Abstract
PD-L1 (programmed cell death 1 ligand 1) is a key driver of tumor-mediated immune suppression, and targeting with antibodies induces therapeutic responses. Given the costs and associated toxicity, alternative therapeutic strategies are needed. Using reverse-phase protein arrays to assess drugs in use or likely to enter trials, we performed a candidate drug screen for inhibitors of PD-L1 expression, identifying verteporfin as a lead small molecule inhibitor. Verteporfin suppressed basal and interferon (IFN)-induced PD-L1 expression in vitro and in vivo through Golgi-related autophagy and disruption of the STAT1-IRF1-TRIM28 signaling cascade, while not affecting the proinflammatory CIITA-MHC II cascade. Within the syngeneic tumor microenvironment, verteporfin inhibited PD-L1 expression, which was associated with enhanced T-lymphocyte infiltration. Moreover, chromatin-associated enzyme poly (ADP-ribose) polymerase 1 (PARP1) inhibition with previously documented immune modulatory effects, induces PD-L1 expression in high endothelial venules (HEVs) in tumors and when combined with verteporfin further enhanced therapeutic efficacy. Thus, verteporfin targets PD-L1 effectively through transcriptional and posttranslational mechanisms and represents an alternative therapeutic strategy for targeting PD-L1.
STATEMENT OF IMPACT
Immunotherapy has revolutionized the treatment of cancer, but immune checkpoint inhibitors are associated with significant costs and toxicities. As an alternative to antibody treatments, we used high-throughput reverse-phase protein arrays to identify agents in use or likely to enter clinical trials for their potential to inhibit PD-L1 expression. There was only one significant hit—verteporfin. We show that verteporfin is a potent inhibitor of PD-L1 expression in vitro and in vivo. Mechanistically, we demonstrated that verteporfin activates Golgi-related PD-L1 autophagy and inhibits the STAT1-IRF1-TRIM28 signaling pathway but not the proinflammatory CIITA-MHC transcriptional cascade, thereby allowing for antitumor immunological responses. In vivo treatment with verteporfin in established models of malignancy was associated with enhanced T-lymphocyte infiltration.
INTRODUCTION
Discovered first as a B7 family ligand of the programmed cell death 1 (PD1) encoded by the CD274 gene (1), PD-L1 has emerged as a major checkpoint molecule that initiates an inhibitory pathway, suppressing cytotoxic T-lymphocytes upon PD-1 engagement. This mechanism that normally serves to prevent over activation of immune responses is frequently co-opted by cancer cells to evade immune surveillance (2), representing one of the major immune checkpoint pathways. Subsequent studies have provided compelling evidence that the PD-L1/PD1 cascade is a highly effective therapeutic target for immune checkpoint blockade therapy that yields durable anticancer efficacy and prolongs patient survival with durable responses (3).
PD-L1 is expressed in a variety of cancer types in either a constitutive (or intrinsic) or IFN-induced manner, and the mechanisms controlling the levels of PD-L1 mRNA and protein have been a subject of numerous studies. The STAT1-IRF1 axis plays a central role in mediating IFN-induced PD-L1 transcription in both cancer and non-cancer cells (4). In addition to transcription regulation, epigenetic mechanisms, 3’ UTR variations, and microRNAs have additional roles in fine-tuning the relative levels of PD-L1 in context-dependent settings (5). Other mechanisms have been reported to control PD-L1 degradation, ubiquitination, autophagy, glycosylation, and recycling from plasma membrane and intracellular compartments (6,7). In contrast to IFN-dependent PD-L1 transcription, the mechanisms underlying intrinsic PD-L1 expression in cancer are poorly understood.
Toxicity and cost are two key issues, among others, that limit the use of antibody approaches to immune checkpoints. Non-antibody approaches to immune checkpoint blockade have not yet been described. Clinically applicable alternatives would probably require a high potency, due to the efficiency of immune checkpoint mechanisms that are sensitive to low levels of ligand signals, and broad cell lineage spectrum coverage, given the high level of cellular heterogeneity typical of the tumor microenvironment (8). The complexity of the mechanisms controlling PD-L1 expression poses another major challenge for the development of small molecule PD-L1 inhibitors. For example, targeting one of the mechanisms controlling either intrinsic or IFN-induced PD-L1 expression may not be sufficient. Further, the pathway controls the expression levels of >7000 genes in addition to PD-L1, and many of the genes are essential for immune responses, especially those along the CIITA-MHC cascade, which is crucial for cancer immunogenicity (9). Thus, it is imperative to preserve the CIITA-MHC cascade and immune responses while targeting PD-L1 expression.
In mammalian cells, the endoplasmic reticulum (ER)-Golgi network plays an important role in autophagy and is a major source of membrane structures contributing to formation of autophagosomes (10,11). PD-L1 is one of a large number of glycosylated proteins, including most of the MHC antigens, that are processed through the Golgi apparatus. The conserved oligomeric Golgi proteins (COGs) in the Golgi apparatus play an important role in posttranslational processing and transport of secreted peptides and proteins that are glycosylated and targeted to the plasma membranes. The oligomeric Golgi complex includes 8 members (COG1-8), with the central COG1 surrounded by a COG2-4 forming lobe A and COG5-8 forming lobe B. Interestingly, PD-L1 has been found to be associated with 7 of the 8 conserved (COGs) (12), whereas the MHC molecules instead associate with the coatomers (12,13), indicating distinct routes of processing. It has been suggested that an essential role for the COGs is maintaining posttranslational homeostasis of either protein glycosylation or glycosylated proteins (14). However, it remains unclear whether PD-L1 expression can be targeted selectively because the central mechanisms controlling PD-L1 levels, e.g., IRF1-dependent transcription and posttranslational processing of PD-L1 through the ER-Golgi network, remain insufficiently understood. Indeed, despite the essential role of IRF1 in PD-L1 mRNA expression, little is known regarding how this mechanism is regulated at the PD-L1 gene promoter and whether PD-L1 transactivation can be dissected from other IRF1 downstream genes. As such, the goal of this study was to identify potential therapeutic agents that could inhibit PD-L1 while maintaining antitumor immune responses.
Materials and Methods
Cell Culture
CRF-SB, EFE184, ETN1, KLE, HOC7, UNP251, OVCAR3, OVCAR8, and MCF7 cells were maintained in RPMI1640 medium supplemented with 5% fetal bovine serum (FBS) and incubated at 37°C in a humidified incubator in an atmosphere of 95% air/5% CO2. All cell lines obtained from the American Type Culture Collection (ATCC) were curated by and redistributed through The University of Texas MD Anderson Cancer Center (MD Anderson) Characterized Cell Line Core from 2015 to 2019. Cell line identities were validated by short tandem repeat (STR) DNA fingerprinting using the AmpF STR identifier kit according to the manufacturer’s instructions (Applied Biosystems, catalogue no. 4322288). MDA-MB231 demonstrates reliable cell surface expression of PD-L1 for flow cytometery (15). The STR profiles were compared with known American Type Culture Collection fingerprints; with the Cell Line Integrated Molecular Authentication database, version 0.1.200808 (Nucleic Acids Research 2009; 37:D925-D932); and with the MD Anderson fingerprint database. The STR profiles matched known DNA fingerprints or were unique. The ID8 mouse ovarian surface epithelial cells, obtained from Vahid Afshar-Kharghan (MD Anderson), were maintained in DMEM (high-glucose, Cellgro) supplemented with 4% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 5 μg/ml insulin, 5 μg/ml transferrin, and 5 ng/ml sodium selenite. All lines were tested for mycoplasma every 6 months.
Antibodies, Immunoprecipitation, Western Blotting, and Immunohistochemistry
Antibodies against PD-L1 (E1L3N-#13684), CIITA (#3793), IDH1 (#8137), mTOR (7C10-#2983), Ub (P4D1-#3936), p62, AKT (40D4-2920), LDHB, LC3 (D11-#3868), ATG5 (2630), GM130 (D6B1-#12480), HSP60, VPS34 (D9A5-4263), β-actin (8H10D10-#3700), ATG16L1 (D6D5-8089), BECN1 (#3738), IFIH1 (#5321), IRF1 (D5E4-#8478), IRF2 (#4943), IRF3 (#11904), STAT1 (D1K9Y-#14994), YAP1 (#15117), TRIM28 (#4123), and protein A-HRP (#12991) from Cell Signaling Technology (CST, Danvers, MA), p62/SQSTM1 (610832) from BD Biosciences (San Jose, CA), HLA-Ds (sc-53302), and ERK2 from Santa Cruz Biotechnology (Dallas, TX), to COG3 (11130-1-AP) from Proteintech group (Rosemont, IL), and to COG7 (EPR9942-ab168362) from Abcam (Cambridge, MA). Cell lysis and immunoblotting were performed using 50 μg of protein lysates and equal protein loading was verified by blotting ERK2 (tumor lines), LDHB, β-actin, mTOR or pan AKT. COG3 and IRF1 immunoprecipitation was performed using 1 mg protein lysate. Mouse specific PD-L1 (#64988) and CD8a (#98941) antibodies from CST, and CD3e (sc-20047) from Santa Cruz were used for immunohistochemistry (IHC) using the CST protocol and SignalStain reagents (#8112, #14746, and #8114 or #8125).
Flow Cytometry
Cell surface PD-L1 was stained with PE-conjugated PD-L1 antibody (clone MIH1; Cat# 12-5983-42) from ThermoFisher for 30 min at 4° C. Control staining was performed using manufacturer recommended mouse IgG1 isotype and treatment-matched samples.
RNAi
ON-TARGETplus siRNAs targeting human COG2, COG3, ATG5, YAP/TAZ, IRF13, IFIH1, IRF1, and IRF2 were from Dharmacon (Lafayette, CO). RNAi silencing was performed using Lipofectamine RNAiMAX transfection reagent (Thermo Fisher) according to the manufacturer's protocol. Briefly, 20 nM siRNA was transfected and control cells were transfected with 20 nM non-targeting siRNA (Dharmacon).
Quantitative real-time PCR assay
Total RNA was isolated with RNeasy Plus Mini Kit (Qiagen, #74136, Gaithersburg, MD) according to the protocols of the manufacturer, and the first strand of cDNA was synthesized using the High Capacity cDNA Reverse Transcription kit (Invitrogen, #4368814, Grand Island, NY). Real-time PCR reactions were performed in a total volume of 20 μl in twin.tec real-time PCR Plates (Eppendorf, #0030132718, Hauppauge, NY), covered with Masterclear® real-time PCR Film (Eppendorf, #0030132947, Hauppauge, NY). TaqMan Gene Expression Assays (Invitrogen, Grand Island, NY) with TaqMan Probes CD274 (Hs01125301_m1) and ACTB (Hs01060665_g1). Each reaction contained 40 ng of cDNA, 10 μl of TaqMan® Fast Universal PCR Master Mix (2X), no AmpErase® UNG (Invitrogen, #4366073, Grand Island, NY). TaqMan reactions were run on Mastercycler® ep realplex (Eppendorf, Hauppauge, NY) with the following thermal cycling protocol: 95 °C for 2 min followed by 40 cycles of 95 °C for 15 s, 55 °C for 15 s and 68 °C for 20 s. Gene expression was quantified by the comparative CT method, by normalizing CT values to the housekeeping gene (ACTB).
Cell Fractionation
Subcellular fractionation was performed using the FractionPREP cell fractionation kit from Biovision (Milpitas, CA) according to the manufacturer’s instructions.
Cell Viability Assay
Cell viability was assayed using the CellTiter-Blue Cell Viability Assay kit from Promega (Madison, WI) according to the manufacturer’s protocol.
Reverse-phase Protein Microarray Analysis (RPPA) and High-throughput Drug Screen
RPPA assays were performed in the MD Anderson CCSG core as described at http://www.mdanderson.org/education-and-research/resources-for-professionals/scientific-resources/core-facilities-and-services/functional-proteomics-rppa-core/index.html. Briefly, serially diluted lysates were spotted onto FAST slides (Schleicher & Schuell BioSciences, USA, Keene, NH) using a robotic GeneTAC arrayer (Genomic Solutions, Inc., Ann Arbor, MI). After printing, the slides were blotted sequentially with Re-Blot (Chemicon,), I-Block, and a biotin blocking system (Dako), probed with primary antibodies, and incubated with biotin-conjugated secondary antibodies. The signals were then amplified using a Catalyzed Signal Amplification kit (DakoCytomation, Carpinteria, CA) according to the manufacturer’s instructions. The processed slides were scanned and quantitated using the Microvigene software (VigeneTech Inc., North Billerica, MA) and the quantitative values of 5 consistently expressed proteins (p38, JNK, ERK, mTor, GSK) as internal controls. To screen for the effects on PD-L1 protein levels, a total of 40 drugs or drug combinations targeting cancer pathways that are entering or likely to enter clinical trials were surveyed across established cancer cell lines and RPPA data sets (Supplementary Table 1).
Mouse study and in vivo tumor models
All animal work and protocols were supervised and approved by the Institutional Animal Care and Use Committee of MD Anderson. ID8 cells (1 x 106) were intraperitoneally injected into C57BL/6 mice (female, 6–8 weeks old, CRL/NCI). After transplantation, cells were allowed to grow in vivo for 1 week, and then mice with established tumors were randomly sorted into different treatment groups with 5 mice/group. Based on our sample size predetermination experiments and statistical analysis, 5 mice per group is sufficient to identify the expected effects with 90% power. ID8-bearing mice were then treated with isotype control IgG or anti-PD-L1 antibody (200 μg/mouse, B7-H1, clone 10F.9G2, Bio X Cell) (i.p. injection) every three days, verteorfin 60 mg/kg daily (intraperitoneal injection), BMN 673 0.33 mg/Kg daily (oral gavage), or the combination of BMN 673 with either anti-PD-L1 or verteporfin. Tumor progression was monitored once a week using a Xenogen IVIS Spectrum in vivo bioluminescence imaging system (PerkinElmer, New Haven, CT). Tumor volume was determined on the basis of the total flux (photons per second). Mice reaching a humane endpoint or weighing more than 35 g as a result of tumor growth and/or ascites were euthanized.
Lewis lung carcinoma cells (LLCs, 5 x 105 in 100 μl phosphate-buffered saline) were inoculated subcutaneously into C57BL/6 mice (male, 8–12 weeks) in both flanks. Five days after inoculation, mice were randomly divided into 4 groups and treated with vehicle, BMN 673 0.33 mg/Kg daily (oral gavage), verteporfin 30 mg/kg daily (intraperitoneal injection) and the combination of BMN 673 with verteporfin for 16 days. Tumors size were measured every 2 days by digital calipers to determine tumor volume using the formula [length/2] × [width2].
Statistical Analysis
All statistical analyses were done in GraphPad Prism 6 software. Correlations between TRIM28 and CIITA-MHC II gene expression levels were analyzed using the linear regression test. Overall survival of various treatment groups was analyzed using the Cox regression model, and the Log-rank test was used to determine the P values. Otherwise, unpaired t tests were used to generate two-tailed P values.
RESULTS
Identification of verteporfin as a potent inhibitor of PD-L1 expression
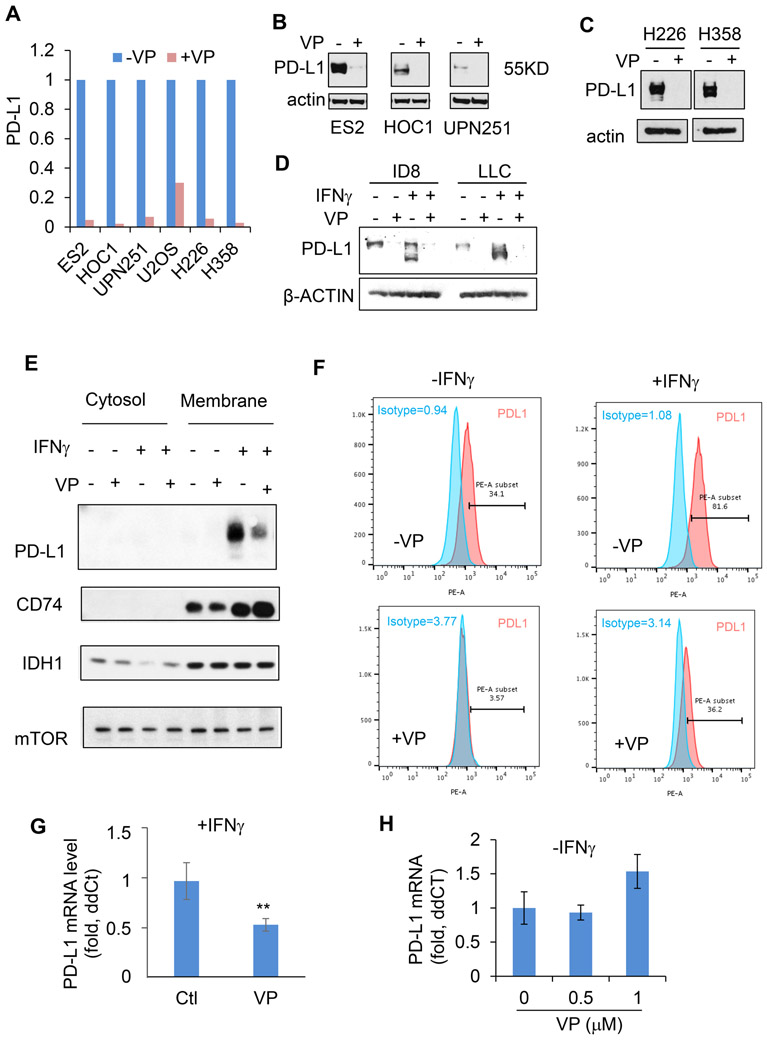
Using downregulation of PD-L1 as a read-out by reverse-phase protein arrays to assess drugs in use or likely to enter trials, we performed a candidate drug screen for inhibitors (n=40) of PD-L1 expression, discarding low-potency, narrow-spectrum, and up-regulating candidates, which identified a single drug candidate - verteporfin. Verteporfin suppressed PD-L1 expression effectively in all 6 cell lines (T cell leukemia; B cell leukemia; ovarian; endometrium n=3) (Fig. 1A, Supplemental Table 1, Supplemental Fig. S1). In an additional panel of 8 human cancer cell lines (ovarian, n=5; osteoblastoma, n=1; and lung cancers, n=2) and 2 murine cancer cell lines (ovarian and lung), verteporfin abolished basal PD-L1 protein expression, including differential glycosylated states as reflected by the double bands on Western Blots (7), regardless of genetic background, lineage specificity, and basal (intrinsic) PD-L1 levels (Fig. 1A-D). Cell fractionation revealed that verteporfin decreased membrane-associated PD-L1 (functionally relevant PD-L1) in EFE184 cells (endometrial cancer) (Fig. 1E) and flow cytometry showed that verteporfin reduced PD-L1 expression on both the surface of cancer cells (Fig. 1F) and on antigen presenting cells (Supplementary Fig. S1D). Verteporfin suppressed both IFN-induced PD-L1 protein expression (Supplemental Fig. S1B, C, D) and mRNA expression (Fig. 1G). However, in contrast to the marked loss of PD-L1 protein, verteporfin had little effect on intrinsic PD-L1 mRNA expression in the absence of IFN-γ (Fig. 1H). Thus, verteporfin engages at least two independent mechanisms to down-regulate PD-L1 expression.
Figure 1: Verteporfin decreases intrinsic and interferon-induced PD-L1 expression.
A, PD-L1 protein levels (fold change) as determined by Western blot analysis in human ovarian (ES2, HOC1, and UPN251), osteoblastoma (U2OS), and lung (H226 and H358) cancer cells treated with (+; blue) or without (−; pink) verteporfin (VP, 1 μM, 24 h). Western blots for PD-L1 levels in human ovarian cancer cells B and lung cancer cells C. D, Murine cancer cells treated with or without verteporfin and interferon. E, Western blots of PD-L1, a loading control (mTOR), membrane fractionation (CD74), and cytosolic fraction (IDH1) of EFE184 cells (endometrial carcinoma) treated with (+) or without (−) interferon-γ (IFN-γ, 10 ng/ml) and verteporfin (1 μM-24 h). F, Flow cytometry analysis of PD-L1 expression on the surface of MDA-MB231 (breast carcinoma) cells treated with (+) or without (−) verteporfin (2 μM) for 24 h. G and H, Relative PD-L1 mRNA levels in EFE184 cells treated with or without verteporfin for 24 h, either in the presence G or absence H of interferon gamma (5 ng/ml). Similar data was also obtained for the HOC1 cell line. Data represent mean+/SD of 3 independent experiments. **, P<0.001 (two-tailed t test).
Verteporfin activates Golgi-related PD-L1 autophagy
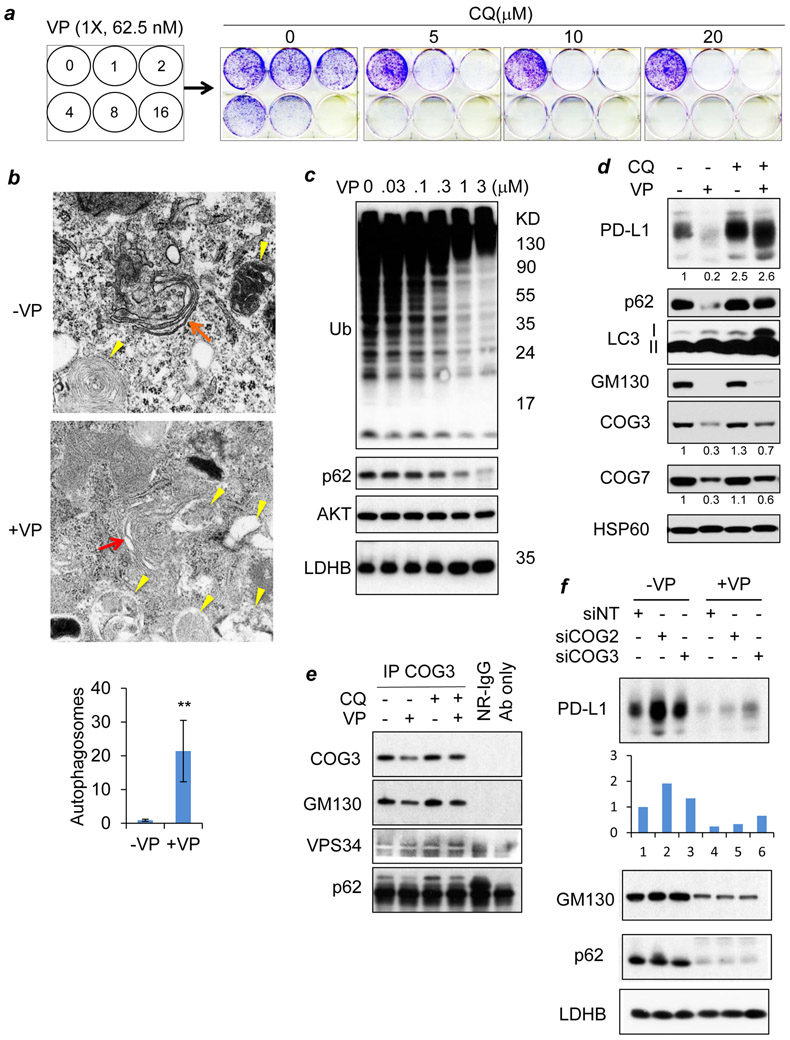
Because verteporfin has been shown to inhibit autophagy (16), we evaluated whether autophagy is required for verteporfin-induced loss of PD-L1. Although verteporfin exhibited a modest dose-dependent growth inhibitory effect on EFE184 cells, cotreatment with chloroquine, which inhibits the final step of autophagy, led to an approximately 16-fold increase in verteporfin-induced growth suppression and a marked loss of cell viability (Fig. 2A), consistent with a crucial role of autophagy in maintenance of viability of cells treated with verteporfin. Transmission electron microscopy (TEM) revealed a marked increase in autophagosomes in the cells treated with verteporfin, and altered Golgi apparatus with swollen and disrupted structures (Fig. 2B). The morphologically disrupted Golgi networks were found to be in proximity to autophagosomes. Consistent with the TEM data, verteporfin treatment led to progressive loss of high molecular weight ubiquitinated proteins in a dose-dependent manner, increases in LC3 lipidation (LC3 1 and II), and loss of the selective autophagy substrate and adaptor p62/SQSTM1 (Fig. 2C-D), indicating active autophagy consistent with a prior report (17).
Figure 2: Autophagy is required for cell survival and verteporfin-induced loss of PD-L1.
A, Cell density after treatment with either verteporfin (VP, 0~1000 nM) or chloroquine (CQ, 0~20 μM) alone or the combination of the two drugs for 6 days. B, Autophagosomes as revealed by TEM (transmission electron microscopy) in EFE184 cells treated with (+) or without (−) verteporfin (VP, 1 μM) for 24 h. Red arrows, Golgi; yellow arrows, autophagosomes. Data represent mean+/− SD from 30 cells. **p<0.001 (two-tailed t test). C, Western blots of ubiquitin and p62/SQSTM1 (p62) in EFE184 cells treated with verteporfin (VP, 0~3 μM). AKT and LDHB were blotted for loading control. D, Western analyses of protein levels in EFE184 cells treated with (+) or without (−) vertepoprfin (0.5 μM) and chloroquine (CQ, 10 μM) for 96 h. Quantification data (densitometry values normalized to lane 1) were provided below respective blots. E, Western blots for proteins present in COG3 immunoprecipitates from EFE184 cells treated with or without verteporfin (VP, 1 μM) and chloroquine (CQ, 10 μM) for 24 h. F, Western blots for protein levels in EFE184 cells treated with or without verteporfin for 24 h after transfection of non-targeting (siNT), COG2, or COG3 siRNA for 48 h. Densitometry data (fold change) of PD-L levels are presented below the blot. HSP60, AKT, and LDHB were blotted for loading control.
In addition to loss of p62 and PD-L1, verteporfin also induced a marked decrease in the Golgi proteins COG3 (the most highly evolutionarily conserved subunit), COG7, and the Golgi matrix protein GM130 (Fig. 2D). Remarkably, chloroquine treatment abrogated verteporfin-induced loss of PD-L1, p62/SQSTM1, and Golgi proteins along with the altered Golgi structures suggesting that verteporfin induces autophagy-mediated degradation of the Golgi apparatus, which is likely a consequence of verteporfin-induced organelle damage. Indeed, we found COG3 physically associated with VPS34 (a crucial membrane component of the autophagy machinery) and the autophagy adaptor molecule p62, in addition to GM130 (Fig. 2E). Next, we interrogated whether the lobe A subunits might have a role in autophagy-dependent PD-L1 removal in cancer cells. RNAi-mediated gene silencing was performed for COG2 and COG3. The COG2 RNAi led to cross depletion of COG3, consistent with the finding that both COG3 and COG2 are required for stabilizing lobe A complexes (18). Nonetheless, RNAi of either COG2 or COG3 increased intrinsic PD-L1 levels, with COG2 depletion having a stronger effect (Fig. 2F), consistent with a role for the oligomeric Golgi complex in regulating PD-L1 levels as a quality control mechanism. However, depletion of COG3 but not COG2 attenuated verteporfin-induced loss of PD-L1, suggesting a crucial role for COG3, but not COG2, in verteporfin-induced PD-L1 removal.
Verteporfin inhibits IRF1-dependent PD-L1 transcription
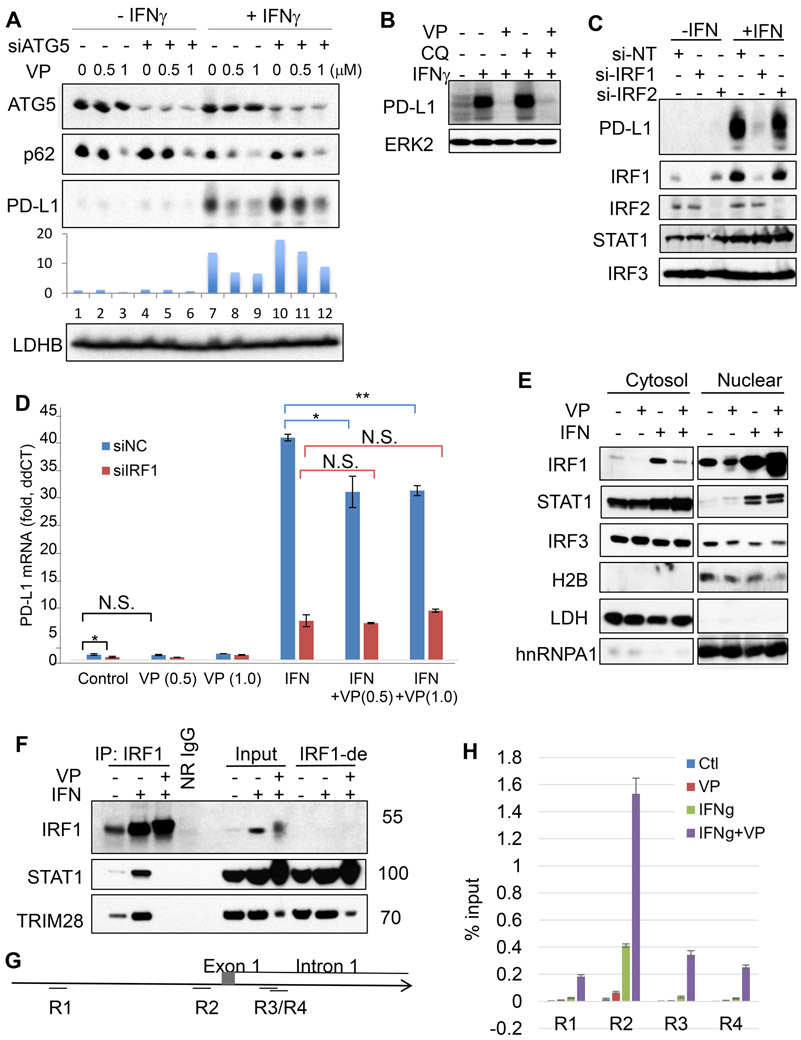
Further supporting a role for autophagy in mediating verteporfin-induced loss of PD-L1, RNAi-mediated knockdown of ATG5, a gene that is essential for canonical autophagy, mitigated the effect of verteporfin on PD-L1 down regulation (Fig. 3A). Despite a role for autophagy in down regulation of PD-L1 protein levels, at increased concentrations (1μM versus 0.5 μM), verteporfin was sufficient to decrease PD-L1 expression in ATG5 depleted cells. Strikingly, at higher drug concentrations verteporfin abolish IFN-induced PD-L1 expression in the presence of chloroquine (Fig. 3B), suggesting that chloroquine-sensitive autophagy is dispensable for loss of PD-L1 in this setting. Because verteporfin treatment also resulted in significant down regulation of PD-L1 mRNA expression (Fig. 1G), we explored mechanisms controlling PD-L1 gene expression.
Figure 3: IRF1 is essential for interferon-induced PD-L1, and verteporfin disrupts IRF1 complex.
A, Western blots for protein levels in EFE184 cells treated with or without verteporfin for 24 h after transfection of non-targeting (siNT) or ATG5 siRNA for 48 h. B, Western analysis of protein levels in EFE184 cells treated with or without verteporfin (VP, 2 μM), chloroquine (CQ, 20 μM), and interferon-γ (IFNy, 5 ng/ml) for 24 h. C, Western blots for protein levels in EFE184 cells treated with or without interferon-γ (IFNγ, 5 ng/ml) for 24 h after transfection of non-targeting, IRF1, or IRF2 siRNA for 48 h. D, qPCR for PD-L1 mRNA levels in EFE184 cells treated with or without verteporfin (μM) and interferon-γ for 24 h after transfection with non-targeting or IRF1 siRNA for 48 h. Data represent mean+/SD (n=6). *, p<0.05; **, p<0.001; N.S., not significant (two-tailed t test). E, Western blots for proteins after sub-cellular fractionation in EFE184 cells treated with or without verteporfin (VP, 1 μM) and interferon-γ(IFNγ, 5 ng/ml) for 24 h. F, Western blots for the level of proteins present in IRF1 immunoprecipitates (IP: IRF1) and mock immunoprecipitation with normal rabbit IgG (NR-IgG) from EFE184 cells treated with or without interferon-γ (IFNγ) and verteporfin (VP) for 24 h. Blots for input and IRF1-immunodepleted (IRF1-de) lysates were shown on the right. G, PD-L1 gene (CD274) promoter regions (R1~4) and H CHIP assays for IRF1 (H) and STAT1 (Supplementary Fig. 4) recruitment in EFE184 cells treated with or without interferon and verteporfin for 24 h.
Previously, verteporfin has been shown to inhibit YAP/TAZ function and YAP1 has been shown to inhibit IRF3 signaling (19). Thus, RNAi was performed to interrogate the roles of the YAP1, TAZ (WWTR1), IFIH1, and IRF3 cascade in association with PD-L1 expression. Knockdown of these genes had minimal effects on both the basal levels of PD-L1 or interferon-induced PD-L1 expression, with depletion of YAP1 and TAZ (WWTR1) leading to increased rather than decreased PD-L1 levels in the presence of IFN-γ (Supplemental Fig. S2). Thus, the effect of verteporfin on PD-L1 expression is unlikely to be attributed to the YAP/TAZ signaling cascade.
Given the ability of verteporfin to abolish IFN-induced PD-L1 expression, we next sought to determine whether verteporfin acts to disrupt PD-L1 gene transcription. Both the signal transducer and activator of transcription 1 (STAT1) and the main transcription factor IRF1, downstream of the IFNγ receptor, are essential for IFN-induced PD-L1 gene expression (20). Indeed, IFN induced marked increases in IRF1 expression in a panel of cancer cell lines, and IRF1 protein levels directly correlated with PD-L1 levels (Supplemental Fig. S3). Further, IRF1 depletion abolished IFN-induced expression of PD-L1 at the protein level (Fig. 3C). Whereas IRF2 knockdown had no effect despite a possible role of IRF2 in either opposing or complimenting that of IRF1 (21). Thus we examined the effect of verteporfin on PD-L1 mRNA in the context of IRF1 depletion. Verteporfin markedly decreased PD-L1 mRNA levels, and IRF1 depletion had a stronger effect, whereas verteporfin did not cause further decreases in PD-L1 mRNA levels in the setting of IRF1 depletion (Fig. 3D), suggesting that verteporfin acts via inhibiting IRF1-dependent PD-L1 transcription. However, IFN-induced STAT1 and IRF1, including their nuclear localization, were not affected by verteporfin (Fig. 3E), suggesting intact IFN-STAT1 signaling upstream of IRF1.
To determine where in the STAT1 pathway verteporfin might be acting, we performed IRF1 immunoprecipitation and found that whereas IFN-γ induced a marked increase in IRF1-STAT1 association, verteporfin treatment led to complete disruption of the IRF1-STAT1 complex (Fig. 3F). We then performed ChIP assays to determine the effect of verteporfin on IFN-induced IRF1-binding to PD-L1 gene promoter regions (Fig. 3G). Indeed, while IRF1-binding could not be detected in unstimulated cells, IFN-γ induced marked increases in the binding of IRF1 to the PD-L1 promoter (Fig. 3H). Strikingly, despite inhibiting PD-L1 expression, verteporfin treatment led to a modest increase in IRF1 binding in the absence of IFN, and a marked increase was detected in the presence of IFN. The paradoxical increase in IRF1 binding to the PD-L1 promoter after verteporfin treatment appeared to suggest futile recruitment of IRF1, probably as a result of disruption of the IRF1-STAT1 protein complex. Indeed, additional ChIP assays showed that verteporfin led to decreases in the recruitment of STAT1 to the PD-L1 promoter in either the absence or presence of IFN (Supplemental Fig. S4). Taken together, our data suggest that verteporfin disrupts IFN-induced IRF1-STAT1 interaction, leading to suppression of PD-L1 transcription with nonproductive trapping of IRF1 to the PD-L1 promoter.
Differential effects of verteporfin on IRF1-dependent transcription
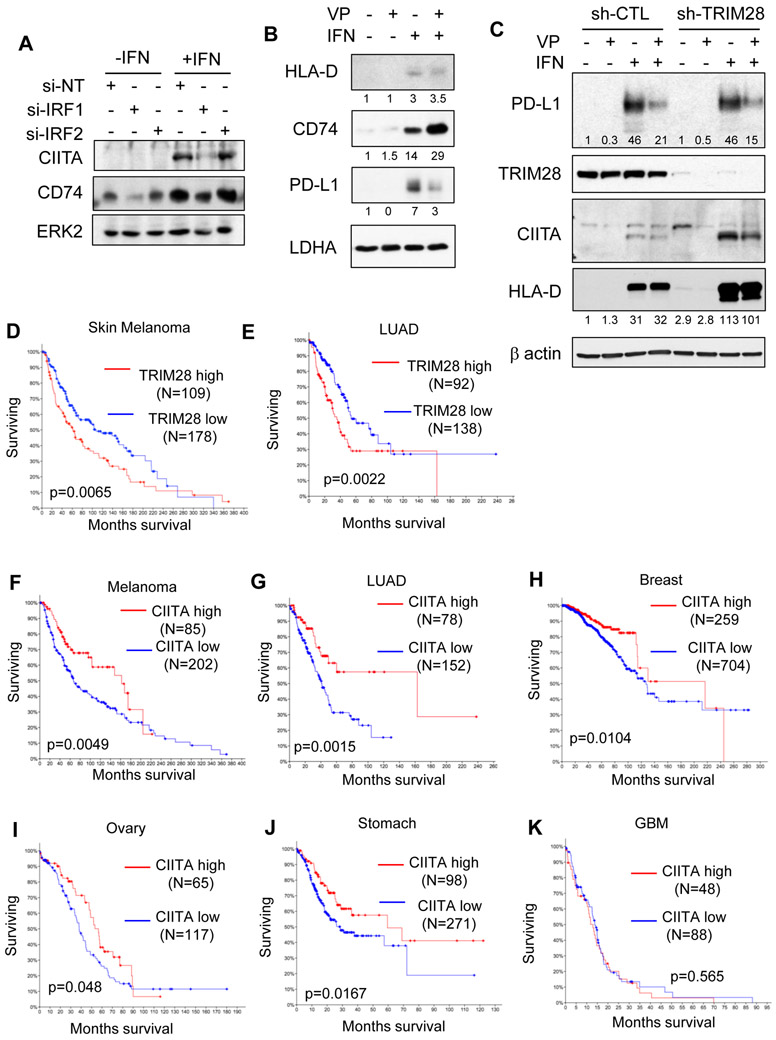
In addition to loss of PD-L1 expression, IRF1 knockdown also led to a marked decrease in the CIITA transcription factor as well as its downstream gene the CD74 protein (Fig. 4A). The MHC II cascade branch of IRF1 signaling was not inhibited in verteporfin-treated cells in the presence of IFN (Fig. 4B) suggesting that verteporfin blocks the IFN-IRF1-PDL1 axis specifically while having little effect on IRF1-CIITA. Such pathway specification would allow the PD-L1 immune checkpoint pathway to be blocked without collateral suppression of CIITA-dependent tumor immunogenicity. We next sought to understand the underlying mechanism. TRIM28 is a cofactor acting either to activate or to repress target gene transcription (22). Although previously reported in association with IRF1 (23), the role of TRIM28 in regulating IRF1 signaling remains unclear. We found in parallel to STAT1, that the TRIM28-IRF1 interaction was also markedly increased in response to IFN and that the interaction was abrogated by verteporfin (Fig. 3F). Strikingly, shRNA-mediated TRIM28 depletion led to marked increases in CIITA and HLA-D (Fig. 4C), suggesting TRIM28 normally acts to inhibit the CIITA-MHC II cascade in a feed-forward manner that is responsive to interferon.
Figure 4: TRIM28 suppresses the CIITA-MHC II cascade downstream of interferon signaling.
A, Western blots for interferon target proteins in EFE184 cells treated with or without interferon gamma (IFN, 5 ng/ml) for 24 h after transfection of non-targeting, IRF1, or IRF2 siRNA for 48 h. B, Western blots for proteins in EFE184 cells treated with (+) or without (−) interferon γ (5 ng/ml) and verteporfin (0.5 μM) for 24 h. Quantification data (densitometry values normalized to lane 1) were provided below respective blots. C, Western blot analyses of protein levels in control (sh-CTL) and TRIM28 knockdown (sh-TRIM28) MCF7 cells treated with or without verteporfin (VP, 1 μM) and interferon gamma (IFN, 5 ng/ml) for 24 h. Quantification data (densitometry values normalized to lane 1) were provided below respective blots. D-K, Kaplan-Meier estimates of overall patient survival of melanoma (D) and lung cancer (E) expressing high and low levels of TRIM28 mRNA and melanoma (F), lung adenocarcinoma (G), breast cancer (H), ovarian cancer (I), stomach cancer (J), and glioblastoma multiforme (GBM) (K) expressing high and low levels of CIITA mRNA.
Indeed, analyses of TCGA data revealed that TRIM28 levels inversely correlate with CIITA-MHC II gene expression and CD74 in the majority of human cancer types that span immunological reactivity (Supplementary Fig. 5A, B). Further, gene expression profiling revealed two distinct tumor clusters in lung cancer, conforming to high and low CIITA signature classes of gene expression, respectively (Supplementary Fig. 5C), which in turn exhibited a very strong (p = 9.27 X 10−16) inverse association with TRIM28 mRNA levels (Supplementary Fig. 5D-F). Thus, the role of TRIM28 in suppressing the IRF1-CIITA pathway appears to be generalizable.
Importantly, higher TRIM28 levels are associated with statistically significant poorer patient outcomes in melanoma and lung cancer (Fig. 4D, E), which are considered immunologically reactive, with a demonstrated benefit to immune checkpoint inhibition (24). To further determine whether the statistical association between TRIM28 levels and patient outcomes might simply reflect a non-causal correlation, we analyzed the effect of a downstream event, i.e., the expression of CIITA. We found that the mRNA levels of CIITA were indeed positively associated with more favorable patient outcomes, with high statistical power across a broad spectrum of immunologically reactive human cancers albeit not necessarily in immunologically non-reactive tumors such as GBM (Fig. 4G-K), suggesting that the effects of TRIM28 and CIITA on patient outcomes are likely to be mechanistically linked in specific cancer lineages.
Verteporfin mediates PD-L1 blockade in syngeneic tumor microenvironments
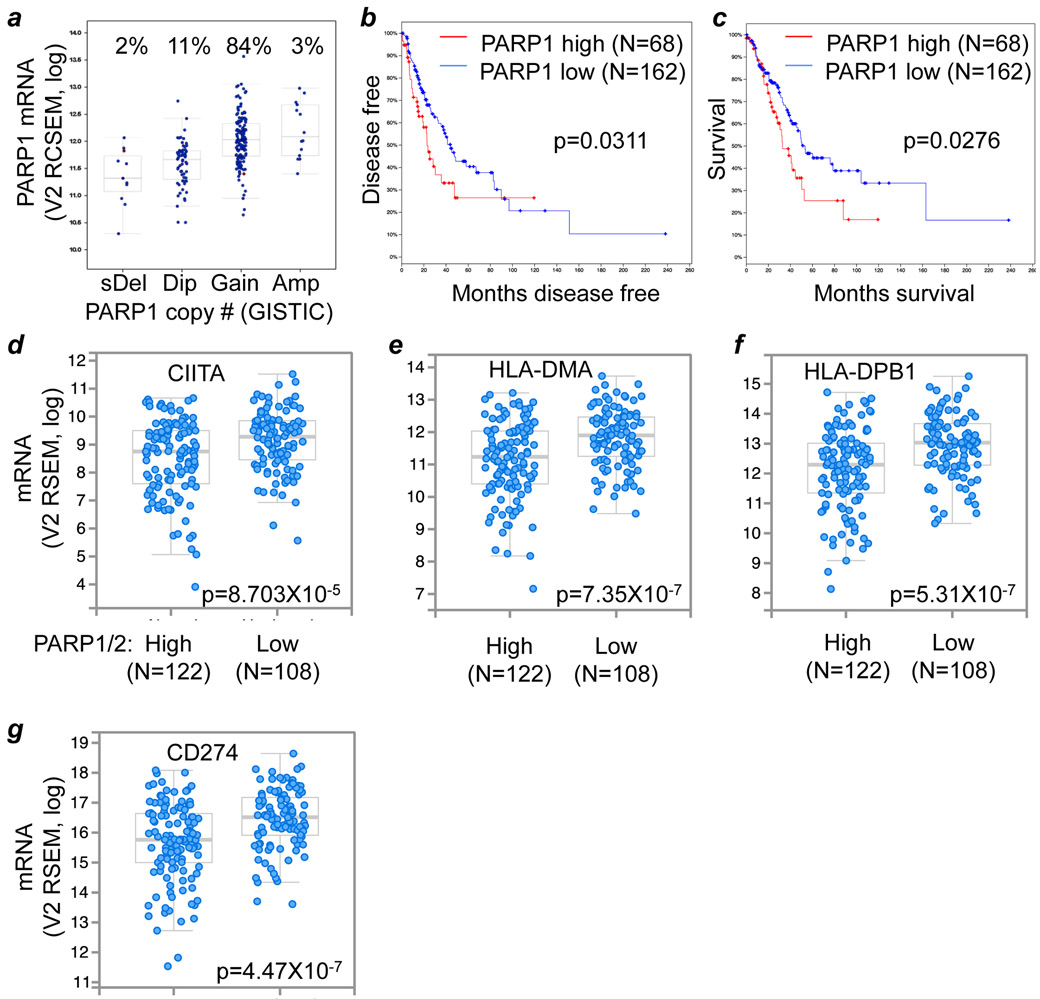
We next hypothesized that therapeutic interventions that activate the CIITA-MHC immunogenicity cascade, which usually also drive PD-L1 expression, would synergize with verteporfin. Because PARP inhibitors were found to synergize with immune checkpoint blockade (25), we sought to determine whether PARP inhibition might affect the CIITA-MHC cascade. We found that PARP1 copy number increases in over 85% of lung cancers (adenocarcinomas), which correlates with PARP1 mRNA expression (Fig. 5A; Pearson’s r=0.511). Higher PARP1/2 mRNA levels were associated with significantly poorer overall and disease-free survival in lung cancer patients as well as with decreased mRNA levels of CIITA-MHC and PD-L1 genes (Fig. 5B-G).
Figure 5: PARP1 expression is associated with PD-L1 levels and the CIITA cascade.
A-C, correlation of PARP1 mRNA levels with putative PARP1 gene copy number (A), disease free (B) and overall survival (C) of lung adenocarcinoma patients (TCGA data). D-G, enrichment analysis for the association of CIITA (D), MHC (E and F), and PD-L1 (G) levels with high and low levels of PARP1/2 mRNA expression in lung adenocarcinoma (TCGA data).
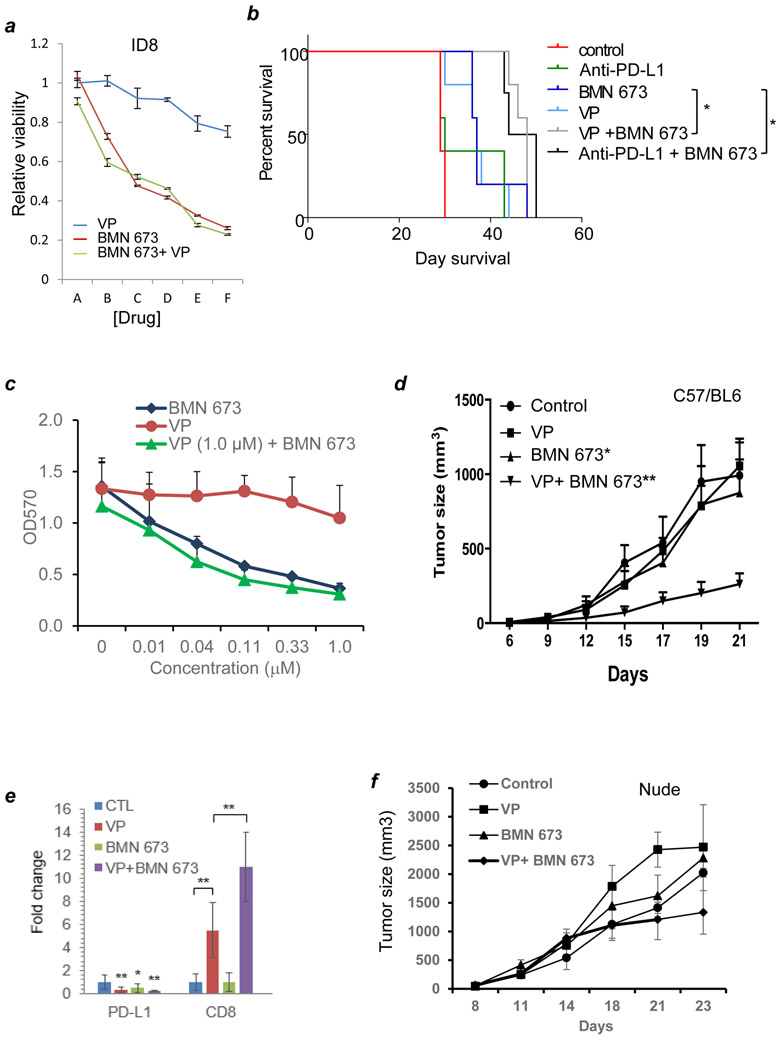
PARP inhibitors have been shown to have activity across a spectrum of cancers and have been approved for first-line treatment of BRCA mutant ovarian cancers (26). Notably, murine ID8 ovarian carcinoma cells, which are commonly used for syngeneic studies, exhibited little sensitivity to verteporfin in cell culture and no synergism with the BMN 673 PARP inhibitor (Fig. 6A). Given these properties, we then tested the in vivo therapeutic effects of verteporfin monotherapy or in combination with BMN 673 in established immune competent mice bearing ID8 cells intraperitoneally including in direct comparison with anti-PD-L1 antibody (Fig. 6B). Monotherapy with either verteporfin or anti-PD-L1 had a modest effect on the survival of ID8-burdened mice, consistent with previous studies showing these cells to have modest intrinsic immunogenicity (25,26). In contrast, the combination of verteporfin and BMN 673 produced a statistically significant improved outcome compared to either monotherapy that was equivalent to the combination of anti-PD-L1 and BMN 673 (Fig. 6B). Independent of BRCA mutations, PARP inhibition induces immune responses in cancer models including that of lung cancer, which has been exploited for therapeutic intervention (27,28). Similar to ID8 cells, murine LLC cells exhibited limited sensitivity to verteporfin and no synergism with the BMN 673 PARP inhibitor in vitro (Fig. 6C). Monotherapy with either verteporfin or BMN 673 had a modest effect on tumor growth in a syngeneic setting; whereas the combination of verteporfin and BMN 673 produced a statistically significantly reduction in tumor volume (Fig. 6D). In LLC tumors for which we could harvest tumors for immune analysis, verteporfin treatment led to marked decreases in PD-L1 expression and increases in CD8 T cells especially in the combinatorial group of vereporfin and BMN 673 (Fig. 6E, D; Supplemental Fig. S6A, B). In contrast, verteporfin and BMN 673, either administrated alone or in combination, were not effective in immunodeficient nude mice relative to control (p>0.1 one-way ANOVA) (Fig. 6F), emphasizing the role of immune responses in the drug action. The difference in tumor size on day 21 of the verteporfin treated mice was not statistically significant.
Figure 6: Verteporfin and PARPi inhibited syngeneic ovarian cancer and Lewis lung carcinoma cell growth.
A, Dose-response curves for relative viability of mouse ovarian cancer cells (ID8) treated with or without verteporfin, BMN 673, or the combination of both for 72 h in standard cell culture. A, B, C, D, E, and F = 0, 0.012, 0.04, 0.12, 0.37, and 1.1 μM respectively. Data represent mean+/− SD (n=9). B, Kaplan-Meier survival estimates of C57/BL6 mice intraperitoneally implanted with ID8 cells (N=5). Established tumors were treated with vehicle (control), anti-PD-L1 antibody (PD-L1), BMN 673, verteporfin (VP60) and the combination of BMN 673 with either anti-PD-L1 antibody (PD-L1+BMN) or verteporfin (VP60+BMN). *, p<0.05; **, p<0.01 (compared with control) (Log-rank test). C, Dose-response curves of verteporfin (VP) and in combination with BMN 673 for relative viability of Lewis lung carcinoma (LLC) cells. D, growth curves of Lewis lung cancer (LLC) in C57BL/6 mice treated with or without verteporfin (VP, 30 mg/Kg), BMN 673, and the combination of the two drugs. *, p<0.05; **, p<0.01 (two-tailed t test). N = 8 tumors per group. The experiment was repeated with similar results. E, immunohistochemical (IHC) staining (and summarized data)of PD-L1 and CD8A, in LCC tumors treated with or without verteporfin (VP), BMN 673, and the combination of the two drugs. Quantitative data were obtained with ImageJ analysis of positive cells per random area. Data represent average +/− SD of 13 areas of 300 nuclear events. scale bar, 50 μm. *, p<0.05; **, p<0.01 (two-tailed t test). The control was used as the baseline and set at 1. Supplementary Figure 6 shows representative immunohistochemistry images. F, growth curves of LLC in nude mice treated with or without verteporfin (VP, 30 mg/Kg), BMN 673, and the combination of the two drugs.
PARP inhibition induces PD-L1 expression on high endothelial venules (HEVs)
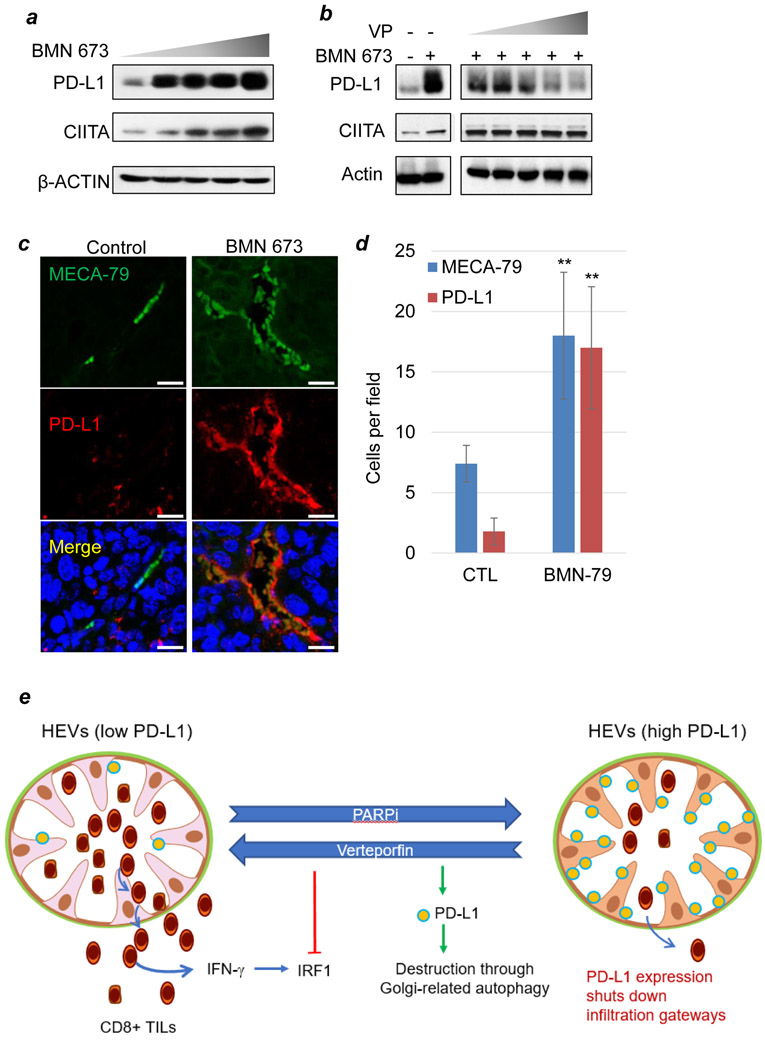
While T-cell infiltration is consistent with the overall outcome of PD-L1 suppression, we observed that PD-L1 expression was highly heterogeneous in the LCC tumor microenvironment. Notably, the strongest PD-L1 signal was detected in the tumor vasculature in BMN 673-treated tumors (Supplemental Fig. S6A). Although PARP inhibition did not affect tumor cell expression of PD-L1 (Supplementary Table 1), it did have a strong inhibitory effect on endothelial expression of PD-L1 (Fig. 7A). We then sought to determine whether tumor-associated endothelial cells are the sources of PD-L1 expression in the BMN 673-treated tumors. Although PD-L1 expression appeared to be associated with LYVE1-positive lymphatic vasculature (Supplemental Fig. S6C), closer examination only revealed a relatively weak expression of PD-L1 in LYVE1 positive cells.
Figure 7: PD-L1 expression on high endothelial venules and the effects of PARPi and verteporfin.
A and B, Western blots for CIITA and PD-L1 protein levels in SVEC4.10 (murine lymphatic endothelial) cells treated with BMN 673 (0 to 100 nM for 96 h) (A) or BMN 673 (100 nM for 96 h) and verteporfin (0-2 μM for 48 h) (B). C and D, Immunofluorescent staining for MECA-79, a selective marker for HEVs, and PD-L1 in syngeneic LCC tumor treated without (−) or with (+) the PARP inhibitor BMN 673. Scale bars, 20 μM. E, Schematic of the mechanism underlying the synergy between PARP inhibition and verteporfin-mediated PD-L1 blockade. PARP inhibition promotes HEV neogenesis and PD-L1 expression, including on HEVs, whereas verteporfin abrogates PD-L1 expression through Golgi-related autophagy and suppression of IRF1-dependent transcription. The combination of PARP inhibition and PD-L1 blockade produces a therapeutic synergy.
Because high endothelial venules (HEVs), which are specialized post-capillary structures, are the only known routes of tumor infiltration of lymphocytes (29,30), we then tested the hypothesis that PARP inhibition drives PD-L1 expression in HEVs. PARP inhibition with BMN 673 on lymphatic endothelial cells led to increases in the protein levels of CIITA and PD-L1 (Fig. 7A). Further, while PARPi-induced PD-L1 expression was suppressed by verteporfin, CIITA levels were not significantly altered even at supraphysiological levels (i.e. =>1 μM) (Fig. 7B). Ex vivo analysis of tumors stained for the selective HEV marker MECA-79 formed poorly developed vascular-like structures in control tumors but highly organized structures were detected in BMN-673-treated tumors with strong luminal MECA-79 staining (Fig. 7C). Strikingly, despite a weak coexpression of PD-L1 and MECA-79 in control tumors, strong PD-L1 expression was detected in MECA-79-positive cells in BMN 673-treated tumors (Fig. 7D), suggesting that PARP inhibition induces PD-L1 expression in HEVs. PD-L1 expression on the HEVs could inhibit effector and proliferating T cells as they exit from the peripheral circulation into the tumor microenvironment (Fig. 7E).
DISCUSSION
High-throughput screening identified verteporfin as a lead candidate for inhibiting PD-L1, suggesting the potential repurposing of the FDA-approved ophthalmological drug verteporfin for oncological therapeutic benefit. Verteporfin is a photosensitizer commonly prescribed for retinopathy but has also shown antitumor properties in various experimental settings (31,32). Further, verteporfin possesses intrinsic antitumor activities through targeting YAP1 and other mechanisms (31,32). Given the patient safety profile and relative low cost ($1700 per injection, wholesale), verteporfin could be considered as an alternative approach to anti-PD-L1 blockade, in effect mimicking the neutralizing antibodies currently used in the clinic. PD-L1 blockade has been shown to have a therapeutic benefit in non-small cell lung carcinoma in a randomized phase III study (33). Notably, the lung cancer patients with the highest levels of PD-L1 derived the greatest benefit from anti-PD-L1, although all did to some degree. We have shown that PARP inhibitors such as BMN 673 can upregulate PD-L1 on HEVs and that the combination of PARP inhibitors and PD-L1 blockade increases the therapeutic effect in vivo. Our preclinical data may provide a strong rationale for the combinatorial use of verteporin and a PARP inhibitor in lung cancer patients, especially given the high frequency of PARP1 gain in lung cancer. In many other cancers such as GBM, PD-L1 is low (34) and the lack of immune effector responses in the tumor microenvironment may not necessarily benefit from this type of strategy.
Our study reveals two unique mechanisms of action of verteporin on PD-L1 expression, which includes an autophagy-dependent mechanism mediated by the oligomeric Golgi complex of the COG proteins (specifically COG2 and COG3) for the removal of intrinsically expressed PD-L1, and suppression of interferon-induced PD-L1 expression. Importantly, this selective downregulation of PD-L1 with verteporfin maintains the proinflammatory IRF1-CIITA-MHC II signaling cascade. IRF1 is a key regulatory factor for interferon, TLR, and TNF (35) (36,37); thus, IRF1 plays an important role in inflammatory responses. Indeed, IRF1 knockout in mice leads to a multifaceted defect in immune surveillance (38). Depending on the specific target gene, IRF1-mediated transactivation can occur with or without its cofactor STAT1 (39). In the case of PD-L1 expression, both IRF1 and STAT1 are essential (4). We show that verteporfin disrupts the IRF1-STAT1 interaction, thereby suppressing interferon-induced PD-L1 transcription, but it increases IRF1 binding to the PD-L1 promoter in response to IFN- γ. It remains unclear as to whether or not this promoter-trapping effect is required for suppression of PD-L1 transcription in addition to loss of STAT1 binding. Supporting this concept, IRF1 has been shown to be involved in target gene silencing (35). Our study also unveiled a previously unrecognized role for TRIM28 in inhibiting the IRF1-CIITA-MHC II axis but not PD-L1 expression. Decommissioning of this role of TRIM28 upon verteporfin treatment provides another layer of selectivity, allowing precision targeting of the IRF1-PD-L1 axis. More studies are needed to determine whether TRIM28 contributes to the development of resistance to adaptive immune surveillance in human cancer and whether TRIM28 represents a therapeutic target for reversing the immunoediting process associated with suppression of the CIITA-MHC genes (40).
We also found that the PARP inhibitors are inducing PD-L1 expression on specialized tumor-associated endothelial structures known as the high endothelial venules (HEVs), which are known to be the gateway for tumor infiltrating lymphocytes (30). The presence of HEVs has been shown to correlate with more favorable patient outcomes in multiple cancer types (41). As shown in other tumor-associated endothelial cells (42,43), HEV expression of PD-L1 is probably modulating the effector and proliferative function of the T cells as they efflux out of circulation into the tumor microenvironment. This mechanism would be in addition to the previously documented role of PARP inhibition, causing the accumulation of double-stranded DNA breaks in cancer cells with increases in mutation loads or the presence of damaged chromosomal DNA in the cytoplasm triggering the cGAS-innate immune response pathway. STING pathway activation in this context also likely drives the robust T cell infiltration we observed in the tumors (27). Importantly, we found a strong inverse correlation between the mRNA levels of PARP1/2 and the proinflammatory CIITA-MHC cascade genes in multiple human cancers. Thus, PARP inhibition is likely to activate the CIITA-MHC cascade, immunogenicity, and T-cell priming through multiple pathways, which consequently elicit immune responses and adaptive PD-L1 expression.
Despite the strong induction of both HEV neogenesis and PD-L1 in response to PARP inhibition, different mechanisms might be involved in these processes. In addition to DNA repair, PARP1 regulates gene expression through multiple mechanisms (44) and can act as a cofactor of multiple transcriptional regulators including NF-κB and TNF-α (45,46). Indeed, the expression of a number of NF-κB-dependent genes has been shown to be impaired in PARP1−/− cells as well as in response to PARP inhibition (47). Intriguingly, while the lymphotoxin-β-LTBR pathway is essential for HEV neogenesis, the signaling of the closely related cytokine TNF-α was shown to have the opposite role in experimental melanoma (48). Future studies will be directed at ascertaining the applicability of modulating HEV PD-L1 expression across cancer lineages and its relevance as a biomarker for response to anti-PD-L1 strategies.
Enrichment markers for response to anti-PD-L1 strategies have not had high predictive value for therapeutic response, and this is also likely to be the scenario for responses to verteporfin, especially because this agent may also have direct antitumor activities through targeting YAP1 (31,32). Yet, at the dosage range that abrogated PD-L1 expression used in our study, verteporfin had little effect on either YAP1 levels or its phosphorylation and exhibited minimal cell-autonomous cytotoxicity on cancer cells. More importantly, verteporfin in combination with PARP inhibition produced a strong synergistic effect in syngeneic models of both ovarian and lung cancers but not in cell culture, suggesting a therapeutic efficacy independent of cell autonomous anticancer activity. Nevertheless, verteporfin may prove uniquely effective in cancers that rely on oncogenic signals from YAP1. In addition, the striking coexpression pattern of MECA-79 and PD-L1 suggests that HEV staining could be considered as a potential biomarker for PD-L1 blockade, especially with a combination of PARP inhibitors and verteporfin within the context of a clinical trial.
Supplementary Material
Acknowledgements
We thank Kenneth Dunner Jr at the High Resolution Electron Microscopy Facility for assistance in performing electron microscopy, David M. Wildrick, Ph.D., for editorial assistance, and Audria Patrick for assisting in manuscript preparation.
Role of the Funding Source
The study was designed by the investigators, who oversaw all data collection and interpretations. The funding agencies had no role in the data analysis, interpretation of the results, or writing of the manuscript. The project was supported by the Gynecologic SPORE (5P50CA098258, NIH/NCI), the Provost Retention Fund and the Brockman Foundation to ABH. This research was performed in the Flow Cytometry & Cellular Imaging Facility, which is supported in part by the National Institutes of Health through M.D. Anderson’s Cancer Center Support Grant CA016672.
Footnotes
Authors’ Disclosures of Potential Conflicts of Interest: None related to the work
REFERENCES
- 1.Dong H, Zhu G, Tamada K, Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med 1999;5(12):1365–9. [DOI] [PubMed] [Google Scholar]
- 2.Drake CG, Jaffee E, Pardoll DM. Mechanisms of immune evasion by tumors. Adv Immunol 2006;90:51–81. [DOI] [PubMed] [Google Scholar]
- 3.Sharma P, Allison JP. The future of immune checkpoint therapy. Science 2015;348(6230):56–61. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep 2017;19(6):1189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, et al. Aberrant PD-L1 expression through 3'-UTR disruption in multiple cancers. Nature 2016;534(7607):402–6. [DOI] [PubMed] [Google Scholar]
- 6.Lim SO, Li CW, Xia W, Cha JH, Chan LC, Wu Y, et al. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell 2016;30(6):925–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun 2016;7:12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pitt JM, Vetizou M, Daillere R, Roberti MP, Yamazaki T, Routy B, et al. Resistance Mechanisms to Immune-Checkpoint Blockade in Cancer: Tumor-Intrinsic and -Extrinsic Factors. Immunity 2016;44(6):1255–69. [DOI] [PubMed] [Google Scholar]
- 9.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A 1998;95(26):15623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geng J, Klionsky DJ. The Golgi as a potential membrane source for autophagy. Autophagy 2010;6(7):950–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge L, Wilz L, Schekman R. Biogenesis of autophagosomal precursors for LC3 lipidation from the ER-Golgi intermediate compartment. Autophagy 2015;11(12):2372–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015;162(2):425–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paulsson KM, Kleijmeer MJ, Griffith J, Jevon M, Chen S, Anderson PO, et al. Association of tapasin and COPI provides a mechanism for the retrograde transport of major histocompatibility complex (MHC) class I molecules from the Golgi complex to the endoplasmic reticulum. J Biol Chem 2002;277(21):18266–71. [DOI] [PubMed] [Google Scholar]
- 14.Foulquier F, Vasile E, Schollen E, Callewaert N, Raemaekers T, Quelhas D, et al. Conserved oligomeric Golgi complex subunit 1 deficiency reveals a previously uncharacterized congenital disorder of glycosylation type II. Proc Natl Acad Sci U S A 2006;103(10):3764–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan H, Lan Y, Wan Y, Wang Q, Wang C, Xu L, et al. PD-L1 mediated the differentiation of tumor-infiltrating CD19(+) B lymphocytes and T cells in Invasive breast cancer. Oncoimmunology 2016;5(2):e1075112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donohue E, Tovey A, Vogl AW, Arns S, Sternberg E, Young RN, et al. Inhibition of autophagosome formation by the benzoporphyrin derivative verteporfin. J Biol Chem 2011;286(9):7290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 2005;1(2):84–91. [DOI] [PubMed] [Google Scholar]
- 18.Blackburn JB, Lupashin VV. Creating Knockouts of Conserved Oligomeric Golgi Complex Subunits Using CRISPR-Mediated Gene Editing Paired with a Selection Strategy Based on Glycosylation Defects Associated with Impaired COG Complex Function. Methods Mol Biol 2016;1496:145–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q, Meng F, Chen S, Plouffe SW, Wu S, Liu S, et al. Hippo signalling governs cytosolic nucleic acid sensing through YAP/TAZ-mediated TBK1 blockade. Nat Cell Biol 2017;19(4):362–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SJ, Jang BC, Lee SW, Yang YI, Suh SI, Park YM, et al. Interferon regulatory factor-1 is prerequisite to the constitutive expression and IFN-gamma-induced upregulation of B7-H1 (CD274). FEBS Lett 2006;580(3):755–62. [DOI] [PubMed] [Google Scholar]
- 21.Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, et al. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell 1989;58(4):729–39. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Xi Y, Li W, McCarthy RL, Stratton SA, Zou W, et al. TRIM28 interacts with EZH2 and SWI/SNF to activate genes that promote mammosphere formation. Oncogene 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narayan V, Pion E, Landre V, Muller P, Ball KL. Docking-dependent ubiquitination of the interferon regulatory factor-1 tumor suppressor protein by the ubiquitin ligase CHIP. J Biol Chem 2011;286(1):607–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012;366(26):2455–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higuchi T, Flies DB, Marjon NA, Mantia-Smaldone G, Ronner L, Gimotty PA, et al. CTLA-4 Blockade Synergizes Therapeutically with PARP Inhibition in BRCA1-Deficient Ovarian Cancer. Cancer Immunol Res 2015;3(11):1257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore K, Colombo N, Scambia G, Kim BG, Oaknin A, Friedlander M, et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N Engl J Med 2018;379(26):2495–505. [DOI] [PubMed] [Google Scholar]
- 27.Shen J, Zhao W, Ju Z, Wang L, Peng Y, Labrie M, et al. PARPi Triggers the STING-Dependent Immune Response and Enhances the Therapeutic Efficacy of Immune Checkpoint Blockade Independent of BRCAness. Cancer Res 2019;79(2):311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chabanon RM, Muirhead G, Krastev DB, Adam J, Morel D, Garrido M, et al. PARP inhibition enhances tumor cell-intrinsic immunity in ERCC1-deficient non-small cell lung cancer. J Clin Invest 2019;129(3):1211–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinet L, Garrido I, Girard JP. Tumor high endothelial venules (HEVs) predict lymphocyte infiltration and favorable prognosis in breast cancer. Oncoimmunology 2012;1(5):789–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinet L, Le Guellec S, Filleron T, Lamant L, Meyer N, Rochaix P, et al. High endothelial venules (HEVs) in human melanoma lesions: Major gateways for tumor-infiltrating lymphocytes. Oncoimmunology 2012;1(6):829–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev 2012;26(12):1300–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Ramakrishnan SK, Triner D, Centofanti B, Maitra D, Gyorffy B, et al. Tumor-selective proteotoxicity of verteporfin inhibits colon cancer progression independently of YAP1. Sci Signal 2015;8(397):ra98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, von Pawel J, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017;389(10066):255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nduom EK, Wei J, Yaghi NK, Huang N, Kong LY, Gabrusiewicz K, et al. PD-L1 expression and prognostic impact in glioblastoma. Neuro Oncol 2016;18(2):195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujita T, Sakakibara J, Sudo Y, Miyamoto M, Kimura Y, Taniguchi T. Evidence for a nuclear factor(s), IRF-1, mediating induction and silencing properties to human IFN-beta gene regulatory elements. EMBO J 1988;7(11):3397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Remoli ME, Gafa V, Giacomini E, Severa M, Lande R, Coccia EM. IFN-beta modulates the response to TLR stimulation in human DC: involvement of IFN regulatory factor-1 (IRF-1) in IL-27 gene expression. Eur J Immunol 2007;37(12):3499–508. [DOI] [PubMed] [Google Scholar]
- 37.Yarilina A, Park-Min KH, Antoniv T, Hu X, Ivashkiv LB. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat Immunol 2008;9(4):378–87. [DOI] [PubMed] [Google Scholar]
- 38.Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell 1993;75(1):83–97. [PubMed] [Google Scholar]
- 39.Ramana CV, Gil MP, Schreiber RD, Stark GR. Stat1-dependent and -independent pathways in IFN-gamma-dependent signaling. Trends Immunol 2002;23(2):96–101. [DOI] [PubMed] [Google Scholar]
- 40.Chikuma S, Suita N, Okazaki IM, Shibayama S, Honjo T. TRIM28 prevents autoinflammatory T cell development in vivo. Nat Immunol 2012;13(6):596–603. [DOI] [PubMed] [Google Scholar]
- 41.Wirsing AM, Ervik IK, Seppola M, Uhlin-Hansen L, Steigen SE, Hadler-Olsen E. Presence of high-endothelial venules correlates with a favorable immune microenvironment in oral squamous cell carcinoma. Mod Pathol 2018. [DOI] [PubMed] [Google Scholar]
- 42.Dieterich LC, Ikenberg K, Cetintas T, Kapaklikaya K, Hutmacher C, Detmar M. Tumor-Associated Lymphatic Vessels Upregulate PDL1 to Inhibit T-Cell Activation. Front Immunol 2017;8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hida K, Hida Y, Amin DN, Flint AF, Panigrahy D, Morton CC, et al. Tumor-associated endothelial cells with cytogenetic abnormalities. Cancer Res 2004;64(22):8249–55. [DOI] [PubMed] [Google Scholar]
- 44.Feng FY, de Bono JS, Rubin MA, Knudsen KE. Chromatin to Clinic: The Molecular Rationale for PARP1 Inhibitor Function. Mol Cell 2015;58(6):925–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hassa PO, Hottiger MO. The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-kappaB in inflammatory disorders. Cell Mol Life Sci 2002;59(9):1534–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amir RE, Haecker H, Karin M, Ciechanover A. Mechanism of processing of the NF-kappa B2 p100 precursor: identification of the specific polyubiquitin chain-anchoring lysine residue and analysis of the role of NEDD8-modification on the SCF(beta-TrCP) ubiquitin ligase. Oncogene 2004;23(14):2540–7. [DOI] [PubMed] [Google Scholar]
- 47.Petrilli V, Herceg Z, Hassa PO, Patel NS, Di Paola R, Cortes U, et al. Noncleavable poly(ADP-ribose) polymerase-1 regulates the inflammation response in mice. J Clin Invest 2004;114(8):1072–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bertrand F, Rochotte J, Colacios C, Montfort A, Tilkin-Mariame AF, Touriol C, et al. Blocking Tumor Necrosis Factor alpha Enhances CD8 T-cell-Dependent Immunity in Experimental Melanoma. Cancer Res 2015;75(13):2619–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.