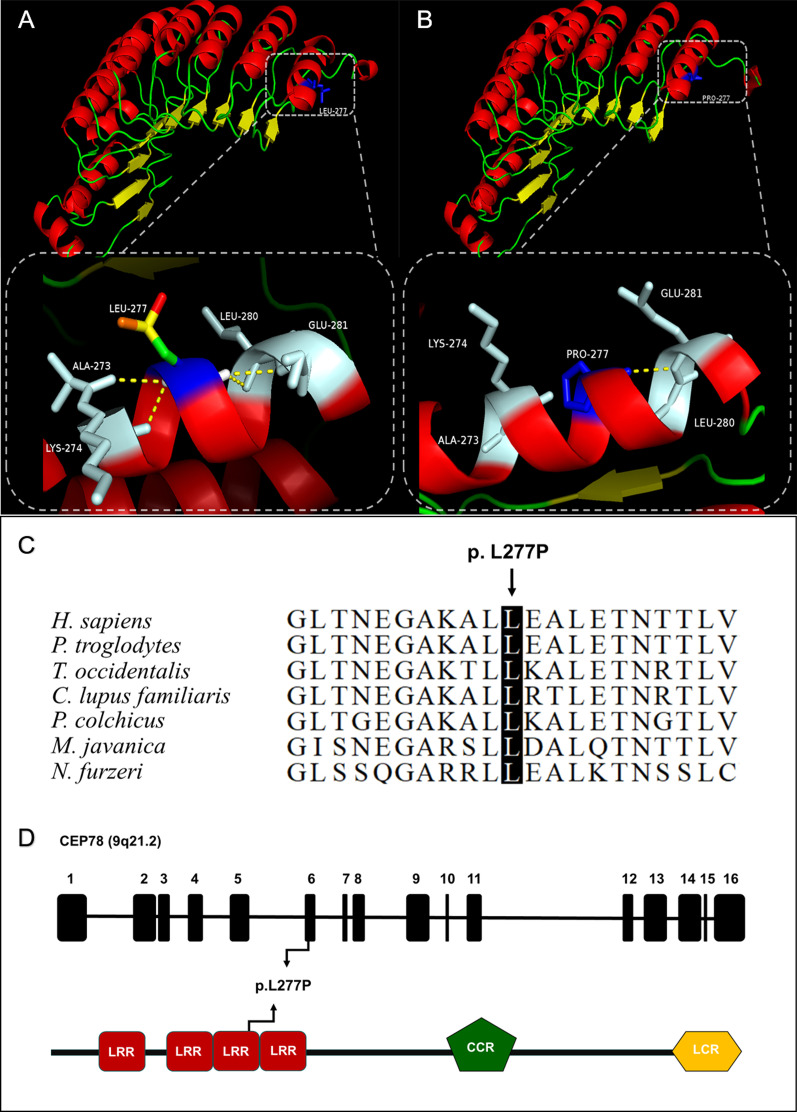
F8-III:5 was found to harbor a homozygous novel missense mutation, c.830T>C (p.L277P) in CEP78. Leucine acid (LEU) at residue 277 is located in a region of a highly conserved domain called leucine-rich repeat (LRR), which presents in a number of proteins with diverse functions. Although Mutation Taster defines the mutation as polymorphism, Sift, Poly-Phen 2, Mutation Assessor and CADD all define it as pathogenic. And the variant had an extremely low allele frequency in healthy control cohort (Table 2). The three-dimensional protein crystal model of CEP78 was constructed to verify the structure impact of p.L277P with PyMOL. As the model showed, the substitution of LEU for proline acid (PRO) resulted in the loss of three hydrogen bonds between the wildtype LEU at residue 277 and Alanine acid (ALA) at residue 273, Lysine acid (LYS) at residue 274, and LEU at residue 280 (Fig. 4). Considering the above, c.830T>C (p.L277P) in CEP78 is categorized as likely pathogenic according to the ACMG guideline (PM2+PM3+PP3+PP4).
Fig. 4.
Evaluation of L277P mutation in CEP78. A Presentation of wildtype CEP78 protein 3D structure. B Mutant status of CEP78 protein. The amino acid at position 277 was mutated from leucine (LEU) to proline acid (PRO), leading to the loss of three hydrogen bonds between the wildtype LEU at residue 277 and Alanine acid (ALA) at residue 273, Lysine acid (LYS) at residue 274, and LEU at residue 280. C Alignment of CEP78 protein sequence from amino acid 267 to 287 to its orthologous protein sequences in different species indicated evolutionary conservation of leucine at position 277 in human CEP78. D Representations of relative linear locations of the L277P mutation in genome structure (top) and protein domains (bottom)
Discussion
Consanguineous marriage is prevalent in rural China, which could potentially increase the risk of suffering from hereditary diseases. In consanguineous families, genetic defects that are caused by homozygous mutations inherited respectively from both parents who are carriers of the heterozygous mutation often manifest in the offspring. Incidence of IRDs, for example, could lead to devastating visual disability and negative livelihood impact. However, few studies focused on Chinese consanguineous families with IRDs. To broaden the knowledge on this population, deep exome sequencing and consanguinity-based analysis was performed in order to elucidate the genetic etiology in patients with IRD.
In this study, 10 homozygous disease-causing mutations were successfully identified. As for the genetic pathogenesis of four typical RP families, USH2A and EYS mutations accounted for 50%, which was consistent with our previous study showing that USH2A and EYS are among the most common disease-causing genes of ARRP in Chinese population [14].
F1-III:4 was diagnosed with RP and high myopia and was identified to harbor a novel homozygous mutation c.1654_1655delAG (p. R552Afs*5) within FAM161A. To date, FAM161A mutations have only been linked to ARRP, and mostly are in the form of null mutations [41]. A previous study did observe that RP patients associated with FAM161A mutation manifested moderate-high myopia, which was consistent with our observation [42]. The protein encoded by FAM161A is evolutionarily conserved in vertebrates such as Pan troglodytes, Macaca mulatta, Equus asinus and Sarcophilus harrisii. It localizes to photoreceptor connecting cilia in human and ciliary basal body in mammalian ciliated cells. Recent research has demonstrated that this protein is involved in microtubule acetylation and stabilization by maintaining transport processes in photoreceptors as a component of the cilia-basal body complex [43, 44]. FAM161A directly interacts with cilia proteins CEP290, SDCCAG8, OFD1 and lebercilin, all of which are implicated in heterogeneous IRDs [45].
CEP78 has been reported to be related to the pathogenesis of Usher syndrome or cone dystrophy along with sensory hearing loss in recent studies [46, 47]. Patient F8-III:5 in our study had a homozygous missense mutation c.830T > C (p.L277P) in CEP78 and presented cone and rod dystrophy without hearing impairment. The mutation is very likely to be the genetic cause based on the evidences above, but the phenotype is unusual as all patients reported to harbor CEP78 mutations have hearing impairment. Our report of the clinical manifestation indicates that CEP78-related IRDs may have a high clinical heterogeneity. However, further research and clinical observation are needed.
A frameshift mutation c.445-448delCTCT (p. Leu149Serfs*31) in gene IQCB1 was determined in F10-III:1, a patient who was diagnosed with LCA. This mutation was pathogenic according to ACMG guidelines, and has been reported in two unrelated German families to be related to Senior-Loken syndrome [40]. Senior-Loken syndrome is a rare autosomal recessive ciliopathy characterized by the association of nephronophthisis (NPHP) and early onset retinal dystrophy. Age of onset, severity and progression of symptoms can vary greatly among affected individuals. Patient F10-III:1 suffered from severe vision impairment in the first year of her life and no kidney disorder was found in the following 30 years. This may be a novel correlation between IQCB1 mutation and complex clinical manifestations. However, regular follow-up at nephrology department is needed.
F2-III:3 was characterized by a rare type of RP associated with macular and choroid coloboma, and had a homozygous mutation c.437T > A (p.V146D) in RDH12. RDH12 coded protein is predominantly expressed in the inner segment of rod and cone photoreceptors as an NADPH-dependent reductase involved in the conversion of all-trans-retinal and 11-cis-retinal to the corresponding retinols [48]. This p.V146D mutation was previously reported in severe early-onset ARRP family [34]. In contrast, the proband in this current study exhibited additional phenotypes. A prior study also indicated that RDH12 mutation could lead to LCA with macular coloboma [49]. However, in our study, characteristic features of LCA, including nystagmus, photophobia, amaurotic pupils and flat or nondetectable ERG, were all absent [50].
F7-III:4 was diagnosed with CORD and harbored a homozygous mutation c.3991C > T (p.R1331C) in CRB1. CRB1 localizes to the outer limiting membrane of the retina and is expressed in adherens junction in Muller glia cells, which may control retinogenesis via signaling pathway such as mTOR, Hippo, Wnt and Notch1 [51].The p.R1331C mutation was reported to have caused unusual maculopathy in an Irish family [38]. In contrast, F7-III:4 had a definite diagnosis of CORD with a wider range of fundus atrophy and evident ERG performance. Furthermore, the patient suffered from more severe binocular glaucoma and cataract. Our findings have verified the implication of the RP or LCA-related CRB1 mutation in maculopathy in a Chinese individual.
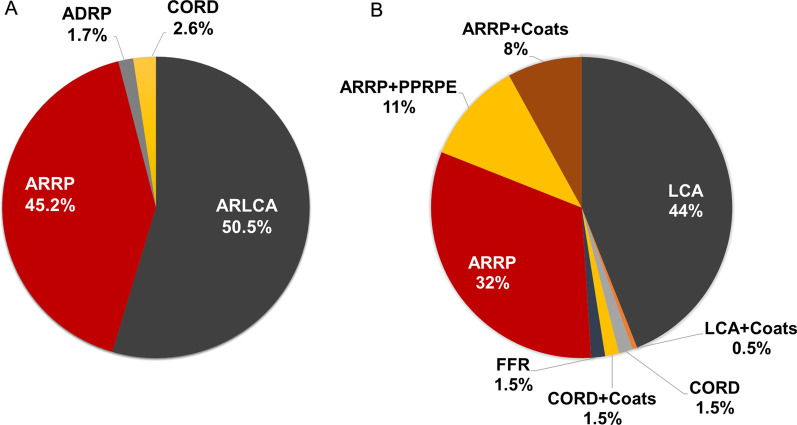
To date, more than 110 mutations within RDH12 have been classified to be responsible for either RP, LCA or CORD. And more than 150 CRB1 mutations are shown to be associated with specific fundus features other than RP or LCA, such as preserved para-arteriole retinal pigment epithelium (PPRPE) and retinal telangiectasia with exudation (Coats-like exudative vasculopathy) [52, 53]. Moreover, some mutations within CRB1 were also causative of Familial foveal Retinoschisis (FFR) [54]. We reviewed RDH12 and CRB1-related disorders from public databases such as ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and HGMD (http://www.hgmd.cf.ac.uk), and summarized the proportion of heterogeneous phenotypes correlated with RDH12 and CRB1 mutations in Fig. 5. Some researches indicate that additional modifying factors could be responsible for the absence of clear correlation between mutations and phenotypes. For example, a previous study has demonstrated an earlier onset and more severe retinal phenotype when Mthfr and rd8 mutations coexist in mice, indicating that Mthfr is a modify factors of Crb1 [55].
Fig. 5.
Proportion of correlated phenotypes of mutated RDH12 and CRB1. Pie charts illustrate the proportion of heterogeneous phenotypes correlated with RDH12 mutations (A) and CRB1 mutations (B). ARRP: autosomal recessive retinitis pigmentosa. ADRP: autosomal dominant retinitis pigmentosa. ARLCA: autosomal recessive Leber congenital amaurosis. CORD: cone and rod dystrophy. PPRPE: preserved para-arteriole retinal pigment epithelium. FFR: familial foveal retinoschisis
The strategy we use in this study would help achieving quick and accurate localization of the possible disease-causing mutations, thus identifying the genetic cause of diseases efficiently and cost-effectively. However, there are some limitations. For example, focusing only on homozygous variations may lead to missing of de novo mutations, as well as the underlying disease-causing mutation in an AD or X linked pattern of hereditary. And due to the extreme genetic and clinical heterogeneity of IRD, as well as the existence of de novo mutation, copy number variation and structural variation, this approach is not 100% successful. In order to solve the genetic causes of patients with negative results, further mining is needed. Homozygosity mapping or detection of runs of homozygosity (ROH) through WES data can be performed to detect novel candidate genes using several practical and reliable tools. AutoMap, for example, was conducted and tested to be one of the best homozygosity mapping tools by a similar recent study in Iranian families. With AutoMap, the researchers predicted ROHs from WES data with high specificity and sensitivity, and also obtain a high detection rate in their IRD cohort [56]. De novo mutations should arouse great attention and whole genome sequencing (WGS) and long-read sequencing can play reliable roles in detecting large structural variations.
Conclusion
Our study employed a consanguinity-based exome sequencing analysis approach to dissect the genetic causes of IRDs in ten consanguineous Chinese families. We revealed two novel mutations and genotype–phenotype correlations, thereby expanding our understanding of the underlying heterogeneous mechanism of IRDs. Moreover, this current study attested to the value of molecular genetic testing using a consanguinity-based approach in the diagnosis of recessive genetic defects, providing a perspective for determining novel pathogenic genes in a more cost-effective and efficient way.
Supplementary Information
Additional file 1: Table S1. List of IRD genes.
Acknowledgements
We would like to thank all the patients and family members for their participation in this study.
Abbreviations
- IRD
Inherited retinal dystrophy
- RP
Retinitis pigmentosa
- CORD
Cone and rod dystrophy
- LCA
Leber congenital amaurosis
- BCD
Bietti crystalline dystrophy
- AR
Autosomal recessive
- AD
Autosomal dominant
- XL
X-linked
- NGS
Next-generation sequencing
- WES
Whole exome sequencing
- OCT
Optical coherence tomography
- ERG
Electroretinogram
- POAG
Primary open-angle glaucoma
- RPE
Retinal pigment epithelium
- ACMG
American College of Medical Genetics and Genomics
- LEU
Leucine acid
- LRR
Leucine-rich repeat
- PRO
Proline acid
- ALA
Alanine acid
- LYS
Lysine acid
- PPRPE
Para-arteriole retinal pigment epithelium
- FFR
Familial foveal retinoschisis
Authors' contributions
ZBJ designed the study, ZBJ and YL recruited patients and recorded clinical data, RJS performed experiments and ophthalmic examinations, RJS and JGW performed the genetic analysis and interpreted the data, RJS wrote the manuscript, ZBJ revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (81970838) and Beijing Natural Science Foundation (Z200014).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Beijing Tongren Hospital.
Informed consent
Informed consent was acquired from all participating subjects.
Consent for publication
Written consent was obtained from all subjects.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Verbakel SK, van Huet RAC, Boon CJF, den Hollander AI, Collin RWJ, Klaver CCW, et al. Non-syndromic retinitis pigmentosa. Prog Retin Eye Res. 2018;66:157–186. doi: 10.1016/j.preteyeres.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Khan NW, Falsini B, Kondo M, Robson AG. Inherited retinal degeneration: genetics, disease characterization, and outcome measures. J Ophthalmol. 2017;2017:2109014. doi: 10.1155/2017/2109014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao ML, Lei XL, Han F, He KW, Jin SQ, Zhang YY, et al. Patient-specific retinal organoids recapitulate disease features of late-onset retinitis pigmentosa. Front Cell Dev Biol. 2020;8:128. doi: 10.3389/fcell.2020.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 5.Xu S, Coku A, Muraleedharan CK, Harajli A, Mishulin E, Dahabra C, et al. Mutation screening in the miR-183/96/182 cluster in patients with inherited retinal dystrophy. Front Cell Dev Biol. 2020;8:619641. doi: 10.3389/fcell.2020.619641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang XF, Huang ZQ, Fang XL, Chen ZJ, Cheng W, Jin ZB. Retinal miRNAs variations in a large cohort of inherited retinal disease. Ophthalmic Genet. 2018;39:175–179. doi: 10.1080/13816810.2017.1329448. [DOI] [PubMed] [Google Scholar]
- 7.Jin ZB, Gu F, Matsuda H, Yukawa N, Ma X, Nao-i N. Somatic and gonadal mosaicism in X-linked retinitis pigmentosa. Am J Med Genet A. 2007;143:2544–2548. doi: 10.1002/ajmg.a.31984. [DOI] [PubMed] [Google Scholar]
- 8.Clark GR, Crowe P, Muszynska D, O’Prey D, O'Neill J, Alexander S, et al. Development of a diagnostic genetic test for simplex and autosomal recessive retinitis pigmentosa. Ophthalmology. 2010;117:2169–2177e3. doi: 10.1016/j.ophtha.2010.02.029. [DOI] [PubMed] [Google Scholar]
- 9.Jin ZB, Huang XF, Lv JN, Xiang L, Li DQ, Chen J, et al. SLC7A14 linked to autosomal recessive retinitis pigmentosa. Nat Commun. 2014;5:3517. doi: 10.1038/ncomms4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roosing S, Thiadens AA, Hoyng CB, Klaver CC, den Hollander AI, Cremers FP. Causes and consequences of inherited cone disorders. Prog Retin Eye Res. 2014;42:1–26. doi: 10.1016/j.preteyeres.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Wang X, Zou X, Xu S, Li H, Soens ZT, et al. Comprehensive molecular diagnosis of a large Chinese Leber congenital amaurosis cohort. Invest Ophthalmol Vis Sci. 2015;56:3642–3655. doi: 10.1167/iovs.14-15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J, Huang XF, Tong Y, Jin ZB. Targeted exome sequencing identified two novel truncation mutations in GPR98 causing Usher syndrome. Clin Exp Ophthalmol. 2016;44:97–199. doi: 10.1111/ceo.12664. [DOI] [PubMed] [Google Scholar]
- 13.Xing DJ, Zhang HX, Huang N, Wu KC, Huang XF, Huang F, et al. Comprehensive molecular diagnosis of Bardet-Biedl syndrome by high-throughput targeted exome sequencing. PLoS ONE. 2014;9:e90599. doi: 10.1371/journal.pone.0090599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang XF, Huang F, Wu KC, Wu J, Chen J, Pang CP, et al. Genotype-phenotype correlation and mutation spectrum in a large cohort of patients with inherited retinal dystrophy revealed by next-generation sequencing. Genet Med. 2015;17:271–278. doi: 10.1038/gim.2014.138. [DOI] [PubMed] [Google Scholar]
- 15.Wang F, Wang H, Tuan HF, Nguyen DH, Sun V, Keser V, et al. Next generation sequencing-based molecular diagnosis of retinitis pigmentosa: identification of a novel genotype-phenotype correlation and clinical refinements. Hum Genet. 2014;133:331–345. doi: 10.1007/s00439-013-1381-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Liu X, Sheng X, Gao X, Zhang X, Li Z, et al. Targeted next-generation sequencing reveals novel EYS mutations in Chinese families with autosomal recessive retinitis pigmentosa. Sci Rep. 2015;5:8927. doi: 10.1038/srep08927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bittles AH, Black ML. Evolution in health and medicine Sackler colloquium: consanguinity, human evolution, and complex diseases. Proc Natl Acad Sci USA. 2010;107(Suppl 1):1779–1786. doi: 10.1073/pnas.0906079106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du RF, Zhao ZL, Xu MJ. Zhongguo butong minzu de jinqin jiehun lv yu leixing. [Types and rates of consanguineous marrige in different ethnic population of China.] Chin Med J. 1981;61(12):723–728. [Google Scholar]
- 19.Hamamy H, Antonarakis SE, Cavalli-Sforza LL, Temtamy S, Romeo G, Kate LP, et al. Consanguineous marriages, pearls and perils: Geneva International Consanguinity Workshop Report. Genet Med. 2011;13:841–847. doi: 10.1097/GIM.0b013e318217477f. [DOI] [PubMed] [Google Scholar]
- 20.Teeuw ME, Loukili G, Bartels EA, ten Kate LP, Cornel MC, Henneman L. Consanguineous marriage and reproductive risk: attitudes and understanding of ethnic groups practising consanguinity in Western society. Eur J Hum Genet. 2014;22:452–457. doi: 10.1038/ejhg.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng Y, Huang H, Wang Y, Liu Z, Li N, Chen Y, et al. A novel missense NMNAT1 mutation identified in a consanguineous family with Leber congenital amaurosis by targeted next generation sequencing. Gene. 2015;569:104–108. doi: 10.1016/j.gene.2015.05.038. [DOI] [PubMed] [Google Scholar]
- 22.Riazuddin SA, Zulfiqar F, Zhang Q, Sergeev YV, Qazi ZA, Husnain T, et al. Autosomal recessive retinitis pigmentosa is associated with mutations in RP1 in three consanguineous Pakistani families. Invest Ophthalmol Vis Sci. 2005;46:2264–2270. doi: 10.1167/iovs.04-1280. [DOI] [PubMed] [Google Scholar]
- 23.Dias MS, Hamel CP, Meunier I, Varin J, Blanchard S, Boyard F, et al. Novel splice-site mutation in TTLL5 causes cone dystrophy in a consanguineous family. Mol Vis. 2017;23:131–139. [PMC free article] [PubMed] [Google Scholar]
- 24.Feldhaus B, Kohl S, Hortnagel K, Weisschuh N, Zobor D. Novel homozygous mutation in the SPATA7 gene causes autosomal recessive retinal degeneration in a consanguineous German family. Ophthalmic Genet. 2018;39:131–134. doi: 10.1080/13816810.2017.1318925. [DOI] [PubMed] [Google Scholar]
- 25.Korf BR, Rehm HL. New approaches to molecular diagnosis. JAMA. 2013;309:1511–1521. doi: 10.1001/jama.2013.3239. [DOI] [PubMed] [Google Scholar]
- 26.Huang XF, Mao JY, Huang ZQ, Rao FQ, Cheng FF, Li FF, et al. Genome-wide detection of copy number variations in unsolved inherited retinal disease. Invest Ophthalmol Vis Sci. 2017;58:424–429. doi: 10.1167/iovs.16-20705. [DOI] [PubMed] [Google Scholar]
- 27.Huang XF, Wu J, Lv JN, Zhang X, Jin ZB. Identification of false-negative mutations missed by next-generation sequencing in retinitis pigmentosa patients: a complementary approach to clinical genetic diagnostic testing. Genet Med. 2015;17:307–311. doi: 10.1038/gim.2014.193. [DOI] [PubMed] [Google Scholar]
- 28.Jin ZB, Li Z, Liu Z, Jiang Y, Cai XB, Wu J. Identification of de novo germline mutations and causal genes for sporadic diseases using trio-based whole-exome/genome sequencing. Biol Rev Camb Philos Soc. 2018;93:1014–1031. doi: 10.1111/brv.12383. [DOI] [PubMed] [Google Scholar]
- 29.Huang XF, Huang ZQ, Lin D, Dai ML, Wang QF, Chen ZJ, et al. Unraveling the genetic cause of a consanguineous family with unilateral coloboma and retinoschisis: expanding the phenotypic variability of RAX mutations. Sci Rep. 2017;7:9064. doi: 10.1038/s41598-017-09276-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin ZB, Mandai M, Homma K, Ishigami C, Hirami Y, Nao-I N, et al. Allelic copy number variation in FSCN2 detected using allele-specific genotyping and multiplex real-time PCRs. Invest Ophthalmol Vis Sci. 2008;49:3799–3805. doi: 10.1167/iovs.07-1656. [DOI] [PubMed] [Google Scholar]
- 31.Chen ZJ, Lin KH, Lee SH, Shen RJ, Feng ZK, Wang XF, et al. Mutation spectrum and genotype-phenotype correlation of inherited retinal dystrophy in Taiwan. Clin Exp Ophthalmol. 2020;48:486–499. doi: 10.1111/ceo.13708. [DOI] [PubMed] [Google Scholar]
- 32.Daiger SP, Rossiter BJF, Greenberg J, Christoffels A, Hide W. Data services and software for identifying genes and mutations causing retinal degeneration. Invest Ophthalmol Vis Sci. 1988;39:S295. [Google Scholar]
- 33.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong B, Wei B, Huang L, Hao J, Li X, Yang Y, et al. Exome sequencing identified a recessive RDH12 mutation in a family with severe early-onset. Retinitis Pigmentosa J Ophthalmol. 2015;2015:942740. doi: 10.1155/2015/942740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang L, Liang X, Li Y, Wang J, Zaneveld JE, Wang H, et al. Comprehensive molecular diagnosis of 67 Chinese Usher syndrome probands: high rate of ethnicity specific mutations in Chinese USH patients. Orphane J Rare Dis. 2015;10:110. doi: 10.1186/s13023-015-0329-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu S, Tian Y, Chen X, Zhao C. Targeted next-generation sequencing extends the phenotypic and mutational spectrums for EYS mutations. Mol Vis. 2016;22:646–657. [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Y, Guan L, Shen T, Zhang J, Xiao X, Jiang H, et al. Mutations of 60 known causative genes in 157 families with retinitis pigmentosa based on exome sequencing. Hum Genet. 2014;133:1255–1271. doi: 10.1007/s00439-014-1460-2. [DOI] [PubMed] [Google Scholar]
- 38.Tsang SH, Burke T, Oll M, Yzer S, Lee W, Xie YA, Allikmets R. Whole exome sequencing identifies CRB1 defect in an unusual maculopathy phenotype. Ophthalmology. 2014;121:1773–1782. doi: 10.1016/j.ophtha.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin X, Yang L, Chen N, Cui H, Zhao L, Feng L, et al. Identification of CYP4V2 mutation in 36 Chinese families with Bietti crystalline corneoretinal dystrophy. Exp Eye Res. 2016;146:154–162. doi: 10.1016/j.exer.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 40.Otto EA, Loeys B, Khanna H, Hellemans J, Sudbrak R, Fan S, et al. Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nat Genet. 2005;37:282–288. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- 41.Langmann T, Di Gioia SA, Rau I, Stöhr H, Maksimovic NS, Corbo JC, et al. Nonsense mutations in FAM161A cause RP28-associated recessive retinitis pigmentosa. Am J Hum Genet. 2010;87:376–381. doi: 10.1016/j.ajhg.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yücel-Yılmaz D, Tarlan B, Kıratlı H, Ozgül R. Genome-wide homozygosity mapping in families with leber congenital amaurosis identifies mutations in AIPL1 and RDH12 genes. DNA Cell Biol. 2014;33:876–883. doi: 10.1089/dna.2014.2554. [DOI] [PubMed] [Google Scholar]
- 43.den Hollander A, Roepman R, Koenekoop R, Cremers F. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog Retin Eye Res. 2008;27:391–419. doi: 10.1016/j.preteyeres.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 44.Di Gioia S, Farinelli P, Letteboer S, Arsenijevic Y, Sharon D, Roepman R, et al. Interactome analysis reveals that FAM161A, deficient in recessive retinitis pigmentosa, is a component of the Golgi-centrosomal network. Hum Mol Genet. 2015;24:3359–3371. doi: 10.1093/hmg/ddv085. [DOI] [PubMed] [Google Scholar]
- 45.Zach F, Grassmann F, Langmann T, Sorusch N, Wolfrum U, Stöhr H. The retinitis pigmentosa 28 protein FAM161A is a novel ciliary protein involved in intermolecular protein interaction and microtubule association. Hum Mol Genet. 2012;21:4573–4586. doi: 10.1093/hmg/dds268. [DOI] [PubMed] [Google Scholar]
- 46.Fu Q, Xu M, Chen X, Sheng X, Yuan Z, Liu Y, et al. CEP78 is mutated in a distinct type of Usher syndrome. J Med Genet. 2017;54:190–195. doi: 10.1136/jmedgenet-2016-104166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ascari G, Peelman F, Farinelli P, Rosseel T, Lambrechts N, Wunderlich KA, et al. Functional characterization of the first missense variant in CEP78, a founder allele associated with cone-rod dystrophy, hearing loss, and reduced male fertility. Hum Mutat. 2020;41:998–1011. doi: 10.1002/humu.23993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C, Thompson D, Koutalos Y. Reduction of all-trans-retinal in vertebrate rod photoreceptors requires the combined action of RDH8 and RDH12. J Biol Chem. 2012;287:24662–24670. doi: 10.1074/jbc.M112.354514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beryozkin A, Khateb S, Idrobo-Robalino CA, Khan MI, Cremers FPM, Obolensky A, et al. Unique combination of clinical features in a large cohort of 100 patients with retinitis pigmentosa caused by FAM161A mutations. Sci Rep. 2020;10:15156. doi: 10.1038/s41598-020-72028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zach F, Stöhr H. FAM161A, a novel centrosomal-ciliary protein implicated in autosomal recessive retinitis pigmentosa. Adv Exp Med Biol. 2014;801:185–190. doi: 10.1007/978-1-4614-3209-8_24. [DOI] [PubMed] [Google Scholar]
- 51.Alves C, Pellissier L, Wijnholds J. The CRB1 and adherens junction complex proteins in retinal development and maintenance. Prog Retin Eye Res. 2014;40:35–52. doi: 10.1016/j.preteyeres.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 52.Bujakowska K, Audo I, Mohand-Saïd S, Lancelot M, Antonio A, Germain A, et al. CRB1 mutations in inherited retinal dystrophies. Hum Mutat. 2012;33:306–315. doi: 10.1002/humu.21653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.den Hollander A, Davis J, van der Velde-Visser S, Zonneveld M, Pierrottet C, Koenekoop R, et al. CRB1 mutation spectrum in inherited retinal dystrophies. Hum Mutat. 2004;24:355–369. doi: 10.1002/humu.20093. [DOI] [PubMed] [Google Scholar]
- 54.Vincent A, Ng J, Gerth-Kahlert C, Tavares E, Maynes J, Wright T, et al. Biallelic mutations in CRB1 underlie autosomal recessive familial foveal retinoschisis. Invest Ophthalmol Vis Sci. 2016;57:2637–2646. doi: 10.1167/iovs.15-18281. [DOI] [PubMed] [Google Scholar]
- 55.Markand S, Saul A, Tawfik A, Cui X, Rozen R, Smith S. Mthfr as a modifier of the retinal phenotype of Crb1(rd8/rd8) mice. Exp Eye Res. 2016;145:164–172. doi: 10.1016/j.exer.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quinodoz M, Peter VG, Bedoni N, Royer Bertrand B, Cisarova K, Salmaninejad A, et al. AutoMap is a high performance homozygosity mapping tool using next-generation sequencing data. Nat Commun. 2021;12:518. doi: 10.1038/s41467-020-20584-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. List of IRD genes.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.