Abstract
Mechanistic (or mammalian) target of rapamycin complex 1 (mTORC1) is a major signalling kinase in cells that regulates proliferation and metabolism and is controlled by extrinsic and intrinsic signals. The lysosome has received considerable attention as a major hub of mTORC1 activation. However, mTOR has also been located to a variety of other intracellular sites, indicating the possibility of spatial regulation of mTORC1 signalling within cells. In particular, there have been numerous recent reports of mTORC1 activation associated with the Golgi apparatus. Here, we review the evidence for the regulation of mTORC1 signalling at the Golgi in mammalian cells. mTORC1 signalling is closely linked to the morphology of the Golgi architecture; a number of Golgi membrane tethers/scaffolds that influence Golgi architecture in mammalian cells that directly or indirectly regulate mTORC1 activation have been identified. Perturbation of the Golgi mTORC1 pathway arising from fragmentation of the Golgi has been shown to promote oncogenesis. Here, we highlight the potential mechanisms for the activation mTORC1 at the Golgi, which is emerging as a major site for mTORC1 signalling.
Keywords: mTORC1, Golgi architecture, trans-Golgi network, actin, Rab1A, GOLPH3, GCC88, Arf1, GAT4, signalling
Background
A major signalling pathway in eukaryotic cells is the mechanistic (or mammalian) target of rapamycin (mTOR) pathway. mTOR is a Ser/Thr kinase that exists in two distinct complexes: mTORC1 and mTORC2. mTORC1 controls cell growth and metabolism, whereas mTORC2 regulates cytoskeletal dynamics1. mTORC1 senses the nutritional and energy status of a cell and initiates a variety of downstream responses by the phosphorylation of target substrates. Activation of mTORC1 promotes ribosome biogenesis, protein translation and nutrient import and also inhibits autophagy and stress-responsive transcription2. For example, active mTORC1 phosphorylates the downstream targets ribosomal S6 kinase and ribosome-associated eukaryotic translation initiation factor 4E-binding protein (4E-BP1) to drive protein translation3.
A number of inputs have been implicated in mTORC1 signalling, namely growth factors, nutrients, oxygen levels, energy levels and stress4. Surface receptors, such as insulin receptor, mediate activation of mTORC1 via phosphoinositide-3 kinase (PI3K)4. Nutrients, especially amino acids such as leucine, arginine and glutamine, activate mTORC15. In addition, mTORC1 senses the energy status of a cell and is inhibited under conditions of energy deprivation via AMP-dependent kinase (AMPK)6. mTORC1 is also inhibited by hypoxia and other signals, including DNA damage2,4. Collectively, inputs regulate the activation and inhibition of mTORC1 and a variety of downstream targets regulate cellular processes. Understanding the regulation of this complex network of pathways is a key issue in cell biology.
Hubs for mTOR
The lysosome is a well-known site/hub for amino acid sensing and activation of mTORC17. The mechanism for lysosomal amino acid sensing involves the heterodimeric, small G protein, Rag GTPase, which binds to the surface of lysosomes via the pentameric Ragulator complex8. Amino acid availability leads to GTP loading of RagA or B which heterodimerises with GDP-bound RagC or D to form an active Rag complex which recruits mTORC1 to the lysosomal membrane surface and is activated in a Rheb GTPase–dependent manner7.
A remaining question in the field is whether the lysosomal hub controls the full repertoire of mTOR signalling outputs. It remains unknown whether the lysosomal hub controls all the downstream signalling responses. Indeed, evidence has emerged that mTOR is localised at a number of different intracellular sites in addition to lysosomes, such as the plasma membrane, secretory pathway, endosomes, mitochondria, peroxisomes and the nucleus9,10. The extensive distribution of mTOR on various organelles suggests that mTORC1 signalling may be regulated at multiple intracellular locations in a spatially specific manner. On a theoretical level, a multi-hub model would provide a greater capacity to fine-tune the complex network of mTOR signalling pathways, as is the case for other signalling pathways such as Toll-like receptor 4 (TLR4) activation11, where the cell surface and endosomes mediate different downstream outputs from the same receptor. Indeed, recent reports have provided evidence for spatially and functionally distinct pools of TORC1 at the vacuoles and endosomes in yeast12 and for activation of mTORC1 at the cell surface of mammalian cells independent of lysosomes13. Notably, there is also growing evidence for a role of the secretory pathway in the regulation of mTORC1 signalling, in particular the Golgi apparatus.
Golgi as a general signalling hub
Many components of mTORC1 were initially identified by genetic screens in yeast1, an organism that lacks the detailed architecture of the vertebrate Golgi apparatus. In vertebrates, individual stacks of Golgi cisternae are fused together into a compact ribbon structure located in close proximity to the microtubule-organising centre14. The vertebrate Golgi was recently recognised to be associated with a range of higher-order cell functions in addition to the classic functions of glycosylation and membrane trafficking. The Golgi architecture is highly dynamic and the Golgi ribbon can undergo rapid changes in morphology15,16. Of relevance to the discussion here on mTORC1, modulation of the Golgi architecture is associated with the regulation of a number of signalling pathways17. It is now clear that there is an intimate relationship between the molecular pathways that regulate the dynamics of the architecture and morphology of the Golgi and signalling (see reviews 17,18). Studies have revealed that the Golgi provides a platform for the regulation of a range of cellular processes, including cell polarisation19, directed migration, stress20, mitosis, metabolism21, pro-inflammatory responses22 and autophagy23. Morphological changes of the Golgi are associated with the regulation of these cell processes (see review 17) and some of these are linked to mTOR signalling.
Location of mTORC1 components on the endoplasmic reticulum/Golgi
There are a number of reports identifying mTORC1, and other components associated with activation of mTORC1, at the Golgi. The mTOR polypeptide, the kinase subunit of the mTOR complexes, has been located at the Golgi24–26. Our study in particular26 used an extensive range of organelle markers and fixation conditions and provided strong evidence for a functional pool of mTOR at the Golgi and particularly enriched at the trans-Golgi network (TGN). The specificity of staining was demonstrated by knockdown of mTOR. Phosphorylated mTOR, representing the active mTOR, was also detected on Golgi membranes, and the phosphorylation was inhibited by the selective mTORC1 inhibitor, rapamycin, indicating that active mTORC1 was present on Golgi membranes26.
Other components associated with the activation of mTORC1 have been located at the endoplasmic reticulum (ER) and Golgi. The GTPase Rheb is an essential and immediate upstream activator of mTORC127. Rheb is recruited from the cytosol to membranes, mediated in part by farnesylation of the Rheb C-terminal CaaX motif28,29. Inhibition of active GTP-bound Rheb results in suppression of mTORC1 activity30. Given the requirement of Rheb to activate mTORC1, the location of this small GTPase is also relevant. Rheb has been reported to be associated with a number of endomembranes, including the Golgi in different cell types29,31,32. It is currently unclear whether Rheb has specific membrane-targeting motifs or whether Rheb binds to membranes non-selectively31,33.
The small GTPase, Rab1A, a component of the ER-to-Golgi trafficking machinery, has also been identified as a regulator of mTORC124. Rab1A overexpression promotes mTORC1 signalling and cell growth24. Using transfections systems and knockdowns, Rab1A was shown to promote Rheb–mTORC1 interactions at the Golgi apparatus in response to amino acid stimulation24. Notably, transporters at the Golgi, in particular the Golgi glutamine transporter PAT4, have also been shown to contribute to mTORC1 activation34. The small GTPase, Arf1, has also been implicated in the regulation of mTORC1 signalling by a Rag-independent mechanism35,36 and, although the study did not provide direct evidence of mTORC1 associated with the Golgi, Arf1 is a well-characterised Golgi-localised small GTPase, located at both the TGN and cis-Golgi/ER-Golgi intermediate compartment37. Hence, mTOR and components of the activation pathway are present at the Golgi or Golgi/ER interface (or both), as illustrated in Figure 1.
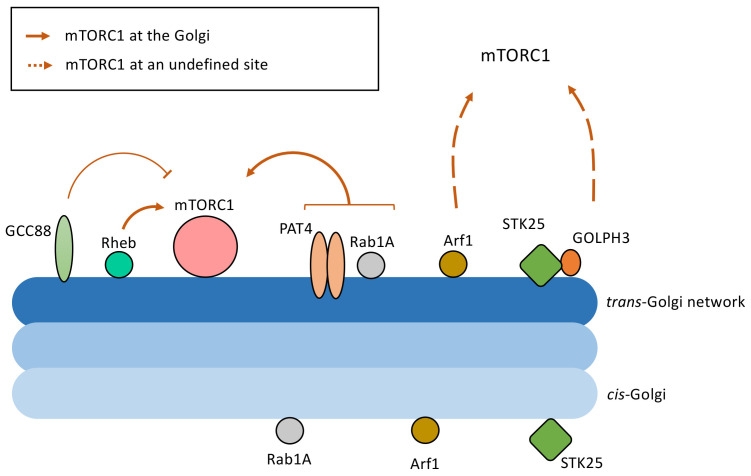
Figure 1. Golgi-localised components that mediate mechanistic (or mammalian) target of rapamycin complex 1 (mTORC1) signalling.
Shown are components of the TORC1 pathway located at the Golgi. All of the components are located at the trans-Golgi network. Some components are also located at the cis-Golgi, as indicated. The components that have been reported to regulate mTORC1 activity at the Golgi are indicated by solid lines, whereas other components that activate mTORC1 at a location yet to be defined are indicated by broken lines. The mechanisms by which the Golgi components regulate mTORC1 remain poorly understood.
Evidence that mTORC1 signalling is mediated by Golgi-localised components
Identification of the components of the mTORC1 pathway at the Golgi demonstrates a contribution by this organelle to the mTOR pathway. However, localisation per se does not demonstrate that activation of mTORC1 occurs on Golgi membranes. In light of the localisation studies alone, it is formally possible that the Golgi represents a reservoir of mTORC1 pathway components and that activation occurs at other sites. However, a growing body of work demonstrates the ability of the Golgi apparatus to directly regulate mTOR activity, independently of lysosomes.
First, the activation of mTORC1 by Rab1A was demonstrated to be independent of lysosomes. Knockdown of Rab1A abrogates the Rheb–mTORC1 interaction at the Golgi but does not interfere with the interaction of mTORC1 with Rag on lysosomes24. Moreover, mTORC1 activation at the Golgi via Rab1A could rescue defects in mTORC1 activation at the lysosomes and vice versa, which argues that the Rab1A and Rag small GTPases are independently regulating mTORC1 at different locations24.
Second, our work found that the organisation of the Golgi as a ribbon structure is essential for regulating the mTORC1 pathway and autophagy. Conversion of the Golgi ribbon into Golgi mini-stacks, mediated by the increased expression of the Golgi membrane tether GCC88, which links Golgi membranes to the actin cytoskeleton at the TGN, resulted in an increase in LC3-positive autophagosomes26. The level of phosphorylated ribosomal S6 in HeLa cells which lack a Golgi ribbon is reduced compared with wild-type HeLa cells26. In addition, there was a significant reduction of both mTOR and p-mTOR26 on the scattered Golgi mini-stacks of HeLa cells lacking a Golgi ribbon compared with wild-type HeLa cells. Using balifomycin A1, which interferes with mTOR recruitment to lysosomes, we also demonstrated that mTOR could be recruited to Golgi membranes in the absence of a lysosomal mTOR pool, indicating that the Golgi and lysosomes have independent mTOR pools.
Third, in situ proximity ligation analysis yielded a positive signal between the glutamine transporter PAT4 and both mTOR and Rab1A, revealing that mTORC1 is located on the Golgi34. In addition, glutamine has been reported to stimulate mTORC1 by a Rag-independent mechanism35, indicating additional pathways of activation. Glutamine and serine activation of mTORC1 by the SLC36 transporter PAT4 shows preference for the phosphorylation of the downstream target 4E-BP1 compared with ribosomal S6, indicating that the Golgi activation pathway may be selective34. These findings suggest that mTORC1 may regulate different downstream pathways at different intracellular locations.
Fourth, mTOR signalling is modulated by Golgi phosphoprotein 3 (GOLPH3), another Golgi membrane tether that links the Golgi membranes to actin cytoskeleton and that has a role in the maintenance of the Golgi structure38,39. Increased levels of GOLPH3 result in enhanced mTORC1 activity and reduced autophagy40,41. Alterations in the level and phosphorylation of GOLPH3 result in changes in Golgi morphology, from the dispersal of Golgi fragments and the loss of the Golgi ribbon mediated by elevated GOLPH3 levels to an enhanced compaction of the Golgi morphology where levels of GOLPH3 are reduced38,40,41. The mechanism whereby GOLPH3 activates mTORC1, by a rapamycin-sensitive manner, is unclear, and a more extensive analysis of the GOLPH3 binding partners is required to resolve this issue.
Fifth, other Golgi proteins have been reported to modulate mTOR activity. Clem16A is a Golgi protein, which was recently reported to negatively regulate autophagy by activating mTOR42, and GOLPH2 (gp73, GOLM1) has been reported to promote mTOR signalling via the PI3K/AKT pathway43. In addition, the Ser/Thr protein kinase, STK25, a Golgi-localised kinase that regulates Golgi morphology and interacts with GOLPH3, downregulates mTORC1 activity and suppresses cell proliferation44. Members of the SLC38 amino acid transporters, namely SNAT2 and SNAT10, are Golgi-localised under certain conditions and have been reported to influence mTORC1 activity45–47. However, the intracellular location of mTORC1 activation via these transporters is not clear.
Collectively, the above indicates that there may be multiple pathways by which the Golgi can regulate mTOR signalling, independent of lysosomes. There is now substantial evidence for a functional pool of mTOR at the Golgi and the architecture of the Golgi influences mTOR activation.
Golgi–organelle membrane contact sites and mTORC1
As discussed above, different morphological states of the Golgi architecture influence mTOR activation. The membrane tethers of the Golgi which influence Golgi architecture are known to interact with a number of components, and we have previously suggested that these membrane tethers may be akin to scaffold molecules to recruit a diverse set of components, including signalling machinery48. Indeed, many of the Golgi membrane tethers bind to small G proteins, which are essential regulators of signalling pathways. Another important consideration which could influence not only mTORC1 signalling but also other signalling pathways is the integrity of membrane contact sites between the Golgi and other intracellular organelles. Golgi membranes, and in particular the TGN, are known to make contacts with the ER and late endosomes/lysosomes49–51. Membrane contact sites between organelles mediate a number of processes, including the transfer of lipids, which in turn regulates the recruitment of cytolosic proteins to membranes. Some reports indicate that the majority of the small GTPase, Rheb, is localised to the Golgi32,52 and that membrane contact sites between the Golgi and lysosomes may be required for lysosomal activation of mTORC132,51. Therefore, the Golgi should also be considered in the context of its membrane contacts with other organelles. An important issue to be investigated is whether the changes in the dynamics of the Golgi morphology modulate the membrane–membrane contacts sites between the Golgi and the ER or lysosomes and, if so, how the dynamics of these membrane contacts affect the recruitment and activation of mTORC1. At this stage, it remains unclear whether activation of mTORC1 is a co-ordinated process between the Golgi and lysosomes and whether changes in Golgi morphology have an indirect effect on lysosomal mTORC1 activation.
Golgi, oncogenes, mTOR and cancer
A number of the Golgi components that influence mTORC1 signalling promote cell growth and tumourigenesis. Rab1A and GOLPH3 have been demonstrated to be oncogenes and are upregulated in a number of human cancers, including colorectal, prostate and gastric cancers24,40,41,53–56. Rab1A overexpression promotes mTORC1 signalling and also oncogenic growth in in vitro and in vivo model systems24. Overexpression of GOLPH3 has been shown to enhance cell survival following DNA damage40. The DNA damage response triggered by clinical therapies, which protects tumour cells from apoptosis, is regulated by GOLPH3. The nuclear kinase, DNA-Pk, which is activated following therapies, phosphorylates GOLPH3 and promotes loss of the compact Golgi ribbon, enhanced Akt1-mTOR signalling and cell survival40,57. In contrast, depletion of GOLPH3 maintains a compact Golgi following treatment with DNA damaging agents, increases apoptosis and reduces cell survival40. Recently, small non-coding RNAs, microRNAs (miRNAs), which have a protective role in preventing Golgi fragmentation mediated by GOLPH3 have been identified. These miRNAs, namely 3135b and 3150b-3p, have been shown to regulate the expression of GOLPH358,59. For example, transfection of miRNA-3135b in HCT-15 cancer cells sensitised the cells to DNA damage by downregulating the expression of GOLPH3, resulting in reduced levels of mTORC1 signalling58. Hence, miRNA-3135b is a tumour suppressor regulating mTORC1 signalling via GOLPH3-mediated changes in Golgi architecture. Notably, miRNA-3135b and miRNA-3150b-3p are downregulated in colorectal cancer cell lines58,59. These findings are important as they indicate that the regulation of mTOR signalling via Golgi pathways is fundamental for normal cellular homeostasis and that aberrant Golgi-mediated signalling promotes tumourigenesis and also escape of cancer cells from treatment by chemotherapy.
Conclusions
The evidence presented here strongly indicates a direct role for the Golgi apparatus in regulation of mTORC1 signalling in a diverse range of mammalian cells, including cultured epithelial and neuronal cells, in vitro tumour cells and in vivo colorectal cancers. Perturbation of these Golgi-mediated mTORC1 pathways has been demonstrated to be relevant for tumourigenesis. The dynamic nature of the structure of the Golgi and the ability to recruit peripheral membrane proteins to specific subdomains of the Golgi membrane60 make the Golgi an ideal template to rapidly modulate the strength of mTOR signalling. In addition, the Golgi has a direct role in regulating the biogenesis of autophagosomes61 and, given the negative regulation of autophagy by mTORC1 signalling, the Golgi represents a site for the co-ordination of both pathways. Currently, the information on mTORC1 activation at the Golgi has been obtained from biochemical, cell biological and microscopic techniques. Biophysical approaches now need to be applied to visualise the activation of mTORC1 in real time at specific intracellular locations and in a range of different primary cells. This approach will also provide the ability to determine which inputs/sensors activate the Golgi mTORC1 signalling pathway. Do both intrinsic and extrinsic sensors, or only intrinsic sensors, activate Golgi mTORC1, and which downstream pathways are activated by Golgi mTORC1? In addition, further consideration should be given to whether mTOR is activated at intracellular locations in addition to lysosomes and the Golgi.
The peer reviewers who approve this article are:
Seth Field, Harrington Discovery Institute, University Hospitals Cleveland Medical Center, and Department of Medicine, Case Western Reserve University, Cleveland, OH, USA
Deborah C I Goberdhan, Department of Physiology, Anatomy and Genetics, University of Oxford, OX1 3QX Oxford, United Kingdom
Michael N. Hall, Biozentrum, University of Basel, CH-4056 Basel, Switzerland
Don Gary Benjamin, Biozentrum, University of Basel, CH-4056 Basel, Switzerland
Carolyn Machamer, Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA
Funding Statement
This work was supported by funding from the Australian Research Council (DP160102394).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wullschleger S, Loewith R, Hall MN: TOR signaling in growth and metabolism. Cell. 2006; 124(3): 471–84. 10.1016/j.cell.2006.01.016 [DOI] [PubMed] [Google Scholar]
- 2. Saxton RA, Sabatini DM: mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017; 169(2): 361–371. 10.1016/j.cell.2017.03.035 [DOI] [PubMed] [Google Scholar]
- 3. Magnuson B, Ekim B, Fingar DC: Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J. 2012; 441(1): 1–21. 10.1042/BJ20110892 [DOI] [PubMed] [Google Scholar]
- 4. Dibble CC, Manning BD: Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol. 2013; 15(6): 555–64. 10.1038/ncb2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takahara T, Amemiya Y, Sugiyama R, et al. : Amino acid-dependent control of mTORC1 signaling: A variety of regulatory modes. J Biomed Sci. 2020; 27(1): 87. 10.1186/s12929-020-00679-2 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 6. Gwinn DM, Shackelford DB, Egan DF, et al. : AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008; 30(2): 214–26. 10.1016/j.molcel.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 7. Sancak Y, Bar-Peled L, Zoncu R, et al. : Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010; 141(2): 290–303. 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 8. Bar-Peled L, Schweitzer LD, Zoncu R, et al. : Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012; 150(6): 1196–208. 10.1016/j.cell.2012.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 9. Betz C, Hall MN: Where is mTOR and what is it doing there? J Cell Biol. 2013; 203(4): 563–74. 10.1083/jcb.201306041 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 10. Carroll B: Spatial regulation of mTORC1 signalling: Beyond the Rag GTPases. Semin Cell Dev Biol. 2020; 107: 103–11. 10.1016/j.semcdb.2020.02.007 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 11. Kagan JC, Su T, Horng T, et al. : TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol. 2008; 9(4): 361–8. 10.1038/ni1569 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 12. Hatakeyama R, Péli-Gulli MP, Hu Z, et al. : Spatially Distinct Pools of TORC1 Balance Protein Homeostasis. Mol Cell. 2019; 73(2): 325–338.e8. 10.1016/j.molcel.2018.10.040 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 13. Rabanal-Ruiz Y, Byron A, Wirth A, et al. : mTORC1 activity is supported by spatial association with focal adhesions. J Cell Biol. 2021; 220(5): e202004010. 10.1083/jcb.202004010 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 14. Wei JH, Seemann J: Golgi ribbon disassembly during mitosis, differentiation and disease progression. Curr Opin Cell Biol. 2017; 47: 43–51. 10.1016/j.ceb.2017.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kulkarni-Gosavi P, Makhoul C, Gleeson PA: Form and function of the Golgi apparatus: Scaffolds, cytoskeleton and signalling. FEBS Lett. 2019; 593(17): 2289–305. 10.1002/1873-3468.13567 [DOI] [PubMed] [Google Scholar]
- 16. Ravichandran Y, Goud B, Manneville JB: The Golgi apparatus and cell polarity: Roles of the cytoskeleton, the Golgi matrix, and Golgi membranes. Curr Opin Cell Biol. 2020; 62: 104–13. 10.1016/j.ceb.2019.10.003 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 17. Makhoul C, Gosavi P, Gleeson PA: The Golgi architecture and cell sensing. Biochem Soc Trans. 2018; 46(5): 1063–72. 10.1042/BST20180323 [DOI] [PubMed] [Google Scholar]
- 18. Zappa F, Failli M, de Matteis MA: The Golgi complex in disease and therapy. Curr Opin Cell Biol. 2018; 50: 102–16. 10.1016/j.ceb.2018.03.005 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 19. Kupfer A, Dennert G, Singer SJ: Polarization of the Golgi apparatus and the microtubule-organizing center within cloned natural killer cells bound to their targets. Proc Natl Acad Sci U S A. 1983; 80(23): 7224–8. 10.1073/pnas.80.23.7224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sasaki K, Yoshida H: Organelle autoregulation-stress responses in the ER, Golgi, mitochondria and lysosome. J Biochem. 2015; 157(4): 185–95. 10.1093/jb/mvv010 [DOI] [PubMed] [Google Scholar]
- 21. Abdel Rahman AM, Ryczko M, Nakano M, et al. : Golgi N-glycan branching N-acetylglucosaminyltransferases I,V and VI promote nutrient uptake and metabolism. Glycobiology. 2015; 25(2): 225–40. 10.1093/glycob/cwu105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen J, Chen ZJ: PtdIns4P on dispersed trans-Golgi network mediates NLRP3 inflammasome activation. Nature. 2018; 564(7734): 71–6. 10.1038/s41586-018-0761-3 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 23. Lamb CA, Yoshimori T, Tooze SA: The autophagosome: Origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013; 14(12): 759–74. 10.1038/nrm3696 [DOI] [PubMed] [Google Scholar]
- 24. Thomas JD, Zhang YJ, Wei YH, et al. : Rab1A is an mTORC1 activator and a colorectal oncogene. Cancer Cell. 2014; 26(5): 754–69. 10.1016/j.ccell.2014.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu X, Zheng XFS: Endoplasmic reticulum and Golgi localization sequences for mammalian target of rapamycin. Mol Biol Cell. 2007; 18(3): 1073–82. 10.1091/mbc.e06-05-0406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gosavi P, Houghton FJ, McMillan PJ, et al. : The Golgi ribbon in mammalian cells negatively regulates autophagy by modulating mTOR activity. J Cell Sci. 2018; 131(3): jcs211987. 10.1242/jcs.211987 [DOI] [PubMed] [Google Scholar]
- 27. Long X, Lin Y, Ortiz-Vega S, et al. : Rheb binds and regulates the mTOR kinase. Curr Biol. 2005; 15(8): 702–13. 10.1016/j.cub.2005.02.053 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 28. Clark GJ, Kinch MS, Rogers-Graham K, et al. : The Ras-related protein Rheb is farnesylated and antagonizes Ras signaling and transformation. J Biol Chem. 1997; 272(16): 10608–15. 10.1074/jbc.272.16.10608 [DOI] [PubMed] [Google Scholar]
- 29. Hanker AB, Mitin N, Wilder RS, et al. : Differential requirement of CAAX-mediated posttranslational processing for Rheb localization and signaling. Oncogene. 2010; 29(3): 380–91. 10.1038/onc.2009.336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garami A, Zwartkruis FJT, Nobukuni T, et al. : Insulin Activation of Rheb, a Mediator of mTOR/S6K/4E-BP Signaling, Is Inhibited by TSC1 and 2. Mol Cell. 2003; 11(6): 1457–66. 10.1016/s1097-2765(03)00220-x [DOI] [PubMed] [Google Scholar]
- 31. Angarola B, Ferguson SM: Weak membrane interactions allow Rheb to activate mTORC1 signaling without major lysosome enrichment. Mol Biol Cell. 2019; 30(22): 2750–60. 10.1091/mbc.E19-03-0146 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 32. Hao F, Kondo K, Itoh T, et al. : Rheb localized on the Golgi membrane activates lysosome-localized mTORC1 at the Golgi-lysosome contact site. J Cell Sci. 2018; 131(3): jcs208017. 10.1242/jcs.208017 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 33. Angarola B, Ferguson SM: Coordination of Rheb lysosomal membrane interactions with mTORC1 activation [version 1; peer review: 4 approved]. F1000Res. 2020; 9: F1000 Faculty Rev-450. 10.12688/f1000research.22367.1 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 34. Fan SJ, Snell C, Turley H, et al. : PAT4 levels control amino-acid sensitivity of rapamycin-resistant mTORC1 from the Golgi and affect clinical outcome in colorectal cancer. Oncogene. 2016; 35(23): 3004–15. 10.1038/onc.2015.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jewell JL, Kim YC, Russell RC, et al. : Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science. 2015; 347(6218): 194–8. 10.1126/science.1259472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li L, Kim E, Yuan H, et al. : Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem. 2010; 285(26): 19705–9. 10.1074/jbc.C110.102483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Donaldson JG, Jackson CL: ARF family G proteins and their regulators: Roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011; 12(6): 362–75. 10.1038/nrm3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dippold HC, Ng MM, Farber-Katz SE, et al. : GOLPH3 bridges phosphatidylinositol-4- phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009; 139(2): 337–51. 10.1016/j.cell.2009.07.052 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 39. Wood CS, Schmitz KR, Bessman NJ, et al. : PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking. J Cell Biol. 2009; 187(7): 967–75. 10.1083/jcb.200909063 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 40. Farber-Katz SE, Dippold HC, Buschman MD, et al. : DNA damage triggers Golgi dispersal via DNA-PK and GOLPH3. Cell. 2014; 156(3): 413–27. 10.1016/j.cell.2013.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 41. Scott KL, Kabbarah O, Liang MC, et al. : GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature. 2009; 459(7250): 1085–90. 10.1038/nature08109 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 42. Tam RCY, Li MWM, Gao YP, et al. : Human CLEC16A regulates autophagy through modulating mTOR activity. Exp Cell Res. 2017; 352(2): 304–12. 10.1016/j.yexcr.2017.02.017 [DOI] [PubMed] [Google Scholar]
- 43. Yan G, Ru Y, Wu K, et al. : GOLM1 promotes prostate cancer progression through activating PI3K-AKT-mTOR signaling. Prostate. 2018; 78(3): 166–77. 10.1002/pros.23461 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 44. Wu F, Gao P, Wu W, et al. : STK25-induced inhibition of aerobic glycolysis via GOLPH3-mTOR pathway suppresses cell proliferation in colorectal cancer. J Exp Clin Cancer Res. 2018; 37(1): 144. 10.1186/s13046-018-0808-1 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 45. Tripathi R, Hosseini K, Arapi V, et al. : SLC38A10 (SNAT10) is Located in ER and Golgi Compartments and Has a Role in Regulating Nascent Protein Synthesis. Int J Mol Sci. 2019; 20(24): 6265. 10.3390/ijms20246265 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 46. Krokowski D, Guan BJ, Wu J, et al. : GADD34 Function in Protein Trafficking Promotes Adaptation to Hyperosmotic Stress in Human Corneal Cells. Cell Rep. 2017; 21(10): 2895–910. 10.1016/j.celrep.2017.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Morotti M, Zois CE, El-Ansari R, et al. : Increased expression of glutamine transporter SNAT2/SLC38A2 promotes glutamine dependence and oxidative stress resistance, and is associated with worse prognosis in triple-negative breast cancer. Br J Cancer. 2021; 124(2): 494–505. 10.1038/s41416-020-01113-y [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 48. Gosavi P, Gleeson PA: The Function of the Golgi Ribbon Structure - An Enduring Mystery Unfolds! Bioessays. 2017; 39(11). 10.1002/bies.201700063 [DOI] [PubMed] [Google Scholar]
- 49. Venditti R, Masone MC, de Matteis MA: ER-Golgi membrane contact sites. Biochem Soc Trans. 2020; 48(1): 187–97. 10.1042/BST20190537 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 50. Wang T, Hong W: Interorganellar regulation of lysosome positioning by the Golgi apparatus through Rab34 interaction with Rab-interacting lysosomal protein. Mol Biol Cell. 2002; 13(12): 4317–32. 10.1091/mbc.e02-05-0280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Starling GP, Yip YY, Sanger A, et al. : Folliculin directs the formation of a Rab34-RILP complex to control the nutrient-dependent dynamic distribution of lysosomes. EMBO Rep. 2016; 17(6): 823–41. 10.15252/embr.201541382 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 52. Yadav RB, Burgos P, Parker AW, et al. : mTOR direct interactions with Rheb-GTPase and raptor: Sub-cellular localization using fluorescence lifetime imaging. BMC Cell Biol. 2013; 14: 3. 10.1186/1471-2121-14-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hu BS, Hu H, Zhu CY, et al. : Overexpression of GOLPH3 is associated with poor clinical outcome in gastric cancer. Tumour Biol. 2013; 34(1): 515–20. 10.1007/s13277-012-0576-z [DOI] [PubMed] [Google Scholar]
- 54. Li W, Guo F, Gu M, et al. : Increased Expression of GOLPH3 is Associated with the Proliferation of Prostate Cancer. J Cancer. 2015; 6(5): 420–9. 10.7150/jca.11228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zeng Z, Lin H, Zhao X, et al. : Overexpression of GOLPH3 promotes proliferation and tumorigenicity in breast cancer via suppression of the FOXO1 transcription factor. Clin Cancer Res. 2012; 18(15): 4059–69. 10.1158/1078-0432.CCR-11-3156 [DOI] [PubMed] [Google Scholar]
- 56. Zhou B, Wang G, Gao S, et al. : Expression of GOLPH3 protein in colon cancer tissues and its association with the prognosis of patients. Oncol Lett. 2016; 12(5): 3936–40. 10.3892/ol.2016.5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Buschman MD, Rahajeng J, Field SJ: GOLPH3 links the Golgi, DNA damage, and cancer. Cancer Res. 2015; 75(4): 624–7. 10.1158/0008-5472.CAN-14-3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Núñez-Olvera SI, Chávez-Munguía B, del Rocío Terrones-Gurrola MC, et al. : A novel protective role for microRNA-3135b in Golgi apparatus fragmentation induced by chemotherapy via GOLPH3/AKT1/mTOR axis in colorectal cancer cells. Sci Rep. 2020; 10(1): 10555. 10.1038/s41598-020-67550-0 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 59. Zhang W, Chen X, Jia J: MiR-3150b-3p inhibits the progression of colorectal cancer cells via targeting GOLPH3. J Investig Med. 2020; 68(2): 425–9. 10.1136/jim-2019-001124 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 60. Goud B, Gleeson PA: TGN golgins, Rabs and cytoskeleton: Regulating the Golgi trafficking highways. Trends Cell Biol. 2010; 20(6): 329–36. 10.1016/j.tcb.2010.02.006 [DOI] [PubMed] [Google Scholar]
- 61. Nakatogawa H: Mechanisms governing autophagosome biogenesis. Nat Rev Mol Cell Biol. 2020; 21(8): 439–58. 10.1038/s41580-020-0241-0 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation