Abstract
Despite a considerable expenditure of time and resources and significant advances in experimental models of disease, cancer research continues to suffer from extremely low success rates in translating preclinical discoveries into clinical practice. The continued failure of cancer drug development, particularly late in the course of human testing, not only impacts patient outcomes, but also drives up the cost for those therapies that do succeed. It is clear that a paradigm shift is necessary if improvements in this process are to occur. One promising direction for increasing translational success is comparative oncology—the study of cancer across species, often involving veterinary patients that develop naturally-occurring cancers. Comparative oncology leverages the power of cross-species analyses to understand the fundamental drivers of cancer protective mechanisms, as well as factors contributing to cancer initiation and progression. Clinical trials in veterinary patients with cancer provide an opportunity to evaluate novel therapeutics in a setting that recapitulates many of the key features of human cancers, including genomic aberrations that underly tumor development, response and resistance to treatment, and the presence of comorbidities that can affect outcomes. With a concerted effort from basic scientists, human physicians and veterinarians, comparative oncology has the potential to enhance the cost-effectiveness and efficiency of pipelines for cancer drug discovery and other cancer treatments.
Keywords: veterinary oncology, cross-species studies, cancer drug discovery, evolutionary biology
Cancer Drug Discovery at a Crossroads
Despite decades of research, cancer ranks as the second-leading cause of mortality globally (GBD 2017 Causes of Death Collaborators 2017). Surgery and radiation therapy are common treatment modalities for local control of primary tumors while systemic therapy is mostly used to treat metastatic disease and as an essential adjuvant for some cancer types. Systemic options include 1) cytotoxic chemotherapies that eradicate rapidly–dividing cells, 2) targeted agents, such as small molecule inhibitors of receptor tyrosine kinases, 3) biologics, such as monoclonal antibodies, as well as combinations of these general classes. In addition to these systemic options, immunotherapies act by reawakening the immune system to target and eradicate cancer cells, often leading to long-term benefits.
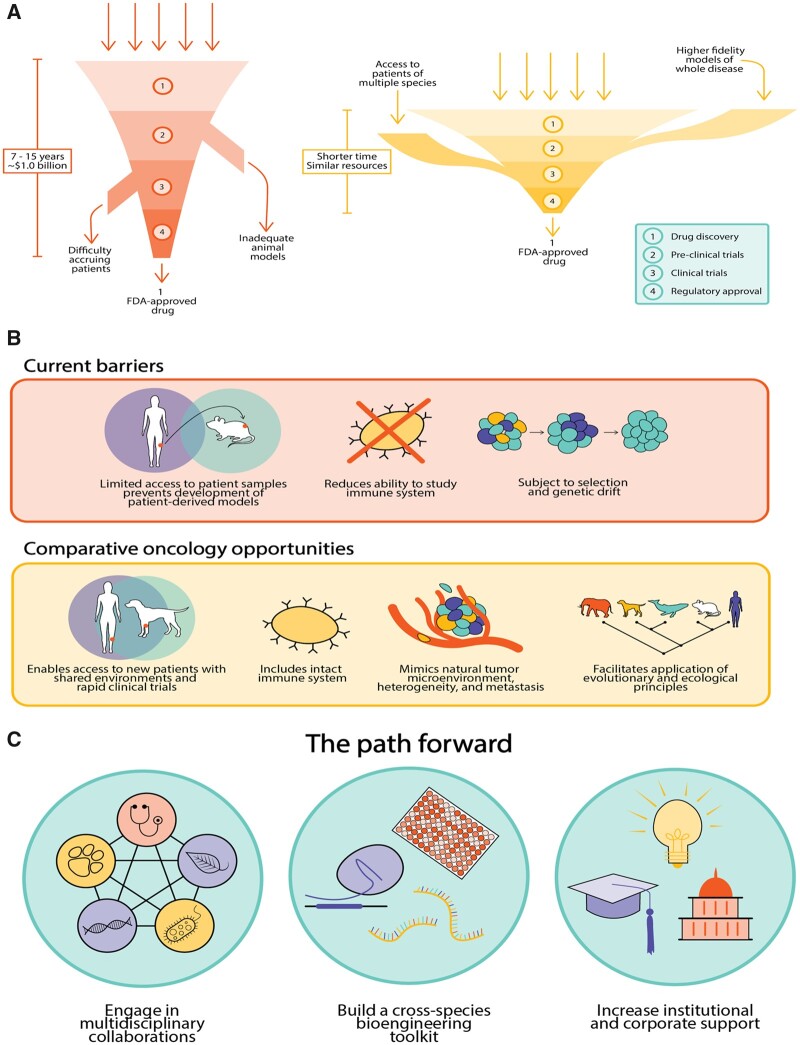
Almost all of these therapies undergo extensive preclinical testing in laboratories, costing millions to billions of dollars while taking years to complete before their evaluation in clinical trials. Although the exact numbers are subject to debate and reanalysis, recent studies suggest that it costs ∼$1.0 billion and from 7 to 15 years for a new drug to pass through the translational pipeline from the bench to the bedside (Paul et al. 2010; Prasad and Mailankody 2017) (fig. 1A). Even for therapies that reach clinical trials, failure rates for new cancer drugs are >80% in Phase II and 50% in Phase III clinical trials, and these failure have been rising since 2001 (Arrowsmith 2011a, 2011b). Clearly, despite extensive efforts, the protocols for bringing new anticancer treatments to the clinic are largely failing: New drugs are not delivered as quickly, as cheaply, or as effectively as required. The reasons for this failure are numerous, encompassing limitations across the entire drug discovery pipeline, from preclinical discovery and validation to clinical evaluation (fig. 1B).
Fig. 1.
Comparative oncology helps overcome current barriers to drug discovery. (A) Challenges to cancer drug discovery include limited access to patient samples for testing, inadequate models to study the immune system and tumor microenvironment, and an absence of models that fully recapitulate inter- and intrapatient tumor heterogeneity. Integrating comparative oncology into the drug discovery process can overcome these challenges and may speed efforts to design and test therapies and treatment paradigms. (B) The current drug discovery process is highly inefficient, taking almost a decade and costing ∼$1 billion to reach an FDA approval. Adopting comparative approaches adds additional inputs to the pipeline in the form of improved models and more diverse patients, leading to more rapid results. (C) Robust implementation of comparative approaches will require 1) institutions to foster cross-disciplinary efforts in oncology; 2) a concerted effort to build tools to study cancer across species; and 3) collaborative partnerships with academia, government, and industry.
First, despite the fact that virtually all cancer patients with solid tumors die from metastatic disease, nearly all studies evaluate a drug’s effectiveness by quantifying the change in growth of a subcutaneously-implanted cancer cell line into an immunocompromised mouse as mouse xenografts. These studies often pay no attention to the critical contributions of the tumor microenvironment, the influence of the immune system, and how the drug performs in the context of both macroscopic and microscopic metastatic disease. To address some of these limitations, researchers have increasingly employed patient-derived models of cancer, in which patient tumors or early passage cell lines are implanted into immunocompromised mice as patient-derived xenografts or grown as organoids to preserve the tumor microenvironment (Day et al. 2015). However, overall, these studies suffer from 1) a lack of access to patient samples, 2) selection and genetic drift over successive passages (Ben-David et al. 2017), and 3) inability to study the contributions of the immune system.
Second, the vast majority of preclinical studies do not consider both the extensive genetic (Wu et al. 2018; Guo et al. 2019; Hai et al. 2019; Hoffman et al. 2019; Zhang et al. 2019) and nongenetic (Jia et al. 2018; Risom et al. 2018; Ryser et al. 2018) heterogeneity within a single patient and the evolutionary forces at play during metastasis that may alter the genomic (Yates et al. 2017; Reiter et al. 2018; Wang et al. 2019) and epigenetic (McDonald et al. 2017) landscapes of metastases.
Finally, another limitation of the current drug discovery pipeline is the near-universal reliance on just two species—humans and mice—to understand cancer’s fundamental mechanisms and identify new treatments. Traditional efforts have almost exclusively relied on 1) human cancer cells in vitro, 2) human cancer cells implanted into immunocompromised mice, and/or 3) genetically engineered mouse models of cancer. Although these systems have provided important insights, particularly with respect to the fundamental molecular and genomic drivers of cancer, a nonreductionist approach to modeling the complexities of cancer development and treatment is necessary to effectively improve translational outcomes. Building off the notion that no single modeling system faithfully recapitulates human cancers, studying cancer across a diverse array of species provides a unique opportunity to interrogate factors that underpin both resistance and susceptibility to cancer, provides access to larger sample sizes from multiple species, and permits the study of new treatments in the setting of spontaneous metastasis complicated by comorbidities.
Can Comparative Oncology Approaches Revolutionize Cancer Drug Discovery?
Comparative oncology is grounded in the One Health concept that animal, environmental, and human health is inexorably linked (Atlas 2013). The core idea of One Health is as old as many of our principles of modern medicine. It is born from the minds of some of the most influential scientists and physicians in medicine, such as Louis Pasteur, Robert Koch, William Osler, and Rudolph Virchow (Atlas 2013). As a subspecialty of One Health that focuses specifically on cancer, comparative oncology bridges the human and veterinary world to study cancer across species, to understand its causes and consequences, and to identify and test new therapies.
Comparative oncology has historically turned to pet dogs with naturally-occurring cancers as unique “models” that closely resemble human cancer. The study of pet dogs with cancer, in contrast to mouse models, offers a high incidence of spontaneous tumors that mirror the biology (Scott et al. 2018), genetics (Edidin 1986; Sakthikumar et al. 2018; Gardner et al. 2019), and both inter- and intratumoral heterogeneity (https://doi.org/10.1101/517086) of humans. Pet dogs share environmental factors with humans, have an intact immune system similar to the human immune system, and possess similar clinical disease presentation, including progression, resistance, recurrence, and metastasis. Moreover, the larger size of dogs compared with rodents permits treatment with therapeutic regimens and interventions that more closely match those used in people. In addition to these similarities, an advantage to studying cancer in dogs is that they tend to have a shorter course of disease progression than human patients (i.e., timelines are somewhat compressed), meaning that therapeutic discoveries could be made more quickly if we developed a platform for integrating canine cancer into our disease models.
Nature’s Solution to Cancer Prevention
Although comparative oncology has traditionally relied on dogs and other companion animals (e.g., cats, horses) with cancer, other species have the potential to enhance our understanding of this highly-complex disease. Cancer has likely been an important selective force in multicellular organismal evolution (Aktipis et al. 2015). Over evolutionary time, natural selection has provided species with mechanisms of tumor suppression to maintain the ability to renew somatic tissue without the risk to the organism that attends neoplastic transformation. Although multicellularity depends upon such mechanisms, very little multicellular life on the planet lacks cancer or cancer-like phenomena (Aktipis et al. 2015). Employing comparative genomics as a tool, cancer biologists have been uncovering both the novel and the shared mechanisms of cancer protection across the tree of life. Shared mechanisms can provide insights into cancer protection universals, such as highly-conserved tumor suppressor genes whereas novel cancer suppression mechanisms can potentially guide new therapeutic strategies.
Recent comparative genomics studies of cancer tend to focus on species with exaggerated life history traits, such as an extended lifespan or large body size. This is because cancer is most often a product of somatic evolution, characterized by the accumulation of mutations in a system where every cell division in a multicellular organism increases the chance that a cancer-causing somatic mutation could occur. According to these principles, the risk of developing cancer should be a function of the number of cell divisions in an organism’s lifetime. However, bigger or longer-lived organisms do not necessarily get more cancer (Abegglen et al. 2015; Caulin et al. 2015). This phenomenon is known as Peto’s Paradox (Peto et al. 1975). Large body size and longevity have evolved independently multiple times during the evolution of complex multicellularity, suggesting there may be many solutions to cancer resistance waiting to be uncovered.
Consistent with this, a few mechanisms of cancer suppression have recently been elucidated. Redundancy of tumor suppressor genes due to gene duplication has been shown to enhance cancer protection. Examples of duplications that increase cancer protection include the expansion of TP53 in elephants (Abegglen et al. 2015; Sulak et al. 2016) and FBX031 in bats (Seim et al. 2013; Caulin et al. 2015) as well as large segmental duplications in dolphins and whales that are enriched in apoptosis and tumor suppression genes (Tollis et al. 2019). Beyond gene duplications, cancer comparative genomic studies have uncovered evidence for positive selection of various tumor suppressor genes in large and/or long-lived species (reviewed in Tollis et al. 2017). These comparative genomics studies will be useful for providing an evolutionarily-informed list of molecular targets that may be exploitable in the service of novel strategies for human cancer prevention and treatment.
In addition to differences in cancer initiation across species, differences also exist in the metastatic potential across species (D’Souza and Wagner 2014). Some of these differences in malignancy across species are associated with their varying degrees of fetal placental invasion (D’Souza and Wagner 2014). Indeed, studies of placental genes have pinpointed novel mechanisms of metastasis in human cancer. For example, the placental invasion gene, PEG10, promotes invasion and metastasis across multiple cancer types (Akamatsu et al. 2015; Li et al. 2016; Peng et al. 2017; Zhao et al. 2017). Further studies of the relationships between placental invasion and metastasis may reveal novel mechanisms to prevent or delay metastasis.
Harnessing comparative oncology to improve drug discovery is still in its nascent stages. Naturally-occurring cancers in dogs and other species offer an unparalleled opportunity to understand cancer genomes, to learn about disease progression, and to trial new investigational drugs in settings where human trials would take too long to complete. Another advantage within the veterinary medicine setting is the ability to offer trials with novel therapies that would be impractical in human medicine. For example, new therapies are most often tested in human patients who have already failed standard-of-care treatments. Such studies typically take years to design and implement and are already based upon years of failure to respond to standard therapies. Many of these patients already have multidrug-resistant disease at the start of the trial. In the veterinary setting, however, veterinarians can test new therapies in earlier disease stages, providing critical data on pharmacodynamics, pharmacokinetics, and evidence of efficacy that may inform future studies and help optimize drug dosing and schedule (Gardner et al. 2016). In addition, integrating the unique access to patients afforded by comparative oncology (for instance, the relatively high numbers of sarcomas in pet dogs) with patient-derived models of cancer could also provide more robust and higher powered preclinical platforms for testing new drugs. Despite the promise of comparative oncology approaches, much work needs to be done to address key barriers to effectively apply the comparative oncology paradigm in a robust, consistent, and truly meaningful way (fig. 1).
Building a Comparative Oncology Toolkit
Much of what we know about cancer comes from decades of study of just a few model species: fruit flies, yeast, zebrafish, nematodes, and mice. Early work on these models relied on inbreeding to reduce genetic variability and thereby facilitate dissection of genetic traits (Ericsson et al. 2013). Later efforts saw most of these model systems being among the first with fully sequenced genomes (C. elegans Sequencing Consortium 1998; Adams et al. 2000; Mouse Genome Sequencing Consortium et al. 2002). Availability of these genomes has led to the development of critical tools with which to study the molecular mechanisms of both normal and cancerous cells.
A similar effort is needed to broaden the toolkit for use in companion animal species. Development of cost-efficient cross-species platforms and reagents, including antibodies, RNA interference reagents, CRISPR/Cas9 reagents, and genomics platforms (e.g., exome-sequencing capture kits) would greatly enhance the ability of researchers to apply comparative oncology approaches. Early pioneering work relied on custom-made genomics platforms (Dunn et al. 2000; Elvers et al. 2015) and the use of cross-reacting therapeutic antibodies from other species to demonstrate efficacy (Maekawa et al. 2014). Further investment is sorely needed to extend the toolkit for studying cancer in companion animals.
Increasing demand will help fuel the creation of these reagents by biotech companies. Ongoing, large-scale, multispecies genome-sequencing efforts (i5K Consortium 2013; GIGA Community of Scientists et al. 2014; Koepfli et al. 2015; Lewin et al. 2018) will facilitate development of genomic, immunologic, and therapeutic small molecule tools. In addition, inclusion of diverse genomes into current analysis toolkits and web portals will enhance researchers’ abilities to design molecular biology and immuno-oncology reagents. The cumulative impact of these collective efforts from multiple groups has the potential to greatly impact how we diagnose and treat cancer by integrating the comparative oncology paradigm into drug discovery pipelines.
Bridging Veterinary and Human Medicine
A major barrier for incorporation of comparative oncology into the current drug discovery pipeline has been the relative lack of communication between veterinarians, zoologists, evolutionary biologists, and human physicians. Veterinarians hold a wealth of knowledge about medicine across species. The establishment of formal relationships between veterinary schools and medical schools can help bring together and incentivize veterinarian/human physician relationships. Several of these partnerships have emerged, including the Clinical and Translational Science Award One Health Alliance (COHA) (https://ctsaonehealthalliance.org/), the National Cancer Institute’s (NCI) Comparative Oncology Program (https://ccr.cancer.gov/Comparative-Oncology-Program), the NCI Comparative Oncology Trials Consortium (https://ccr.cancer.gov/Comparative-Oncology-Program/sponsors/consortium), and the Duke/North Carolina State University/V Foundation joint Consortium for Canine Comparative Oncology (https://c3oncology.org/).
In addition to these relationships, training in oncology should include as standard practice training in evolutionary biology. Comparative approaches and evolutionary biology should be part of the curriculum at medical schools. Cancer biology can play a more prominent role in undergraduate education in the biological sciences. The somatic evolution of cancer can be usefully compared and contrasted with the evolution across organismal generations, which is unfortunately sometimes the sole focus of evolutionary biology in undergraduate curricula.
Cancer and multicellularity are intimately connected (Aktipis et al. 2015). Studying cancer in only one or two species impairs our ability to fully and broadly understand oncogenesis and cancer prevention strategies as basic components of multicellular life. A better appreciation for evolutionary biology and comparative medicine will also help train physicians who can more fully understand the essential processes of disease. Engagement with ecologists, phylogeneticists, comparative physiologists, and evolutionary biologists will illuminate how evolutionary, environmental, and life history factors contribute to cancer (Somarelli et al. 2019).
Acceptance from biotech and pharma of the value of comparative oncology can spur innovation and facilitate evaluation and testing of novel agents. For example, novel COX2 inhibitors were tested in canines with transitional cell carcinoma prior to being evaluated in humans (reviewed in Gardner et al. 2016). Similarly, a study of mifamurtide in canine osteosarcoma showed improvement in overall survival and led to trials in humans that showed similar survival benefits (reviewed in Gardner et al. 2016). Although it remains an investigational drug in United States, mifamurtide was subsequently approved in Europe for the treatment of human osteosarcoma (reviewed in Gardner et al. 2016).
Establishment of comparative oncology approaches as a formal part of the Food and Drug Administration’s drug approval pipeline has the potential to speed testing of new therapies. In this way, even fast failures can be an enormous success. In one recent example of this, Somarelli JA, Ruprecht G, Altunel E, Rao S, Lazarides A, Hoskinson SM, Sheth MU, Cheng S, Kim SY, Ware KE, Agarwal A, Selmic LE, Eward C, Eward WC, Hsu SD. (unpublished data) identified proteasome inhibitors as a promising candidate therapy for osteosarcoma. The team verified the efficacy of proteasome inhibitors both in vitro and in vivo using patient-derived models of cancer. Based on these encouraging results, a pilot canine clinical trial was initiated in 10 dogs with osteosarcoma. The trial met the patient accrual target in <1 year, but the proteasome inhibitor was ineffective in prolonging metastasis-free survival. Within 1 year, the trial showed proteasome inhibitors to be ineffective in a specific clinical setting. Conversely, the same trial in humans took 7 years before it closed due to low patient accrual, and the drug could not be evaluated (Maki et al. 2005). The accelerated oncologic progression of canine cancer enabled a prompt and cost-effective evaluation of a new drug candidate, whereas clinicians were unable to evaluate the effects of the drug in humans. These “fast failures” allow a more nimble assessment of novel therapeutic strategies and a more rapid shift to assessment of the next candidate. Such studies do not preclude additional testing in humans, but complement and inform efforts in the human setting.
The Path Ahead
Despite significant and continuing advances in the understanding of cancer, there remains an urgent, unmet need to improve cancer drug discovery pipelines. The translation from bench to bedside is slow, inefficient, and extremely costly to patients, their families, and the healthcare system (fig. 1A). Studying cancer across the tree of life has the potential to alleviate impediments to new drug discovery. As described earlier, a greater breadth of comparison across species yields better insights into the fundamental processes of cancer; there is an immense opportunity to learn about biology and mechanism from comparative approaches (fig. 1C). Naturally-occurring cancer in animals provides an opportunity to improve the understanding of drivers of cancer, guiding cancer drug development and pinpointing the most promising therapies for human trials. Broader application of these advantages will require hard work in tool development and engagement across disciplines. With continued effort, the field of comparative oncology has the potential to become an essential component of translational cancer drug discovery that will improve accurate development of new treatments for cancer patients and their families.
Acknowledgments
JAS acknowledges funding from the Society for Molecular Biology and Evolution and the Triangle Center for Evolutionary Medicine. JAS and WCE acknowledge support from the Consortium for Canine Comparative Oncology.
References
- Abegglen LM, Caulin AF, Chan A, Lee K, Robinson R, Campbell MS, Kiso WK, Schmitt DL, Waddell PJ, Bhaskara S, et al. 2015. Potential mechanisms for cancer resistance in elephants and comparative cellular response to DNA damage in humans. JAMA 314(17):1850–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287(5461):2185–2195. [DOI] [PubMed] [Google Scholar]
- Akamatsu S, Wyatt AW, Lin D, Lysakowski S, Zhang F, Kim S, Tse C, Wang K, Mo F, Haegert A, et al. 2015. The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Rep. 12(6):922–936. [DOI] [PubMed] [Google Scholar]
- Aktipis CA, Boddy AM, Jansen G, Hibner U, Hochberg ME, Maley CC, Wilkinson GS. 2015. Cancer across the tree of life: cooperation and cheating in multicellularity. Philos Trans R Soc Lond B Biol Sci. 370:1673–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowsmith J. 2011a. Trial watch: phase II failures: 2008–2010. Nat Rev Drug Discov. 10(5):328–329. [DOI] [PubMed] [Google Scholar]
- Arrowsmith J. 2011b. Trial watch: phase III and submission failures: 2007–2010. Nat Rev Drug Discov. 10(2):87.. [DOI] [PubMed] [Google Scholar]
- Atlas RM. 2013. One health: its origins and future. Curr Top Microbiol Immunol. 365:1–13. [DOI] [PubMed] [Google Scholar]
- Ben-David U, Ha G, Tseng YY, Greenwald NF, Oh C, Shih J, McFarland JM, Wong B, Boehm JS, Beroukhim R, et al. 2017. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet. 49(11):1567–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282:2012–2018. [DOI] [PubMed] [Google Scholar]
- Caulin AF, Graham TA, Wang LS, Maley CC. 2015. Solutions to Peto’s paradox revealed by mathematical modelling and cross-species cancer gene analysis. Philos Trans R Soc Lond B Biol Sci. 370:1850–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CP, Merlino G, Van Dyke T. 2015. Preclinical mouse cancer models: a maze of opportunities and challenges. Cell 163(1):39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza AW, Wagner GP. 2014. Malignant cancer and invasive placentation: a case for positive pleiotropy between endometrial and malignancy phenotypes. Evol Med Public Health. 2014:136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn KA, Thomas R, Binns MM, Breen M. 2000. Comparative genomic hybridization (CGH) in dogs–application to the study of a canine glial tumour cell line. Vet J. 160(1):77–82. [DOI] [PubMed] [Google Scholar]
- Edidin M. 1986. Major histocompatibility complex haplotypes and the cell physiology of peptide hormones. Hum Immunol. 15(4):357–365. [DOI] [PubMed] [Google Scholar]
- Elvers I, Turner-Maier J, Swofford R, Koltookian M, Johnson J, Stewart C, Zhang CZ, Schumacher SE, Beroukhim R, Rosenberg M, et al. 2015. Exome sequencing of lymphomas from three dog breeds reveals somatic mutation patterns reflecting genetic background. Genome Res. 25(11):1634–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson AC, Crim MJ, Franklin CL. 2013. A brief history of animal modeling. Mo Med. 110(3):201–205. [PMC free article] [PubMed] [Google Scholar]
- Gardner HL, Fenger JM, London CA. 2016. Dogs as a model for cancer. Annu Rev Anim Biosci. 4(1):199–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner HL, Sivaprakasam K, Briones N, Zismann V, Perdigones N, Drenner K, Facista S, Richholt R, Liang W, Aldrich J, et al. 2019. Canine osteosarcoma genome sequencing identifies recurrent mutations in DMD and the histone methyltransferase gene SETD2. Commun Biol. 2(1):266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD 2017 Causes of Death Collaborators. 2017. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390:1151–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIGA Community of Scientists, Bracken-Grissom H, Collins AG, Collins T, Crandall K, Distel D, Dunn C, Giribet G, Haddock S, Knowlton N, et al. 2014. The Global Invertebrate Genomics Alliance (GIGA): developing community resources to study diverse invertebrate genomes. J Hered. 105:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Chen Z, Xu C, Zhang X, Yan H, Su J, Yang J, Xie Z, Guo W, Li F, et al. 2019. Intratumoral heterogeneity of EGFR-activating mutations in advanced NSCLC patients at the single-cell level. BMC Cancer. 19(1):369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai P, Imai T, Xu S, Zhang R, Aft RL, Zou J, Wang LV. 2019. High-throughput, label-free, single-cell photoacoustic microscopy of intratumoral metabolic heterogeneity. Nat Biomed Eng. 3(5):381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman M, Gillmor AH, Kunz DJ, Johnston MJ, Nikolic A, Narta K, Zarrei M, King J, Ellestad K, Dang NH, et al. 2019. Intratumoral genetic and functional heterogeneity in pediatric glioblastoma. Cancer Res. 79(9):2111–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- i5K Consortium. 2013. The i5K Initiative: advancing arthropod genomics for knowledge, human health, agriculture, and the environment. J Hered. 104:595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Q, Wu W, Wang Y, Alexander PB, Sun C, Gong Z, Cheng JN, Sun H, Guan Y, Xia X, et al. 2018. Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat Commun. 9(1):5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepfli KP, Paten B, Genome K, O’Brien SJ. 2015. The Genome 10K Project: a way forward. Annu Rev Anim Biosci. 3(1):57–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin HA, Robinson GE, Kress WJ, Baker WJ, Coddington J, Crandall KA, Durbin R, Edwards SV, Forest F, Gilbert MTP, et al. 2018. Earth BioGenome Project: sequencing life for the future of life. Proc Natl Acad Sci U S A. 115(17):4325–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xiao R, Tembo K, Hao L, Xiong M, Pan S, Yang X, Yuan W, Xiong J, Zhang Q. 2016. PEG10 promotes human breast cancer cell proliferation, migration and invasion. Int J Oncol. 48(5):1933–1942. [DOI] [PubMed] [Google Scholar]
- Maekawa N, Konnai S, Ikebuchi R, Okagawa T, Adachi M, Takagi S, Kagawa Y, Nakajima C, Suzuki Y, Murata S, et al. 2014. Expression of PD-L1 on canine tumor cells and enhancement of IFN-gamma production from tumor-infiltrating cells by PD-L1 blockade. PLoS One 9(6):e98415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki RG, Kraft AS, Scheu K, Yamada J, Wadler S, Antonescu CR, Wright JJ, Schwartz GK. 2005. A multicenter Phase II study of bortezomib in recurrent or metastatic sarcomas. Cancer 103(7):1431–1438. [DOI] [PubMed] [Google Scholar]
- McDonald OG, Li X, Saunders T, Tryggvadottir R, Mentch SJ, Warmoes MO, Word AE, Carrer A, Salz TH, Natsume S, et al. 2017. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet. 49(3):367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing ConsortiumWaterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420(6915):520–562. [DOI] [PubMed] [Google Scholar]
- Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL. 2010. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat Rev Drug Discov. 9:203–214. [DOI] [PubMed] [Google Scholar]
- Peng YP, Zhu Y, Yin LD, Zhang JJ, Wei JS, Liu X, Liu XC, Gao WT, Jiang KR, Miao Y. 2017. PEG10 overexpression induced by E2F-1 promotes cell proliferation, migration, and invasion in pancreatic cancer. J Exp Clin Cancer Res. 36(1):30.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peto R, Roe FJ, Lee PN, Levy L, Clack J. 1975. Cancer and ageing in mice and men. Br J Cancer. 32(4):411–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad V, Mailankody S. 2017. Research and development spending to bring a single cancer drug to market and revenues after approval. JAMA Intern Med. 177(11):1569–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JG, Makohon-Moore AP, Gerold JM, Heyde A, Attiyeh MA, Kohutek ZA, Tokheim CJ, Brown A, DeBlasio RM, Niyazov J, et al. 2018. Minimal functional driver gene heterogeneity among untreated metastases. Science 361(6406):1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risom T, Langer EM, Chapman MP, Rantala J, Fields AJ, Boniface C, Alvarez MJ, Kendsersky ND, Pelz CR, Johnson-Camacho K, et al. 2018. Differentiation-state plasticity is a targetable resistance mechanism in basal-like breast cancer. Nat Commun. 9(1):3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryser MD, Yu M, Grady W, Siegmund K, Shibata D. 2018. Epigenetic heterogeneity in human colorectal tumors reveals preferential conservation and evidence of immune surveillance. Sci Rep. 8(1):17292.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakthikumar S, Elvers I, Kim J, Arendt ML, Thomas R, Turner-Maier J, Swofford R, Johnson J, Schumacher SE, Alfoldi J, et al. 2018. SETD2 is recurrently mutated in whole-exome sequenced canine osteosarcoma. Cancer Res. 78(13):3421–3431. [DOI] [PubMed] [Google Scholar]
- Scott MC, Temiz NA, Sarver AE, LaRue RS, Rathe SK, Varshney J, Wolf NK, Moriarity BS, O’Brien TD, Spector LG, et al. 2018. Comparative transcriptome analysis quantifies immune cell transcript levels, metastatic progression, and survival in osteosarcoma. Cancer Res. 78(2):326–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seim I, Fang X, Xiong Z, Lobanov AV, Huang Z, Ma S, Feng Y, Turanov AA, Zhu Y, Lenz TL, et al. 2013. Genome analysis reveals insights into physiology and longevity of the Brandt’s bat Myotis brandtii. Nat Commun. 4(1):2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somarelli JA, Gardner H, Cannataro VL, Gunady EF, Boddy AM, Johnson NA, Fisk JN, Gaffney SG, Chuang JH, Li S, et al. 2019. Molecular biology and evolution of cancer: from discovery to action. Mol Biol Evol pii: msz242. doi: 10.1093/molbev/msz242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulak M, Fong L, Mika K, Chigurupati S, Yon L, Mongan NP, Emes RD, Lynch VJ. 2016. TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M, Robbins J, Webb AE, Kuderna LFK, Caulin AF, Garcia JD, Berube M, Pourmand N, Marques-Bonet T, O’Connell MJ, et al. 2019. Return to the sea, get huge, beat cancer: an analysis of cetacean genomes including an assembly for the humpback whale (Megaptera novaeangliae). Mol Biol Evol. 36(8):1746–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M, Schiffman JD, Boddy AM. 2017. Evolution of cancer suppression as revealed by mammalian comparative genomics. Curr Opin Genet Dev. 42:40–47. [DOI] [PubMed] [Google Scholar]
- Wang D, Niu X, Wang Z, Song CL, Huang Z, Chen KN, Duan J, Bai H, Xu J, Zhao J, et al. 2019. Multiregion sequencing reveals the genetic heterogeneity and evolutionary history of osteosarcoma and matched pulmonary metastases. Cancer Res. 79(1):7–20. [DOI] [PubMed] [Google Scholar]
- Wu H, Yu J, Li Y, Hou Q, Zhou R, Zhang N, Jing Z, Jiang M, Li Z, Hua Y, et al. 2018. Single-cell RNA sequencing reveals diverse intratumoral heterogeneities and gene signatures of two types of esophageal cancers. Cancer Lett. 438:133–143. [DOI] [PubMed] [Google Scholar]
- Yates LR, Knappskog S, Wedge D, Farmery JHR, Gonzalez S, Martincorena I, Alexandrov LB, Van Loo P, Haugland HK, Lilleng PK, et al. 2017. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell 32(2):169–184 e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Peng Y, Li C, Li Q, Yu Z, Pang Y, Wu AR, Huang Y, Li H. 2019. Genomic heterogeneity and branched evolution of early stage primary acral melanoma shown by multiregional microdissection sequencing. J Invest Dermatol. 139(7):1526–1534. [DOI] [PubMed] [Google Scholar]
- Zhao M, Sun D, Li X, Xu Y, Zhang H, Qin Y, Xia M. 2017. Overexpression of long noncoding RNA PEG10 promotes proliferation, invasion and metastasis of hypopharyngeal squamous cell carcinoma. Oncol Lett. 14(3):2919–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]