Abstract
Objectives
Rapid rate-of-kill (RoK) is a key parameter in the target candidate profile 1 (TCP1) for the next-generation antimalarial drugs for uncomplicated malaria, termed Single Encounter Radical Cure and Prophylaxis (SERCaP). TCP1 aims to rapidly eliminate the initial parasite burden, ideally as fast as artesunate, but minimally as fast as chloroquine. Here we explore whether the relative RoK of the Medicine for Malaria Venture (MMV) Malaria Box compounds is linked to their mode of action (MoA) and identify scaffolds of medicinal chemistry interest.
Methods
We used a bioluminescence relative RoK (BRRoK) assay over 6 and 48 h, with exposure to equipotent IC50 concentrations, to compare the cytocidal effects of Malaria Box compounds with those of benchmark antimalarials.
Results
BRRoK assay data demonstrate the following relative RoKs, from fast to slow: inhibitors of PfATP4>parasite haemoglobin catabolism>dihydrofolate reductase-thymidylate synthase (DHFR-TS)>dihydroorotate dehydrogenase (DHODH)>bc1 complex. Core-scaffold clustering analyses revealed intrinsic rapid cytocidal action for diamino-glycerols and 2-(aminomethyl)phenol, but slow action for 2-phenylbenz-imidazoles, 8-hydroxyquinolines and triazolopyrimidines.
Conclusions
This study provides proof of principle that a compound’s RoK is related to its MoA and that the target’s intrinsic RoK is also modified by factors affecting a drug’s access to it. Our findings highlight that as we use medicinal chemistry to improve potency, we can also improve the RoK for some scaffolds. Our BRRoK assay provides the necessary throughput for drug discovery and a critical decision-making tool to support development campaigns. Finally, two scaffolds, diamino-glycerols and 2-phenylbenzimidazoles, exhibit fast cytocidal action, inviting medicinal chemistry improvements towards TCP1 candidates.
Introduction
Resistance of Plasmodium falciparum to front-line therapeutic agents necessitates new drugs with a novel mode of action (MoA) to circumvent parasite resistance mechanisms.1 , 2 This need was initially met by the identification of 20 000 hits with submicromolar potency against P. falciparum intraerythrocytic stages from an extensive screening campaign of around four million compounds from the libraries of St. Jude Children’s Research Hospital, TN, USA, Novartis and GSK.2–6 Triaging these hits to establish development priorities requires additional pharmacodynamic information, key amongst which is their rate of kill (RoK).7 Rapid RoK is specifically identified by the Medicine for Malaria Venture (MMV) as a key requirement within a future Single Encounter Radical Cure and Prophylaxis (SERCaP) to treat malaria.1 , 8 The target candidate profile 1 (TCP1) requires an immediate effect to rapidly eliminate parasites, minimally as fast as chloroquine and ideally as fast as artesunate. If resistance renders artemisinin ineffective, TCP1 candidates will ideally replace it.1 , 8
Antimalarial RoK is currently determined in vivo with mouse models or Phase IIa clinical trials.9 It is defined by: (i) the parasite reduction ratio (PRR), the fold-reduction from starting parasitaemia after 48 h (one erythrocyte-stage cycle) of treatment; and (ii) parasite clearance time (PCT), the time until parasites are no longer detectable in peripheral blood films.9 The only in vitro RoK assay that provides the PRR and PCT parameters is the recrudescence assay at GSK, Tres Cantos,10 representing the gold standard for RoK determination in vitro. However, its challenging technical aspects, such as requirements for parasite recrudescence over 21–28 days, limit applicability to small-scale lead validation.7 , 11–13 To address this assay bottleneck, we reported a microplate-based bioluminescence relative RoK (BRRoK) assay that discriminates between minimum essential and ideal TCP1 candidates within 6 h (BRRoK6 h)10 and published RoK data for 370 open-access MMV Malaria Box compounds relative to a panel of known antimalarial benchmarks.2 , 7 In this study, we extend the previous study of RoK for the Malaria Box compounds to demonstrate the following proofs of principle. First, to show that compounds with similar BRRoKs have similar MoAs, we compared the BRRoK6 h of Malaria Box compounds with their predicted MoA. Five clusters emerged, with each representing distinct relative RoKs correlating with different MoAs. Second, to demonstrate that Malaria Box compounds with related scaffolds have similar rates of antimalarial killing, BRRoK6 h values were compared based on compounds’ core scaffold, with five clusters emerging that we then correlated with what we know about potential MoAs. Third, we had previously identified 178 Malaria Box compounds that showed little cytocidal activity within 6 h.10 Therefore, we extended the assay over 48 h (BBRoK48 h) to ensure completion of one intraerythrocytic cycle. Most compounds without activity in the BRRoK6 h assay showed activity in the BRRoK48 h assay, providing links to their MoAs. Our data demonstrate that a revised BRRoK assay at two timepoints, 6 and 48 h, provides a critical decision-making tool for antimalarial drug discovery and development campaigns.
Methods
The transgenic Dd2 P. falciparum clone (Dd2luc)14 , 15 was cultured as described previously.7 The antimalarial drugs and the Malaria Box compounds were prepared as shown in Table S1 (available as Supplementary data at JAC Online). Malaria Box IC50 values were measured in Dd2luc and deposited in the ChEMBL-NTD open-access repository (ChEMBL3392923, see Van-Voorhis et al.16).
The BRRoK48 h assay was carried out as described previously.7 Briefly, compounds were serially diluted (9× IC50, 3× IC50, 1× IC50 and 0.3× IC50 concentrations from a determination of IC50 at 48 h) in 96-multiwell plates, trophozoite-stage (20–26 h post-infection) cultures of Dd2luc were added and mixed by pipetting to give a final 200 μL volume in each well with 3-fold IC50 dilution series of drugs, 1% parasitaemia and 2% haematocrit. To estimate the BRRoK48 h, the plates were incubated continuously at 37°C in the presence of the compounds for 48 h prior to assay. As described previously,7 , 17 40 μL of P. falciparum culture was transferred to a white 96-multiwell plate (Greiner, UK) and lysed with 10 μL of passive lysis buffer (Promega, UK). An equal volume, 50 μL, of the supplied luminogenic substrate was mixed with the lysed parasites and the bioluminescence was measured for 2 s in a GloMax-Multi Detection System (Promega, UK). Experiments were carried out as technical triplicates on the same plate, with three independent biological repeats of each plate performed. Controls in each biological replicate consisted of trophozoite-stage culture with no drug added (100%) or uninfected erythrocytes (0%). The mean and SD of bioluminescence data from three independent biological repeats were expressed as a proportion of the untreated control (100%) and calculated as follows: 100 × [μ(S) − μ(−)/μ(+) − μ(−)], where μ(S), μ(+) and μ(−) represent the means for the sample in question and 100% and 0% controls, respectively. The Z′ score of the BRRoK48 h assay was calculated as follows: Z′=1 − [(3σ (+) + 3σ (−))/μ (+) − μ (−)], where μ (+)and σ (+) are the mean and SD of the no-drug (untreated) positive control, respectively, and μ (−) and σ (−) are the mean and SD from uninfected erythrocytes (negative control), respectively.18 The signal/background (S/B) ratio was calculated as follows: (μ (+) − μ (−))/σ (−).
As previously described,7 a principle components analysis (PCA) was performed on the BRRoK assay data for the MMV Malaria Box compounds (48 h assays using a 9× IC50, 3× IC50, 1× IC50 and 0.3× IC50 series) using the KNIME analytics platform, to reduce the dimensionality of these datasets,19 allowing the concentration–rate relationship to be captured in one parameter. The first principle component (PC1) accounted for 78% of the total variance of the data (see Supplementary data). A zero-meaned PC1 value is used to provide a description of the RoK relative to known antimalarial benchmark controls (see Table S1).7
Results
BRRoK6 h for the Malaria Box identifies compound clusters linked by common modes of antimalarial action
That antiplasmodial in vitro RoK correlates with MoA has been established for a small number of antimalarial drugs, predominantly within classes that have been or are currently used.10 We have previously described a determination of the rates of initial cytocidal kill (over 6 h) using the BRRoK assay for 370 compounds from the Malaria Box open-access drug discovery resource relative to a range of benchmark antimalarials for which both in vitro and in vivo RoK data were available.7 This determination used a P. falciparum strain genetically modified to express a bioluminescent luciferase reporter protein, with cytocidal action determined by loss of bioluminescent signal following exposure to increasing concentrations of test compound. Analysis of the normalized concentration-dependent bioluminescent signals by PCA provides a rank of initial cytocidal action that enables RoK relative to known controls to be described. PC1 values are presented as zero-meaned data where low values such as −97.4 relate to the extremely rapidly acting dihydroartemisinin and higher values, such as 55.4, to the slow-acting atovaquone.7
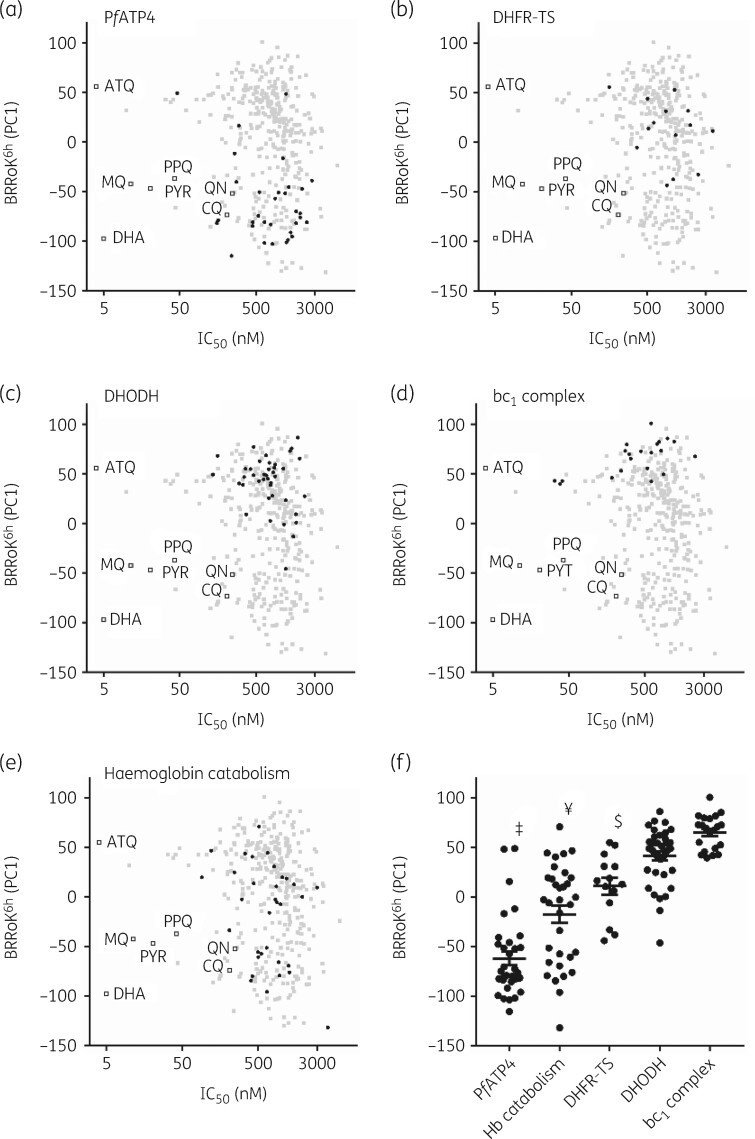
With BRRoK6 h data for 370 Malaria Box compounds, we correlated these with MoA data made available as part of this open-source drug discovery project (Figure 1).20–34 PC1s were plotted against their IC50 (ChEMBL3392923, see Van-Voorhis et al. 16) and mapped against benchmark antimalarials. Compounds with RoK ≥ dihydroartemisinin (PC1 = −97.4) and ≥ chloroquine (PC1 = −73.7, log PRR=4.5, 99% PCT=32 h) meet the TCP1 ideal and minimum essential criteria, respectively. Generally, compounds with RoK ≥ chloroquine are considered fast-acting, those with RoK ≥ quinine (PC1 = −52), mefloquine (PC1=−42.4, log PRR=3.7 and 99.9% PCT=43 h) or piperaquine (PC1 = −37, log PRR=4.6, 99% PCT=33 h) are considered moderate-acting and those with RoK ≥ atovaquone (PC1 = 55.4, log PRR=2.9 and 99.9% PCT=90 h) are slow-acting (Figure 1, Table S1). Thus, compounds with an initial rapid RoK and nanomolar potency, such as artemisinins, occupy the bottom-left quadrant and those such as atovaquone, which although potent is slow-acting, occupy the upper left-hand quadrant (Figure 1).
Figure 1.
Correlating mode of drug action with the BRRoK6 h in the MMV Malaria Box compounds. Zero-meaned PC1 data for known antimalarial drugs (open squares), all Malaria Box compounds (grey-filled squares) and Malaria Box compounds predicted to use the indicated MoA (black-filled circles) for (a) PfATP4, (b) DHFR-TS, (c) DHODH, (d) bc 1 complex and (e) parasite haemoglobin catabolism are plotted against their IC50. Faster initial rates of cytocidal activity are represented by lower PC1 values. See Table S2 for PC1, IC50 and predicted MoA data for individual compounds. (f) One-way ANOVA with post hoc Tukey test comparing the BRRoK6 h data7 for each MoA group (whisker plots represent the mean and SD).16 , 20 , 29 , 30 ‡, PfATP4 cluster, significantly different from all clusters (P<0.01). ¥, parasite haemoglobin catabolism (Hb catabolism), significantly different to DHODH and bc1 complex (P<0.01). $, DHFR-TS, significantly different to the bc1 complex (P<0.01). ATQ, atovaquone; CQ, chloroquine; DHA, dihydroartemisinin; MQ, mefloquine; PPQ, piperaquine; PYR, pyronaridine; QN, quinine.
MoA data were sourced from specific activity assays (e.g. in vitro enzyme inhibition assays) to comparative metabolomic profiling and, as such, the MoA associations for the Malaria Box are often tentative. We hypothesized that compounds with a shared MoA would exhibit similar BRRoK6 h data. Five MoAs, including those of compounds targeting: (i) PfATP4, an Na+-ATPase in the parasite’s plasma membrane; (ii) bifunctional Plasmodium enzyme dihydrofolate reductase-thymidylate synthase (DHFR-TS); (iii) dihydroorotate dehydrogenase (DHODH); (iv) the bc1 complex of the mitochondrial electron transport chain; and (v) parasite haemoglobin catabolism (Figure 1a–e), were clustered. These MoAs were selected because in vitro PRR data are available for ≥10 compounds (Table S2) in each class.16 , 20 , 21 , 27 , 28 , 35 RoKs were identified from fast to slow: PfATP4 > parasite haemoglobin catabolism > DHFR-TS > DHODH > bc1 complex. Using one-way ANOVA with a post hoc Tukey test,7 , 16 , 20 , 29 , 30 we found that compounds targeting PfATP4 exhibit the fastest RoKs and are significantly faster than other clusters (Figure 1f). Compounds targeting parasite haemoglobin catabolism are significantly faster than those targeting DHFR-TS, DHODH and bc1 complex, and compounds targeting DHFR-TS are faster than DHODH and bc1 complex inhibitors (all P<0.01), while other pairwise comparisons were not significant.
BRRoK6 h highlights rapid cytocidal activity for diamino-glycerols and 2-(aminomethyl)phenol scaffolds in the Malaria Box
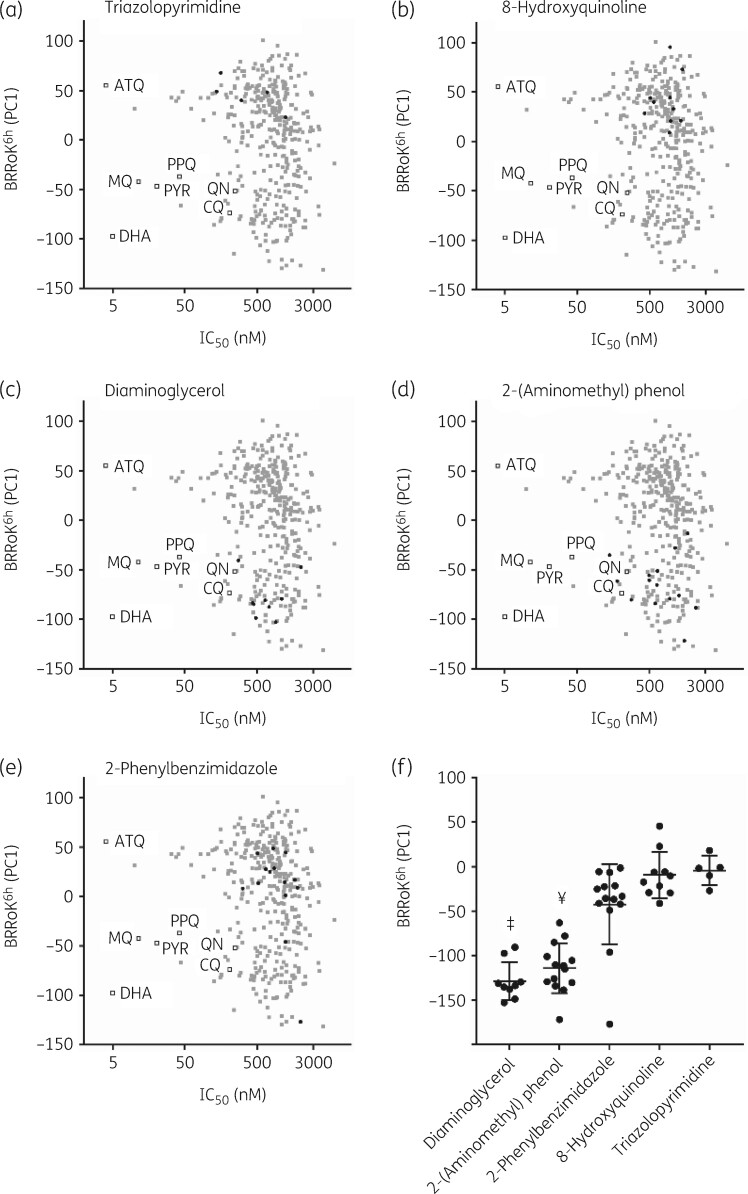
The Malaria Box compounds were selected to be structurally diverse.2 We wished to determine whether substructure analysis of these novel Malaria Box compounds revealed novel core scaffolds with shared RoK activity and thus potentially with new MoAs. BRRoK6 h data were overlaid with five distinct scaffolds: diamino-glycerols; 2-(aminomethyl)phenol; 2-phenylbenzimidazole; 8-hydroxyquinolines; and triazolopyrimidine (Figure 2). Table S3 shows full structures and the core-scaffold substructures, with five or more compounds for each scaffold annotated. We found a fast BRRoK for diamino-glycerols and 2-(aminomethyl)phenols, and a slow BRRoK for 2-phenylbenzimidazoles, 8-hydroxyquinolines and triazolopyrimidines (Figure 2a–e). The five core scaffolds identified BRRoK ranking from fast to slow: diamino-glycerols>2-(aminomethyl)phenol > 2-phenylbenzimidazoles>8-hydroxyquinolines > triazolopyrimidine. The diamino-glycerol scaffold exhibited the fastest cytocidal action among the group [P<0.01 for all, except 2-(aminomethyl)phenol where P>0.05 by ANOVA] (Figure 2f). Similarly, compounds in the 2-(aminomethyl)phenol scaffold exhibited significantly faster action (P<0.01) than the 2-phenylbenzimidazole, 8-hydroxyquinoline and triazolopyrimidine scaffolds (Figure 2f).
Figure 2.
BRRoK6 h data illustrate related compounds in the MMV Malaria Box that share a similar relative RoK. Zero-meaned PC1 data for known antimalarial drugs (open squares), all Malaria Box compounds (grey-filled squares) and Malaria Box compounds sharing the indicated related core scaffolds (black-filled circles) of (a) triazolopyrimidine, (b) 8-hydroxyquinolines, (c) diaminoglycerols, (d) 2-(aminomethyl)phenol and (e) 2-phenylbenzimidazole are plotted against their IC50. Faster initial rates of cytocidal activity are represented by lower PC1 values. See Table S3 for PC1, IC50 and structures for individual compounds. (f) One-way ANOVA with post hoc Tukey test comparing the BRRoK6 h data7 for each group (whisker plots represent the mean and SD) based on the indicated related core scaffold. ‡, Diamino-glycerols, significantly different to 2-Ph-Bz, 8-hydroxyquinolines and triazolopyrimidine scaffolds (P<0.01). ¥, 2-(aminomethyl)phenol, significantly different to 2-phenylbenzimidazoles, 8-hydroxyquinolines and triazolopyrimidine scaffolds (P<0.01). ATQ, atovaquone; CQ, chloroquine; DHA, dihydroartemisinin; MQ, mefloquine; PPQ, piperaquine; PYR, pyronaridine; QN, quinine.
BRRoK48 h confirms slow cytocidal action for a subset of compounds in the Malaria Box
The BRRoK6 h assay identified fast-acting Malaria Box compounds as TCP1 candidates, including the fastest-acting PfATP4 inhibitor spiroindolone MMV396749 (Table S2). However, almost half of the Malaria Box compounds showed little cytocidal activity against intraerythrocytic trophozoites over 6 h. We predicted that these compounds might have a lag phase in their cytocidal action, such as shown by the antimalarial atovaquone with a 48 h lag in cytocidal action.10 We therefore employed a revised BRRoK assay over 48 h (BRRoK48 h) to ensure completion of one full intraerythrocytic cycle. For validation, we selected different benchmark antimalarials, which covered multiple MoAs.7 Dd2luc parasites were exposed to a 3-fold serial dilution (9–0.33× IC50) for 48 h, the resulting bioluminescence signal normalized to an untreated control and the normalized bioluminescent signal plotted against drug concentration (Figure S1). We found the identical relative ranking order of benchmark antimalarial drugs (i.e. artemisinin>chloroquine>4-methanolquinolines>atovaquone) to BRRoK6 h,7 which is identical to both the in vivo and in vitro RoKs.31–33 , 35
We had sufficient material available for 178 slow-acting Malaria Box compounds. Along with the benchmark antimalarial drugs atovaquone, chloroquine, dihydroartemisinin, mefloquine, piperaquine, pyronaridine and quinine, we subjected them to a BRRoK48 h assay (Table S4; Figures S2, S3). The 95% CI for the Z′ score (0.85–0.95), maximum coefficient of variation (0.9%–2.84%) and S/B ratio (2580–5001) indicate a robust and sensitive microplate-based assay of the BRRoK48 h data. Using mean ± SD for each IC50-fold BRRoK48 h normalized bioluminescent signal, a PCA was carried out for concentration-dependent effects (Figure S4; Tables S5–S6). PC1 accounts for 78% of the variance at 48 h, with most contributions provided by the 3× IC50 data.
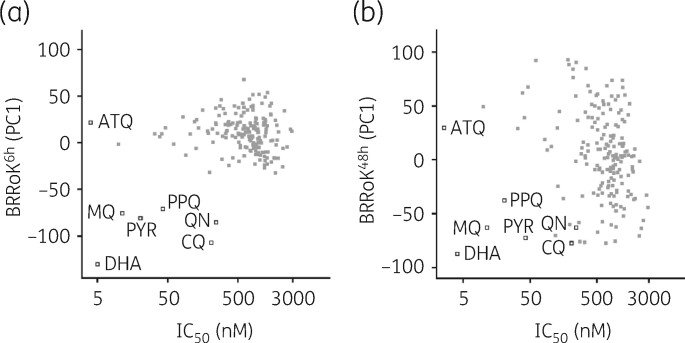
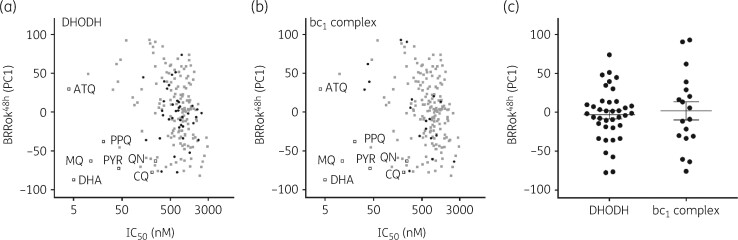
We next plotted BRRoK6 h and BRRoK48 h PC1 against IC50 data (Figure 3, Table S7). Figure 3(a) highlights these compounds’ slow action over 6 h, with compounds clustering adjacent to the slow-acting atovaquone. Plotting BRRoK48 h data against IC50 results in a wide distribution of 48 h RoKs for these compounds (Figure 3b). Interestingly, a number of initially slow-acting compounds now showed a 48 h RoK within the TCP1 target range (>chloroquine) and presumably reflect a shorter lag phase in their action, such as the 24 h lag phase reported for pyrimethamine.10 The majority of compounds, however, still showed a BRRoK PC1 more similar to that of atovaquone, and thus potentially a longer lag phase. To explore this, compounds with two predicted slow-acting MoAs16 , 20 , 22–26 , 29 , 30 , 34 , 36–39 were correlated with BRRoK48 h; 38 were DHODH inhibitors (Figure 4a, Table S7) and 18 were bc1 complex inhibitors (Figure 4b, Table S7). Unfortunately, due to the small sample size, one-way ANOVA did not indicate statistical significance (P>0.05) (Figure 4c) between these different MoAs but did indicate that their longer lag phases resulted in higher BRRoK48 h PC1 scores (Figure 4c).
Figure 3.
Distribution of BRRoK (PC1) against IC50 for the MMV Malaria Box compounds. Zero-meaned PC1 data for 178 compounds in the MMV Malaria Box (grey-filled squares) and 7 benchmark antimalarial drugs (open squares) are plotted against their IC50 for (a) 6 h and (b) 48 h. See Table S2 for PC1 and IC50 data for individual compounds. ATQ, atovaquone; CQ, chloroquine; DHA, dihydroartemisinin; MQ, mefloquine; PPQ, piperaquine; PYR, pyronaridine; QN, quinine.
Figure 4.
Correlating mode of drug action with the BRRoK48 h in the MMV Malaria Box compounds. Zero-meaned PC1 data for known antimalarial drugs (open squares), all Malaria Box compounds (grey-filled squares) and Malaria Box compounds with a predicted MoA (black-filled circles) that target (a) DHODH or (b) the bc1 complex are plotted against their IC50. See Table S2 for PC1, IC50 and predicted MoA data for individual compounds. (c) One-way ANOVA with post hoc Tukey test comparing the BRRoK48 h data (whisker plots represent the mean and SD) for both groups clustered based on the indicated predicted MoA data.16 , 20 , 29 , 30 No significant difference was found (P=0.6). ATQ, atovaquone; CQ, chloroquine; DHA, dihydroartemisinin; MQ, mefloquine; PPQ, piperaquine; PYR, pyronaridine; QN, quinine.
Discussion
The next-generation antimalarial drugs should rapidly eliminate parasite burden, ideally as fast as artesunate, but at least as fast as chloroquine.1 Whilst we have previously used the BRRoK6 h assay to measure the relative RoK for 370 Malaria Box compounds, here we show that BRRoK6 h data provide links to the antimalarial MoAs (with PfATP4 > parasite haemoglobin catabolism > DHFR-TS > DHODH > bc1 complex) and that comparison with scaffold substructures identified five core scaffolds with the relative RoKs: diamino-glycerols>2-(aminomethyl)phenol>2-phenylbenzimidazoles > 8-hydroxyquinolines > triazolopyrimidine. We also predicted that compounds with minimal activity at 6 h might have a lag phase, similar to that seen with atovaquone and DSM265.7 , 10 , 35 Thus, we determined the RoK of apparently slow-acting compounds using a BRRoK48 h assay and showed that many of the slow-acting compounds are likely DHODH and bc1 complex inhibitors. In short, compounds in the Malaria Box with similar targets and chemical core substructure exhibit similar time-dependent RoK dynamics. Although our study is limited to a library of 400 compounds that lack a full biochemical target validation, it provides the proof of principle that BRRoK data offer an opportunity to rapidly prioritize compounds in the TCAMS library, or other libraries, by informing predictions of structure–activity and MoA. Moreover, we note that using the BRRoK assay at two timepoints, 6 and 48 h, we not only have the potential to rapidly identify and discriminate between compounds that meet the ideal and minimum TCP1 criteria, but also identify compounds that likely exhibit a lag time in drug action between 6 and 48 h. This BRRoK assay format, however, does not provide a reliable assessment of the extent and timing of this lag time, as would be reported by a recrudescence assay.10
A compound’s immediate cytocidal activity likely results from the nature of the target and the ease of access to the target. The first aspect considers how quickly a deficit in this target’s function will lead to cell death, i.e. its MoA. In vitro assays of RoK report that antimalarial drugs with a similar MoA result in a similar RoK.7 , 10 We have significantly extended this observation here for the open-source Malaria Box, a critical collection of antimalarial drug discovery compounds. Whilst an important caveat is that for most compounds described the target association is tentative, this library is still the best-described and investigated resource in this endeavour.21 Nonetheless, here we were able to consider five MoA groups due to availability of in vitro PRR data and at least 10 MMV compounds annotated for each MoA from a range of sources.16 , 20 , 21 , 27 , 28 , 35 Specifically: (i) PfATP4 (Figure 1a): 33 compounds are annotated as PfATP4 inhibitors (Figure 1a).20 , 29 In vitro PRR data are available for exemplar PfATP4 inhibitors, (+)-SJ73336,21 a dihydroisoquinoline with a slow-to-moderate RoK, and KAE609/NITD609,27 a spiroindolone with a moderate-to-fast RoK. Most potential Malaria Box PfATP4 inhibitors were reported as having a BRRoK6 h between that of the moderately acting mefloquine10 (comparable to the PRR reference pyrimethamine10) and the rapidly acting dihydroartemisinin. The fastest-acting PfATP4 inhibitor was the spiroindolone MMV396749, with several studies reporting fast-to-moderate cytocidal activity for PfATP4 inhibitors.7 , 16 , 20 , 21 , 27 , 29 The Malaria Box also contains five structural analogues of the slower-acting PfATP4 inhibitor (+)-SJ733; two have PC1s falling between the fast-acting dihydroartemisinin and chloroquine, with the remaining three between mefloquine and atovaquone, supporting the prediction of a moderate-to-slow RoK of the dihydroisoquinolines. (ii) Plasmodium DHFR-TS (Figure 1b): 14 compounds are annotated as DHFR-TS inhibitors, clustering with known antifolate antimalarial drugs that target DHFR-TS, such as P218, pyrimethamine and WR99210.29 , 40–42 Pyrimethamine has a lag phase of 24 h, which is the slowest RoK after atovaquone.10 The BRRoK6 h confirms slow cytocidal activity for this cluster, between that of slow-acting atovaquone and moderate-acting pyronaridine. (iii) DHODH (Figure 1c): 43 compounds are annotated as DHODH inhibitors and BRRoK6 h data show that they share a slow initial cytocidal action. This slow RoK correlates with the atovaquone-like in vitro PRR data for DSM265,35 due to its 24–48 h lag phase. One outlier, MMV666102, falls between pyronaridine and mefloquine, showing a slow-to-moderate cytocidal action. Whilst no additional target information is available, we predict that this compound may have additional targets. (iv) bc1 complex inhibitors: 18 compounds are annotated as bc1 complex inhibitors and were slowly cytocidal in the BRRoK6 h assay; they are comparable to atovaquone, which shares the same MoA.10 Comparing BRRoK with the predicted MoA for all four groups indicates that compounds with a similar MoA have a similar RoK. The predicted MoA used here was primarily obtained through metabolomics16 , 29 , 30 and readily highlights the potential for BRRoK to complement such studies. (v) Parasite haemoglobin catabolism: Allman et al. 29 report a compound group in the Malaria Box that perturb parasite haemoglobin catabolism. Parasite haemoglobin catabolism compounds formed our second fastest-acting cluster (Figure 1e). However, as expected, the BRRoK6 h data reveal a broad RoK range for these compounds, which agrees with metabolomics data, as these compounds have a range of predicted targets. For example, chloroquine, known for accumulation within the digestive vacuole of Plasmodium, clusters with this group but the resulting metaprint is divergent, due to the overall lack of significant metabolic changes or dysregulation induced by chloroquine.29 , 43 MMV390048, which inhibits the phosphatidylinositol 4-kinase (PI4K), and AZ412, which inhibits the putative vacuolar ATPase,12 , 29 , 44 also cluster with this group. Interestingly, as expected from compounds with different targets, the BRRoK6 h appears to form subclusters within this group. Upon additional target data availability, we would predict that this currently broad class of compounds could be further categorized into slow-, moderate- and fast-acting groups.
A second means to classify compounds for comparison with the BRRoK6 h data is through their chemical structure (Figure 2). Our analysis suggests that structurally similar compounds exert similar RoKs. This is not surprising if they share the same target, and our analysis suggests that medicinal chemistry may not only improve IC50 potency for candidates but may also help improve RoK within a chemical class and a well-defined MoA. For example, all five triazolopyrimidine scaffold members (Figure 2a) inhibit DHODH and are structural analogues of DSM265, a known slow-acting compound in clinical trials.35 However, their PC1 varies between 23 and 67, highlighting room to influence the initial cytocidal action within the limits of this chemical class and the intrinsic limits of the MoA. A range of slow cytocidal activity is also reported for 8-hydroxyquinolines (PC1 of 8.8–95.4) (Figure 2b), with one annotated as a DHODH inhibitor. We also report two fast-acting scaffolds: diamino-glycerols and 2-(aminomethyl)phenol (Figure 2c and d). The diamino-glycerol is the fastest scaffold described here, which agrees with a predicted MoA as four of these nine compounds are PfATP4 inhibitors.16 , 20 , 29 It would be interesting to investigate whether the remaining five compounds also affect PfATP4. Three of these five compounds are designated as probe-like and were not characterized in metabolomic studies that focused on drug-like compounds in the Malaria Box.29 Furthermore, five compounds are structurally related to the amino alcohol-carbazoles, which have demonstrated long-lasting and fast-acting antimalarial activity in vivo,45 in agreement with BRRoK6 h measurements here. The next most fast-acting compound cluster was the 2-(aminomethyl)phenol scaffold. Interestingly, BRRoK6 h indicated 5 of 14 compounds with this scaffold are likely inhibitors of parasite haemoglobin catabolism (PC1 between −79 and −51),29 which is the second fastest-acting compound cluster according to MoA comparisons and agrees with our chemical clustering analyses. Eight compounds are probe-like, so metabolomic data are not available; however, Creek et al.30 have shown an artemisinin-like metabolomic signature for three of these compounds, thus confirming the relatively fast action of this scaffold. These data illustrate how BRRoK data can be effectively employed alongside other datasets to inform how decisions are made regarding the selection of targets for further study and/or development.
Given the short time frame of the BRRoK6 h assay, a second attribute that may influence RoK is ease of target access. Within our in vitro assay, compounds must migrate through up to four membranes to access a target within an infected erythrocyte and the biophysical parameters of size, hydrophobicity, hydrogen-bonding capabilities and charge may contribute to how easily access occurs. Another consideration for compounds with a basic charge at physiological pH is that of access/accumulation within the digestive vacuole in the trophozoite, irrespective of the final target site. Biophysical properties span charge type, lipophilicity, polarity, size, 3D shape, flexibility and H-bond properties.46 To investigate what influence molecular properties have on BRRoK6 h, we calculated key biophysical properties for Malaria Box compounds (PC1) (Table S8) and compared compounds with relative RoK faster than dihydroartemisinin and slower than atovaquone to see whether extremes of RoK are associated with significantly different molecular properties. We expanded analyses to include compounds reported to have a common fast MoA (PfATP4), a common slow MoA (bc1 complex), a common fast core [2-(methylamino)-phenols (2-MAP)] and a common slow core (2-phenylbenzimidazoles) (Table S8 and S9). These analyses do not reveal molecular property differences associated with BRRoK, although an important limitation here is the number of compounds in each group. Finally, we compared individual compounds with the fastest BRRoK6 h and slowest BRRoK6 h in the five MoA clusters investigated here and found some small differences (Table S10). The fastest compound in each MoA group often has a lower molecular weight, fewer rotatable bonds and is more aromatic in nature compared with the slowest, suggesting that careful biophysical property control may allow compound design to achieve improvements in RoK within a well-defined MoA/chemical class.
Perhaps the key benefit of RoK analysis considering both the MoA and chemical substructure is that outliers emerge that would appear to warrant additional validation or follow-up. An example from this study are the three 1,2-diaza-9-fluorenones (MMV666021, MMV6666026 and MMV665934) for which the proposed MoA is the bc1 complex, which would imply a very slow RoK. However, we instead found that two of these have a very fast BRRoK. Also, of interest is the structural singleton MMV142383, which has the fastest RoK (PC1 = −131.5) and is categorized as acting by haemoglobin catabolism. Exceptions found using substructure analysis may have either a miscategorized MoA or, alternatively, they may have more than one MoA. The latter would be of particular interest as it would theoretically lead to less resistance if more than one target is involved.
In summary, we provide a demonstration for leading antimalarial drug discovery compounds that their RoK is related to their MoA, and that a compound’s RoK is also likely modified by factors that affect target access. Thus, as we use medicinal chemistry to improve compound potency, we could also influence the RoK for some scaffolds. Our modified BRRoK assay provides the necessary throughput for drug discovery and a critical decision-making tool to support development campaigns. Although our study was performed on a small pool of compounds, the scaffolds we identified provide a strong basis for prioritization of discovery antimalarials. Our core analysis approach has identified two scaffolds, diamino-glycerols and 2-(aminomethyl) phenol, that exhibit fast cytocidal action, inviting medicinal chemistry improvements towards possible TCP1 candidates. Some less-represented scaffolds have also been identified with fast cytocidal action, and medicinal chemistry may allow discovery of compounds that meet the TCP1 profile. Further insights might be gained from our data, as targets are defined for additional MMV compounds.
Supplementary Material
Acknowledgements
We thank MMV for the assembly and supply of the Malaria Box, Dr Leah Imlay for review of the manuscript and Dr Sneha Ray for her medicinal chemistry advice.
Funding
This study was supported by Keele University [ACORN PhD scholarship award and the Charles Wallace Trust (to I.U.) and The Biochemical Society (to P.D.H.)]. G.A.B. is supported, in part, by the Medical Research Council (MR/L000644/1, MC_PC_13069 and MC_PC_14111). D.M.W. was supported by NIH K08 AI103106, a Children’s Clinical Research Advisory Committee (CCRAC) Junior Investigator Award, a CCRAC Early Investigator Award, a 2019 Harrington Scholar-Innovator Award, and funds from the UT Southwestern Department of Pediatrics.
Transparency declarations
None to declare.
References
- 1. Burrows JN, Hooft Van Huijsduijnen R, Möhrle JJ et al. Designing the next generation of medicines for malaria control and eradication. Malar J 2013; 12: 187.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Spangenberg T, Burrows JN, Kowalczyk P et al. The open access Malaria Box: a drug discovery catalyst for neglected diseases. PLoS One 2013; 8: e62926.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gamo F-J, Sanz LM, Vidal J et al. Thousands of chemical starting points for antimalarial lead identification. Nature 2010; 465: 305–10. [DOI] [PubMed] [Google Scholar]
- 4. Guiguemde WA, Shelat AA, Bouck D et al. Chemical genetics of Plasmodium falciparum. Nature 2010; 465: 311–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Plouffe D, Brinker A, McNamara C et al. In silico activity profiling reveals the mechanism of action of antimalarials discovered in a high-throughput screen. Proc Natl Acad Sci USA 2008; 105: 9059–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Meister S, Plouffe DM, Kuhen KL et al. Imaging of Plasmodium liver stages to drive next-generation antimalarial drug discovery. Science 2011; 334: 1372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ullah I, Sharma R, Biagini GA et al. A validated bioluminescence-based assay for the rapid determination of the initial rate of kill for discovery antimalarials. J Antimicrob Chemother 2017; 72: 717–26. [DOI] [PubMed] [Google Scholar]
- 8. Burrows JN, Duparc S, Gutteridge WE et al. New developments in anti-malarial target candidate and product profiles. Malaria J 2017; 16: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. White NJ. Assessment of the pharmacodynamic properties of antimalarial drugs in vivo. Antimicrob Agents Chemother 1997; 41: 1413–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sanz LM, Crespo B, De-Cózar C et al. P. falciparum in vitro killing rates allow to discriminate between different antimalarial mode-of-action. PLoS One 2012; 7: e30949.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baragaña B, Hallyburton I, Lee MCS et al. A novel multiple-stage antimalarial agent that inhibits protein synthesis. Nature 2015; 522: 315–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hameed P S, Solapure S, Patil V et al. Triaminopyrimidine is a fast-killing and long-acting antimalarial clinical candidate. Nat Commun 2015; 6: 6715.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McConville M, Fernández J, Angulo-Barturen Í et al. Carbamoyl triazoles, known serine protease inhibitors, are a potent new class of antimalarials. J Med Chem 2015; 58: 6448–55. [DOI] [PubMed] [Google Scholar]
- 14. Wong EH, Hasenkamp S, Horrocks P. Analysis of the molecular mechanisms governing the stage-specific expression of a prototypical housekeeping gene during intraerythrocytic development of P. falciparum. J Mol Biol 2011; 408: 205–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hasenkamp S, Sidaway A, Devine O et al. Evaluation of bioluminescence-based assays of anti-malarial drug activity. Malar J 2013; 12: 58.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van Voorhis WC, Adams JH, Adelfio R et al. Open source drug discovery with the Malaria Box compound collection for neglected diseases and beyond. PLoS Pathog 2016; 12: e1005763.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hasenkamp S, Wong EH, Horrocks P. An improved single-step lysis protocol to measure luciferase bioluminescence in Plasmodium falciparum. Malar J 2012; 11: 42.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang J-H, Chung TDY, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 1999; 4: 67–73. [DOI] [PubMed] [Google Scholar]
- 19. Berthold MR, Cebron N, Dill F et al. KNIME - the Konstanz information miner. In: Preisach C, Berhardt H, Schmidt-Theime L et al. , eds. Data Analysis, Machine Learning and Applications. Germany: Springer, 2007; 319–26. [Google Scholar]
- 20. Lehane AM, Ridgway MC, Baker E et al. Diverse chemotypes disrupt ion homeostasis in the malaria parasite. Mol Microbiol 2014; 94: 327–39. [DOI] [PubMed] [Google Scholar]
- 21. Fedewa G, Clark JA, Kirk K et al. (+)-SJ733, a clinical candidate for malaria that acts through ATP4 to induce rapid host-mediated clearance of Plasmodium. Proc Natl Acad Sci USA 2014; 111: E5455–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fong KY, Sandlin RD, Wright DW. Identification of β-hematin inhibitors in the MMV Malaria Box. Int J Parasitol Drugs Drug Resist 2015; 5: 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu L, Richard J, Kim S et al. Small molecule screen for candidate antimalarials targeting Plasmodium kinesin-5. J Biol Chem 2014; 289: 16601–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dickerman BK, Elsworth B, Cobbold SA et al. Identification of inhibitors that dually target the new permeability pathway and dihydroorotate dehydrogenase in the blood stage of Plasmodium falciparum. Sci Rep 2016; 6: 37502.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tiwari NK, Reynolds PJ, Calderón AI. Preliminary LC-MS based screening for inhibitors of Plasmodium falciparum thioredoxin reductase (PfTrxR) among a set of antimalarials from the Malaria Box. Molecules 2016; 21: 424.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paiardini A, Bamert RS, Kannan-Sivaraman K et al. Screening the Medicines for Malaria Venture ‘Malaria Box’ against the Plasmodium falciparum aminopeptidases, M1, M17 and M18. PLoS One 2015; 10: e0115859.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rottmann M, McNamara C, Yeung BKS et al. Spiroindolones, a potent compound class for the treatment of malaria. Science 2010; 329: 1175–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coteron JM, Marco M, Esquivias J et al. Structure-guided lead optimization of triazolopyrimidine-ring substituents identifies potent Plasmodium falciparum dihydroorotate dehydrogenase inhibitors with clinical candidate potential. J Med Chem 2011; 54: 5540–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Allman EL, Painter HJ, Samra J et al. Metabolomic profiling of the Malaria Box reveals antimalarial target pathways. Antimicrob Agents Chemother 2016; 60: 6635–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Creek DJ, Chua HH, Cobbold SA et al. Metabolomics-based screening of the Malaria Box reveals both novel and established mechanisms of action. Antimicrob Agents Chemother 2016; 60: 6650–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bahamontes-Rosa N, Rodríguez-Alejandre A, González-Del-Rio R et al. A new molecular approach for cidal vs static antimalarial determination by quantifying mRNA levels. Mol Biochem Parasitol 2012; 181: 171–7. [DOI] [PubMed] [Google Scholar]
- 32. Linares M, Viera S, Crespo B et al. Identifying rapidly parasiticidal anti-malarial drugs using a simple and reliable in vitro parasite viability fast assay. Malar J 2015; 14: 441.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pukrittayakamee S, Chantra A, Simpson JA et al. Therapeutic responses to different antimalarial drugs in vivax malaria. Antimicrob Agents Chemother 2000; 44: 1680–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ahyong V, Sheridan CM, Leon KE et al. Identification of Plasmodium falciparum specific translation inhibitors from the MMV Malaria Box using a high throughput in vitro translation screen. Malar J 2016; 15: 173.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Phillips MA, Lotharius J, Marsh K et al. A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria. Sci Transl Med 2015; 7: 296ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aroonsri A, Akinola O, Posayapisit N et al. Identifying antimalarial compounds targeting dihydrofolate reductase-thymidylate synthase (DHFR-TS) by chemogenomic profiling. Int J Parasitol 2016; 46: 527–35. [DOI] [PubMed] [Google Scholar]
- 37. Von Koschitzky I, Gerhardt H, Lämmerhofer M et al. New insights into novel inhibitors against deoxyhypusine hydroxylase from Plasmodium falciparum: compounds with an iron chelating potential. Amino Acids 2015; 47: 1155–66. [DOI] [PubMed] [Google Scholar]
- 38. Hain AUP, Bartee D, Sanders NG et al. Identification of an Atg8-Atg3 protein-protein interaction inhibitor from the Medicines for Malaria Venture Malaria Box active in blood and liver stage Plasmodium falciparum parasites. J Med Chem 2014; 57: 4521–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vallières C, Avery SV. The candidate antimalarial drug MMV665909 causes oxygen-dependent mRNA mistranslation and synergizes with quinoline-derived antimalarials. Antimicrob Agents Chemother 2017; 61: e00459-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Basco LK, de Pécoulas PE, Wilson CM et al. Point mutations in the dihydrofolate reductase-thymidylate synthase gene and pyrimethamine and cycloguanil resistance in Plasmodium falciparum. Mol Biochem Parasitol 1995; 69: 135–8. [DOI] [PubMed] [Google Scholar]
- 41. Canfield CJ, Milhous WK, Ager AL et al. PS-15: a potent, orally active antimalarial from a new class of folic acid antagonists. Am J Trop Med Hyg 1993; 49: 121–6. [DOI] [PubMed] [Google Scholar]
- 42. Yuthavong Y, Tarnchompoo B, Vilaivan T et al. Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target. Proc Natl Acad Sci USA 2012; 109: 16823–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cobbold SA, Chua HH, Nijagal B et al. Metabolic dysregulation induced in Plasmodium falciparum by dihydroartemisinin and other front-line antimalarial drugs. J Infect Dis 2016; 213: 276–86. [DOI] [PubMed] [Google Scholar]
- 44. Younis Y, Douelle F, Feng T-S et al. 3,5-Diaryl-2-aminopyridines as a novel class of orally active antimalarials demonstrating single dose cure in mice and clinical candidate potential. J Med Chem 2012; 55: 3479–87. [DOI] [PubMed] [Google Scholar]
- 45. Charman SA, Greco B, Bashyam S et al. Long-lasting and fast-acting in vivo efficacious antiplasmodial azepanylcarbazole amino alcohol. ACS Med Chem Lett 2017; 8: 1304–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Veber DF, Johnson SR, Cheng HY et al. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 2002; 45: 2615–23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.