Abstract
The spike glycoprotein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the first point of contact for the virus to recognize and bind to host receptors, is the focus of biomedical research seeking to effectively prevent and treat coronavirus disease (COVID-19). The mass production of spike glycoproteins is usually carried out in different cell systems. Studies have been shown that different expression cell systems alter protein glycosylation of hemagglutinin and neuraminidase in the influenza virus. However, it is not clear whether the cellular system affects the spike protein glycosylation. In this work, we investigated the effect of an expression system on the glycosylation of the spike glycoprotein and its receptor-binding domain. We found that there are significant differences in the glycosylation and glycans attached at each glycosite of the spike glycoprotein obtained from different expression cells. Since glycosylation at the binding site and adjacent amino acids affects the interaction between the spike glycoprotein and the host cell receptor, we recognize that caution should be taken when selecting an expression system to develop inhibitors, antibodies, and vaccines.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a strain of novel coronavirus that caused the 2019 pandemic disease (COVID-19). SARS-CoV-2 has close genetic similarity to bat coronavirus. Since its first appearance in Wuhan, China, in December 2019, SARS-CoV-2 has spread globally in a few months.1 It was confirmed by January 20, 2020, that SARS-CoV-2 can be transmitted from person to person through direct or indirect contact, such as respiratory droplets (coughs or sneezes), airborne, fomite, and urine or feces. As of March 2021, the SARS-CoV-2 virus has caused 2.7 million deaths and 123.2 million cases worldwide.
Similar to the earlier coronavirus strains middle east respiratory syndrome (MERS)-CoV and SARS-CoV that transmits to humans, SARS-CoV-2 consists of four structural proteins, called spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. The S, E, and M proteins together form the viral envelope, while the N protein retains the RNA genome.2,3 The entry of SARS-CoV-2 cells depends on the binding of the viral S protein to the cell receptors and the S protein triggered by the host cell proteases. Studies have shown that the cell entry process engages angiotensin-converting enzyme 2 (ACE2) to bind the S protein and uses transmembrane protease serine 2 (TMPRSS2) to trigger the S protein.4,5 TMPRSS2 not only cleaves and activates the spike glycoprotein for membrane fusion but also splits ACE2 into two to enhance viral infectivity.6 Because ACE2 and TMPRSS2 are highly present in the respiratory system, digestive tract, and gastrointestinal tract (such as human airway epithelium),7,8 bronchial transient secretory cells,9 nasal epithelial cells,10 human ocular surface,11 and small intestine,12 various routes of infection may occur when SARS-CoV-2 comes into contact with humans through any of these organs. Furin, another receptor highly expressed in the lungs, binds to the spike and cleaves the furin cleavage site (FCS) of SARS-CoV-2.13 The presence of ACE2, TMPRSS2, and furin in these cells and tissues may indicate that there are multiple routes of transmission through their respective viral infections.
The spike glycoprotein of coronavirus plays a key role in virus infection, mediates virus entry, and is a primary determinant of cell tropism and pathogenesis.14 The spike S1 is the first possible point of contact for recognition and binding of host receptors (ACE2, Furin, or GRP78 via CD14715), allowing subsequently conformational changes in S2, thereby promoting the fusion between the viral envelope and the host cell membrane. According to reports, the binding affinity of the SARS-CoV-2 S1 receptor-binding domain (RBD) to ACE2 is considerably higher than that of SARS-CoV,16,17 leading to severe infection and the widespread of the SARS-CoV-2 virus. Although the mortality rate has decreased from ∼10% of SARS-CoV to 1–5% of SARS-CoV-2, the number of deaths caused by SARS-CoV-2 is substantially higher than that of SARS-CoV, e.g., over 2.7 million for the former and 812 for the latter globally to date. Therefore, it is important to understand the structure of the spike glycoprotein and the mechanism of infection.
The spike glycoprotein deploys S1 for attachment to the host cell and S2 for fusion. Obviously, the high affinity promotes the attachment of S1 to the host cell and increases the spread of the virus. The detailed structural comparison of S1 between SARS-CoV and SARS-CoV-2 shows that 10 regions in the S1 domain play critical roles in ACE2 binding; mutations in certain amino acid residues in these regions result in low affinity of S1 to ACE2.4 In contrast, SARS-CoV-2 mutations on some amino acids may help enhance affinity, such as Y442 in SARS-CoV to L457 in SARS-CoV-2, N479 to Q493, Y484 to Q498.4 Thus, any mutations in amino acid residues or post-translational modification (PTM) of amino acids may affect the attachment of the spike S1 to the host cell receptors. Because the spike is a glycoprotein, its glycosylated variants have a profound effect on the affinity and infectivity of SARS-CoV-2. Recent studies have identified 22 N-glycosites in the protomer of the trimeric spike and have a high-density N-glycan mask on the surface of the viral protein, similar to the S1 subunit of MERS-CoV.18,19 Several studies have also detected trace levels of O-glycosites at T323 and S325 of the spike glycoprotein19,20 and T678 near FCS occupied by core-1 and core-2 structures.21 Recent studies have identified 25 O-glycosites in the S1 of the spike glycoprotein expressed from HEK293 cells, of which 16 O-glycosites are located within the three amino acids from the N-glycosites.22 These results are consistent with our predictions using ISOGlyP, indicating that S1 RBD is highly O-glycosylated in SARS-CoV-2. On the other hand, as observed in the influenza viruses, when viral glycoproteins are expressed in different cell systems, their glycosylation can change.23,24 Yet, it has not been studied whether the expression cell system has an impact on the O-glycosylation of SARS-CoV-2 S1 RBD.
In this study, we intend to comprehensively characterize N-linked and O-linked glycosylation of the spike S1 subunit of SARS-CoV and SARS-CoV-2 produced by different expression host cells. We recognize that host expression may alter the glycosylation pattern of spike glycoproteins. HEK293 cells and baculovirus-insect system Hi5 cells are used for virus production and recombinant spike glycoprotein production in our work. The effect of host cell lines on viral protein glycosylation has been reported. The influenza A virus glycoprotein can contain structures of paucimannose (Sf9 cells), core-fucosylated bisected N-GlcNAc (embryonated hen egg), or sialylated biantennary glycans (HEK293).23 baculovirus-insect cells, already used in influenza and human papillomavirus (HPV), is an ideal baculovirus expression system for the production of recombinant spike glycoproteins and vaccines.25 baculovirus-insect cells can synthesize glycans with one or two core fucoses. There is a report of glucuronic acid (GlcA) in the cells,26 even though other insect cells may have GlcA residues.27 It should be investigated whether baculovirus-insect cells have GlcA and other glycans to analyze the glycosylation of the spike glycoprotein.
To reveal these uncertainties, we compared the S1 subunits of Spike expressed in HEK293 cells and baculovirus-insect Hi5 cells (Table 1). The spike S1 was digested with trypsin, and then glycopeptides were enriched using hydrophilic interaction liquid chromatography (HILIC). The enriched glycopeptides were analyzed by liquid chromatography–mass spectrometry (LC-MS/MS) using electron-transfer/higher-energy collision dissociation (EThcD) fragments. In another experiment, N-glycans and O-glycans were released from spike S1 and evaluated using a Bruker Autoflex Matrix-Assisted Laser Desorption/Ionization (MALDI)-MS.
Table 1. Recombinant Spike S1 Expressed in Different Expression Cellsa.
| sample | catalog | description | species | expression host | sequence |
|---|---|---|---|---|---|
| BIC1 | 40150-V08B1 | spike S1 | SARS-CoV | baculovirus-insect | M7-R667 |
| BIC2 | 40591-V08B1 | spike S1 | SARS-CoV-2 | baculovirus-insect | V16-R685 |
| HEK2 | 40591-V08H | spike S1 | SARS-CoV-2 | HEK293 | V16-R685 |
Samples were purchased from Sino Biological.
Results and Discussion
Most Diverse Mutations of Amino Acids Occurred in the S1 Domain of the Spike Glycoprotein
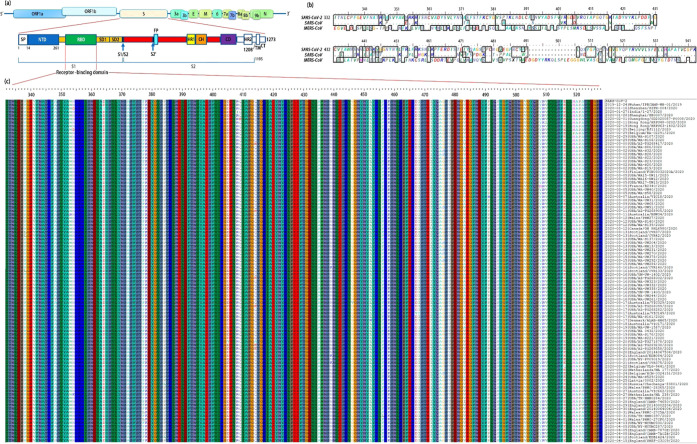
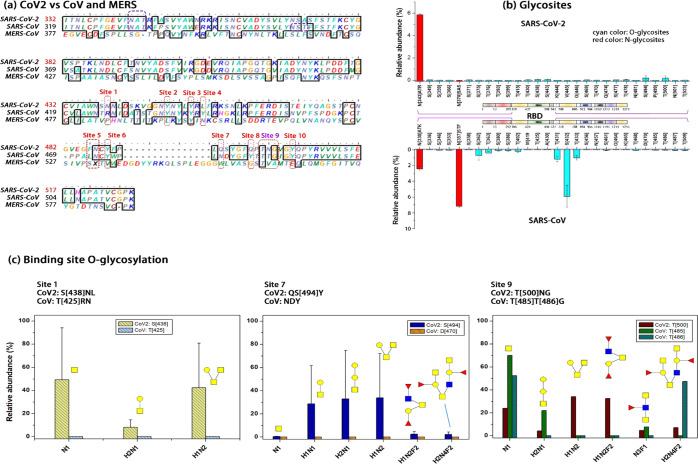
The global initiative on sharing all influenza data (GISAID) has updated the SARS-CoV-2 genome and the spike glycoprotein sequence based on data submitted by laboratories and research institutes around the world. As of February 2021, we have downloaded more than 200 000 protein sequences of the spike glycoproteins. After removing redundant and incomplete sequences, we found that there are 98 unique spike glycoproteins, most of which have mutations in the receptor-binding domain (RBD) of spike S1 (Figure 1). The sequences are arranged according to their submission date (strain list is given in Table S1). Figure 1a illustrates the schematic structure of SARS-CoV-2 and its spike glycoprotein, and Figure 1b compares the alignment of SARS-CoV-2, SARS-CoV, and MERS-CoV. Genetic analysis showed 79% similarity between SARS-CoV and SARS-CoV-2, and the amino acid sequence identity was 76.47%;28 the sequence alignment between MERS-CoV and SARS-CoV showed significant differences.29 There were 51 amino acid changes between SARS-CoV and SARS-CoV-2, or 25.8% variation. Importantly, the variation falls in several sites that are critical for binding affinity to the host cell receptors.4 From 12/2019 to 05/2020, amino acid mutations were observed at 19.3% positions within the RBD domain (Figure 1c). This result indicates that the diversity of SARS-CoV-2 is caused by its frequent mutation on the spike RBD. Thus, it is essential to clarify the spike RBD domain variation to provide necessary information for the development of inhibitors, antibodies, and vaccines.
Figure 1.
Amino acid mutation predominantly occurred on the receptor-binding domain of the SARS-CoV-2 spike glycoprotein from 12/2019 to 05/2020. (a) Domains of SARS-CoV-2 virion include ORF1a&b, spike (S), 3a, 3b, envelope (E), membrane (M), 6, 7a, 7b, 8a, 8b, 9b, and nucleocapsid (N). The spike S1 domain consists of an N-terminal domain (NTD), a receptor-binding domain (RBD), a subdomain 1 (SD1), and a SD2; the other domains are S2, heptad repeat 1 (HR1), central helix (CH), connector domain (CD), HR2, transmembrane (TM), and cytoplasmic tail (CT). S1/S2 is the protease cleavage site, FP is the fusion peptide, and S2′ is the protease cleavage site. (b) Spike RBD sequence alignment between SARS-CoV-2, SARS-CoV, and MERS-CoV. (c) Alignment on RBD of SARS-CoV-2 strains from 12/2019 to 05/2020. The 98 complete and unique sequences are listed, most of which are conserved. The amino acid mutations are highlighted with white bars, while few mutations are observed in other domains of spike glycoproteins.
N-Glycosylation of SARS-CoV-2 Regulated by the Expression System
The purified recombinant S1 proteins expressed in HEK293 cells (HEK2) and baculovirus-insect cells (BIC2 and BIC1) (Table 1) were purchased from Sino Biological. The analysis of each sample was performed in triplicate. Each N-glycosite was plotted using relative abundance related to all N-glycosites.
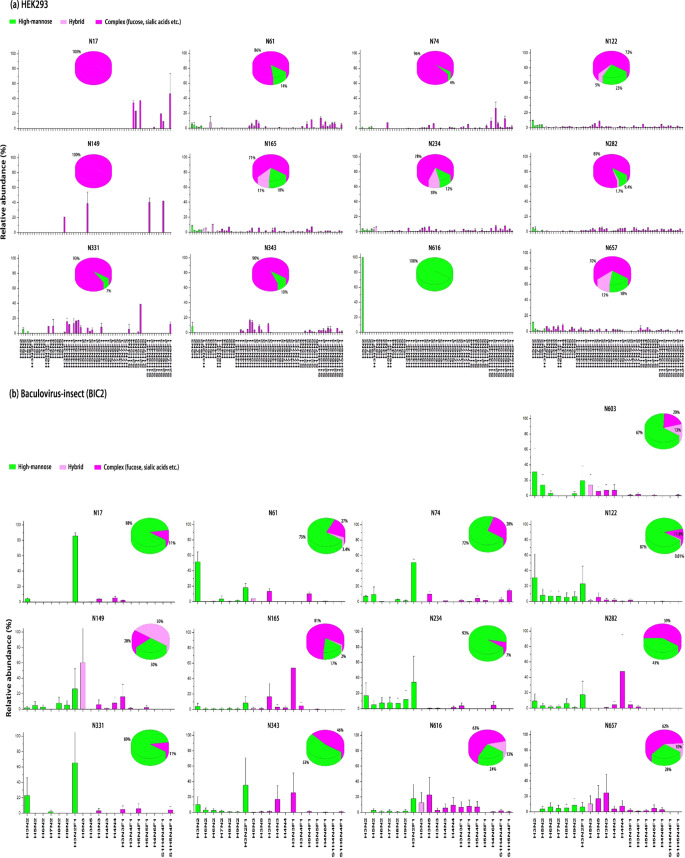
The expression system impacts N-glycosylation and the types of N-glycans at each site. As shown in Figure 2a,b, the spike S1 expressed in HEK293 cells has 12 N-glycosites. When expressed in baculovirus-insect cells, it will carry an additional N-glycosite N603. N-glycans show distinct patterns between the proteins expressed by HEK2 and BIC2. For example, N17 only exhibits complex N-glycans in HEK2, and N17 in BIC2 predominantly contains complex N-glycans with 4% high mannose. A similar observation was also found in N149 of HEK2. On the other hand, HEK2 N616 only has the Man5 (Man5GlcNAc2: Man = Mannose, GlcNAc = N-acetylglucosamine), while N616 from BIC2 mainly contains complex N-glycans, a small amount of hybrid and high-mannose N-glycans. These results indicate that the N616 site from HEK2 cells can be accessed by α1,2-mannosidases, but not as much as GlcNAcT-I.18 Other sites containing complex and high-mannose N-glycans, such as N61, N74, N331, and N343 in HEK2, or N74, N234, N282, and N331 in BIC2, are good substrates for GlcNAcT-I when forming complex N-glycans. N122, N165, N234, N282, and N657 in HEK2 show hybrid N-glycans; N-61, N122, N149, N165, N603, N616, and N657 in BIC2 also have hybrid N-glycans, indicating that the N-glycan process of SARS-CoV-2 depends on the expression system. Moreover, the sialylation distribution of N-glycans is strikingly different between HEK2 and BIC2. Except for N616, all other N-glycosites in HEK2 contain large amounts of sialylated N-glycans. Further linkage analysis by matrix-assisted laser desorption ionization-MS (MALDI-MS) showed that these sialic acids have α2,3 or α2,6 linkage (Figure 2d and Table S2a),30 suggesting that these peptide substrates may be processed by sialyltransferases (e.g., ST3Gal4 or ST6Gal1). Our results are consistent with previous studies on the SARS-CoV-2 spike proteins recombinantly expressed on the HEK293 supernatant,20,31 except for the identification of N-glycans in N17 and N603 in our study, even though the number of N-glycans observed in these N-glycosites is limited.
Figure 2.
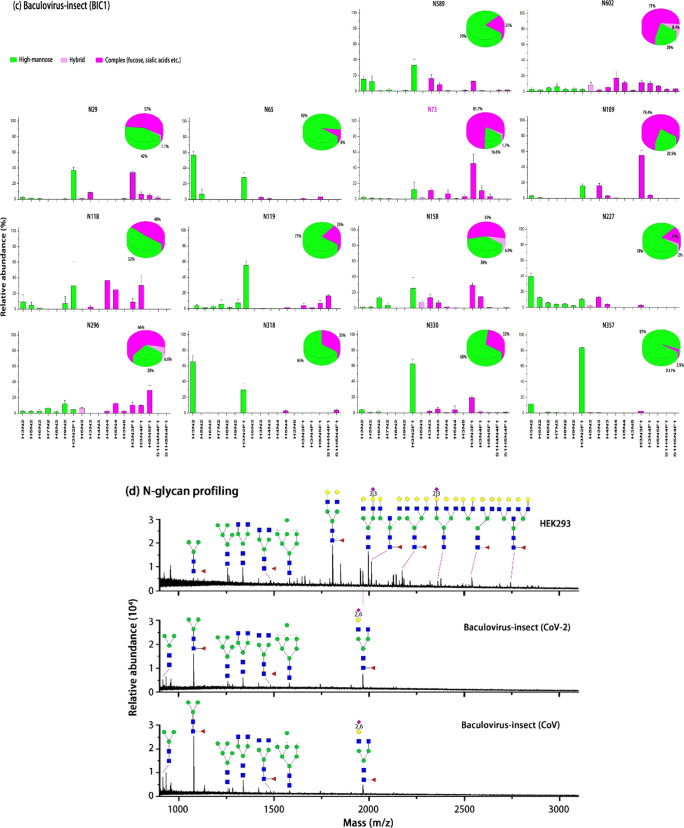
Site-specific characterization of N-glycosylation of the S1 domain of SARS-CoV and SARS-CoV-2 spike glycoprotein. (a) SARS-CoV-2 virus expressed in HEK293 cells. Twelve N-glycosites in S1 were identified by LC-MS/MS. N-glycans are divided into high-mannose (green), hybrid (light purple), and complex (purple). N-glycosites, N17 and N149, are attached by complex N-glycans, N616 only has high-mannose (Man5), and other sites are predominantly complex types. Among these sites, N165, N234, and N657 have more than 10% hybrid N-glycans. (b) SARS-CoV-2 virus expressed in baculovirus-insect. In addition to 12 N-glycosites similar to HEK293 cells, another N-glycosite N603 was detected. High-mannose and complex N-glycans are present in all N-glycosites, while hybrid N-glycans are present in N61, N122, N149, N165, N343, N616, N657, and N603. (c) SARS-CoV virus expressed in baculovirus-insect cells. There are 14 N-glycosites in SARS-CoV. High-mannoses are predominantly present in N65, N227, and N318. Complex N-glycans are highly abundant in N29, N73, N109, N118, N119, N158, N296, N330, N357, N589, and N602. (d) MALDI-MS profiling of N-glycans released from SARS-CoV and SARS-CoV-2. Spike S1 was immobilized on AminoLink plus resins and derivatized by ethyl esterification/ethylenediamine amidation. The most abundant N-glycans are represented, and complete N-glycans for HEK293, CoV-2, and CoV are listed in Table S2. Data are given as mean ± standard deviation.
SARS-CoV expressed in baculovirus-insect (BIC1) has 14 N-glycosites, and SARS-CoV-2 expressed in baculovirus-insect (BIC2) has 13 N-glycosites. BIC1 and BIC2 produce high-mannose and complex N-glycans, and all of these N-glycans contain fucosylated complex types such as Man3GlcNAc2Fuc1, or known as paucimannose specific to the insect. These results demonstrate the synthesis of core fucose in the presence of α1,3-fucosyltransferase in the baculovirus-insect cells.32 Conversely, almost no sialylated N-glycans were identified in the BIC1 or BIC2, although treatment of baculovirus-insect with a β-N-acetylglucosaminidase inhibitor may produce terminally sialylated N-glycans.33 N-glycosylation primarily glycosylated by high-mannoses is located at N61, N122, and N234 in BIC2 and N65, N227, and N318 in BIC1. The subtle difference in N-glycosylation may be attributed to the change in the amino acid sequence between BIC2 and BIC1. Generally, the N-glycan profile is highly conserved between BIC2 and BIC1 (Figure 2d).
Differential Pattern of O-Glycosites of SARS-CoV-2 in Host Cells
Table S3 shows the potential O-glycosylation on the SARS-CoV-2 spike glycoprotein predicted by ISOGlyP.34,35 In this study, T or S sites marked as “high” were reported in the literature and detected in our work, and our method also detected other O-glycosites marked as “medium” (Table S3). It is worth noting that the detected O-glycosites are mainly in the peptide substrate cluster, e.g., T22, T29, S31, and T33 are in the peptide cluster of T[22]QLPPAYT[29]NS[31]FT[33]R. This is consistent with the finding that an amino acid substrate containing P (proline) is beneficial to GalNAcTs (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases (E.C. E.C. 2.4.1.41)) accessible to the T or S residues.36,37 The charge state surrounding T or S may be a factor because the polarity of the GalNAcTs lectin domain affects glycosylation.38
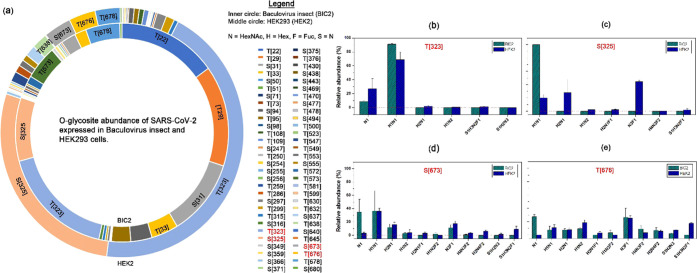
According to N-glycopeptide analysis, spike S1 also showed different O-glycosylation expressed in HEK293 and baculovirus-insect cells. In the HEK293 cells, T323 and T325 are O-glycosylated by GalNAc and GalGalNAc mucin-type O-glycans. S637, T676, and T638 are more abundant than BIC2. In BIC2, T22, T29, S31, T33, S94, T95, T323, and T325 are the most abundant O-glycosites; T572 and T573 are only present in BIC2. These results may imply that the types of GalNAcTs are different in HEK2 and BIC2 because the glycopeptide substrate preferences of GalNAcTs may cause distinct O-glycosylation.39 It is expected that for the same peptide substrates, such as S323, S325, T676, and T678, there will be some O-glycosites with similar glycosylation. A comparison of the site-specific O-glycan profiles on these O-glycosites is given in Figure 3b–e. We noticed that T323 has O-glycans similar to GalNAc (N1) and GalGalNAc (H1N1). The other three main O-glycosites have divergent O-glycans. For example, BIC2 has H1N1 at S325, N1, H1N1, H2N1, and H3F1 (F = Fucose) at S673, H1, H1N1, H2N1, H1N2, H1N2F2, H3F1, H4N3F2, and H2N4F2 at T676; HEK2 has H1N1, H2N1, and N3F1 at S325, H1N1, H2N1, H3F1, H2N4F2, and S1H3N2F1 (S = NeuAc) at S673, H1N1, H2N1, H1N2, N3F1, H2N4F2, S1N2N3, and S1H3N2F1 at T676. This demonstrates the combination of the availability of branched glycoenzyme and the preference for GalNAcTs on peptide substrates.40
Figure 3.
Differential O-glycosylation in spike S1 expressed in baculovirus-insect and HEK293. (a) Relative abundance of O-glycosites identified in the Spike S1 domain. The most abundance O-glycosites are labeled in the ring, and the complete list of all O-glycosites are described in the legend. (b) Most abundant O-glycosite, T323, is present in both BIC2 and HEK2. This O-glycosite consists of GalNAc (N1) and GalGalNAc (H1N1). (c) S325 in BIC2 is mainly H1N1, while S325 in HEK2 is more diverse. O-glycosites (d) S673 and (e) T676 reveal more diverse O-glycans in HEK2, including several sialylated species.
O-Glycosylation in the BIC2 RBD Domain
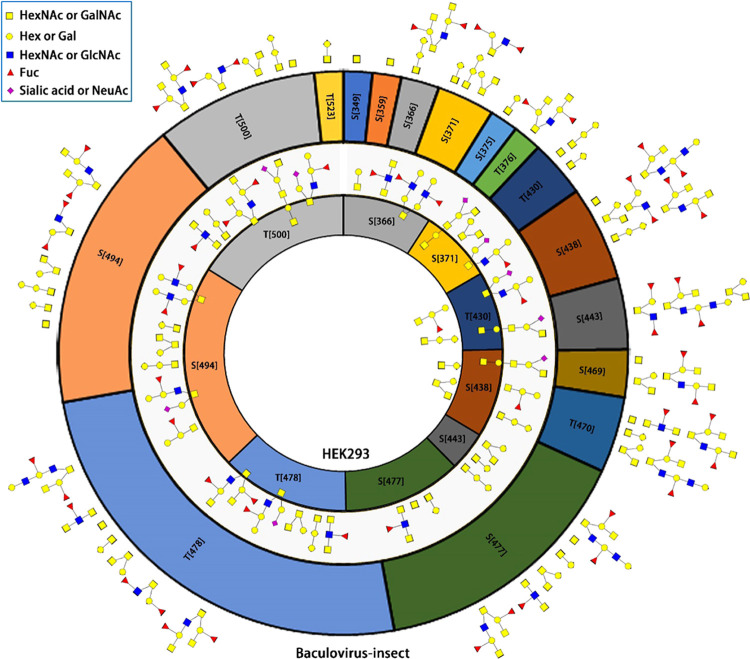
The location of the O-glycosites is different between HEK293 and baculovirus-insect (Figure 3a). We paid special attention to the RBD domain of SARS-CoV-2 expressed in two cell lines. RBD has 197 amino acids starting from I332 to K528. When SARS-CoV-2 was expressed in HEK293 cells, nine O-glyosites were found including S366, T371, T430, S438, S443, S477, T478, S494, and T500 (Figure 4). GalNAc, GalGalNAc, and GalGalNAc2 are the main O-glycans in all O-glycosites, and the abundance of S494 and T500 is high (the area inside the circle in HEK293 represents the relative abundance). These two O-glycosites and T438 are the key positions that may affect the binding affinity of RDB to the ACE2 receptor.4
Figure 4.
Site-specific O-glycan profiling of SARS-CoV-2 receptor-binding domain expressed in baculovirus-insect and HEK293 cells. The outer ring, 16 O-glycosites within RBD of baculovirus-insect cells expressed SARS-CoV-2. Conversely, HEK293 cells expressing RBD have nine O-glycosites. The area within the ring denotes the relative abundance of the O-glycosite, while the ring color illustrates the same O-glycosites between baculovirus-insect and HEK293, e.g., S371 in yellow for both BIC2 and HEK2. Baculovirus-insect has fucosylated O-glycans in most O-glycosites, and HEK293 produces sialylated O-glycans in several O-glycosites, including T500.
Compared with HEK293 cells, the O-glycosylation of SARS-CoV-2 S1 RBD domain expressed in Baculovirus-insect cells is more diverse and complex. Besides the nine O-glycosites identified in HEK2, six additional O-glycosites were found in BIC2, revealing that the density of O-glycosites is higher in BIC2. Additionally, no sialylated O-glycans were found in BIC2, while HEK2 showed sialic acid at S371, T430, S438, T478, S494, and T500. This is consistent with previous reports that insect cells lack sialyltransferases, rarely produce sialylated glycans, and often require metabolic engineering to make terminal sialic acid.41,42 It is worth noting that terminal sialic acid plays an important role in viral infection by attaching to the surface of host cells (such as influenza virus hemagglutinin or receptor determinants for coronaviruses).43,44
Potential Impact of Glycosite Differentiation on the RBD–ACE2 Binding
We further compared how RBD amino acid mutations change RBD glycosylation (Figure 5). Although the lengths of the RBD domains of SARS-CoV-2, SARS-CoV, and MERS-CoV are different, they contain a receptor-binding motif (RBM) in which 10 sites directly interact with the ACE2 receptor.4,45Figure 5a shows 10 sites (red dotted circle) across their coronavirus strains. We emphasized whether the amino acid inside, before or after each binding site is T or S (e.g., S438 before site 1 in SARS-CoV-2, T425 before site 1 in SARS-CoV). The reason is that glycosylation changes at these sites may impact the binding affinity between the spike S1 and ACE2. Figure 5b compares N- and O-glycosites of the spike S1 RBD between SARS-CoV-2 and SARS-CoV. The red bars indicate the relative abundance of N-glycosites, while the cyan bars indicate O-glycosites (note: the purple dotted line is a value equal to 0). There is one N-glycosite located within the RBD domain in SARS-CoV-2 and two N-glycosites in SARS-CoV; however, these N-glycosites are not in the RBM domain. There are several O-glycosites highly abundant in SARS-CoV than that in CoV-2, such as S362, T363, T431, S432, and T433. These O-glycosites are in the secondary structures of SARS-CoV-2 RBD.45 The O-glycosites at S438, S494, and T500 are ACE2 contact residues or adjacent to them (Figure 5c). The high abundance of these O-glycosites in SARS-CoV-2 may be the determinant of the attachment of spike S1 to ACE2.
Figure 5.
O-glycosites in or nearby key ACE2–RBD binding sites. (a) Ten binding sites that are crucial in the ACE2–RBM interaction. These sites are aligned for SARS-CoV-2, SARS-CoV, and MERS. (b) N- and O-glycosites in the RBD domain of the SARS-CoV-2 and SARS-CoV. The red bar is the relative abundance of N-glycosites, and the cyan bar is that of O-glycosites. Each amino acid is aligned based on the sequence described in (a). (c) Three ACE2–RBM binding sites (1, 7, and 9) overlapping with O-glycosites. SARS-CoV-2 has S438, S494, and T500; SARS-CoV has T485 and T486. The RBM, receptor-binding motif, starts from S438 to Q506.
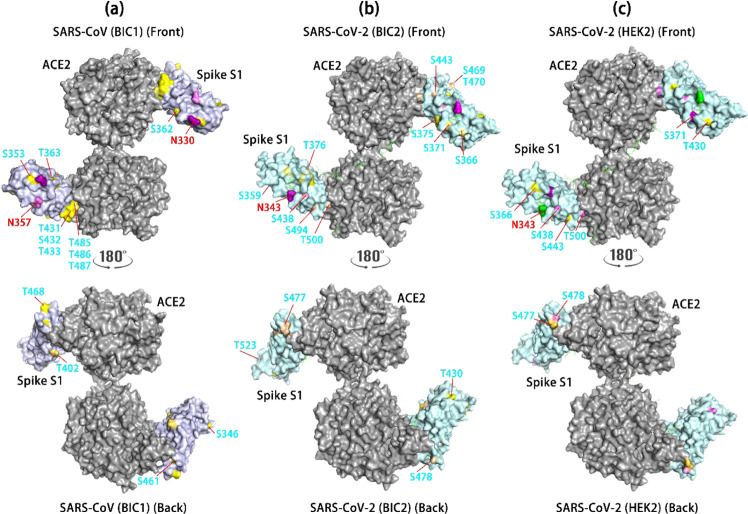
Based on PDB 6VW1 (dimer) for SARS-CoV-2 and PDB 3D0H for SARS-CoV, we mapped S1 glycosites using receptor-binding domain (RBD) in complex with ACE2.46,47Figure 6 shows the site-specific glycosylation mapping of SARS-CoV in baculovirus-insect (BIC1) (Figure 6 a), SARS-CoV-2 in baculovirus-insect (BIC2) (Figure 6b), and SARS-CoV-2 in HEK293 cells (HEK2) (Figure 6c). Compared with BIC2, BIC1 has less glycosites on the spike S1. The latter has O-glycosite at T500 in the RBM domain and may affect the affinity of the spike S1 and ACE2. BIC1 retains complex N-glycans and GalGalNAc or Gal O-glycans; in contrast, BIC2 also carries complex O-glycans and a higher number of O-glycosites. HEK2 revealed a similar location of glycosylation but showed different high-mannose N-glycans at N343 and fewer O-glycosites of spike S1. The spike S1 glycosylation in RBM and secondary structure may interact with ACE2 receptors, whose glycosylation adds another factor in S1 attachment and virus fusion into host cells.48,49 Further studies on the stoichiometric structure of RBD and ACE2 can provide valuable insights into the interaction between RBD and ACE2.
Figure 6.
Mapping glycosites of the Spike S1 RBD domain and its human receptor ACE2. N-glycosites are labeled in red and O-glycosites in cyan. The site mapping color represents different types of glycans: yellow = Gal (H1), GalGalNAc (H1N1) without or with minimal fucosylation or sialylation; light yellow = H1 or H1N1 with fucosylation or sialylation; pink = fucosylation and/or sialylation; green = high-mannose; purple = sialylated complex N-glycans; light purple = other types of complex N-glycans. SARS-CoV is based on 3D0H47 and SARS-CoV-2 on 6WV1.46 (a) SARS-CoV Spike S1 RBD domain glycosites include T485, T486, and T487 near or within the binding sites between ACE2 and RBD. These sites are H1 and H1N1. The front and back sides of the S1 are illustrated for glycosites. (b) Glycosites on the SARS-CoV-2 spike S1 RBD domain expressed in baculovirus-insect cells. (c) Glycosites on the SARS-CoV-2 spike S1 RBD domain expressed in HEK293 cells.
Summary and Perspectives
In this study, we investigated the effect of host expression cells on the glycosylation of the SARS-CoV-2 spike S1 protein. SARS-CoV-2 virus particles infect host cells through S1 attachment to cells and S2 fusion. The affinity between S1 and host cell receptors plays a critical role in viral infection and transmission. The receptor-binding domain of spike S1 has a specific receptor-binding motif (RBM), which may directly interact with the receptor through hydrogen bonds and salt bridges.45 From S438 to Q506, the RBM domain has 10 sites that directly interact with the ACE2 receptor. The binding kinetics between RBM and ACE2 receptor may be affected by glycosylation on these two proteins,50 which has been similarly manifested by influenza A virus hemagglutinin51 and HIV-1 whose encapsulated glycan moieties determine viral propagation.52 The glycosylation of spike S depends on the host cell line, which can express varying glycoenzymes and transporters, resulting in specificity and heterogeneity.53 Differential glycosylation not only impacts the infectivity of the virus but also changes the clinical effectiveness of therapeutic products. Thus, we intend to explore how the expression system regulates the glycosylation of spike S1 RBM and secondary structure and compare the glycosylation distribution between SARS-CoV and SARS-CoV-2.
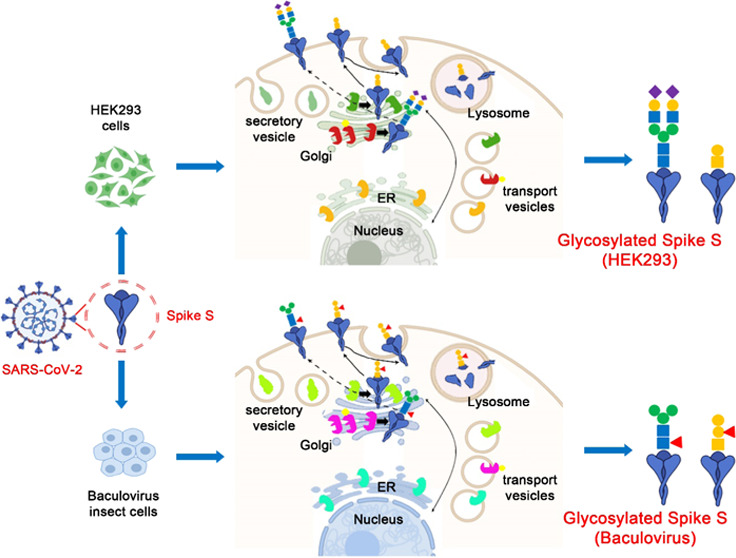
HEK293 and baculovirus-insect cell expression system is used for non-mRNA COVID-19 vaccine development.54,55 Our results show that the expression cell determines glycosylation of the spike S1 and the type of attached glycans. SARS-CoV-2 derived from baculovirus-insect cells contains high-mannose and fucosylated complex N-glycans and fucosylated mucin-type O-glycans. SARS-CoV-2 in HEK293 cells constructs hybrid and sialylated complex N-glycans and sialylated O-glycans. MALDI-MS analysis found that SARS-CoV-2 in HEK293 contains α2,3- and α2,6-linked sialic acids. These observations are consistent with the glycan biosynthesis of the expression system. The known glycan biosynthetic pathways of insects can form Man3GlcNAc2Fuc through GlcNAcMan5GlcNAc2 with α-mannosidase II, core α1,3-fucosyltransferase, and N-Acetylglucosaminidase. Complex glycans are further extended by additional glycoenzymes.56 HEK293 follows the general mammalian glycosylation pathways, forming biantennary, triantennary, or tetraantennary complex glycans in the presence of sialic acid or fucose residues.57 As expected, we found that SARS-CoV-2 expressed by HEK293 has bisected, fucosylated, and sialylated N-glycans and fucosylated/sialylated O-glycans. When using the same expression host cell, similar glycosylation was still detected in BIC2 and BIC1 despite the different strains.
Glycosite mapping of spike S1 suggests the potential influence of host cells on the binding affinity to the ACE2 receptor. Eight O-glycosites in the RBM domain were identified in the baculovirus-insect and six O-glycosites in HEK293. The difference in glycosylation and the three-dimensional (3D) conformation of spike S1 can improve the interaction with the ACE2 receptor. It is very important to systematically study glycosylation, since the RBD (especially RBM) in the SARS-CoV-2 spike glycoprotein may be the target for the development of virus attachment inhibitors, neutralizing antibodies, and vaccines.58 Given that SARS-CoV-2 can be infected and transmitted through many media (lungs, oral, eyes, intestine, etc.), consideration should be given to selecting suitable host cell lines for diagnostic applications and the development of inhibitors, antibodies, or vaccines.
Methods
Sample Preparation of SARS-CoV and SARS-CoV-2
Recombinant spike S1 was purchased from Sino Biological (HEK2, BIC2, and BIC1) (Table 1). The amino acid sequences of HEK2 and BIC2 were from V16 to K685 and that of BIC1 from M1 to R667. The sample preparation followed the procedure described in Figure S1. Each sample was performed in technical triplicate. First, 40 μg of the protein was denatured in high-performance liquid chromatography (HPLC) water at 90 °C/10 min, and half of which was reduced in 12 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP)/37 °C/1 h and alkylated in 16 mM iodoacetamide (IAA)/room temperature/1 h. The sample was then digested with trypsin (1:25) (Promega, Madison, WI) at 37 °C/overnight. The digest solution was acidified with 30 μL of 100% trifluoroacetic acid (TFA) prior to solid-phase extraction (SPE) cartridge C18 cleanup (Waters, Milford, MA). An in-house packed Amide-80 (Tosoh Bioscience LLC, King of Prussia, PA) HILIC SPE column was used to further enrich glycopeptides.59 The glycopeptides and flow-through peptides after HILIC were analyzed using LC-MS/MS.
The remaining 20 μg of the protein after denaturation was conjugated with an Aminolink plus coupling resin (Thermo Fisher Scientific, Waltham, MA) for glycan analysis. The solid-phase method is called glycoprotein immobilization for glycan extraction (GIG),60 in which α2,6-linked sialic acid underwent an ethyl esterification reaction (0.5 M N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC·HCl) and 0.5 M 1-hydroxybenzotriazole hydrate (HBot), 250 μL) and α2,3-linked sialic acid through a carbodiimide coupling (1 M p-toluidine in the presence of EDC (pH 4–6)).59 First, we used 1 μL of PNGase F (New England BioLabs, Ipswich, MA) in 25 mM ammonium bicarbonate to release N-glycans; the remaining sample on the Aminolink resin was further processed to release O-glycans through β-elimination (0.1 M NaOH) and permethylation. The permethylated O-glycans were purified using a C18 SPE cartridge and eluted with 300 μL of 60% ACN in 0.1% TFA. Glycans were analyzed by MALDI-time-of-flight/TOF-MS (MALDI-TOF/TOF-MS) (Bruker Autoflex).
MALDI-TOF/TOF-MS Identification of Glycans
The eluted glycans in 60% ACN (0.1% TFA) were spotted onto a μFocus MALDI plate (384 circles; Hudson Surface Technology, West New York, NJ), together with 1 μL of 10 mg/mL dihydroxybenzoic acid (DHB) matrix in the presence of 2% N,N-dimethylaniline (DMA) (50% ACN in 0.1 mM NaCl). The plate was dried on the top of a 50–60 °C hot plate. Each MALDI-MS test was performed in triplicate for 8000 shots. The mass (m/z) was searched against the glycan database in GlycoWorkBench.61 For N-glycans, the mass range was set between 900 and 6000 Da, while it was set between 300 and 3000 Da for O-glycans.
LC-MS/MS Analysis of Glycopeptides
The samples were analyzed using a Dionex U3000 nanoHPLC system connected to a Thermo Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). Glycopeptides (1 μg) were injected and desalted with an Acclaim PepMap C18 Nano trap column (3 μm, 100 Å, 75 μm × 2 cm) at 5 μL/min with 100% solvent A (0.1% formic acid in HPLC water) for 5 min. Then, glycopeptides were separated by an Acclaim PepMap 100 nano column (3 μm, 100 Å, 75 μm × 250 mm) using a linear gradient of 2.5–37.5% solvent B (80% ACN, 0.1% formic acid) over 85 min, with a wash at 90% B for 5 min. The column was equilibrated at 2.5% B for 10 min before the next injection. Data-dependent analysis (DDA) was carried out with a duty cycle of 2 s. Precursor masses were detected in the orbitrap at a resolution (R) of 120 000 (at m/z 200) with internal calibration (Easy IC). Stepped HCD spectra (HCD energy at 15, 25, and 35%) were acquired for precursors with charges between 2 and 8 and intensities over 5.0 × 104 at R = 30 000. Dynamic exclusion was set at 20 s. When at least one glycan oxonium fragment ion (m/z 138.0545, 204.0867, 366.1396 Da) was observed within the top 20 most abundant fragments and within 15 ppm mass accuracy, an EThcD spectrum was acquired in the orbitrap at R = 30 000. The electron-transfer dissociation (ETD) reagent target was 2.0 × 105, with supplemental collision energy at 15%. The ETD reaction time was dependent on the precursor charge state: 125 ms (ETD reaction time) for charge 2, 100 ms for 3, 75 ms for 4, and 50 ms for ≥5.
Data Analysis
Through precursor and MS/MS fragmentation matching, the glycan composition analysis was performed in GlycoWorkBench, which uses glycan databases from a consortium for functional glycomics (CFG), Carbbank, GlycomeDB, and Glycosciences. The derivatization of the sialic acid linkages added a mass tag to its residues, namely, 28.031301 on α2,6-link or 42.058183 on α2,3-link. The identified N-glycans and O-glycans were used as the glycan database for glycopeptide analysis (Tables S2 and S4).
MS/MS spectra were searched using Byos (Protein Metrics, San Carlos, CA) against a spiked protein database compiled in-house. The identified glycans in the MALDI-TOF were used as the glycan database. Search parameters include precursor mass tolerance (15 ppm), HCD fragment mass tolerance (20 ppm), EThcD fragment mass tolerance (20 ppm), missed cleavage (3), oxidation (+15.994915, variable), carbamidomethyl (+57.021464, fixed), common modification (≤2), rare modification (1), maximum precursor mass (30 000), protein FDR (2%), and missed cleavage (3). The identified glycopeptides were manually verified according to oxonium ions, pep-HexNAc, and y and b ions with fragments surrounding an O-glycosite. An example of glycopeptide tandem MS is shown in Figure S2. For a peptide that has multiple glycosites, such as N-glycosite and T/S O-glycosites, we use a fragment ion calculator (http://db.systemsbiology.net:8080/proteomicsToolkit/FragIonServlet.html) to check the fragmentation mass of glycopeptides.
The quantification of glycopeptides was performed as follows. After searching the LC-MS/MS spectra against Byonic, Byologic further analyzed the Byonic output files. The total area under the curve (AUC) of each glycopeptide was extracted from LC-MS/MS by Byologics. The AUC of the same glycopeptide was summed up, and the relative abundance was estimated by dividing the AUC (single glycopeptide) by the total AUC (all glycopeptides). To quantify glycans on each glycosite, we used the AUC of each glycoform divided by the total AUC of all glycoforms.
Acknowledgments
This work was supported by the Intramural Research Program of the NIDCR, National Institutes of Health (ZIA-000751-02 to Y.W. and 1-ZIA-DE000739-05 to L.A.T.) and Soochow University Start-up funding.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c01785.
List of 98 unique sequences of the SARS-CoV-2 spike receptor-binding domain (Table S1); quantitative profiling of N-glycans from SARS-CoV and SARS-CoV-2 expressed in HEK293 cells and baculovirus-insect cells (Table S2); O-glycosite prediction based on the amino acid sequence of SARS-CoV-2 using ISOGlyP (Table S3); O-glycans for Byonic O-glycopeptide search, O-glycans were prepared by the solid-phase glycan extraction, including β-elimination and permethylation (Table S4); schematic workflow of spike glycoprotein analysis (Figure S1); typical HCD fragmentation of the O-glycopeptide precursor (Figure S2); and MALDI-MS of O-glycans of SARS-CoV-2 in BIC1, BIC2, and HEK293 (Figure S3) (PDF)
Author Contributions
S.Y. conducted the experiments, analyzed data, and wrote the paper. S.Y. and Y.W. designed the experiments. Y.W., Z.W., W.W., and P.H. conducted LC-MS and MALDI-MS. Y.W. conducted part of data analysis using Byos. S.Y. revised the paper.
The authors declare no competing financial interest.
Supplementary Material
References
- Li R.; Pei S.; Chen B.; Song Y.; Zhang T.; Yang W.; Shaman J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 2020, 368, 489–493. 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A. C.; Park Y.-J.; Tortorici M. A.; Wall A.; McGuire A. T.; Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.; Li W.; Farzan M.; Harrison S. C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005, 309, 1864–1868. 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Hoffmann M.; Kleine-Weber H.; Schroeder S.; Krüger N.; Herrler T.; Erichsen S.; Schiergens T. S.; Herrler G.; Wu N.-H.; Nitsche A.; Müller M. A.; Drosten C.; Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y.; Shang J.; Graham R.; Baric R. S.; Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020, 94, e00127-20 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurich A.; Hofmann-Winkler H.; Gierer S.; Liepold T.; Jahn O.; Pöhlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014, 88, 1293–1307. 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Rostami M. R.; Leopold P. L.; Mezey J. G.; O’Beirne S. L.; Strulovici-Barel Y.; Crystal R. G. Expression of the SARS-CoV-2 ACE2 receptor in the human airway epithelium. Am. J. Respir. Crit. Care Med. 2020, 202, 219–229. 10.1164/rccm.202003-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase M.; Shirato K.; van der Hoek L.; Taguchi F.; Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 2012, 86, 6537–6545. 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukassen S.; Chua R. L.; Trefzer T.; Kahn N. C.; Schneider M. A.; Muley T.; Winter H.; Meister M.; Veith C.; Boots A. W.; Hennig B. P.; Kreuter M.; Conrad C.; Eils R. SARS-CoV-2 receptor ACE 2 and TMPRSS 2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020, 39, e105114 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W.; Huang N.; Bécavin C.; Berg M.; Queen R.; Litvinukova M.; Talavera-López C.; Maatz H.; Reichart D.; Sampaziotis F.; Worlock K. B.; Yoshida M.; Barnes J. L. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020, 26, 681–687. 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L.; Xu Z.; Castiglione G. M.; Soiberman U. S.; Eberhart C. G.; Duh E. J. ACE2 and TMPRSS2 are expressed on the human ocular surface, suggesting susceptibility to SARS-CoV-2 infection. Ocul. Surf. 2020, 18, 537–544. 10.1016/j.jtos.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R.; Castro M. F. G.; McCune B. T.; Zeng Q.; Rothlauf P. W.; Sonnek N. M.; Liu Z.; Brulois K. F.; Wang X.; Greenberg H. B.; Diamond M. S.; Ciorba M. A.; Whelan S. P. J.; Ding S. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020, 5, eabc3582 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A.; Paray B. A.; Hussain A.; Qadir F. A.; Attar F.; Aziz F. M.; Sharifi M.; Derakhshankhah H.; Rasti B.; Mehrabi M.; Shahpasand K.; Saboury A. A.; Falahati M. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. J. Biomol. Struct. Dyn. 2021, 39, 3025–3033. 10.1080/07391102.2020.1754293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S.; Millet J. K.; Licitra B. N.; Whittaker G. R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 2012, 4, 1011–1033. 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim I. M.; Abdelmalek D. H.; Elshahat M. E.; Elfiky A. A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020, 80, 554–562. 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Zhang Y.; Wu L.; Niu S.; Song C.; Zhang Z.; Lu G.; Qiao C.; Hu Y.; Yuen K.-Y.; Wang Q.; Zhou H.; Yan J.; Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 2020, 181, 894–904. 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A.; Peng Y.; Huang B.; Ding X.; Wang X.; Niu P.; Meng J.; Zhu Z.; Zhang Z.; Wang J.; Sheng J.; Quan L.; Xia Z.; Tan W.; Cheng G.; Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 2020, 27, 325–328. 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y.; Allen J. D.; Wrapp D.; McLellan J. S.; Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y.; Berndsen Z. T.; Raghwani J.; Seabright G. E.; Allen J. D.; Pybus O. G.; McLellan J. S.; Wilson I. A.; Bowden T. A.; Ward A. B. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat. Commun. 2020, 11, 2688 10.1038/s41467-020-16567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajahan A.; Supekar N. T.; Gleinich A. S.; Azadi P. Deducing the N-and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology 2020, 30, 981–988. 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanda M.; Morrison L.; Goldman R. N and O glycosylation of the SARS-CoV-2 spike protein. Anal. Chem. 2021, 93, 2003–2009. 10.1021/acs.analchem.0c03173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdonaite I.; Thompson A. J.; Wang X.; Søgaard M.; Fougeroux C.; Frank M.; Diedrich J. K.; Yates J. R.; Salanti A.; Vakhrushev S. Y.; Paulson J. C.; Wandall H. H. Site-specific O-glycosylation analysis of SARS-CoV-2 spike protein produced in insect and human cells. Viruses 2021, 13, 551 10.3390/v13040551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Y.; Rininger J. A.; Jarvis D. L.; Jing X.; Ye Z.; Aumiller J. J.; Eichelberger M.; Cipollo J. F. Comparative glycomics analysis of influenza hemagglutinin (H5N1) produced in vaccine relevant cell platforms. J. Proteome Res. 2013, 12, 3707–3720. 10.1021/pr400329k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X.; Jayaraman A.; Maniprasad P.; Raman R.; Houser K. V.; Pappas C.; Zeng H.; Sasisekharan R.; Katz J. M.; Tumpey T. M. N-linked glycosylation of the hemagglutinin protein influences virulence and antigenicity of the 1918 pandemic and seasonal H1N1 influenza A viruses. J. Virol. 2013, 87, 8756–8766. 10.1128/JVI.00593-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F.; Krammer F. SARS-CoV-2 vaccines: status report. Immunity 2020, 52, 583–589. 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmberger D.; Wilson I. B. H.; Berger I.; Grabherr R.; Rendic D. SweetBac: a new approach for the production of mammalianised glycoproteins in insect cells. PLoS One 2012, 7, e34226 10.1371/journal.pone.0034226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.; De Schutter K.; Van Damme E. J. M.; Smagghe G. Synthesis and biological roles of O-glycans in insects. Glycoconjugate J. 2020, 37, 47–56. 10.1007/s10719-019-09867-1. [DOI] [PubMed] [Google Scholar]
- Wang H.; Li X.; Li T.; Zhang S.; Wang L.; Wu X.; Liu J. The genetic sequence, origin, and diagnosis of SARS-CoV-2. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1629–1635. 10.1007/s10096-020-03899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Rajashankar K. R.; Yang Y.; Agnihothram S. S.; Liu C.; Lin Y.-L.; Baric R. S.; Li F. Crystal structure of the receptor-binding domain from newly emerged Middle East respiratory syndrome coronavirus. J. Virol. 2013, 87, 10777–10783. 10.1128/JVI.01756-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.; Jankowska E.; Kosikova M.; Xie H.; Cipollo J. Solid-phase chemical modification for sialic acid linkage analysis: application to glycoproteins of host cells used in influenza virus propagation. Anal. Chem. 2017, 89, 9508–9517. 10.1021/acs.analchem.7b02514. [DOI] [PubMed] [Google Scholar]
- Zhou D.; Tian X.; Qi R.; Peng C.; Zhang W. Identification of 22 N-glycosites on spike glycoprotein of SARS-CoV-2 and accessible surface glycopeptide motifs: Implications for vaccination and antibody therapeutics. Glycobiology 2020, 31, 69–80. 10.1093/glycob/cwaa052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis D. L. Developing baculovirus-insect cell expression systems for humanized recombinant glycoprotein production. Virology 2003, 310, 1–7. 10.1016/S0042-6822(03)00120-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S.; Kokuho T.; Takahashi H.; Takahashi M.; Kubota T.; Inumaru S. Sialylation of N-glycans on the recombinant proteins expressed by a baculovirus-insect cell system under β-N-acetylglucosaminidase inhibition. J. Biol. Chem. 2002, 277, 5090–5093. 10.1074/jbc.M110548200. [DOI] [PubMed] [Google Scholar]
- Kong Y.; Joshi H. J.; Schjoldager K. T.-B. G.; Madsen T. D.; Gerken T. A.; Vester-Christensen M. B.; Wandall H. H.; Bennett E. P.; Levery S. B.; Vakhrushev S. Y.; Clausen H. Probing polypeptide GalNAc-transferase isoform substrate specificities by in vitro analysis. Glycobiology 2015, 25, 55–65. 10.1093/glycob/cwu089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohl J. E.; Gerken T. A.; Leung M.-Y. ISOGlyP: de novo prediction of isoform specific mucin-type O-glycosylation. Glycobiology 2021, 31, 168–172. 10.1093/glycob/cwaa067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hema Thanka Christlet T.; Veluraja K. Database analysis of O-glycosylation sites in proteins. Biophys. J. 2001, 80, 952–960. 10.1016/S0006-3495(01)76074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett E. P.; Mandel U.; Clausen H.; Gerken T. A.; Fritz T. A.; Tabak L. A. Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology 2012, 22, 736–756. 10.1093/glycob/cwr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May C.; Ji S.; Syed Z. A.; Revoredo L.; Daniel E. J. P.; Gerken T. A.; Tabak L. A.; Samara N. L.; Ten Hagen K. G. Differential splicing of the lectin domain of an O-glycosyltransferase modulates both peptide and glycopeptide preferences. J. Biol. Chem. 2020, 295, 12525–12536. 10.1074/jbc.RA120.014700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt M. R.; Hang H. C.; Ten Hagen K. G.; Rarick J.; Gerken T. A.; Tabak L. A.; Bertozzi C. R. Deconvoluting the functions of polypeptide N-α-acetylgalactosaminyltransferase family members by glycopeptide substrate profiling. Chem. Biol. 2004, 11, 1009–1016. 10.1016/j.chembiol.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Moremen K. W.; Ramiah A.; Stuart M.; Steel J.; Meng L.; Forouhar F.; Moniz H. A.; Gahlay G.; Gao Z.; Chapla D.; et al. Human glycosylation enzymes for enzymatic, structural and functional studies. Nat. Chem. Biol. 2018, 14, 156–162. 10.1038/nchembio.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal I.; Jarvis D. L.; Cacan R.; Verbert A. Glycoproteins from insect cells: sialylated or not?. Biol. Chem. 2001, 382, 151–159. 10.1515/BC.2001.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubelka V.; Altmann F.; Kornfeld G.; Marz L. Structures of the N-linked oligosaccharides of the membrane glycoproteins from three lepidopteran cell lines (Sf-21, IZD-Mb-0503, Bm-N). Arch. Biochem. Biophys. 1994, 308, 148–157. 10.1006/abbi.1994.1021. [DOI] [PubMed] [Google Scholar]
- Stencel-Baerenwald J. E.; Reiss K.; Reiter D. M.; Stehle T.; Dermody T. S. The sweet spot: defining virus–sialic acid interactions. Nat. Rev. Microbiol. 2014, 12, 739–749. 10.1038/nrmicro3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwegmann-Weßels C.; Herrler G. Sialic acids as receptor determinants for coronaviruses. Glycoconjugate J. 2006, 23, 51–58. 10.1007/s10719-006-5437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J.; Ge J.; Yu J.; Shan S.; Zhou H.; Fan S.; Zhang Q.; Shi X.; Wang Q.; Zhang L.; Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Shang J.; Ye G.; Shi K.; Wan Y.; Luo C.; Aihara H.; Geng Q.; Auerbach A.; Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. J. Virol. 2008, 82, 6984–6991. 10.1128/JVI.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P.; Praissman J. L.; Grant O. C.; Cai Y.; Xiao T.; Rosenbalm K. E.; Aoki K.; Kellman B. P.; Bridger R.; Barouch D. H.; Brindley M. A.; Lewis N. E.; Tiemeyer M.; Chen B.; Woods R. J.; Wells L. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe 2020, 28, 586–601. 10.1016/j.chom.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D. P.; Lohani M.; Cho M. W. Specific asparagine-linked glycosylation sites are critical for DC-SIGN-and L-SIGN-mediated severe acute respiratory syndrome coronavirus entry. J. Virol. 2007, 81, 12029–12039. 10.1128/JVI.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi A.; Huang Y.; Harris B.; Xiong Y.; Nandi S.; McDonald K. A.; Faller R. Development and simulation of fully glycosylated molecular models of ACE2-Fc fusion proteins and their interaction with the SARS-CoV-2 spike protein binding domain. PLoS One 2020, 15, e0237295 10.1371/journal.pone.0237295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishin V. P.; Novikov D.; Hayden F. G.; Gubareva L. V. Effect of hemagglutinin glycosylation on influenza virus susceptibility to neuraminidase inhibitors. J. Virol. 2005, 79, 12416–12424. 10.1128/JVI.79.19.12416-12424.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R.; Raska M.; Bimczok D.; Novak J.; Smith P. D. HIV-1 envelope glycan moieties modulate HIV-1 transmission. J. Virol. 2014, 88, 14258–14267. 10.1128/JVI.02164-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh J. B.; Ng S. K. Impact of host cell line choice on glycan profile. Crit. Rev. Biotechnol. 2018, 38, 851–867. 10.1080/07388551.2017.1416577. [DOI] [PubMed] [Google Scholar]
- Le Ru A.; Jacob D.; Transfiguracion J.; Ansorge S.; Henry O.; Kamen A. A. Scalable production of influenza virus in HEK-293 cells for efficient vaccine manufacturing. Vaccine 2010, 28, 3661–3671. 10.1016/j.vaccine.2010.03.029. [DOI] [PubMed] [Google Scholar]
- Tian J.-H.; Patel N.; Haupt R.; Zhou H.; Weston S.; Hammond H.; Logue J.; Portnoff A. D.; Norton J.; Guebre-Xabier M.; Zhou B.; Jacobson K.; Maciejewski S.; Khatoon R.; Wisniewska M.; Moffitt W.; Kluepfel-Stahl S.; Ekechukwu B.; Papin J.; Boddapati S.; Jason Wong C.; Piedra P. A.; Frieman M. B.; Massare M. J.; Fries L.; Bengtsson K. L.; Stertman L.; Ellingsworth L.; Glenn G.; Smith G. SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat. Commun. 2021, 12, 372 10.1038/s41467-020-20653-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R. L.; Jarvis D. L. Protein N-glycosylation in the baculovirus–insect cell expression system and engineering of insect cells to produce “mammalianized” recombinant glycoproteins. Adv. Virol. Res. 2006, 68, 159–191. [DOI] [PubMed] [Google Scholar]
- Wang Q.; Stuczynski M.; Gao Y.; Betenbaugh M. J.. Strategies for Engineering Protein N-Glycosylation Pathways in Mammalian Cells. In Methods in Molecular Biology; Springer, 2015; Vol. 1321, pp 287–305. [DOI] [PubMed] [Google Scholar]
- Tai W.; He L.; Zhang X.; Pu J.; Voronin D.; Jiang S.; Zhou Y.; Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.; Wu W. W.; Shen R.-F.; Bern M.; Cipollo J. Identification of sialic acid linkages on intact glycopeptides via differential chemical modification using IntactGIG-HILIC.. J. Am. Soc. Mass Spectrom. 2018, 29, 1273–1283. 10.1007/s13361-018-1931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.; Li Y.; Shah P.; Zhang H. Glycomic analysis using glycoprotein immobilization for glycan extraction. Anal. Chem. 2013, 85, 5555–5561. 10.1021/ac400761e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceroni A.; Maass K.; Geyer H.; Geyer R.; Dell A.; Haslam S. M. GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 2008, 7, 1650–1659. 10.1021/pr7008252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.